Abstract
In mammals, the first cell-fate decision occurs during preimplantation embryo development when the inner cell mass (ICM) and trophectoderm (TE) lineages are established. The ICM develops into the embryo proper, while the TE lineage forms the placenta. The underlying molecular mechanisms that govern lineage formation involve cell-to-cell interactions, cell polarization, cell signaling and transcriptional regulation. In this review, we will discuss the current understanding regarding the cellular and molecular events that regulate lineage formation in mouse preimplantation embryos with an emphasis on cell polarity and the Hippo signaling pathway. Moreover, we will provide an overview on some of the molecular tools that are used to manipulate the Hippo pathway and study cell-fate decisions in early embryos. Lastly, we will provide exciting future perspectives on transcriptional regulatory mechanisms that modulate the activity of the Hippo pathway in preimplantation embryos to ensure robust lineage segregation.
Keywords: mouse, preimplantation embryos, Hippo signaling, transcription factors, lineage formation
Introduction
In placental mammals, life begins as a totipotent one-cell embryo that has the capacity to transform into a differentiated multi-cellular organism. A central question in developmental biology is how do totipotent cells in the early embryo become specialized tissues and organs. During preimplantation embryo development totipotent cells must undergo the first cell-fate decision to become the pluripotent inner cell mass (ICM) or multi-potent trophectoderm (TE) lineages. These two cellular lineages develop into the fetus and the placenta, respectively. Proper specification of the ICM and TE is absolutely crucial for subsequent development. For example, disruption of the ICM lineage in human preimplantation embryos may result in fetal malformations and congenital defects (Ferrer-Vaquer and Hadjantonakis, 2013), whereas perturbations in the TE lineage can lead to defects in placentation and pregnancy-associated problems such as preeclampsia and preterm birth (Norwitz, 2007; Faye-Petersen, 2008; Chaiworapongsa et al., 2014). In the subsequent sections, we will provide an overview of mouse preimplantation embryo development, a model organism for investigating the etiology of early embryonic loss in humans. We will give background information on the cellular and transcriptional events that are required for lineage formation and discuss the importance of the Hippo signaling pathway in early embryo development.
A synopsis of mouse preimplantation embryo development
The window of preimplantation embryo development encompasses a series of cellular and morphological events that culminate in blastocyst formation. Preimplantation development begins immediately after sperm and oocyte fusion when the metaphase II arrested oocyte undergoes resumption of meiosis. The newly formed zygote contains one maternal and paternal haploid genome, both of which undergo DNA synthesis and will coalescence in preparation for the first mitotic cleavage. Beginning at the late one-cell to two-cell stage, the embryo will transition from utilizing maternal transcripts and proteins stored in the oocyte, to actively transcribing its own embryonic genome (i.e. zygotic genome activation) (Latham and Schultz, 2001; Schultz, 2002). Between the one-cell stage and eight-cell stage, the embryonic cells (i.e. blastomeres) undergo three symmetrical cell divisions.
At the eight-cell stage, the embryo begins to exhibit the first obvious signs of differentiation when the blastomeres compact and undergo polarization forming the apical and basolateral domains. Compaction is mediated through the expression and localization of E-cadherin on the basal lateral cell membranes (Ducibella et al., 1977; Larue et al., 1994; De Vries et al., 2004). Concomitant with compaction, core cell polarity complexes, consisting of Par-6 family cell polarity regulator beta (PARD6B), Par-3 family cell polarity regulator and atypical PKC (e.g. PKC zeta or delta), assemble on the apical membranes (outer region) of each blastomere, while on the inside of the embryo, MAP/microtubule affinity-regulating kinase 2, scribbled homolog and lethal giant larvae homolog 1 assemble at the basal lateral membrane of each blastomere (Vinot et al., 2005; Dard et al., 2009a; Cockburn and Rossant, 2010). The asymmetrical localization of these protein complexes generates the apical–basal axis, which is critical for subsequent blastomere differentiation into polar and apolar cells (Cockburn and Rossant, 2010).
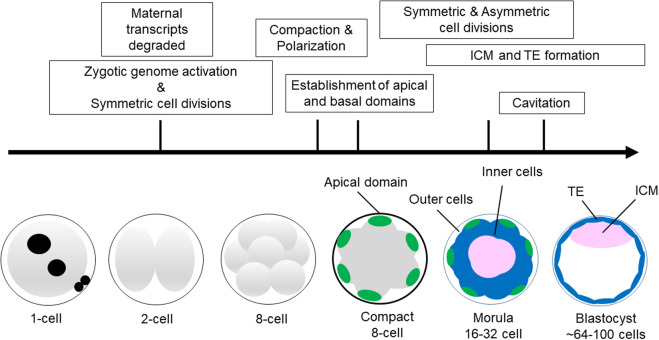
Next, at the 8–16 cell stage, there is a switch from all symmetrical to combined symmetrical and asymmetrical cell divisions that generate populations of polar outside cells and apolar inside cells, respectively (Johnson and McConnell, 2004; Cockburn and Rossant, 2010). This is a critical stage in development because the embryo must allocate its blastomeres into either the TE or ICM. How this is precisely accomplished remains elusive, but upon the fourth mitotic division the placement of the mitotic spindle is random resulting in either asymmetric or symmetric cell divisions (Dard et al., 2009b). Accompanying these differential cell divisions, a number of different tight junction proteins, Na/K-ATPases and aquaporins are expressed and localized to the apical membranes on the outside TE cells (Barcroft et al., 2003; Madan et al., 2007; Moriwaki et al., 2007; Eckert and Fleming, 2008; Katsuno et al., 2008; Sheth et al., 2008). These molecules are critical for establishment of paracellular sealing and fluid accumulation (i.e. blastocoel formation). In Fig. 1, we provide a schematic illustrating the basic cellular and morphological events that are associated with preimplantation development. The proper execution of these events is essential for blastocyst formation and serves as a prerequisite for implantation, placentation and subsequent fetal development.
Figure 1.
Overview of the basic cellular and morphological events which occur during mouse preimplantation embryo development. Key events are emphasized in the boxes. Lineage formation is thought to begin around the time of compaction when the apical (green) and basolateral domains (not shown) are established. Subsequent symmetric and asymmetric cell divisions generate outside (blue) and inside (pink) cells at the morula stage. During the morula to blastocyst transition, the trophectoderm (TE) (blue) and inner cell mass (ICM) (pink) lineages are formed.
Transcriptional regulation of the first cell-fate decision in mouse preimplantation embryos
Along with the cellular and morphological events that mediate lineage formation in preimplantation embryos, there is a cohort of transcription factors that promote specification of the ICM and TE lineages via molecular mechanisms. These transcription factors can be separated into three subgroups based on their expression pattern in blastocysts. The first group consists of transcription factors that regulate pluripotency and inhibit cellular differentiation in the ICM lineage. Examples of these include octamer-binding transcription factor 4 (OCT4), NANOG and SRY-box transcription factor 2 (SOX2) (Palmieri et al., 1994; Nichols et al., 1998; Mitsui et al., 2003; Chen et al., 2009; Keramari et al., 2010; Thomson et al., 2011; Bessonnard et al., 2014; Mulas et al., 2018; Heurtier et al., 2019). The second group of transcription factors is enriched in the TE lineage and is important for implantation and subsequent placental development. Examples of these include caudal type homeobox 2 (CDX2), GATA binding protein 3 (GATA3), eomesodermin and transcription factor AP2 gamma (TFAP2C) (Auman et al., 2002; Werling and Schorle, 2002; Strumpf et al., 2005; Winger et al., 2006; Home et al., 2009; Choi et al., 2012). Lastly, the third group of transcription factors is expressed in both the ICM and TE lineages. Examples, of these include TEA domain family member 4 (TEAD4), as well as other transcriptional regulators (e.g. epigenetic modifiers) that have been extensively reviewed elsewhere by our lab and others (Yagi et al., 2007; Nishioka et al., 2008; Home et al., 2012; Knott and Paul, 2014; Paul and Knott, 2014; Miller and Hendrich, 2018). In Fig. 2, we provide a general overview on the importance of SOX2, OCT4, NANOG, TFAP2C, TEAD4, GATA3 and CDX2 during lineage formation. Throughout the remainder of this review, we will focus specifically on the regulation and/or function of OCT4, SOX2, TEAD4, CDX2 and TFAP2C in the context of the first cell-fate decision in preimplantation embryos.
