Abstract
The gut and the liver have a bidirectional communication via the biliary system and the portal vein. The intestinal microbiota and microbial products play an important role for modulating liver diseases such as alcohol-associated liver disease, non-alcoholic fatty liver disease and steatohepatitis, and cholestatic liver diseases. Here, we review the role of the gut microbiota and its products for the pathogenesis and therapy of chronic liver diseases.
Keywords: microbiome, bile acids, short-chain fatty acids, non-alcoholic fatty liver disease, alcoholic liver disease
Introduction
Interactions between the gut microbiota and host are important for maintaining homeostasis and human health. Microbiota contributes to carbohydrate digestion (110), bile acid metabolism (56), maintenance of gut barrier integrity against pathogen infection (78), and vitamin synthesis (173). On the other hand, the intestinal microbiota, including bacteria, fungi, archaea, and viruses, contributes to the pathogenesis of many diseases (87). Changes in gut microbial composition have been associated with a variety of diseases, such as inflammatory bowel disease (IBD) (85), cardiovascular disease (89), colorectal carcinoma (91), and obesity (154). Whether changes in microbial composition are the cause or consequences of disease states is not fully understood (157). There are also instances where the complete absence of the microbiota does not affect the disease course. Germ-free mice show no difference in the extent of acetaminophen-induced liver damage compared with conventional mice (129).
To characterize the microbial community, different approaches have been developed, including metagenomics (genes), metatranscriptomics (RNA), metaproteomics (proteins), and metabolomics (metabolites) (34, 92, 113, 145). Disturbance of essential microbial functions results in microbial dysbiosis, which can promote both intestinal and extra-intestinal diseases. Various liver disorders such as alcohol-associated liver disease (ALD) (141), nonalcoholic fatty liver disease (NAFLD) (17), nonalcoholic steatohepatitis (NASH) (22), hepatocellular carcinoma (HCC) (60), primary biliary cholangitis (PBC) (108, 132), primary sclerosing cholangitis (PSC) (67), and liver cirrhosis (131) have been associated with altered gut microbiota. The liver and the microbiota have a bidirectional communication (148). The venous blood from the intestine reaches the liver directly via the portal vein. Nutrients, microbial products and metabolites, and intestinal toxins reach the liver as the first organ in the human body. Nutrients and microbial products are also drained into lymphatic vessels and reach the liver via the thoracic duct and systemic circulation (5). The liver is connected to the intestine via the biliary system. Bile, bile constituents, and bile acids are secreted from the liver and reach the small intestine via bile ducts (FIGURE 1). In this review, we will focus on the interactions between microbiota and liver, describing the role of microbiota and its products for the pathogenesis and treatment of liver diseases. By summarizing the current literature and pointing out the limitations of common approaches in microbiomics, we provide a comprehensive review for what we know and discuss gaps about the contribution of the gut microbiota to liver disease pathogenesis. We also focus on human microbiome studies, which mostly rely on a cross-sectional, observational study design. As such, these studies are predominantly associative in nature and cannot establish causality.
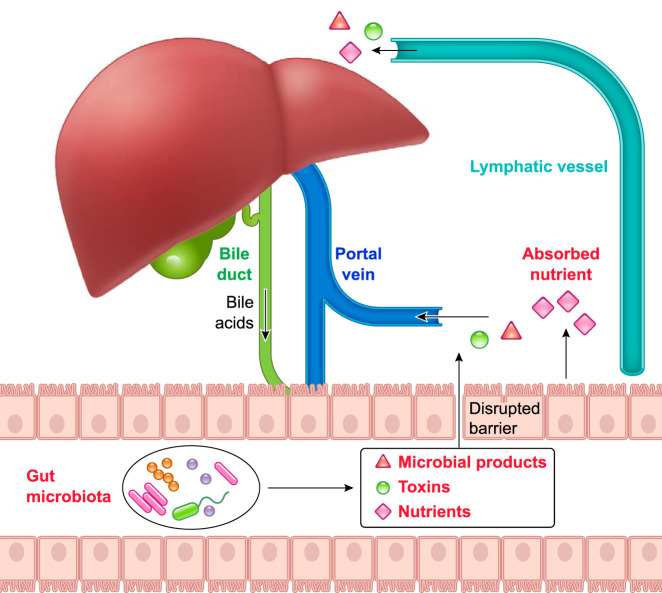
FIGURE 1.
The interplay between liver and gut
Liver secretes bile acids to the intestinal lumen through bile ducts. When gut barrier is intact, the intestine absorbs nutrients through enterocytes and transports to the liver. When gut barrier is disrupted, microbial-derived toxins and products translocate to liver from intestine through portal vein and influence liver functions. Nutrients and microbial products can reach the systemic circulation via lymph vessels and the thoracic duct.
Gut Microbiota-Derived Products
Bile Acids
Bile acids are synthesized in the liver from cholesterol, and they function by facilitating digestion and lipid absorption in the intestine (97). In addition to their role in digestion, bile acids also function as signaling molecules by activating farnesoid X receptor (FXR) and the G-protein-coupled bile acid receptor 1 (GPBAR1/TGR5) (111, 142). In humans, primary bile acids cholic acid (CA) and chenodeoxycholic acid (CDCA) are both endogenous FXR agonists (159). Upon metabolism by gut microbiota, primary bile acids CA and CDCA are converted to secondary bile acids deoxycholic acid (DCA) and lithocholic acid (LCA), respectively (46, 126). DCA is reabsorbed in the intestine and transported back to the liver, whereas only a small portion of LCA is reabsorbed and conjugated in the liver for reentry into the enterohepatic circulation (30, 66). The gut microbiota plays an important role in maintaining the bile acid pool by modulating bile acids through deconjugation, dehydroxylation, esterification, oxidation, and desulfation (29). Conjugated bile acids are deconjugated in the colon by the microbial enzyme bile salt hydrolase; elevating its activity reduced body weight, plasma cholesterol, and liver triglycerides in the host (74). Patients with NAFLD and NASH have an altered systemic bile acid composition, and increased key bile acids are associated with higher grades of steatosis (taurocholate), hepatocyte ballooning (taurocholate), lobular (glycocholate), and portal inflammation (taurolithocholate) (130). In addition, chronic ethanol administration in rats decreased hydrophilic taurine-conjugated bile acids and increased unconjugated bile acids in the liver and intestinal contents (duodenum, ileum, cecum, and rectum) (169). Cirrhotic patients who are actively drinking have higher fecal secondary bile acids than patients with cirrhosis due to a different liver disease etiology, and the increase in secondary bile acids is accompanied by increased inflammatory cytokines in colonic mucosa (75). Rats subjected to chronic ethanol together with CDCA, a hydrophobic bile salt, showed aggravated hepatic steatosis, lipid peroxidation, and liver injury (120).
FXR is not only highly expressed in liver and intestine, but also in kidneys and adrenal cortex (51). Activation of FXR in hepatocytes induces the transcription of an orphan nuclear receptor, small heterodimer protein (SHP), which inhibits the transcription of cytochrome P450 family 7 subfamily A member 1 (CYP7A1) and bile acid synthesis (59, 105). The rate-limiting enzyme CYP7A1 is regulated by negative feedback mechanism via fibroblast growth factor (FGF) 15/19, which is produced and secreted from enterocytes in the terminal ileum upon FXR activation (73). In addition, FXR signaling is also critical for lipid and glucose metabolism. FXR−/− mice exhibit decreased basal expression of SHP, increased hepatic triglyceride level (164), glucose intolerance, and insulin resistance (182). Bile acid-induced FXR activity can have anti-bacterial effects to protect the small intestine from bacterial overgrowth and barrier disruption, and mice lacking FXR have a compromised gut-barrier function (70).
TGR5 is distributed in a wide range of tissues, such as cholangiocytes, gallbladder epithelial cells and enterocytes (mainly in the ileum and colon), smooth muscle, brown adipose tissue, immune cells, and enteroendocrine L-cells (114). Endogenous agonists for TGR5 include LCA, DCA, CDCA, and CA, among which LCA is the most potent activator (80). Upon stimulation by bile acids, enteroendocrine L-cells can secrete glucagon-like peptide-1 (GLP-1) to regulate glucose homeostasis (23). Induction of GLP-1 release by INT-777 (a TGR5 agonist) increases energy expenditure, reduces hepatic steatosis, and improves insulin sensitivity in obese mice (152), suggesting TGR5 may be a promising therapeutic target for metabolic diseases. TGR5 activation also exhibits anti-inflammatory effects. TGR5 on Kupffer cells can be activated by bile acids, which reduces LPS-induced inflammatory cytokine production (81). TGR5 is also detected in sinusoidal endothelial cells; the activation of which induces expression of endothelial nitric oxide synthase (eNOS) and nitric oxide production. This suggests a possible protective mechanism from oxidative stress (82). TGR5 is also expressed on cholangiocytes, which protects them from bile acid toxicity under cholestatic conditions, whereas it may promote cholangiocarcinoma by stimulating cholangiocyte proliferation (137).
Bile acids and microbiota can regulate each other. To examine how bile acids affect gut microbial composition, rats were fed with CA-supplemented diet. CA-fed rats exhibited a significant reduction in total bacteria number and variety, probably due to the direct anti-microbial activity of CA. Furthermore, gut microbiota was altered at phylum level with CA administration compared with chow diet. More specifically, Firmicutes predominated, and several bacteria in the classes of Clostridia and Erysipelotrichi increased in the CA group (72). In addition, Kakiyama et al. found a connection between fecal bile acid concentration, liver cirrhosis, and microbiota composition. As chronic liver disease progresses to cirrhosis in patients, there is bacterial dysbiosis paralleled by a decreased bile flow and low bile acid levels in the intestine, which are also accompanied by an increase of the inflammatory taxa Enterobacteriaceae (76). Compared with conventional mice, germ-free mice have a strikingly increased bile acid pool. Downregulation of apical sodium-dependent bile acid transporter (ASBT) through FXR signaling might contribute to this increase (69, 147). Interestingly, administration of antibiotics to wild-type mice increased bile acid pool through antagonizing FXR-signaling and reducing DCA (69). Gut microbiota can affect bile acid synthesis through modulating FXR-FGF15/19 signaling in the ileum (140). Overall, bile acids are important signal molecules in the bidirectional communication between gut and liver. Targeting bile acid signaling or related microbiota may represent an excellent treatment strategy for liver diseases that are associated with disturbances in bile acid pool and/or bile acid composition.
Short-Chain Fatty Acids
Short-chain fatty acids (SCFAs) are major products of bacterial fermentation of carbohydrates in the gastrointestinal tract (16). In the human body, the most abundant SCFAs in the colon are acetate, propionate, and butyrate. Other SCFAs are produced in lesser amounts, including formate, valerate, and caproate (109). Among all the SCFAs, acetate and propionate are mainly produced by Bacteroidetes, whereas butyrate is mainly synthesized by Firmicutes (47, 121). SCFAs can be rapidly absorbed in the colon through passive diffusion (136), monocarboxylate transporters (119, 138), or exchange with bicarbonate (HCO3−) via Solute Carrier Family 26 Member 3 (SLC26A3) (149). Absorbed SCFAs enter the tricarboxylic acid cycle to generate ATP and energy (143). A small proportion of SCFAs that are not metabolized can be released into the liver via portal vein, where they can be used as energy substrates for hepatocytes (37, 143).
SCFAs are ligands for a number of deorphaned G-protein-coupled receptors (21, 86). Free fatty acids receptors (FFAR) 2 (GPR43) and 3 (GPR41) are predominantly activated by acetate, propionate, and butyrate (25, 95). FFA2 and FFA3 are highly expressed in small and large intestines (153), pancreas, adipose tissue, neurons, and immune cells (25). In the intestine, butyrate has beneficial effects through regulating transepithelial fluid transport, maintaining the intestinal barrier, ameliorating mucosal inflammation, and modulating intestinal motility (27). Butyrate also inhibits intestinal cholesterol biosynthesis gene expression (8) and prevents diet-induced obesity in mice (55).
