Casein kinase 1 regulates cross-kingdom infections of a cytorhabdovirus in host plants and insect vectors by phosphorylating an intrinsically disordered region of viral phosphoprotein.
Abstract
Casein kinase 1 (CK1) family members are conserved Ser/Thr protein kinases that regulate important developmental processes in all eukaryotic organisms. However, the functions of CK1 in plant immunity remain largely unknown. Barley yellow striate mosaic virus (BYSMV), a plant cytorhabdovirus, infects cereal crops and is obligately transmitted by the small brown planthopper (SBPH; Laodelphax striatellus). The BYSMV phosphoprotein (P) exists as two forms with different mobilities corresponding to 42 kD (P42) and 44 kD (P44) in SDS-PAGE gels. Mass spectrometric analyses revealed a highly phosphorylated serine-rich (SR) motif at the C-terminal intrinsically disordered region of the P protein. The Ala-substitution mutant (PS5A) in the SR motif stimulated virus replication, whereas the phosphorylation-mimic mutant (PS5D) facilitated virus transcription. Furthermore, PS5A and PS5D associated preferentially with nucleocapsid protein–RNA templates and the large polymerase protein to provide optimal replication and transcription complexes, respectively. Biochemistry assays demonstrated that plant and insect CK1 protein kinases could phosphorylate the SR motif and induce conformational changes from P42 to P44. Moreover, overexpression of CK1 or a dominant-negative mutant impaired the balance between P42 and P44, thereby compromising virus infections. Our results demonstrate that BYSMV recruits the conserved CK1 kinases to achieve its cross-kingdom infection in host plants and insect vectors.
INTRODUCTION
Phosphorylation is one of the most important posttranslational modifications in eukaryotic cells and has important roles in enzyme activation, transcription, and biotic/abiotic stress responses (Friso and van Wijk, 2015; Millar et al., 2019). As a reversible process, phosphorylation is catalyzed by protein kinases, while dephosphorylation is achieved by protein phosphatases (Friso and van Wijk, 2015; Millar et al., 2019). Casein kinase 1 (CK1) proteins are highly conserved Ser/Thr protein kinases in eukaryotes. CK1-type kinases have been extensively studied in their regulatory role of divergent biological processes in yeast and animals (Gross and Anderson, 1998; Knippschild et al., 2005). By contrast, only a few studies have focused on the biological roles of plant CK1 proteins. Arabidopsis (Arabidopsis thaliana) CK1.3 and CK1.4 phosphorylate the photoreceptor cryptochrome 2 to regulate blue light signaling (Tan et al., 2013). Arabidopsis CK1.8 regulates ethylene biosynthesis by phosphorylating ACC SYNTHASE5 (ACS5), a key enzyme in ethylene biosynthesis, thereby promoting ACS5 turnover (Tan and Xue, 2014). Arabidopsis CASEIN KINASE 1-LIKE6 (CKL6) mainly plays a role in macromolecular trafficking at plasmodesmata sites (Lee et al., 2005). A rice (Oryza sativa) CK1 has been shown to be involved in signal transduction and hormone metabolism (Liu et al., 2003). However, the function of CK1 in plant immunity has not yet been elucidated.

The virus family Rhabdoviridae contains bacilliform or bullet-shaped, enveloped viruses that have monopartite single-stranded, negative-sense RNA genomes (Ammar et al., 2009). More than 160 rhabdoviruses infect a broad range of hosts from human, wildlife, livestock, to plant. Plant rhabdoviruses infect various monocot and dicot plants, including agriculturally significant crops such as wheat (Triticum aestivum), rice, barley (Hordeum vulgare), maize (Zea mays), potato (Solanum tuberosum), and tomato (Solanum lycopersicum). Plant rhabdoviruses are taxonomically divided into six genera: Cytorhabdovirus, Dichorhavirus, Varicosavirus, Alphanucleorhabdovirus, Betanucleorhabdovirus, and Gammanucleorhabdovirus (Dietzgen et al., 2020). Previous work on plant rhabdoviruses has largely focused on biochemical and molecular analyses of viral proteins during ectopic expression in plant cells, but not in the context of authentic virus infections due to the lack of available reverse genetic systems. Recently, Wang and colleagues developed the first reverse genetics system of plant negative-stranded RNA viruses in Nicotiana benthamiana plants, which facilitates investigation of the biological functions of viral proteins (Jackson et al., 2005; Wang et al., 2015; Jackson and Li, 2016).
A typical rhabdovirus genome encodes five conserved structural proteins: the nucleocapsid protein (N), a phosphoprotein (P), a matrix protein (M), a glycoprotein (G), and the large subunit (L) of the RNA-dependent RNA polymerase complex (RdRp). These genes are organized in the conserved order 3′−N−P−M−G−L−5′ (Jackson et al., 2005; Mann and Dietzgen, 2014). In addition, plant rhabdoviruses encode one or more ancillary proteins in the N−P, P−M, or G−L gene junctions (Walker et al., 2011). The rhabdovirus nucleocapsid is a minimal infectious unit that comprises the genomic RNA (gRNA) associated with the N, P, and L core proteins. During replication, the nascent rhabdovirus gRNAs and antigenomic RNAs (agRNAs) are rapidly encapsidated by the N proteins to form a long helical N–gRNA complex that serves as a template for RdRp-mediated transcription and genome replication (Moyer et al., 1991). The rhabdovirus P protein provides a noncatalytic cofactor that facilitates access of the L protein to N–RNA template. In addition, the P protein forms N0-P complexes with nascent RNA-free N (N0) proteins to solubilize N0 and to encapsulate gRNAs and agRNAs generated during replication (Emerson and Schubert, 1987; Moyer et al., 1991; Green and Luo, 2009; Rahmeh et al., 2012). Rhabdovirus transcription and replication are regulated in a dynamic balance by the polymerase complex, and an intriguing open question is how the viral RdRp fine-tunes the balance between transcription and replication.
Barley yellow striate mosaic virus (BYSMV) is a member of the genus Cytorhabdovirus and elicits chlorotic striations and mosaic symptoms in cereal plant species. BYSMV is transmitted in a persistent-propagative manner by the small brown planthopper (SBPH; Laodelphax striatellus; Cao et al., 2018). The BYSMV genome consists of 12,706 nucleotides and encodes five conserved structural proteins and five ancillary proteins in the order 3′−N−P−P3−P4/P5−P6−M−G−P9−L−5′ (Yan et al., 2015). We recently developed a BYSMV antigenomic minireplicon (BYSMV-agMR) system in N. benthamiana leaves (Fang et al., 2019). We subsequently rescued BYSMV (i.e., reconstituted a functional and infectious genome) from full-length cDNA clones and developed BYSMV vectors as versatile delivery platforms in monocot plants and planthoppers (Gao et al., 2019). Using these genetic systems, we recently demonstrated that the BYSMV P protein hijacks the host deadenylation factor CARBON CATABOLITE REPRESSION4 (CCR4) to promote viral replication (Zhang et al., 2020). Here, we explored the regulation of BYSMV P phosphorylation in viral transcription and replication.
Accumulating evidence suggests that many host protein kinases play pivotal roles in viral life cycles by phosphorylating both viral and host cellular proteins (Jakubiec and Jupin, 2007; Hu et al., 2015; Wang, 2015; Zhao et al., 2015; Hoover and Kao, 2016; Zhong et al., 2017; Mei et al., 2018). However, the role of phosphorylation during plant negative-stranded RNA virus infections has not been described. Here, we first undertook mass spectrometric analyses of BYSMV P proteins to identify physiologically relevant phosphorylation sites. Intriguingly, we identified a highly phosphorylated serine-rich (SR) motif at the C-terminal domain intrinsically disordered region (IDRCTD) of BYSMV P. We then used reverse genetic approaches to investigate the effects of these phosphorylation events on BYSMV infections. We present several lines of evidence showing that the SR motif of BYSMV P is phosphorylated by the conserved CK1 family of plant and insect hosts to finely regulate the balance of BYSMV transcription and replication.
RESULTS
Identification of BYSMV P Phosphorylated Residues by Mass Spectrometry
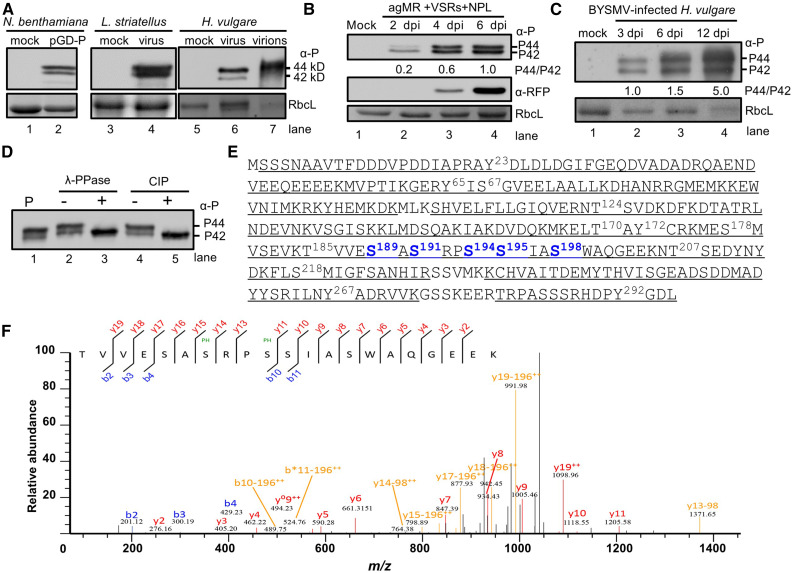
To investigate the phosphorylated forms of BYSMV P in vivo, we performed immunoblot analyses to detect BYSMV P proteins isolated from N. benthamiana leaves infiltrated with the BYSMV agMR (Fang et al., 2019) via Agrobacterium (Agrobacterium tumefasciens), from BYSMV-infected SBPHs, and from barley plants infiltrated with BYSMV agMR, using purified BYSMV virions from infected barley plants as a positive control. We detected two main bands for BYSMV P in SDS-PAGE gels with slightly different mobilities, corresponding to 42 kD (designated P42) and 44 kD (designated P44; Figure 1A). Notably, P44 was the principal form in BYSMV virions (Figure 1A, lane 7). In BYSMV-agMR assays, BYSMV P mainly existed as the P42 form 2 d postinfiltration (2 dpi; Figure 1B). Subsequently, the appearance of P44 was accompanied by red fluorescent protein (RFP) accumulation and agMR replication at 4 and 6 dpi (Figure 1B). The RFP coding sequence was inserted in between the virus N and P genes in the infectious clone and provides a positive control for transcription of the viral genome. The P44:P42 ratio gradually increased during BYSMV infections in barley plants (Figure 1C). Collectively, these results suggest that P44 production is involved in BYSMV infections.
Figure 1.
Identification of Phosphorylated Residues of BYSMV P by Mass Spectrometry.
(A) Immunoblot analysis showing two forms of the P protein expressed in N. benthamiana, L. striatellus, and H. vulgare, and purified virions, using specific antibodies against the P protein. Note the presence of two BYSMV P protein bands (P42 and P44) with molecular masses of 42 and 44 kD in gels.
(B) Accumulation of BYSMV P and RFP in agMR-inoculated N. benthamiana leaves at 2, 4, and 6 dpi. Total protein extracts from agMR-inoculated N. benthamiana leaves were probed with anti-P and anti-RFP antibodies. The P44:P42 ratios were calculated from band intensities.
(C) Accumulation of BYSMV P in BYSMV-infected H. vulgare plants at 3, 6, and 12 dpi. Coomassie Brilliant blue (CBB) staining of Rubisco complex large subunit (RbcL) was used as protein loading controls in (A) to (C).
(D) Treatment of BYSMV P proteins with phosphatase. P-Flag proteins were isolated from N. benthamiana leaves infiltrated with the plasmid pMDC32-P by immunoprecipitation with anti-Flag beads. P-Flag proteins were treated with lambda protein phosphatase (λ-PPase) or CIP. Mock treatment (–) was incubated with buffer without phosphatases.
(E) Mass spectrometric identification of BYSMV protein phosphorylation sites. Underlined peptides (bold) were identified by mass spectrometry. The bold and numbered residues represent phosphorylation sites. Highly phosphorylated Ser residues of the BYSMV P protein SR motif are shown in blue.
(F) MS/MS spectrum of the peptide 185TVVESASRPSSIASWAQGEEK205 containing the Ser-191 and Ser-194 phosphorylated residues. The b and y ions are highlighted in the sequence and corresponding labels in the spectrum.
To determine whether the two BYSMV P forms resulted from phosphorylation, we immunoprecipitated BYSMV P proteins from N. benthamiana leaves overexpressing P and treated the immunoprecipitates with either λ-protein phosphatase or calf intestinal phosphatase (CIP). As expected, we failed to detect P44 after treatment with either phosphatase in vitro (Figure 1D), indicating that P42 and P44 represent hypo-phosphorylated and hyper-phosphorylated forms, respectively. In agreement with previous studies about the accumulation of animal rhabdovirus P proteins as two forms (Barik and Banerjee, 1992; Gupta et al., 2000), our results indicate that phosphorylation of BYSMV P similarly affects the mobility of P and phosphorylated P in SDS-PAGE gels.
To further identify phosphorylated residues in BYSMV P during replication and transcription, we immunoprecipitated the BYSMV P protein from BYSMV-agMR–infiltrated leaves and then subjected the protein samples to SDS-PAGE gel electrophoresis. We excised the P42 and P44 bands and treated them with trypsin for in-gel digestion. We analyzed the resulting products by liquid chromatography–tandem mass spectrometry (LC-MS/MS). The results showed that 95% of the BYSMV P protein was covered in the LC-MS/MS results from the P42 and P44 bands (Figure 1E), indicating that P42 is a full-length product rather than a truncated derivative. Notably, both P42 and P44 were characterized by different degrees of phosphorylation on a series of Ser, Thr, and Tyr residues (Figure 1E; Supplemental Table). Our LC-MS/MS results showed 17 phosphorylated Ser, Thr, and Tyr residues in 15 peptide fragments (Figure 1E; Supplemental Table). Notably, five Ser residues (amino acids 189, 191, 194, 195, and 198) in an SR motif (189SASRPSSIAS198) were highly phosphorylated in different peptides (Figures 1E and 1F; Supplemental Figure 1).
Collectively, the BYSMV P protein in plants and insect vectors exists as hyper-phosphorylation (P44) and hypo-phosphorylation (P42) forms in vivo. Moreover, the C terminus of BYSMV P contains a highly phosphorylated SR motif (Ser residues 189, 191, 194, 195, and 198).
