Abstract
CRISPR-mediated DNA base editors, which include cytosine base editors (CBEs) and adenine base editors (ABEs), are promising tools that can induce point mutations at desired sites in a targeted manner to correct or disrupt gene expression. Their high editing efficiency, coupled with their ability to generate a targeted mutation without generating a DNA double-strand break (DSB) or requiring a donor DNA template, suggests that DNA base editors will be useful for treating genetic diseases, among other applications. However, this hope has recently been challenged by the discovery of DNA base editor shortcomings, including off-target DNA editing, the generation of bystander mutations, and promiscuous deamination effects in both DNA and RNA, which arise from the main DNA base editor constituents, a Cas nuclease variant and a deaminase. In this review, we summarize information about the DNA base editors that have been developed to date, introduce their associated potential challenges, and describe current efforts to minimize or mitigate those issues of DNA base editors.
Keywords: CRISPR-Cas, cytosine base editor, adenine base editor, off-target effects, genome-wide DNA deamination, transcriptome-wide RNA deamination, high-fidelity DNA base editors
Graphical Abstract
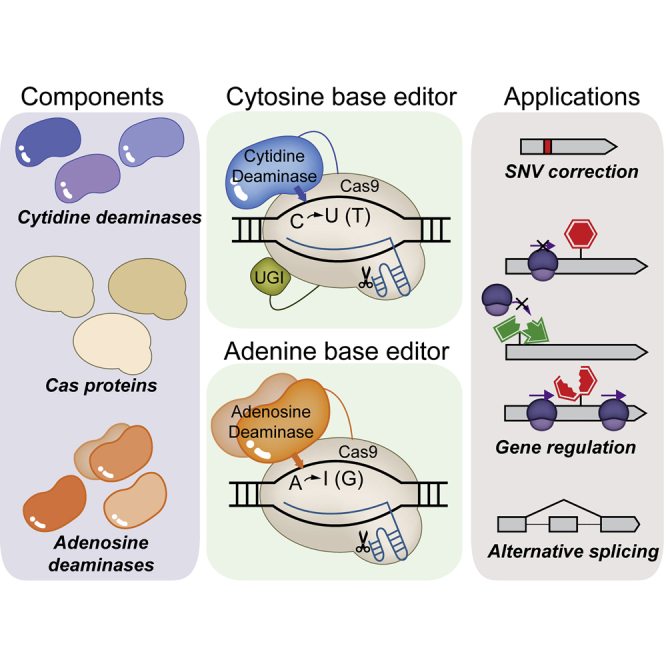
DNA base editors, which can induce point mutations at desired sites in a targeted manner, are useful to correct or disrupt gene expression. In this review, we introduce their associated potential challenges and describe current efforts to minimize or mitigate those issues of DNA base editors.
Main Text
Since CRISPR-Cas9 nucleases were first harnessed for site-specific DNA editing in the human genome,1, 2, 3, 4, 5 the gene-editing field has rapidly advanced, and many CRISPR-associated tools have been developed that allow targeted gene disruption, recovery, and regulation. Despite their versatility, conventional CRISPR-based tools are associated with fundamental problems. For example, these tools inevitably generate DNA double-strand breaks (DSBs), which can potentially disturb efficient and safe gene editing by inducing the p53-mediated DNA damage response6,7 and by causing unexpected large chromosomal deletions or genomic rearrangements.8 In addition, the DSB repair processes that are key to conventional CRISPR-based methods are error-prone in the case of non-homologous end joining (NHEJ), which generates small insertions and deletions (indels) at the DSB site, or operate only in a specific cell cycle phase, S and G2 phases, for homology-directed repair (HDR).9 Recently, newly developed base editing tools, cytosine base editors (CBEs) and adenine base editors (ABEs), have attracted a great deal of attention as alternatives to conventional CRISPR-associated tools. CBEs and ABEs deaminate DNA pyrimidine and purine (cytosine and adenine, respectively) with accompanying little DSB generation, maintaining their high editing efficiency. Additionally, they can be used in both dividing and non-dividing cells because they are associated with base excision repair (BER) or mismatch repair (MMR), which occurs extensively in most cell-cycle phases rather than NHEJ or HDR. Furthermore, because DNA base editors do not require donor DNAs, they are relatively easy to use.
Nevertheless, base editing technologies do not yet represent a complete “Swiss army knife” for gene editing. For instance, not all genes are targetable by DNA base editors because of protospacer adjacent motif (PAM) sequence preferences.10,11 Moreover, their editing specificity is restricted by the width of the editing window.12 Even more seriously, DNA base editors can induce genome-wide off-target deamination on both DNA and RNA, as well as unexpected nucleotide conversions, both of which could have serious consequences in medical applications.13, 14, 15, 16, 17, 18, 19 In this review, we introduce the current status and challenges of base editing tools and describe recent approaches for addressing their limitations.
The Construction and Mechanism of CBEs
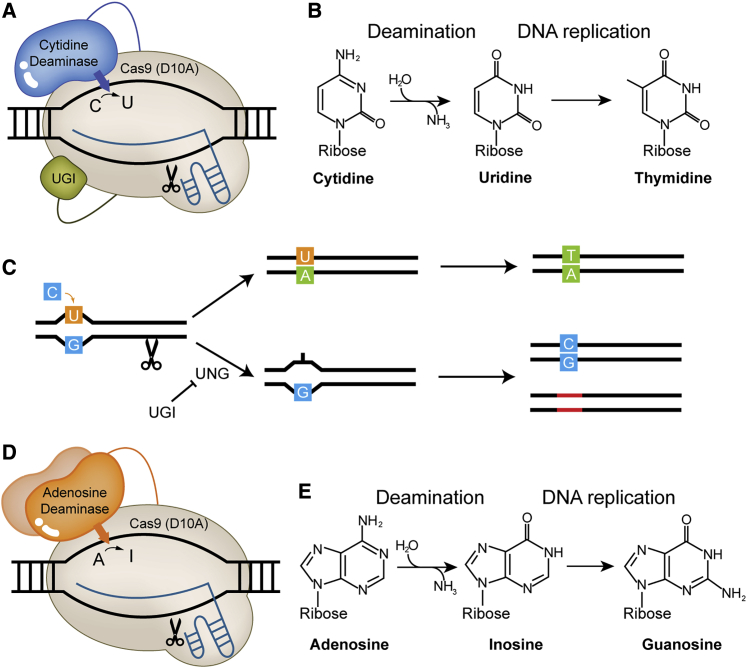
A CBE is basically composed of three fused elements: a cytidine deaminase, a uracil DNA glycosylase inhibitor (UGI), and a Cas nuclease that is either catalytically inactive (dCas) or partially inactive (Cas nickase or nCas). Such Cas9 variants contain mutations that prevent the generation of DSBs. An associated single-guide RNA (sgRNA) confers target sequence specificity (Figure 1A).10 Once a CBE complex has been recruited to the target DNA by the Cas protein and sgRNA, the cytidine deaminase recognizes the single-stranded DNA (ssDNA) in the R-loop structure formed by the pairing between the sgRNA and the non-edited DNA strand and converts a cytosine into a uracil (Figure 1B), generating a U/G pair. The mismatched U/G pair is then sequentially converted into a U/A pair and a T/A pair by the MMR pathway (Figure 1C).
Figure 1.
DNA Base Editor Mechanisms
(A) Schematic diagram of a cytosine base editor. A Cas9-sgRNA complex forms an R-loop at the target site in the DNA. The linked cytidine deaminase converts the exposed cytidine into a uridine. An additional linked protein, UGI, protects uracil from uracil-DNA glycosylase (UDG). (B) After deamination, the resulting uridine is read as thymidine by DNA polymerase. (C) The artificial uracil deoxynucleotide created by the CBE system is repaired by two major pathways. In the upper pathway, DNA polymerase reads uracil as thymine, pairing the uracil with adenine. Ultimately, a T:A pair is made. In the bottom pathway, the UDG protein removes uracil from the DNA. The resulting deoxyribonucleotide, which lacks a base, is repaired to cytosine, which is complementary to the guanine on the opposite strand. Alternatively, the uracil is excised, which induces the formation of insertions, deletions, or substitutions. The UGI protein inhibits the bottom pathway. (D) Schematic diagram of an adenine base editor. (E) After deamination converts an adenosine into an inosine, the inosine is read as a guanosine during DNA replication.
Two groups, employing different cytidine deaminases, developed CBEs at about the same time. The Liu group chose rat APOBEC1 as a cytidine deaminase and fused it to the N terminus of dCas9 (D10A and H840A) through a 16-residue linker protein (XTEN).10 On the other hand, the Kondo group employed an activation-induced cytidine deaminase (AID) from sea lamprey (PmCDA1), which is fused to the C terminus of dCas9.20 Each primitive CBE was improved in subsequent engineering, generating BE3 and Target-AID, in which the UGI was introduced to prevent the restoration of mismatched U/G pairs to C/G pairs by BER. Furthermore, the catalytic activity of dCas9 was partially restored (by correcting the H840A mutation) to create nCas9 (D10A), which generates nicks in the non-edited DNA strand and thereby enables MMR to convert guanosine into adenosine preferentially. The differences between BEs and Target-AID in temperature-sensitiveness and sequence preference suggest that the two types of CBEs should complement each other.
