The phosphoregulatory region of citrus MAF1 not only contributes to, but also regulates the binding of MAF1 to the C34 subunit of RNA polymerase III.
Abstract
MAF1 is a phosphoprotein that plays a critical role in cell growth control as the central regulator of RNA polymerase (Pol) III activity. Citrus MAF1 (CsMAF1) was identified as a direct target of PthA4, a bacterial effector protein required to induce tumors in citrus. CsMAF1 binds to Pol III to restrict transcription; however, exactly how CsMAF1 interacts with the polymerase and how phosphorylation modulates this interaction is unknown. Moreover, how CsMAF1 binds PthA4 is also obscure. Here we show that CsMAF1 binds predominantly to the WH1 domain of the citrus Pol III subunit C34 (CsC34) and that its phosphoregulatory region, comprising loop-3 and α-helix-2, contributes to this interaction. We also show that phosphorylation of this region decreases CsMAF1 affinity to CsC34, leading to Pol III derepression, and that Ser 45, found only in plant MAF1 proteins, is critical for CsC34 interaction and is phosphorylated by a new citrus AGC1 kinase. Additionally, we show that the C-terminal region of the citrus TFIIIB component BRF1 competes with CsMAF1 for CsC34 interaction, whereas the C-terminal region of CsMAF1 is essential for PthA4 binding. Based on CsMAF1 structural data, we propose a mechanism for how CsMAF1 represses Pol III transcription and how phosphorylation controls this process.
INTRODUCTION
MAF1 plays an essential role in eukaryotes as the central regulator of RNA polymerase (Pol) III activity. Through its direct interaction with components of the Pol III machinery, MAF1 represses Pol III transcription in response to stress and nutrient availability, thus optimizing the cellular resources needed for ribosome biogenesis, protein synthesis and cell growth (Desai et al., 2005; Reina et al., 2006; Cieśla et al., 2007; Rollins et al., 2007; Willis and Moir, 2007; Rideout et al., 2012; Leśniewska and Boguta, 2017).
Since its discovery as a Pol III repressor in yeast (Saccharomyces cerevisiae; Boguta et al., 1997), MAF1 has been studied in several organisms and shown to play many physiological roles associated with cell growth that go beyond the regulation of tRNA and 5S rRNA synthesis (Khanna et al., 2015; Zhang et al., 2018). Loss of MAF1 in Drosophila led, for instance, to an increase in body size, whereas in mice it conferred obesity resistance and a lean phenotype due to changes in energy metabolism (Rideout et al., 2012; Bonhoure et al., 2015; Willis, 2018). MAF1 also differentially affected the lifespan in yeast and animals (Bonhoure et al., 2015; Cai and Wei, 2016; Shetty et al., 2020), and induced adipocyte differentiation (Chen et al., 2018), mediated T cell priming (Reverendo et al., 2019), and ameliorated cardiac hypertrophy (Sun et al., 2019) in mice. Additionally, MAF1 displayed tumor-suppressor activity in several mammalian cells and its down-regulation has been linked to liver carcinomas (Palian et al., 2014; Li et al., 2016).
The role of MAF1 in plants in response to stress and pathogen attack has also been investigated (Soprano et al., 2013, 2017; Ahn et al., 2019). The Citrus sinensis MAF1 (CsMAF1) was originally identified as an interactor of PthA4, a transcription activator-like (TAL) effector from the citrus canker pathogen, Xanthomonas citri, that is essential to induce cankers on citrus (Soprano et al., 2013; Abe and Benedetti, 2016). Silencing of CsMAF1 increased tRNA transcription and cell proliferation during citrus canker development triggered by PthA4, whereas CsMAF1 overexpression impaired seedling growth, inhibited tRNA synthesis, and attenuated canker pustule formation in sweet orange plants (Soprano et al., 2013). These findings show that CsMAF1 also acts as a Pol III repressor and suppressor of cell growth in plants. However, how PthA4 interacts with CsMAF1 to promote disease in citrus is presently unknown.
MAF1 is a phosphoprotein whose activity is regulated by reversible phosphorylation reactions. Yeast cells employ several protein kinases, including Protein kinase A (PKA), Target of rapamycin complex 1, and Ser/threonine-protein kinase SCH9 (SCH9), to regulate MAF1 subcellular localization and activity, whereas mammalian cells inactivate MAF1 through mechanistic target of rapamycin (mTOR)-dependent phosphorylation. Nonetheless, in both yeast and mammalian cells, the underlying mechanism of Pol III repression is that MAF1 binds to Pol III in its dephosphorylated or hypophosphorylated state (Moir et al., 2006; Oficjalska-Pham et al., 2006; Roberts et al., 2006; Towpik et al., 2008; Kantidakis et al., 2010; Michels et al., 2010; Shor et al., 2010; Michels, 2011; Oler and Cairns, 2012).
The TOR kinase has also been implicated in the regulation of MAF1 activity in plants in response to biotic and abiotic stresses (Soprano et al., 2017; Ahn et al., 2019). In citrus, the mTOR inhibitor AZD8055 blocked canker development induced by X. citri in a CsMAF1-dependent manner, suggesting that CsMAF1 is regulated by a citrus mTOR homologue, an idea that is supported by the fact that recombinant mTOR phosphorylates CsMAF1 at sites that are conserved between human MAF1 (hMAF1) and CsMAF1 (Soprano et al., 2017, 2018). Moreover, CsMAF1 was also shown to be phosphorylated in vitro by murine PKA at Ser 45 (S45), a residue that is conserved only in plant MAF1 proteins and which controls CsMAF1 nucleolar localization (Soprano et al., 2017, 2018).