Phosphorylation of the BYSMV P SR Motif Elicits P44 Production
IDRs have been described in numerous proteins required for various biological functions (Uversky et al., 2005). IDRs usually serve as flexible scaffolds for multi-component complex assemblies and exhibit different binding affinities to their partners (Spolar and Record, 1994; Wright and Dyson, 2009). Based on meta-prediction of IDRs from 15 independent predictors, we inferred that BYSMV P contained three structured domains, including an N-terminal domain (amino acids 9 to 33), a central domain (amino acids 60 to 183, and a C-terminal domain (amino acids 208 to 270), separated by two IDR regions, IDRN (amino acids 34 to 59) and IDRC (amino acids 184 to 207; Figure 2A). Interestingly, the highly phosphorylated SR motif (189SASRPSSIAS198) was located in the IDRC region (amino acids 184 to 207; Figure 2A).
Figure 2.
Effects of P Phosphorylation on Minigenome Transcription and Replication.
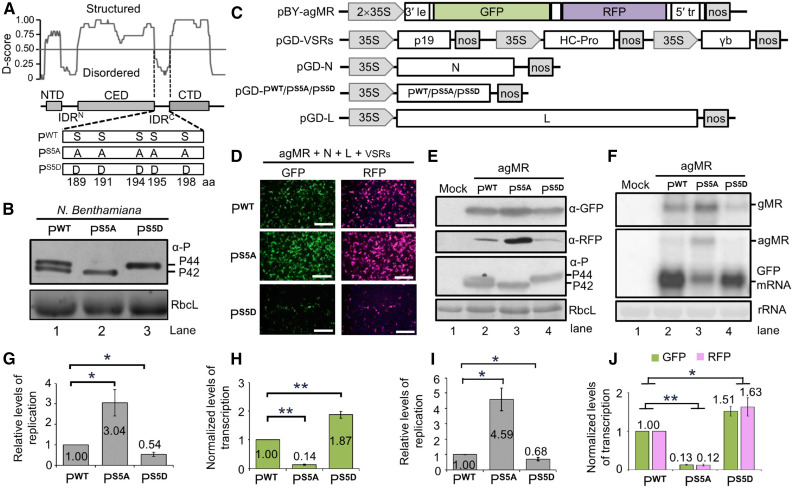
(A) Modular organization of the BYSMV P protein and schematic representation of its phosphorylation mutants. Normalized scores for disorder (D-score) were calculated from 15 different predictors as described in Methods. Structured regions and disordered regions are indicated by gray boxes and lines, respectively. Five Ser residues (amino acids 189, 191, 194, 195, and 198) in the SR motif of PWT were mutated to either Ala (PS5A) or to Asp (PS5D).
(B) Immunoblot analyses of PWT, PS5A, and PS5D levels in infiltrated leaves.
(C) Illustration of the binary vectors used for generating antigenomic-sense minireplicon RNA (pBY-agMR) and pGD constructs for expression of the BYSMV N, P/PS5A/PS5D, and L proteins in Agrobacterium-infiltrated N. benthamiana leaves. The pGD-VSRs plasmid contains three expression cassettes for simultaneous expression of the viral suppressors of RNA silencing TBSV p19, TEV HC-Pro, and BSMV γb.
(D) GFP and RFP foci in N. benthamiana leaves at 5 dpi with Agrobacterium cultures containing pBY-agMR, pGD-N, pGD-L, pGD-VSRs, and pGD-P/PS5A/PS5D. GFP and RFP was photographed with a fluorescence microscope. Bar = 1 mm.
(E) Immunoblot analysis showing GFP, RFP, and P expression in Agrobacterium-infiltrated leaves of (D) with anti-GFP (α-GFP), anti-RFP (α-RFP), and anti-P (α-P) polyclonal antibodies. Mock buffer-infiltrated leaves were used as negative controls.
(F) RNA gel blot analysis of MR replication and transcription supported by PWT, PS5A, or PS5D proteins in Agrobacterium-infiltrated leaves. rRNAs were used as loading controls.
(G) Relative levels of minigenome RNA replication calculated from signal intensities of gMR bands of (F).
(H) Normalized levels of GFP mRNA transcription supported by the agMR plasmid in combination with the PWT, PS5A, or PS5D proteins. Note: Normalized levels of transcription refer to the ratio between GFP mRNA accumulation and gMR accumulation.
(I) Quantification by qPCR of the relative levels of minigenome replication supported by the PWT, PS5A, or PS5D proteins.
(J) GFP and RFP mRNA in the same agMR samples of (I) were analyzed by qPCR.
Note: All the results in (G) to (J) were obtained from three biologically independent experiments. The values in PWT samples were set to 1. The EF-1A gene served as internal control in (I) and (J). Error bars indicate sd. Data points above columns are mean values of three independent experiments. Statistically significant differences were determined by Student’s t test. *P-value < 0.05; **P- < 0.01.
To examine whether phosphorylation of the BYSMV P SR motif affected conformational changes, we generated two BYSMV P mutants in which the five Ser residues at positions 189, 191, 194, 195, and 198 were substituted with Ala (designated as PS5A) to mimic the nonphosphorylated state or with Asp (designated as PS5D) to mimic the phosphorylated state (Figure 2A). We then expressed the wide-type P protein (PWT), PS5A, and PS5D in N. benthamiana leaves by agro-infiltration. As described above, PWT presented as both basally phosphorylated (P42) and hyper-phosphorylated (P44) forms (Figure 2B). By contrast, PS5A and PS5D migrated on SDS-PAGE gels only as P42 and P44, respectively (Figure 2B). These results indicate that BYSMV P phosphorylation (PS5D) and nonphosphorylation (PS5A) in the SR motif result in conformational changes that affect electrophoretic mobility of BYSMV P.
Effects of BYSMV P Phosphorylation on Minigenome Transcription and Replication
We recently developed a BYSMV-agMR system using a coinfiltration strategy in N. benthamiana leaves with Agrobacterium-harboring plasmids driving the accumulation of core proteins and a BYSMV anti-minigenome derivative cDNA (Fang et al., 2019). In the pBYSMV-agMR plasmid, we substituted the BYSMV N and P open reading frames (ORFs) from the antigenome strand with those of green fluorescent protein (GFP) and RFP flanked by the BYSMV 5′ and 3′ terminal sequences (Figure 2C). In addition, the infiltration system consisted of the pGD vector carrying the N, P, or L ORF from BYSMV (pGD-N, pGD-P, and pGD-L, respectively) and a pGD-VSRs plasmid that encodes the viral suppressors of RNA silencing p19 from tomato bushy stunt virus, γb from barley stripe mosaic virus, and P1/HC-Pro from tobacco etch virus (Figure 2C).
We next determined the effects of P mutants on BYSMV minigenome replication and transcription in vivo. We infiltrated Agrobacterium cultures containing plasmids for expression of PWT, PS5A, or PS5D in N. benthamiana leaves along with mixtures of bacterial cultures harboring the pGD-N, pGD-L, pGD-VSRs, and agMR plasmids (Figure 2C). Compared to PWT, the phosphorylation-deficient PS5A mutant resulted in more GFP and RFP fluorescent foci, whereas the phospho-mimic variant PS5D had the opposite effect (Figure 2D). In agreement, immunoblot analyses showed that PS5A supported enhanced accumulation of GFP and RFP, whereas PS5D caused a reduced accumulation of GFP and RFP, compared to PWT (Figure 2E). As described above (Figure 2B), PWT existed as the two forms P42 and P44, whereas PS5A and PS5D migrated with P42 and P44, respectively (Figure 2E).
We also performed a RNA gel blot analysis to measure BYSMV minigenome replication and associated transcription products. The RNAs corresponding to the genome- (gMR) and antigenomic-stranded (agMR) minigenome were the most abundant in PS5A-infiltrated leaves, while they accumulated to intermediate levels in PWT-infiltrated leaves and were the lowest in PS5D-infiltrated leaves (Figure 2F). These results indicate that the replication of the minigenome is upregulated by PS5A and downregulated by PS5D. Quantification of band intensities from three independent experiments showed that accumulation of gMR in PS5A samples increased threefold, whereas gMR in PS5D only accumulated to 54% of PWT levels (Figure 2G).
Next, we compared the transcriptional activities of PWT, PS5A, and PS5D by detecting GFP mRNA. Since replicating minigenomes are templates for viral transcription, a rise in viral replication should positively affect viral transcription. We had therefore expected to see higher transcription in the PS5A samples than in PWT or PS5D. However, PS5A supported much lower GFP transcription than either PS5D or PWT (Figure 2F, GFP mRNA). To compare the transcriptional activities of these mutants, we normalized GFP transcript levels relative to the gMR template. Our results showed that PS5A decreased transcriptional activity to only 14% of that observed with PWT, whereas viral transcription with PS5D increased by 87% over that seen with PWT (Figure 2H).
To further verify the RNA gel blot results, we performed a real-time quantitative RT-PCR (RT-qPCR) analysis to quantify the levels of the minigenome RNA and GFP/RFP transcripts. Minigenome RNA levels were significantly upregulated in the PS5A samples but were reduced in the PS5D samples compared to the PWT samples (Figure 2I). GFP and RFP transcript levels were significantly downregulated in PS5A samples but were upregulated in PS5D samples (Figure 2J), providing further support to our RNA gel blot results (Figures 2E to 2H).
We next mutated each Ser residue within the SR motif individually to Ala to determine the relative contribution of each Ser residue during viral RNA synthesis. The Ser-191 (PS191A), Ser-195 (PS195A), and Ser-198 (PS198A) single mutants resulted in similar numbers of fluorescent foci as PS5A (Supplemental Figure 2A). By contrast, the PS189A and PS194A mutants had fewer fluorescent foci (Supplemental Figure 2A). Immunoblot analysis confirmed that the P variants PS191A, PS195A, PS198A, and PS5A all supported higher RFP accumulation than that seen with PS189A or PS194A (Supplemental Figure 2B, top). In addition, PS191A, PS195A, PS198A, and PS5A coelectrophoresed with the P42 form, while PS189A and PS194A existed as both P42 and P44 forms (Supplemental Figure 2B, middle). These results demonstrate that the PS191A, PS195A, or PS198A single mutants each affect P protein conformation and functionally similar to PS5A.
Taken together, our results clearly demonstrate that the nonphosphorylated form (PS5A) of BYSMV P enhances virus replication, whereas the phosphorylated form (PS5D) facilitates virus transcription. These results thus provide evidence that phosphorylation of the SR motif of BYSMV P modulates the balance between RNA replication and transcription.
Effects of BYSMV P Phosphorylation on Full-Length Virus Infection in N. benthamiana Leaves
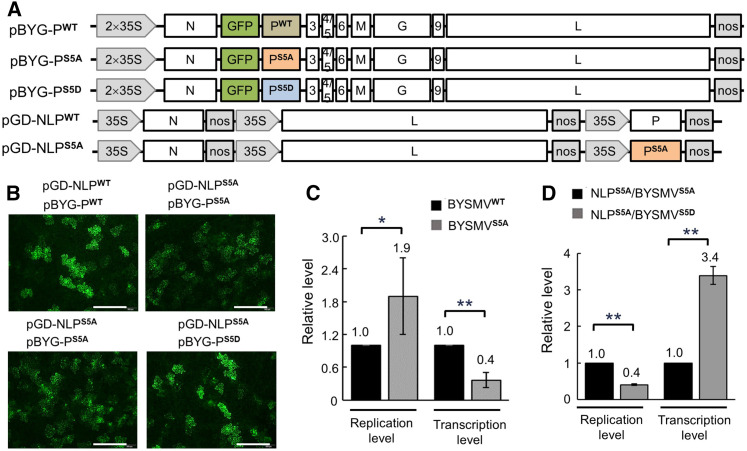
We have previously demonstrated the rescue of infectious BYSMV from cloned full-length cDNAs in SBPHs and cereal plants (Gao et al., 2019). To examine the effects of BYSMV P phosphorylation on virus infections, we generated a pGD-NLPS5A mutant, based on the pGD-NLPWT plasmid, that contains expression cassettes for the BYSMV N, PS5A, and L core proteins (Figure 3A). In addition, we inserted point mutations into BYSMV-GFP (BYG) carried by the pBYG-PWT plasmid to create pBYG-PS5A and pBYG-PS5D mutant versions of the full-length antigenome BYSMV cDNA containing a GFP gene insertion, as described previously by Gao et al. (2019; Figure 3A).
Figure 3.
Effects of P Protein Phosphorylation on Full-Length BYSMV Transcription and Replication in N. benthamiana Leaves.
(A) Illustration of BYSMV genome organization and the binary Agrobacterium vectors harboring pBYG-PWT, pBYG-PS5A, pBYG-PS5D, pGD-NLPWT, and pGD-NLPS5A.
(B) GFP foci in N. benthamiana leaves at 10 dpi with Agrobacterium cultures containing the indicated plasmids. GFP fluorescence was observed and photographed with a fluorescence microscope. Bar = 0.5 mm.
(C) Relative replication and transcription levels of recombinant BYSMVWT and BYSMVS5A as quantified by RT-qPCR. BYSMVWT was rescued by coinfiltration of pGD-NLPWT and pBYG-PWT, while BYSMVS5A was rescued by coinfiltration of pGD-NLPS5A and pBYG-PS5A. The values of viral replication and transcription supported by BYSMVWT were set to 1.
(D) Relative replication and transcription levels of recombinant BYSMVS5A and BYSMVS5D as quantified by RT-qPCR. BYSMVS5A and BYSMVS5D were rescued by coinfiltration of pBYG-PS5A and pBYG-PS5D under the support of pGD-NLPS5A. The values of viral replication and transcription supported by BYSMVS5A were set to 1.
In (C) and (D), error bars indicate sd. Data points above the columns are the mean values of three independent experiments. Statistically significant differences were determined by Student’s t test. *P-value < 0.05; **P-value < 0.01.
We achieved BYG-PS5A infection by infiltration with Agrobacterium cultures harboring the pGD-VSRs, pGD-NLPS5A, and pBYG-PS5A binary vectors into N. benthamiana leaves. Similarly, we coexpressed the pGD-VSRs, pGD-NLPWT, and pBYG-PWT plasmids to rescue the wild-type BYG-PWT. Compared to BYG-PWT, the BYG-PS5A mutant resulted in more fluorescent foci in infiltrated leaves, although their fluorescence intensity was much weaker (Figure 3B, top). RT-qPCR analysis showed that accumulation of the BYG-PS5A gRNA was 1.9-fold higher than that of BYG-PWT, whereas the BYG-PS5A transcription levels were ∼60% lower than those seen for BYG-PWT (Figure 3C). These results indicate that PS5A facilitates full-length virus replication but inhibits virus transcription.