Interestingly, it has been reported that CBEs employing rAPOBEC1 frequently induce C-to-G/A conversion rather than C-to-T conversion depending on the cell types,21,22 which is mediated by BER.23 To reduce the unwanted C-to-G/A conversion, a second UGI with a longer linker sequence was added in BE423 or rAPOBEC1 was replaced with CDA1 or AID, generating CDA1-BE3 and AID-BE3, respectively.23 The editing efficiency of BE4 was further enhanced by codon optimization and ancestral reconstruction, generating BE4max and AncBE4max, respectively.24 In addition, unwanted indels via CBEs could be reduced by fusing the Gam protein of bacteriophage Mu to the N terminus of CBEs, which can bind to the ends of DSBs reduced indel frequency of CBEs.23
The Construction and Mechanism of ABEs
A considerable fraction (∼47%) of disease-related point mutations are conversions of a G/C pair to an A/T pair,25 a type of mutation that cannot be rescued by CBEs; instead, ABEs would be required. Following their work on CBEs, the Liu group developed ABEs, which resemble CBEs in both structure and base editing mechanisms, except that an adenosine deaminase replaces the cytidine deaminase (Figure 1D). Because no natural adenosine deaminase that operates on DNA has been discovered, the tRNA-specific adenosine deaminase TadA from Escherichia coli (E. coli) was evolved to generate a version (eTadA∗) with such activity for use as an adenosine deaminase in ABEs.11 The ABE complex is recruited to the target DNA in a process similar to that used by CBEs, after which the adenosine deaminase converts an adenosine into an inosine (Figure 1E), generating an I/T pair. MMR then sequentially converts the mismatched I/T pair into an I/C pair and a G/C pair.
The initially developed ABE (ABE1.2) was based on BE3. Because TadAs operate as homodimers, in which one monomer carries out deamination and the other interacts with the substrate,26 a wild-type TadA (wtTadA) was additionally tethered in a subsequent ABE variant (ABE2.9).11 In addition, several mutations were introduced into eTadA∗ to achieve high editing efficiency, generating ABE7.9 and ABE7.10. The activity of ABE7.10 was further enhanced by addition of modified NLS and codon-optimization (ABEmax).24 Recently, ABE8 variants (ABE8e and ABE8s) were developed by removing wtTadA and introducing additional mutations into eTadA∗.27,28
Expanding Targetable Sites of DNA Base Editors by Using Engineered Cas9 Variants or Cas Orthologs
Initially, both CBEs and ABEs were constructed using variants of the most popular Cas9 nuclease in the genome editing field, SpCas9, which originates from Streptococcus pyogenes. However, SpCas9 prefers specific PAM sequences (NGG) in the DNA target, and this preference limits the number of genes that can be targeted by DNA base editors. The replacement of SpCas9 with variants derived from other Cas nucleases, which exhibit different PAM sequence preferences, has expanded the targeting capability of DNA base editors.
In the case of CBEs, the adoption of engineered SpCas9 variants having different PAM sequences (NG/GAA/GAT, NGA, NGAG, NGCG, and NG) in BE3 or Target-AID resulted in new targetable sites.12,29, 30, 31 The PAM sequence preference can also be altered by replacing the PAM-interacting region of SpCas9 with that of Streptococcus macacae Cas9 (Smac Cas9); this manipulation resulted in the generation of Spy-mac BE4max, which recognizes NAA PAMs.32 Cas9 orthologs have also been used as SpCas9 alternatives. CBEs employing Cas9 from Staphylococcus aureus (SaCas9), its variant (KKH-SaCas9), and Cas9 from Staphylococcus auricularis (SauriCas9) target sites with NNGRRT, NNNRRT, and NNGG PAMs, respectively.12,33 Likewise, replacement of SpCas9 with Cas12a (also known as Cpf1) provides a different PAM preference (TTTV). In particular, CBEs that are constructed with a catalytically inactive Cpf1 that originates from L. bacterium (dLbCpf1-BE)34 or an enhanced ortholog from Acidaminococcus sp. (enAsCas12a-BE)35 reveal higher C-to-T conversion frequencies with a lower rate of unwanted indel generation compared with the SpCas9-based CBEs (Table 1).
Table 1.
Current Versions of High-Fidelity CBEs
 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cytidine Deaminase |
Cas Nuclease |
PAM | Level of DNA Deamination | Level of RNA Deamination | Notes | Reference | ||||
| CBE Name | Type | Mutation | Type | Mutation | Editing Window | |||||
| BE1 | rAPOBEC1 | WT | SpCas9 | D10A/H840A | 4th–8th | NGG | unknown (maybe high) | unknown (maybe high) | no UGI | 10 |
| BE2 | WT | SpCas9 | D10A/H840A | 4th–8th | NGG | unknown (maybe high) | unknown (maybe high) | one UGI with 4 aa linker | 10 | |
| BE3 | WT | SpCas9 | D10A | 4th–8th | NGG | high | high | one UGI with 4 aa linker | 10,13,14 | |
| BE4 | WT | SpCas9 | D10A | 4th–8th | NGG | high | high | two UGI with 9 aa linker | 23 | |
| BE4max | WT | SpCas9 | D10A | 4th–8th | NGG | unknown (maybe high) | unknown (maybe high) | codon-opimization, bpNLS at both N and C termini | 24 | |
| AncBE4max | Anc689 | SpCas9 | D10A | 4th–8th | NGG | unknown (maybe high) | unknown (maybe high) | Anc689 from ancestral sequence reconstruction of rAPOBEC1 | 24 | |
| xCas9(3.7)-BE3 | WT | SpCas9 | D10A + 7 mutations (A262T/R324L/S409I/E480K/E543D/M694I/E1219V) | 4th–8th | NG/GAA/GAT | unknown (maybe high) | unknown (maybe high) | 29,36 | ||
| VQR-BE3 | WT | SpCas9 | D10A + D1135V/R1335Q/T1337R | 4th–8th | NGA | unknown (maybe high) | unknown (maybe high) | 12 | ||
| EQR-BE3 | WT | SpCas9 | D10A + D1135E/R1335Q/T1337R | 4th–8th | NGAG | unknown (maybe high) | unknown (maybe high) | 12 | ||
| VRER-BE3 | WT | SpCas9 | D10A + D1135V/G1218R/R1335E/T1337R | 4th–8th | NGCG | unknown (maybe high) | unknown (maybe high) | 12 | ||
| CBE-NG | WT | SpCas9 | D10A + 7 mutations (R1335V/L1111R/D1135V/G1218R/E1219F/A1322R/T1337R) | 4th–8th | NG | unknown (maybe high) | unknown (maybe high) | 36 | ||
| Spy-mac BE4max | WT | Spy-mac Cas9 | D10A | 4th–8th | NAA | unknown (maybe high) | unknown (maybe high) | 32 | ||
| Sa-BE3 | WT | SaCas9 | D10A | 3th–12th | NNGRRT | unknown (maybe high) | unknown (maybe high) | 12 | ||
| SaKKH-BE3 | WT | SaCas9 | D10A + E782K/N968K/R1015H | 3th–12th | NNNRRT | unknown (maybe high) | unknown (maybe high) | 12 | ||
| SauriBE4max | WT | SauriCas9 | D15A | 6th–9th | NNGG | unknown (maybe high) | unknown (maybe high) | 33 | ||
| dLbCpf1-BE | WT | LbCas12a | D832A | 8th–13th | TTTV | unknown (maybe high) | unknown (maybe high) | 34 | ||
| enAsCas12a-BE | WT | AsCas12a | D908A + E174R/S542R/K548R | 8th–13th | TTTV | unknown (maybe high) | unknown (maybe high) | 35 | ||
| ScCas9-BE3 | WT | ScCas9 | D10A | 4th–8th | NNG | unknown (maybe high) | unknown (maybe high) | 37 | ||
| YE1-BE3 or YE1-BE4 | W90Y/R126E | SpCas9 | D10A | 5th–6th | NGG | reduced | reduced | 12,16,38,39 | ||
| EE-BE3 | R126E/R132E | SpCas9 | D10A | 5th–6th | NGG | unknown (maybe low) | unknown | 12,38 | ||
| YE2-BE3 | W90Y/R132E | SpCas9 | D10A | 5th–6th | NGG | unknown (maybe low) | unknown | 12,38 | ||
| YEE-BE3 | W90Y/R126E/R132E | SpCas9 | D10A | 5th–6th | NGG | unknown (maybe low) | reduced | 12,38 | ||
| YFE-BE4max | W90Y/Y120F/R126E | SpCas9 | D10A | 4th–6th | NGG | unknown (maybe low) | unknown (maybe low) | 40 | ||
| dCpf1-BE-YE | W90Y/R126E | LbCas12a | D832A | 10th–12th | TTTV | unknown (maybe low) | unknown (maybe low) | 34 | ||
| CP1012-CBE | WT | SpCas9 | CP1012 | 4th–11th | NGG | unknown (maybe high) | unknown (maybe high) | 41 | ||
| CP1028-CBE | WT | SpCas9 | CP1028 | 4th–11th | NGG | unknown (maybe high) | unknown (maybe high) | 41 | ||
| HF-BE3 | WT | SpCas9 | D10A + (N497A/R661A/Q695A/Q926A) | 4th–8th | NGG | high | unknown (maybe high) | 14,42 | ||
| Sniper-BE3 | WT | SpCas9 | D10A + (F539S/M763I/K890N) | 4th–8th | NGG | unknown (maybe high) | unknown (maybe high) | 43 | ||
| SECURE-BE3 | R33A | SpCas9 | D10A | 5th–7th | NGG | unknown (maybe low) | reduced | 15,38 | ||
| SECURE-BE3 | R33A/K34A | SpCas9 | D10A | 5th–6th | NGG | unknown (maybe low) | reduced | 15,38 | ||
| BE3-R132E | R132E | SpCas9 | D10A | 4th–8th | NGG | reduced | reduced | 39 | ||
| BE3-(W90F+R126E) | W90F/R126E | SpCas9 | D10A | 4th–8th | NGG | reduced | reduced | 39 | ||