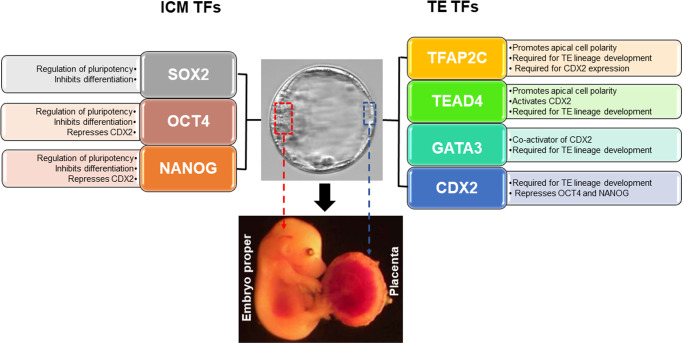
Figure 2.
Role and regulation of key lineage transcription factors during the first-cell fate decision in mouse preimplantation embryos. On the left side are SRY-box transcription factor 2 (SOX2), octamer-binding transcription factor 4 (OCT4) and NANOG. These transcription factors (TFs) are important for regulation of pluripotency and inhibition of differentiation in the ICM lineage. In the center is an expanded mouse blastocyst with the ICM and TE lineages highlighted in red and blue boxes. Below the blastocyst, there is a representative mouse fetus and placenta that developed from the ICM and TE lineages, respectively. On the right side are transcription factor AP2 gamma (TFAP2C), TEA domain family member 4 (TEAD4), GATA binding protein 3 (GATA3) and caudal type homeobox 2 (CDX2). These TFs are listed in a hierarchy from top to bottom. TFAP2C, TEAD4 and GATA3 are required for activation of CDX2 expression and proper TE lineage development. Mechanistically, CDX2 can negatively regulate Oct4 and Nanog expression in the TE lineage, while OCT4 and NANOG can repress Cdx2 expression in the ICM lineage.
The molecular mechanisms by which early embryonic transcription factors become exclusively expressed in the ICM and TE to promote lineage development have been extensively investigated. Several studies have shown that at the eight-cell stage, OCT4 and CDX2 are initially ubiquitously expressed (Strumpf et al., 2005; Dietrich and Hiiragi, 2007). However, during the morula-to-blastocyst transition, OCT4 becomes restricted to the ICM, while CDX2 becomes enriched in the TE lineage (Strumpf et al., 2005; Dietrich and Hiiragi, 2007). This is accomplished by a combination of transcriptional and epigenetic mechanisms. For example, OCT4 and CDX2 exhibit reciprocal regulation of one another by binding and repressing each other’s gene promoters (Niwa et al., 2005; Wang et al., 2010). Moreover, the Oct4 and Cdx2 promoters acquire specific active and repressive epigenetic marks that modulate their transcriptional activity in the ICM and TE, respectively (Yuan et al., 2009; Saha et al., 2013). For more information on the role of epigenetic modifications in lineage formation, refer to an excellent review (Paul and Knott, 2014).
Of particular interest, there is a subset of transcription factors that exhibit a unique developmental expression pattern that insinuates important roles in lineage formation. Examples of these transcription factors include TFAP2C and SOX2. The TE regulator TFAP2C is one of the earliest transcription factors expressed during preimplantation development. In the mouse, it is expressed both maternally and zygotically (Winger et al., 2006; Choi et al., 2012). During preimplantation development, the expression and nuclear localization of TFAP2C precedes CDX2 and other TE transcription factors such as GATA3 (Dietrich and Hiiragi, 2007; Home et al., 2009; Ralston et al., 2010; Cao et al., 2015). Research in our laboratory demonstrated that TFAP2C acts upstream of CDX2 and is required for transcriptional activation of the Cdx2 gene in two-cell embryos (Cao et al., 2015). Likewise, the pluripotency transcription factor SOX2 exhibits a unique expression pattern in preimplantation development. Unlike OCT4 which is ubiquitously expressed at the eight-cell and morula stages, SOX2 is only expressed in a subset of inside cells at the morula stage that develop into the ICM (Wicklow et al., 2014; Frum et al., 2018). Functional studies from our laboratory and others have demonstrated that TFAP2C and SOX2 are critical for blastocyst formation and/or proper lineage specification (Keramari et al., 2010; Kuckenberg et al., 2010; Choi et al., 2012; Wicklow et al., 2014). Altogether, these experimental findings highlight the importance of lineage transcription factors in preimplantation development.
Discovery of the evolutionarily conserved Hippo signaling pathway
A long-standing fundamental question in mammalian development is what regulatory pathways orchestrate lineage formation and promote subsequent blastocyst development. For example, what signaling pathways act upstream of CDX2 and SOX2 to govern the first cell-fate decision? In 2009, researchers in Japan revealed that the Hippo signaling pathway is crucial for lineage formation and specification of the ICM and TE lineages (Nishioka et al., 2009). Originally discovered in Drosophila melanogaster in 1995, the Hippo pathway is essential for regulation of organ growth and prevention of tumorigenesis (Justice et al., 1995; Xu et al., 1995; Udan et al., 2003). It contains several key components such as Warts (WTS), Salvador (SAV) and Hippo (HPO) (Justice et al., 1995; Tapon et al., 2002; Udan et al., 2003; Bennett and Harvey, 2006). Characterization of these molecules demonstrated that they function as protein kinases that are part of a key regulatory pathway that controls organ and tissue growth. Over the next ten years, a number of other Hippo signaling pathway components were identified in Drosophila. These included Merlin (MER), Mob as a tumor suppressor, the effector protein Yorkie (YKI) and the transcription factor Scalloped (SD) (LaJeunesse et al., 1998; Huang et al., 2005; Lai et al., 2005; Wu et al., 2008; Kim and Jho, 2018). Notably, there are multiple orthologs in mammals plus additional regulatory molecules that are not present in Drosophila. Table 1 contains a list of the Hippo signaling gene names for both mammals and Drosophila. In mammals, this pathway plays numerous roles in development and adult life including organ growth, apoptosis, cellular differentiation and tumor suppression (Pan, 2010; Kim and Jho, 2018). In the ensuing sections, we will focus on the role of the Hippo signaling pathway in lineage formation (i.e. the first cell-fate decision) as it relates to regulation of Cdx2 and Sox2 expression, as well as the molecular mechanisms that negatively and positively control Hippo signaling during preimplantation development. We will focus exclusively on the Hippo signaling pathway and we will not discuss other signaling pathways that are involved with lineage formation.
Table I.