The role of SCFAs has been implicated in multiple liver diseases. For example, higher fecal SCFAs (propionate and acetate) and increased abundance of SCFA-producing bacteria, such as Fusobacteriaceae and Prevotellaceae, were observed in NAFLD patients compared with healthy controls (135). SCFA-supplemented diet prevents and reverses high-fat diet (HFD)-induced obesity and insulin resistance in mice through downregulating peroxisome proliferator-activated receptor-γ (PPARγ) in liver and white adipose tissue, but not in muscle (40). Furthermore, butyrate also shows protective effects in a murine model of NASH. Dietary supplementation of butyrate not only alleviated methionine-choline-deficient-induced liver injury, inflammation, and fibrosis, but also improved gut dysbiosis. Interestingly, butyrate shifted the microbial composition by reducing the abundance of LPS-producing bacteria Bilophila and promoting the abundance of probiotic genus Akkermansia (174). Synbiotic supplementation comprised of butyrate-producing bacteria (Faecalibacterium prausnitzii) and a butyrate-yielding prebiotic (potato starch) showed protective effects against chronic-binge ethanol-induced liver steatosis, liver injury, and gut injury (139). Inulin treatment increased intestinal SCFAs (propionate, butyrate, and valerate) and ameliorated hepatic inflammation by reducing M1 and increasing M2 macrophages in mice subjected to chronic ethanol feeding (163). Although SCFAs are considered to be health-promoting in most cases, they should be approached with caution. A recent study observed detrimental effects of inulin in promoting HCC in dysbiotic mice. Inulin is a soluble fiber that can be metabolized to SCFAs. Consumption of inulin-enriched high-fat diet induces dysbiosis and HCC in wild-type mice via early onset of cholestasis (146). Thus manipulating SCFAs directly by dietary supplementation or indirectly by SCFA-producing bacteria may prevent or treat liver diseases. However, it is hard to draw an accurate generalizable conclusion about the benefit of SCFAs, so the specific context should be considered.
Choline and Tryptophan Metabolites
Choline is a water-soluble nutrient essential for human life that can be metabolized by the gut microbiota to trimethylamine (TMA) (179). TMA-producing bacteria in fecal samples from subjects without known atherosclerosis or severe cardiovascular disease include various taxa Clostridium XIVa strains, Eubacterium sp., and Gammaproteobacteria (primarily from Escherichia coli) (134). TMA can be absorbed by the host and be further oxidized to generate trimethylamine-N-oxide (TMAO) in the liver (13). Although previous studies suggest that choline is beneficial to humans (161), TMAO can be detrimental. In cardiovascular disease, metabolomic analysis showed a dose-dependent association between plasma phosphatidylcholine metabolites (choline, TMAO, and betaine) and cardiovascular disease risk. Choline- or TMAO-supplemented diet promoted atherosclerosis in atherosclerosis-prone mice (C57BL/6J Apoe−/−), whereas reducing the commensal microbiota with antibiotics inhibited choline-induced atherosclerosis (161). This study emphasizes the role of dietary microbiota-dependent phosphatidylcholine metabolism and cardiovascular disease pathogenesis. Studies in chronic kidney disease further demonstrated that plasma TMAO level is increased in patients with chronic kidney disease and is significantly correlated with 5-year mortality rate. Wild-type mice fed with choline- or TMAO-supplemented diet showed tubulointerstitital fibrosis and renal injury, suggesting that TMAO may contribute to chronic kidney disease progression (151). Inhibition of microbial TMA production with 3,3-dimethyl-1-butanol attenuated choline-diet-enhanced macrophage foam cell formation and atherosclerosis in mice fed a high-choline or L-carnitine diet, indicating that microbial production of TMA may serve as a therapeutic approach for cardiometabolic diseases (162). Another approach to inhibit TMA biosynthesis is by inhibiting the enzymes that convert choline to TMA, CutC and CutD (178). Alternatively, inhibiting the hepatic enzyme that produces TMAO can be another option. For example, knockdown of the enzyme flavin-containing monooxygenase 3 reduced TMAO level, and prevented the development of hyperglycemia, hyperlipidemia, and atherosclerosis in Liver Insulin Receptor Knockout (LIRKO) mice (117).
Tryptophan is an essential amino acid derived from diet (77). It plays an important role in protein synthesis, metabolic functions, and immune homeostasis (94, 156). Most of the absorbed tryptophan is metabolized in the kynurenine pathway, and only a small percentage is metabolized into serotonin or degraded by the gut microbiota to indole and derivatives (10). Tryptophan metabolism has great impact on maintaining intestinal tract immunity, gut eubiosis, and intestinal barrier (54). Gut microbiota not only can directly utilize tryptophan but also can regulate its availability for the host and can metabolize tryptophan metabolites (3). Several tryptophan metabolites exhibit health-promoting and disease-protective effects (181). Metabolites of tryptophan, such as indole-3-acetic acid, act as ligands for aryl hydrocarbon receptor (AhR) to stimulate interleukin 22 (IL-22) production and expression of regenerating islet-derived 3 gamma (REG3G), and reduce bacterial translocation to the liver in a mouse model of ethanol-induced liver disease (65). This microbiota-AhR axis activation provides microbial symbiosis and protects gut mucosal surfaces during conditions of inflammation (181). The role of tryptophan as anti-inflammatory modulator is further supported by feeding a tryptophan-free diet to mice, which are subjected to the chemical dextran sodium sulfate-induced colitis model. A tryptophan-free diet resulted in decreased antimicrobial peptide synthesis, altered microbiota composition, and increased susceptibility to colitis, providing a link between dietary amino acids, microbiota, and innate immunity (64). Analysis of serum samples from IBD patients showed a negative correlation between disease severity and tryptophan level. Furthermore, levels of tryptophan metabolites such as quinolinic acid were also found increased in active IBD patients, suggesting a possible role of tryptophan and its metabolites as biomarkers (125). In patients with metabolic syndrome, a significant increase of kynurenine-to-tryptophan ratio was reported, and the increased ratio correlates with obesity, body mass index, and serum triglyceride (112). Furthermore, tryptophan metabolism is altered in several other diseases, such as irritable bowel syndrome (33), obesity (45), and infectious diseases (180, 183), suggesting the use of tryptophan or its metabolites as biomarkers for diagnosis. In addition, tryptophan metabolism can serve as therapeutic targets, which can be achieved by supplementation with tryptophan metabolites, targeting AhR receptor, or enhancing metabolic pathways in the gut microbiota.
Other Microbial Products
Bacterial lipopolysaccharide (LPS) is the major component of outer cell membrane in Gram-negative bacteria (4). LPS is anchored in the outer membrane by a lipophilic, carbohydrate lipid moiety termed lipid A (165). The lipid A portion is responsible for immunological activities of LPS and is detected by CD14/Toll-like receptor 4 (TLR4) (128, 167). Activation of CD14/TLR4 triggers downstream signaling through nuclear factor-κB (NF-κB) or Jun NH2-terminal kinase/stress-activated protein kinase (JNK/SAPK) pathway to release inflammatory cytokines and Type I interferon genes (106). A previous study found that Bacteroides LPS is structurally distinct from E. coli LPS. It inhibits innate immune signaling and endotoxin tolerance, whereas E. coli LPS can decrease incidence of autoimmune diabetes in non-obese diabetic (NOD) mice (model of Type 1 diabetes) by inducing endotoxin tolerance. This suggests that exposure to immunostimulatory LPS may be protective in immune-mediated diseases (155). LPS has been well-studied for its role in alcohol-related liver diseases. For example, circulating LPS level is increased in animals and a subset of human patients with alcohol-use disorder (20, 83). Treating rats with antibiotics protects the liver from ethanol-induced injury, suggesting that alcohol consumption leads to increased endotoxemia, which contributes to pathogenesis of alcohol-related liver disease (2). In NAFLD, the importance of LPS is suggested by the fact that systemic LPS level is significantly increased in animals and a subset of patients, and that a combination of disaccharides-rich diet and LPS accelerates hepatic steatosis (15, 52, 63).
Bacterial flagellum is a whip-like complex apparatus that facilitates bacterial motility and bacterial invasion. One of the major subunits, flagellin, plays an important role in innate and adaptive immune response via activating its receptor TLR5 (61, 62). In liver, TLR5 is expressed in hepatocytes, Kupffer cells, and hepatic dendritic cells (168). Mice lacking TLR5 in hepatocytes are predisposed to HFD-induced hepatic steatosis, and they exhibited increased expression of proinflammatory cytokines and fibrosis-related genes in models of NASH (43). This suggests a protective role of TLR5 against circulating bacteria and against NAFLD and NASH.
Gut Microbiome and Liver Diseases
Gut Microbiome and Liver Health
Colonization of gut microbial communities is different in various regions of the human gut. Normal gut microbiota benefits human health by contributing nutrients and energy through digestion of carbohydrates and absorption (50). Liver is the organ exposed to a large number of microbial components and metabolites; thus it is critical to maintain a healthy gut environment for liver health. According to the World Health Organization’s definition of “health” as “the absence of diseases,” although one might think of “healthy gut” as healthy upper and lower gastrointestinal tract without indication of bowel diseases, the “healthy” microbiome has not been defined yet (18, 115). Changes in the gut microbiota composition could have an adverse effect since detrimental bacterial toxins and products are drained to the liver via the portal vein (186). LPS is important for induction of tolerance in the liver during homeostasis (36) but can activate Kupffer cells via binding to TLR4 during disease conditions. Upon activation, inflammatory cytokines are secreted, resulting in liver injury and inflammation (118). In addition, SCFAs, the major fermentation product of microbiota, exhibit opposing effects in liver health depending on the balance between caloric intake and expenditure (186). In the following section, we will discuss how gut microbiota and host interact with regard to liver diseases (FIGURE 2).
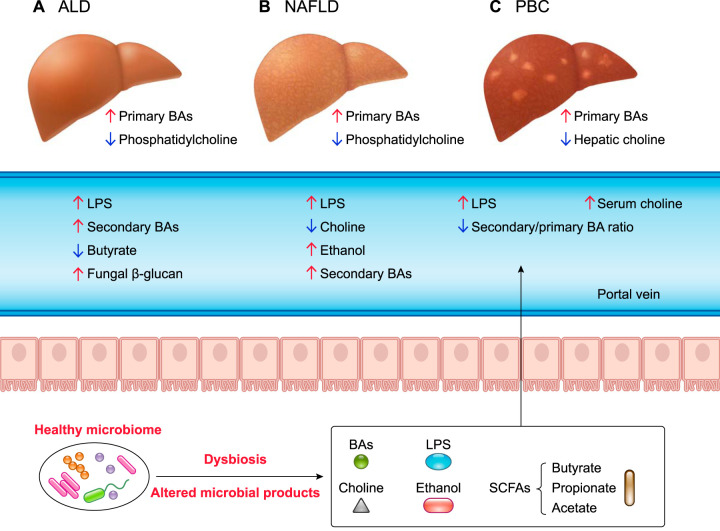
FIGURE 2.
The role of microbial products in different liver diseases
Intestinal dysbiosis is observed in alcohol-associated liver disease (ALD), nonalcoholic fatty liver disease (NAFLD), and primary biliary cholangitis (PBC). A and B: ALD- and NAFLD-associated dysbiosis increases lipopolysaccharide (LPS) and secondary bile acids, leading to an increase of primary bile acids. ALD also increases fungal β-glucan and decreases butyrate level. In NAFLD, an increase of ethanol and a decrease of choline are also observed. C: in PBC, increases of serum choline and LPS are observed in addition to decreases in hepatic choline and secondary-to-primary bile acid ratio.