| A3A-BE3 | hAPOBEC3A | WT | SpCas9 | D10A | 4th–8th | NGG | high | high | 22,39 | |
| eA3A-BE3 | N57G | SpCas9 | D10A | 4th–8th | NGG | unknown | reduced | 18 | ||
| eA3A-HF1-BE3-2xUGI | N57G | SpCas9 | D10A + (N497A/R661A/Q695A/Q926A) | 4th–8th | NGG | unknown | unknown (maybe low) | 22 | ||
| eA3A-Hypa-BE3-2xUGI | N57G | SpCas9 | D10A + (N692A/M694A/Q695A/H698A) | 4th–8th | NGG | unknown | unknown (maybe low) | 22 | ||
| BE3(hA3A-Y130F) | Y130F | SpCas9 | D10A | 4th–8th | NGG | high | reduced | 16,39 | ||
| BE3(hA3A-R128A) | R128A | SpCas9 | D10A | 4th–8th | NGG | unknown | reduced | 16 | ||
| Target-AID | PmCDA1 | WT | SpCas9 | D10A | 2nd–4th | NGG | unknown | low | deaminase linked with C-terminal of Cas nuclease | 18,20 |
| nCDA1-BE3 | truncated | SpCas9 | D10A | 2nd–8th | NGG | unknown | unknown (maybe low) | 31 | ||
| Target-AID-NG | WT | SpCas9 | D10A + 7 mutations (R1335V/L1111R/D1135V/G1218R/E1219F/A1322R/T1337R) | 2nd–4th | NG | unknown | unknown (maybe low) | deaminase linked with C-terminal of Cas nuclease | 30 | |
| hAID-BE3 | hAID | WT | SpCas9 | D10A | 3rd–8th | NGG | unknown | low | 18,44 | |
| BE4-PpAPOBEC1 [R33A] | PpAPOBEC1 | R33A | SpCas9 | D10A | 6th–10th | NGG | low | low | 45 | |
| BE4-PpAPOBEC1 [H122A] | PpAPOBEC1 | H122A | SpCas9 | D10A | 4th–9th | NGG | low | low | 45 | |
| BE4-RrA3F | RrA3F | WT | SpCas9 | D10A | 2nd–12th | NGG | low | low | 45 | |
| BE4-AmAPOBEC1 | AmAPOBEC1 | WT | SpCas9 | D10A | 4th–12th | NGG | low | low | 45 | |
| BE4-SsAPOBEC3B | SsAPOBEC3B | WT | SpCas9 | D10A | 2nd–15th | NGG | low | low | 45 | |
As with CBEs, genes that can be targeted by ABEs are also limited by the PAM sequence preference of the Cas9 protein, and targetable sites can be varied by replacing it with variants or orthologs with different PAM preferences.29,36,41,46,47 Adoption of SpCas9 variants, Spy-mac Cas9, or SaCas9 or its variant KKH-SaCas9 in ABEs expanded the ABE target range.32,46 A newly characterized Cas9 from Streptococcus canis (ScCas9) is another alternative to SpCas9, providing an NNG PAM sequence preference (Table 1).37
However, unlike these newer CBE versions, ABEs built with non-SpCas9 proteins showed lower base editing efficiencies compared to the canonical ABE, because the TadA protein was initially evolved based on a fusion with SpCas9. To solve this problem, recently the Liu group further evolved the TadA protein and developed a new version (TadA-8e) with faster deamination kinetics.27 The TadA-8e protein exhibits high editing efficiencies when it is combined with non-SpCas9 proteins, such as SaCas9, SaCas9-KKH, LbCas12a, or enAsCas12a, as well as in combination with SpCas9 (ABE8e). Moreover, the Gaudelli group developed another set of eTadA∗ variants (TadA8s; variants with different mutations with TadA-8e) starting from TadA7.10 and using directed evolution. TadA8s also showed high editing efficiencies when combined with SaCas9 and SpCas9-NG.28
The range of sites targetable by the DNA base editors can be further expanded by the use of newly developed SpCas9 variants. For example, the Liu group recently introduced several SpCas9 variants, which respectively recognize NRRH, NRTH, and NRCH PAM sequences, thereby enabling DNA base editors to operate on targets with NR PAM sequences.48 Another SpCas9 variant developed by the Kleinstiver group, named SpRY, revealed a preference for both NRN and NYN PAM sequences but to a lesser extent, suggesting that DNA base editors employing SpRY could target genes almost independently of a PAM.49
Efforts to Reduce DNA Base Editor-Induced Bystander Mutations
DNA base editors operate in a confined editing window (several nucleotides [nts] in length), which sometimes limits targetable bases, but at the same time, if multiple Cs or As exist within or nearby the editing window, they can undergo accompanying unwanted base conversions (bystander mutations). One way to reduce the frequency of bystander mutations is to narrow the editing window. The width of the editing window is determined by the DNA base editor deaminase, and the introduction of several mutations in the deaminase can reduce the size of the editing window with little effect on the deaminase activity. For example, rAPOBEC1, the cytidine deaminase in BE3, initially had a 5-nt editing window (4th–8th nts, Table 1 counting from 5′ end of sgRNA-binding region);10 the introduction of double mutations in rAPOBEC1 reduced the width of the editing window to 2 nts (5th–6th) with little activity loss (YE1-BE3 and EE-BE3) or a modest activity decrease (YE2-BE3), whereas a triple mutation (YEE-BE3) reduced the editing window to 1–2 nts (5th–6th) with a drastic decrease in editing activity.12 Another triple mutations (YFE-BE4max) also reduced editing window to 3 nts (4th–6th) with enhanced editing efficiency.40 Similarly, introducing YE mutations into dCpf1-BE, generating dCpf1-BE-YE, resulted in narrowing the editing window from 6 to 3 nts.34
Another way to reduce the frequency of bystander mutations is to use an alternative deaminase that requires a specific motif for its operation. For example, human APOBEC3A (hA3A) is known to preferentially deaminate Cs in TCR motifs in vitro,50, 51, 52 although this editing specificity was not observed in human U2OS cells.22 The introduction of a mutation into A3A (N57G) to create eA3A restored the TCR motif preference in vivo; eA3A-BE3 was associated with the higher cognate-to-bystander editing ratio than YE1-BE3, YE2-BE3, and YEE-BE3.22 In addition to modifying A3A, it has been reported that exploiting a shorter linker sequence and removing a non-essential part from CDA1 (nCDA1-BE3) could selectively edit a single cytosine at a specific position, resulting in precise base editing.31
On the other hand, the use of circularly permuted SpCas9 (CP-SpCas9) variants in CBEs expanded base editing windows while maintaining the NGG PAM sequence. CP-CBE variants, which incorporated the CP-SpCas9 variants (CP1012-Cas9, CP1028-Cas9, and CP1041-Cas9), positioned the cytidine deaminase adjacent to different SpCas9 N-terminal residues (the number after “CP” indicates the original position of the SpCas9 amino acid that is newly positioned at the N terminus) and broadened the editing window. CP1012-CBE and CP1028-CBE showed a wide base editing window (4th–11th) compared with that of BE4 (4th–8th). Furthermore, CP1012-CBE, CP1028-CBE, and CP1041-CBE additionally edited bases upstream of the protospacer on both the target and non-target stands.41 Likewise, the use of the CP-Cas9 variants (CP1012-Cas9, CP1028-Cas9, CP1041-Cas9, and CP1249-Cas9) in ABEs also resulted in a broader editing window (4th–12th) compared to that of ABE7.10 (4th–7th).41
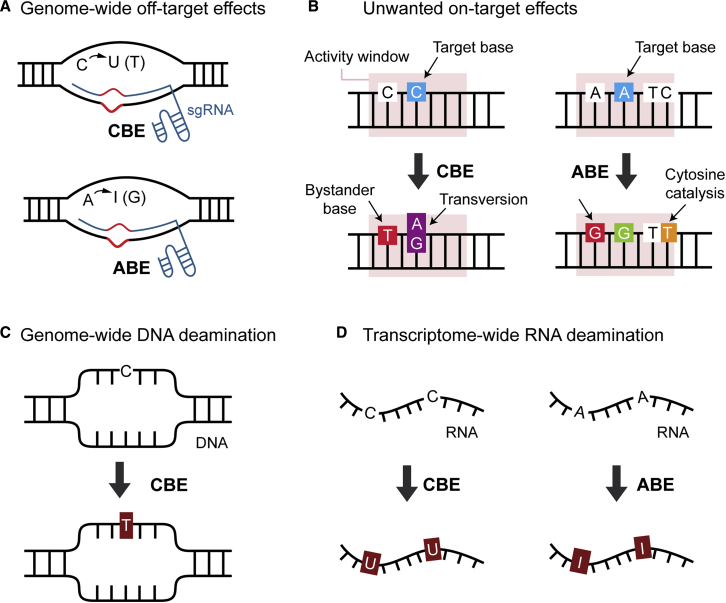
Genome-wide sgRNA-Dependent Off-Target DNA Editing by CBEs and ABEs
It is well known that Cas nucleases exhibit sgRNA-dependent, genome-wide off-target DNA editing, and it is reasonable to suppose that both CBEs and ABEs also display such behavior (Figure 2A). However, genome-wide profiling of DNA base editor off-target sites is difficult because DNA base editors do not generate DSBs, which are relatively easy to detect. The first breakthrough for CBE-mediated, genome-wide profiling of off-target sites was made by the Kim group.53,54 They employed BE3ΔUGI, which cleaves the non-edited strand during the editing process, and a uracil-specific excision reagent (USER), a mixture of uracil-DNA glycosylase (UDG) and DNA glycosylase-lyase endonuclease VII, which cleaves the edited strand, thereby generating DSBs. Then, off-target editing sites were detected at the genome level using an in vitro nuclease-digested whole-genome sequencing (Digenome-seq) method. The results showed that BE3ΔUGI exhibits less sgRNA-dependent off-target editing compared to the conventional Cas9 nucleases. For ABEs, the Kim group and the Songyang group independently used a restriction EndoV enzyme, which cleaves the edited strand, to induce DSBs.55,56 Analysis using Digenome-seq and EndoV-seq revealed that sgRNA-dependent off-target editing by ABE7.10 is also reduced compared to that of the conventional Cas9 nucleases and is comparable to that of CBEs.