Key Hippo signaling proteins in Drosophila and mammals.
| Drosophila | Mammalian orthologues | Function | References |
|---|---|---|---|
| Mer | NF2 | Tumor suppressor; interacts with LATS | LaJeunesse et al. (1998) |
| Merlin | Neurofibromatosis 2 | ||
| Hpo | MST1/2 | Upstream activator of Hippo signaling; protein kinase | Udan et al. (2003) |
| Hippo | Macrophage stimulating 1/2 | ||
| Sav | SAV1 | Promotes exit of cell cycle and apoptosis; scaffold protein that interacts with Hippo | Tapon et al. (2002) |
| Salvador | Salvador 1 | ||
| Wts | LATS1/2 | Regulates cell proliferation, differentiation and apoptosis; protein kinase that targets YAP1 | Justice et al. (1995), Nishioka et al. (2009), Xu et al. (1995) |
| Warts | Large tumor suppressor 1/2 | ||
| Mats | MOBKL1A/1B | Growth inhibitor and tumor suppressor; LATS interacting protein | Lai et al. (2005) |
| Mob as a tumor suppressor | MOB as tumor suppressor | ||
| Yki | YAP1 | Hippo effector protein; transcriptional co-activator of TEAD4; promotes cell proliferation and differentiation; inhibits apoptosis | Huang et al. (2005), Nishioka et al. (2009) |
| Yorkie | Yes-associated protein 1 | ||
| Sd | TEAD4 | Mediates YAP1 activity and is required for YAP1-induced cell proliferation and differentiation | Wu et al. (2008) |
| Scalloped | TEA domain transcription factor 4 | ||
| NA | AMOT | Regulates the localization of Hippo signaling in cells; inhibits YAP1 activity via activation of LATS | Zhao et al. (2011) |
| Angiomotin |
Cellular and molecular mechanisms that regulate Hippo signaling during mouse preimplantation development
Foundational work conducted by Nishioka et al. (2008) and others (Yagi et al., 2007; Nishioka et al., 2009) elegantly showed that the activity of the Hippo signaling pathway and TEAD4 are crucial for formation of the ICM and TE lineages in mice. They demonstrated that beginning at the 8- to 16-cell stage, the Hippo signaling pathway is exclusively active in the inside cells of the embryo and inactive in the outside cells. The working model in preimplantation embryos proposes that the Hippo signaling is position-dependent, regulated by cell polarity and cell-to-cell contact (Nishioka et al., 2009; Anani et al., 2014; Hirate et al., 2015). For example, on the outside of the embryo where there is low cell contact, apical cell polarity complexes can suppress Hippo signaling by inhibiting large tumor suppressor kinase 1/2 (LATS1/2). Consequently, yes-associated protein 1 (YAP1) and WW domain containing transcription regulator 1 (WWTR1), two effector proteins that share redundant functions, enter the nucleus and interact with TEAD4 to selectively activate TE-specific genes such as Cdx2 (Nishioka et al., 2009; Alarcon, 2010; Anani et al., 2014). In contrast, on the inside of the embryo where there is high cell contact and the presence of basolateral cell polarity complexes, LATS phosphorylates YAP1/WWTR1, preventing YAP1/WWTR1 from entering the nucleus (Nishioka et al., 2009; Anani et al., 2014; Hirate et al., 2015). This results in the downregulation of Cdx2 and the upregulation of Sox2 (Nishioka et al., 2009; Wicklow et al., 2014). Furthermore, an exciting recent study asserts that YAP1/WWTR1 and TEAD4 may directly repress Sox2 expression in the outside cells (Frum et al., 2019). Future research will help elucidate whether YAP1/WWTR1 and TEAD4 recruit additional co-repressors and/or epigenetic modifiers to silence Sox2 transcription.
In addition to LATS1/2, a second regulatory protein known as Angiomotin (AMOT) is required for activation of the Hippo pathway in mammals (Zhao et al., 2011). AMOT is a junction-associated binding protein that interacts with adherens junctions on the inside of the embryo. There, it forms a regulatory complex with Neurofibromatosis 2 (NF2), LATS and YAP1/WWTR1 (Hirate et al., 2013; Leung and Zernicka-Goetz, 2013; Hirate and Sasaki, 2014). The working model proposes that phospho-AMOT interacts with YAP1/WWTR1 allowing LATS-dependent phosphorylation of YAP1/WWTR1. This keeps YAP1/WWTR1 localized exclusively in the cytoplasm where it is degraded by multiple mechanisms (Hirate et al., 2013; Leung and Zernicka-Goetz, 2013; Hirate and Sasaki, 2014). Conversely, in the outside cells, AMOT is sequestered away from adherens junctions by F-actin and the apical cell polarity complex, preventing it from binding to the LATS-NF2 complex (Hirate et al., 2013; Leung and Zernicka-Goetz, 2013; Hirate and Sasaki, 2014). This inactivates the Hippo pathway allowing YAP1/WWTR1 to enter the nucleus and interact with TEAD4 to activate Cdx2 expression (Hirate et al., 2013; Leung and Zernicka-Goetz, 2013; Hirate and Sasaki, 2014). A graphical overview of the Hippo signaling pathway and its key effector proteins in preimplantation embryos is shown in Fig. 3.
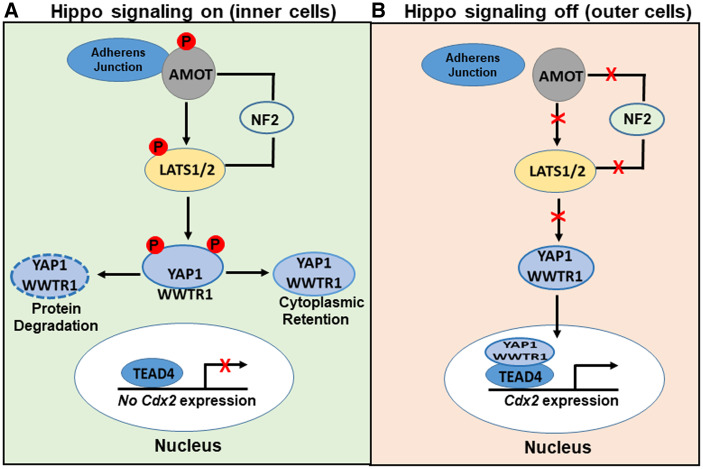
Figure 3.
Schematic overview of the Hippo signaling pathway in mouse preimplantation embryos. (A) When Hippo signaling is on, angiomotin (AMOT) binds to adherens junctions and forms a complex with Neurofibromatosis 2 (NF2) and tumor suppressor kinases 1/2 (LATS1/2). This complex phosphorylates yes-associated protein/WW domain containing transcription regulator 1 (YAP/WWTR1) causing it to either undergo degradation or cytoplasmic retention. As a result, TE-specific genes are repressed. (B) When Hippo signaling is off, YAP/WWTR1 accumulates in the nucleus and interacts with TEAD4, resulting in the upregulation of TE-specific genes.
