Abstract
Transcriptional bursting is a major source of noise in gene expression. The telegraph model of gene expression, whereby transcription switches between on and off states, is the dominant model for bursting. Recently, it was shown that the telegraph model cannot explain a number of experimental observations from perturbation data. Here, we study an alternative model that is consistent with the data and which explicitly describes RNA polymerase recruitment and polymerase pause release, two steps necessary for messenger RNA (mRNA) production. We derive the exact steady-state distribution of mRNA numbers and an approximate steady-state distribution of protein numbers, which are given by generalized hypergeometric functions. The theory is used to calculate the relative sensitivity of the coefficient of variation of mRNA fluctuations for thousands of genes in mouse fibroblasts. This indicates that the size of fluctuations is mostly sensitive to the rate of burst initiation and the mRNA degradation rate. Furthermore, we show that 1) the time-dependent distribution of mRNA numbers is accurately approximated by a modified telegraph model with a Michaelis-Menten like dependence of the effective transcription rate on RNA polymerase abundance, and 2) the model predicts that if the polymerase recruitment rate is comparable or less than the pause release rate, then upon gene replication, the mean number of RNA per cell remains approximately constant. This gene dosage compensation property has been experimentally observed and cannot be explained by the telegraph model with constant rates.
Significance
The random nature of gene expression is well established experimentally. Mathematical modeling provides a means of understanding the factors leading to the observed stochasticity. There is evidence that the classical two-state model of stochastic messenger RNA (mRNA) dynamics (the telegraph model) cannot describe perturbation experiments, and a new model that includes polymerase dynamics has been proposed. In this article, we present the first detailed study of this model, deriving an exact solution for the mRNA distribution in steady-state conditions and an approximate time-dependent solution and showing that the model can explain gene dosage compensation. As well, we use the theory together with transcriptomic data to deduce which parameters when perturbed lead to a maximal change in the size of mRNA fluctuations.
Introduction
There is widespread evidence that mammalian genes are expressed in bursts: infrequent periods of transcriptional activity that produce a large number of messenger RNA (mRNA) transcripts within a short period of time (1, 2, 3). This is in contrast to constitutive expression in which mRNAs are produced in random, uncorrelated events, with a time-independent probability (4). The size and frequency of transcriptional bursts affect the magnitude of temporal fluctuations in mRNA and the protein content of a cell and thus constitute an important source of intracellular noise (5).
A large number of studies have sought to elucidate the mechanisms leading to bursting by constructing simple stochastic models that can explain the data. The simplest of these models is the telegraph model whereby 1) a gene is in two states, an ON state where mRNA is expressed, and an OFF state where there is no expression, and 2) mRNA degrades in the cytoplasm. These first-order reactions are effective because each encapsulates the effect of a large number of underlying biochemical reactions. The chemical master equation of this model has been solved exactly to obtain the probability distribution of mRNA numbers as a function of time (6). For parameter conditions consistent with bursty expression, the steady-state distribution is well approximated by a negative binomial that fits some of the experimental data (7).
Recent studies have extended the telegraph model in various directions (see (8,9) for a recent review). Mammalian cells have been shown to display complex promoter dynamics during the switch from transcriptionally inactive to active states. Such dynamics cannot be described by a single reaction step whose time is exponentially distributed (2), as assumed by the telegraph model. In (10), this complexity is accounted for by deriving analytical expressions linking the Fano factor of mRNA distributions to the general waiting-time distribution of the time to switch from inactive to active states. In contrast, other works (11, 12, 13) have sought to describe promoter dynamics with transitions between a number of discrete promoter states, only some of which are active; in special cases of such models, the steady-state distribution of mRNA fluctuations can be derived analytically. Moreover, dynamic regulation of eve stripe 2 expression in living Drosophila (14) suggests the occurrence of multiple rates of RNA polymerase II (Pol II) loading, which argues in favor of the multistate model rather than the simpler telegraph model. Another study, based on live cell imaging of the amoeba Dictyostelium, postulates a continuum of transcriptional states (15) rather than discrete states. All these models share a common property with the telegraph model, namely that when a transcript is produced, the gene state is unchanged.
Bartman et al. (16) recently argued that it is unclear how polymerase recruitment and pause release, two well-known steps in mRNA production, map onto the active and inactive states assumed by the telegraph model. This argument also applies to the various multistate variants of the telegraph model. In particular, in these models, one cannot tell whether the initiation of a burst permits polymerase recruitment to occur or whether it permits release from the paused state. In (16), the telegraph model and several possible models of transcription were considered that incorporated bursting (burst initiation and termination steps) together with polymerase recruitment and pause release steps. Using stochastic simulations in conjunction with RNA fluorescence in situ hybridization and Pol II chromatin immunoprecipitation sequencing measurements, they showed that the only model compatible with the data is one in which 1) polymerase recruitment follows after burst initiation, and 2) only one polymerase is permitted to bind each promoter-proximal region at a time, and this bound polymerase has to undergo pause release before a second polymerase can be recruited to a gene copy (in line with the findings in (17,18)). We emphasize that although this model has three effective gene states, it is not a special case of the multistate gene models studied in (11, 12, 13). These models assume that the gene state does not change upon production of mRNA because they model the production of a mature transcript without detailed modeling of the steps between transcriptional initiation and termination. However, the model expounded in (16) models transcription at a finer level of detail, which requires that the production of nascent mRNA results in a change of gene state, a property that is crucial to capture the second property above. Note the number of nascent mRNA molecules, irrespective of their length, is equal to the number of polymerases currently transcribing the gene (19). An interesting recent review discussing the assumptions behind common gene expression models including those with polymerase dynamics can be found in (20).