Figure 2.
Possible DNA Base Editor Side Effects
(A) Schematic diagram of the mechanism behind CBE- and ABE-induced genome-wide off-target DNA mutations. The Cas9-sgRNA complex binds with mismatched DNA targets in the genome, allowing the linked deaminase to alter bases in the exposed ssDNA. (B) Unwanted on-target effects including the generation of bystander mutations within activity windows of base editors, inaccurate transversion mutation of target base via a CBE, and cytosine conversions mediated by the catalysis activity of an ABE. (C) Cytidine deaminases exhibit sgRNA-independent, genome-wide deamination effects. (D) Both cytidine and adenosine deaminases exhibit sgRNA-independent, transcriptome-wide deamination effects.
Although sgRNA-dependent off-target editing occurs relatively infrequently for both CBEs and ABEs, it represents a potential obstacle to their medical application. The simplest way to reduce sgRNA-dependent off-target editing is to use a high-fidelity SpCas9 variant instead of WT SpCas9 in CBEs and ABEs. SpCas9-HF1, xCas9(3.7), Sniper-Cas9, and eSpCas9(1.1), and evoCas9 are known to have low levels of off-target binding; the first three have been combined with BE3 to generate the high-specificity DNA base editors, HF-BE3, xCas(3.7)-BE3, and Sniper-BE3, respectively.29,42,43 In addition, SpCas9-HF1 and HypaCas9, other high fidelity SpCas9 variants, were introduced into eA3A-BE3 (generating eA3A-HF1-BE3-2xUGI and eA3A-Hypa-BE3-2xUGI, respectively) to make high-specificity DNA base editors with low level of bystander mutations.22
In addition to replacing SpCas9, changing how DNA base editors are delivered into the cell can also reduce off-target editing. Most previous studies were conducted with transfection of DNA-based constructs such as plasmids or viral vectors.21,57, 58, 59 While the delivery of plasmids exploits non-viral methods such as electroporation or cationic lipid nanoparticles, viral vector transduction is mediated by viruses such as lentivirus, adeno virus, or adeno-associated virus. Although DNA-based constructs show efficient expression and high editing efficacy, their long-term expression frequently increases off-target effects.60 Alternatively, DNA base editors can be delivered as a form of ribonucleoprotein38,42,61,62 (base editor protein and sgRNA) or mRNA,48,63,64 which can reduce off-target editing because both RNP and mRNA are rapidly degraded in cell cytoplasm.28
Recently, a new type of off-target editing, induced by the deaminase rather than by Cas9, was found. The Bae group reported that ABE can catalyze cytosine conversions at the target site (Figure 2B).19 This ABE-mediated cytosine conversion is sgRNA-dependent, similar to Cas9-mediated off-target editing, and occurs in sgRNA binding regions containing a TCN motif that spans positions 5–7, counting from the 5′ end of the target site.
Genome-wide sgRNA-Independent DNA Deamination by CBEs
In a surprising development, two groups reported that CBEs can induce genome-wide cytosine mutations regardless of the presence of sgRNAs (Figure 2C).13,14 It is difficult to detect DNA base editor-mediated deamination effects at the genome level because of the presence of numerous natural single nucleotide variants (SNVs). To address this issue, the Yang group developed a genome-wide off-target analysis by two-cell embryo injection (GOTI) method in which one of the two blastomeres in the embryo is exposed to a DNA base editor. Whole-genome sequence analysis of progeny cells of both blastomeres is performed to identify off-target SNVs. Because there are few SNVs in two-cell mouse embryos, the SNVs that are found are regarded to be induced by the DNA base editor. Using this technique, abundant sgRNA-independent deamination by the CBE (BE3) was detected dominantly at transcription sites.13,14 On the other hand, SNVs were barely found when ABE was used, suggesting that the level of ABE-induced sgRNA-independent deamination is very low.
Consistently, the Gao group found that unexpected SNVs were induced by BE3 and HF1-BE3 (a plant version of HF-BE3), but not by ABE, in rice seeds. The finding that a significant number of unwanted SNVs are generated by high-fidelity BE3 variants such as HF1-BE3 suggests that the cytidine deaminase, rather than Cas9, causes the sgRNA-independent deamination. Therefore, to reduce this effect, further cytidine deaminase modifications will be required.
Transcriptome-wide sgRNA-Independent RNA Deamination by CBEs and ABEs
DNA base editor-mediated, sgRNA-independent deamination of RNA has also been reported. The Joung group and the Yang group independently reported that CBEs can also induce transcriptome-wide mutations in cytosines in RNAs, which were detected through whole RNA sequencing data (transcriptome data; Figure 2D).15,16 Additionally, three groups further reported that ABEs can also deaminate adenosine in RNA on a transcriptome-wide level, whereas ABEs showed less genome-wide deamination of adenosines in DNA.17,18 Considering that ABEs initially contained dual adenosine deaminases (wtTadA and eTadA∗), it is believed that the sgRNA-independent deamination is caused by excess wtTadA protein, because the engineered TadA∗ preferentially interacts with ssDNA rather than RNA. The CBE- and ABE-mediated RNA deamination effects are a direct result of the DNA base editor deaminase activity; thus, further modification of the deaminases will be required to reduce these effects although it is uncertain how adverse the RNA deamination effects are due to the short lifetime of RNA.
Efforts to Reduce sgRNA-Independent CBE-Mediated RNA and DNA Deamination
Among the various candidates for improvement, CBE variants with reduced transcriptome-wide RNA deamination effects received the highest priority for development. The Joung group found that the introduction of mutations into rAPOBEC1 (rAPOBEC1-R33A and rAPOBEC1-R33A/K34A) in BE3, generating SECURE-BE3, reduced sgRNA-independent deamination in RNA.15 They also found that the RNA deamination effect is low when eA3A is used as a deaminase.18 In particular, hAID-BE3 and Target-AID exhibited negligible transcriptome-wide RNA deamination effects compared to other CBEs employing rAPOBEC1.44 The Yang group found that the double mutations in rAPOBEC1 of YE1-BE3, which was previously developed by the Liu group to have a narrower base-editing window, reduced the binding affinity of the cytidine deaminase to RNA, resulting in decreased transcriptome-wide RNA deamination.16 Furthermore, they identified A3A variants (hA3A-R128A and hA3A-Y130F) with reduced RNA deamination effects.
To reduce CBE-mediated genome-wide DNA deamination effects, several groups have tried to develop highly specific versions of CBEs. The Liu group screened several versions of BE4 carrying various mutations in the cytidine deaminase by using a reporter assay (in bacteria) or an R-loop assay (in human cells).38 In the reporter assay with E. coli, an increase in the survival rate of E. coli under rifampin by the activation of the rifampin resistance gene (rpoB) reflects sgRNA-independent deamination, whereas an increase in the survival rate of E. coli under chloramphenicol is regarded as on-target activity of CBEs. Therefore, the activity and specificity of CBEs can be evaluated by comparing the resistance to rifampin and to chloramphenicol in the reporter assay. In the R-loop assay with human cells, an additional inactive CRISPR ortholog (dSaCas9) and its sgRNA are exploited to expose ssDNA by forming the R-loop, and sgRNA-independent DNA deamination can be introduced in the ssDNA via CBEs, which can be confirmed through high-throughput sequencing. By using these two assay methods, the Liu group revealed that YE1-BE4 has lower genome-wide DNA deamination effects, compared to BE4.
On the other hand, using the GOTI method, the Yang group found that BE3 containing rAPOBEC1-R132E, YE1-BE3, and BE3 containing rAPOBEC1-W90F/R126E showed high specificity.39 In another study, the Gaudelli group found several cytidine deaminase orthologs that showed lower levels of DNA and RNA deamination by using the R-loop assay in human cells.45 They showed that BE3 containing PpAPOBEC1, RrA3F, AmAPOBEC1, and SsAPOBEC3B were more specific than that containing rAPOBEC1. Further engineering of PpAPOBEC1 (H122A or R33A) enhanced the on-target activity of the CBE and reduced DNA and RNA deamination. Current versions of high-fidelity CBEs are summarized in Table 1.