Several studies have also shown that Rho and Rho-associated coiled-coil kinases 1 and 2 (ROCK1 and ROCK2) are necessary for lineage formation by negatively regulating the Hippo pathway (Kono et al., 2014; Shi et al., 2017; Alarcon and Marikawa, 2018). ROCK is activated by Rho small GTPases and then it phosphorylates a variety of targets involved in modulation of cellular processes such as cell polarity and gene expression (Amano et al., 2010; Julian and Olson, 2014). In preimplantation embryos, ROCK functions in opposition to LATS to negatively regulate the Hippo signaling pathway (Kono et al., 2014; Mihajlovic and Bruce, 2016). Rho and/or ROCK negatively regulates Hippo signaling in the outside cells by at least two mechanisms. Firstly, Rho and ROCK can interfere with activators of LATS, such as NF2 and AMOT, by controlling their subcellular localization (Mihajlovic and Bruce, 2016; Shi et al., 2017). This allows YAP1/WWTR1 to enter the nucleus and activate TE-specific genes such as Cdx2. Also, ROCK can inactivate the Hippo signaling pathway indirectly by regulating the localization of apical and basolateral cell polarity complexes (Kono et al., 2014; Cao et al., 2015). In support of this model, inhibition of ROCK via a pharmacological approach (Y-27632) disrupts apical and basolateral cell polarity resulting in global activation of the Hippo signaling pathway and the upregulation of pluripotency genes in both the inside and outside of the embryo (Kono et al., 2014; Cao et al., 2015). Altogether, these studies demonstrate that the Hippo signaling pathway is a highly regulated molecular circuit that is crucial for proper formation of the ICM and TE lineages in mouse preimplantation embryos.
Molecular tools for manipulating the Hippo pathway and studying cell-fate decisions
Our current knowledge about the role of the Hippo signaling pathway in lineage formation in mouse preimplantation embryos was attained by using various molecular tools and gene knockout (KO) models. This experimental tool kit can be divided into three groups. In the first group, one-cell embryos or two-cell embryos are microinjected with synthetic RNAs (RNAs) encoding either wild-type or mutant versions of specific Hippo signaling proteins to modulate the activity of the pathway. In addition, small interfering RNAs (siRNAs) can be injected to assess the function of a particular Hippo signaling protein. In the second group, mutant mice are generated by various gene targeting approaches and the males and females are mated to produce heterozygous and homozygous preimplantation embryos for phenotypic analysis. In the third group, pharmacological methods are employed to alter the activity of the Hippo pathway. In Table 2, we list a subset of tools that are frequently used to manipulate the Hippo pathway in mouse preimplantation embryos. In the following section, we provide specific examples of how these tools were used to experimentally control the Hippo signaling pathway and study cell-fate.
Table II.
Molecular tools for manipulating the Hippo signaling pathway and altering cell-fate in mouse preimplantation embryos
| Experimental approach | Hippo signaling component | Effect on Hippo signaling | Mutant phenotype/effect on cell-fate | References |
|---|---|---|---|---|
| Microinjection of zygotes or two-cell embryos |
|
|
|
|
| Lats2 RNA | YAP1 Phosphorylation | Decreased CDX2; increased SOX2; enhanced ICM features | Frum et al. (2018), Nishioka et al. (2009) | |
| KD-Lats2 RNA | Inhibition of YAP1 phosphorylation; increase in nuclear YAP1 | Loss of Hippo signaling | Nishioka et al. (2009) | |
| Lats1/2 siRNA | Loss of LATS function; Increase in nuclear YAP1 | Misspecification of ICM; gastrulation failure | Lorthongpanich et al. (2013) | |
| Gene KO |
|
|
|
|
| Lats1/2−/− | Loss of LATS function; Increase in nuclear YAP1 | Increased CDX2 in the inside cells | Nishioka et al. (2009) | |
| Pharmacological inhibitors |
|
|
|
|
| Verteporfin | Inhibition of YAP/TEAD4 interactions | Decreased CDX2; embryonic arrest | Menchero et al. (2019) |
In the seminal study by Nishioka et al. (2009), the authors used a vast combination of gene KOs and synthetic RNA microinjection approaches to elucidate the role of specific Hippo signaling pathway members and effectors in lineage formation. For instance, in a subset of experiments, the authors microinjected wild-type Lats2 RNA, a kinase dead (KD) Lats2 or a constitutively activate (CA)-Yap1 RNA into early mouse embryos. Overexpression of LATS in early embryos suppressed Cdx2 expression in the outer cells of morulae via inactivation of YAP1/WWTR1. Conversely, overexpression of KD-LATS2 or KO of LATS1/2 inactivated Hippo signaling, as inferred by accumulation of nuclear YAP1/WWTR1 and upregulation of Cdx2 in the inside of the embryo. Likewise, microinjection of CA-Yap1 RNA into early embryos caused ectopic expression of Cdx2 in the inside of embryos through increased YAP1 accumulation in the nucleus.
Furthermore, in a recent study (Frum et al., 2018), the authors investigated the role of Hippo signaling in cell-fate decisions in preimplantation embryos. They co-injected CA-Yap1 and GFP RNA into a single blastomere at the two-cell stage and through lineage tracing showed that YAP could repress Sox2 expression within the GFP labeled cells. Consequently, there was a decrease in SOX2 positive cells in the ICM of blastocysts. Likewise, they also co-injected Lats2 and GFP RNA into a single blastomere and showed that LATS2 could induce Sox2 expression in the GFP labeled cells. The cellular progeny of these injected blastomeres localized to the ICM. Further genetic studies using gene KO mice demonstrated that maternal and zygotic Yap1/Wwtr1 gene dosage (e.g. +/− and −/−) had differential effects on Cdx2 and Sox2 expression resulting in alternate cell-fates in the preimplantation embryo (Frum et al., 2018).
In another study, Lats1/2 siRNAs were injected into zygotes to transiently reduce Lats1/2 expression during the window of preimplantation embryo development (Lorthongpanich et al., 2013). A temporary reduction in LATS1/2 resulted in accumulation of nuclear YAP on the inside of the embryo and misspecification of the blastocyst ICM. For example, in the ICM both pluripotency and TE markers were co-expressed. Transfer of these blastocysts into surrogate female mice resulted in early post-implantation embryo arrest and failure to undergo gastrulation (Lorthongpanich et al., 2013). These data indicate that transient perturbations in Hippo signaling in early embryos causes detrimental effects later during post-implantation development.
Pharmacological approaches can also be quite useful for studying Hippo signaling and cell-fate specification in preimplantation embryos. Two chemical inhibitors that are frequently utilized in preimplantation embryos are Y-27632 and verteporfin. As discussed in the previous section Y-27632 inhibits ROCK1/2 resulting in changes in apical and basal cell polarity (Kono et al., 2014). Consequently, Hippo signaling is no longer position dependent as inferred by global LATS activation and loss of nuclear YAP in the outside cells. This leads to misexpression of Sox2 on the outside of the embryo (Frum et al., 2018). Verteporfin is used to disrupt nuclear YAP1 and TEAD4 interactions in cells (Liu-Chittenden et al., 2012). Treatment of embryos with verteporfin during the morula-to-blastocyst transition impairs blastocyst development by repressing Cdx2 expression and altering TE characteristics (Menchero et al., 2019). One advantage of using chemical inhibitors is that embryos can be treated in a stage-specific manner. For example, embryos can be cultured in the presence of the inhibitor during certain periods of development to elucidate when the Hippo signaling pathway is functionally relevant. This information is harder to obtain when using mutant embryos generated by gene KO or RNAi because the target protein is absent throughout most of development. All in all, several molecular approaches can effectively be employed to investigate the role of the Hippo signaling pathway in lineage formation and blastocyst development. The implementation of these tools in mice has led to numerous discoveries on the regulation and role of Hippo signaling in preimplantation embryos.