We further compared the replication and transcription levels of BYG-PS5A and BYG-PS5D when coexpressed with NLPS5A. BYG-PS5A infection resulted in increased numbers of foci but lower fluorescence intensity for GFP compared to BYG-PS5D infection (Figure 3B, bottom). In addition, BYG-PS5D infections led to an ∼60% reduction in replication but a twofold increase in transcription relative to BYG-PS5A (Figure 3D). Immunoblot analyses revealed that NLPS5A/BYG-PS5A only produced the P42 form, whereas the NLPWT/BYG-PWT and NLPS5A/BYG-PS5D combinations resulted in the production of both P42 and P44 forms (Supplemental Figure 3). The detection of the BYSMV M protein accumulation by immunoblot indicates that all combinations supported virus infection and were able to rescue recombinant BYSMV-GFP in N. benthamiana leaves (Supplemental Figure 3).
Collectively, these results indicate that the PS5A mutant facilitates replication, while the PS5D mutant assists in transcription during infections with full-length BYSMV virus genome, which is consistent with the agMR assays (Figure 2).
Effects of BYSMV P Phosphorylation on BYSMV Transmission by SBPHs
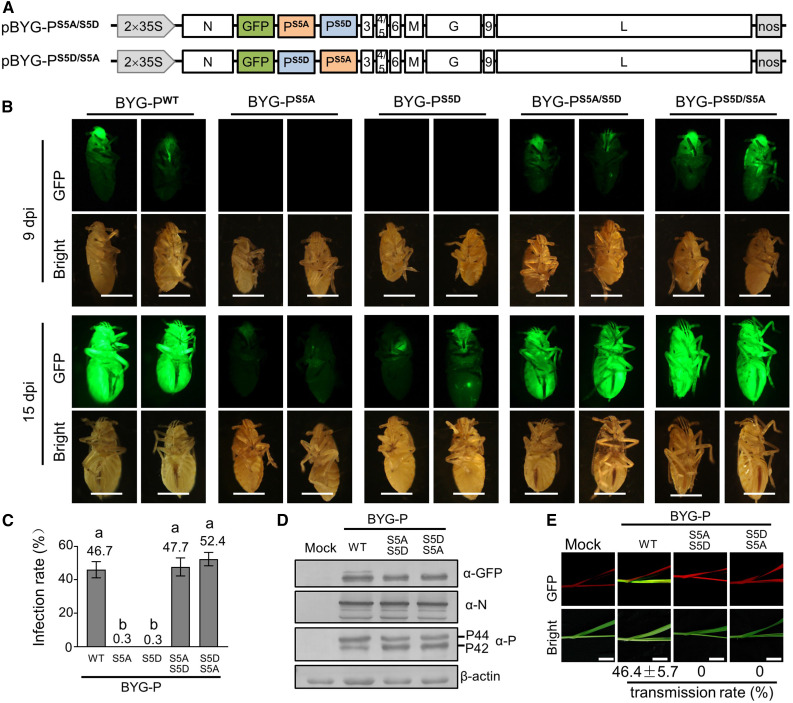
BYSMV is transmitted to cereal plants by SBPHs in a persistent-propagative manner (Cao et al., 2018). Our recent studies have demonstrated the successful infection of SBPHs by injection with crude extracts of N. benthamiana leaves containing recombinant BYSMV-GFP (Gao et al., 2019). Since PWT can produce both P42 and P44 in vivo, we generated pBYG-PS5A/S5D and pBYG-PS5D/S5A constructs to coexpress PS5A and PS5D in full-length virus infections (Figure 4A).
Figure 4.
Effects of P Phosphorylation on Full-Length Virus Transcription and Replication in Planthopper Vectors.
(A) Illustration of the genome organization of the pBYG-PS5A/S5D and pBYG-PS5D/S5A binary vectors harbored by the Agrobacterium cultures.
(B) GFP fluorescence of SBPHs microinjected with crude extracts of N. benthamiana leaves infected by BYG-PWT, BYG-PS5A, BYG-PS5D, BYG-PS5A/S5D, and BYG-PS5D/S5A at 9 and 15 dpi. Bar = 1 mm.
(C) Infection rates (n ≥ 30) of BYG-PWT, BYG-PS5A, BYG-PS5D, BYG-PS5A/S5D, and BYG-PS5D/S5A in SBPHs. Data are based on three independent experiments. Error bars indicate sd. Data points above the columns represent mean values of three independent experiments. Different letters indicate significant differences (ANOVA, P < 0.001).
(D) Immunoblot analysis of GFP, N, and P accumulation in SBPHs infected by BYG-PWT, BYG-PS5A/S5D, and BYG-PS5D/S5A with anti-GFP (α-GFP), anti-N (α-N), and anti-P (α-P) polyclonal antibodies. The SBPH β-actin protein served as a loading control.
(E) GFP fluorescence in systemically infected barley leaves at 15 d after feeding with SBPHs infected by BYG-PWT, BYG-PS5A/S5D, or BYG-PS5D/S5A. Photographs were taken with a digital camera under a hand-held UV lamp. The values under the photos represent the mean infection rates of three independent experiments. Error bars indicate sd. Bar = 1 cm.
To determine the effects of P phosphorylation on SBPH infection and transmission, we injected SBPHs with crude extracts of the N. benthamiana leaves infiltrated with BYG-PWT, BYG-PS5A, BYG-PS5D, BYG-PS5A/S5D, or BYG-PS5D/S5A (Supplemental Figure 4A). At 9 dpi, we observed faint GFP fluorescence from SBPHs infected with BYG-PWT, BYG-PS5A/S5D, or BYG-PS5D/S5A leaf extracts, but not in SBPHs injected with BYG-PS5A or BYG-PS5D extracts (Figure 4B). By 15 dpi, more than 45% of the SBPHs infected with BYG-PWT, BYG-PS5A/S5D, or BYG-PS5D/S5A exhibited intense GFP fluorescence. By contrast, only 0.30% of all SBPHs injected with BYG-PS5A or BYG-PS5D extracts exhibited very faint GFP fluorescence (Figures 4B and 4C). Immunoblot analyses revealed that BYG-PWT, BYG-PS5A/S5D, and BYG-PS5D/S5A infections resulted in similar accumulation of GFP, N, and P proteins (Figure 4D). These results demonstrate that both PS5A and PS5D are required for efficient BYSMV infections in SBPHs.
Next, we transferred SBPHs that had been infected with BYG-PWT, BYG-PS5A/S5D, and BYG-PS5D/S5A to healthy barley plants (five insects per plant) for a 2-d inoculation access period (IAP). Approximately 15 d after the inoculation access period, we detected GFP fluorescence in ∼46% of newly emerging leaves of BYG-PWT–infected barley plants (Figure 4E). However, none of the barley plants exhibited GFP fluorescence after being fed to BYG-PS5A/S5D– or BYG-PS5D/S5A–infected SBPHs (Figure 4E), indicating that neither BYG-PS5A/S5D nor BYG-PS5D/S5A in SBPHs were transmitted to barley plants by SBPHs.
Collectively, these results suggest that both PS5A and PS5D are essential for the dissemination of BYSMV in SBPHs. Nonetheless, coexpression of PS5A and PS5D in SBPHs is not sufficient for BYSMV transmission to barley plants. Therefore, the transmission of BYG-PWT by SBPHs may require spatial- and temporal-specific expression of P42 and P44, rather than coexpression of P42 and P44.
BYSMV P Phosphorylation Mutants Have Different Association for the N–RNA Complex and L Protein
In rhabdovirus replication and transcription complexes, the P protein is involved in increasing (1) the solubility of the N protein and (2) the attachment of the L protein subunit to the N–RNA template. Because the BYSMV P SR motif is located in a highly disordered region, we hypothesized that the phosphorylation state within the SR motif might affect the binding of P protein to the N–RNA template and/or the L protein. To test this hypothesis, we performed biomolecular fluorescence complementation (BiFC) assays and coimmunoprecipitation (Co-IP) assays to investigate the binding of P mutants with N–RNA templates and the L protein.
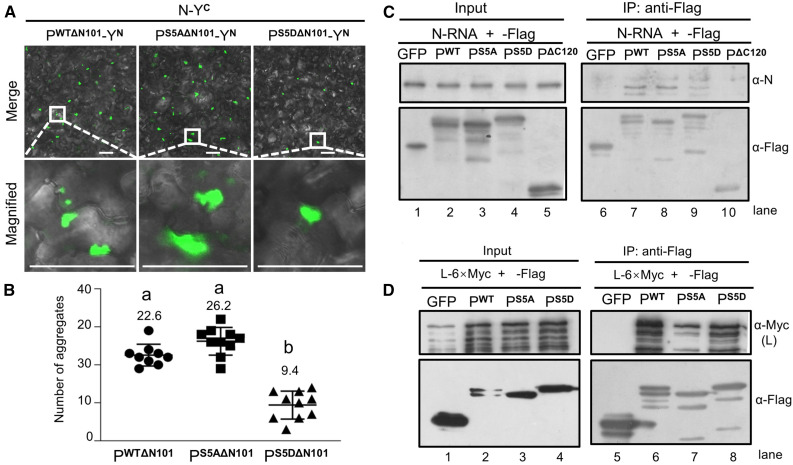
BiFC and Co-IP assays demonstrated that full length PWT, PS5A, and PS5D exhibited similar interaction with the BYSMV N0 protein (Supplemental Figures 5A and 5B). In addition, the point mutations did not affect the self-interactions of full-length PWT, PS5A, and PS5D (Supplemental Figures 5C and 5D). The N and C termini of rhabdovirus P proteins affect how P binds to RNA-free N0 and to N–RNA templates (Leyrat et al., 2011). Thus, to prevent the binding of the P protein with N0, we deleted the N-terminal 101 amino acids of PWT, PS5A, and PS5D, while the remaining C termini were tagged with the N-terminal half (YN) of yellow fluorescent protein (YFP). We tagged the BYSMV N protein with the C-terminal half (YC) of YFP. In addition, as a negative control, we fused ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco) with YC and YN individually. We observed no BiFC fluorescence in Rubisco control samples that were coexpressed with PWTΔN101-YN, PS5AΔN101-YN, PS5DΔN101-YN, or N-YC (Supplemental Figure 6). However, after coexpression of N-YC with PWTΔN101-YN, PS5AΔN101-YN, or PS5DΔN101-YN, we detected many punctate granules in the cytoplasm (Figure 5A). Leaves coinfiltrated with PS5DΔN101-YN and N-YC had far fewer granules than in leaves coinfiltrated with N-YC and either PWTΔN101-YN or PS5AΔN101-YN (Figures 5A and 5B), suggesting that PS5D has a lower binding affinity to the N–RNA template than PWT and PS5A.
Figure 5.
Association of BYSMV PS5A and PS5D With N–RNA Templates and the L Protein.
(A) BiFC detection of interactions between PWT/PS5A/PS5D and the N–RNA complexes. N. benthamiana leaves were photographed at 3 dpi, after infiltration with Agrobacterium cultures harboring plasmids encoding N-YC with PWTΔN101-YN, PS5AΔN101-YN, or PS5DΔN101-YN. YN and YC represent the N-terminal and C-terminal halves of YFP, respectively. Bar = 50 μm.
(B) Quantification of the number of fluorescent aggregates in (A). Each data point represents the number of fluorescent aggregates in a single photograph. Error bars indicate sd. Letters indicate significant differences (ANOVA, P < 0.001).
(C) Co-IP assays showing in vivo interactions between the PWT/PS5A/PS5D proteins and purified N–RNA complexes. Flag-tagged PWT/PS5A/PS5D proteins were expressed in and precipitated from N. benthamiana leaves with Flag M2 beads at 3 dpi. The N–RNA complexes were isolated from purified virions. The N (α-N) and Flag (α-Flag) antibodies were used to detect accumulation of N–RNA or P, respectively. GFP-Flag or PΔC120-Flag served as negative controls.
(D) Co-IP assays showing PWT/PS5A/PS5D protein interacting with L. Flag-tagged PWT/PS5A/PS5D and L-6×Myc were coexpressed in N. benthamiana leaves and Co-IPed with anti-Flag beads at 3 dpi. Protein levels was detected by immunoblot using α-Flag and α-Myc antibodies. GFP-Flag and L-Myc were coexpressed as negative controls.
To further examine the association of P mutants with N–RNA in vivo, we performed Co-IP assays using N–RNA complexes and Agrobacterium-infiltrated leaves expressing Flag-tagged PWT, PS5A, or PS5D and anti-Flag beads. A mutant bearing a deletion of the C-terminal 120 residues (PΔC120) that does not contain the N–RNA complex binding domain served as a negative control. We isolated the BYSMV N–RNA complexes from detergent-treated purified virus using a standard protocol of high-salt incubation and double isolation on a cesium chloride (CsCl) gradient (Emerson and Yu, 1975). Immunoblot and RT-PCR analyses showed that BYSMV N protein and gRNA were present in the purified N–RNA complexes within the nucleocapsid complexes (Supplemental Figure 7). Flag-PWT and Flag-PS5A efficiently immunoprecipitated BYSMV N proteins from the N–RNA complexes (Figure 5C, lanes 7 and 8). By contrast, Flag-PS5D immunoprecipitated much less N protein from the purified N–RNA complexes (Figure 5C, lane 9). Flag-GFP and Flag-PΔC120 were unable to immunoprecipitate N from the purified N–RNA complexes (Figure 5C, lane 10). Thus, these results show that the PS5D mutant exhibits compromised association with the N–RNA complex.
We next examined the binding between the P protein mutants and the L protein in Co-IP assays with Agrobacterium-infiltrated N. benthamiana leaves coexpressing L-6×Myc and Flag-tagged GFP, PWT, PS5A, or PS5D. Both the Flag-PWT and Flag-PS5D proteins, but not the Flag-GFP control, efficiently immunoprecipitated L-6×Myc (Figure 5D, lanes 6 and 8). By contrast, the Flag-PS5A protein immunoprecipitated L-6×Myc with a reduced efficiency despite equal protein amounts in the starting samples (Figure 5D, lanes 3 and 7).
Collectively, we conclude that the PS5A variant mimicking the nonphosphorylated state of P associates strongly with N–RNA templates and weakly with the L protein. Conversely, the phosphorylation mimic PS5D mutant is compromised in the N–RNA template interaction. Therefore, the nonphosphorylated and phosphorylated states of the SR motif drive P protein conformational changes, thereby forming different complexes to initiate either transcription or replication.
The SR Motif of BYSMV P Is Phosphorylated by Plant and Insect CK1 Proteins
Phosphorylation is a universal posttranslational modification in all eukaryotes, including yeasts, animals, and plants. Therefore, we expressed the BYSMV P protein in yeast (Saccharomyces cerevisiae, strain BY4741) to determine whether BYSMV P could be phosphorylated by yeast protein kinases. Indeed, yeast-expressed BYSMV P also electrophoresed as P42 and P44, which match the migration pattern of PS5A and PS5D, respectively (Supplemental Figure 8). These results indicate that one or more conserved protein kinase(s) might be responsible for phosphorylating BYSMV P in host both plants and insect vectors. Therefore, we explored conserved protein kinases and constructed a phylogenetic tree using multiple sequence alignment tools (Supplemental Figure 9). In the yeast proteome, we identified 47 protein kinases that were highly conserved in barley plants and planthoppers (Supplemental Figure 9, labeled in red). We cloned 16 protein kinase genes from yeast to test their kinase activity against the BYSMV P protein.