Alcohol-Associated Liver Disease
Alcohol-related liver disease is a leading cause of morbidity and mortality worldwide. Among deaths caused by liver cirrhosis, 48% of these are alcohol-related in the U.S. (53). Alcohol consumption is associated with intestinal bacterial overgrowth and gut dysbiosis, which leads to bacterial translocation into the liver through the portal vein (100). Forty-three percent of alcohol-dependent subjects had increased intestinal permeability compared with control subjects, whereas most patients with cirrhosis show increased intestinal permeability (84, 96). Ethanol-fed mice exhibited bacterial translocation before obvious changes observed in the microbiota; intestinal bacterial overgrowth was seen after 3 wk of ethanol feeding (160, 171). To examine bacterial taxonomic changes, 16s rDNA sequencing was performed using colon samples from alcohol-use disorder patients with and without liver disease. Alcohol-use disorder patients with dysbiosis had a decreased proportion of Bacteroidetes and increased Proteobacteria compared with healthy controls (123). In a study culturing stool samples from 66 patients with alcohol-use disorder and 24 healthy controls, a significant decrease in beneficial bacteria such as bifidobacteria and lactobacilli were found in patients with alcohol-use disorder (88).
Since changes in microbial composition, function, and products have been shown to contribute to the pathogenesis of alcohol-related liver diseases (FIGURE 2A) (11), modulating intestinal microbiota could be important for effective therapy. Indeed, reducing intestinal bacteria with antibiotics prevented bacterial translocation and ethanol-induced liver injury in rats (2, 48). Furthermore, TLR4−/− mice displayed less liver injury, inflammation, and hepatic steatosis after ethanol administration, suggesting a contribution of TLR4 signaling for ethanol-induced liver disease (71). Ethanol feeding downregulated gene and protein expression of bactericidal C-type lectins Reg3b and Reg3g in the small intestine. Prebiotics treatment with fructooligosaccharides (FOS) partially restored Reg3g level, decreased bacterial overgrowth, and alleviated ethanol-induced steatohepatitis in mice (171). Fecal samples from patients with alcoholic hepatitis have 2,700-fold more Enterococcus faecalis than non-alcoholic controls. Some E. faecalis strains secrete the exotoxin cytolysin. Importantly, cytolysin-positive E. faecalis correlates with the severity of liver disease and mortality in alcoholic hepatitis patients. Targeting cytolysin with bacteriophages attenuated ethanol-induced liver disease in humanized mice colonized with feces from cytolysin-positive alcoholic hepatitis patients. These data suggest that fecal cytolysin can be used as a biomarker in patients with alcoholic hepatitis; targeting cytolysin may be used as therapy for patients with alcoholic hepatitis (41). In addition to bacterial dysbiosis, patients with alcohol-use disorder displayed reduced intestinal fungal diversity and Candida overgrowth. Alcohol increases fungal β-glucan translocation to the systemic circulation, and it induced liver inflammation via the C-type lectin domain family 7 member A (Clec7A) in mice. Treating mice with antifungal agents reduced intestinal fungal overgrowth and ethanol-induced liver disease, implying a contributing role of the fungal mycobiome in alcohol-associated liver disease (171). Recent studies have been focusing on targeting the microbiota with prebiotics, probiotics, antibiotics, or fecal microbiota transplantation (FMT). Probiotics are live organisms that confer beneficial effects to the host body supplied from outside in adequate dosage. Species of Lactobacillus and Bifidobacterium are commonly used probiotics. Prebiotics (such as inulin-type fructans) are substances fermented by the microbiota that change the gastrointestinal microbial composition in favor of host health and promote the growth of beneficial bacteria (14, 32). Supplementing mice with Lactobacillus rhamnosus GG restored intestinal occludin level and gut-barrier function, and protected against ethanol-induced liver disease (185). Probiotic treatment also exhibits protective effects in acute ethanol-induced liver injury. Mice binged with ethanol and treated with probiotics Bifidobacterium animalis subsp. lactis KV9 showed reduced hepatic inflammation and liver injury, and increased ethanol metabolism (184). These examples show the beneficial effects of probiotics for both acute and chronic ethanol-induced liver disease in mice.
Nonalcoholic Fatty Liver Disease and Steatohepatitis (NASH)
As the most common chronic liver disease, nonalcoholic fatty liver disease (NAFLD) affects over 64 million Americans, with an annual direct medical cost of $103 billion (175). NAFLD is the liver manifestation of the metabolic syndrome. It often occurs with Type 2 diabetes, obesity, hypertriglyceridemia, and hypertension (176). Obesity is associated with an altered gut microbiome. Germ-free mice colonized with microbiota from obese mice exhibit a greater percentage of body fat compared with germ-free mice colonized with microbiota from lean mice, linking the microbiota causatively to obesity development (9, 99, 154). Enteric microbiome can affect the host through metabolites. For example, choline deficiency has been implicated in the pathogenesis of experimental NASH for decades (19). After conversion to phosphatidylcholine, it prevents hepatic triglyceride accumulation by facilitating VLDL excretion from the liver (116). Interestingly, commensal bacteria such as E.coli and Desulfovibrio desulfuricans can also convert choline into methylamines (179). The critical role of gut microbiota in NAFLD (FIGURE 2B) and NASH has been corroborated by several studies. Mice susceptible to high-fat-diet-induced NAFLD are associated with lower levels of plasma phosphatidylcholine and a high urinary methylamines, suggesting a reduction of choline bioavailability (42). Similarly, dysbiosis increases choline conversion, which can lead to choline deficiency and contribute to development of NASH in experimental rodent models (90) (Table 1).
TABLE 1.
The role of microbiota in nonalcoholic fatty liver disease
| Factor | Species | Effect | Reference(s) |
|---|---|---|---|
| Fecal microbiota | Human | Family: ↑Kiloniellaceae, Pasteurellaceae, Lactobacillaceae, Lachnospiraceae, Veillonellaceae, ↓Ruminococcaceae, Porphyromonadaceae Genus: ↑Lactobacillus, Robinsoniella, Roseburia, Dorea, ↓Oscillibacter |
133 |
| Fecal microbiota | Human | ↓Firmicutes (phylum) ↑Proteobacteria (phylum) |
104 |
| Bile acids | Human | ↑Serum bile acids ↑Total fecal bile acids, primary-to-secondary bile acid ratio |
49, 122 |
| Short-chain fatty acids | Human | ↑Fecal acetate ↑Fecal propionate |
135 |
| Ethanol | Human | ↑Blood ethanol | 187 |
| Choline | Mice | ↓Phosphatidylcholine in plasma ↑Urinary trimethylamine |
42 |
| Tryptophan | Human | ↑Serum tryptophan | 57 |
Comparisons were made relative to controls or milder NAFLD.
In addition to choline, other microbial products such as bile acids, ethanol (187), and tryptophan (57) are also implicated in NAFLD/NASH progression. Patients with NASH have increased fecal bile acids and an altered profile, and the ratio of primary to secondary bile acids was also increased (122). Patients with NAFLD and NASH have elevated levels of bile acids in liver, serum, and urine. Bile acid pool from NASH patients included more hydrophobic and cytotoxic species compared with healthy controls (31, 49). The implication of gut microbiota in NASH is further supported by a study showing an elevated abundance of ethanol-producing bacteria such as Escherichia in NASH patients, accompanied by increased blood ethanol concentrations (187). In a study comparing serum endotoxin level between NASH and healthy subjects, only 8 of 19 patients exhibited elevated endotoxin (177). Similarly, a meta-analysis showed 39.1% of NAFLD patients had increased intestinal permeability compared with 6.8% healthy controls, suggesting that only a subset of NAFLD patients have increased intestinal permeability (107). In a cohort of children with NAFLD, endotoxin and plasminogen activator inhibitor-1 (PAI-1) were found to be significantly higher than in control subjects. Endotoxin and PAI-1 were also significantly associated with NAFLD activity score (6). Finally, genetically obese mice are more susceptible to LPS-induced liver injury through two possible mechanisms: altered Kupffer cell function and increased sensitivity to tumor necrosis factor-α (TNF-α) (172).
Given the intense interest and understanding between altered microbiota and NAFLD/NASH, a number of studies characterized the microbial composition by high-throughput sequencing. Deep sequencing of fecal samples from a cohort of NAFLD patients revealed an increased abundance of Lactobacillus and several members of phylum Firmicutes (Lachnospiraceae) compared with healthy controls. They also found that Ruminococcaceae and Oscillibacter were decreased in NAFLD patients (133). In another cohort of NAFLD patients, Proteobacteria and Escherichia coli were found increased in fecal samples of patients with advanced fibrosis, whereas Firmicutes were reduced. By using selected bacterial species combined with Shannon diversity, age, and body mass index as input, the random forest model had a robust and statistically significant diagnostic accuracy for predicting advanced fibrosis in patients with NASH (104). In the same cohort, changes in serum metabolites were also measured. In NAFLD patients with mild/moderate fibrosis (stage 0–2), inosine and hypoxanthine were enriched, whereas nine metabolites associated with carbon and amino acid metabolism (succinate, malate, α-ketoglutarate, glutamine, serine, fumarate, α-ketobutyrate, glutamate, and lactate) were enriched in advanced fibrosis (104). In particular, succinate was found increased in serum and fecal samples from NAFLD patients (104), and it was suggested to promote liver fibrosis by activating hepatic stellate cells through G-protein-coupled receptor 91 (101). Succinate can be produced by intestinal microbes such as Prevotella to serve as substrate for intestinal gluconeogenesis. Dietary succinate has been shown to improve glucose and insulin tolerance in wild-type mice fed with high-fat/high-sucrose diet. Wild-type mice colonized with the succinate producer Prevotella copri showed increased cecal succinate and reduced hepatic glucose production, suggesting metabolic benefits of succinate as a microbial product (39). In addition, plasma phenylacetic acid, a microbial product, showed a positive correlation with liver steatosis in obese patients. Chronic treatment with phenylacetic acid triggers liver steatosis in primary human hepatocytes and mice (68). The 3-(4-hydroxyphenyl) lactate, produced by bacterial enzymes and intestinal microbes, is also increased in serum samples from NAFLD patients and has a shared gene effect with hepatic steatosis and fibrosis (28). In summary, succinate, phenylacetic acid, and 3-(4-hydroxyphenyl) lactate are metabolites produced by intestinal microbes that can serve as biomarkers for NAFLD. Their value as therapeutic target requires mechanistic studies in rodent models in the future.
Given the connection between gut microbiota and NAFLD/NASH, targeting microbiota could be a promising therapeutic approach. Prebiotics and probiotics showed beneficial results in fatty liver disease. Mice fed with high-fat diet were treated with probiotic Lactobacillus johnsonii BS15 for a total period of 17 wk. BS15 treatment modulated gut microbiota composition by increasing Lactobacillus spp. and Lactobacillus johnsonii, and decreasing Enterobacteriaceae family. It improved gut-barrier function, and protected against hepatic steatosis and insulin resistance compared with PBS control (170). Similarly, high-fat-diet-fed mice treated with a multispecies probiotic formulation showed that tight-junction proteins zonula occludens (ZO)-1 and ZO-2 were restored and that hepatic triglyceride and inflammation were reduced compared with non-probiotic treated mice (24). In patients with NAFLD and NASH, probiotic treatment also exhibited beneficial effects. Administration of a mixture of Lactobacillus bulgaricus and Streptococcus thermophilus in adult patients with NAFLD reduced liver aminotransferases (AST, ALT, GGT) levels compared with the placebo group (starch) (7). In a cohort of patients with NASH, probiotic plus metformin showed better improvement on liver enzyme aminotransferases than metformin alone (144). In another NASH patient cohort, long-time probiotic treatment with Lepicol (a mixture of five probiotic strains) for 6 mo reduced intrahepatic triglyceride content and AST but didn't affect body mass index, glucose, and lipid levels (166). In summary, changes in the microbial composition contribute to the onset and progression of NAFLD and NASH. Future studies are needed to confirm the efficacy and long-term effect of probiotics in independent patient cohorts with NAFLD. Since the gut microbiome is a robust system, long-term effects of probiotics should be evaluated to examine whether the gut microbiota reverts back after treatment is terminated. Although no safety issues have been reported in this patient population, the dose, treatment combination, and treatment length should be harmonized to reproduce and validate results across clinical trials.
Cholestatic Liver Disease
Cholestatic liver disease is a diverse cluster of diseases that can affect both children and adults. It ranges from acute reversible cholestasis to chronic, progressive diseases such as primary biliary cholangitis (PBC) and primary sclerosing cholangitis (PSC) (93). In this section, we will provide an overview of available data about preclinical and clinical studies targeting selected microbiota from a treatment perspective.