Future perspectives
Even though we understand the basic mechanisms by which Hippo signaling governs the first cell-fate decision in the preimplantation embryo, there are significant gaps in our knowledge on how Hippo signaling is precisely regulated at the cellular and molecular level. For example, what role do lineage transcription factors play in modulation of position-dependent Hippo signaling? Do TFAP2C, TEAD4 and SOX2 exert negative and/or positive feedback on the activity of the Hippo signaling pathway on the inside and outside of the embryo? Two earlier studies from our laboratory demonstrated that TFAP2C can function as a master regulator of TE lineage development in preimplantation embryos (Choi et al., 2012; Cao et al., 2015). Loss-of-function studies combined with binding motif analysis, chromatin immunoprecipitation and gene expression analysis revealed that TFAP2C regulates a number of different genes involved in apical cell polarity, ROCK signaling and tight junction biogenesis (Choi et al., 2012). As described earlier in this review, PARD6B and ROCK proteins play critical roles in position-dependent Hippo signaling by regulating formation and maintenance of the apical domain. Interestingly, loss of maternal and zygotic TFAP2C downregulates Pard6b and Rock1/2 transcription resulting in global activation of LATS1/2, as inferred by phosphorylation of YAP1 on the inside and outside of embryos (Cao et al., 2015). Consequently, YAP1 is prevented from entering the nucleus in the outside cells resulting in downregulation of Cdx2 expression. Intriguingly, the results of this study indicate that TFAP2C can negatively control the activity of the Hippo signaling pathway in the outside cells by positively regulating Pard6b and Rock1/2 expression. In support of this, Wang et al. (2018) showed that TFAP2C can negatively regulate the activity of the Hippo signaling pathway in cancer cells via transcriptional regulation of Rock1/2, indicating that TFAP2C may have a conserved role in early development and in disease.
Consistent with the established role of TFAP2C in apical cell polarity (Cao et al., 2015), a recent exciting study demonstrated that both TFAP2C and TEAD4 can work together to regulate the assembly of the apical domain via transcriptional regulation of key genes that encode for actin regulators such as ARP2/3, MARCKSL1 and CDC42 (Zhu et al., 2020). The authors showed that these proteins along with RhoA are required for actin polymerization and proper assembly of the apical domain (Zhu et al., 2020). Figure 4 is a working model illustrating how together TFAP2C and TEAD4 positively regulate the formation of the apical domain via transcriptional regulation of Pard6b, Rock1/2 and Arp2/3. This mechanism, in return, negatively affects LATS1/2 activity in the outside cells during the eight-cell to morula transition. In future studies, it will be noteworthy to test whether ICM lineage TFs such as SOX2 have opposite transcriptional effects (i.e. repressive) on genes that encode for cell polarity, ROCK signaling and actin proteins. Even more so, it will be enticing to ascertain whether SOX2 can positively or negatively regulate the expression and/or activity of key Hippo signaling proteins that promote ICM lineage development. In support of this notion, SOX2 can antagonize NF2 and other Hippo signaling components to enhance YAP1 activity in some SOX2-dependent cancers (Basu-Roy et al., 2015). These results indicate that SOX2 can regulate key Hippo signaling proteins in other cellular contexts.
Figure 4.
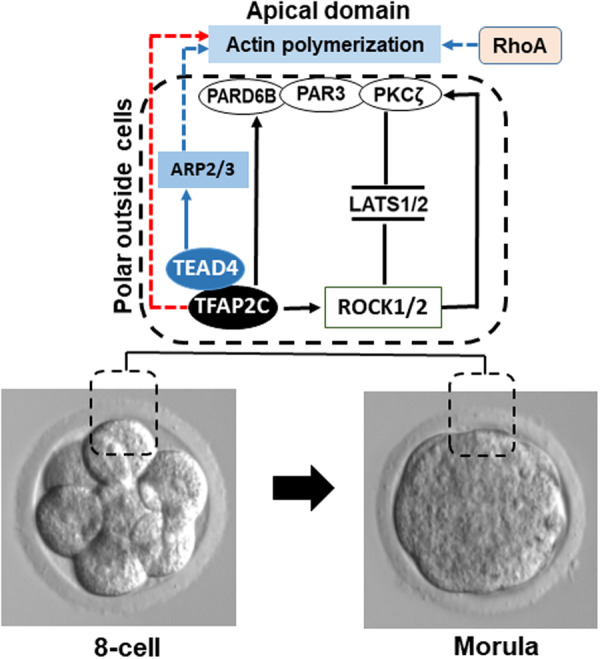
Working model proposing how TFAP2C and TEAD4 promote formation of the apical domain, which in turn, negatively regulate LATS1/2 activity in the outside polar cells. Between the eight-cell to morula transition TFAP2C positively regulates the expression of Pard6b and Rock1/2 genes. TEAD4 positively regulates the expression of key actin regulators such as ARP2/3 to promote actin polymerization. Par-6 family cell polarity regulator beta (PARD6B) contributes to formation of the apical domain by forming a complex with PAR3 and PKCζ. The TEAD4-ARP2/3 axis promotes localization of apical domain proteins to the outside membrane. Rho and Rho-associated coiled-coil kinases 1 and 2 (ROCK1/2) reinforces the apical localization of cell polarity proteins and represses LATS1/2 activity. Depletion of TFAP2C, TEAD4, PARD6B or ARP2/3 or inhibition of ROCK1/2 activity disrupts apical cell polarity and triggers the activation of LATS1/2 in the outside cells.
A second exciting possibility is that TFAP2C may also act downstream of the Hippo signaling pathway and converge with YAP1 and TEAD4 to upregulate Cdx2 transcription. As mentioned earlier in this review, TFAP2C can bind and regulate the expression of Cdx2 in preimplantation embryos. Work in our laboratory using an immunofluorescence proximity ligation assay revealed that TFAP2C can form a nuclear complex with YAP1 in the outside cells during the morula to blastocyst transition (Fig. 5A). Importantly, the localization of TFAP2C-YAP1 is consistent with the normal expression pattern of YAP1 in the outside cells (Nishioka et al., 2009). These results suggest that TFAP2C may regulate Cdx2 expression in collaboration with YAP1 and TEAD4. In support of these findings, an exciting new study revealed that glycolysis-independent glucose metabolism regulates Cdx2 expression via formation of a functional TFAP2C/YAP1/TEAD4 transcriptional complex in the outside cells (Chi et al., 2020). Glucose metabolism is a key biochemical process required for TE lineage formation (Brown and Whittingham, 1991; Leppens-Luisier and Sakkas, 1997). This TFAP2C-Hippo signaling mechanism involves the hexosamine biosynthetic pathway which allows nuclear localization of YAP1 (Chi et al., 2020). As part of this mechanism, TFAP2C translation is regulated by nucleotide synthesis by the pentose phosphate pathway (PPP) and sphingolipid signaling (Chi et al., 2020). Depletion of glucose and/or inhibition of the PPP blocked translation of TFAP2C and downregulated TEAD4 expression, preventing both proteins from forming a functional nuclear complex with YAP1 (Chi et al., 2020). Additional research is necessary to elucidate the precise molecular mechanisms by which TFAP2C regulates Hippo signaling and activates Cdx2 and other TE-specific genes. In Fig. 5B, we provide a working model illustrating how TFAP2C acts downstream of the Hippo signaling pathway during the morula to blastocyst transition to regulate Cdx2 expression in outside cells.
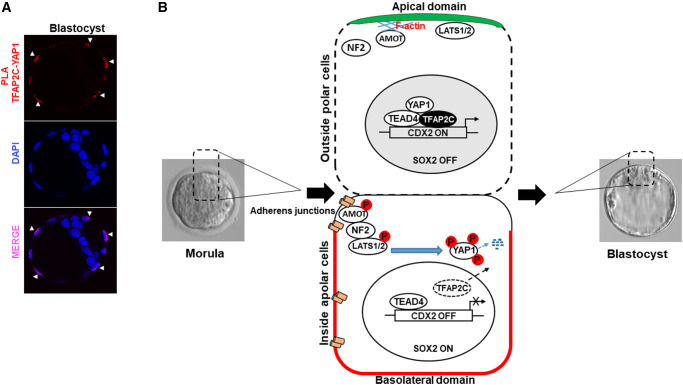
Figure 5.