In this article, we present the first detailed study of the model proposed by Bartman et al. (16). The article is organized as follows. In Model, we introduce the chemical master equation formulation of the model. In Exact Solution, we obtain an exact steady-state solution of this model, and in Sensitivity Analysis, we use the theoretical results and transcriptomic data to investigate the sensitivity of the size of mRNA fluctuations to the five parameters. In Effective Telegraph Model, we show that by mapping the model onto an effective telegraph model, we can obtain an approximate time-dependent solution. In Connection to the Refractory Model, we show that although our model has three effective promoters states, it is not the same as the refractory model of gene expression devised by Naef and co-workers (2). In Protein Dynamics, we show that the protein number distribution can also be obtained in the limit of fast mRNA decay and that this is generally different than that obtained using the conventional three-stage model of gene expression (21). We finish with a discussion of the biological implications of our results in Conclusions.
Results and Discussion
Model
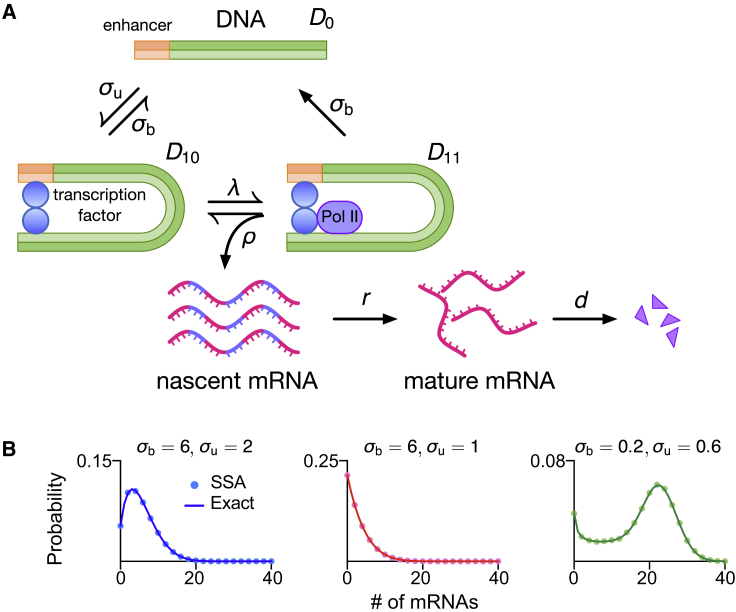
We consider a stochastic transcriptional bursting model (recently introduced in (16) and henceforth referred to as the multiscale model; see Fig. 1 A), whereby a gene fluctuates between three states: two permissive states (D10 and D11) and a nonpermissive state (D0).
Figure 1.
(A) Schematic of the stochastic multiscale transcriptional bursting model. (B) Analytical distribution for mature mRNA numbers (under the assumption of short-lived nascent mRNA) is given by Eq. 6 and agrees with stochastic simulations using the SSA. The kinetic parameters are ρ = 60, λ = 40, and d = 1; other parameters are indicated in each panel. To see this figure in color, go online.
The transition from D0 to D10 (burst initiation) is mediated by transcription factor binding with rate constant σu, which is reversible with rate constant σb (this transition may alternatively represent other processes such as nucleosome remodeling). Subsequently, the binding of Pol II to D10 with rate constant λ (which is proportional to Pol II abundance) leads to D11. This represents a state in which Pol II is paused and models the experimental observation that Pol II pauses downstream of the transcription initiation site preceding productive elongation (18). The polymerase is released from this state with rate constant ρ, leading to two simultaneous processes: 1) because now the polymerase can actively transcribe RNA, it implies the production of nascent mRNA (denoted as N) with rate ρ; and 2) the gene state changes from D11 to D10. This step models the experimental observation that unless the polymerase is unpaused, there is no binding of new Pol II (17,18). In the paused state D11, both the polymerase and the transcription factor can unbind from the gene and lead to the nonpermissive state D0 (burst termination). Both reversible switches operate at different timescales (hours versus minutes) with max{σb, σu} ≪ min {ρ, λ}, leading to multiscale transcriptional bursting (16,22). After termination, the nascent mRNA becomes a mature mRNA (denoted by M); this occurs with rate r. Subsequently, the mature mRNA decays with rate constant d. Note that we assume all reactions to be first order, characterized by exponentially distributed waiting times between successive reactions.
In what follows, for simplicity, we assume that the lifetime of nascent mRNA is very short, i.e., r is large, such that the reaction D11 → D10 + N, N → M can be approximated by the single reaction step D11 → D10 + M. In the next section, we derive the steady-state distribution of mature mRNA (simply called mRNA henceforth).
Exact solution
Let Pθ (n, t) (θ = 0, 10, 11) denote the probability of a cell being in state Dθ with n mRNAs at time t (arguments n and t are hereafter omitted for brevity). The dynamics of probability Pθ are described by the set of coupled master equations
| (1) |
where the step operator acts on a general function g(n) as (23). To solve Eq. 1, we use the generating function method and define for θ = 0, 10, 11 so that Eq. 1 can be recast as a set of coupled partial differential equations
| (2a) |
| (2b) |
| (2c) |
wherein the variable z is dropped for brevity. By setting z = 1 and the time derivatives to zero (considering steady-state conditions), we can deduce that the probability of being in the nonpermissive state D0 is G0(1) = σb/(σu + σb) and the probability of being in one of the two permissive states D10 or D11 is G10(1) + G11(1) = σu/(σu + σb).