PBC is an autoimmune liver disease characterized by portal inflammation and destruction of intrahepatic bile ducts, leading to fibrosis and potentially cirrhosis (79). Recent progress has been made to assess the microbial composition in PBC and how altered gut microbiota can be used as biomarker and therapeutic target (FIGURE 2C) (108). Metagenomic sequencing of fecal samples from PBC patients showed that the microbiota was depleted of Lachnobacterium sp., Bacteroides eggerthii, Ruminococcus bromii, and Acidobacteria, but was enriched in some of the pathobiont taxa, such as Enterobacteriaceae, Neisseriaceae, Streptococcus, and Proteobacteria. Interestingly, several altered gut bacteria exhibited association with metabolic functions, immunity, and liver functions (108). Gut microbial alteration was confirmed in a larger cohort of PBC patients, showing increases in Eubacterium and Veillonella and a significant decrease in Fusobacterium in the oral microbiota compared with heathy controls (1). In the fecal microbiota of patients with PSC, Klebsiella pneumoniae was identified to disrupt the intestinal epithelial barrier, translocate to the mesenteric lymph nodes, and initiate a Th17 inflammatory response in the liver. Gnotobiotic mice colonized with fecal material from PSC patients exhibited T-helper 17 cell response and increased susceptibility to hepatobiliary injuries (124). Currently, ursodeoxycholic acid (UDCA) and obeticholic acid (as second line therapy) are the only two drugs approved for the treatment of PBC (44). UDCA has been shown to inhibit cholangiocyte proliferation and to have choleretic effects (35, 58, 103). UDCA therapy of PBC patients for 6 mo modulated the fecal microbiota and reduced the abundance of three PBC-enriched genera (Hemophilus spp, Streptococcus spp, and Pseudomonas spp) (150).
In addition to bacteria, a recent study found fungal microbiota changes in PSC patients compared with IBD patients and healthy subjects, characterized by an increased biodiversity and composition alteration. Specifically, an increased proportion of Exophiala and decreased Saccharomyces cerevisiae were observed. By building an abundance correlation network between bacterial and fungi microbiota, Lemoinne et al. were able to detect that PSC was associated with disruption of the bacterial-fungi network (98).
In line with human studies, animal model of cholestasis (bile duct ligation) showed changes in microbial composition as early as 3 days after the procedure (26). Another mouse model of human PSC, the Mdr2−/− mouse, also exhibited an increased bacterial translocation and altered gut microbiota composition compared with wild-type mice. Transferring microbiota from Mdr2−/− mice to wild-type mice induced significant intestinal dysbiosis and liver injury, suggesting a link between gut microbiota and disease progression in Mdr2−/− mice (102). In conclusion, accumulating evidence from preclinical and clinical research efforts continues to strengthen the association between microbiota and cholestatic liver diseases. With the development in high-throughput sequencing, more novel mechanisms and therapeutic targets should be revealed.
Conclusions
In the past decade, the interplay between gut microbiome and liver diseases has been increasingly recognized. Microbial products regulate bile acid synthesis, and glucose and lipid metabolism in the liver through the portal vein. Gut dysbiosis and changes in microbial taxonomy are observed in patients within a wide variety of liver diseases (ALD, NAFLD, NASH, and cholestatic liver disease). With animal models mimicking human disease, we are able to elucidate many important mechanistic pathways and apply findings to humans. However, commonly used approaches in the field of microbiota research have limitations to establish causality between microbiome changes and liver diseases. For example, FMT in mice is commonly used for investigating a causal role of microbiota (38). However, it may fail to alter the microbiome if not severely altered in diseases, and colonization of mice with a human microbiota is not always stable (158). In chronic diseases, FMTs are impractical to assess the causative role of gut microbiota, considering the need for long prospective studies (157). In addition, as we characterize the microbial community, technical platforms should be standardized so that results can be replicated and compared across the laboratories. Since we use animal models extensively, it is critical that the animal models recapitulate human diseases as close as possible. Therefore, selectively targeting microbiota within a controlled environment could be more therapeutically relevant. It is encouraging that manipulating microbiota through FMTs and antibiotic/probiotic treatment provides us microbiota-based therapeutic modalities for diseases that can easily be translated into clinical practice. In a pilot study, patients with alcoholic hepatitis who received 1 wk of FMT from healthy donors showed improved liver function and survival (127). Moreover, patients with cirrhosis with recurrent hepatic encephalopathy received FMT from a rationally selected donor. FMT recipients showed lower hospitalizations, improved cognitive tests, and increased microbial diversity and beneficial taxa (12). Several clinical trials are still ongoing to investigate the effect of FMT on NASH (NCT02469272), chronic hepatitis B (NCT03429439), obesity (NCT02741518), and Type 2 diabetes mellitus (NCT02346669). Since the majority of human studies cannot provide mechanistic insights, more preclinical effort is required to identify the underlying “causal component” of gut microbiota changes. We still have a long way to go, but with the development of new technologies, we are closer to more accurate diagnostic tools and effective treatments to improve liver diseases in patients.
Acknowledgments
This study was supported in part by a Biocodex Microbiota Foundation Grant, National Institutes of Health Grants R01 AA-24726, R01 AA-020703, and U01 AA-026939, by Award No. BX004594 from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development (to B.S.), and services provided by P30 DK-120515 and P50 AA-011999.
Dr. Bernd Schnabl has been consulting for Ferring Research Institute, Intercept Pharmaceuticals, HOST Therabiomics, and Patara Pharmaceuticals. Dr. Bernd Schnabl's institution (UC San Diego) has received grant support from BiomX, NGM Biopharmaceuticals, CymaBay Therapeutics, and Synlogic Operating Company.
L.J. prepared figures; L.J. drafted manuscript; B.S. conceived and designed research; B.S. edited and revised manuscript; B.S. approved final version of manuscript.
References
- 1.Abe K, Takahashi A, Fujita M, Imaizumi H, Hayashi M, Okai K, Ohira H. Dysbiosis of oral microbiota and its association with salivary immunological biomarkers in autoimmune liver disease. PLoS One 13: e0198757, 2018. doi: 10.1371/journal.pone.0198757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adachi Y, Moore LE, Bradford BU, Gao W, Thurman RG. Antibiotics prevent liver injury in rats following long-term exposure to ethanol. Gastroenterology 108: 218–224, 1995. doi: 10.1016/0016-5085(95)90027-6. [DOI] [PubMed] [Google Scholar]
- 3.Agus A, Planchais J, Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe 23: 716–724, 2018. doi: 10.1016/j.chom.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Alexander C, Rietschel ET. Bacterial lipopolysaccharides and innate immunity. J Endotoxin Res 7: 167–202, 2001. [PubMed] [Google Scholar]
- 5.Alexander JS, Ganta VC, Jordan PA, Witte MH. Gastrointestinal lymphatics in health and disease. Pathophysiology 17: 315–335, 2010. doi: 10.1016/j.pathophys.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alisi A, Manco M, Devito R, Piemonte F, Nobili V. Endotoxin and plasminogen activator inhibitor-1 serum levels associated with nonalcoholic steatohepatitis in children. J Pediatr Gastroenterol Nutr 50: 645–649, 2010. doi: 10.1097/MPG.0b013e3181c7bdf1. [DOI] [PubMed] [Google Scholar]
- 7.Aller R, De Luis DA, Izaola O, Conde R, Gonzalez Sagrado M, Primo D, De La Fuente B, Gonzalez J. Effect of a probiotic on liver aminotransferases in nonalcoholic fatty liver disease patients: a double blind randomized clinical trial. Eur Rev Med Pharmacol Sci 15: 1090–1095, 2011. [PubMed] [Google Scholar]
- 8.Alvaro A, Solà R, Rosales R, Ribalta J, Anguera A, Masana L, Vallvé JC. Gene expression analysis of a human enterocyte cell line reveals downregulation of cholesterol biosynthesis in response to short-chain fatty acids. IUBMB Life 60: 757–764, 2008. doi: 10.1002/iub.110. [DOI] [PubMed] [Google Scholar]
- 9.Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci USA 104: 979–984, 2007. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Badawy AA. Tryptophan availability for kynurenine pathway metabolism across the life span: Control mechanisms and focus on aging, exercise, diet and nutritional supplements. Neuropharmacology 112: 248–263, 2017. doi: 10.1016/j.neuropharm.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 11.Bajaj JS. Alcohol, liver disease and the gut microbiota. Nat Rev Gastroenterol Hepatol 16: 235–246, 2019. doi: 10.1038/s41575-018-0099-1. [DOI] [PubMed] [Google Scholar]
- 12.Bajaj JS, Kassam Z, Fagan A, Gavis EA, Liu E, Cox IJ, Kheradman R, Heuman D, Wang J, Gurry T, Williams R, Sikaroodi M, Fuchs M, Alm E, John B, Thacker LR, Riva A, Smith M, Taylor-Robinson SD, Gillevet PM. Fecal microbiota transplant from a rational stool donor improves hepatic encephalopathy: A randomized clinical trial. Hepatology 66: 1727–1738, 2017. doi: 10.1002/hep.29306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker JR, Chaykin S. The biosynthesis of trimethylamine-N-oxide. J Biol Chem 237: 1309–1313, 1962. [PubMed] [Google Scholar]
- 14.Bengmark S. Bioecologic control of the gastrointestinal tract: the role of flora and supplemented probiotics and synbiotics. Gastroenterol Clin North Am 34: 413–436, 2005. doi: 10.1016/j.gtc.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 15.Bergheim I, Weber S, Vos M, Krämer S, Volynets V, Kaserouni S, McClain CJ, Bischoff SC. Antibiotics protect against fructose-induced hepatic lipid accumulation in mice: role of endotoxin. J Hepatol 48: 983–992, 2008. doi: 10.1016/j.jhep.2008.01.035. [DOI] [PubMed] [Google Scholar]
- 16.Bergman EN. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol Rev 70: 567–590, 1990. doi: 10.1152/physrev.1990.70.2.567. [DOI] [PubMed] [Google Scholar]
- 17.Betrapally NS, Gillevet PM, Bajaj JS. Changes in the intestinal microbiome and alcoholic and nonalcoholic liver diseases: causes or effects? Gastroenterology 150: 1745–1755e3, 2016. doi: 10.1053/j.gastro.2016.02.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bischoff SC. ‘Gut health’: a new objective in medicine? BMC Med 9: 24, 2011. doi: 10.1186/1741-7015-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blumberg H, McCollum EV. The prevention by choline of liver cirrhosis in rats on high fat, low protein diets. Science 93: 598–599, 1941. doi: 10.1126/science.93.2425.598. [DOI] [PubMed] [Google Scholar]