The crystal structures of hMAF1 and CsMAF1 show that these proteins share the same fold characterized by a single globular α-β sandwich domain (Vannini et al., 2010; Soprano et al., 2017). However, these structures lack two critical regulatory regions: the phosphorylation region that includes the phospholoop, where the mTOR sites are located, and the C-terminal region (Vannini et al., 2010; Lee et al., 2015; Pradhan et al., 2017; Soprano et al., 2017). A recently published structure of yeast MAF1 (ScMAF1) also lacks these structural elements (Vorländer et al., 2020). Consistent with the fact that neither hMAF1 nor CsMAF1 crystallized with these regions, hydrogen-deuterium exchange experiments combined with limited proteolysis and SEC-MALS analysis revealed that the phospholoop region and C-terminal region of CsMAF1 and hMAF1 are flexible, further suggesting that MAF1 proteins are structurally conserved and similarly regulated between plants and mammals (Soprano et al., 2017).
The precise molecular mechanism by which MAF1 represses Pol III transcription is not fully understood. Early genetic and biochemical studies revealed that ScMAF1 interacts with the Pol III subunits C160, C82, AC40, and C34 and with the TFIIIB component BRF1 (Pluta et al., 2001; Desai et al., 2005). Human MAF1 also physically associates with BRF1 and with the largest Pol III subunit RPC1 in glutathione S-transferase (GST)-pulldown and coimmunoprecipitation assays (Reina et al., 2006; Rollins et al., 2007; Goodfellow et al., 2008). Crystallographic studies of hMAF1 combined with low-resolution Cryo-EM structures of yeast Pol III in the presence and absence of MAF1 suggested that MAF1 binds to the Pol III clamp domain and rearranges the C82/34/31 subcomplex close to the active site cleft (Vannini et al., 2010). More recently, a higher resolution Cryo-EM structure of yeast Pol III in complex with ScMAF1 revealed that ScMAF1 interacts with the C34 subunit precluding the binding of TFIIIB (Vorländer et al., 2020). However, because the structure of ScMAF1 bound to Pol III also lacks the critical phosphoregulatory region, the mechanism by which phosphorylation of MAF1 affects its interaction with Pol III is still a mystery.
Here we show that CsMAF1 binds mostly to the WH1 domain of the citrus Pol III subunit C34 (CsC34) and that its phospholoop region and adjacent α-helix 2 contributes to this interaction. In addition, we show that the conserved S45 residue is critical for CsMAF1-CsC34 interaction and is specifically phosphorylated by a new citrus AGC1 kinase. This residue sits in the α-helix-2, which harbors other phosphosites, and which, according to our structural models, approaches the CsC34 WH1 domain. Moreover, we show that the C-terminal region of citrus BRF1 (CsBRF1) competes with CsMAF1 for CsC34 interaction. Based on these results and on the crystal structures of CsMAF1, we propose a model in which progressive phosphorylation of the phospholoop and α-helix-2 disrupts the interaction of CsMAF1 with CsC34. Furthermore, our data showing that CsMAF1 binds PthA4 with high affinity supports the idea that PthA4 targets CsMAF1 to promote cell growth in citrus.
RESULTS
The C-terminal Domain of CsMAF1 Is Essential for PthA4 Interaction
Prior studies showed that CsMAF1 interacted specifically with the TAL effector PthA4 in yeast two-hybrid assays (Soprano et al., 2013). To map the CsMAF1 and PthA4 regions responsible for this interaction, we performed far-protein gel blot (FWB) analysis using truncated forms of CsMAF1 and PthA4 (Figure 1A). In line with yeast two-hybrid and GST-pulldown assays (Soprano et al., 2013), full-length CsMAF1 interacted with PthA4 but not with PthA2, thus validating our FWB assays (Figure 1B).
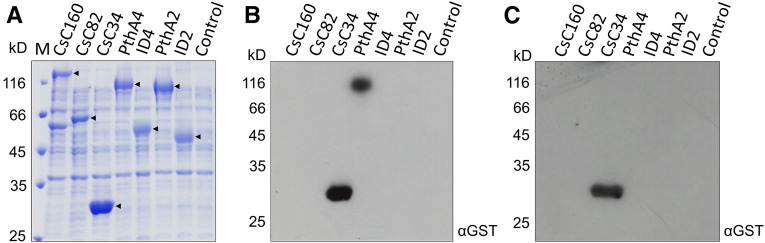
Figure 1.
CsMAF1 Interacts with the TAL Effector PthA4 and with the Citrus RNA Pol III C34 Subunit.
(A) SDS-PAGE gel of IPTG-induced E. coli cell extracts showing major bands corresponding to the expressed proteins CsC160, CsC82, CsC34, PthA4, ID4, PthA2, and ID2 (arrowheads), compared with a control cell extract with no expression plasmid.
(B) FWB analysis showing that full-length CsMAF1 interacted with CsC34 and PthA4, but not with PthA2 or with the internal DNA binding domains of PthA4 (ID4) and PthA2 (ID2).
(C) FWB analysis showing that the C-terminal region of CsMAF1 is required for PthA4 but not CsC34 interaction. Blots were probed with an anti-GST serum (α-GST) to detect the GST-CsMAF1 (B) and GST-CsMAF1_dCT (C) fusions.