Working model proposing how TFAP2C functions downstream of the Hippo signaling pathway to regulate Cdx2 expression in the outside polar cells. (A) Proximity ligation assay (PLA) in blastocysts demonstrating nuclear interactions between TFAP2C and YAP1. Embryos (n = 3 biological replicates, 5/embryos/group/replicate). Red fluorescence dots (white arrow heads) indicate interactions between TCFAP2C and YAP1 in the outside cells. (B) During the morula to blastocyst transition, Hippo signaling is position dependent. During this period, TFAP2C is enriched in the polar outside cells and downregulated in the inside apolar cells. In the polar outside cells, AMOT is sequestered at the apical domain by F-actin preventing it from activating LATS1/2. Consequently, YAP can enter the nucleus and activates Cdx2 expression via TEAD4 and TFAP2C. Sox2 is repressed via YAP1 and TEAD4. In the apolar inside cells, AMOT becomes phosphorylated and is associated with adherens junctions. There it forms a complex with NF2 and LATS1/2. Activated LATS1/2 subsequently phosphorylates YAP1 causing it to be degraded in the cytoplasm. As a result, Cdx2 is repressed and Sox2 is expressed allowing ICM lineage development.
Conclusion
In conclusion, the Hippo signaling pathway is essential for lineage formation in mouse preimplantation embryos. Disruption of Hippo signaling, or its downstream lineage transcription factors, results in misspecification of the ICM and TE lineages resulting in either pre- or post-implantation embryo arrest. There are many things we do not fully understand with regard to how Hippo signaling is regulated and how lineage transcription factors such as TFAP2C negatively and/or positively regulate the pathway. Furthermore, the clinical relevance of the Hippo signaling pathway in human preimplantation development and early human pregnancy is not fully established. Interestingly, YAP1 is expressed in the human placenta and its levels are downregulated in patients with preeclampsia (Sun et al., 2018), indicating that Hippo signaling may be important for human trophoblast lineage development. In support of this notion, a recent study using human cell models revealed that YAP1 and TEAD4 promote self-renewal of cytotrophoblast progenitor cells, while inhibiting formation of differentiated syncytiotrophoblast cells (Meinhardt et al., 2020). Based on the function of the Hippo signaling pathway in mouse preimplantation embryos and embryos from other large animal species such as cattle and pigs (Negron-Perez and Hansen, 2018; Cao et al., 2019; Emura et al., 2020; Sharma and Madan, 2020), it is easy to postulate that Hippo signaling plays a fundamental role in human preimplantation embryo development and early lineage formation.
Acknowledgements
We thank Ms. Catherine A. Wilson for critically reading our manuscript.
Authors’ roles
J.G.K., C.K. and M.A. wrote the manuscript. J.G.K., C.K. and M.A. created the figures. J.G.K. M.A., C.K. and C.S.D. edited and approved the final version.
Funding
This work was supported by a grant from the National Institute of Child Health and Development of the National Institutes of Health (HD095371 to J.G.K.), MSU AgBio Research, and a T32 fellowship (T32HD087166 to M.A.). The funding agencies had no role in writing of the manuscript or in the decision to submit the paper for publication.
Conflict of interest
None declared.
References
- Alarcon VB. Cell polarity regulator PARD6B is essential for trophectoderm formation in the preimplantation mouse embryo. Biol Reprod 2010;83:347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon VB, Marikawa Y.. ROCK and RHO playlist for preimplantation development: streaming to HIPPO pathway and apicobasal polarity in the first cell differentiation. Adv Anat Embryol Cell Biol 2018;229:47–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano M, Nakayama M, Kaibuchi K.. Rho-kinase/ROCK: a key regulator of the cytoskeleton and cell polarity. Cytoskeleton (Hoboken) 2010;67:545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anani S, Bhat S, Honma-Yamanaka N, Krawchuk D, Yamanaka Y.. Initiation of Hippo signaling is linked to polarity rather than to cell position in the pre-implantation mouse embryo. Development 2014;141:2813–2824. [DOI] [PubMed] [Google Scholar]
- Auman HJ, Nottoli T, Lakiza O, Winger Q, Donaldson S, Williams T.. Transcription factor AP-2gamma is essential in the extra-embryonic lineages for early postimplantation development. Development 2002;129:2733–2747. [DOI] [PubMed] [Google Scholar]
- Barcroft LC, Offenberg H, Thomsen P, Watson AJ.. Aquaporin proteins in murine trophectoderm mediate transepithelial water movements during cavitation. Dev Biol 2003;256:342–354. [DOI] [PubMed] [Google Scholar]
- Basu-Roy U, Bayin NS, Rattanakorn K, Han E, Placantonakis DG, Mansukhani A, Basilico C.. SOX2 antagonizes the Hippo pathway to maintain stemness in cancer cells. Nat Commun 2015;6:6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett FC, Harvey KF.. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr Biol 2006;16:2101–2110. [DOI] [PubMed] [Google Scholar]
- Bessonnard S, De Mot L, Gonze D, Barriol M, Dennis C, Goldbeter A, Dupont G, Chazaud C.. Gata6, Nanog and Erk signaling control cell fate in the inner cell mass through a tristable regulatory network. Development 2014;141:3637–3648. [DOI] [PubMed] [Google Scholar]
- Brown JJ, Whittingham DG.. The roles of pyruvate, lactate and glucose during preimplantation development of embryos from F1 hybrid mice in vitro. Development 1991;112:99–105. [DOI] [PubMed] [Google Scholar]
- Cao Z, Carey TS, Ganguly A, Wilson CA, Paul S, Knott JG.. Transcription factor AP-2gamma induces early Cdx2 expression and represses HIPPO signaling to specify the trophectoderm lineage. Development 2015;142:1606–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Xu T, Tong X, Wang Y, Zhang D, Gao D, Zhang L, Ning W, Qi X, Ma Y. et al. Maternal yes-associated protein participates in porcine blastocyst development via modulation of trophectoderm epithelium barrier function. Cells 2019;8:1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaiworapongsa T, Chaemsaithong P, Yeo L, Romero R.. Pre-eclampsia part 1: current understanding of its pathophysiology. Nat Rev Nephrol 2014;10:466–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Yabuuchi A, Eminli S, Takeuchi A, Lu CW, Hochedlinger K, Daley GQ.. Cross-regulation of the Nanog and Cdx2 promoters. Cell Res 2009;19:1052–1061. [DOI] [PubMed] [Google Scholar]
- Chi F, Sharpley MS, Nagaraj R, Roy SS, Banerjee U.. Glycolysis-independent glucose metabolism distinguishes TE from ICM fate during mammalian embryogenesis. Dev Cell 2020;53:9–26.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi I, Carey TS, Wilson CA, Knott JG.. Transcription factor AP-2gamma is a core regulator of tight junction biogenesis and cavity formation during mouse early embryogenesis. Development 2012;139:4623–4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockburn K, Rossant J.. Making the blastocyst: lessons from the mouse. J Clin Invest 2010;120:995–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dard N, Le T, Maro B, Louvet-Vallee S.. Inactivation of aPKClambda reveals a context dependent allocation of cell lineages in preimplantation mouse embryos. PLoS One 2009. a;4:e7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dard N, Louvet-Vallee S, Maro B.. Orientation of mitotic spindles during the 8- to 16-cell stage transition in mouse embryos. PLoS One 2009. b;4:e8171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries WN, Evsikov AV, Haac BE, Fancher KS, Holbrook AE, Kemler R, Solter D, Knowles BB.. Maternal beta-catenin and E-cadherin in mouse development. Development 2004;131:4435–4445. [DOI] [PubMed] [Google Scholar]