To solve (2a), (2b), (2c) for G0(z), G10(z), and G11(z) in steady-state conditions, we set , solve G10 from Eq. 2c as a function of G11, and combine the yielded result to solve G0 from Eq. 2b as a function of G11 so that Eq. 2a consequently becomes a differential equation with G11 being the only variable
| (3) |
with u = z − 1, γ1 = σb + σu, and γ2 = ρ + λ + σb. By defining a new variable x = ρλu/d2, Eq. 3 can be further simplified to
which is in the canonical form of the differential equation for the generalized hypergeometric function
admitting the solution f(x) = C1F2(a1, b1, b2, x), with C being an integration constant. Hence, the solution for G11 is in terms of the generalized hypergeometric function
| (4) |
On the other hand, summing Eqs. 2a, 2b, and 2c and denoting , one can get , which together with Eq. 4 leads to
Note that in the last step, we made use of the general relation . The integration constant C2 is found to be 1 by using the normalization condition G(0) = 1. Hence, the exact solution for the generating function is
| (5) |
Hence, it follows that the marginal probability of finding n mRNAs in a cell is
| (6) |
where is the Pochhammer symbol. In Fig. 1 B, we show that distributions obtained from Eq. 6 as well as the corresponding modality (a phenotypic signature (24)) are indistinguishable from distributions produced using the stochastic simulation algorithm (SSA) (25). Note that here, we have solved for the mature mRNA distribution under the assumption that nascent mRNA is short lived. In cases in which this assumption is not physiologically meaningful and one is interested in the nascent mRNA distribution, then the latter is given by Eq. 6 with d replaced by r (the rate at which nascent mRNA changes to mature mRNA because of the termination of transcription).
Special case of bursty transcription
It can be further shown by perturbation theory in Appendix A that when ρ, λ, and σb are much greater than the rest of the parameters, the exact solution Eq. 6 reduces to the negative binomial distribution with α = σbγ2/λ. Note the constraint on the parameters leads to a time series with large- and short-lived bursts of transcription (because ρ, λ, and σb are large), separated by long silent intervals (because σu is small). Such bursty trancription is common in mammalian cells (3).
Relationship to the telegraph model
It can also be shown that that in the limit of large ρ, the exact solution of Eq. 6 reduces to the confluent hypergeometric solution of the telegraph model (see Appendix B). This is equivalent to the steady-state solution of the two-state system . The reduction to a two-state model results from genes spending a short time in state D11 because of the large value of ρ. The production of an mRNA molecule involves the slow reaction step from D10 to D11 with rate λ followed by a very fast reverse step with rate ρ. Hence, the rate of mRNA production is determined by the reaction rate of the slowest reaction, i.e., it is equal to λ. By similar reasoning, we can deduce that in the limit of large λ, the gene spends a short time in the state D10, and the multiscale model reduces to the two-state telegraph model with a rate of mRNA production equal to ρ.
Sensitivity analysis
The exact solution in Eq. 5 allows us to examine the stochastic properties of the multiscale model over large swathes of parameter space. We investigate the relative sensitivity of the coefficient of variation of mRNA fluctuations, , which is typically employed as a measure of the magnitude of transcriptional noise. To this end, we calculate the first two central moments, ( and ), from Eq. 5 using and . The mean and CV are then given by
| (7a) |
| (7b) |
Note that because the parameters ρ and λ appear symmetrically in (7a), (7b), for simplicity, we enforce the constraint ρ = λ (we will relax this constraint later). Hence, the relative sensitivity of the quantity , which can serve as a gauge of transcriptional noise, is insightful to study and defined as for a model parameter p, meaning that 1% change in p leads to a Λp% change in . The parameter values for the sensitivity analysis were sampled from experimental distributions recently inferred for 3575 genes of CAST allele in mouse fibroblasts (3) using the telegraph model. To obtain values for ρ and λ, we equate the mean of the telegraph model (with ON switching rate σb, OFF switching rate σu, transcription rate ρu, and degradation rate d) with the mean of the multiscale model (Eq. 7a) under the constraint ρ = λ, giving
| (8) |
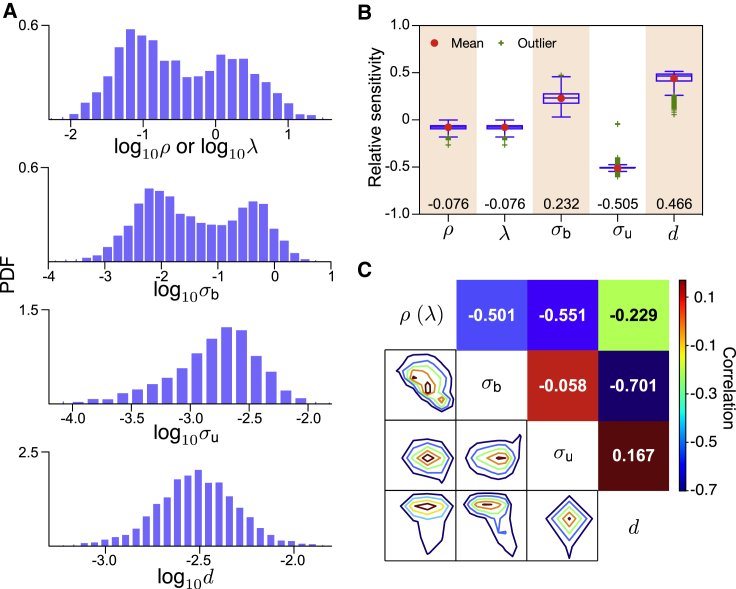
Distributions for each parameter in the data set are presented in Fig. 2 A, and the box plots in Fig. 2 B show the relative sensitivity for each parameter. The parameters in order of most sensitive first are σu, d, σb, and ρ = λ. This order is the same as obtained by ranking parameters according to the inverse of their mean experimental values (the mean of the distributions in Fig. 2 A), implying that changes to the CV are most easily accomplished by perturbations to the slowest reactions. Given the vectors and for any pair and p1, p2 in the set {ρ, λ, σb, σu, d} where each entry is a different gene, in Fig. 2 C, we calculate the Pearson correlation coefficient between the vectors and the corresponding joint distributions. This shows that (σu, σb) is the least dependent pairing, and hence, they constitute a quasiorthogonal decomposition of the sensitivity. In other words, a change in the CV due to a change in σu is practically uncorrelated with a change in the CV due to a change in σb, and hence, these two parameters can be seen as independent “control knobs” to change the CV; this is of interest in synthetic biology, in which an engineering design approach is taken to modify a biological system for improved functionality (26,27). The same set of parameters ranked by sensitivity are obtained, if instead of setting λ = ρ, we consider ρ ≫ λ or λ ≫ ρ, and hence, it appears that our results in this section are robust and invariant with respect to the ratio λ/ρ.