Efforts to Reduce sgRNA-Independent ABE-Mediated RNA Deamination
Because wtTadA, which is included in ABE7.10 and ABEmax, operates on RNA rather than DNA, it has been presumed to be the culprit responsible for ABE-mediated, sgRNA-independent RNA deamination, and it is expected that an appropriate mutation in wtTadA could reduce levels of RNA deamination. Indeed, the Liu group showed that ABEmax-AW, in which the wtTadA catalytic site is silenced (wtTadA-E59A) and eTadA∗ is further engineered (eTadA-V106W), exhibits less RNA deamination activity than ABEmax.65 In addition, the Liu group reduced RNA deamination by introducing the same mutation (V106W) into ABE8e, which has faster kinetics than ABEmax.27 Similarly, a V106W mutation in the ABE8.17 m variant (one of the set of ABE8s developed by the Gaudelli group) reduced RNA deamination effects.28
In a different approach, the Joung group completely removed wtTadA to reduce RNA deamination activity; they found that an ABE containing eTadA∗ only, named miniABEmax, retains on-target activity but exhibits reduced RNA deamination activity compared to the version with wtTadA.18 The Joung group further engineered TadA-7.10 and developed high-fidelity versions of ABE containing eTadA-K20A/R21A or eTadA-V82G, referred to as SECURE-ABE variants. The Yang group independently suggested improving ABE7.10 by incorporating wtTadA-F148A and eTadA-F148A based on the previous result that an F148A substitution in E. coli TadA abolishes TadA activity.16
To date, however, ABE variants with reduced levels of cytosine conversion have not yet been reported. Current versions of high-fidelity ABEs are summarized in Table 2.
Table 2.
Current Versions of High-Fidelity ABEs
 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| ABE Name | Adenosine Deaminase |
Cas Nuclease |
PAM | Level of DNA Deamination | Level of RNA Deamination | Reference | |||
| wtTadA (Mutation) | eTadA (Mutation) | Type | Mutation | Editing Window | |||||
| ABE7.10 | WT | TadA7.10 | SpCas9 | D10A | 4th–7th | NGG | low | high | 11,13,14 |
| ABEmax | WT | TadA7.10 | SpCas9 | D10A | 4t–7th | NGG | low | high | 14,24 |
| VQR-ABE | WT | TadA7.10 | SpCas9 | D10A + D1135V/R1335Q/T1337R | 4th–7th | NGA | unknown (maybe low) | unknown (maybe high) | 41,46,47 |
| VRQR-ABE | WT | TadA7.10 | SpCas9 | D10A + (D1135V/G1218R/R1335Q/T1337R) | 4th–7th | NGA | unknown (maybe low) | unknown (maybe high) | 41 |
| VRER-ABE | WT | TadA7.10 | SpCas9 | D10A + D1135V/G1218R/R1335E/T1337R | 4th–7th | NGCG | unknown (maybe low) | unknown (maybe high) | 41,47 |
| NG-ABE | WT | TadA7.10 | SpCas9 | D10A + 7 mutations (R1335V/L1111R/D1135V/G1218R/E1219F/A1322R/T1337R) | 4th–7th | NG | unknown (maybe low) | unknown (maybe high) | 36,41,47 |
| xCas9(3.7)-ABE | WT | TadA7.10 | SpCas9 | D10A + 7 mutations (A262T/R324L/S409I/E480K/E543D/M694I/E1219V) | 4th–7th | NG/GAA/GAT | unknown (maybe low) | unknown (maybe high) | 29,36,41 |
| Spy-mac ABEmax | WT | TadA7.10 | Spy-mac Cas9 | D10A | 4th–7th | NAA | unknown (maybe low) | unknown (maybe high) | 32 |
| Sa-ABE | WT | TadA7.10 | saCas9 | D10A | 4th–14th | NNGRRT | unknown (maybe low) | unknown (maybe high) | 41 |
| SaKKH-ABE | WT | TadA7.10 | SaCas9 | D10A + E782K/N968K/R1015H | 4th–14th | NNNRRT | unknown (maybe low) | unknown (maybe high) | 41,46 |
| ScCas9-ABE | WT | TadA7.10 | ScCas9 | D10A | 4th–7th | NNG | unknown (maybe low) | unknown (maybe high) | 37 |
| SauriABEmax | WT | TadA7.10 | SauriCas9 | D15A | 6th–14th | NNGG | unknown (maybe low) | unknown (maybe high) | 33 |
| ABE8e | Del | TadA-8e | SpCas9 | D10A | 4th–8th | NGG | high | high | 27 |
| SaABE8e | Del | TadA-8e | SaCas9 | D10A | 3rd–14th | NNGRRT | high | high | 27 |
| LbABE8e | Del | TadA-8e | LbCas12a | D832A | 8th–14th | TTTV | high | high | 27 |
| enAsABE8e | Del | TadA-8e | AsCas12a | D908A + E174R/S542R/K548R | 8th–14th | TTTV | high | high | 27 |
| ABE8s | Del | TadA-8 s | SpCas9 | D10A | 3rd–9th | NGG | unknown | high | 28 |
| NG-ABE8s | Del | TadA-8 s | SpCas9 | D10A + 7 mutations (R1335V/L1111R/D1135V/G1218R/E1219F/A1322R/T1337R) | 3rd–9th | NG | unknown | high | 28 |
| Sa-ABE8s | Del | TadA-8 s | SaCas9 | D10A | 5th–14th | NNGRRT | unknown | high | 28 |
| CP1012-ABE | WT | TadA7.10 | SpCas9 | CP1012 | 4th–12th | NGG | unknown (maybe low) | unknown (maybe high) | 41 |
| CP1028-ABE | WT | TadA7.10 | SpCas9 | CP1028 | 4th–12th | NGG | unknown (maybe low) | unknown (maybe high) | 41 |
| CP1041-ABE | WT | TadA7.10 | SpCas9 | CP1041 | 4th–12th | NGG | unknown (maybe low) | unknown (maybe high) | 41 |
| CP1249-ABE | WT | TadA7.10 | SpCas9 | CP1249 | 4th–12th | NGG | unknown (maybe low) | unknown (maybe high) | 41 |
| ABEmax-AW | E59A | TadA7.10 (V106W) | SpCas9 | D10A | 4th–7th | NGG | low | reduced | 65 |
| ABE8e(TadA-8e V106W) | Del | TadA-8e (V106W) | SpCas9 | D10A | 4th–8th | NGG | reduced | reduced | 27 |
| ABE8.17-m | Del | TadA8.17 (V106W) | SpCas9 | D10A | 3rd–9th | NGG | low | reduced | 28 |
| SECURE-ABE | Del | TadA7.10 (K20A+R21A) | SpCas9 | D10A | 4th–7th | NGG | low | reduced | 18 |
| SECURE-ABE | Del | TadA7.10 (V82G) | SpCas9 | D10A | 4th–7th | NGG | low | reduced | 18 |
| ABE7.10-F148A | TadA (F148A) | TadA7.10 (F148A) | SpCas9 | D10A | 4th–6th | NGG | low | reduced | 16 |
Therapeutic Applications of DNA Base Editors
A single nucleotide mutation generating either a protein with a wrong amino acid (missense mutation) or a truncated protein (nonsense mutation) is the main cause of genetic diseases (>58% of the entries in the ClinVar database).25 The advantages of DNA base editors—high editing efficiency and not generating DSB—suggest a bright outlook of DNA base editors as a therapeutic tool. Indeed, CBE variants efficiently corrected the mutated genes or their promoters associated with diseases such as p53 (cancer),10 HBB (β-thalassemia),66,67 and FBN1 (Marfan syndrome).68 Likewise, ABE variants were effective in correcting the genes such as HFE (hereditary haemochromatosis),11 DMD (Duchenne muscular dystrophy),64 Fah (hereditary tyrosinemia type I),69 COL7A1 (recessive dystrophic epidermolysis bullosa),70,71 and TRET (brain tumor).72 Conversely, introducing mutations into genes relevant to diseases by DNA base editors can be another strategy to prevent or treat the diseases. APOE4 (Alzheimer’s disease) could be converted into less risky APE3r by BE3-induced mutation.10 Fetal hemoglobin upregulation, which alleviates the symptoms of β-globin-related blood diseases, could be achieved by introducing the mutations into the enhancer of BCL11A or the promoter of HBG1 and HBG2 through ABE7.10, ABEmax, or ABE8e.11,24,27 Furthermore, multiplex editing ability of DNA base editor brightens the prospect for its medical application. Disruption of the BCL11A erythroid enhancer with correction of the HBB promoter mutation could be achieved in primary human hematopoietic stem and progenitor cells, suggesting an effective way for curing blood disorders,67 and multiplex gene modification in primary human T cells showed the potential to improve chimeric antigen receptor engineered T cell immunotherapy minimizing risks from DSB generation.28,73
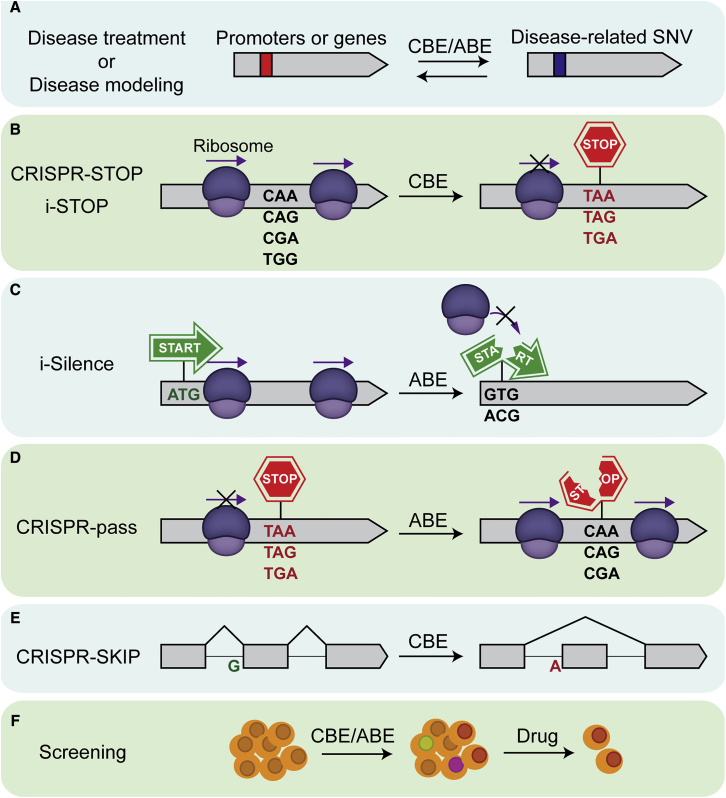
Versatile Applications of DNA Base Editors
The base conversion ability of DNA base editors has been harnessed not only for correcting mutated genes but for versatile applications in various ways (Figure 3). One idea is that CBEs could be used for introducing a premature termination codon (PTC) in the middle of a target gene to disrupt its expression. Two independent approaches, named CRISPR-STOP and iSTOP, were used for showing that BE3 can create in-frame stop codons by converting CAA, CAG, CGA, and/or TGG codons into TAA, TAG, and TGA stop codons in multiple genes in the human genome.74,75 Another idea for gene disruption is to use an ABE to mutate the start codon (ATG), thereby abolishing gene expression. A new strategy, named i-Silence, involves conversion of an ATG to GTG or ACG using ABEmax, resulting in silencing of a gene of interest.76 Unlike CRISPR-STOP and iSTOP, gene disruption by i-Silence does not generate truncated protein variants because the start of transcription is blocked. Conversely, pre-existing PTCs can be bypassed by ABEs to avoid truncated protein generation. A strategy named CRISPR-pass successfully changed PTCs to glutamine (CAA or CAG) or arginine (CGA) codons through A-to-G or T-to-C conversion to allow transcription to proceed.77
Figure 3.