Because the C-terminal region of CsMAF1 is negatively charged and unfolded in solution, we postulated that this region might interact with the internal DNA binding domain of PthA4 (ID4; Soprano et al., 2017, 2018). The results show, nevertheless, that CsMAF1 does not interact with ID4 or with the internal DNA binding domain of PthA2 (ID2), used as negative control (Figure 1B). However, when the C-terminal region of CsMAF1 was deleted (CsMAF1_dCT mutant), no interaction with PthA4 was observed, indicating that CsMAF1 does bind PthA4 through its flexible C-terminal domain (Figure 1C).
The interaction of CsMAF1 with PthA4 was further examined by NMR heteronuclear single quantum coherence (HSQC) experiments using 15N-labeled CsMAF1. As shown in Figure 2A, several amino acid residues in CsMAF1 showed substantial chemical shift changes or a decrease in the NMR signal in the presence of PthA4, indicating that the two proteins interact in solution at low micromolar concentrations. Conversely, no significant chemical shift differences or signal intensity in the HSQC spectra were observed when CsMAF1_dCT was titrated with PthA4 (Figure 2B), thus confirming that the C-terminal domain of CsMAF1 is required for PthA4 binding.
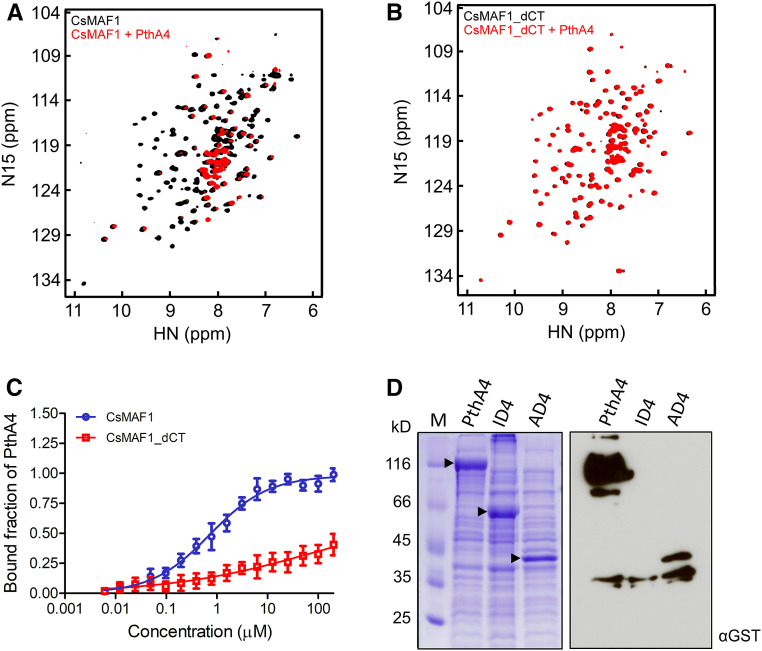
Figure 2.
The C-terminal Domain of CsMAF1 Is Required for PthA4 Interaction.
(A) NMR HSQC spectrum of 15N-labeled CsMAF1 showing the interaction of CsMAF1 with PthA4 in solution, as revealed by the NMR signal depletion and changes in the chemical shifts of several CsMAF1 residues in the presence of PthA4 (compare black signals of CsMAF1 alone with red signals of CsMAF1 plus PthA4).
(B) NMR HSQC spectrum of 15N-labeled CsMAF1_dCT showing that the interaction of CsMAF1_dCT with PthA4 is lost when the C-terminal domain of CsMAF1 is removed, as indicated by the restoration of the NMR signals and chemical shifts of CsMAF1.
(C) MST assays using NHS-labeled PthA4 showing that full-length CsMAF1 has a PthA4 binding affinity of ∼0.7 µM ± 80 nM (blue curve). Error bars denote standard deviations from the mean of three measurements. No binding affinity could be measured when PthA4 was titrated against CsMAF1 lacking its C-terminal domain (red curve).
(D) SDS-PAGE gel of IPTG-induced E. coli cell extracts expressing full-length PthA4, or the respective ID4 and AD4 domains (arrowheads), coupled with an FWB assay showing that the activation domain of PthA4 is sufficient to interact with CsMAF1. The blot was probed with an anti-GST serum to detect GST-CsMAF1.
To determine the binding affinity of CsMAF1 to PthA4, CsMAF1 was titrated against N-hydroxysuccinimide (NHS)–labeled PthA4 in microscale thermophoresis (MST) assays. The binding curves showed that while full-length CsMAF1 has a PthA4 binding affinity of ∼0.7 µM ± 80 nM, no binding affinity could be measured when PthA4 was titrated against the CsMAF1_dCT mutant (Figure 2C).
To finely map the PthA4 region responsible for CsMAF1 binding, the C-terminal region of PthA4 (residues 884–1163), referred to here as the transcriptional activation domain (AD4), was probed against CsMAF1 in FWB assays in comparison with ID4 and the full-length protein. In line with our yeast-two hybrid assays (Soprano et al., 2013), the AD4 region was sufficient to interact with CsMAF1; nevertheless, this interaction was significantly weaker than that observed with the full-length protein (Figure 2D). Thus, despite the existence of polymorphic residues in the C-terminal region of PthAs, particularly between PthAs 2 and 4, which could contribute to the specific interaction of PthA4 with CsMAF1 (Supplemental Figure 1), our data suggest that full-length PthA4 is required for a strong interaction with CsMAF1.
