Cancer-associated gene isoforms, arising from aberrant RNA splicing and/or processing, can play a functional role in tumor pathogenesis1 and are attractive as biomarkers and targets for cancer therapy. To date, the prevalence and significance of such alternative transcript isoforms in esophageal adenocarcinoma (EAC), an increasingly prevalent and lethal malignancy,2 remain unknown. Here, using an agnostic genome-scale approach, we sought to identify and characterize aberrant cancer-associated transcript-variants in EAC.
Whole transcriptome sequencing (RNAseq) was performed on a discovery sample set of 49 treatment-naive EAC and 40 normal/premalignant fresh-frozen biopsy tissues (Supplementary Table 1 and Supplementary Methods), followed by de novo transcriptome analysis to specifically identify novel/unannotated gene transcript-variants primarily induced in EACs but not in normal/premalignant tissues. Following stringent and orthogonal evaluation using transcript-variant specific polymerase chain reaction (PCR) in respective primary EAC tumors, we identified 7 novel candidate EAC-associated transcript-variants (Supplementary Figure 1, Supplementary Table 2). Together, the 7 candidate transcript-variants accounted for 71% of EACs tested, with each of the transcript-variants being induced in 10%–30% of EACs in the RNAseq discovery cohort.
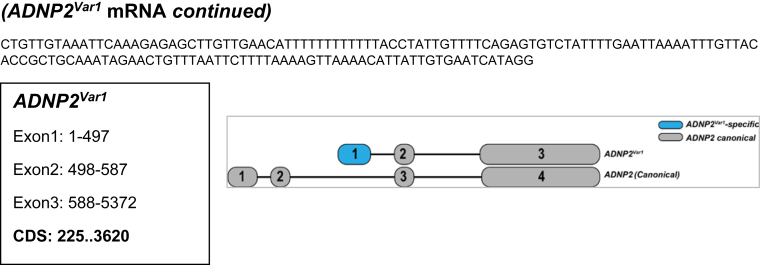
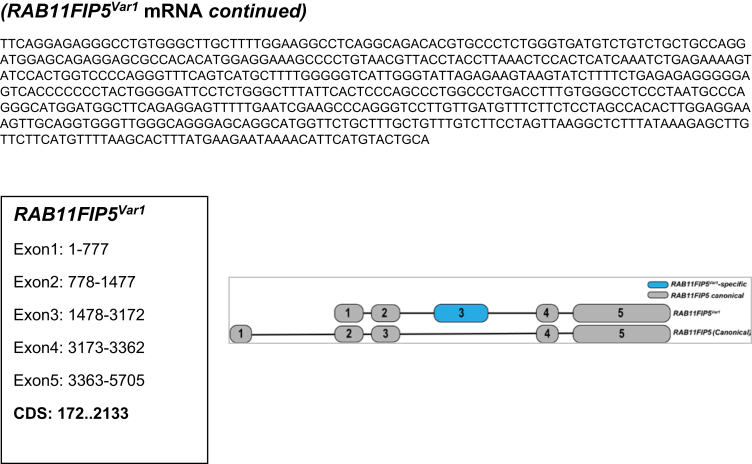
Supplementary Figure 1.
Full-length structure of novel transcript-variants identified in EACs. Shown are the complete mRNA sequences (5′ to 3′) of the respective candidate transcript-variants discovered in EACs. For each of the 7 candidates, variant-specific sequences are highlighted in blue font. Shown below each of the sequences are positions of individual exons and coding sequence. For each of the variants and their corresponding canonical genes, exon-intron structures along with their relative sizes-distances are illustrated on the right.