Figure 2.
Relative sensitivity analysis of the coefficient variation of mRNA noise over five kinetic parameters for 3575 genes of CAST allele data for mouse fibroblasts. (A) Distributions of the kinetic parameters in the dataset (obtained from (3)); values of ρ or λ are calculated using Eq. 8. (B) Box plots indicate the median (values shown at bottom), the 25% and 75% quantiles, and mean and outliers of relative sensitivity. (C) Joint distributions and Pearson correlation between the relative sensitivity vectors for each pair of parameters suggest that (σb, σu) and (σu, d) are the least-dependent pairs. To see this figure in color, go online.
Effective telegraph model
Earlier, we showed that in the limit of large ρ or large λ, the solution of the multiscale model tends to the solution of the telegraph model. Next, we use the first-passage time method to reduce the multiscale model into an effective telegraph model, without making the aforementioned assumptions. To this end, we consider the transcription motif of the multiscale model, , whose corresponding master equations for producing newborn mRNA starting from state D10 are
| (9) |
where P10, P11, and PM represent the probability of staying in states D10, D11, or producing a new mRNA, respectively. We remark that the reaction D11 → D0 is absent from the motif because of its relatively small reaction rate σb compared to ρ and λ. The initial conditions for Eq. 9 are and . Solving for PM in Eq. 9, we can calculate the mean first-passage time for mRNA production
| (10) |
where is the first-passage time distribution (28). Because the effective transcription rate is the inverse of the mean first-passage time, it immediately follows that the effective telegraph model is
| (11) |
Alternatively, one can obtain this result by equating the means of our model Eq. 7a and of the telegraph model and solving for the effective production rate ρu, giving because, typically, ρ, λ ≫ σb.
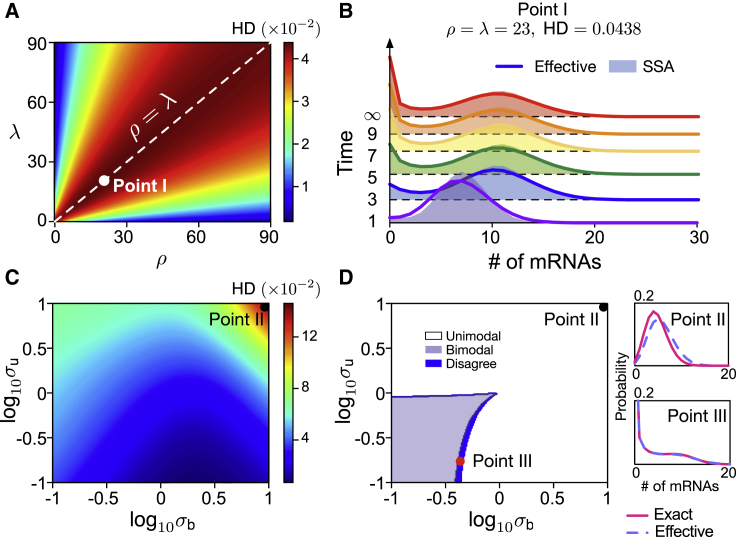
In Fig. 3, we show the high accuracy of the effective telegraph model approximation from Eq. 11. In particular, Fig. 3 A shows a heatmap of the distance between the distributions of mRNA numbers predicted by the effective telegraph model and the multiscale model. As a distance measure, we use the Hellinger distance (HD), a Euclidean distance-based metric normalized to the interval between 0 and 1. The effective telegraph model is naturally a more accurate description to the multiscale model when there is one rate-limiting step (large difference between ρ and λ) rather than when there are two rate-limiting steps (ρ = λ).
Figure 3.
An effective telegraph model (given by reaction scheme (11)) approximates the distribution of mRNA numbers of the multiscale model. (A) Hellinger distance (HD) between steady-state distributions of mRNA numbers for the effective telegraph model and the multiscale model as a function of ρ and λ with σu = 0.2, σb = 0.1, and d = 1. The discrepancy between the two distributions grows as ρ and λ approach the line ρ = λ. (B) Shown is the time-dependent distributions for Point I in (A) (the point with the largest HD) predicted by the effective model compared to those computed by the SSA for the multiscale model. (C) Heat map of HD between both distributions as a function of σb and σu with ρ = λ = 23 and d = 1. (D) Stochastic bifurcation diagram for the number of modes of the steady-state distributions predicted by the two models. The small dark blue region is where modality of both models disagree. Insets show distributions corresponding to the points marked in (C and D). To see this figure in color, go online.