Altogether, the results show that the C-terminal domain of CsMAF1 is essential for PthA4 binding and that the C-terminal domain of PthA4 contributes to this binding.
CsMAF1 Predominantly Binds the WH1 Domain of the Citrus Pol III C34 Subunit
Genetic, biochemical, and structural studies have shown that yeast and mammalian MAF1 interact with several Pol III components including the C160, C82, and C34 subunits (Pluta et al., 2001; Reina et al., 2006; Vannini et al., 2010; Vorländer et al., 2020). Thus, to test whether CsMAF1 could also interact with any of these Pol III subunits, the corresponding Citrus sinensis CsC160, CsC82, and CsC34 proteins were expressed in bacteria and probed in FWB assays (Figure 1). The results show that CsMAF1 interacts only with the CsC34 subunit of the citrus RNA Pol III (Figure 1B) and that its flexible C-terminal domain is not required for this interaction (Figure 1C).
To further characterize the CsMAF1-CsC34 interaction, HSQC experiments were performed using the 15N-labled CsMAF1_dCT mutant protein. CsMAF1_dCT was used instead of full-length CsMAF1 because the C-terminal region of CsMAF1 was not required for CsC34 interaction (Figure 1C). In addition, since the C-terminal region of CsMAF1 is unfolded in solution (Soprano et al., 2017), the CsMAF1_dCT protein showed a better distribution of the NMR signals in the HSQC spectra compared with full-length CsMAF1 (Supplemental Figure 2).
The HSQC experiments revealed that several residues of CsMAF1_dCT undergo substantial chemical shift changes or a decrease in signal intensity in the presence of increasing amounts of CsC34 (Figure 3A). Such changes were, however, not observed when CsMAF1_dCT was titrated with NusA used as a tag for expression of soluble CsC34 (Figure 3B). These results thus confirm that CsMAF1 binds the C34 subunit of the citrus RNA Pol III at low micromolar concentrations.
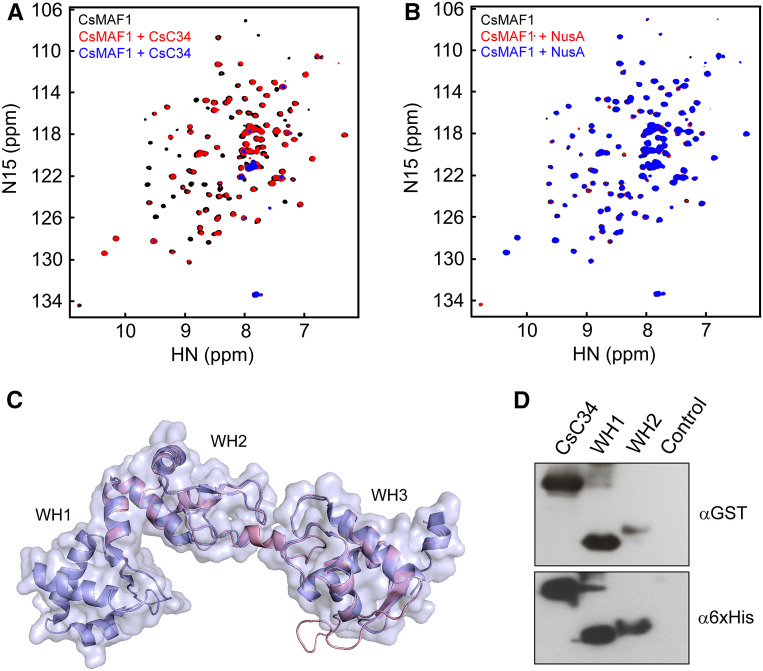
Figure 3.
CsC34 Interacts with CsMAF1 via its WH1 Domain.
(A) NMR HSQC spectrum of 15N-labeled-CsMAF1 (150 µM) in the presence of 15 µM (red signals) or 30 µM CsC34 (blue signals), compared with CsMAF1 alone (black signals), showing that CsMAF1 binds CsC34 in solution, as revealed by the NMR signal depletion and changes in the chemical shifts of several CsMAF1 residues in the presence of CsC34.
(B) NMR HSQC spectrum of 15N-labeled CsMAF1 in the presence of 15 µM (red signals) or 30 µM NusA (blue signals), compared with CsMAF1 alone (black signals). The absence of significant changes in the chemical shifts or signal intensities of the NMR peaks indicates lack of interaction between CsMAF1 and NusA, used as a tag for CsC34 expression.
(C) Diagrammatic representation of the structural alignment between the cryo-EM structure of the yeast RNA Pol III C34 subunit (PDB ID 5FJ8, light blue) and the CsC34 model (light pink) showing that the WH1 and WH2 domains of CsC34 align with the WH2 and WH3 domains of yeast C34.
(D) FWB assay showing that CsC34 interacts with CsMAF1 mainly via its WH1 domain. The FWB was probed with the anti-GST serum (α-GST) to detect the GST-CsMAF1 protein, whereas the loading control blot was probed with the anti-6xHis (α-6xHis) serum. A bacterial cell extract with no expression plasmid was used as control.