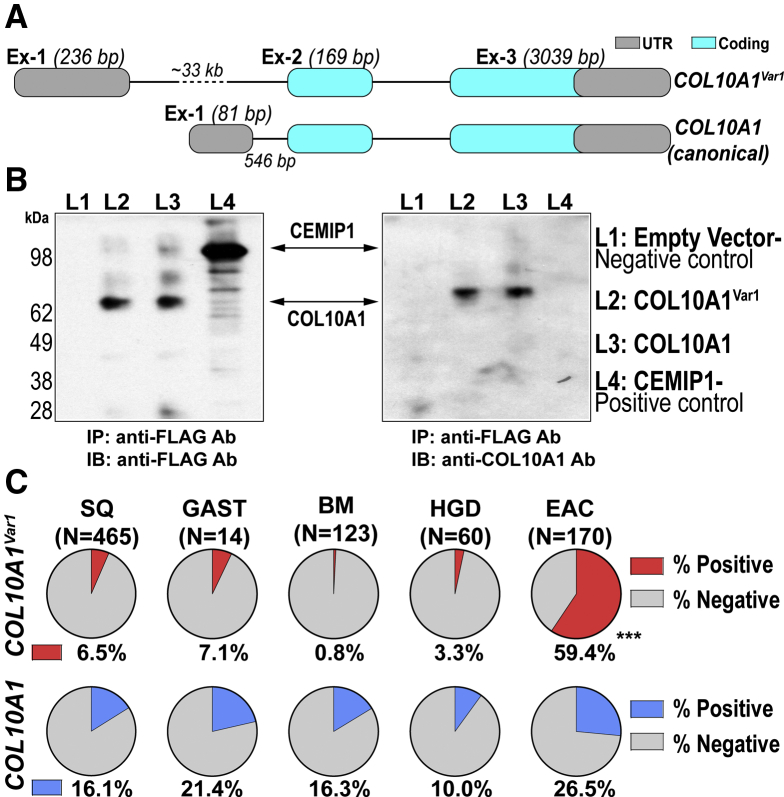
We subsequently prioritized a novel transcript-variant of the collagen X alpha 1 chain precursor (COL10A1) gene for further studies, on the basis of the recognized pro-tumorigenic role of COL10A1 pathway network in other tumor contexts.3, 4, 5, 6, 7, 8 Using bidirectional rapid amplification of cDNA ends (RACE) analysis, we first characterized the full-length transcript structure of this novel COL10A1-variant, hereafter referred to as COL10A1Var1 (deposited in GenBank: MN308081). COL10A1Var1 is a 3-exon transcript (3444 base pairs [bp]), containing a longer and distinct 5′ exon compared with the canonical (NM_000493.4) transcript (Figure 1A, Supplementary Figure 1). In silico analyses (NCBI ORFfinder) predicted COL10A1Var1 to encode for a ∼66 kDa (680 aa) protein, identical in size to the secreted canonical COL10A1 protein, which we confirmed by using orthogonal immunoprecipitation and Western blot analyses upon transfecting HEK293T cells with full-length COL10A1Var1 transcript (3444 bp), or the coding sequence of canonical COL10A1 transcript (Figure 1B).
Figure 1.
Characterization of COL10A1Var1. (A) Shown are the 5′ to 3′ exon (Ex)-introns (thin line) structures of COL10A1Var1 and canonical COL10A1. UTR, untranslated region. (B) Western blot analyses depicting COL10A1Var1 and COL10A1 proteins. IB, immunoblotting; IP, immunoprecipitation. CEMIP1 was used as positive control for secreted protein and Empty vector as a negative control. (C) Pie charts demonstrating the proportion (%) of samples positive for COL10A1Var1 transcript (top, red color) or canonical COL10A1 (bottom, blue color) in respective SQ, GAST, BM, HGD, and malignant (EAC) tissue biopsies. ∗∗∗P< .0001 indicates significant difference in the proportion COL10A1Var1 positivity between malignant (EAC) vs any of the respective non-EAC tissue groups, estimated by using a one-tailed Fisher exact test.
Using a robust quantitative real-time PCR (qPCR) assay that specifically detects COL10A1Var1 but not the canonical transcript, we next evaluated the generality and frequency of COL10A1Var1 expression in a validation cohort (N = 832) consisting of treatment-naive EAC (N = 170), Barrett’s metaplasia (BM) (N = 123), Barrett’s with high grade dysplasia (HGD) (N = 60), normal esophageal squamous (SQ) (N = 465), and normal gastric (GAST) (N = 14) biopsy tissues (Supplementary Table 1). Our orthogonal analysis demonstrated COL10A1Var1 to be robustly induced in the majority (∼60%) of EACs (Figure 1C, Supplementary Table 3). In striking contrast to EAC, only a minority of BM, HGD, SQ, and GAST samples tested positive for COL10A1Var1 (Fisher exact test, P < .0001; Figure 1C, Supplementary Table 3). We also note that COL10A1Var1 is a more frequently detected isoform in EACs, as compared with the canonical COL10A1 transcript that was detected in approximately one-fourth of EAC samples with no marked differences between EAC and normal/premalignant tissues (Figure 1C, Supplementary Table 3). Taken together, these findings strongly point to COL10A1Var1 as a recurrently induced transcript-variant in advanced stages of EAC development.