Applications of DNA Base Editors
(A) DNA base editors can be used for versatile therapeutic tools for disease treatment or disease modeling. (B) CBEs can be used for converting a CAA, CAG, CGA, or TGG codon into a premature stop codon (TAA, TAG, or TGA), which abolishes protein synthesis and results in a knockout of gene function. (C) ABE can be used for converting the adenine in a start codon (ATG) into a guanine to abolish protein synthesis. (D) ABE can be used for converting a premature stop codon into a codon for an amino acid. Although the resulting codon is not always the same as the unmutated original, whole protein synthesis is no longer abolished. (E) CBEs can be used for converting Gs within splicing acceptor sites into As by editing Cs in the complementary strand of the target site. (F) BEs can be used for generating various substitutions for screening experiments.
In addition to gene expression and disruption, isoform-specific gene expression can also be controlled by DNA base editors. Most proteins have multiple isoforms, and the alternative splicing of the pre-mRNA is a key step to determine the isoform type by which some exons are excluded from the mature transcripts.78 CRISPR-SKIP is a method based on the fact that most introns end with G.79 Cytidine deaminases of CRISPR-SKIP convert Gs within splice acceptor sites into As by editing Cs in the complementary strand of the target site. As a result, the corresponding exons fail to be incorporated into the mature transcripts while the other exons are expressed normally. DNA base editors can be used for functional screening of single-nucleotide variants as well, thanks to their accuracy and simplicity in single-nucleotide editing. Indeed, recent studies revealed that DNA base editors can be a powerful tool for functional genetic screenings.80, 81, 82, 83
Conclusions
It is undoubted that base editing technologies have had a large impact on the gene-editing field. Their remarkable properties, including their high editing efficiency, acceptance for non-dividing cells, and ability to generate targeted mutations without generating DSBs, make DNA base editors particularly compelling among current gene-editing tools. On the other hand, as described above, DNA base editors have several drawbacks, such as limitations in their targetable sites, the generation of bystander mutations, and unexpected off-target effects in both DNA and RNA, that raise concerns about potential future therapeutic applications of these tools. However, a high level of research is currently ongoing to cross these hurdles, and evidence from these studies suggests that we will be able to utilize the potential of DNA base editors fully in the near future.
Designing proper sgRNAs is another critical issue of DNA base editors because the design and optimization of sgRNA for DNA base editors is more complicated compared to those for CRISPR-based methods. Currently, typical CRISPR sgRNA design programs such as CRISPOR,84 CHOPCHOP,85, 86, 87 and Cas-Designer88 can be used for sgRNAs of DNA base editors. In particular, DNA base editor-dedicated tools such as BE-Designer,89 sgSTOPs,75 beditor,90 SNP-CRISPR,91 BE-FF,92 and Benchling are available as a command-line program or at the website. Recently, a machine learning-based sgRNA design tool, BE-Hive,93 was newly developed, which provides predicted editing efficacy, as well as genotype outcomes for each target according to 11 different CBEs and ABEs and 2 different cell lines.
Despite the intense efforts to improve DNA base editors, there are fundamental boundaries in usage; DNA base editing is confined to transition mutations (incapable of transversion mutations) and is not competent in inducing indel mutations. As an alternative, the Liu group developed a prime editing (PE) system,94 which enables generating small insertion and deletion in addition to substitution of several nucleotides at target sites. With these properties, PEs are expected to complement DNA base editors. Nevertheless, DNA base editors are still attractive because they have higher on-target activities and generate lower unwanted indel frequencies. Furthermore, variable developments and applications of DNA base editors have plentifully been reported, possibly suggesting that DNA base editors will be applied to DNA base-editor-capable targets with a priority.
Author Contributions
All authors have contributed to the writing of the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This research was supported by grants from the National Research Foundation of Korea (NRF; grant number 2018M3A9H3022412) and the Korea Healthcare Technology R&D Project (HI16C1012) to S.B.
References
- 1.Mali P., Esvelt K.M., Church G.M. Cas9 as a versatile tool for engineering biology. Nat. Methods. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sander J.D., Joung J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim H., Kim J.S. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- 4.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 5.Koonin E.V., Makarova K.S., Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017;37:67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haapaniemi E., Botla S., Persson J., Schmierer B., Taipale J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 2018;24:927–930. doi: 10.1038/s41591-018-0049-z. [DOI] [PubMed] [Google Scholar]
- 7.Ihry R.J., Worringer K.A., Salick M.R., Frias E., Ho D., Theriault K., Kommineni S., Chen J., Sondey M., Ye C. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat. Med. 2018;24:939–946. doi: 10.1038/s41591-018-0050-6. [DOI] [PubMed] [Google Scholar]
- 8.Kosicki M., Tomberg K., Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018;36:765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J.P., Li X.L., Li G.H., Chen W., Arakaki C., Botimer G.D., Baylink D., Zhang L., Wen W., Fu Y.W. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol. 2017;18:35. doi: 10.1186/s13059-017-1164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I., Liu D.R. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim Y.B., Komor A.C., Levy J.M., Packer M.S., Zhao K.T., Liu D.R. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol. 2017;35:371–376. doi: 10.1038/nbt.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuo E., Sun Y., Wei W., Yuan T., Ying W., Sun H., Yuan L., Steinmetz L.M., Li Y., Yang H. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science. 2019;364:289–292. doi: 10.1126/science.aav9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin S., Zong Y., Gao Q., Zhu Z., Wang Y., Qin P., Liang C., Wang D., Qiu J.L., Zhang F., Gao C. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science. 2019;364:292–295. doi: 10.1126/science.aaw7166. [DOI] [PubMed] [Google Scholar]
- 15.Grünewald J., Zhou R., Garcia S.P., Iyer S., Lareau C.A., Aryee M.J., Joung J.K. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature. 2019;569:433–437. doi: 10.1038/s41586-019-1161-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou C., Sun Y., Yan R., Liu Y., Zuo E., Gu C., Han L., Wei Y., Hu X., Zeng R. Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature. 2019;571:275–278. doi: 10.1038/s41586-019-1314-0. [DOI] [PubMed] [Google Scholar]
- 17.Rees H.A., Wilson C., Doman J.L., Liu D.R. Analysis and minimization of cellular RNA editing by DNA adenine base editors. Sci. Adv. 2019;5:eaax5717. doi: 10.1126/sciadv.aax5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grünewald J., Zhou R., Iyer S., Lareau C.A., Garcia S.P., Aryee M.J., Joung J.K. CRISPR DNA base editors with reduced RNA off-target and self-editing activities. Nat. Biotechnol. 2019;37:1041–1048. doi: 10.1038/s41587-019-0236-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim H.S., Jeong Y.K., Hur J.K., Kim J.S., Bae S. Adenine base editors catalyze cytosine conversions in human cells. Nat. Biotechnol. 2019;37:1145–1148. doi: 10.1038/s41587-019-0254-4. [DOI] [PubMed] [Google Scholar]
- 20.Nishida K., Arazoe T., Yachie N., Banno S., Kakimoto M., Tabata M., Mochizuki M., Miyabe A., Araki M., Hara K.Y. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016;353:aaf8729. doi: 10.1126/science.aaf8729. [DOI] [PubMed] [Google Scholar]
- 21.Zafra M.P., Schatoff E.M., Katti A., Foronda M., Breinig M., Schweitzer A.Y., Simon A., Han T., Goswami S., Montgomery E. Optimized base editors enable efficient editing in cells, organoids and mice. Nat. Biotechnol. 2018;36:888–893. doi: 10.1038/nbt.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gehrke J.M., Cervantes O., Clement M.K., Wu Y., Zeng J., Bauer D.E., Pinello L., Joung J.K. An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities. Nat. Biotechnol. 2018;36:977–982. doi: 10.1038/nbt.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komor A.C., Zhao K.T., Packer M.S., Gaudelli N.M., Waterbury A.L., Koblan L.W., Kim Y.B., Badran A.H., Liu D.R. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci. Adv. 2017;3:eaao4774. doi: 10.1126/sciadv.aao4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koblan L.W., Doman J.L., Wilson C., Levy J.M., Tay T., Newby G.A., Maianti J.P., Raguram A., Liu D.R. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 2018;36:843–846. doi: 10.1038/nbt.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rees H.A., Liu D.R. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018;19:770–788. doi: 10.1038/s41576-018-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Losey H.C., Ruthenburg A.J., Verdine G.L. Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA. Nat. Struct. Mol. Biol. 2006;13:153–159. doi: 10.1038/nsmb1047. [DOI] [PubMed] [Google Scholar]