Since eukaryotic-type protein kinases are absent in Escherichia coli, we modified a bacterial coexpression system to screen kinase candidates easily through the coexpression of individual protein kinases and their candidate target in E. coli (Heger-Stevic et al., 2018). The BYSMV P protein expressed alone in E. coli only produced the P42 form, indicating that protein kinases targeting BYSMV P are absent in E. coli (Supplemental Figure 10A, lane 1). We then coexpressed BYSMV P with our 16 yeast protein kinase candidates in E. coli. Immunoblot analyses revealed that BYSMV P migrated as both the P42 and P44 forms only when coexpressed with the yeast kinase HO and Radiation Repair25 (HRR25; Supplemental Figure 10A, lane 13). By contrast, coexpression of the glutathione S-transferase (GST) fusion protein GST-HRR25 did not change the mobility of PS5A, indicating that the Ser residues of the SR motif were phosphorylated by HRR25 (Supplemental Figure 10B). Furthermore, the GST-HRR25–generated P44 form disappeared after treatment with CIP, confirming that P44 was the hyper-phosphorylated form (Supplemental Figure 10C).
The yeast kinase HRR25 belongs to the CK1 family. BLAST searches showed that the N. benthamiana, SBPHs, and barley genomes encode 14, 5, and 14 yeast CK1 orthologs, respectively (Supplemental Figure 11). We next evaluated the effects of representative probable CK1 orthologs from N. benthamiana (NbCK1), barley (HvCK1), and planthoppers (LsCK1) on the phosphorylation of BYSMV P in E. coli. Coexpression of NbCK1.3, NbCK1.4, NbCK1.7, and NbCK1.13 resulted in the conversion of P42 to P44 (Supplemental Figure 12), but these kinases had no effect on PS5A mobility, as expected (Supplemental Figure 12). Using a similar strategy, we established that LsCK1.3, LsCK1.4, HvCK1.2, HvCK1.3, and HvCK1.12 phosphorylated PWT but had no effect on PS5A in E. coli (Supplemental Figure 12). Therefore, these results suggest that CK1 orthologs in plants and insect hosts contribute to phosphorylation of the BYSMV P SR motif.
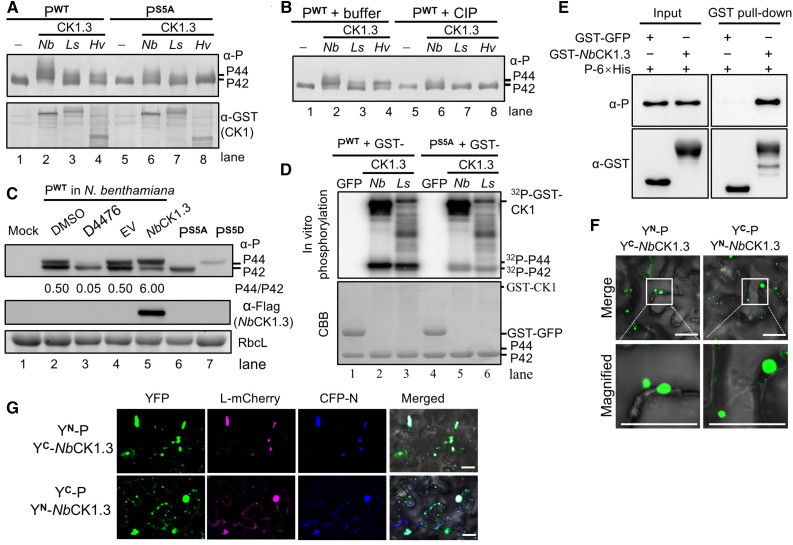
We selected NbCK1.3, LsCK1.3, and HvCK1.3 for further analysis. NbCK1.3, LsCK1.3, and HvCK1.3 resulted in the accumulation of the hyper-phosphorylated P44 form of P when coexpressed with BYSMV PWT (Figure 6A, lanes 2 to 4), but had no effect on the mobility of BYSMV PS5A (Figure 6A, lanes 6 to 8). Moreover, the P44 derivatives produced by NbCK1.3, LsCK1.3, and HvCK1.3 disappeared after CIP treatment (Figure 6B, lanes 6 to 8), indicating that P44 is hyper-phosphorylated. To further confirm that CK1-type kinases are responsible for BYSMV P phosphorylation in plants, we transiently expressed BYSMV P in N. benthamiana leaves and then treated the Agrobacterium-infiltrated leaves with 50 μM D4476, a specific CK1 inhibitor, or with DMSO alone as solvent control (Rena et al., 2004). Immunoblot analyses revealed that the D4476 treatment significantly inhibited the accumulation of P44 compared to the DMSO control treatment (Figure 6C, cf. lanes 2 and 3). Moreover, P44 production was significantly induced by overexpression of NbCK1.3 relative to the empty vector control (Figure 6C, cf. lanes 4 and 5). These results indicate that CK1 phosphorylates the SR motif of BYSMV P in vivo.
Figure 6.
CK1 Phosphorylates the SR Motif of BYSMV P.
(A) Detection of the phosphorylation forms of PWT and PS5A expressed alone (–) or with NbCK1.3, LsCK1.3, and HvCK1.3 in E. coli. The anti-P and anti-GST polyclonal antibodies were used to detect accumulation of P and protein kinases, respectively.
(B) Treatment with CIP affects the forms of BYSMV P. Purified PWT-6×His proteins were treated with buffer or CIP and then analyzed by immunoblot analysis.
(C) Accumulation of the two forms of BYSMV PWT after transient expression in N. benthamiana leaves. Infiltrated leaves were treated with DMSO buffer (lane 2) or the CK1-specific inhibitor D4476 (lane 3) at 2 dpi and collected for P protein detection at 3 dpi. Infiltrated leaves coexpressing PWT with empty vector (EV) or NbCK1.3 were collected for P protein detection at 3 dpi. PS5A and PS5D from Agrobacterium-infiltrated N. benthamiana leaves served as P protein phosphorylation markers.
(D) Autoradiograph showing in vitro [γ-32P]ATP phosphorylation of the SR motif of BYSMV P by NbCK1.3 and LsCK1.3. Recombinant GST-NbCK1.3 and GST-LsCK1.3 proteins purified from E. coli were used as protein kinases. GFP-GST acted as a negative control. The purified PWT-6×His and PS5A-6×His proteins from E. coli served as substrates. Autophosphorylated CK1 and phosphorylated P are indicated. Coomassie Brilliant blue (CBB)–stained gels served as protein loading controls.
(E) Pull-down assays showing physical interactions between BYSMV P-6×His and GST-NbCK1.3 in vitro. GST-GFP was used as a negative control.
(F) BiFC assays showing the association between BYSMV P and NbCK1.3 in N. benthamiana leaves. The photographs were taken at 3 dpi. The N-terminal and C-terminal halves of YFP are designated YN and YC. Bar = 20 μm.
(G) Colocalization of BYSMV L and N with punctate particles formed by BiFC assay of P and NbCK1.3. L-mCherry and CFP-N were coexpressed with YN-P and YC-NbCK1.3 or YC-P and YN-NbCK1.3 combinations in N. benthamiana leaves, respectively. Fluorescence images were photographed at 3 dpi. Bar = 20 μm.
To verify BYSMV P phosphorylation in vitro, we purified P-6×His, PS5A-6×His, GST-GFP, GST-NbCK1.3, and GST-LsCK1.3 recombinant proteins from E. coli for in vitro phosphorylation assays with [γ-32P]ATP. In control experiments, we detected no radiolabel after incubation of GST-GFP with P-6×His or PS5A-6×His (Figure 6D, lanes 1 and 4). The incubation of P-6×His with GST-NbCK1.3 or GST-LsCK1.3 generated radiolabeled phosphorylated P-6×His (Figure 6D, lanes 2 and 3). By contrast, radiolabeled phosphorylated PS5A-6×His products significantly decreased in the presence of GST-NbCK1.3 or GST-LsCK1.3, as expected (Figure 6D, lanes 5 and 6). Collectively, these results indicate that the Ser residues (Ser-189, Ser-191, Ser-194, Ser-195, and Ser-198) of the SR motif are phosphorylated by host CK1 kinases in vivo and in vitro.
BYSMV P Recruits NbCK1.3 into Viroplasm-Like Bodies
Since CK1 can phosphorylate the BYSMV P protein, we hypothesized that BYSMV P may interact with CK1 within the viroplasm-like bodies that include the N, P, and L core proteins. To test this hypothesis, we performed GST pull-down assays to determine whether P and NbCK1.3 interact. Indeed, GST-NbCK1.3, but not GST-GFP, was immunoprecipitated with P-6×His in vitro (Figure 6E). To visualize the association of P with NbCK1.3 in living cells, we conducted BiFC assays in N. benthamiana leaves. In agreement with the pull-down assays, coexpression of both YN-P/YC-NbCK1.3 and YC-P/YN-NbCK1.3 resulted in fluorescent granules in the cytoplasm of infiltrated cells (Figure 6F). Notably, these granules trafficked rapidly throughout the cytoplasm (Supplemental Movies 1 and 2). By contrast, we observed no BiFC fluorescence in the control samples, in which Rubisco was coexpressed with P or NbCK1.3 fused with different YN and YC (Supplemental Figure 13).
BiFC of YN-P/YC-NbCK1.3 and YC-P/YN-NbCK1.3 resulted in trafficking granules that were reminiscent of the BYSMV P-mediated viroplasm-like bodies seen in our recent work (Fang et al., 2019). We therefore coexpressed L-mCherry and CFP-N with the BiFC combinations of YN-P/YC-NbCK1.3 and YC-P/YN-NbCK1.3. Satisfyingly, L-mCherry and CFP-N colocalized with the trafficking granules formed by the interaction of YN-P/YC-NbCK1.3 with YC-P/YN-NbCK1.3 (Figure 6G; Supplemental Movies 3 and 4). These results suggest that NbCK1.3 is recruited by BYSMV P to form trafficking viroplasm-like bodies in which BYSMV P can be efficiently phosphorylated by NbCK1.3.
RNA Interference against CK1 Inhibits Efficient Infection of BYSMV in SBPHs
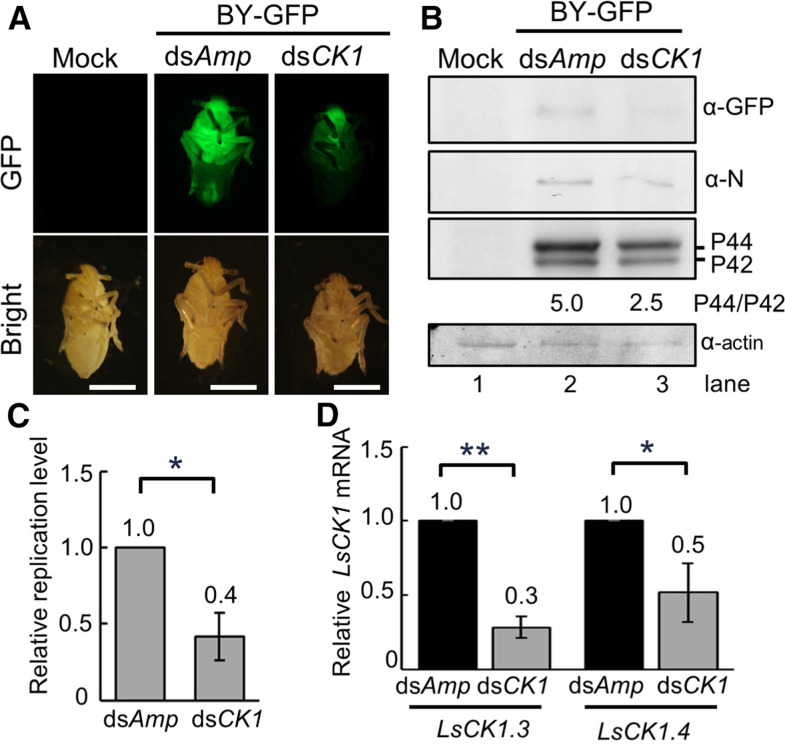
Our results thus far show that host CK1 interacts with BYSMV P and phosphorylates the SR motif of BYSMV P (Figure 6), indicating that CK1 plays important roles in BYSMV infections. To obtain genetic confirmation of this hypothesis, we synthetized a 600-bp double RNA fragment (dsLsCK1) corresponding to the homologous region between LsCK1.3 and LsCK1.4 in vitro to reduce the expression of LsCK1.3 and LsCK1.4 in SBPHs. We coinjected the thoraxes of L. striatellus with crude extracts of BY-GFP–infected barley plants and either dsLsCK1 or dsAmp (ampicillin, 70 ng per insect) as a negative control. At 5 dpi, dsLsCK1-treated insects exhibited much lower GFP fluorescence signal than dsAmp-injected insects (Figure 7A). In agreement, immunoblot analyses revealed that GFP, N, and P proteins also accumulated to a lower level in dsLsCK1-treated insects than in dsAmp-injected insects (Figure 7B, compare lanes 2 and 3).
Figure 7.
Knocking Down CK1 Expression Suppresses BYSMV Infection of SBPHs.
(A) GFP fluorescence in SBPHs microinjected with crude extracts of BYG-P–infected barley leaves with the dsAmp or dsCK1 RNAs at 5 dpi. Bar = 1 mm.
(B) Immunoblot analysis showing accumulation of the GFP, N, and P proteins in the samples of (A). The SBPH β-actin protein served as a loading control.
(C) RT-qPCR analysis showing relative replication levels of BYSMV in the samples of (A).
(D) RT-qPCR analysis showing accumulation of LsCK1.3 and LsCK1.4 in the samples shown in (A). The viral replication values with LsCK1.3 and LsCK1.4 in the dsAmp-treated SBPHs were set to 1. In (C) and (D), error bars indicate sd. Data points are mean value of three independent experiments. Statistically significant differences were determined by the Student’s t test. *P-value < 0.05; **P-value < 0.01.
RT-qPCR analyses further revealed that BY-GFP gRNA levels decreased by ∼58% in dsLsCK1-treated SBPHs compared with dsAmp-injected SBPHs (Figure 7C). Moreover, LsCK1.3 and LsCK1.4 in the dsLsCK1-treated insects decreased by ∼72 and 48%, respectively (Figure 7D). Collectively, these results suggest that knocking down LsCK1 transcript levels negatively affects virus infections in SBPHs.