CsC34 is a 25.9-kD protein that is homologous to many plant proteins annotated as ‘DNA-directed RNA Pol III subunit RPC6’, which have not been functionally or structurally characterized yet (Supplemental Figure 3). When compared with the yeast and human C34 subunits that have three winged-helix (WH) domains, CsC34 has only two WH domains, a feature that appears to be conserved among plant C34 proteins (Supplemental Figure 3). Protein sequence alignments and structural modeling studies using the yeast C34 structure as a template model (PDB ID 5FJ8; Hoffmann et al., 2015) indicate that the WH1 and WH2 domains of CsC34 correspond to the WH2 and WH3 domains of yeast C34, respectively (Figure 3C; Supplemental Figure 3). In addition, we found that CsC34 also carries the four C-terminal Cys residues corresponding to the zinc-finger motif commonly found in C34 proteins (Supplemental Figure 3; Blombach et al., 2009).
To determine the contribution of the CsC34 WH domains to the CsMAF1 interaction, FWB assays were performed with each of the domains expressed separately. We found that CsMAF1 predominantly interacted with the WH1 domain of CsC34 (Figure 3D).
The CsMAF1 Phospholoop Contributes to CsC34 Interaction
The well-resolved 15N-HSQC spectra obtained for CsMAF1_dCT (Figure 3) prompted us to assign the amino acid residues involved in CsC34 binding. Surprisingly, however, when CsMAF1_dCT was triple-labeled with 2H, 15N, and 13C, many of the peaks that showed chemical shift changes or a decrease in signal in the presence of CsC34 (Figure 3A) were undetected in the NMR spectra in the absence of CsC34 (Supplemental Figure 4). Despite several attempts to improve the production of CsMAF1_dCT in deuterated medium, the problem with specific NMR signal depletion with the triple-labeled protein persisted. Moreover, the low stability of CsMAF1_dCT over time under the NMR experimental conditions also precluded data collection from experiments lasting more than 36 h.
To overcome these limitations and gain insights into the CsMAF1 region responsible for CsC34 binding, a competition approach was used to identify peptides that could prevent CsMAF1 from interacting with CsC34. Based on our previous work, the flexible phospholoop, comprising residues GKSLDTDSSSPAEFLLSRSS (Figure 4A), was a candidate region for CsC34 interaction because this region carries the conserved mTOR phosphorylation sites (S59, T62, and S66) shared with hMAF1 (Kantidakis et al., 2010; Shor et al., 2010; Michels, 2011; Soprano et al., 2017, 2018). In fact, in addition to S59, previously identified as an mTOR site (Soprano et al., 2017), we show here that S66 and S47 are also phosphorylated by mTOR in vitro (Supplemental Figure 5).
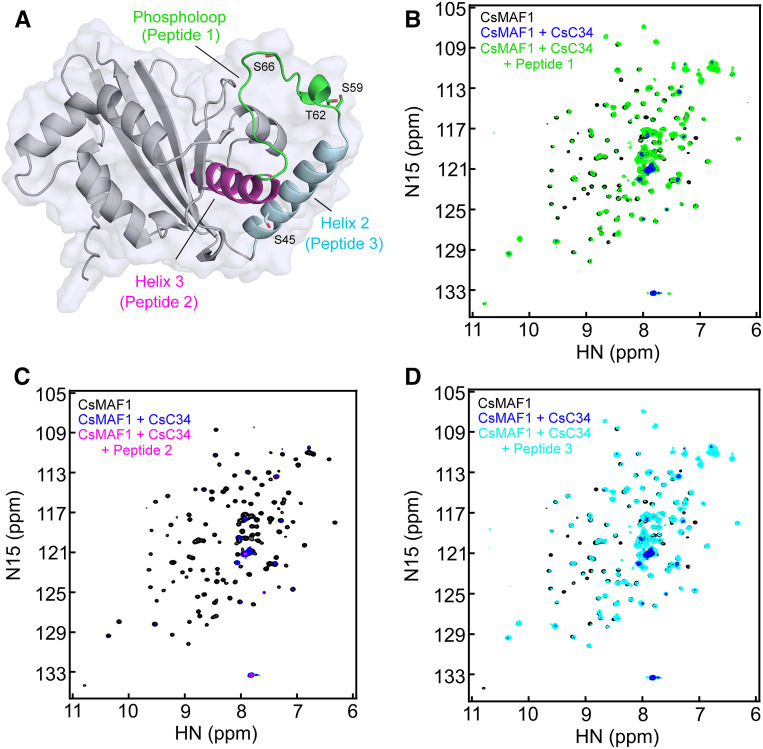
Figure 4.
The Phospholoop and α-Helix-2 of CsMAF1 Are Important for CsC34 Binding.
(A) Diagrammatic representation of the CsMAF1 crystal structure (PDB ID 5U4Z) depicting the α-helices 2 (cyan) and 3 (magenta) flanking the phospholoop (green), which was modeled in this representation. The phospholoop residues mapped as mTOR sites in both hMAF1 and CsMAF1, as well as S45, identified as a PKA and CsPK3 site, are shown as sticks.
(B) NMR HSQC spectrum of 15N-labeled CsMAF1 in the presence of CsC34 (blue signals), or CsC34 plus peptide 1 corresponding to the CsMAF1 phospholoop (green), compared to CsMAF1 alone (black signals). Peptide 1 significantly restored the CsMAF1 signals depleted by CsC34, indicating that the phospholoop competes with CsMAF1 for CsC34 binding.
(C) HSQC spectrum of 15N-labeled CsMAF1 (black signals) in the presence of CsC34 (blue signals) showing that peptide 2 corresponding to the α-helix-3 does not compete with CsMAF1 for CsC34 binding (magenta signals).