Because the time-dependent distribution of the telegraph model is known in closed form (6,29), it follows that by the effective model in Eq. 11 we have an approximation for the time-dependent distribution of the multiscale model too. The accuracy of this approximation is shown in Fig. 3 B, where it is compared to the time-dependent distributions computed using the SSA for the multiscale model. The parameters here correspond to those of Point I in Fig. 3 A (the largest HD). Differences between the distributions of the two models are negligible except near time t = 0. We further investigate how burst initiation and termination rates (σu, σb) affect the approximation error with a heatmap of HD as a function of σu and σb (Fig. 3 C) and a stochastic bifurcation diagram for the number of modes of the effective telegraph and multiscale model distributions (Fig. 3 D) at steady state. The point of maximal HD in Fig. 3 C (Point II) displays distributions that are not that different from each other; see upper right inset of Fig. 3 D. The two models display the same number of modes in all regions of parameter space except for a narrow region in which modality detection is challenging because the distributions have a broad plateau; see lower right inset of Fig. 3 D (Point III). This again confirms the high accuracy of the effective telegraph model approximation. The biological implications of the Michaelis-Menten dependence of the transcription rate ρu in Eq. 11 on λ and ρ is discussed in Conclusions; in particular, there we argue how this special feature of our model can explain gene dosage compensation observed in experiments.
Connection to the refractory model
Besides the telegraph model, another prevalent stochastic transcriptional model is the refractory model (2) (a three-state model, see Fig. 4 A, left), wherein the burst initiation requires two steps. This model was devised to explain the experimental observation that the distribution of “off” intervals is not exponential but rather has a peak at a nonzero value. To understand the connection between our model and the refractory model, we first exactly solve the refractory model for the steady-state distribution of mRNA numbers.
Figure 4.
Effective telegraph model approximation for the refractory model. (A) Schematics of both models. (B) Hellinger distance between the steady-state distributions of mRNA numbers predicted by both models and a bifurcation diagram of their number of modes (black lines) as a function of σu and λ with σb = 0.8, ρu = 30, and d = 1. (C) Distributions for Points I and II in (B), showing significant disagreement in the height of the zero mode (insets show a zoom at the mode at zero). To see this figure in color, go online.
Given the reaction scheme illustrated in Fig. 4 A, it follows that the temporal evolution of probability Pθ(n) of finding n mRNAs and gene state Dθ (θ = 0, 1, or 2) can be described by the following master equations:
The corresponding generating function equations are given by
| (12a) |
| (12b) |
| (12c) |
where . We intend to solve Eqs. 12a, 12b, and 12c at steady state and thus set . Then, we solve G1 as a function of G2 from Eq. 12c, subsequently substitute it into Eq. 12b, and solve G0 as a function of G2. After that, Eq. 12a becomes an ordinary differential equation with G2 being the only variable to be solved
| (13) |
where , , , and are the kinetic parameters normalized with respect to d and u = z − 1. Eq. 13 is the canonical form of the differential equation for the generalized hypergeometric function 2F2, admitting the solution
| (14) |
where C is an integration constant, and β1 and β2 denote
Summing Eqs. 12a, 12b, and 12c leads to , one can obtain G from Eq. 14 in the form of the generalized hypergeometric function
| (15) |
and C2 is found to be 1 by the normalization condition G(0) = 1. Eq. 15 together with defines the distribution of mRNA numbers for the refractory model in steady-state conditions. A similar solution is also known for a generalization of the refractory model (11).
The next step is to map the refractory model onto an effective telegraph model by matching the mean mRNA numbers
leading to an effective burst initiation rate and the corresponding effective model shown in Fig. 4 A (right). Note that whereas the multiscale model is approximately equivalent to an effective telegraph model with a renormalized mRNA production rate, the refractory model’s telegraph approximation leads to a renormalized rate of switching to the active state.
We then compare the steady-state distributions of the refractory model and its effective telegraph model. A heatmap of HD quantifying their distributional difference and a modality diagram (marked as black lines) of the two distributions are illustrated in Fig. 4 B. Both the regions of high HD and Region 2 where only the telegraph model predicts bimodality are significantly large, and Region 1 where both predict bimodality is small. This shows that the refractory model, in general, is not well approximated by the telegraph model, particularly the latter’s probability for low mRNA numbers is not accurate (see Fig. 4 C). Given the telegraph model’s excellent approximation to the multiscale model, it is clear that the multiscale model and refractory model can be distinguished.
Protein dynamics
Finally, for completeness, we extend the multiscale model to provide analytic steady-state distributions of protein numbers. This allows interpretations of single-cell data of protein expression (see, for example, (30)). We consider the network in Fig. 1 A with two additional reactions: 1) a first-order reaction modeling the translation of mRNA to proteins with rate constant k and 2) a first-order reaction modeling the decay of protein with rate constant dp. It is shown in Appendix C that under the classic short-lived mRNA assumption (d ≫ dp) (21), the generating function corresponding to the steady-state distribution of protein numbers is given by
| (16) |
with b1 = (σb + σu)/dp, b2 = (σb + λ + ρ)/dp, the mean translational burst size b = k/d, and the parameters a1, a2, and a3 being solutions of the equations
In the limit of large λ or ρ, we show in Appendix C that Eq. 16 reduces to the Gaussian hypergeometric function (2F1), which was reported in (21), for the classical three-stage model of gene expression in the limit of fast mRNA decay.
Conclusions
Here, we performed the first detailed analytical study of a multiscale model of bursty gene expression based on recent experimental data from mammalian cells (16). The conventional telegraph model does not include an independently regulated pause release step and hence cannot differentiate the effects of changing polymerase pause release versus polymerase recruitment rates, whereas the multiscale model studied here can distinguish these effects. Although our model has three effective gene states (one of which regulates pause release), it is not a special case of existing multistate models because in our model, the gene state changes upon production of new nascent mRNAs to model the experimental observation that unless the polymerase is unpaused (and nascent mRNA starts being actively transcribed by this polymerase), there can be no binding of new Pol II. In contrast, current models assume the gene state does not change upon production of mRNA because they model the production of a mature transcript without detailed modeling of the steps between transcriptional initiation and termination.