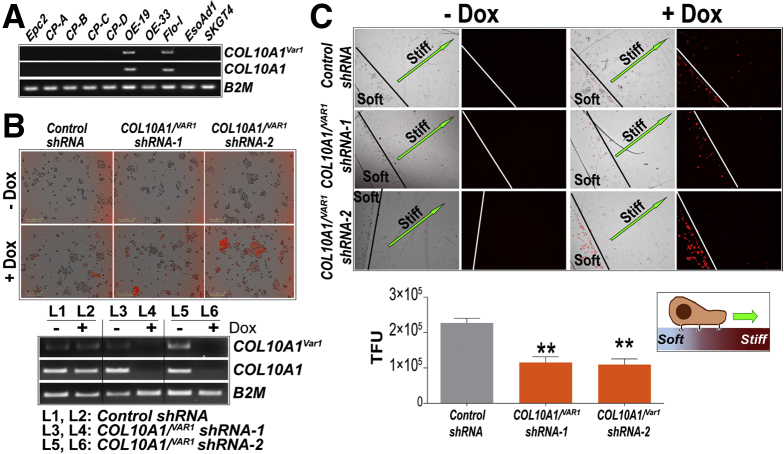
Because fibrillary protein networks (collagen, elastin) and glycoproteins (fibronectin) play a vital role in facilitating migration and invasion of cancer cells,9 we next evaluated the impact of COL10A1Var1 knockdown on the migratory potential of EAC cells in a durotaxis10 assay. We note that the EAC cell lines positive for COL10A1Var1 also expressed canonical COL10A1 transcript (Figure 2A), and repeated attempts to specifically knockdown COL10A1Var1 with custom short hairpin RNAs (shRNAs) proved technically unsuccessful. Nonetheless, because both COL10A1Var1 and canonical COL10A1 transcripts code for identical protein (Figure 1B) and consequently may exhibit similar function, as an alternative approach we used well-characterized COL10A1 shRNAs that also target COL10A1Var1 for subsequent studies. OE19 EAC cells (Figure 2A), stably expressing control or COL10A1 shRNAs under the control of doxycycline (Figure 2B), were seeded onto one-half of a glass coverslip coated with fibronectin alone (representing soft surface). Migration (durotaxis) of cells from the soft surface to an adjacent fibronectin-coated hydrogel (stiffer, 12 kPa) surface was monitored over time in the presence of doxycycline. Loss of COL10A1Var1/COL10A1 indeed significantly impeded the durotactic ability of EAC cells (P < .004) (Figure 2C), suggesting COL10A1 isoforms as potential regulators of mechanosensing ability of EAC cells.
Figure 2.
Impact of COL10A1/Var1on durotaxis of EAC cells. (A) PCR-based analysis showing COL10A1Var1 and canonical COL10A1 expression in normal esophageal squamous (Epc2), non-dysplastic BE (CP-A), dysplastic BE (CP-B, CP-C, CP-D), and EAC (OE19, OE33, FLO-1, EsoAd1, SKGT4) cell lines. B2M was used as the internal RNA control. BE, Barrett’s esophagus. (B) Representative images (left) demonstrating shRNA induction on doxycycline (Dox) treatment in stable OE19 cells, carrying either non-targeting control shRNA or shRNAs targeting both COL10A1Var1 and canonical COL10A1 transcripts (depicted as COL10A1/Var1). Note the specific induction of TurboRFP, a red fluorescent reporter of shRNA induction, on doxycyline treatment in these cells. PCR analysis (right) demonstrating knockdown of COL10A1/Var1 RNA on doxycycline treatment of the stable OE19 cells. B2M was used as an internal RNA control. (C) Representative images of durotaxis assay in stable OE19 cells. Quantitative analysis of cell migration (bar graph), measured as total fluorescence units (TFU, Y-axis) of TurboRFP-positive cells in the stiffer surface. All data are plotted as mean ± standard error of the mean, obtained from 3 replicate experiments. ∗∗P < .004 indicates significant differences in COL10A1/Var1 knockdown vs control shRNA cells, estimated by using a Student t test assuming unequal variances.