(D) HSQC spectrum of 15N-labeled CsMAF1 (black signals) in the presence of CsC34 (blue signals) showing that peptide 3 also restored the CsMAF1 signals depleted by CsC34 (cyan signals), indicating that α-helix-2 is also important for CsC34 binding.
We found that the phospholoop peptide prevented the interaction of CsMAF1_dCT with CsC34, as most of the CsMAF1_dCT peaks remained unchanged in the HSQC spectra in the presence of this peptide (Figure 4B). On the other hand, the RKALIYLVLTLYHM peptide corresponding to the α-helix-3 (Figure 4A; Soprano et al., 2017) did not compete with CsMAF1_dCT for CsC34 binding (Figure 4C).
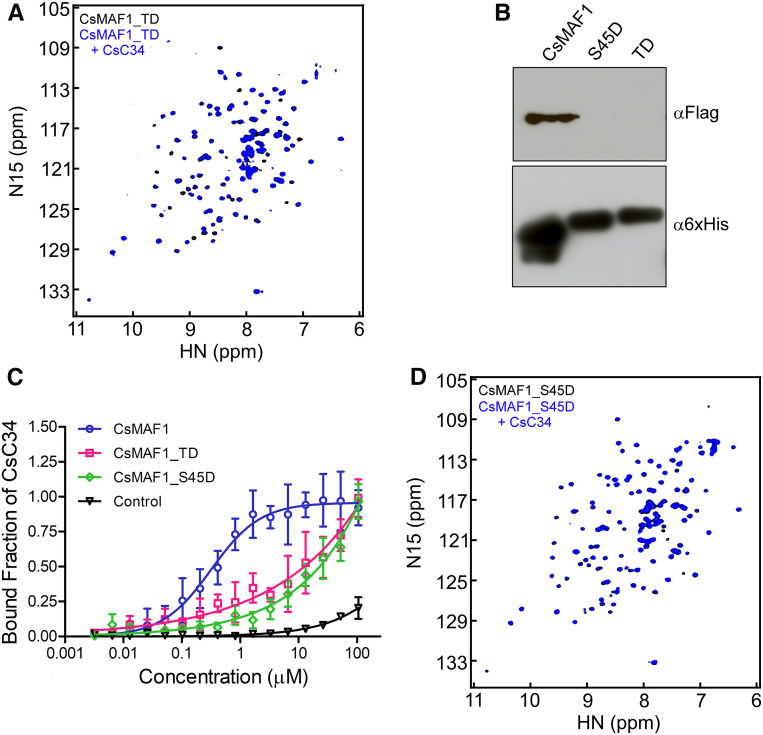
To further investigate the role of the phospholoop in CsMAF1-CsC34 interaction, we performed HSQC and FWB experiments using the phosphomimetic CsMAF1 triple D mutant (CsMAF1_TD), where the mTOR phosphosites S59, T62, and S66 were replaced by Asp (Soprano et al., 2017). We found that the CsMAF1_TD mutant did not interact with CsC34, as revealed by HSQC experiments (Figure 5A) and FWB assays (Figure 5B), thus indicating that phosphorylation of the phospholoop residues reduces the binding of CsMAF1 to CsC34. Accordingly, MST assays showed that while CsMAF1 has a binding affinity of ∼0.3 µM ± 50 nM to CsC34, no binding affinity could be measured between CsC34 and the phosphomimetic CsMAF1_TD mutant (Figure 5C). Although the HSQC spectra of CsMAF1_TD differ slightly from those of wild-type CsMAF1 (Supplemental Figure 6), which could indicate a change in protein conformation as a result of the amino acid substitutions, they clearly show that CsMAF1_TD is well-folded in solution. Thus, loss of interaction with CsC34 cannot be attributed to a misfolded protein.
Figure 5.
The Phosphomimetic Mutants CsMAF1_TD (TD) and CsMAF1_S45D do not Bind CsC34.
(A) NMR HSQC spectrum of 15N-labeled-CsMAF1_TD in the absence (black signals) and presence (blue signals) of CsC34 showing that the phosphomimetic CsMAF1_TD mutant did not bind CsC34 in solution.
(B) FWB assays showing that CsMAF1, but not the CsMAF1_TD or the CsMAF1_S45D mutant, interacted with CsC34. The FWB was probed with the anti-FLAG and anti-6xHis sera to detect CsC34 and the CsMAF1 proteins, respectively.
(C) MST assays showing that while CsMAF1 has a binding affinity of ∼0.3 µM ± 50 nM to CsC34, no binding affinity could be measured between CsC34 and the phosphomimetic mutants CsMAF1_TD and CsMAF1_S45D. EGFP was used as a negative control. Error bars denote standard deviations from the mean of three measurements.
(D) NMR HSQC spectrum of 15N-labeled-CsMAF1_S45 in the absence (black signals) and presence (blue signals) of CsC34 showing that CsMAF1_S45D did not also bind CsC34 in solution.
Together, the results show that the phospholoop is involved in CsC34 interaction and indicate that phosphorylation of S59, T62, and S66 is sufficient to substantially decrease the binding affinity of CsMAF1 to CsC34.