Overexpression of CK1 and a Dominant-Negative Mutant Impairs BYSMV Infections in Plants
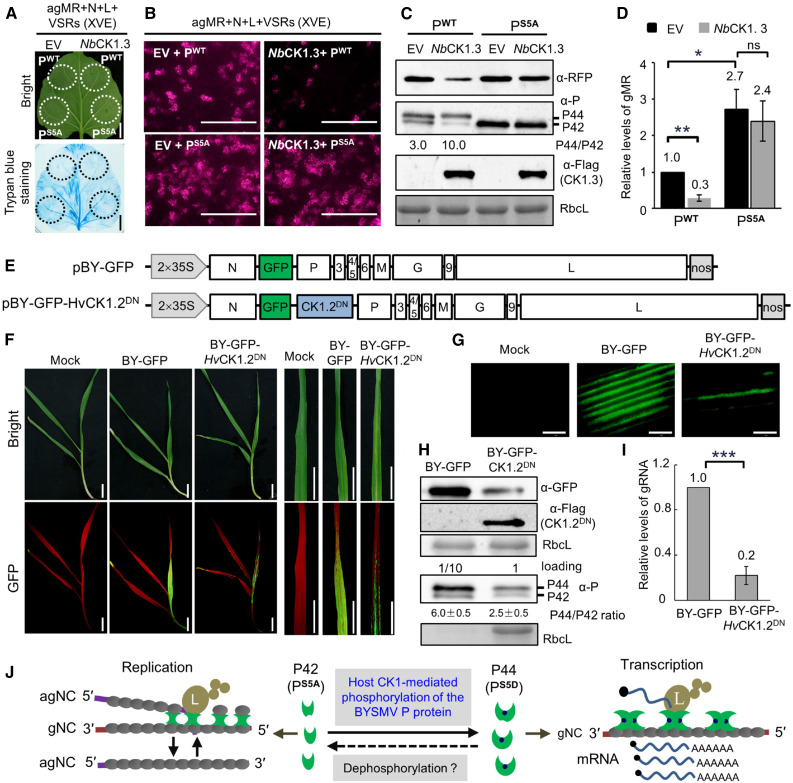
To investigate whether CK1 affects BYSMV infections in plants, we overexpressed NbCK1 proteins by placing their coding regions under the control of the cauliflower mosaic virus 35S promoter with the agMR systems. Unfortunately, coinfiltrated leaves exhibited cell death in the infiltrated patches (Supplemental Figure 14A), which would affect the agMR replication. To avoid the appearance of cell death, we used an estrogen-inducible promoter to drive the expression of plant CK1 genes in N. benthamiana leaves. In these experiments, we treated coinfiltrated leaves with 2 μM estradiol at 3 dpi, before subjecting them to trypan blue staining and observation for RFP foci 2 d later. Estradiol-induced expression of CK1 did not cause cell death in the infiltrated patches (Figure 8A). Compared to the empty vector control, coexpression of NbCK1.3 with PWT greatly decreased the numbers of RFP foci (Figure 8B, top). Notably, coexpression of NbCK1.3 also reduced the accumulation of the P42 form (Figure 8C), indicating that NbCK1.3 phosphorylated the PWT protein in vivo. Accumulation of the RFP protein and genomic minireplicon RNA (gMR) decreased in the samples overexpressing NbCK1.3 (Figures 8C and 8D). By contrast, coexpression of NbCK1.3 did not affect the PS5A-mediated improvement in agMR replication (Figures 8A and 8B). In these assays, NbCK1.13 was also coexpressed with PWT, which improved P44 production and inhibited agMR replication in N. benthamiana leaves (Supplemental Figure 15). Collectively, these results indicate that overexpression of CK1 negatively regulates BYSMV infections through phosphorylation of the P SR motif.
Figure 8.
Overexpression of CK1 Suppresses BYSMV Infections in Plants.
(A) Trypan blue staining of N. benthamiana leaves at 5 dpi after infiltration with Agrobacterium cultures containing pBY-agMR, pGD-N, pGD-L, pGD-VSRs, and pGD-PWT (wild type)/PS5A (phosphorylation defective mutant) with empty vector (EV) or NbCK1.3 under the control of the estradiol-inducible promoter. Estradiol (2 μM) was infiltrated at 3 dpi, and the leaves were stained at 5 dpi. Bar = 2 cm.
(B) RFP foci in N. benthamiana leaves from (A) at 5 dpi. Bar = 1 mm.
(C) Immunoblot analysis showing accumulation of the RFP, P, and NbCK1.3 proteins in the samples of (B). RbcL served as a loading control.
(D) RT-qPCR analysis showing accumulation of gMR in the samples shown in (B).
(E) Illustration of the genome organization of the pBY-GFP and pBY-GFP-HvCK1.2DN binary vectors harbored by the Agrobacterium cultures.
(F) Disease symptoms and GFP fluorescence of barley plants infected with BY-GFP and BY-GFP-HvCK1.2DN at 15 dpi. Bar = 2 cm.
(G) GFP foci in barley leaves of (F) observed by confocal microscope. Bar = 1 mm.
(H) Immunoblot analysis showing accumulation of the GFP and HvCK1.2DN proteins in the samples of (F). For detection of the BYSMV P protein, the total protein samples of BY-GFP were diluted 10 times for immunoblot analysis. RbcL served as a loading control.
(I) RT-qPCR analysis showing accumulation of BYSMV gRNA in the samples shown in (F).
(J) Proposed model for host CK1-mediated phosphorylation of the BYSMV P SR motif regulating virus transcription and replication. The BYSMV P proteins exist as a basally phosphorylated form (P42) and a hyper-phosphorylated form (P44). Viral replication and transcription of the N protein–encapsidated gRNA is accomplished by the polymerase L, P protein, and N proteins. The P protein dimer provides a physical link to attach the L protein to the N–RNA complex. Plant and insect CK1 proteins can phosphorylate the BYSMV P42 form to produce p44, which promotes the switch from replication to transcription.
In (D) and (I), error bars indicate sd. Data points are mean value of three independent experiments. Statistically significant differences were determined by the Student’s t test. *P < 0.05; **P < 0.01; ***P < 0.001.
Dominant-negative approaches have been used to inhibit endogenous CK1 activity in mammalian cells (Zhu et al., 1998; Lee et al., 2009). The point mutants CKIε (K38R) and CKIα (D136N) were used as dominant-negative mutants in previous studies by Zhu et al. (1998) and Lee et al. (2009). Protein blast showed that the K38 of CKIε and D136 of CKIα are also conserved in plant CK1 homologs (K38 and D128; Supplemental Figure 14B). Accordingly, we generated the point mutant (K38R and D128N) in HvCK1.2 to serve as a dominant-negative mutant, HvCK1.2DN (Supplemental Figure 14B). Unlike the wild-type HvCK1.2, HvCK1.2DN did not induce cell death in N. benthamiana leaves due to its lack of kinase activity (Supplemental Figure 14A). Moreover, coexpression of HvCK1.2DN interfered with its phosphorylation of the BYSMV P protein (Supplemental Figure 14C).
We next generated a recombinant BYSMV plasmid containing HvCK1.2DN (pBY-GFP-HvCK1.2DN) based on pBY-GFP, which would drive the accumulation of GFP to act as a visual marker of systemic BYSMV infections in barley plants (Figure 8E). Following rescue in N. benthamiana leaves and transmission by SBPHs, BY-GFP and BY-GFP-HvCK1.2DN established systemic infections in barley plants at 10 dpi (Figure 8F). BY-GFP spread extensively in the first fully expanded leaves of infected plants, whereas BY-GFP-HvCK1.2DN was limited to several veins (Figures 8F and 8G). Immunoblot analysis showed that GFP accumulated to much lower levels in BY-GFP-HvCK1.2DN–infected leaves than BY-GFP (Figure 8H). To ensure the clear detection and differentiation of the P42 and P44 bands, we diluted BY-GFP total protein samples 10 times for immunoblot analysis. As hoped, overexpression of the dominant-negative mutant (HvCK1.2DN) reduced the ratio of P44 to P42 compared with that of BY-GFP (Figure 8H). In agreement, RT-qPCR results showed that the accumulation of gRNA in BY-GFP-HvCK1.2DN–infected leaves decreased to 22% of that seen with BY-GFP (Figure 8I). As a control for infection assays, we replaced HvK1.2DN from BY-GFP-HvCK1.2DN with the β-GLUCURONIDASE (GUS) gene to generate BY-GFP-GUS. Similarly to BY-GFP, BY-GFP-GUS exhibited higher pathogenicity than BY-GFP-HvCK1.2DN in barley plants (Supplemental Figure 16). Collectively, these results indicate that overexpression of CK1.2DN inhibits BYSMV infections in barley plants.
In summary, our results demonstrate that CK1 proteins from plant and insect vectors can phosphorylate the BYSMV P protein and induce transition from virus replication to transcription (Figure 8J). The phosphorylated P44 form facilitates virus transcription, whereas the nonphosphorylated P42 form improves virus replication (Figure 8J). Since both virus replication and transcription are required for effective virus infection, either inhibition or overexpression of host CK1 genes impair the balance between virus replication and transcription, thereby inhibiting BYSMV infections (Figure 8).
DISCUSSION
Phosphorylation of cellular proteins is a widespread posttranslational modification that regulates signal transduction, molecular interactions, and subcellular localization phenomena (Peck, 2006; Bond et al., 2011). Viral proteins are usually phosphorylated by host protein kinases that play pivotal roles in viral life cycles (Jakubiec and Jupin, 2007; Wang, 2015). Here, we have identified two phosphorylated states of the BYSMV P protein that represent the hypo-phosphorylated (P42) and the hyper-phosphorylated (P44) forms (Figure 1). In this respect, the BYSMV P protein is similar to the Rabies virus (RV) P protein, which has both a basally phosphorylated and a hyper-phosphorylated form (Tuffereau et al., 1985; Takamatsu et al., 1998). However, the protein kinases catalyzing the phosphorylation of the RV P protein remain elusive. Here, we discovered that host CK1 protein kinases are responsible for BYSMV P phosphorylation and P44 production (Figure 6). Furthermore, multiple CK1 orthologs phosphorylated BYSMV P protein in N. benthamiana, planthoppers (L. striatellus), and barley (Supplemental Figure 12). BYSMV infections of barley plants did not induce expression of these CK1 genes (Supplemental Figure 17), possibly because normal CK1 levels are sufficient for the phosphorylation of BYSMV P. It is important to note that BYSMV is a cross-kingdom virus infecting both plants and insects. Correspondingly, CK1 is a well-conserved kinase family in eukaryotes, including dicot and monocot plants, as well as in SBPHs (Knippschild et al., 2005); thus, it is not surprising that CK1 plays an important role in the cross-kingdom life cycle of BYSMV in plants and insect vectors.
Another interesting finding in our study is that the Ser-rich region (SR motif) of the BYSMV P protein was highly phosphorylated (Figure 1). Using site-directed mutants and phosphorylation assays in E. coli and in vitro, we clearly show that the SR motif is phosphorylated by host CK1 (Figure 6). Furthermore, the nonphosphorylated PS5A mutant of the SR motif only migrates as P42, whereas the phospho-mimic mutant PS5D exists as P44 in SDS-PAGE gels (Figure 2B), suggesting that the phosphorylated and nonphosphorylated SR motifs drive conformational changes between the two forms (P42 and P44). More interestingly, the SR motif is located in the C-terminal IDRs of the BYSMV P protein. Many IDRs with increased flexibility usually serve as scaffolds for the assembly of functional complexes involved in cell signaling, molecular recognition, and regulation processes (Uversky et al., 2005). Thus, our results strongly suggest that host CK1-mediated SR phosphorylation changes the flexibility of the BYSMV P C-terminal IDR to mediate conformational changes from P42 to P44. Moreover, the conformational changes between P42 and P44 result in different complexes for optimal replication or transcription. We have therefore provided evidence showing that a host kinase can target the C-terminal IDR region of a rhabdovirus P protein and affect viral RNA replication and transcription.
Rhabdovirus transcription and replication are executed by the same polymerase complexes and are finely regulated in a dynamic balance (Ivanov et al., 2011). An intriguing question is how viral polymerase complexes are finely regulated to initiate either transcription or replication. Using our recent BYSMV reverse genetics systems, we showed that the nonphosphorylated mutant (PS5A) of the SR motif increased replication but reduced transcription. Conversely, the PS5D variant, which mimics the P-phosphorylated form, stimulates transcription and inhibits replication (Figure 2). It should be noted that PS5D-induced transcripts were based on significantly reduced gMR templates, which probably led to aberrant transcripts with no coding capacity. Therefore, it is not surprising that PS5D supported higher accumulation of GFP mRNA but reduced the accumulation of GFP protein than PS5A (Figures 2E and 2F). Collectively, our results demonstrate that the phosphorylation states of the SR motif modulate the balance between viral replication and transcription in vivo. Several pioneering studies using Vesicular stomatitis virus (VSV) minigenome systems have demonstrated that N- or C-terminal phosphorylation of the VSV P protein affects RNA replication or transcription (Hwang et al., 1999; Chen et al., 2007, 2013; Mondal et al., 2014). For instance, CK2 phosphorylates the N terminus of the VSV P protein and affects P oligomerization, which is required for virus transcription rather than replication (Pattnaik et al., 1997; Qanungo et al., 2004). Here, the IDRC motif of the rhabdovirus P protein is a new phosphorylated domain, which affects both virus replication and transcription. It is striking to note that the P proteins of animal rhabdovirus RV and VSV, as well as plant cytorhabdovirus, such as Northern cereal mosaic virus, also contain SR motifs in their C-terminal IDR regions (Supplemental Figure 18; Gerard et al., 2009). Broader implications of our finding await investigations into whether the CK1 family is a common modulator of other plant and animal rhabdoviruses.
In the field, BYSMV is transmitted by SBPHs in a persistent-propagative manner (Cao et al., 2018), meaning that BYSMV is disseminated to plants solely by these insects, and its survival depends entirely on replication in SBPHs. Our results show that CK1 in SBPHs can phosphorylate the SR motif of the BYSMV P protein (Figures 1 and 6). Thus, the SBPH CK1 protein is instrumental in BYSMV infections. Indeed, knockdown of LsCK1 by RNA interference significantly affected virus accumulation and dissemination (Figure 7). Using our full-length infectious clones, we clearly show that highly efficient replication and transcription supported by PS5A and PS5D, respectively, are essential for virus infection in SBPHs. Nonetheless, BYG-PS5A/S5D and BYG-PS5D/S5A failed to be transmitted to barley plants by the infected SBPHs. Thus, it remains to be determined whether spatial- and temporal-specific accumulation of PS5A and PS5D or other factors play a role during the different infection stages of BYSMV in SBPHs and barley plants.