Taken in toto, we identify COL10A1Var1 as a novel and recurrent EAC-associated transcript-variant with a potential pro-tumorigenic function. On a broader scale, our study represents the first genome-wide analysis identifying novel transcript-variants induced in EAC. Further comprehensive studies are warranted to decipher the biologic role of the identified candidates and to evaluate their utility as biomarkers and therapeutic targets in this increasingly prevalent and lethal malignancy.
CRediT Authorship Contributions
Biswa Pratim Das Purkayastha (Formal analysis: Lead; Investigation: Lead; Writing – original draft: Lead; Writing – review & editing: Lead),
E. Ricky Chan (Data curation: Lead; Formal analysis: Lead; Software: Lead),
Durgadevi Ravillah (Formal analysis: Equal; Methodology: Equal),
Lakshmeswari Ravi (Methodology: Supporting),
Rajesh Gupta (Methodology: Supporting; Resources: Supporting),
Marcia I. Canto (Resources: Equal)
Jean S. Wang (Resources: Equal)
Nicholas J. Shaheen (Resources: Equal)
Joseph E. Willis (Resources: Equal)
Amitabh Chak (Data curation: Lead; Funding acquisition: Lead; Project administration: Equal; Resources: Lead; Supervision: Equal; Writing – review & editing: Equal),
Vinay Varadan (Data curation: Equal; Formal analysis: Lead; Funding acquisition: Supporting; Investigation: Equal; Methodology: Lead; Supervision: Equal; Writing – review & editing: Lead),
Kishore Guda (Conceptualization: Lead; Formal analysis: Lead; Funding acquisition: Lead; Investigation: Lead; Methodology: Lead; Supervision: Lead; Validation: Lead; Writing – original draft: Lead; Writing – review & editing: Equal)
Footnotes
Conflicts of interest This author discloses the following: V. Varadan is a consultant/advisory board member for Curis, Inc. The remaining authors disclose no conflicts.
Funding Supported by PHS awards: R01 CA204549 (K. Guda), U01 CA152756 (K. Guda), Case BETRNetU54 CA163060 (J. E. Willis, A. Chak, K. Guda), Case GI SPOREP50 CA150964 (J. E. Willis, A. Chak, K. Guda), K25 DK115904 (V. Varadan), P30 CA043703 (V. Varadan, K. Guda), K24 DK100548 (N. J. Shaheen), P30 DK034987 (N. J. Shaheen), P30 DK89502 (M. I. Canto), and by the DeGregorio Family Foundation, the Savone Family, and the Esophageal Cancer Awareness Association (K. Guda).
Supplementary Methods
Patient Samples
We compiled an in-house whole-transcriptome RNA sequencing (RNAseq) dataset previously generated by our group1,2 for discovery studies and an independent validation cohort (N = 832) consisting of treatment-naive malignant, premalignant, and nonmalignant biopsy tissues (Supplementary Table 1). All samples were accrued with informed consent under an institutional review board approved protocol (UHCMC IRB, #CC301) as previously described.2
Identification of Novel Transcript-Variants Using Whole-Transcriptome RNA Sequencing
Briefly, RNAseq reads that passed quality control were aligned to the human reference genome (GRCh37p13) using the STAR aligner v2.5.1. The resulting bam files were sorted for de novo transcriptome assembly using Cufflinks in addition to the Gencode transcriptome annotation for GRCh37 version 19 as a guide. The resulting final merged transcriptome assembly was compared with the reference annotation from Gencode using Cuffcompare. Of note, the detection of any transcript, including novel transcripts, in the de novo transcriptome assembly was required to be supported by a minimum of 10 paired-reads. Novel transcripts were further examined visually to confirm the presence of supporting reads spanning the novel junctions using the Integrative Genomics Viewer (IGV). Subsequent experimental validations of these junctions were performed by using PCR analysis with custom intron-spanning primer sets.
Quantitative Real-time Polymerase Chain Reaction
One microgram of total RNA was reverse-transcribed by using Superscript III First-Strand Synthesis (Life Technologies, Carlsbad, CA; #18080). Quantitative PCR analysis was performed by using iQ SYBR Green Supermix system (Bio-Rad Laboratories, Hercules, CA; #170-8887) with custom intron-spanning primer set for COL10A1Var1 (Supplementary Table 2) or commercially available primer set for canonical COL10A1 transcript (Qiagen, Hilden, Germany). B2M was used as an endogenous RNA control as previously described by our group.2 Each qPCR reaction was carried out in triplicate in a 25 μL volume for 50 cycles using a Bio-Rad CFX96 Real-Time PCR machine. Samples were designated positive for COL10A1 transcript isoforms using respective melt-curve signals in the qPCR assay. Representative qPCR products were further subjected to direct Sanger sequencing for additional confirmation of transcript isoforms. A negative sample indicates no signal in a 50-cycle qPCR assay.