- 20.Bode C, Kugler V, Bode JC. Endotoxemia in patients with alcoholic and non-alcoholic cirrhosis and in subjects with no evidence of chronic liver disease following acute alcohol excess. J Hepatol 4: 8–14, 1987. doi: 10.1016/S0168-8278(87)80003-X. [DOI] [PubMed] [Google Scholar]
- 21.Bolognini D, Tobin AB, Milligan G, Moss CE. The pharmacology and function of receptors for short-chain fatty acids. Mol Pharmacol 89: 388–398, 2016. doi: 10.1124/mol.115.102301. [DOI] [PubMed] [Google Scholar]
- 22.Brandl K, Schnabl B. Intestinal microbiota and nonalcoholic steatohepatitis. Curr Opin Gastroenterol 33: 128–133, 2017. doi: 10.1097/MOG.0000000000000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brighton CA, Rievaj J, Kuhre RE, Glass LL, Schoonjans K, Holst JJ, Gribble FM, Reimann F. Bile acids trigger GLP-1 release predominantly by accessing basolaterally located G protein-coupled bile acid receptors. Endocrinology 156: 3961–3970, 2015. doi: 10.1210/en.2015-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Briskey D, Heritage M, Jaskowski LA, Peake J, Gobe G, Subramaniam VN, Crawford D, Campbell C, Vitetta L. Probiotics modify tight-junction proteins in an animal model of nonalcoholic fatty liver disease. Therap Adv Gastroenterol 9: 463–472, 2016. doi: 10.1177/1756283X16645055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D, Muir AI, Wigglesworth MJ, Kinghorn I, Fraser NJ, Pike NB, Strum JC, Steplewski KM, Murdock PR, Holder JC, Marshall FH, Szekeres PG, Wilson S, Ignar DM, Foord SM, Wise A, Dowell SJ. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem 278: 11312–11319, 2003. doi: 10.1074/jbc.M211609200. [DOI] [PubMed] [Google Scholar]
- 26.Cabrera-Rubio R, Patterson AM, Cotter PD, Beraza N. Cholestasis induced by bile duct ligation promotes changes in the intestinal microbiome in mice. Sci Rep 9: 12324, 2019. doi: 10.1038/s41598-019-48784-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Canani RB, Costanzo MD, Leone L, Pedata M, Meli R, Calignano A. Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J Gastroenterol 17: 1519–1528, 2011. doi: 10.3748/wjg.v17.i12.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caussy C, Hsu C, Min-Tzu L, Amy L, Bettencourt R, Ajmera V, Shirin B, Hooker J, Ethan S, Richards L, Schork N, Schnabl B, David B, Sirlin C, Chi-Hua C, Loomba R. Novel link between gut-microbiome derived metabolite and shared gene-effects with hepatic steatosis and fibrosis in NAFLD. Hepatology 68: 918–932, 2018. doi: 10.1016/S0168-8278(18)30275-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J, Thomsen M, Vitetta L. Interaction of gut microbiota with dysregulation of bile acids in the pathogenesis of nonalcoholic fatty liver disease and potential therapeutic implications of probiotics. J Cell Biochem 120: 2713–2720, 2019. doi: 10.1002/jcb.27635. [DOI] [PubMed] [Google Scholar]
- 30.Chiang JY. Bile acids: regulation of synthesis. J Lipid Res 50: 1955–1966, 2009. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chow MD, Lee YH, Guo GL. The role of bile acids in nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Mol Aspects Med 56: 34–44, 2017. doi: 10.1016/j.mam.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ciorba MA. A gastroenterologist’s guide to probiotics. Clin Gastroenterol Hepatol 10: 960–968, 2012. doi: 10.1016/j.cgh.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clarke G, Fitzgerald P, Cryan JF, Cassidy EM, Quigley EM, Dinan TG. Tryptophan degradation in irritable bowel syndrome: evidence of indoleamine 2,3-dioxygenase activation in a male cohort. BMC Gastroenterol 9: 6, 2009. doi: 10.1186/1471-230X-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci USA 106: 14728–14733, 2009. doi: 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Copaci I, Micu L, Iliescu L, Voiculescu M. New therapeutical indications of ursodeoxycholic acid. Rom J Gastroenterol 14: 259–266, 2005. [PubMed] [Google Scholar]
- 36.Crispe IN. Immune tolerance in liver disease. Hepatology 60: 2109–2117, 2014. doi: 10.1002/hep.27254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dalile B, Van Oudenhove L, Vervliet B, Verbeke K. The role of short-chain fatty acids in microbiota-gut-brain communication. Nat Rev Gastroenterol Hepatol 16: 461–478, 2019. doi: 10.1038/s41575-019-0157-3. [DOI] [PubMed] [Google Scholar]
- 38.de Groot PF, Frissen MN, de Clercq NC, Nieuwdorp M. Fecal microbiota transplantation in metabolic syndrome: history, present and future. Gut Microbes 8: 253–267, 2017. doi: 10.1080/19490976.2017.1293224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Vadder F, Kovatcheva-Datchary P, Zitoun C, Duchampt A, Bäckhed F, Mithieux G. Microbiota-produced succinate improves glucose homeostasis via intestinal gluconeogenesis. Cell Metab 24: 151–157, 2016. doi: 10.1016/j.cmet.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 40.den Besten G, Bleeker A, Gerding A, van Eunen K, Havinga R, van Dijk TH, Oosterveer MH, Jonker JW, Groen AK, Reijngoud DJ, Bakker BM. Short-chain fatty acids protect against high-fat diet-induced obesity via a PPARγ-dependent switch from lipogenesis to fat oxidation. Diabetes 64: 2398–2408, 2015. doi: 10.2337/db14-1213. [DOI] [PubMed] [Google Scholar]
- 41.Duan Y, Llorente C, Lang S, Brandl K, Chu H, Jiang L, White RC, Clarke TH, Nguyen K, Torralba M, Shao Y, Liu J, Hernandez-Morales A, Lessor L, Rahman IR, Miyamoto Y, Ly M, Gao B, Sun W, Kiesel R, Hutmacher F, Lee S, Ventura-Cots M, Bosques-Padilla F, Verna EC, Abraldes JG, Brown RS Jr, Vargas V, Altamirano J, Caballería J, Shawcross DL, Ho SB, Louvet A, Lucey MR, Mathurin P, Garcia-Tsao G, Bataller R, Tu XM, Eckmann L, van der Donk WA, Young R, Lawley TD, Stärkel P, Pride D, Fouts DE, Schnabl B. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature 575: 505–511, 2019. doi: 10.1038/s41586-019-1742-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dumas ME, Barton RH, Toye A, Cloarec O, Blancher C, Rothwell A, Fearnside J, Tatoud R, Blanc V, Lindon JC, Mitchell SC, Holmes E, McCarthy MI, Scott J, Gauguier D, Nicholson JK. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci USA 103: 12511–12516, 2006. doi: 10.1073/pnas.0601056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Etienne-Mesmin L, Vijay-Kumar M, Gewirtz AT, Chassaing B. Hepatocyte Toll-like receptor 5 promotes bacterial clearance and protects mice against high-fat diet-induced liver disease. Cell Mol Gastroenterol Hepatol 2: 584–604, 2016. doi: 10.1016/j.jcmgh.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.European Association for the Study of the Liver EASL Clinical Practice Guidelines: management of cholestatic liver diseases. J Hepatol 51: 237–267, 2009. doi: 10.1016/j.jhep.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 45.Favennec M, Hennart B, Caiazzo R, Leloire A, Yengo L, Verbanck M, Arredouani A, Marre M, Pigeyre M, Bessede A, Guillemin GJ, Chinetti G, Staels B, Pattou F, Balkau B, Allorge D, Froguel P, Poulain-Godefroy O. The kynurenine pathway is activated in human obesity and shifted toward kynurenine monooxygenase activation. Obesity (Silver Spring) 23: 2066–2074, 2015. doi: 10.1002/oby.21199. [DOI] [PubMed] [Google Scholar]
- 46.Feng HY, Chen YC. Role of bile acids in carcinogenesis of pancreatic cancer: An old topic with new perspective. World J Gastroenterol 22: 7463–7477, 2016. doi: 10.3748/wjg.v22.i33.7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng W, Ao H, Peng C. Gut microbiota, short-chain fatty acids, and herbal medicines. Front Pharmacol 9: 1354, 2018. doi: 10.3389/fphar.2018.01354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferrier L, Bérard F, Debrauwer L, Chabo C, Langella P, Buéno L, Fioramonti J. Impairment of the intestinal barrier by ethanol involves enteric microflora and mast cell activation in rodents. Am J Pathol 168: 1148–1154, 2006. doi: 10.2353/ajpath.2006.050617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferslew BC, Xie G, Johnston CK, Su M, Stewart PW, Jia W, Brouwer KL, Barritt AS IV. Altered bile acid metabolome in patients with nonalcoholic steatohepatitis. Dig Dis Sci 60: 3318–3328, 2015. doi: 10.1007/s10620-015-3776-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flint HJ, Scott KP, Louis P, Duncan SH. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol 9: 577–589, 2012. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- 51.Forman BM, Goode E, Chen J, Oro AE, Bradley DJ, Perlmann T, Noonan DJ, Burka LT, McMorris T, Lamph WW, Evans RM, Weinberger C. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell 81: 687–693, 1995. doi: 10.1016/0092-8674(95)90530-8. [DOI] [PubMed] [Google Scholar]
- 52.Fukunishi S, Sujishi T, Takeshita A, Ohama H, Tsuchimoto Y, Asai A, Tsuda Y, Higuchi K. Lipopolysaccharides accelerate hepatic steatosis in the development of nonalcoholic fatty liver disease in Zucker rats. J Clin Biochem Nutr 54: 39–44, 2014. doi: 10.3164/jcbn.13-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao B, Bataller R. Alcoholic liver disease: pathogenesis and new therapeutic targets. Gastroenterology 141: 1572–1585, 2011. doi: 10.1053/j.gastro.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao J, Xu K, Liu H, Liu G, Bai M, Peng C, Li T, Yin Y. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front Cell Infect Microbiol 8: 13, 2018. doi: 10.3389/fcimb.2018.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao Z, Yin J, Zhang J, Ward RE, Martin RJ, Lefevre M, Cefalu WT, Ye J. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes 58: 1509–1517, 2009. doi: 10.2337/db08-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gérard P. Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens 3: 14–24, 2013. doi: 10.3390/pathogens3010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goffredo M, Santoro N, Tricò D, Giannini C, D’Adamo E, Zhao H, Peng G, Yu X, Lam TT, Pierpont B, Caprio S, Herzog RI. A branched-chain amino acid-related metabolic signature characterizes obese adolescents with non-alcoholic fatty liver disease. Nutrients 9: E642, 2017. doi: 10.3390/nu9070642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gong Y, Huang Z, Christensen E, Gluud C. Ursodeoxycholic acid for patients with primary biliary cirrhosis: an updated systematic review and meta-analysis of randomized clinical trials using Bayesian approach as sensitivity analyses. Am J Gastroenterol 102: 1799–1807, 2007. doi: 10.1111/j.1572-0241.2007.01235.x. [DOI] [PubMed] [Google Scholar]