- 27.Richter M.F., Zhao K.T., Eton E., Lapinaite A., Newby G.A., Thuronyi B.W. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol. 2020;38:883–891. doi: 10.1038/s41587-020-0453-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaudelli N.M., Lam D.K., Rees H.A., Solá-Esteves N.M., Barrera L.A., Born D.A., Edwards A., Gehrke J.M., Lee S.J., Liquori A.J. Directed evolution of adenine base editors with increased activity and therapeutic application. Nat. Biotechnol. 2020;38:892–900. doi: 10.1038/s41587-020-0491-6. [DOI] [PubMed] [Google Scholar]
- 29.Hu J.H., Miller S.M., Geurts M.H., Tang W., Chen L., Sun N., Zeina C.M., Gao X., Rees H.A., Lin Z., Liu D.R. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556:57–63. doi: 10.1038/nature26155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishimasu H., Shi X., Ishiguro S., Gao L., Hirano S., Okazaki S., Noda T., Abudayyeh O.O., Gootenberg J.S., Mori H. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science. 2018;361:1259–1262. doi: 10.1126/science.aas9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan J., Zhang F., Karcher D., Bock R. Engineering of high-precision base editors for site-specific single nucleotide replacement. Nat. Commun. 2019;10:439. doi: 10.1038/s41467-018-08034-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Z., Shan H., Chen S., Chen M., Song Y., Lai L., Li Z. Efficient base editing with expanded targeting scope using an engineered Spy-mac Cas9 variant. Cell Discov. 2019;5:58. doi: 10.1038/s41421-019-0128-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu Z., Wang S., Zhang C., Gao N., Li M., Wang D., Wang D., Liu D., Liu H., Ong S.G. A compact Cas9 ortholog from Staphylococcus Auricularis (SauriCas9) expands the DNA targeting scope. PLoS Biol. 2020;18:e3000686. doi: 10.1371/journal.pbio.3000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X., Wang Y., Liu Y., Yang B., Wang X., Wei J., Lu Z., Zhang Y., Wu J., Huang X. Base editing with a Cpf1-cytidine deaminase fusion. Nat. Biotechnol. 2018;36:324–327. doi: 10.1038/nbt.4102. [DOI] [PubMed] [Google Scholar]
- 35.Kleinstiver B.P., Sousa A.A., Walton R.T., Tak Y.E., Hsu J.Y., Clement K., Welch M.M., Horng J.E., Malagon-Lopez J., Scarfò I. Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 2019;37:276–282. doi: 10.1038/s41587-018-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hua K., Tao X., Han P., Wang R., Zhu J.K. Genome Engineering in Rice Using Cas9 Variants that Recognize NG PAM Sequences. Mol. Plant. 2019;12:1003–1014. doi: 10.1016/j.molp.2019.03.009. [DOI] [PubMed] [Google Scholar]
- 37.Chatterjee P., Jakimo N., Jacobson J.M. Minimal PAM specificity of a highly similar SpCas9 ortholog. Sci. Adv. 2018;4:eaau0766. doi: 10.1126/sciadv.aau0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doman J.L., Raguram A., Newby G.A., Liu D.R. Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat. Biotechnol. 2020;38:620–628. doi: 10.1038/s41587-020-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zuo E., Sun Y., Yuan T., He B., Zhou C., Ying Y. High-fidelity base editor with no detectable genome-wide off-target effects. bioRxiv. 2020 doi: 10.1101/2020.02.07.939074. [DOI] [Google Scholar]
- 40.Liu Z., Chen S., Shan H., Jia Y., Chen M., Song Y., Lai L., Li Z. Efficient base editing with high precision in rabbits using YFE-BE4max. Cell Death Dis. 2020;11:36. doi: 10.1038/s41419-020-2244-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang T.P., Zhao K.T., Miller S.M., Gaudelli N.M., Oakes B.L., Fellmann C., Savage D.F., Liu D.R. Circularly permuted and PAM-modified Cas9 variants broaden the targeting scope of base editors. Nat. Biotechnol. 2019;37:626–631. doi: 10.1038/s41587-019-0134-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rees H.A., Komor A.C., Yeh W.H., Caetano-Lopes J., Warman M., Edge A.S.B., Liu D.R. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat. Commun. 2017;8:15790. doi: 10.1038/ncomms15790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee J.K., Jeong E., Lee J., Jung M., Shin E., Kim Y.H., Lee K., Jung I., Kim D., Kim S., Kim J.S. Directed evolution of CRISPR-Cas9 to increase its specificity. Nat. Commun. 2018;9:3048. doi: 10.1038/s41467-018-05477-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng T.L., Li S., Yuan B., Wang X., Zhou W., Qiu Z. Expanding C-T base editing toolkit with diversified cytidine deaminases. Nat. Commun. 2019;10:3612. doi: 10.1038/s41467-019-11562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu Y., Leete T.C., Born D.A., Young L., Barrera L.A., Lee S.J., Rees H.A., Ciaramella G., Gaudelli N.M. Cytosine base editors with minimized unguided DNA and RNA off-target events and high on-target activity. Nat. Commun. 2020;11:2052. doi: 10.1038/s41467-020-15887-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang L., Zhang X., Wang L., Yin S., Zhu B., Xie L., Duan Q., Hu H., Zheng R., Wei Y. Increasing targeting scope of adenosine base editors in mouse and rat embryos through fusion of TadA deaminase with Cas9 variants. Protein Cell. 2018;9:814–819. doi: 10.1007/s13238-018-0568-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeong Y.K., Yu J., Bae S. Construction of non-canonical PAM-targeting adenosine base editors by restriction enzyme-free DNA cloning using CRISPR-Cas9. Sci. Rep. 2019;9:4939. doi: 10.1038/s41598-019-41356-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller S.M., Wang T., Randolph P.B., Arbab M., Shen M.W., Huang T.P., Matuszek Z., Newby G.A., Rees H.A., Liu D.R. Continuous evolution of SpCas9 variants compatible with non-G PAMs. Nat. Biotechnol. 2020;38:471–481. doi: 10.1038/s41587-020-0412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walton R.T., Christie K.A., Whittaker M.N., Kleinstiver B.P. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science. 2020;368:290–296. doi: 10.1126/science.aba8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Logue E.C., Bloch N., Dhuey E., Zhang R., Cao P., Herate C., Chauveau L., Hubbard S.R., Landau N.R. A DNA sequence recognition loop on APOBEC3A controls substrate specificity. PLoS ONE. 2014;9:e97062. doi: 10.1371/journal.pone.0097062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi K., Carpenter M.A., Banerjee S., Shaban N.M., Kurahashi K., Salamango D.J., McCann J.L., Starrett G.J., Duffy J.V., Demir Ö. Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B. Nat. Struct. Mol. Biol. 2017;24:131–139. doi: 10.1038/nsmb.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kouno T., Silvas T.V., Hilbert B.J., Shandilya S.M.D., Bohn M.F., Kelch B.A., Royer W.E., Somasundaran M., Kurt Yilmaz N., Matsuo H., Schiffer C.A. Crystal structure of APOBEC3A bound to single-stranded DNA reveals structural basis for cytidine deamination and specificity. Nat. Commun. 2017;8:15024. doi: 10.1038/ncomms15024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim D., Bae S., Park J., Kim E., Kim S., Yu H.R., Hwang J., Kim J.I., Kim J.-S. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods. 2015;12:237–243. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- 54.Kim D., Lim K., Kim S.T., Yoon S.H., Kim K., Ryu S.M., Kim J.S. Genome-wide target specificities of CRISPR RNA-guided programmable deaminases. Nat. Biotechnol. 2017;35:475–480. doi: 10.1038/nbt.3852. [DOI] [PubMed] [Google Scholar]
- 55.Kim D., Kim D.E., Lee G., Cho S.I., Kim J.S. Genome-wide target specificity of CRISPR RNA-guided adenine base editors. Nat. Biotechnol. 2019;37:430–435. doi: 10.1038/s41587-019-0050-1. [DOI] [PubMed] [Google Scholar]
- 56.Liang P., Xie X., Zhi S., Sun H., Zhang X., Chen Y., Chen Y., Xiong Y., Ma W., Liu D. Genome-wide profiling of adenine base editor specificity by EndoV-seq. Nat. Commun. 2019;10:67. doi: 10.1038/s41467-018-07988-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Levy J.M., Yeh W.H., Pendse N., Davis J.R., Hennessey E., Butcher R., Koblan L.W., Comander J., Liu Q., Liu D.R. Cytosine and adenine base editing of the brain, liver, retina, heart and skeletal muscle of mice via adeno-associated viruses. Nat. Biomed. Eng. 2020;4:97–110. doi: 10.1038/s41551-019-0501-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Villiger L., Grisch-Chan H.M., Lindsay H., Ringnalda F., Pogliano C.B., Allegri G., Fingerhut R., Häberle J., Matos J., Robinson M.D. Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat. Med. 2018;24:1519–1525. doi: 10.1038/s41591-018-0209-1. [DOI] [PubMed] [Google Scholar]