- Dietrich JE, Hiiragi T.. Stochastic patterning in the mouse pre-implantation embryo. Development 2007;134:4219–4231. [DOI] [PubMed] [Google Scholar]
- Ducibella T, Ukena T, Karnovsky M, Anderson E.. Changes in cell surface and cortical cytoplasmic organization during early embryogenesis in the preimplantation mouse embryo. J Cell Biol 1977;74:153–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert JJ, Fleming TP.. Tight junction biogenesis during early development. Biochim Biophys Acta 2008;1778:717–728. [DOI] [PubMed] [Google Scholar]
- Emura N, Saito Y, Miura R, Sawai K.. Effect of downregulating the Hippo pathway members YAP1 and LATS2 transcripts on early development and gene expression involved in differentiation in porcine embryos. Cell Reprogram 2020;22:62–70. [DOI] [PubMed] [Google Scholar]
- Faye-Petersen OM. The placenta in preterm birth. J Clin Pathol 2008;61:1261–1275. [DOI] [PubMed] [Google Scholar]
- Ferrer-Vaquer A, Hadjantonakis AK.. Birth defects associated with perturbations in preimplantation, gastrulation, and axis extension: from conjoined twinning to caudal dysgenesis. Wiley Interdiscip Rev Dev Biol 2013;2:427–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frum T,, Murphy TM, Ralston A.. HIPPO signaling resolves embryonic cell fate conflicts during establishment of pluripotency in vivo. Elife 2018;7:e42298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frum T, Watts JL, Ralston A.. TEAD4, YAP1 and WWTR1 prevent the premature onset of pluripotency prior to the 16-cell stage. Development 2019;146:dev179861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurtier V, Owens N, Gonzalez I, Mueller F, Proux C, Mornico D, Clerc P, Dubois A, Navarro P.. The molecular logic of Nanog-induced self-renewal in mouse embryonic stem cells. Nat Commun 2019;10:1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirate Y, Hirahara S, Inoue K, Kiyonari H, Niwa H, Sasaki H.. Par-aPKC-dependent and -independent mechanisms cooperatively control cell polarity, Hippo signaling, and cell positioning in 16-cell stage mouse embryos. Dev Growth Differ 2015;57:544–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirate Y, Hirahara S, Inoue K, Suzuki A, Alarcon VB, Akimoto K, Hirai T, Hara T, Adachi M, Chida K. et al. Polarity-dependent distribution of angiomotin localizes Hippo signaling in preimplantation embryos. Curr Biol 2013;23:1181–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirate Y, Sasaki H.. The role of angiomotin phosphorylation in the Hippo pathway during preimplantation mouse development. Tissue Barriers 2014;2:e28127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Home P, Ray S, Dutta D, Bronshteyn I, Larson M, Paul S.. GATA3 is selectively expressed in the trophectoderm of peri-implantation embryo and directly regulates Cdx2 gene expression. J Biol Chem 2009;284:28729–28737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Home P, Saha B, Ray S, Dutta D, Gunewardena S, Yoo B, Pal A, Vivian JL, Larson M, Petroff M. et al. Altered subcellular localization of transcription factor TEAD4 regulates first mammalian cell lineage commitment. Proc Natl Acad Sci USA 2012;109:7362–7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D.. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 2005;122:421–434. [DOI] [PubMed] [Google Scholar]
- Johnson MH, McConnell JM.. Lineage allocation and cell polarity during mouse embryogenesis. Semin Cell Dev Biol 2004;15:583–597. [DOI] [PubMed] [Google Scholar]
- Julian L, Olson MF.. Rho-associated coiled-coil containing kinases (ROCK): structure, regulation, and functions. Small GTPases 2014;5:e29846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ.. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev 1995;9:534–546. [DOI] [PubMed] [Google Scholar]
- Katsuno T, Umeda K, Matsui T, Hata M, Tamura A, Itoh M, Takeuchi K, Fujimori T, Nabeshima Y, Noda T. et al. Deficiency of zonula occludens-1 causes embryonic lethal phenotype associated with defected yolk sac angiogenesis and apoptosis of embryonic cells. Mol Biol Cell 2008;19:2465–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keramari M, Razavi J, Ingman KA, Patsch C, Edenhofer F, Ward CM, Kimber SJ.. SOX2 is essential for formation of trophectoderm in the preimplantation embryo. PLoS One 2010;5:e13952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Jho EH.. The history and regulatory mechanism of the Hippo pathway. BMB Rep 2018;51:106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott JG, Paul S.. Transcriptional regulators of the trophoblast lineage in mammals with hemochorial placentation. Reproduction 2014;148:R121–R136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono K, Tamashiro DA, Alarcon VB.. Inhibition of RHO-ROCK signaling enhances ICM and suppresses TE characteristics through activation of Hippo signaling in the mouse blastocyst. Dev Biol 2014;394:142–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuckenberg P, Buhl S, Woynecki T, van Furden B, Tolkunova E, Seiffe F, Moser M, Tomilin A, Winterhager E, Schorle H.. The transcription factor TCFAP2C/AP-2gamma cooperates with CDX2 to maintain trophectoderm formation. Mol Cell Biol 2010;30:3310–3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai ZC, Wei X, Shimizu T, Ramos E, Rohrbaugh M, Nikolaidis N, Ho LL, Li Y.. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell 2005;120:675–685. [DOI] [PubMed] [Google Scholar]
- LaJeunesse DR, McCartney BM, Fehon RG.. Structural analysis of Drosophila merlin reveals functional domains important for growth control and subcellular localization. J Cell Biol 1998;141:1589–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larue L, Ohsugi M, Hirchenhain J, Kemler R.. E-cadherin null mutant embryos fail to form a trophectoderm epithelium. Proc Natl Acad Sci USA 1994;91:8263–8267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham KE, Schultz RM.. Embryonic genome activation. Front Biosci 2001;6:D748–D759. [DOI] [PubMed] [Google Scholar]
- Leppens-Luisier G, Sakkas D.. Development, glycolytic activity, and viability of preimplantation mouse embryos subjected to different periods of glucose starvation. Biol Reprod 1997;56:589–596. [DOI] [PubMed] [Google Scholar]
- Leung CY, Zernicka-Goetz M.. Angiomotin prevents pluripotent lineage differentiation in mouse embryos via Hippo pathway-dependent and -independent mechanisms. Nat Commun 2013;4:2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Chittenden Y, Huang B, Shim JS, Chen Q, Lee SJ, Anders RA, Liu JO, Pan D.. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev 2012;26:1300–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorthongpanich C, Messerschmidt DM, Chan SW, Hong W, Knowles BB, Solter D.. Temporal reduction of LATS kinases in the early preimplantation embryo prevents ICM lineage differentiation. Genes Dev 2013;27:1441–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan P, Rose K, Watson AJ.. Na/K-ATPase beta1 subunit expression is required for blastocyst formation and normal assembly of trophectoderm tight junction-associated proteins. J Biol Chem 2007;282:12127–12134. [DOI] [PubMed] [Google Scholar]