- 59.Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME, Maloney PR, Willson TM, Kliewer SA. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell 6: 517–526, 2000. doi: 10.1016/S1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 60.Gupta H, Youn GS, Shin MJ, Suk KT. Role of gut microbiota in hepatocarcinogenesis. Microorganisms 7: 121, 2019. doi: 10.3390/microorganisms7050121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haiko J, Westerlund-Wikström B. The role of the bacterial flagellum in adhesion and virulence. Biology (Basel) 2: 1242–1267, 2013. doi: 10.3390/biology2041242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hajam IA, Dar PA, Shahnawaz I, Jaume JC, Lee JH. Bacterial flagellin-a potent immunomodulatory agent. Exp Mol Med 49: e373, 2017. doi: 10.1038/emm.2017.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harte AL, da Silva NF, Creely SJ, McGee KC, Billyard T, Youssef-Elabd EM, Tripathi G, Ashour E, Abdalla MS, Sharada HM, Amin AI, Burt AD, Kumar S, Day CP, McTernan PG. Elevated endotoxin levels in non-alcoholic fatty liver disease. J Inflamm (Lond) 7: 15, 2010. doi: 10.1186/1476-9255-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hashimoto T, Perlot T, Rehman A, Trichereau J, Ishiguro H, Paolino M, Sigl V, Hanada T, Hanada R, Lipinski S, Wild B, Camargo SM, Singer D, Richter A, Kuba K, Fukamizu A, Schreiber S, Clevers H, Verrey F, Rosenstiel P, Penninger JM. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature 487: 477–481, 2012. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hendrikx T, Duan Y, Wang Y, Oh JH, Alexander LM, Huang W, Stärkel P, Ho SB, Gao B, Fiehn O, Emond P, Sokol H, van Pijkeren JP, Schnabl B. Bacteria engineered to produce IL-22 in intestine induce expression of REG3G to reduce ethanol-induced liver disease in mice. Gut 68: 1504–1515, 2019. doi: 10.1136/gutjnl-2018-317232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hofmann AF. Detoxification of lithocholic acid, a toxic bile acid: relevance to drug hepatotoxicity. Drug Metab Rev 36: 703–722, 2004. doi: 10.1081/DMR-200033475. [DOI] [PubMed] [Google Scholar]
- 67.Hov JR, Karlsen TH. The microbiome in primary sclerosing cholangitis: current evidence and potential concepts. Semin Liver Dis 37: 314–331, 2017. doi: 10.1055/s-0037-1608801. [DOI] [PubMed] [Google Scholar]
- 68.Hoyles L, Fernández-Real JM, Federici M, Serino M, Abbott J, Charpentier J, Heymes C, Luque JL, Anthony E, Barton RH, Chilloux J, Myridakis A, Martinez-Gili L, Moreno-Navarrete JM, Benhamed F, Azalbert V, Blasco-Baque V, Puig J, Xifra G, Ricart W, Tomlinson C, Woodbridge M, Cardellini M, Davato F, Cardolini I, Porzio O, Gentileschi P, Lopez F, Foufelle F, Butcher SA, Holmes E, Nicholson JK, Postic C, Burcelin R, Dumas ME. Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat Med 24: 1070–1080, 2018. doi: 10.1038/s41591-018-0061-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu X, Bonde Y, Eggertsen G, Rudling M. Muricholic bile acids are potent regulators of bile acid synthesis via a positive feedback mechanism. J Intern Med 275: 27–38, 2014. doi: 10.1111/joim.12140. [DOI] [PubMed] [Google Scholar]
- 70.Inagaki T, Moschetta A, Lee YK, Peng L, Zhao G, Downes M, Yu RT, Shelton JM, Richardson JA, Repa JJ, Mangelsdorf DJ, Kliewer SA. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci USA 103: 3920–3925, 2006. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Inokuchi S, Tsukamoto H, Park E, Liu ZX, Brenner DA, Seki E. Toll-like receptor 4 mediates alcohol-induced steatohepatitis through bone marrow-derived and endogenous liver cells in mice. Alcohol Clin Exp Res 35: 1509–1518, 2011. doi: 10.1111/j.1530-0277.2011.01487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Islam KBMS, Fukiya S, Hagio M, Fujii N, Ishizuka S, Ooka T, Ogura Y, Hayashi T, Yokota A. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology 141: 1773–1781, 2011. doi: 10.1053/j.gastro.2011.07.046. [DOI] [PubMed] [Google Scholar]
- 73.Jiang C, Xie C, Li F, Zhang L, Nichols RG, Krausz KW, Cai J, Qi Y, Fang ZZ, Takahashi S, Tanaka N, Desai D, Amin SG, Albert I, Patterson AD, Gonzalez FJ. Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. J Clin Invest 125: 386–402, 2015. doi: 10.1172/JCI76738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Joyce SA, MacSharry J, Casey PG, Kinsella M, Murphy EF, Shanahan F, Hill C, Gahan CG. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc Natl Acad Sci USA 111: 7421–7426, 2014. doi: 10.1073/pnas.1323599111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kakiyama G, Hylemon PB, Zhou H, Pandak WM, Heuman DM, Kang DJ, Takei H, Nittono H, Ridlon JM, Fuchs M, Gurley EC, Wang Y, Liu R, Sanyal AJ, Gillevet PM, Bajaj JS. Colonic inflammation and secondary bile acids in alcoholic cirrhosis. Am J Physiol Gastrointest Liver Physiol 306: G929–G937, 2014. doi: 10.1152/ajpgi.00315.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB, Heuman DM, Daita K, Takei H, Muto A, Nittono H, Ridlon JM, White MB, Noble NA, Monteith P, Fuchs M, Thacker LR, Sikaroodi M, Bajaj JS. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol 58: 949–955, 2013. doi: 10.1016/j.jhep.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kałużna-Czaplińska J, Gątarek P, Chirumbolo S, Chartrand MS, Bjørklund G. How important is tryptophan in human health? Crit Rev Food Sci Nutr 59: 72–88, 2019. doi: 10.1080/10408398.2017.1357534. [DOI] [PubMed] [Google Scholar]
- 78.Kamada N, Chen GY, Inohara N, Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol 14: 685–690, 2013. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kaplan MM, Gershwin ME. Primary biliary cirrhosis. N Engl J Med 353: 1261–1273, 2005. doi: 10.1056/NEJMra043898. [DOI] [PubMed] [Google Scholar]
- 80.Kawamata Y, Fujii R, Hosoya M, Harada M, Yoshida H, Miwa M, Fukusumi S, Habata Y, Itoh T, Shintani Y, Hinuma S, Fujisawa Y, Fujino M. A G protein-coupled receptor responsive to bile acids. J Biol Chem 278: 9435–9440, 2003. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- 81.Keitel V, Donner M, Winandy S, Kubitz R, Häussinger D. Expression and function of the bile acid receptor TGR5 in Kupffer cells. Biochem Biophys Res Commun 372: 78–84, 2008. doi: 10.1016/j.bbrc.2008.04.171. [DOI] [PubMed] [Google Scholar]
- 82.Keitel V, Reinehr R, Gatsios P, Rupprecht C, Görg B, Selbach O, Häussinger D, Kubitz R. The G-protein coupled bile salt receptor TGR5 is expressed in liver sinusoidal endothelial cells. Hepatology 45: 695–704, 2007. doi: 10.1002/hep.21458. [DOI] [PubMed] [Google Scholar]
- 83.Keshavarzian A, Farhadi A, Forsyth CB, Rangan J, Jakate S, Shaikh M, Banan A, Fields JZ. Evidence that chronic alcohol exposure promotes intestinal oxidative stress, intestinal hyperpermeability and endotoxemia prior to development of alcoholic steatohepatitis in rats. J Hepatol 50: 538–547, 2009. doi: 10.1016/j.jhep.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Keshavarzian A, Holmes EW, Patel M, Iber F, Fields JZ, Pethkar S. Leaky gut in alcoholic cirrhosis: a possible mechanism for alcohol-induced liver damage. Am J Gastroenterol 94: 200–207, 1999. doi: 10.1111/j.1572-0241.1999.00797.x. [DOI] [PubMed] [Google Scholar]
- 85.Khan I, Ullah N, Zha L, Bai Y, Khan A, Zhao T, Che T, Zhang C. Alteration of gut microbiota in inflammatory bowel disease (IBD): Cause or consequence? IBD treatment targeting the gut microbiome. Pathogens 8: E126, 2019. doi: 10.3390/pathogens8030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kimura I, Inoue D, Hirano K, Tsujimoto G. The SCFA receptor GPR43 and energy metabolism. Front Endocrinol (Lausanne) 5: 85, 2014. doi: 10.3389/fendo.2014.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kinross JM, Darzi AW, Nicholson JK. Gut microbiome-host interactions in health and disease. Genome Med 3: 14, 2011. doi: 10.1186/gm228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kirpich IA, Solovieva NV, Leikhter SN, Shidakova NA, Lebedeva OV, Sidorov PI, Bazhukova TA, Soloviev AG, Barve SS, McClain CJ, Cave M. Probiotics restore bowel flora and improve liver enzymes in human alcohol-induced liver injury: a pilot study. Alcohol 42: 675–682, 2008. doi: 10.1016/j.alcohol.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, DiDonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WH, Bushman FD, Lusis AJ, Hazen SL. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med 19: 576–585, 2013. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kolodziejczyk AA, Zheng D, Shibolet O, Elinav E. The role of the microbiome in NAFLD and NASH. EMBO Mol Med 11: e9302, 2019. doi: 10.15252/emmm.201809302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, Baselga J, Liu C, Shivdasani RA, Ogino S, Birren BW, Huttenhower C, Garrett WS, Meyerson M. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 22: 292–298, 2012. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kuczynski J, Lauber CL, Walters WA, Parfrey LW, Clemente JC, Gevers D, Knight R. Experimental and analytical tools for studying the human microbiome. Nat Rev Genet 13: 47–58, 2011. doi: 10.1038/nrg3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kummen M, Hov JR. The gut microbial influence on cholestatic liver disease. Liver Int 39: 1186–1196, 2019. doi: 10.1111/liv.14153. [DOI] [PubMed] [Google Scholar]
- 94.Le Floc’h N, Otten W, Merlot E. Tryptophan metabolism, from nutrition to potential therapeutic applications. Amino Acids 41: 1195–1205, 2011. doi: 10.1007/s00726-010-0752-7. [DOI] [PubMed] [Google Scholar]
- 95.Le Poul E, Loison C, Struyf S, Springael JY, Lannoy V, Decobecq ME, Brezillon S, Dupriez V, Vassart G, Van Damme J, Parmentier M, Detheux M. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J Biol Chem 278: 25481–25489, 2003. doi: 10.1074/jbc.M301403200. [DOI] [PubMed] [Google Scholar]
- 96.Leclercq S, Matamoros S, Cani PD, Neyrinck AM, Jamar F, Stärkel P, Windey K, Tremaroli V, Bäckhed F, Verbeke K, de Timary P, Delzenne NM. Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc Natl Acad Sci USA 111: E4485–E4493, 2014. doi: 10.1073/pnas.1415174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev 89: 147–191, 2009. doi: 10.1152/physrev.00010.2008. [DOI] [PubMed] [Google Scholar]
- 98.Lemoinne S, Kemgang A, Ben Belkacem K, Straube M, Jegou S, Corpechot C, Saint-Antoine IBDN, Chazouilleres O, Housset C, Sokol H. Fungi participate in the dysbiosis of gut microbiota in patients with primary sclerosing cholangitis. Gut 69: 92–102, 2020. doi: 10.1136/gutjnl-2018-317791. [DOI] [PubMed] [Google Scholar]
- 99.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA 102: 11070–11075, 2005. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li F, Duan K, Wang C, McClain C, Feng W. Probiotics and alcoholic liver disease: treatment and potential mechanisms. Gastroenterol Res Pract 2016: 5491465, 2016. doi: 10.1155/2016/5491465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li YH, Woo SH, Choi DH, Cho EH. Succinate causes α-SMA production through GPR91 activation in hepatic stellate cells. Biochem Biophys Res Commun 463: 853–858, 2015. doi: 10.1016/j.bbrc.2015.06.023. [DOI] [PubMed] [Google Scholar]
- 102.Liao L, Schneider KM, Galvez EJC, Frissen M, Marschall HU, Su H, Hatting M, Wahlström A, Haybaeck J, Puchas P, Mohs A, Peng J, Bergheim I, Nier A, Hennings J, Reißing J, Zimmermann HW, Longerich T, Strowig T, Liedtke C, Cubero FJ, Trautwein C. Intestinal dysbiosis augments liver disease progression via NLRP3 in a murine model of primary sclerosing cholangitis. Gut 68: 1477–1492, 2019. doi: 10.1136/gutjnl-2018-316670. [DOI] [PubMed] [Google Scholar]