- 59.Chadwick A.C., Wang X., Musunuru K. In Vivo Base Editing of PCSK9 (Proprotein Convertase Subtilisin/Kexin Type 9) as a Therapeutic Alternative to Genome Editing. Arterioscler. Thromb. Vasc. Biol. 2017;37:1741–1747. doi: 10.1161/ATVBAHA.117.309881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim S., Kim D., Cho S.W., Kim J., Kim J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yeh W.H., Chiang H., Rees H.A., Edge A.S.B., Liu D.R. In vivo base editing of post-mitotic sensory cells. Nat. Commun. 2018;9:2184. doi: 10.1038/s41467-018-04580-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gao X., Tao Y., Lamas V., Huang M., Yeh W.H., Pan B., Hu Y.J., Hu J.H., Thompson D.B., Shu Y. Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents. Nature. 2018;553:217–221. doi: 10.1038/nature25164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hendel A., Bak R.O., Clark J.T., Kennedy A.B., Ryan D.E., Roy S., Steinfeld I., Lunstad B.D., Kaiser R.J., Wilkens A.B. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 2015;33:985–989. doi: 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ryu S.M., Koo T., Kim K., Lim K., Baek G., Kim S.T., Kim H.S., Kim D.E., Lee H., Chung E., Kim J.S. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat. Biotechnol. 2018;36:536–539. doi: 10.1038/nbt.4148. [DOI] [PubMed] [Google Scholar]
- 65.Rees H.A., Wilson C., Doman J.L., Liu D.R. Analysis and minimization of cellular RNA editing by DNA adenine base editors. Sci. Adv. 2019;5:eaax5717. doi: 10.1126/sciadv.aax5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liang P., Ding C., Sun H., Xie X., Xu Y., Zhang X., Sun Y., Xiong Y., Ma W., Liu Y. Correction of β-thalassemia mutant by base editor in human embryos. Protein Cell. 2017;8:811–822. doi: 10.1007/s13238-017-0475-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zeng J., Wu Y., Ren C., Bonanno J., Shen A.H., Shea D., Gehrke J.M., Clement K., Luk K., Yao Q. Therapeutic base editing of human hematopoietic stem cells. Nat. Med. 2020;26:535–541. doi: 10.1038/s41591-020-0790-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zeng Y., Li J., Li G., Huang S., Yu W., Zhang Y., Chen D., Chen J., Liu J., Huang X. Correction of the Marfan Syndrome Pathogenic FBN1 Mutation by Base Editing in Human Cells and Heterozygous Embryos. Mol. Ther. 2018;26:2631–2637. doi: 10.1016/j.ymthe.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Song C.Q., Jiang T., Richter M., Rhym L.H., Koblan L.W., Zafra M.P., Schatoff E.M., Doman J.L., Cao Y., Dow L.E. Adenine base editing in an adult mouse model of tyrosinaemia. Nat. Biomed. Eng. 2020;4:125–130. doi: 10.1038/s41551-019-0357-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mittapalli V.R., Madl J., Löffek S., Kiritsi D., Kern J.S., Römer W., Nyström A., Bruckner-Tuderman L. Injury-Driven Stiffening of the Dermis Expedites Skin Carcinoma Progression. Cancer Res. 2016;76:940–951. doi: 10.1158/0008-5472.CAN-15-1348. [DOI] [PubMed] [Google Scholar]
- 71.Rashidghamat E., McGrath J.A. Novel and emerging therapies in the treatment of recessive dystrophic epidermolysis bullosa. Intractable Rare Dis. Res. 2017;6:6–20. doi: 10.5582/irdr.2017.01005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li J.-X., Cao Z., Guo S., Tian Z., Liu W., Zhu F. Molecular characterization and functional analysis of two trehalose transporter genes in the cabbage beetle, Colaphellus bowringi. J. Asia Pac. Entomol. 2020;23:627–633. [Google Scholar]
- 73.Webber B.R., Lonetree C.L., Kluesner M.G., Johnson M.J., Pomeroy E.J., Diers M.D. Highly efficient multiplex human T cell engineering without double-strand breaks using Cas9 base editors. Nat. Commun. 2019;10:5222. doi: 10.1038/s41467-019-13007-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuscu C., Parlak M., Tufan T., Yang J., Szlachta K., Wei X., Mammadov R., Adli M. CRISPR-STOP: gene silencing through base-editing-induced nonsense mutations. Nat. Methods. 2017;14:710–712. doi: 10.1038/nmeth.4327. [DOI] [PubMed] [Google Scholar]
- 75.Billon P., Bryant E.E., Joseph S.A., Nambiar T.S., Hayward S.B., Rothstein R., Ciccia A. CRISPR-Mediated Base Editing Enables Efficient Disruption of Eukaryotic Genes through Induction of STOP Codons. Mol Cell. 2017;67:1068–1079. doi: 10.1016/j.molcel.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang X., Liu Z., Li G., Dang L., Huang S., He L., Ma Y., Li C., Liu M., Yang G. Efficient Gene Silencing by Adenine Base Editor-Mediated Start Codon Mutation. Mol. Ther. 2020;28:431–440. doi: 10.1016/j.ymthe.2019.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee C., Hyun Jo D., Hwang G.H., Yu J., Kim J.H., Park S.E., Kim J.S., Kim J.H., Bae S. CRISPR-Pass: Gene Rescue of Nonsense Mutations Using Adenine Base Editors. Mol. Ther. 2019;27:1364–1371. doi: 10.1016/j.ymthe.2019.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Graveley B.R. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 79.Gapinske M., Luu A., Winter J., Woods W.S., Kostan K.A., Shiva N., Song J.S., Perez-Pinera P. CRISPR-SKIP: programmable gene splicing with single base editors. Genome Biol. 2018;19:107. doi: 10.1186/s13059-018-1482-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kuscu C., Adli M. CRISPR-Cas9-AID base editor is a powerful gain-of-function screening tool. Nat. Methods. 2016;13:983–984. doi: 10.1038/nmeth.4076. [DOI] [PubMed] [Google Scholar]
- 81.Jun S., Lim H., Chun H., Lee J.H., Bang D. Single-cell analysis of a mutant library generated using CRISPR-guided deaminase in human melanoma cells. Commun. Biol. 2020;3:154. doi: 10.1038/s42003-020-0888-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kweon J., Jang A.H., Shin H.R., See J.E., Lee W., Lee J.W., Chang S., Kim K., Kim Y. A CRISPR-based base-editing screen for the functional assessment of BRCA1 variants. Oncogene. 2020;39:30–35. doi: 10.1038/s41388-019-0968-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hanna R.E., Hegde M., Fagre C.R., DeWeirdt P.C., Sangree A.K., Szegletes Z. Massively parallel assessment of human variants with base editor screens. bioRxiv. 2020 doi: 10.1101/2020.05.17.100818. [DOI] [PubMed] [Google Scholar]
- 84.Haeussler M., Schönig K., Eckert H., Eschstruth A., Mianné J., Renaud J.B., Schneider-Maunoury S., Shkumatava A., Teboul L., Kent J. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17:148. doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Montague T.G., Cruz J.M., Gagnon J.A., Church G.M., Valen E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014;42:W401-7. doi: 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Labun K., Montague T.G., Gagnon J.A., Thyme S.B., Valen E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016;44 doi: 10.1093/nar/gkw398. W272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Labun K., Montague T.G., Krause M., Torres Cleuren Y.N., Tjeldnes H., Valen E. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019;47(W1):W171–W174. doi: 10.1093/nar/gkz365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Park J., Bae S., Kim J.S. Cas-Designer: a web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics. 2015;31:4014–4016. doi: 10.1093/bioinformatics/btv537. [DOI] [PubMed] [Google Scholar]
- 89.Hwang G.H., Park J., Lim K., Kim S., Yu J., Yu E., Kim S.T., Eils R., Kim J.S., Bae S. Web-based design and analysis tools for CRISPR base editing. BMC Bioinformatics. 2018;19:542. doi: 10.1186/s12859-018-2585-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dandage R., Després P.C., Yachie N., Landry C.R. beditor: A Computational Workflow for Designing Libraries of Guide RNAs for CRISPR-Mediated Base Editing. Genetics. 2019;212:377–385. doi: 10.1534/genetics.119.302089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen C.L., Rodiger J., Chung V., Viswanatha R., Mohr S.E., Hu Y., Perrimon N. SNP-CRISPR: A Web Tool for SNP-Specific Genome Editing. G3 (Bethesda) 2020;10:489–494. doi: 10.1534/g3.119.400904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rabinowitz R., Abadi S., Almog S., Offen D. Prediction of synonymous corrections by the BE-FF computational tool expands the targeting scope of base editing. Nucleic Acids Res. 2020;48(W1):W340–W347. doi: 10.1093/nar/gkaa215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Arbab M., Shen M.W., Mok B., Wilson C., Matuszek Ż., Cassa C.A., Liu D.R. Determinants of Base Editing Outcomes from Target Library Analysis and Machine Learning. Cell. 2020 doi: 10.1016/j.cell.2020.05.037. Published online June 9, 2020. 10.1016/j.cell.2020.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Anzalone A.V., Randolph P.B., Davis J.R., Sousa A.A., Koblan L.W., Levy J.M., Chen P.J., Wilson C., Newby G.A., Raguram A., Liu D.R. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]