The Phosphomimetic CsMAF1_TD does not Repress Pol III Transcription in Yeast
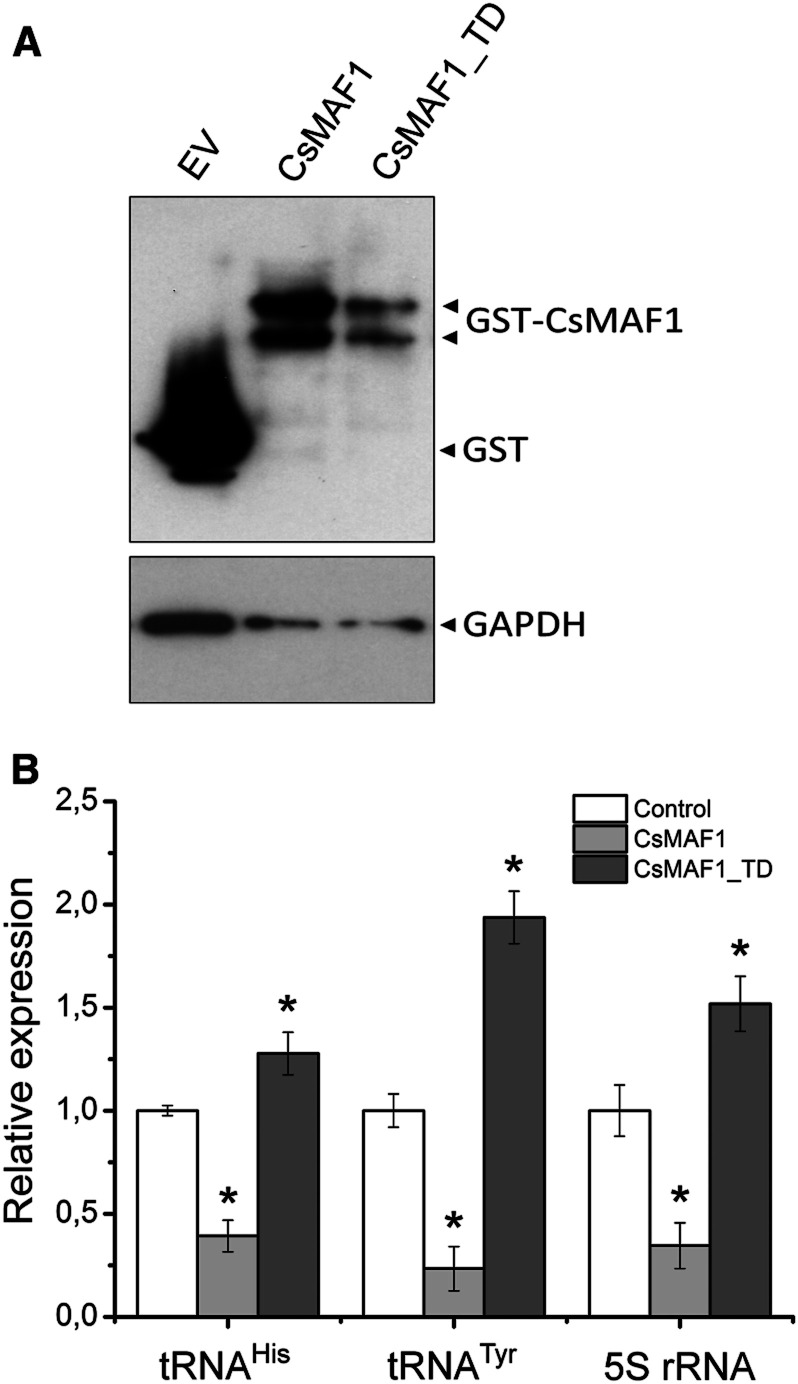
Considering the results shown above and the fact that CsMAF1 complements the maf1 mutation in yeast (Soprano et al., 2013), we tested whether the phosphomimetic CsMAF1_TD protein would repress RNA Pol III transcription in vivo. Thus, the wild-type CsMAF1 and CsMAF1_TD mutant proteins were each expressed in the yeast maf1- mutant (Figure 6A) and the accumulation of tRNAHis, tRNATyr, and 5S rRNA transcripts were quantified by RT-qPCR; Figure 6B). Contrary to the wild-type CsMAF1, CsMAF1_TD did not repress tRNAHis, tRNATyr, or 5S rRNA transcription in the yeast maf1- mutant (Figure 6B). In fact, we noticed that transcription of the tRNAHis, tRNATyr, and 5S rRNA genes was enhanced in the yeast cells expressing CsMAF1_TD relative to cells carrying the empty vector, used as a control (Figure 6B). These results thus indicate that phosphorylation of the phospholoop prevents CsMAF1 from binding to yeast Pol III.
Figure 6.
The CsMAF1_TD Mutant does not Repress Pol III Transcription in Yeast.
(A) Immunoblot analysis of the wild-type CsMAF1 and CsMAF1_TD proteins expressed in the yeast maf1− mutant as GST fusions, showing the accumulation of the fusion proteins (GST-MAF1) relative to GST alone derived from the empty vector (EV). The blot was probed with the anti-GST serum to detect the GST-MAF1 fusions, and with the anti-GAPDH serum for sample loading control.
(B) RT-qPCR analysis showing the relative expression levels of tRNAHis, tRNATyr, and 5S rRNA transcripts in yeast cells expressing the wild-type CsMAF1 or the CsMAF1_TD mutant, relative to GST alone (control). The plot shows that contrary to wild-type CsMAF1, the phosphomimetic CsMAF1_TD mutant did not repress tRNAHis, tRNATyr, or 5S rRNA transcription in the yeast maf1− mutant. Indeed, an increase in transcription of tRNAHis, tRNATyr, or 5S rRNA was observed in the yeast cells expressing CsMAF1_TD mutant. Error bars represent standard deviations, and asterisks denote statistically significant differences between the means at the 0.05 level, relative to control samples.