Rapid Amplification of cDNA Ends
We obtained the full-length sequence of the novel transcript-variant, COL10A1Var1, through RACE in OE19 EAC cell line using the SMARTer RACE cDNA kit (Takara Bio, Kusatsu, Shiga, Japan; #634860). The RACE products were purified, cloned into TOPO TA vector (ThermoFisher Scientific, Waltham, MA), and subsequently confirmed by Sanger sequencing.
Cell Culture and Transfection
EAC and premalignant Barrett’s esophagus cell lines were cultured as previously described by our group.1,2 HEK293T cells were transfected with pcDNA3.1 vector containing either FLAG-tagged canonical COL10A1 ORF (GenScript USA Inc, Piscataway, NJ; #OHU18227D), full-length COL10A1Var1, empty vector (negative control), or with CEMIP 3 (positive control for secreted protein) using Lipofectamine 2000 (Life Technologies; #11668019).
Immunoprecipitation and Immunoblotting
Cell culture supernatants were transferred to an Amicon Ultra-4 10K filter column (Millipore, Burlington, MA; #UFC801024), concentrated by centrifugation at 4000g for 15 minutes, immunoadsorbed overnight at 4°C using anti-FLAG antibody conjugated agarose beads (Sigma-Aldrich, St. Louis, MO; #A2220), and washed with RIPA buffer (150 mmol/L NaCl, 25 mmol/L Tris [pH 7.4], 0.1% sodium dodecyl sulfate, 1% NP-40). The immunoprecipitated proteins were subjected to electrophoresis on 4%–12% polyacrylamide gel (Life Technologies; #0321) and transferred to Hybond–C Extra nitrocellulose membrane (GE Healthcare, Chicago, IL; #10600016). Membranes were blocked with 5% milk in TBST (0.05% Tween-20 in Tris buffered saline) and incubated overnight at 4°C with either horseradish peroxidase–conjugated anti-FLAG antibody (Cell Signaling Technology, Danvers, MA; #2044S) or anti-COL10A1 (Abcam, Cambridge, UK; #ab182563) primary antibody at 1:1000 dilution. For COL10A1, blots were incubated with anti-rabbit horseradish peroxidase secondary antibody (Cell Signaling Technology; #7074) at 1:5000 in 5% milk in TBST. Chemiluminescence was visualized by using ECL-Plus Western Blotting Detection Kit (GE Healthcare; #RPN2232).
Stable OE19 Cell Line Generation With Conditional COL10A1Var1 Knockdown
Doxycycline-regulated TurboRFP lentiviral vectors, containing nonoverlapping shRNAs targeting different regions of COL10A1/Var1 transcript (Dharmacon, Lafayette, CO; #V3SH11252-227571902, #V3SH11252-228435149) or non-targeting shRNA (Dharmacon; #VSC11655), were produced in HEK293T cells using standard procedures, and viral titers were analyzed by using a 24-gag ELISA kit (Takara; #632200). OE19 EAC cells were infected with the viral particles and treated with puromycin (500 ng/mL) for subsequent stable cell line generation. Induction of shRNAs on doxycyline (0.6 μg/mL) treatment was confirmed by TurboRFP signal under fluorescent microscope, and knockdown of COL10A1/Var1 was confirmed by qPCR with isoform-specific primers. At least 3 independently derived clones per shRNA were used for the study. These lentiviral-based shRNAs were used in the durotaxis assay as described below.