- 103.Lindor KD, Dickson ER, Baldus WP, Jorgensen RA, Ludwig J, Murtaugh PA, Harrison JM, Wiesner RH, Anderson ML, Lange SM, Lesage G, Rossi SS, Hofmann AF. Ursodeoxycholic acid in the treatment of primary biliary cirrhosis. Gastroenterology 106: 1284–1290, 1994. doi: 10.1016/0016-5085(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 104.Loomba R, Seguritan V, Li W, Long T, Klitgord N, Bhatt A, Dulai PS, Caussy C, Bettencourt R, Highlander SK, Jones MB, Sirlin CB, Schnabl B, Brinkac L, Schork N, Chen CH, Brenner DA, Biggs W, Yooseph S, Venter JC, Nelson KE. Gut microbiome-based metagenomic signature for non-invasive detection of advanced fibrosis in human nonalcoholic fatty liver disease. Cell Metab 25: 1054–1062e5, 2017. doi: 10.1016/j.cmet.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, Mangelsdorf DJ. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell 6: 507–515, 2000. doi: 10.1016/S1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- 106.Lu YC, Yeh WC, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine 42: 145–151, 2008. doi: 10.1016/j.cyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 107.Luther J, Garber JJ, Khalili H, Dave M, Bale SS, Jindal R, Motola DL, Luther S, Bohr S, Jeoung SW, Deshpande V, Singh G, Turner JR, Yarmush ML, Chung RT, Patel SJ. Hepatic injury in nonalcoholic steatohepatitis contributes to altered intestinal permeability. Cell Mol Gastroenterol Hepatol 1: 222–232, 2015. doi: 10.1016/j.jcmgh.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lv LX, Fang DQ, Shi D, Chen DY, Yan R, Zhu YX, Chen YF, Shao L, Guo FF, Wu WR, Li A, Shi HY, Jiang XW, Jiang HY, Xiao YH, Zheng SS, Li LJ. Alterations and correlations of the gut microbiome, metabolism and immunity in patients with primary biliary cirrhosis. Environ Microbiol 18: 2272–2286, 2016. doi: 10.1111/1462-2920.13401. [DOI] [PubMed] [Google Scholar]
- 109.Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proc Nutr Soc 62: 67–72, 2003. doi: 10.1079/PNS2002207. [DOI] [PubMed] [Google Scholar]
- 110.Makki K, Deehan EC, Walter J, Bäckhed F. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe 23: 705–715, 2018. doi: 10.1016/j.chom.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 111.Malhi H, Camilleri M. Modulating bile acid pathways and TGR5 receptors for treating liver and GI diseases. Curr Opin Pharmacol 37: 80–86, 2017. doi: 10.1016/j.coph.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mallmann NH, Lima ES, Lalwani P. Dysregulation of tryptophan catabolism in metabolic syndrome. Metab Syndr Relat Disord 16: 135–142, 2018. doi: 10.1089/met.2017.0097. [DOI] [PubMed] [Google Scholar]
- 113.Maron PA, Ranjard L, Mougel C, Lemanceau P. Metaproteomics: a new approach for studying functional microbial ecology. Microb Ecol 53: 486–493, 2007. doi: 10.1007/s00248-006-9196-8. [DOI] [PubMed] [Google Scholar]
- 114.Maruyama T, Miyamoto Y, Nakamura T, Tamai Y, Okada H, Sugiyama E, Nakamura T, Itadani H, Tanaka K. Identification of membrane-type receptor for bile acids (M-BAR). Biochem Biophys Res Commun 298: 714–719, 2002. doi: 10.1016/S0006-291X(02)02550-0. [DOI] [PubMed] [Google Scholar]
- 115.McBurney MI, Davis C, Fraser CM, Schneeman BO, Huttenhower C, Verbeke K, Walter J, Latulippe ME. Establishing what constitutes a healthy human gut microbiome: state of the science, regulatory considerations, and future directions. J Nutr 149: 1882–1895, 2019. doi: 10.1093/jn/nxz154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mehedint MG, Zeisel SH. Choline’s role in maintaining liver function: new evidence for epigenetic mechanisms. Curr Opin Clin Nutr Metab Care 16: 339–345, 2013. doi: 10.1097/MCO.0b013e3283600d46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Miao J, Ling AV, Manthena PV, Gearing ME, Graham MJ, Crooke RM, Croce KJ, Esquejo RM, Clish CB, Vicent D, Biddinger SB; Morbid Obesity Study Group . Flavin-containing monooxygenase 3 as a potential player in diabetes-associated atherosclerosis. Nat Commun 6: 6498, 2015. doi: 10.1038/ncomms7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Minemura M, Shimizu Y. Gut microbiota and liver diseases. World J Gastroenterol 21: 1691–1702, 2015. doi: 10.3748/wjg.v21.i6.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Miyauchi S, Gopal E, Fei YJ, Ganapathy V. Functional identification of SLC5A8, a tumor suppressor down-regulated in colon cancer, as a Na(+)-coupled transporter for short-chain fatty acids. J Biol Chem 279: 13293–13296, 2004. doi: 10.1074/jbc.C400059200. [DOI] [PubMed] [Google Scholar]
- 120.Montet AM, Oliva L, Beaugé F, Montet JC. Bile salts modulate chronic ethanol-induced hepatotoxicity. Alcohol Alcohol 37: 25–29, 2002. doi: 10.1093/alcalc/37.1.25. [DOI] [PubMed] [Google Scholar]
- 121.Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 7: 189–200, 2016. doi: 10.1080/19490976.2015.1134082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mouzaki M, Wang AY, Bandsma R, Comelli EM, Arendt BM, Zhang L, Fung S, Fischer SE, McGilvray IG, Allard JP. Bile acids and dysbiosis in non-alcoholic fatty liver disease. PLoS One 11: e0151829, 2016. doi: 10.1371/journal.pone.0151829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mutlu EA, Gillevet PM, Rangwala H, Sikaroodi M, Naqvi A, Engen PA, Kwasny M, Lau CK, Keshavarzian A. Colonic microbiome is altered in alcoholism. Am J Physiol Gastrointest Liver Physiol 302: G966–G978, 2012. doi: 10.1152/ajpgi.00380.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nakamoto N, Sasaki N, Aoki R, Miyamoto K, Suda W, Teratani T, Suzuki T, Koda Y, Chu PS, Taniki N, Yamaguchi A, Kanamori M, Kamada N, Hattori M, Ashida H, Sakamoto M, Atarashi K, Narushima S, Yoshimura A, Honda K, Sato T, Kanai T. Gut pathobionts underlie intestinal barrier dysfunction and liver T helper 17 cell immune response in primary sclerosing cholangitis. Nat Microbiol 4: 492–503, 2019. doi: 10.1038/s41564-018-0333-1. [DOI] [PubMed] [Google Scholar]
- 125.Nikolaus S, Schulte B, Al-Massad N, Thieme F, Schulte DM, Bethge J, Rehman A, Tran F, Aden K, Hasler R, Moll N, Schutze G, Schwarz MJ, Waetzig GH, Rosenstiel P, Krawczak M, Szymczak S, Schreiber S. Increased tryptophan metabolism is associated with activity of inflammatory bowel diseases. Gastroenterology 153: 1504–1516e2, 2017. doi: 10.1053/j.gastro.2017.08.028. [DOI] [PubMed] [Google Scholar]
- 126.Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, Stimmel JB, Willson TM, Zavacki AM, Moore DD, Lehmann JM. Bile acids: natural ligands for an orphan nuclear receptor. Science 284: 1365–1368, 1999. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- 127.Philips CA, Pande A, Shasthry SM, Jamwal KD, Khillan V, Chandel SS, Kumar G, Sharma MK, Maiwall R, Jindal A, Choudhary A, Hussain MS, Sharma S, Sarin SK. Healthy donor fecal microbiota transplantation in steroid-ineligible severe alcoholic hepatitis: a pilot study. Clin Gastroenterol Hepatol 15: 600–602, 2017. doi: 10.1016/j.cgh.2016.10.029. [DOI] [PubMed] [Google Scholar]
- 128.Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282: 2085–2088, 1998. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 129.Possamai LA, McPhail MJ, Khamri W, Wu B, Concas D, Harrison M, Williams R, Cox RD, Cox IJ, Anstee QM, Thursz MR. The role of intestinal microbiota in murine models of acetaminophen-induced hepatotoxicity. Liver Int 35: 764–773, 2015. doi: 10.1111/liv.12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Puri P, Daita K, Joyce A, Mirshahi F, Santhekadur PK, Cazanave S, Luketic VA, Siddiqui MS, Boyett S, Min HK, Kumar DP, Kohli R, Zhou H, Hylemon PB, Contos MJ, Idowu M, Sanyal AJ. The presence and severity of nonalcoholic steatohepatitis is associated with specific changes in circulating bile acids. Hepatology 67: 534–548, 2018. doi: 10.1002/hep.29359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, Guo J, Le Chatelier E, Yao J, Wu L, Zhou J, Ni S, Liu L, Pons N, Batto JM, Kennedy SP, Leonard P, Yuan C, Ding W, Chen Y, Hu X, Zheng B, Qian G, Xu W, Ehrlich SD, Zheng S, Li L. Alterations of the human gut microbiome in liver cirrhosis. Nature 513: 59–64, 2014. doi: 10.1038/nature13568. [DOI] [PubMed] [Google Scholar]
- 132.Quigley EM. Primary biliary cirrhosis and the microbiome. Semin Liver Dis 36: 349–353, 2016. doi: 10.1055/s-0036-1594006. [DOI] [PubMed] [Google Scholar]
- 133.Raman M, Ahmed I, Gillevet PM, Probert CS, Ratcliffe NM, Smith S, Greenwood R, Sikaroodi M, Lam V, Crotty P, Bailey J, Myers RP, Rioux KP. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol 11: 868–875e1, 2013. doi: 10.1016/j.cgh.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 134.Rath S, Heidrich B, Pieper DH, Vital M. Uncovering the trimethylamine-producing bacteria of the human gut microbiota. Microbiome 5: 54, 2017. doi: 10.1186/s40168-017-0271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rau M, Rehman A, Dittrich M, Groen AK, Hermanns HM, Seyfried F, Beyersdorf N, Dandekar T, Rosenstiel P, Geier A. Fecal SCFAs and SCFA-producing bacteria in gut microbiome of human NAFLD as a putative link to systemic T-cell activation and advanced disease. United European Gastroenterol J 6: 1496–1507, 2018. doi: 10.1177/2050640618804444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rechkemmer G, von Engelhardt W. Concentration- and pH-dependence of short-chain fatty acid absorption in the proximal and distal colon of guinea pig (Cavia porcellus). Comp Biochem Physiol A Comp Physiol 91: 659–663, 1988. doi: 10.1016/0300-9629(88)90944-9. [DOI] [PubMed] [Google Scholar]
- 137.Reich M, Deutschmann K, Sommerfeld A, Klindt C, Kluge S, Kubitz R, Ullmer C, Knoefel WT, Herebian D, Mayatepek E, Häussinger D, Keitel V. TGR5 is essential for bile acid-dependent cholangiocyte proliferation in vivo and in vitro. Gut 65: 487–501, 2016. doi: 10.1136/gutjnl-2015-309458. [DOI] [PubMed] [Google Scholar]
- 138.Ritzhaupt A, Wood IS, Ellis A, Hosie KB, Shirazi-Beechey SP. Identification and characterization of a monocarboxylate transporter (MCT1) in pig and human colon: its potential to transport L-lactate as well as butyrate. J Physiol 513: 719–732, 1998. doi: 10.1111/j.1469-7793.1998.719ba.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Roychowdhury S, Glueck B, Han Y, Mohammad MA, Cresci GAM. A designer synbiotic attenuates chronic-binge ethanol-induced gut-liver injury in mice. Nutrients 11: 97, 2019. doi: 10.3390/nu11010097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall HU, Bamberg K, Angelin B, Hyötyläinen T, Orešič M, Bäckhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab 17: 225–235, 2013. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 141.Schnabl B, Brenner DA. Interactions between the intestinal microbiome and liver diseases. Gastroenterology 146: 1513–1524, 2014. doi: 10.1053/j.gastro.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Schoeler M, Caesar R. Dietary lipids, gut microbiota and lipid metabolism. Rev Endocr Metab Disord 20: 461–472, 2019. doi: 10.1007/s11154-019-09512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Schönfeld P, Wojtczak L. Short- and medium-chain fatty acids in energy metabolism: the cellular perspective. J Lipid Res 57: 943–954, 2016. doi: 10.1194/jlr.R067629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Shavakhi A, Minakari M, Firouzian H, Assali R, Hekmatdoost A, Ferns G. Effect of a probiotic and metformin on liver aminotransferases in non-alcoholic steatohepatitis: a double blind randomized clinical trial. Int J Prev Med 4: 531–537, 2013. [PMC free article] [PubMed] [Google Scholar]
- 145.Shi Y, Tyson GW, DeLong EF. Metatranscriptomics reveals unique microbial small RNAs in the ocean’s water column. Nature 459: 266–269, 2009. doi: 10.1038/nature08055. [DOI] [PubMed] [Google Scholar]