We have derived simple closed-form expressions for the approximate time evolution of the mRNA numbers and used the theory to understand which reactions contribute mostly to fluctuations. We also showed that 1) this model can be distinguished from the refractory model, another three-gene-state model popular in the literature and 2) a number of previous models in the literature are special cases of our model, valid only in certain parameter regimes. Specifically, the mRNA and protein distributions of the conventional three-stage model of gene expression provide a good approximation to the multiscale bursting model in certain regions of parameter space as shown in Appendices B and C.
The simplicity of the equations for the mean and the variance allow the inference of rate parameters from single-cell data using maximal likelihood methods (31). Potential extensions include 1) the impact of cell cycle effects such as binomial partitioning and variability in the cell cycle duration and 2) introducing a detailed description of polymerase movement along the gene during elongation. The use of the recently developed linear mapping approximation (32) appears to be a promising means to extend the analytical solution of this model to include feedback loops via DNA-protein interactions (33,34).
An important result of the article is that the time-dependent mRNA distribution of the multiscale model with polymerase dynamics and three states can be accurately approximated by the two-state telegraph model, modified with a Michaelis-Menten-like dependence of the effective transcription rate on polymerase abundance. Specifically, by Eq. 11, the transcription rate of a gene locus is ρu = λρ/(λ + ρ), where λ is the binding rate of Pol II (see Fig. 1 A), which is proportional to the local number of Pol II molecules at the gene locus with active transcription (35). This equation implies that the transcription rate is proportional to the local number of Pol II molecules if λ is approximately less than ρ, i.e., if the Pol II binding rate is less than or equal to the rate at which Pol II is unpaused. In contrast, if unpausing is the rate-limiting step (ρ ≪ λ), then the transcription rate is practically independent of the local Pol II number.
Now, when the number of gene copies doubles during replication, the local number of Pol II molecules will correspondingly decrease because of increased sharing of Pol II. Hence, if we are in the regime , the transcription rate per gene copy decreases; thus, the total transcription rate for a gene per cell postreplication will be consequently slower than twice the total transcription rate prereplication. This implies that the mean number of RNA per cell is not significantly affected by replication; indeed, this “dosage compensation” has been observed experimentally for some genes in mouse embryonic stem cells (36) though a different explanation than above was suggested. In one study (37), it was estimated that for 6 yeast genes (RPB2, RPB3, TAF5, TAF6, TAF12, and KAP104), the formation of the preinitiation complex at the promoter (λ) is approximately equal to the rate at which the RNA polymerase escapes the promoter (ρ); hence, gene dosage compensation via polymerase sharing, as implied by our model, may be common. In contrast, if we are in the regime ρ ≪ λ, the transcription rate per gene copy before and after replication is the same, and hence, the total transcription rate for a gene per cell postreplication will be twice the total transcription rate prereplication. This is also what is predicted by the telegraph model with constant burst initiation and termination rates and observed experimentally for a reporter gene expressed from a strong synthetic promoter (36). Note that because the mean burst size is the mean number of RNAs transcribed when the gene is on, by our reasoning above, it also follows that when , the mean burst size is altered upon gene replication. The idea that the number of RNA polymerases is the limiting factor in transcription has been recently hypothesized (38) and has implications for the mitigation of burden imposed by gene circuits in synthetic biology (39). Our model here goes one step further by deriving the explicit relationship between the transcription rate and the number of RNA polymerases. Generally, our model supports the observation that there are differences in transcriptional activity between different stages of the cell cycle (40) that cannot be explained by the conventional telegraph model.
Author Contributions
Z.C. formulated the research question, performed the calculations, produced the figures, and wrote an initial draft of the manuscript. T.F. performed some of the calculations for protein distributions. D.O. supervised the research and edited the manuscript. R.G. formulated the research question, supervised the research, and wrote the manuscript with assistance from the co-authors.
Acknowledgments
Z.C. gratefully acknowledges support of the UK Research Councils Synthetic Biology for Growth programme and of the Biotechnology and Biological Sciences Research Council, Engineering and Physical Sciences Research Council, and Medical Research Council (B/M018040/1) and careful proofreading by J. Holehouse. R.G. acknowledges support from Biotechnology and Biological Sciences Research Council grant BB/M025551/1. D.O. acknowledges support from the Human Frontier Science Program (grant RGY076/2015).
Editor: Alexander Berezhkovskii.
Appendix A: Analytic Distribution For Mrna Numbers When , , And Are Large
Given the large values of ρ, λ, and σb, we implement the following parametrization:
where δ is a large real number.
By means of the method of characteristics, solving (2a), (2b), (2c) is tantamount to seeking a solution to the ordinary differential equation system
| (17a) |
| (17b) |
| (17c) |
Dividing δ on both sides of Eqs. 17a, 17b, and 17c, one obtains a singular system consisting of
| (18) |
with . Expanding G, G10, and G11 in Eq. 18 as a series in powers of ,
and matching the orders of , we have
and
Then, we have
where α = σbγ2/λ, and u = z − 1 =reds. Its solution immediately follows as
| (19) |
with C(r) being a function of r to be determined from the initial condition. Suppose that the initial condition for this process is , which is known a priori. For instance, say the initial distribution of n mRNA molecules is P(n) = pn, then g(u) = ∑npn(u + 1)n. Letting s be equal to 0 (or equivalently t = 0), it follows u = r and g(u) = g(r), and we can establish the following relation
from which we can solve C(r) as
Substituting the latter back into Eq. 19 and replacing r = ue−dt, we can calculate the leading-order solution of G from (Eq. 19) as
| (20) |
At steady state, the leading-order solution in (Eq. 20) becomes
and the corresponding distribution of mRNA numbers is a negative binomial distribution .