In summary, the P44 hyper-phosphorylated form of the P protein is the principal product in BYSMV virus nucleocapsids (Figure 1A) and facilitates initiation of virus transcription from ribonucleoproteins (RNPs) released during the infection of cells. Subsequently, the freshly translated P42 form binds N0 to form P–N0 complexes and efficiently delivers the N0 to the nascent RNA chains, which drives the switch from transcription to RNA replication. Upon phosphorylation of the P42 form by host CK1 proteins, the resulting P44 form exhibits compromised binding to N–RNA templates, which results in the dissociation of the complexes from N–RNA templates and the switch from gRNA replication to mRNA transcription (Figure 8J).
METHODS
Preparation of Host Plants and Insect Vectors
Nicotiana benthamiana and barley (Hordeum vulgare cv Golden promise) plants were cultivated in a growth room at 25°C with a 16-h-day/8-h-night cycle. SBPHs were initially isolated from Hebei Province, China, and reared in illumination incubators that were placed inside a climate chamber under a 16-h-d/8-h-night cycle at 26°C.
Virus Inoculation and Transmission
Virus recovery and transmission by SBPHs were performed as previously described by Gao et al. (2019) and Zhang et al. (2020). Briefly, the BYSMV infectious clones were transformed into Agrobacterium (Agrobacterium tumefaciens) strain EHA105. The bacteria were cultivated at 28°C for 12 h, harvested by centrifugation, and suspended in buffer (10 mM MES, 150 μM acetosyringone, and 10 mM MgCl2). After incubation at 25°C for 3 h, bacterial suspensions were mixed and infiltrated in N. benthamiana leaves. At 14 dpi, inoculated leaves were homogenized in buffer (100 mM Tris‐HCl, 40 mM Na2SO3, 10 mM Mg (CH3COO)2, and 1 mM MnCl2, pH 8.4) and centrifuged at 12,000g for 15 min at 4°C. Crude extracts of infected N. benthamiana leaves (13.8 nL per insect) were injected into second‐instar nymphs with a Nanoinject II auto-nanoliter injector (Drummond Scientific). After a 10-d incubation period, injected SBPH nymphs were transferred to healthy barley plants for a 2-d inoculation period. Observation of symptoms and virus analyses were performed at 10 to 15 dpi.
Plasmid Constructions
The pBYSMV-agMR, pGD-VSRs, pGD-N, pGD-P, pGD-L, and pGDG-P plasmids were previously described by Fang et al. (2019) as well as the pGD-NLP, pBY-GFP, and pBY-GR plasmids (Gao et al., 2019). We introduced the full-length P gene and the deletion fragment lacking the C-terminal 120 amino acids into plasmid pMDC32. The plasmids pMDC-PS5A and pMDC-PS5D were constructed by the Quick-Change mutagenesis method. The pGD-NLPS5A was constructed by replacing the P coding sequence with PS5A in the pGD-NLP plasmid. Based on the pBY-GFP construct that was used to design pBYG-PWT (Gao et al., 2019), the fragment including P and P3 was PCR amplified and cloned into the pTOPO-Blunt vector to generate the pTO-BYP-P3 vector. For the recombinant virus plasmids pBYG-PS5A and pBYG-PS5D, the PS5A and PS5D coding sequence was PCR amplified and subcloned into the pTO-BYP-P3 vector, and the fragments including PS5A-P3- or -PS5D-P3 were PCR amplified and then substituted in place of the corresponding region of pBYG-PWT. For pBYG-PS5A/S5D and pBYG-PS5D/S5A, the RFP-P-P3 was PCR amplified from pBY-GR and subcloned into the M5 HiPer pTOPO-Blunt vector (Mei5 Biotechnology) to generate pTORFP-P-P3. The pTORFP-PS5A-P3 and pTORFP-PS5D-P3 plasmids were constructed by site-directed mutagenesis. Next, the RFP ORF of pTORFP-PS5A-P3 and RFP-PS5D-P3 were replaced with the PS5D or PS5A ORFs to generate pTO-PS5D/S5A-P3 and pTO-PS5A/S5D-P3, respectively. Finally, the PS5D/S5A-P3 and PS5A/S5D-P3 fragments were cloned into pBY-GR to generate pBYG-PS5A/S5D and pBYG-PS5D/S5A. All primers used for these manipulations and other cloning experiments are listed in Supplemental Data Set 1.
Immunoblot Analysis
Agrobacterium-infiltrated N. benthamiana leaves were harvested at 2 to 6 dpi, and total proteins were extracted in SDS buffer (100 mM Tris, pH 6.8, 20% [v/v] glycerol, 4% [w/v] SDS, 0.2% [w/v] bromophenol blue, and 10% [v/v] β-mercaptoethanol) for immunoblot analysis. Proteins were separated on SDS-PAGE gels followed either by Coomassie Brilliant Blue R 250 staining or transfer onto nitrocellulose membranes for detection with polyclonal antibodies specific to the BYSMV N (1:5000), P (1:3000), GFP (1:2000), or RFP (1:2000) proteins. The antibodies of the BYSMV N, P, RFP, and GFP were acquired from rabbits immunized with prokaryotic expressed proteins as previously described by Fang et al., (2019) and Gao et al. (2019). Secondary reactions were performed with goat anti-rabbit IgG horseradish peroxidase conjugate (1:3000; catalog no. 1,706,515, Bio-Rad) and Pierce ECL Plus chemiluminescent substrate followed by exposure of the filters to x-ray films (Fujifilm).
Mass Spectrometric Analysis
Immunoprecipitation of Flag-tagged P protein and identification of phosphorylation sites by LC-MS/MS were performed as previously described by Hu et al. (2015). Briefly, soluble proteins were extracted from Agrobacterium-infiltrated N. benthamiana leaves and dissolved in immunoprecipitation (IP) buffer (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% [v/v] glycerol, 0.1% [v/v] Nonidet P-40, 2% [w/v] Polyvinylpyrrolidone 40, 1 mM NaF, 1mM NaVO3, 1 mM EDTA, 10 mM DTT, and 1× protease inhibitor cocktail [Sigma-Aldrich]), followed by centrifugation at 10,000g for 30 min at 4°C. The supernatant was incubated with 20 μL of Flag-specific agarose beads at 4°C for 2 h. The immunoprecipitated Flag-P was washed five times with IP buffer and separated onto 12.5% (v/v) SDS-PAGE gels. After staining with the Plus Silver Stain Kit (Sigma-Aldrich), the Flag-P bands were cut out and subjected to in-gel trypsin digestion for 8 h at 37°C. The digested peptides were analyzed by Q-Exactive LC-MS/MS (Thermo Fisher Scientific) at the Mass Spectrometry Facility of China Agricultural University.
Meta-Prediction of IDRs
Disordered regions within the BYSMV P protein were predicted by submitting the amino acid sequence to 15 different predictors that perform well and are available through web servers. These include DisEMBL (Coils, Hotloops, and Remark465; Linding et al., 2003), DISpro (Obradovic et al., 2005), Spine-D (Zhang et al., 2012), Dispro (Cheng et al., 2005), IUPred (long and short; Dosztányi et al., 2005), Pdisorder (http://linux1.softberry.com/berry.phtml?topic=pdisorderandgroup=programsandsubgroup=propt), Poodle-S (Shimizu et al., 2007), Poodle-L (Hirose et al., 2007), PrDOS (Ishida and Kinoshita, 2007), Spritz (long and short; Vullo et al., 2006), and RONN (Yang et al., 2005). For each predicting algorithm, a score of 1 was attributed to each residue predicted to be in a structured region, whereas the predicted disordered residues were attributed a score of 0.
Phylogenetic Analysis
The CK1 protein sequences for Saccharomyces cerevisiae, H. vulgare, and Laodelphax striatellus were downloaded from different sources: S. cerevisiae kinases were downloaded from KinBase (http://kinase.com/web/current/), H. vulgare kinases from Ensembl Plants (plants.ensembl.org), and L. striatellus kinases from Gigadb (http://gigadb.org/data set/100361). The gene families of these three species were identified by the OrthoMCL v2.0.9 package (Li et al., 2003). We used each protein as a query against the Pfam database to identify their kinase domain. Phylogenetic analysis was conducted using MEGA7.0 (Kumar et al., 2016) followed by annotation with EvolView (He et al., 2016). For broad phylogenetic analysis of CK1 kinases in N. benthamiana, L. striatellus, and H. vulgare, we searched and downloaded yeast protein HRR25 homologs in the three species using BLAST in the N. benthamiana Genome and Transcriptome database (https://sefapps02.qut.edu.au/blast/blast_link2.cgi), the L. striatellus protein databases (http://gigadb.org/data set/100361), and the barley protein database in Ensembl Plant (http://plants.ensembl.org/Multi/Tools/Blast?db=core). The phylogenetic trees were constructed using MEGA7.0.
Kinase Assays in Escherichia coli
We subcloned 16 yeast kinase genes in the prokaryotic expression vector pGEX-KG. Briefly, yeast kinase genes were queried in yeast databases (https://www.yeastgenome.org). The corresponding ORFs were amplified from yeast total RNA by RT-PCR and cloned into the pGEX-KG vector. For PWT and PS5A protein prokaryotic expression, the coding sequence was PCR amplified and cloned between the NdeI and XhoI restriction sites in pET-30a. To ensure coexpression of kinases and P protein in E. coli, the segments containing the -tac promoter-GST tag-kinase ORF- were PCR amplified and then fused with SphI-linearized pET-P and pET-PS5A to generate pET-PWT-GST-Kinase and pET-PS5A-GST-Kinase. The pET-PWT-GST-Kinases and pET-PS5A-GST-Kinases plasmids were transformed into E. coli (Rosetta strain) and induced by isopropyl β-d-1-thiogalactopyranoside (1 mM) for 8 h at 25°C. The E. coli cultures were harvested by centrifugation at 10,000g for 10 min at 25°C and extracted in SDS buffer (100 mM Tris, pH 6.8, 20% [v/v] glycerol, 4% [w/v] SDS, 0.2% [w/v] bromophenol blue, and 10% [v/v] β-mercaptoethanol) for immunoblot analysis with polyclonal antibodies against BYSMV P (1:3000) and GST (1:5000; GenScript, catalog no. A00866), respectively.
In Vitro Phosphorylation Assays
In vitro phosphorylation assays were performed according to previous studies by Hu et al. (2015) and Zhang et al. (2018). The pGEX-NbCK1.3, pGEX-LsCK1.3, pGEX-GFP, pET-PWT, and pET-PS5A plasmids were transformed into E. coli (Rosetta strain) and induced by isopropyl β-d-1-thiogalactopyranoside (0.3 mM) for 16 h at 18°C. GST-NbCK1.3, GST-LsCK1.3, and GST-GFP were purified by glutathione Sepharose 4 Fast Flow medium (GE Healthcare). The PWT-6×His and PS5A-6×His fusion proteins were purified by nickel-affinity agarose chromatography (Bio-Rad) according to the manufacturer’s instructions. For in vitro phosphorylation assays, 0.1 μg of GST-NbCK1.3, GST-LsCK1.3, or GST-GFP was incubated for 30 min at 30°C with 1 μg of PWT-6×His or PS5A-6×His in 30 μL of 1× kinase buffer (25 mM HEPES, pH 7.5, 20 mM MgCl2, 0.1 mg/mL BSA, 0.1 mM DTT, 0.1 mM ATP, and 0.3 μCi of [γ-32P]ATP). Next, 5 μL of 5× SDS sample buffer was added to terminate the reactions. The proteins were separated on a 12.5% (v/v) SDS-PAGE gel, and the gel was dried using a model 583 Gel Dryer (Bio-Rad) for 45 min at 80°C. The phosphorylated P proteins were visualized via autoradiography.
The P-Flag protein immunoprecipitated from Agrobacterium-infiltrated N. benthamiana leaves or PWT purified from E. coli was eluted from the resin by shaking for 1 h with IP buffer containing 0.1 mg/mL Flag peptide. The eluted P-Flag protein was incubated with 2 μL of 10 units/μL CIP (New England Biolabs) or 1 μL of 400 units/μL λ-protein phosphatase (New England Biolabs) in 50-μL reactions with protease inhibitor (1× cocktail; Roche) for 1 h at 37°C. Reactions were stopped by boiling them in gel loading buffer for immunoblot analysis with the anti-P antibody.
GST Pull-Down Assay
GST pull-down assays were performed as previously described by Yang et al. (2018). P-6×His proteins (∼10 μg) were incubated at 4°C for 3 h with GST-GFP and GST-NbCK1.3 proteins (∼10 μg) in 600 μL of binding buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% [v/v] glycerol, 0.2% [v/v] Triton X-100, 1 mM DTT, and 1× protease inhibitor cocktail) with 30 μL of glutathione Sepharose 4B beads (GE Healthcare). The beads were washed six times with binding buffer and eluted with 50 μL of binding buffer containing 10 mM reduced l-glutathione. The eluted fractions were boiled in equal volumes of 2× SDS loading buffer and then the proteins were analyzed by immunoblot with anti-GST (1:5000) and anti-P (1:3000) antibodies.
BYSMV-agMR Assays in N. benthamiana Leaves
BYSMV-agMR assays were performed as previously described by Fang et al. (2019). N. benthamiana plants with six to eight fully expanded leaves were used for BYSMV-agMR assays. Briefly, Agrobacterium harboring the pBYSMV-agMR (OD600 0.3), pGD-VSRs (OD600 0.1), pGD-N (OD600 0.1), pGD-P (OD600 0.1), and pGD-L (OD600 0.3) plasmids were mixed and infiltrated into N. benthamiana leaves. Epidermal cells of infiltrated leaves were observed for GFP and RFP fluorescence at 6 dpi using a fluorescent microscope (model BX53; Olympus) or confocal laser scanning microscopy (model FV1000; Olympus).
RNA Gel Blots
For analysis of RNA synthesis in the BYSMV MR system, total RNA was extracted from Agrobacterium-infiltrated N. benthamiana leaves using TRIzol reagent (Sigma-Aldrich). RNA gel blot detection of BYSMV MR gRNA and agRNA derivatives and GFP mRNA was performed using 5 μg of total RNA as previously described by Dong et al. (2016). For gRNA or agRNA detection, the PCR fragments were obtained using pBYSMV agMR as a template with primers designed with T7 or SP6 promoters in the desired genomic or antigenomic orientations, followed by in vitro transcription with T7 or SP6 RNA polymerase in the presence of [32P]UTP.