Durotaxis Assay
Durotaxis assay was performed following protocol of Wen et al4 with some modification. Cells were seeded onto one-half of 18 mm2 glass coverslip coated with fibronectin alone (representing soft surface), whereas the second-half of the glass coverslip contained a fibronectin-coated polyacrylamide hydrogel, representing the stiffer (12 kPa) surface. Briefly, the coverslip was functionalized by using 3-(trimethoxysilyl) propyl methacrylate (Millipore Sigma; #440159) to facilitate covalent attachment of hydrogel substrates to glass surface. A polymer solution containing acrylamide monomers (Millipore Sigma; #A7802), cross-linker N,N methylene-bis-acrylamide, ammonium persulfate (Millipore Sigma; #A3678), and N,N,N0,N0-tetramethylethylenediamine (TEMED) (Bio-Rad; #1610801) was prepared and allowed to polymerize on one-half of the glass coverslips. The 6.1% acrylamide was used to obtain the 12 kPa of hydrogel stiffness. The gels were sterilized through ultraviolet exposure for 2 × 30 minutes. To allow for cell adhesion and fibrous-protein tethering, substrates were incubated in 1 mmol/L N-sulphosuccinimidyl-6-(40-azido-20-nitrophenylamino) hexanoate (sulpho-SANPAH) (Millipore Sigma; #803332), activated with ultraviolet light exposure for 2 × 5 minutes, followed by 1× phosphate-buffered saline wash for 3 times. The entire glass coverslips were then incubated in fibronectin (ThermoFisher Scientific; #PHE0023) overnight, followed by normalization with cell culture medium for at least 2 hours. The 1 × 104 OE19 EAC cells, expressing COL10A1/Var1 shRNAs or control shRNA (see above), were seeded on one-half of the coverslip with the fibronectin-coated glass surface and allowed them to grow overnight. Subsequently, the cells were treated with 10% (Tet-free) fetal bovine serum supplemented culture media with or without doxycycline (0.6 μg/mL). Experiments were performed in triplicates, and the fluorescent signals were captured and measured over time with Keyence BZ-X800 (Osaka, Japan) fluorescence microscope and analyzed with the Keyence image analyzer.
Supplementary Table 1.
Discovery and Validation Sample Cohorts
| Discovery RNAseq samples | Number of samples | Median age at diagnosis, y (range) | Gender distribution | Cancer stage distribution |
|---|---|---|---|---|
| EACa | 49 | 65 (36 - 88) | 89% (male) 11% (female) | Stage I (17.9%), Stage II (19.6%), Stage III (46.4%), Stage IV (16.1%) |
| Nondysplastic stable Barrett's esophagusb | 18 | 56 (18-84) | 94% (male) 6% (female) | NA |
| Normal esophageal squamous (SQ)c | 11 | 64 (45-83) | 90% (male) 10% (female) | NA |
| Normal gastric (GAST) | 11 | 63 (36-82) | 82% (male) 18% (female) | NA |
| Total | 89 |
| Validation samples | Number of samples | Median age at diagnosis, y (range) | Gender distribution | Cancer stage distribution |
|---|---|---|---|---|
| EACd | 170 | 64 (34–89) | 77% (male) 15% (female) | Stage I (14.1%), Stage II (16.8%), Stage III (52.2%), Stage IV (15.0%) |
| Normal esophageal squamous (SQ) | 465 | 64 (34 –89) | 77% (male) 15% (female) | NA |
| Barrett's metaplasia (BM)e | 123 | 65.5 (36–93) | 71% (male) 34% (female) | NA |
| BM with high-grade dysplasia (HGD) | 60 | 66 (46–80) | 89% (male) 11% (female) | NA |
| Normal gastric (GAST) | 14 | 63 (36–82) | 85% (male) 15% (female) | NA |
| Total | 832 |
11% of EACs were gastroesophageal junctional adenocarcinomas.
Median surveillance of 9 years, ranging from 6 to 22 years.
Each of the 11 normal SQ samples was obtained from respective EAC patients included in the RNA sequencing.
13% of EACs were gastroesophageal junctional adenocarcinomas.
Clinical follow-up information unavailable (progression status unknown) for these patients.
Supplementary Table 2.