Appendix B: Convergence to Telegraph Model for Large
To this end, we parametrize ρ as , where δ is a large real number. As such, (2a), (2b), (2c) can be recast as
| (21a) |
| (21b) |
| (21c) |
Dividing both sides of Eqs. 21b and 21c by δ and setting ϵ = δ−1, we have that
| (22a) |
| (22b) |
Again using the same method as before, we expand G0, G10, and G11 in Eqs. 21a and (22a), (22b) as a series in powers of , collect the terms for ϵ0 and ϵ1, and obtain
| (23) |
and
| (24) |
From Eq. 23, we can solve that , with which we can further get from Eq. 24. Given both results, Eqs. 23 and 24 can be simplified to
which are exactly the generating function equations of the telegraph model (see Eqs. A2 and A3 in (29)), thus showing that the multiscale transcriptional bursting model converges to the telegraph model when ρ → ∞. A similar proof can be constructed to show that the telegraph model is also obtained in the limit λ → ∞.
Appendix C: Analytic Marginal Distribution for Protein Numbers for the Multiscale Model in the Limit of Fast mRNA Decay
To the reaction scheme illustrated in Fig. 1 A, we add two reactions: 1) a first-order reaction modeling the translation of mRNA to proteins with rate constant k and 2) a first-order reaction modeling the decay of protein with rate constant dp. The following coupled master equations describe the time evolution of the probability Pθ(n, m) of finding n mRNAs, m proteins, and gene state Dθ (θ = 0, 10, 11) in a cell:
| (25) |
By defining , solving Eq. 25 is tantamount to seeking solutions to the set of differential equations
| (26) |
By means of the method of characteristics, Eq. 26 is equivalently represented as
and
Assuming that mRNA decays much faster than protein such that ((21a), (21b), (21c)), we get that
| (27) |
and b = k/d is the mean translational burst size. Using Eq. 27, we can reduce Eq. 26 to
| (28a) |
| (28b) |
| (28c) |
where , , , and are kinetic parameters normalized with respect to protein degradation rate dp. It follows from summing (28a), (28b), (28c) that
| (29) |
Using the definitions and and plugging Eq. 29 into Eqs. 28b and 28c, it gives us that
which admits a solution
| (30) |
with a1, a2, and a3 being roots of
Hence, summarizing, Eq. 30 and define the steady-state distribution of protein numbers, which is
given that mRNA is short lived.
Next, we will show the solution Eq. 30 converges to the Gaussian hypergeometric function (2F1) for the three-stage gene expression model (21) when ρ is large. To this end, we parameterize in Eqs. 28b and 28c as , where δ is a large number. Dividing both sides of Eqs. 28b and 28c by δ, we have
| (31a) |
| (31b) |
where . Again similarly, we expand G0, G10, and G11 in Eqs. 28a and (31a), (31b) as a series in powers of , collect the terms for 0 and 1, and obtain
| (32) |
and
| (33) |
From Eq. 32, we get , which is used to reduce Eq. 33 and the first equation in Eq. 32 to
| (34) |
Note that Eq. 34, which is the leading order of (28a), (28b), (28c), is exactly the same as the generating functions of the three-stage gene expression model reported in (21) (see Eqs. 68–69). By means of similar arguments, one can show the reduction of our model when λ is large.
References
- 1.Bahar Halpern K., Tanami S., Itzkovitz S. Bursty gene expression in the intact mammalian liver. Mol. Cell. 2015;58:147–156. doi: 10.1016/j.molcel.2015.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suter D.M., Molina N., Naef F. Mammalian genes are transcribed with widely different bursting kinetics. Science. 2011;332:472–474. doi: 10.1126/science.1198817. [DOI] [PubMed] [Google Scholar]
- 3.Larsson A.J.M., Johnsson P., Sandberg R. Genomic encoding of transcriptional burst kinetics. Nature. 2019;565:251–254. doi: 10.1038/s41586-018-0836-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zenklusen D., Larson D.R., Singer R.H. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat. Struct. Mol. Biol. 2008;15:1263–1271. doi: 10.1038/nsmb.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanchez A., Golding I. Genetic determinants and cellular constraints in noisy gene expression. Science. 2013;342:1188–1193. doi: 10.1126/science.1242975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peccoud J., Ycart B. Markovian modeling of gene-product synthesis. Theor. Popul. Biol. 1995;48:222–234. [Google Scholar]
- 7.Raj A., Peskin C.S., Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tunnacliffe E., Chubb J.R. What is a transcriptional burst? Trends Genet. 2020;36:288–297. doi: 10.1016/j.tig.2020.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Cao Z., Grima R. Analytical distributions for detailed models of stochastic gene expression in eukaryotic cells. Proc. Natl. Acad. Sci. USA. 2020;117:4682–4692. doi: 10.1073/pnas.1910888117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar N., Kulkarni R.V. Constraining the complexity of promoter dynamics using fluctuations in gene expression. Phys. Biol. 2019;17:015001. doi: 10.1088/1478-3975/ab4e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou T., Zhang J. Analytical results for a multistate gene model. SIAM J. Appl. Math. 2012;72:789–818. [Google Scholar]
- 12.Rodriguez J., Ren G., Larson D.R. Intrinsic dynamics of a human gene reveal the basis of expression heterogeneity. Cell. 2019;176:213–226.e18. doi: 10.1016/j.cell.2018.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J.J., Zhou T.S. Stationary moments, distribution conjugation and phenotypic regions in stochastic gene transcription. Math. Biosci. Eng. 2019;16:6134–6166. doi: 10.3934/mbe.2019307. [DOI] [PubMed] [Google Scholar]