RT-qPCR
Total RNA isolated from MR-infiltrated leaves was treated with DNase I (Takara) to eliminate DNA contamination. Next, the treated total RNA (2 μg) was used as template for reverse transcription with oligo(dT) primer to detect mRNA or with B-RT-F and EF1a-R primers to detect minigenome RNA with Moloney Murine leukemia virus reverse transcriptase (Promega). RT-qPCR reactions were performed by amplifying GFP and RFP fragments to detect transcript levels and trailer fragments to monitor replication levels with SsoFast EvaGreen Supermix (Bio-Rad). The primers are listed in Supplemental Data Set 1. All RT-qPCR data were normalized to the EF1a gene. Three independent biological replicates were performed, each in technical triplicates, for statistical analysis as shown in Supplemental Data Set 2.
In Vivo Co-IP Assays
In vivo Co-IP assays were performed as previously described by Zhang et al. (2017b). N. benthamiana plants were infiltrated with Agrobacterium cultures for transient expression of recombinant proteins, and leaf tissues were homogenized in Co-IP buffer (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% [v/v] glycerol, 0.1% [v/v] Nonidet P-40, 2% [w/v] Polyvinylpyrrolidone 40, 1 mM NaF, 1 mM NaVO3, 1 mM EDTA, 10 mM DTT, and 1× protease inhibitor cocktail [Sigma-Aldrich]). The supernatants produced by filtration and centrifugation (10,000g for 20 min at 4°C) were incubated for 2 h with anti-Flag M2 affinity beads (Sigma-Aldrich), followed by five washes with IP buffer. The immunoprecipitated products were boiled in SDS buffer for immunoblot assays with corresponding antibodies.
BiFC Assays
BiFC assays were performed as previously described by Zhang et al. (2017a), with minor modifications. The fragments of the BYSMV N, L, P, and P mutant were cloned into the BiFC vectors pSPYNE-35S and pSPYCE-35S with different primers listed in Supplemental Data Set 1. As control, the Rubisco coding sequence was cloned into the BiFC vectors (Zhang et al., 2017a). N. benthamiana leaves were coinfiltrated with Agrobacterium EHA105 cultures containing BiFC plasmids and the P19 viral suppressor plasmid at a final ratio of 0.2:0.2:0.1 (OD600). The epidermal cells of Agrobacterium-infiltrated leaves were observed for fluorescence (YFP) using confocal laser scanning microscopy (model FV1000; Olympus).
N–RNA Purification by CsCl Gradient Centrifugation and In Vitro Co-IP
BYSMV-infected barley tissues were homogenized in buffer (0.3 M Gly and 0.1 M MgCl2, pH 8.0), and the homogenates were filtered through two layers of gauze and centrifuged at 150g at 4°C for 5 min. The supernatant was mixed with chloroform (20% [v/v] final volume) and centrifuged at 1200g at 4°C for 20 min. The resulting aqueous supernatant was used for BYSMV RNP complex precipitation after addition of polyethylene glycol 6000 and NaCl to final concentrations of 6% and 3% (w/v), respectively, and centrifuged at 3500g at 4°C for 20 min. The pellet was resuspended in extraction buffer (100 mM Gly and 0.01 M MgCl2, pH 7.0) and centrifuged at 160,000g for 90 min at 4°C in a Beckman L-80XP centrifuge, and N–RNA was purified from a portion of the BYSMV RNP as previously described by Zhang et al. (2013). BYSMV RNP pellets were also resuspended in 2 mL of CsCl buffer (10 mM Tris-HCl, pH 7.8, 150 mM NaCl, and 1 mM EDTA), loaded onto a discontinuous 25 to 40% (w/v) CsCl gradient (25%, 1.5 mL; 30%, 1.5 mL; and 40%, 1.5 mL) in dilution buffer (10 mM Tris-HCl, pH 7.8, 150 mM NaCl, and 1 mM EDTA), and centrifuged at 250,000g at 20°C for 1 h. The contents of the visible band were collected from the region between 30 and 40% (w/v) CsCl gradient and subjected to centrifugation at 200,000g at 4°C for 1 h. The resulting pellet was resuspended in 100 μL of 1× PBS for in vitro Co-IP. The purified N–RNA was divided into five equal aliquots and incubated with buffer containing Flag-tagged GFP, PWT, PS5A, PS5D, and PΔC120 immunoprecipitated from infiltrated N. benthamiana leaves with anti-Flag M2 affinity beads. Co-IP and immunoblot analyses were performed as described above.
CK1 Overexpression and Dominant-Negative Assays
The coding sequences of NbCK.3, NbCK1.7, NbCK.13, and HvCK.12 were cloned into the pMDC32 vector for overexpression of CK1 in N. benthamiana. We induced expression of CK1 with estradiol according to a previous study by Zuo et al. (2000). Briefly, The NbCK1.3 and NbCK1.13 coding sequences were cloned into the pER8 vector to generate pER8-NbCK1.3 and pER8-NbCK1.13. Agrobacterium cultures harboring the pER8-NbCK1.3/pER8-NbCK1.13 and BYSMV-agMR systems plasmids were mixed and infiltrated into N. benthamiana leaves. Next, the infiltrated leaves were infiltrated with 2 μM estradiol at 3 dpi, and samples were collected at 5 dpi for RFP fluorescence observation and MR replication analysis.
For CK dominant-negative assays, the RFP ORF of pBY-GR was replaced with HvCK1.2 with two amino acid substitutions (K38R and D128N) or a GUS fragment to generate pBY-GFP-HvCK1.2DN or pBY-GFP-GUS. Next, the recombinant viruses were rescued from N. benthamiana to planthopper and then to barley as previously described by Gao et al. (2019). The accumulation of recombinant viruses in infected barley plants was evaluated with a hand-held blue green lamp (Luyor Instrument) and photographed with a Canon digital camera at 15 dpi.
Virus Inoculation, Double-Stranded RNA Injection, and Insect Transmission
Protocols for crude extraction of BYSMV virions from plant materials and injection into planthopper nymphs have been previously described by Gao et al. (2019). To determine virus transmission by planthoppers, injected nymphs were allowed to feed on healthy barley plants for a 2-d inoculation period, and fluorescence of infected plants was evaluated with a hand-held blue green lamp (Luyor Instrument) at 15 dpi. For RNA interference assays, the 600 bp of LsCK1.3 and the 555 bp of negative control ampicillin resistance gene β-lactamase fragments (Amp) containing T7 promoters at the two termini were amplified. Before injection, second-instar nymphs were anesthetized by incubation on ice for 30 min. Next, ∼70 ng of dsLsCK1 and dsAmp were mixed with crude virus extracts from Agrobacterium-infiltrated regions of N. benthamiana leaves that were exhibiting GFP fluorescence, and 23 nL was injected into the thoraxes of each insect with a Nanoinject II auto-nanoliter injector (Drummond Scientific). RNA interference effects and virus accumulation were detected by RT-qPCR at 5 dpi. All primers are listed in Supplemental Data Set 1.
Trypan Blue Staining
Trypan blue staining was performed as previously reported, with minor modifications, by Zhang et al. (2018). N. benthamiana leaves infiltrated with Agrobacterium cultures were soaked in ethanol for 2 to 3 min and then transferred to staining buffer (10 mL of lactic acid, 10 mL of water phenol, 10 mL of glycerol, and 10 mL of double distilled water with 15 mg of trypan blue), boiled in water for 10 to 15 min, and left to stand for 6 to 8 h at room temperature. Leaves were photographed after destaining three times using chloral hydrate (2.5 g/mL).
Accession Numbers
Sequence data of proteins and genes used this article can be found in Supplemental Data Set 3 and at UniProt protein resource (https://www.uniprot.org/), GenBank database (https://ncbi.nlm.nih.gov), Nicotiana benthamiana Genome & Transcriptome database (http://benthgenome.qut.edu.au/), and Saccharomyces genome database (https://www.yeastgenome.org/).
Supplemental Data
Supplemental Figure 1. LC-MS/MS analysis of the SR motif phosphorylation of BYSMV P.
Supplemental Figure 2. RFP accumulation from agMR after co-expression with SR motif P protein mutants deficient in phosphorylation.
Supplemental Figure 3. Immunoblot analysis showing accumulation of P and M in co-infiltrated N. benthamiana leaves at 10 dpi.
Supplemental Figure 4. GFP foci in N. benthamiana leaves at 10 d post infiltration with Agrobacterium cultures containing the indicated plasmids.
Supplemental Figure 5. BiFC assays showing interaction of P/P mutants with themselves or with the BYSMV N protein.
Supplemental Figure 6. BiFC assays showing the negative controls of Rubisco with P/P mutants and N.
Supplemental Figure 7. The presence of N and genome RNA in the purified N–RNA complexes.
Supplemental Figure 8. Immunoblot analysis showing two phosphorylation forms of the BYSMV P protein in yeast.
Supplemental Figure 9. Phylogenetic analysis of the protein sequences of conserved protein kinases.
Supplemental Figure 10. Yeast HRR25 phosphorylates the SR motif of BYSMV P.
Supplemental Figure 11. Phylogenetic analysis of CK1 orthologs based on their protein sequences.
Supplemental Figure 12. Orthologs of yeast HRR25 phosphorylates the SR motif of BYSMV P.
Supplemental Figure 13. BiFC assays showing the negative controls of Rubisco with P and NbCK1.3.
Supplemental Figure 14. Overexpression of CK1, but not a dominant-negative mutant, results in cell death.
Supplemental Figure 15. Overexpression of NbCK1.13 suppresses BYSMV infections in N. benthamiana plants.
Supplemental Figure 16. Overexpression of a CK1 dominant-negative mutant suppresses BYSMV infections in plants.
Supplemental Figure 17. Transcript levels of CK1 genes in response to BYSMV infections in barley plants.
Supplemental Figure 18. Potential phosphorylation sites within the predicted C- terminal IDRs of P proteins.
Supplemental Table. BYSMV P phosphopeptides identified by mass.
Supplemental Data Set 1. Primers used in this study.
Supplemental Data Set 2. Statistical analysis data in this study.
Supplemental Data Set 3. Accession numbers for all CK1 proteins described in this study.
Supplemental File 1. Alignment of conserved kinases in yeast, barley and planthopper.
Supplemental File 2. The phylogenetic tree of yeast, barley and planthopper conserved kinases (Newick format).
Supplemental File 3. Alignments of NbCK1, LsCK1, and HvCK1 orthologs.
Supplemental File 4. The phylogenetic tree of NbCK1, LsCK1, and HvCK1 orthologs (Newick format).
Supplemental Movie 1. Intracellular movement of the BiFC punctate particles of YN-P and YC-NbCK1.3.
Supplemental Movie 2. Intracellular movement of the BiFC punctate particles of YC-P and YN-NbCK1.3.
Supplemental Movie 3. A representative movie showing trafficking granules formed by L-mCherry and CFP-N with YN-P/YC-NbCK1.3.
Supplemental Movie 4. A representative movie showing trafficking granules formed by L-mCherry and CFP-N with YC-P/YN-NbCK1.3.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
BSMV Gramene: Barley stripe mosaic virus
BSMV Araport: Barley stripe mosaic virus
Acknowledgments
We thank our colleagues Chenggui Han, Dawei Li, and Yongliang Zhang for their helpful suggestions and constructive criticism. We thank our colleague Qun He for his gift of the GST antibody. We thank Andrew O. Jackson (Department of Plant and Microbial Biology, University of California, Berkeley) for his gift of pGD and pGDG and for helpful suggestions. We thank Xiaorong Tao (Department of Plant Pathology, Nanjing Agricultural University) for his gift of the pCB301-2×35S-Nos plasmid. We thank Zhen Li for his assistance with LC-MS/MS analysis at the Mass Spectrometry Facility of China Agricultural University. This work was supported by the National Natural Science Foundation of China (grants 31872920 and 31571978).
AUTHOR CONTRIBUTIONS
X.-B.W., Q.G., T.Y., and Z.-J.Z. conceived and designed the experiments; Q.G., T.Y., X.-D.F., Z.-J.Z., Z.-H.D., Q.C., W.-Y.X., D.-M.G., J.-H.Q., and Y.-Z.Y. performed the experiments; X.-B.W., Q.G., T.Y., and S.-Y.L. analyzed the data and drafted the article; X.-B.W., Y.W., and J.Y. participated in experimental coordination and revision of the article; and X.-B.W., Q.G., and T.Y. proofread and finalized the article.