Candidate Novel Transcript-Variants
| Transcript_variant | CHR | Transcript_variant Genomic START (hg19) | Transcript_variant Genomic END (hg19) | Transcript_variant STRAND | Transcript_variant EXON NUMBER | Transcript_variant EXON SIZE (bp) | Transcript_variant LENGTH (bp) | Transcript_variant PREDICTED CDS START-STOP (bp) a | Transcript_variant PREDICTED PROTEIN LENGTH (AA)a | Transcript_variant– specific Forward_primer (5' to 3') | Transcript_variant-specific Reverse_primer (5' to 3') | PCR_product_size (bp) | Canonical_Gene symbol | Canonical_Gene ID | Canonical_Transcript NUCLEOTIDE ID | Canonical_Transcript LENGTH (bp) | Canonical_Transcript CDS_START-STOP (bp) | Canonical_Transcript PROTEIN ID | Canonical_Transcript PROTEIN LENGTH (AA) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| chr6 | 116440086 | 116443124 | – | 3 | 3038 | AGCAGCCAACAACAAGCATA | GTGGACCAGGAGTACCTTGC | 252 | |||||||||||
| COL10A1Var1 | chr6 | 116446502 | 116446670 | – | 2 | 168 | 3442 | 252..2294 | 680 | COL10A1 | 1300 | NM_000493 | 3302 | 96..2138 | NP_000484 | 680 | |||
| chr6 | 116479777 | 116480013 | – | 1 | 236 | ||||||||||||||
| chr6 | 39063820 | 39073552 | – | 3 | 9732 | CATCCTCCTTCCCACTACCA | TGCCATCATTACATGCACCT | 2241 | |||||||||||
| SAYSD1Var1 | chr6 | 39077090 | 39081496 | – | 2 | 4406 | 14523 | 4788..5138 | 116 | SAYSD1 | 55776 | NM_001304793 | 6425 | 4706..5056 | NP_001291722 | 116 | |||
| chr6 | 39082659 | 39083044 | – | 1 | 385 | ||||||||||||||
| chr5 | 68462688 | 68463110 | + | 1 | 422 | AGAGGCAGACCACGTGAGAG | GCTTAGGAGTTCTGTGGGACA | 1431 | |||||||||||
| chr5 | 68463735 | 68463905 | + | 2 | 170 | ||||||||||||||
| chr5 | 68464000 | 68464170 | + | 3 | 170 | ||||||||||||||
| CCNB1Var1 | chr5 | 68467097 | 68467279 | + | 4 | 182 | 1643 | 403..1512 | 369 | CCNB1 | 891 | NM_031966 | 2029 | 114..1415 | NP_114172 | 433 | |||
| chr5 | 68470078 | 68470236 | + | 5 | 158 | ||||||||||||||
| chr5 | 68470704 | 68470940 | + | 6 | 236 | ||||||||||||||
| chr5 | 68471224 | 68471529 | + | 7 | 305 | ||||||||||||||
| chr2 | 73300510 | 73302852 | – | 5 | 2342 | TTGGCTCTTCAGAGTCAGCA | GGTAGTACTTGGCCGACTGG | 778 | |||||||||||
| chr2 | 73303121 | 73303310 | – | 4 | 189 | ||||||||||||||
| RAB11FIP5Var1 | chr2 | 73306779 | 73308473 | – | 3 | 1694 | 5700 | 144..3566 | 1140 | RAB11FIP5 | 26056 | NM_015470 | 4272 | 172..2133 | NP_056285 | 653 | |||
| chr2 | 73315178 | 73315877 | – | 2 | 699 | ||||||||||||||
| chr2 | 73316007 | 73316783 | – | 1 | 776 | ||||||||||||||
| chr18 | 77889764 | 77890260 | + | 1 | 496 | CCATCAAAACTTGCTGAGAGC | GGCCACAACAGTATGGCTTT | 288 | |||||||||||
| ADNP2Var1 | chr18 | 77890986 | 77891075 | + | 2 | 89 | 5369 | 765..3785 | 1006 | ADNP2 | 22850 | NM_014913 | 5157 | 225..3620 | NP_055728 | 1131 | |||
| chr18 | 77893495 | 77898279 | + | 3 | 4784 | ||||||||||||||
| chr16 | 46989335 | 46989534 | – | 10 | 199 | GGATGCCGCAGTATCGTAAT | TTGTGGGGAAGTAACCTTGG | 503 | |||||||||||
| chr16 | 46990919 | 46991132 | – | 9 | 213 | ||||||||||||||
| chr16 | 46992915 | 46993042 | – | 8 | 127 | ||||||||||||||
| chr16 | 46993187 | 46993331 | – | 7 | 144 | ||||||||||||||
| DNAJA2Var1 | chr16 | 46998523 | 46998719 | – | 6 | 196 | 1857 | 407..1645 | 412 | DNAJA2 | 10294 | NM_005880 | 3008 | 103..1341 | NP_005871 | 412 | |||
| chr16 | 47001425 | 47001558 | – | 5 | 133 | ||||||||||||||
| chr16 | 47001996 | 47002076 | – | 4 | 80 | ||||||||||||||
| chr16 | 47005261 | 47005484 | – | 3 | 223 | ||||||||||||||
| chr16 | 47005808 | 47005867 | – | 2 | 59 | ||||||||||||||
| chr16 | 47007406 | 47007889 | – | 1 | 483 | ||||||||||||||
| chr14 | 54941202 | 54944877 | – | 3 | 3675 | TCCCCAGGTGTTGGTAAAT | GGTCTTCGGTATTTCTTATTTCAA | 250 | |||||||||||
| GMFBVar1 | chr14 | 54946504 | 54946577 | – | 2 | 73 | 3996 | 270..395 | 41 | GMFB | 2764 | NM_004124 | 4085 | 54..482 | NP_004115 | 142 | |||
| chr14 | 54947592 | 54947840 | – | 1 | 248 |
Putative candidate transcript–variant coding regions were predicted using NCBI ORF finder. Listed are only those predicted ORFs for transcript–variants that are in the same reading frame as respective canonical transcripts.