- 14.Bothma J.P., Garcia H.G., Levine M. Dynamic regulation of eve stripe 2 expression reveals transcriptional bursts in living Drosophila embryos. Proc. Natl. Acad. Sci. USA. 2014;111:10598–10603. doi: 10.1073/pnas.1410022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corrigan A.M., Tunnacliffe E., Chubb J.R. A continuum model of transcriptional bursting. eLife. 2016;5:e13051. doi: 10.7554/eLife.13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartman C.R., Hamagami N., Raj A. Transcriptional burst initiation and polymerase pause release are key control points of transcriptional regulation. Mol. Cell. 2019;73:519–532.e4. doi: 10.1016/j.molcel.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shao W., Zeitlinger J. Paused RNA polymerase II inhibits new transcriptional initiation. Nat. Genet. 2017;49:1045–1051. doi: 10.1038/ng.3867. [DOI] [PubMed] [Google Scholar]
- 18.Gressel S., Schwalb B., Cramer P. CDK9-dependent RNA polymerase II pausing controls transcription initiation. eLife. 2017;6:e29736. doi: 10.7554/eLife.29736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu H., Skinner S.O., Golding I. Stochastic kinetics of nascent rna. Phys. Rev. Lett. 2016;117:128101. doi: 10.1103/PhysRevLett.117.128101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phillips R., Belliveau N.M., Scholes C. Figure 1 theory meets figure 2 experiments in the study of gene expression. Annu. Rev. Biophys. 2019;48:121–163. doi: 10.1146/annurev-biophys-052118-115525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shahrezaei V., Swain P.S. Analytical distributions for stochastic gene expression. Proc. Natl. Acad. Sci. USA. 2008;105:17256–17261. doi: 10.1073/pnas.0803850105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tantale K., Mueller F., Bertrand E. A single-molecule view of transcription reveals convoys of RNA polymerases and multi-scale bursting. Nat. Commun. 2016;7:12248. doi: 10.1038/ncomms12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Kampen N.G. Elsevier; Amsterdam, the Netherlands: 1992. Stochastic Processes in Physics and Chemistry. [Google Scholar]
- 24.Thomas P., Popović N., Grima R. Phenotypic switching in gene regulatory networks. Proc. Natl. Acad. Sci. USA. 2014;111:6994–6999. doi: 10.1073/pnas.1400049111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gillespie D.T. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 1977;81:2340–2361. [Google Scholar]
- 26.Mannan A.A., Liu D., Oyarzún D.A. Fundamental design principles for transcription-factor-based metabolite biosensors. ACS Synth. Biol. 2017;6:1851–1859. doi: 10.1021/acssynbio.7b00172. [DOI] [PubMed] [Google Scholar]
- 27.Arpino J.A.J., Hancock E.J., Polizzi K. Tuning the dials of synthetic biology. Microbiology. 2013;159:1236–1253. doi: 10.1099/mic.0.067975-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Redner S. Cambridge University Press; Cambridge, UK: 2001. A Guide to First-Passage Processes. [Google Scholar]
- 29.Iyer-Biswas S., Hayot F., Jayaprakash C. Stochasticity of gene products from transcriptional pulsing. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2009;79:031911. doi: 10.1103/PhysRevE.79.031911. [DOI] [PubMed] [Google Scholar]
- 30.Bothma J.P., Norstad M.R., Garcia H.G. Llamatags: a versatile tool to image transcription factor dynamics in live embryos. Cell. 2018;173:1810–1822.e16. doi: 10.1016/j.cell.2018.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao Z., Grima R. Accuracy of parameter estimation for auto-regulatory transcriptional feedback loops from noisy data. J. R. Soc. Interface. 2019;16:20180967. doi: 10.1098/rsif.2018.0967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao Z., Grima R. Linear mapping approximation of gene regulatory networks with stochastic dynamics. Nat. Commun. 2018;9:3305. doi: 10.1038/s41467-018-05822-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holehouse J., Grima R. Revisiting the reduction of stochastic models of genetic feedback loops with fast promoter switching. Biophys. J. 2019;117:1311–1330. doi: 10.1016/j.bpj.2019.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holehouse J., Cao Z., Grima R. Stochastic modeling of auto-regulatory genetic feedback loops: a review and comparative study. Biophys. J. 2020;118:1517–1525. doi: 10.1016/j.bpj.2020.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cisse I.I., Izeddin I., Darzacq X. Real-time dynamics of RNA polymerase II clustering in live human cells. Science. 2013;341:664–667. doi: 10.1126/science.1239053. [DOI] [PubMed] [Google Scholar]
- 36.Skinner S.O., Xu H., Golding I. Single-cell analysis of transcription kinetics across the cell cycle. eLife. 2016;5:e12175. doi: 10.7554/eLife.12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choubey S., Kondev J., Sanchez A. Deciphering transcriptional dynamics in vivo by counting nascent rna molecules. PLoS Comput. Biol. 2015;11:e1004345. doi: 10.1371/journal.pcbi.1004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin J., Amir A. Homeostasis of protein and mRNA concentrations in growing cells. Nat. Commun. 2018;9:4496. doi: 10.1038/s41467-018-06714-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nikolados E.M., Weiße A.Y., Oyarzún D.A. Growth defects and loss-of-function in synthetic gene circuits. ACS Synth. Biol. 2019;8:1231–1240. doi: 10.1021/acssynbio.8b00531. [DOI] [PubMed] [Google Scholar]
- 40.Zopf C.J., Quinn K., Maheshri N. Cell-cycle dependence of transcription dominates noise in gene expression. PLoS Comput. Biol. 2013;9:e1003161. doi: 10.1371/journal.pcbi.1003161. [DOI] [PMC free article] [PubMed] [Google Scholar]