References
- Ammar D., Tsai C.-W., Whitfield A.E., Redinbaugh M.G., Hogenhout S.A.(2009). Cellular and molecular aspects of rhabdovirus interactions with insect and plant hosts. Annu. Rev. Entomol. 54: 447–468. [DOI] [PubMed] [Google Scholar]
- Barik S., Banerjee A.K.(1992). Sequential phosphorylation of the phosphoprotein of vesicular stomatitis virus by cellular and viral protein kinases is essential for transcription activation. J. Virol. 66: 1109–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond A.E., Row P.E., Dudley E.(2011). Post-translation modification of proteins; methodologies and applications in plant sciences. Phytochemistry 72: 975–996. [DOI] [PubMed] [Google Scholar]
- Cao Q., Xu W.-Y., Gao Q., Jiang Z.-H., Liu S.-Y., Fang X.-D., Gao D.-M., Wang Y., Wang X.-B.(2018). Transmission characteristics of barley yellow striate mosaic virus in its planthopper vector Laodelphax striatellus. Front. Microbiol. 9: 1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Zhang S., Banerjee A.K., Chen M.(2013). N-Terminal phosphorylation of phosphoprotein of vesicular stomatitis virus is required for preventing nucleoprotein from binding to cellular RNAs and for functional template formation. J. Virol. 87: 3177–3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Ogino T., Banerjee A.K.(2007). Interaction of vesicular stomatitis virus P and N proteins: identification of two overlapping domains at the N terminus of P that are involved in N0-P complex formation and encapsidation of viral genome RNA. J. Virol. 81: 13478–13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J.L., Sweredoski M.J., Baldi P.(2005). Accurate prediction of protein disordered regions by mining protein structure data. Data Min. Knowl. Discov. 11: 213–222. [Google Scholar]
- Dietzgen R.G., Bejerman N.E., Goodin M.M., Higgins C.M., Huot O.B., Kondo H., Martin K.M., Whitfield A.E.(2020). Diversity and epidemiology of plant rhabdoviruses. Virus Res. 281: 197942. [DOI] [PubMed] [Google Scholar]
- Dong K., Wang Y., Zhang Z., Chai L.X., Tong X., Xu J., Li D., Wang X.B.(2016). Two amino acids near the N-terminus of Cucumber mosaic virus 2b play critical roles in the suppression of RNA silencing and viral infectivity. Mol. Plant Pathol. 17: 173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosztányi Z., Csizmok V., Tompa P., Simon I.(2005). IUPred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21: 3433–3434. [DOI] [PubMed] [Google Scholar]
- Emerson S.U., Schubert M.(1987). Location of the binding domains for the RNA polymerase L and the ribonucleocapsid template within different halves of the NS phosphoprotein of vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA 84: 5655–5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson S.U., Yu Y.(1975). Both NS and L proteins are required for in vitro RNA synthesis by vesicular stomatitis virus. J. Virol. 15: 1348–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X.D., Yan T., Gao Q., Cao Q., Gao D.M., Xu W.Y., Zhang Z.J., Ding Z.H., Wang X.B.(2019). A cytorhabdovirus phosphoprotein forms mobile inclusions trafficked on the actin/ER network for viral RNA synthesis. J. Exp. Bot. 70: 4049–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso G., van Wijk K.J.(2015). Posttranslational protein modifications in plant metabolism. Plant Physiol. 169: 1469–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Xu W.-Y., Yan T., Fang X.-D., Cao Q., Zhang Z.-J., Ding Z.-H., Wang Y., Wang X.-B.(2019). Rescue of a plant cytorhabdovirus as versatile expression platforms for planthopper and cereal genomic studies. New Phytol. 223: 2120–2133. [DOI] [PubMed] [Google Scholar]
- Gerard F.C., Ribeiro Ede.A. Jr., Leyrat C., Ivanov I., Blondel D., Longhi S., Ruigrok R.W., Jamin M.(2009). Modular organization of rabies virus phosphoprotein. J. Mol. Biol. 388: 978–996. [DOI] [PubMed] [Google Scholar]
- Green T.J., Luo M.(2009). Structure of the vesicular stomatitis virus nucleocapsid in complex with the nucleocapsid-binding domain of the small polymerase cofactor, P. Proc. Natl. Acad. Sci. USA 106: 11713–11718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S.D., Anderson R.A.(1998). Casein kinase I: Spatial organization and positioning of a multifunctional protein kinase family. Cell. Signal. 10: 699–711. [DOI] [PubMed] [Google Scholar]
- Gupta A.K., Blondel D., Choudhary S., Banerjee A.K.(2000). The phosphoprotein of rabies virus is phosphorylated by a unique cellular protein kinase and specific isomers of protein kinase C. J. Virol. 74: 91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z., Zhang H., Gao S., Lercher M.J., Chen W.H., Hu S.(2016). Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 44: W236–W241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger-Stevic J., Zimmermann P., Lecoq L., Böttcher B., Nassal M.(2018). Hepatitis B virus core protein phosphorylation: Identification of the SRPK1 target sites and impact of their occupancy on RNA binding and capsid structure. PLoS Pathog. 14: e1007488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose S., Shimizu K., Kanai S., Kuroda Y., Noguchi T.(2007). POODLE-L: A two-level SVM prediction system for reliably predicting long disordered regions. Bioinformatics 23: 2046–2053. [DOI] [PubMed] [Google Scholar]
- Hoover H.S., Kao C.C.(2016). Phosphorylation of the viral coat protein regulates RNA virus infection. Virus Adapt. Treat. 8: 13–20. [Google Scholar]
- Hu Y., Li Z., Yuan C., Jin X., Yan L., Zhao X., Zhang Y., Jackson A.O., Wang X., Han C., Yu J., Li D.(2015). Phosphorylation of TGB1 by protein kinase CK2 promotes barley stripe mosaic virus movement in monocots and dicots. J. Exp. Bot. 66: 4733–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang L.N., Englund N., Das T., Banerjee A.K., Pattnaik A.K.(1999). Optimal replication activity of vesicular stomatitis virus RNA polymerase requires phosphorylation of a residue(s) at carboxy-terminal domain II of its accessory subunit, phosphoprotein P. J. Virol. 73: 5613–5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T., Kinoshita K.(2007). PrDOS: Prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 35: W460–W464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I., Yabukarski F., Ruigrok R.W.H., Jamin M.(2011). Structural insights into the rhabdovirus transcription/replication complex. Virus Res. 162: 126–137. [DOI] [PubMed] [Google Scholar]
- Jackson A.O., Dietzgen R.G., Goodin M.M., Bragg J.N., Deng M.(2005). Biology of plant rhabdoviruses. Annu. Rev. Phytopathol. 43: 623–660. [DOI] [PubMed] [Google Scholar]
- Jackson A.O., Li Z.(2016). Developments in plant negative-strand RNA virus reverse genetics. Annu. Rev. Phytopathol. 54: 469–498. [DOI] [PubMed] [Google Scholar]
- Jakubiec A., Jupin I.(2007). Regulation of positive-strand RNA virus replication: The emerging role of phosphorylation. Virus Res. 129: 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knippschild U., Gocht A., Wolff S., Huber N., Löhler J., Stöter M.(2005). The casein kinase 1 family: Participation in multiple cellular processes in eukaryotes. Cell. Signal. 17: 675–689. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K.(2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Chen R., Lee Y., Yoo S., Lee C.(2009). Essential roles of CKIdelta and CKIepsilon in the mammalian circadian clock. Proc. Natl. Acad. Sci. USA 106: 21359–21364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Taoka K., Yoo B.C., Ben-Nissan G., Kim D.J., Lucas W.J.(2005). Plasmodesmal-associated protein kinase in tobacco and Arabidopsis recognizes a subset of non-cell-autonomous proteins. Plant Cell 17: 2817–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyrat C., Ribeiro E.A., Gerard F.C.A., Ivanov I., Ruigrok R.W.H., Jamin M.(2011). Structure, interactions with host cell and functions of rhabdovirus phosphoprotein. Future Virol. 6: 465–481. [Google Scholar]
- Li L., Stoeckert C.J. Jr., Roos D.S.(2003). OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 13: 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linding R., Jensen L.J., Diella F., Bork P., Gibson T.J., Russell R.B.(2003). Protein disorder prediction: Implications for structural proteomics. Structure 11: 1453–1459. [DOI] [PubMed] [Google Scholar]
- Liu W., Xu Z.-H., Luo D., Xue H.-W.(2003). Roles of OsCKI1, a rice casein kinase I, in root development and plant hormone sensitivity. Plant J. 36: 189–202. [DOI] [PubMed] [Google Scholar]
- Mann K.S., Dietzgen R.G.(2014). Plant rhabdoviruses: New insights and research needs in the interplay of negative-strand RNA viruses with plant and insect hosts. Arch. Virol. 159: 1889–1900. [DOI] [PubMed] [Google Scholar]
- Mei Y., Wang Y., Hu T., Yang X., Lozano-Duran R., Sunter G., Zhou X.(2018). Nucleocytoplasmic shuttling of Geminivirus C4 protein mediated by phosphorylation and myristoylation is critical for viral pathogenicity. Mol. Plant 11: 1466–1481. [DOI] [PubMed] [Google Scholar]
- Millar A.H., Heazlewood J.L., Giglione C., Holdsworth M.J., Bachmair A., Schulze W.X.(2019). The scope, functions, and dynamics of posttranslational protein modifications. Annu. Rev. Plant Biol. 70: 119–151. [DOI] [PubMed] [Google Scholar]
- Mondal A., Victor K.G., Pudupakam R.S., Lyons C.E., Wertz G.W.(2014). Newly identified phosphorylation site in the vesicular stomatitis virus P protein is required for viral RNA synthesis. J. Virol. 88: 1461–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer S.A., Smallwood-Kentro S., Haddad A., Prevec L.(1991). Assembly and transcription of synthetic vesicular stomatitis virus nucleocapsids. J. Virol. 65: 2170–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obradovic Z., Peng K., Vucetic S., Radivojac P., Dunker A.K.(2005). Exploiting heterogeneous sequence properties improves prediction of protein disorder. Proteins 61: 176–182. [DOI] [PubMed] [Google Scholar]
- Pattnaik A.K., Hwang L., Li T., Englund N., Mathur M., Das T., Banerjee A.K.(1997). Phosphorylation within the amino-terminal acidic domain I of the phosphoprotein of vesicular stomatitis virus is required for transcription but not for replication. J. Virol. 71: 8167–8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck S.C.(2006). Analysis of protein phosphorylation: Methods and strategies for studying kinases and substrates. Plant J. 45: 512–522. [DOI] [PubMed] [Google Scholar]
- Qanungo K.R., Shaji D., Mathur M., Banerjee A.K.J.P.N.A.S.(2004). Two RNA polymerase complexes from vesicular stomatitis virus-infected cells that carry out transcription and replication of genome RNA. Proc. Natl. Acad. Sci. USA 101: 5952–5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmeh A.A., Morin B., Schenk A.D., Liang B., Heinrich B.S., Brusic V., Walz T., Whelan S.P.J.(2012). Critical phosphoprotein elements that regulate polymerase architecture and function in vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA 109: 14628–14633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rena G., Bain J., Elliott M., Cohen P.(2004). D4476, a cell-permeant inhibitor of CK1, suppresses the site-specific phosphorylation and nuclear exclusion of FOXO1a. EMBO Rep. 5: 60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K., Hirose S., Noguchi T.(2007). POODLE-S: Web application for predicting protein disorder by using physicochemical features and reduced amino acid set of a position-specific scoring matrix. Bioinformatics 23: 2337–2338. [DOI] [PubMed] [Google Scholar]
- Spolar R.S., Record M.T. Jr.(1994). Coupling of local folding to site-specific binding of proteins to DNA. Science 263: 777–784. [DOI] [PubMed] [Google Scholar]
- Takamatsu F., Asakawa N., Morimoto K., Takeuchi K., Eriguchi Y., Toriumi H., Kawai A.(1998). Studies on the rabies virus RNA polymerase: 2. Possible relationships between the two forms of the non-catalytic subunit (P protein). Microbiol. Immunol. 42: 761–771. [DOI] [PubMed] [Google Scholar]
- Tan S.T., Dai C., Liu H.-T., Xue H.-W.(2013). Arabidopsis casein kinase1 proteins CK1.3 and CK1.4 phosphorylate cryptochrome2 to regulate blue light signaling. Plant Cell 25: 2618–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S.T., Xue H.W.(2014). Casein kinase 1 regulates ethylene synthesis by phosphorylating and promoting the turnover of ACS5. Cell Rep. 9: 1692–1702. [DOI] [PubMed] [Google Scholar]
- Tuffereau C., Fischer S., Flamand A.(1985). Phosphorylation of the N and M1 proteins of rabies virus. J. Gen. Virol. 66: 2285–2289. [DOI] [PubMed] [Google Scholar]
- Uversky V.N., Oldfield C.J., Dunker A.K.(2005). Showing your ID: Intrinsic disorder as an ID for recognition, regulation and cell signaling. J. Mol. Recognit. 18: 343–384. [DOI] [PubMed] [Google Scholar]
- Vullo A., Bortolami O., Pollastri G., Tosatto S.C.E.(2006). Spritz: A server for the prediction of intrinsically disordered regions in protein sequences using kernel machines. Nucleic Acids Res. 34: W164–W168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker P.J., Dietzgen R.G., Joubert D.A., Blasdell K.R.(2011). Rhabdovirus accessory genes. Virus Res. 162: 110–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A.(2015). Dissecting the molecular network of virus-plant interactions: The complex roles of host factors. Annu. Rev. Phytopathol. 53: 45–66. [DOI] [PubMed] [Google Scholar]
- Wang Q., Ma X., Qian S., Zhou X., Sun K., Chen X., Zhou X., Jackson A.O., Li Z.(2015). Rescue of a plant negative-strand RNA virus from cloned cDNA: Insights into enveloped plant virus movement and morphogenesis. PLoS Pathog. 11: e1005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright P.E., Dyson H.J.(2009). Linking folding and binding. Curr. Opin. Struct. Biol. 19: 31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan T., Zhu J.R., Di D., Gao Q., Zhang Y., Zhang A., Yan C., Miao H., Wang X.B.(2015). Characterization of the complete genome of Barley yellow striate mosaic virus reveals a nested gene encoding a small hydrophobic protein. Virology 478: 112–122. [DOI] [PubMed] [Google Scholar]
- Yang M., Zhang Y., Xie X., Yue N., Li J., Wang X.B., Han C., Yu J., Liu Y., Li D.(2018). Barley stripe mosaic virus γb protein subverts autophagy to promote viral infection by disrupting the ATG7-ATG8 Interaction. Plant Cell 30: 1582–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.R., Thomson R., McNeil P., Esnouf R.M.(2005). RONN: The bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics 21: 3369–3376. [DOI] [PubMed] [Google Scholar]
- Zhang K., Zhang Y., Yang M., Liu S., Li Z., Wang X., Han C., Yu J., Li D.(2017a). The Barley stripe mosaic virus γb protein promotes chloroplast-targeted replication by enhancing unwinding of RNA duplexes. PLoS Pathog. 13: e1006319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Chen L., Zhang G., Yan Q., Yang X., Ding B., Tang Q., Sun S., Hu Z., Chen M.(2013). An amino acid of human parainfluenza virus type 3 nucleoprotein is critical for template function and cytoplasmic inclusion body formation. J. Virol. 87: 12457–12470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Faraggi E., Xue B., Dunker A.K., Uversky V.N., Zhou Y.(2012). SPINE-D: Accurate prediction of short and long disordered regions by a single neural-network based method. J. Biomol. Struct. Dyn. 29: 799–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Dong K., Xu K., Zhang K., Jin X., Yang M., Zhang Y., Wang X., Han C., Yu J., Li D.(2018). Barley stripe mosaic virus infection requires PKA-mediated phosphorylation of γb for suppression of both RNA silencing and the host cell death response. New Phytol. 218: 1570–1585. [DOI] [PubMed] [Google Scholar]
- Zhang X.-P., Liu D.-S., Yan T., Fang X.-D., Dong K., Xu J., Wang Y., Yu J.-L., Wang X.-B.(2017b). Cucumber mosaic virus coat protein modulates the accumulation of 2b protein and antiviral silencing that causes symptom recovery in planta. PLoS Pathog. 13: e1006522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.-J., et al. (2020). CCR4, a RNA decay factor, is hijacked by a plant cytorhabdovirus phosphoprotein to facilitate virus replication. eLife 9: e53753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Wang X., Dong K., Zhang Y., Hu Y., Zhang X., Chen Y., Wang X., Han C., Yu J., Li D.(2015). Phosphorylation of Beet black scorch virus coat protein by PKA is required for assembly and stability of virus particles. Sci. Rep. 5: 11585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Wang Z.Q., Xiao R., Cao L., Wang Y., Xie Y., Zhou X.(2017). Mimic phosphorylation of a βC1 protein encoded by TYLCCNB impairs its functions as a viral suppressor of RNA silencing and a symptom determinant. J. Virol. 91: e00300–e00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Shibasaki F., Price R., Guillemot J.-C., Yano T., Dötsch V., Wagner G., Ferrara P., McKeon F.(1998). Intramolecular masking of nuclear import signal on NF-AT4 by casein kinase I and MEKK1. Cell 93: 851–861. [DOI] [PubMed] [Google Scholar]
- Zuo J., Niu Q.-W., Chua N.-H.(2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265–273. [DOI] [PubMed] [Google Scholar]