- 146.Singh V, Yeoh BS, Chassaing B, Xiao X, Saha P, Olvera RA, Lapek JD, Zhang LM, Wang WB, Hao SJ, Flythe MD, Gonzalez DJ, Cani PD, Conejo-Garcia JR, Xiong N, Kennett MJ, Joe B, Patterson AD, Gewirtz AT, Vijay-Kumar M. Dysregulated microbial fermentation of soluble fiber induces cholestatic liver cancer. Cell 175: 679–694e22, 2018. doi: 10.1016/j.cell.2018.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sinha J, Chen F, Miloh T, Burns RC, Yu Z, Shneider BL. Beta-Klotho and FGF-15/19 inhibit the apical sodium-dependent bile acid transporter in enterocytes and cholangiocytes. Am J Physiol Gastrointest Liver Physiol 295: G996–G1003, 2008. doi: 10.1152/ajpgi.90343.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Stärkel P, Schnabl B. Bidirectional communication between liver and gut during alcoholic liver disease. Semin Liver Dis 36: 331–339, 2016. doi: 10.1055/s-0036-1593882. [DOI] [PubMed] [Google Scholar]
- 149.Stumpff F. A look at the smelly side of physiology: transport of short chain fatty acids. Pflugers Arch 470: 571–598, 2018. doi: 10.1007/s00424-017-2105-9. [DOI] [PubMed] [Google Scholar]
- 150.Tang R, Wei Y, Li Y, Chen W, Chen H, Wang Q, Yang F, Miao Q, Xiao X, Zhang H, Lian M, Jiang X, Zhang J, Cao Q, Fan Z, Wu M, Qiu D, Fang J-Y, Ansari A, Gershwin ME, Ma X. Gut microbial profile is altered in primary biliary cholangitis and partially restored after UDCA therapy. Gut 67: 534–541, 2018. doi: 10.1136/gutjnl-2016-313332. [DOI] [PubMed] [Google Scholar]
- 151.Tang WHW, Wang Z, Kennedy DJ, Wu Y, Buffa JA, Agatisa-Boyle B, Li XS, Levison BS, Hazen SL. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ Res 116: 448–455, 2015. doi: 10.1161/CIRCRESAHA.116.305360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, Rizzo G, Macchiarulo A, Yamamoto H, Mataki C, Pruzanski M, Pellicciari R, Auwerx J, Schoonjans K. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab 10: 167–177, 2009. doi: 10.1016/j.cmet.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tolhurst G, Heffron H, Lam YS, Parker HE, Habib AM, Diakogiannaki E, Cameron J, Grosse J, Reimann F, Gribble FM. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes 61: 364–371, 2012. doi: 10.2337/db11-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444: 1027–1031, 2006. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 155.Vatanen T, Kostic AD, d’Hennezel E, Siljander H, Franzosa EA, Yassour M, Kolde R, Vlamakis H, Arthur TD, Hämäläinen AM, Peet A, Tillmann V, Uibo R, Mokurov S, Dorshakova N, Ilonen J, Virtanen SM, Szabo SJ, Porter JA, Lähdesmäki H, Huttenhower C, Gevers D, Cullen TW, Knip M, Xavier RJ; DIABIMMUNE Study Group . Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 165: 842–853, 2016. doi: 10.1016/j.cell.2016.04.007. A correction for this article is available at https://doi.org/10.1016/j.cell.2016.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Waclawiková B, El Aidy S. Role of microbiota and tryptophan metabolites in the remote effect of intestinal inflammation on brain and depression. Pharmaceuticals (Basel) 11: 63, 2018. doi: 10.3390/ph11030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Walter J, Armet AM, Finlay BB, Shanahan F. Establishing or exaggerating causality for the gut microbiome: lessons from human microbiota-associated rodents. Cell 180: 221–232, 2020. doi: 10.1016/j.cell.2019.12.025. [DOI] [PubMed] [Google Scholar]
- 158.Walter J, Maldonado-Gómez MX, Martínez I. To engraft or not to engraft: an ecological framework for gut microbiome modulation with live microbes. Curr Opin Biotechnol 49: 129–139, 2018. doi: 10.1016/j.copbio.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang H, Chen J, Hollister K, Sowers LC, Forman BM. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol Cell 3: 543–553, 1999. doi: 10.1016/S1097-2765(00)80348-2. [DOI] [PubMed] [Google Scholar]
- 160.Wang L, Fouts DE, Stärkel P, Hartmann P, Chen P, Llorente C, DePew J, Moncera K, Ho SB, Brenner DA, Hooper LV, Schnabl B. Intestinal REG3 lectins protect against alcoholic steatohepatitis by reducing mucosa-associated microbiota and preventing bacterial translocation. Cell Host Microbe 19: 227–239, 2016. doi: 10.1016/j.chom.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WH, DiDonato JA, Lusis AJ, Hazen SL. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472: 57–63, 2011. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wang Z, Roberts AB, Buffa JA, Levison BS, Zhu W, Org E, Gu X, Huang Y, Zamanian-Daryoush M, Culley MK, DiDonato AJ, Fu X, Hazen JE, Krajcik D, DiDonato JA, Lusis AJ, Hazen SL. Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell 163: 1585–1595, 2015. doi: 10.1016/j.cell.2015.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wang Z, Zhang X, Zhu L, Yang X, He F, Wang T, Bao T, Lu H, Wang H, Yang S. Inulin alleviates inflammation of alcoholic liver disease via SCFAs-inducing suppression of M1 and facilitation of M2 macrophages in mice. Int Immunopharmacol 78: 106062, 2020. doi: 10.1016/j.intimp.2019.106062. [DOI] [PubMed] [Google Scholar]
- 164.Watanabe M, Houten SM, Wang L, Moschetta A, Mangelsdorf DJ, Heyman RA, Moore DD, Auwerx J. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J Clin Invest 113: 1408–1418, 2004. doi: 10.1172/JCI21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wisniewski PJ, Dowden RA, Campbell SC. Role of dietary lipids in modulating inflammation through the gut microbiota. Nutrients 11: 117, 2019. doi: 10.3390/nu11010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wong VW, Won GL, Chim AM, Chu WC, Yeung DK, Li KC, Chan HL. Treatment of nonalcoholic steatohepatitis with probiotics. A proof-of-concept study. Ann Hepatol 12: 256–262, 2013. doi: 10.1016/S1665-2681(19)31364-X. [DOI] [PubMed] [Google Scholar]
- 167.Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science 249: 1431–1433, 1990. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 168.Wu J, Meng Z, Jiang M, Zhang E, Trippler M, Broering R, Bucchi A, Krux F, Dittmer U, Yang D, Roggendorf M, Gerken G, Lu M, Schlaak JF. Toll-like receptor-induced innate immune responses in non-parenchymal liver cells are cell type-specific. Immunology 129: 363–374, 2010. doi: 10.1111/j.1365-2567.2009.03179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Xie G, Zhong W, Li H, Li Q, Qiu Y, Zheng X, Chen H, Zhao X, Zhang S, Zhou Z, Zeisel SH, Jia W. Alteration of bile acid metabolism in the rat induced by chronic ethanol consumption. FASEB J 27: 3583–3593, 2013. doi: 10.1096/fj.13-231860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Xin J, Zeng D, Wang H, Ni X, Yi D, Pan K, Jing B. Preventing non-alcoholic fatty liver disease through Lactobacillus johnsonii BS15 by attenuating inflammation and mitochondrial injury and improving gut environment in obese mice. Appl Microbiol Biotechnol 98: 6817–6829, 2014. doi: 10.1007/s00253-014-5752-1. [DOI] [PubMed] [Google Scholar]
- 171.Yan AW, Fouts DE, Brandl J, Stärkel P, Torralba M, Schott E, Tsukamoto H, Nelson KE, Brenner DA, Schnabl B. Enteric dysbiosis associated with a mouse model of alcoholic liver disease. Hepatology 53: 96–105, 2011. doi: 10.1002/hep.24018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yang SQ, Lin HZ, Lane MD, Clemens M, Diehl AM. Obesity increases sensitivity to endotoxin liver injury: implications for the pathogenesis of steatohepatitis. Proc Natl Acad Sci USA 94: 2557–2562, 1997. doi: 10.1073/pnas.94.6.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. Human gut microbiome viewed across age and geography. Nature 486: 222–227, 2012. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ye J, Lv L, Wu W, Li Y, Shi D, Fang D, Guo F, Jiang H, Yan R, Ye W, Li L. Butyrate protects mice against methionine-choline-deficient diet-induced non-alcoholic steatohepatitis by improving gut barrier function, attenuating inflammation and reducing endotoxin levels. Front Microbiol 9: 1967, 2018. doi: 10.3389/fmicb.2018.01967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Younossi ZM, Blissett D, Blissett R, Henry L, Stepanova M, Younossi Y, Racila A, Hunt S, Beckerman R. The economic and clinical burden of nonalcoholic fatty liver disease in the United States and Europe. Hepatology 64: 1577–1586, 2016. doi: 10.1002/hep.28785. [DOI] [PubMed] [Google Scholar]
- 176.Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 64: 73–84, 2016. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 177.Yuan J, Baker SS, Liu W, Alkhouri R, Baker RD, Xie J, Ji G, Zhu L. Endotoxemia unrequired in the pathogenesis of pediatric nonalcoholic steatohepatitis. J Gastroenterol Hepatol 29: 1292–1298, 2014. doi: 10.1111/jgh.12510. [DOI] [PubMed] [Google Scholar]
- 178.Zeisel SH, Warrier M. Trimethylamine N-oxide, the microbiome, and heart and kidney disease. Annu Rev Nutr 37: 157–181, 2017. doi: 10.1146/annurev-nutr-071816-064732. [DOI] [PubMed] [Google Scholar]
- 179.Zeisel SH, Wishnok JS, Blusztajn JK. Formation of methylamines from ingested choline and lecithin. J Pharmacol Exp Ther 225: 320–324, 1983. [PubMed] [Google Scholar]
- 180.Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D’Angelo C, Massi-Benedetti C, Fallarino F, Carvalho A, Puccetti P, Romani L. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 39: 372–385, 2013. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 181.Zelante T, Iannitti RG, Fallarino F, Gargaro M, De Luca A, Moretti S, Bartoli A, Romani L. Tryptophan feeding of the IDO1-AhR axis in host-microbial symbiosis. Front Immunol 5: 640, 2014. doi: 10.3389/fimmu.2014.00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zhang Y, Lee FY, Barrera G, Lee H, Vales C, Gonzalez FJ, Willson TM, Edwards PA. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proc Natl Acad Sci USA 103: 1006–1011, 2006. doi: 10.1073/pnas.0506982103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Zhang YJ, Reddy MC, Ioerger TR, Rothchild AC, Dartois V, Schuster BM, Trauner A, Wallis D, Galaviz S, Huttenhower C, Sacchettini JC, Behar SM, Rubin EJ. Tryptophan biosynthesis protects mycobacteria from CD4 T-cell-mediated killing. Cell 155: 1296–1308, 2013. doi: 10.1016/j.cell.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Zhang Z, Zhou H, Bai L, Lv YY, Yi HX, Zhang LW, Li R. Protective effects of probiotics on acute alcohol-induced liver injury in mice through alcohol metabolizing enzymes activation and hepatic TNF-alpha response reduction. J Funct Foods 59: 234–241, 2019. doi: 10.1016/j.jff.2019.05.018. [DOI] [Google Scholar]
- 185.Zhao H, Zhao C, Dong Y, Zhang M, Wang Y, Li F, Li X, McClain C, Yang S, Feng W. Inhibition of miR122a by Lactobacillus rhamnosus GG culture supernatant increases intestinal occludin expression and protects mice from alcoholic liver disease. Toxicol Lett 234: 194–200, 2015. doi: 10.1016/j.toxlet.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 186.Zhu L, Baker RD, Baker SS. Gut microbiome and nonalcoholic fatty liver diseases. Pediatr Res 77: 245–251, 2015. doi: 10.1038/pr.2014.157. [DOI] [PubMed] [Google Scholar]
- 187.Zhu L, Baker SS, Gill C, Liu W, Alkhouri R, Baker RD, Gill SR. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology 57: 601–609, 2013. doi: 10.1002/hep.26093. [DOI] [PubMed] [Google Scholar]