- Meinhardt G, Haider S, Kunihs V, Saleh L, Pollheimer J, Fiala C, Hetey S, Feher Z, Szilagyi A, Than NG. et al. Pivotal role of the transcriptional co-activator YAP in trophoblast stemness of the developing human placenta. Proc Natl Acad Sci USA 2020;117:13562–13570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menchero S, Rollan I, Lopez-Izquierdo A, Andreu MJ, Sainz de Aja J, Kang M, Adan J, Benedito R, Rayon T, Hadjantonakis AK. et al. Transitions in cell potency during early mouse development are driven by Notch. Elife 2019;8:e42930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihajlovic AI, Bruce AW.. Rho-associated protein kinase regulates subcellular localisation of Angiomotin and Hippo-signalling during preimplantation mouse embryo development. Reprod Biomed Online 2016;33:381–390. [DOI] [PubMed] [Google Scholar]
- Miller A, Hendrich B.. Chromatin remodelling proteins and cell fate decisions in mammalian preimplantation development. Adv Anat Embryol Cell Biol 2018;229:3–14. [DOI] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S.. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 2003;113:631–642. [DOI] [PubMed] [Google Scholar]
- Moriwaki K, Tsukita S, Furuse M.. Tight junctions containing claudin 4 and 6 are essential for blastocyst formation in preimplantation mouse embryos. Dev Biol 2007;312:509–522. [DOI] [PubMed] [Google Scholar]
- Mulas C, Chia G, Jones KA, Hodgson AC, Stirparo GG, Nichols J.. OCT4 regulates the embryonic axis and coordinates exit from pluripotency and germ layer specification in the mouse embryo. Development 2018;145:dev159103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negron-Perez VM, Hansen PJ.. Role of yes-associated protein 1, angiomotin, and mitogen-activated kinase kinase 1/2 in development of the bovine blastocyst. Biol Reprod 2018;98:170–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A.. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor OCT4. Cell 1998;95:379–391. [DOI] [PubMed] [Google Scholar]
- Nishioka N, Inoue K, Adachi K, Kiyonari H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki N. et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell 2009;16:398–410. [DOI] [PubMed] [Google Scholar]
- Nishioka N, Yamamoto S, Kiyonari H, Sato H, Sawada A, Ota M, Nakao K, Sasaki H.. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech Dev 2008;125:270–283. [DOI] [PubMed] [Google Scholar]
- Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J.. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 2005;123:917–929. [DOI] [PubMed] [Google Scholar]
- Norwitz ER. Defective implantation and placentation: laying the blueprint for pregnancy complications. Reprod Biomed Online 2007;14:101–109. [DOI] [PubMed] [Google Scholar]
- Palmieri SL, Peter W, Hess H, Scholer HR.. Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev Biol 1994;166:259–267. [DOI] [PubMed] [Google Scholar]
- Pan D. The hippo signaling pathway in development and cancer. Dev Cell 2010;19:491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Knott JG.. Epigenetic control of cell fate in mouse blastocysts: the role of covalent histone modifications and chromatin remodeling. Mol Reprod Dev 2014;81:171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston A, Cox BJ, Nishioka N, Sasaki H, Chea E, Rugg-Gunn P, Guo G, Robson P, Draper JS, Rossant J.. GATA3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development 2010;137:395–403. [DOI] [PubMed] [Google Scholar]
- Saha B, Home P, Ray S, Larson M, Paul A, Rajendran G, Behr B, Paul S.. EED and KDM6B coordinate the first mammalian cell lineage commitment to ensure embryo implantation. Mol Cell Biol 2013;33:2691–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz RM. The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum Reprod Update 2002;8:323–331. [DOI] [PubMed] [Google Scholar]
- Sharma J, Madan P.. Characterisation of the Hippo signalling pathway during bovine preimplantation embryo development. Reprod Fertil Dev 2020;32:392–401. [DOI] [PubMed] [Google Scholar]
- Sheth B, Nowak RL, Anderson R, Kwong WY, Papenbrock T, Fleming TP.. Tight junction protein ZO-2 expression and relative function of ZO-1 and ZO-2 during mouse blastocyst formation. Exp Cell Res 2008;314:3356–3368. [DOI] [PubMed] [Google Scholar]
- Shi X, Yin Z, Ling B, Wang L, Liu C, Ruan X, Zhang W, Chen L.. Rho differentially regulates the Hippo pathway by modulating the interaction between Amot and Nf2 in the blastocyst. Development 2017;144:3957–3967. [DOI] [PubMed] [Google Scholar]
- Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J.. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 2005;132:2093–2102. [DOI] [PubMed] [Google Scholar]
- Sun M, Na Q, Huang L, Song G, Jin F, Li Y, Hou Y, Kang D, Qiao C.. YAP Is Decreased in Preeclampsia and Regulates Invasion and Apoptosis of HTR-8/SVneo. Reprod Sci 2018;25:1382–1393. [DOI] [PubMed] [Google Scholar]
- Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber D, Hariharan IK.. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 2002;110:467–478. [DOI] [PubMed] [Google Scholar]
- Thomson M, Liu SJ, Zou LN, Smith Z, Meissner A, Ramanathan S.. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell 2011;145:875–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G.. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol 2003;5:914–920. [DOI] [PubMed] [Google Scholar]
- Vinot S, Le T, Ohno S, Pawson T, Maro B, Louvet-Vallée S.. Asymmetric distribution of PAR proteins in the mouse embryo begins at the 8-cell stage during compaction. Dev Biol 2005;282:307–319. [DOI] [PubMed] [Google Scholar]
- Wang K, Sengupta S, Magnani L, Wilson CA, Henry RW, Knott JG.. Brg1 is required for Cdx2-mediated repression of OCT4 expression in mouse blastocysts. PLoS One 2010;5:e10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Sun D, Tai J, Chen S, Yu M, Ren D, Wang L.. TFAP2C promotes stemness and chemotherapeutic resistance in colorectal cancer via inactivating hippo signaling pathway. J Exp Clin Cancer Res 2018;37:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werling U, Schorle H.. Transcription factor gene AP-2 gamma essential for early murine development. Mol Cell Biol 2002;22:3149–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicklow E, Blij S, Frum T, Hirate Y, Lang RA, Sasaki H, Ralston A.. HIPPO pathway members restrict SOX2 to the inner cell mass where it promotes ICM fates in the mouse blastocyst. PLoS Genet 2014;10:e1004618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winger Q, Huang J, Auman HJ, Lewandoski M, Williams T.. Analysis of transcription factor AP-2 expression and function during mouse preimplantation development. Biol Reprod 2006;75:324–333. [DOI] [PubMed] [Google Scholar]
- Wu S, Liu Y, Zheng Y, Dong J, Pan D.. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell 2008;14:388–398. [DOI] [PubMed] [Google Scholar]
- Xu T, Wang W, Zhang S, Stewart RA, Yu W.. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development 1995;121:1053–1063. [DOI] [PubMed] [Google Scholar]
- Yagi R, Kohn MJ, Karavanova I, Kaneko KJ, Vullhorst D, DePamphilis ML, Buonanno A.. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development 2007;134:3827–3836. [DOI] [PubMed] [Google Scholar]
- Yuan P, Han J, Guo G, Orlov YL, Huss M, Loh YH, Yaw LP, Robson P, Lim B, Ng HH.. Eset partners with OCT4 to restrict extraembryonic trophoblast lineage potential in embryonic stem cells. Genes Dev 2009;23:2507–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Lu Q, Wang LH, Liu CY, Lei Q, Guan KL.. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev 2011;25:51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Wang P, Handford CE, Na J, Zernicka-Goetz M.. Transcriptional control of apical protein clustering drives de novo cell polarity establishment in the early mouse embryo. bioRxiv 2020:2020.2002.2010.942201. [Google Scholar]