Supplementary Table 3.
Expression Status of COL10A1Var1 and Canonical COL10A1 Across Lesions
| EAC (N = 219)a |
||
|---|---|---|
| Canonical COL10A1-positive | Canonical COL10A1-negative | |
| COL10A1Var1-positive | 53 (24.2%) | 79 (36.07%) |
| COL10A1Var1-negative | 1 (0.46%) | 86 (39.27%) |
| NDBE (N = 141)a | ||
|---|---|---|
| Canonical COL10A1-positive | Canonical COL10A1-negative | |
| COL10A1Var1-positive | 0 (0%) | 2 (1.42%) |
| COL10A1Var1-negative | 22 (15.6%) | 117 (82.98%) |
| HGD (N = 60)a | ||
|---|---|---|
| Canonical COL10A1-positive | Canonical COL10A1-negative | |
| COL10A1Var1-positive | 1 (1.67%) | 1 (1.67%) |
| COL10A1Var1-negative | 5 (8.33%) | 53 (88.33%) |
| SQ (N = 476)a | ||
|---|---|---|
| Canonical COL10A1-positive | Canonical COL10A1-negative | |
| COL10A1Var1-positive | 9 (1.89%) | 21 (4.41%) |
| COL10A1Var1-negative | 67 (14.08%) | 379 (79.62%) |
| GAST (N= 25)a | ||
|---|---|---|
| Canonical COL10A1-positive | Canonical COL10A1-negative | |
| COL10A1Var1-positive | 0 (0%) | 1 (4%) |
| COL10A1Var1-negative | 9 (36%) | 15 (60%) |
NDBE, nondysplastic Barrett’s esophagus.
Number of samples combined from both Discovery and Validation cohorts.
References
- 1.Chen J. Oncogene. 2015;34:1–14. doi: 10.1038/onc.2013.570. [DOI] [PubMed] [Google Scholar]
- 2.Bray F. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Yuzhalin A.E. Br J Cancer. 2018;118:435–440. doi: 10.1038/bjc.2017.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sole X. PLoS One. 2014;9 [Google Scholar]
- 5.Huang H. Onco Targets Ther. 2018;11:1571–1581. doi: 10.2147/OTT.S160196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li T. Cell Death Dis. 2018;9:849. doi: 10.1038/s41419-018-0877-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naba A. BMC Cancer. 2014;14:518. doi: 10.1186/1471-2407-14-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brodsky A.S. BMC Cancer. 2016;16:274. doi: 10.1186/s12885-016-2302-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hynes R.O. Science. 2009;326:1216–1219. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartman C.D. Proc Natl Acad Sci U S A. 2016;113:11190–11195. doi: 10.1073/pnas.1611324113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplementary References
- 1.Blum A.E. Cancer Res. 2016;76:5628–5633. doi: 10.1158/0008-5472.CAN-16-0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blum A.E. Gastroenterology. 2019;156:1761–1774. doi: 10.1053/j.gastro.2019.01.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fink S.P. Oncotarget. 2015;6:30500–30515. doi: 10.18632/oncotarget.5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wen J.H. Nat Mater. 2014;13:979–987. doi: 10.1038/nmat4051. [DOI] [PMC free article] [PubMed] [Google Scholar]