Ser 45 Is also Required for CsMAF1 Interaction with CsC34
Prior studies showed that CsMAF1 is also phosphorylated in vitro by murine PKA at S45. Because S45 is conserved only in plant MAF1 proteins and the phosphomimetic mutation S45D retarded CsMAF1 accumulation in the nucleolus, S45 phosphorylation is thought to also modulate CsMAF1 interaction with Pol III (Soprano et al., 2017).
To investigate the role of S45 in CsC34 binding, we performed FWB assays using the phosphomimetic CsMAF1_S45D mutant (Soprano et al., 2017). The results show that, like CsMAF1_TD, the CsMAF1_S45D mutant did not interact with CsC34 (Figure 5B). These results were confirmed by NMR experiments showing that many of the CsMAF1_S45D peaks remain in the HSQC spectra in the presence of CsC34 (Figure 5D). Moreover, we noticed that the HSQC profile of CsMAF1_S45D is more closely related to that of CsMAF1_TD than to the wild-type CsMAF1 (Supplemental Figure 6), suggesting that the two phosphomimetic proteins assume a similar conformation that differs from that of wild-type CsMAF1.
In agreement with this idea, the DKRLSISLEHEILDYL peptide corresponding to CsMAF1 α-helix 2 (Figure 4A; Soprano et al., 2017) also prevented the interaction of CsMAF1_dCT with CsC34 (Figure 4D). Moreover, as observed for CsMAF1_TD, the affinity of CsMAF1 to CsC34 was significantly affected by the S45D mutation (Figure 5C), indicating that phosphorylation of S45 also abrogates the binding of CsMAF1 to CsC34.
Identification a New Citrus AGC1 Kinase that Phosphorylates CsMAF1 at S45
Given that S45 is specifically phosphorylated by murine PKA and controls CsMAF1 subcellular localization (Soprano et al., 2017) and CsC34 interaction (Figures 4 and 5), we set out to identify the citrus PKA homologue that could modulate CsMAF1 activity in vivo. To this end, we retrieved all the C. sinensis sequences annotated as “Ser-Thr protein kinase” and selected those that likely belong to the AGC1 subfamily, also referred to as AGCVIII kinases, because members of this subfamily in plants are the closest relatives of mammalian PKAs (Bögre et al., 2003; Rademacher and Offringa, 2012). Candidate proteins with transmembrane domains were discarded and from the remaining set of sequences we selected those carrying one or more canonical nuclear localization signals (NLS). From this group, we chose four proteins named CsPK1–4 for further studies, none of which had been characterized in citrus plants before. Except for CsPK4, which could be classified as a member of the AGC2 subfamily based on its PIF (PDK1-interacting fragment) motif, all the CsPK proteins selected carry the same activation loop signature sequence (DFDx89SFVGTHEYLAPE) and PIF motif (FD/ExF) that characterize the AGC1 subfamily (Supplemental Figure 7; Bögre et al., 2003).
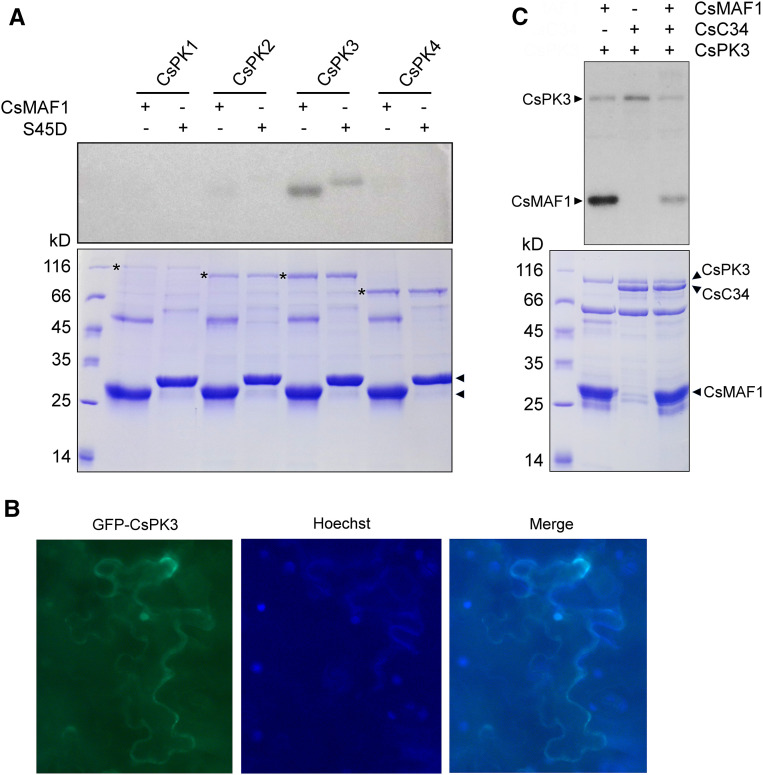
These putative kinases share a high degree of sequence identity within their C-terminal catalytic domain; however, they differ substantially in their N-terminal regions (Supplemental Figure 7), suggesting that they represent distinct functional homologs. Consistent with this, only CsPK3 phosphorylated CsMAF1 in vitro (Figure 7A).
Figure 7.
CsPK3 Phosphorylates CsMAF1 at S45 and Competes with CsC34 for CsMAF1 Binding.
(A) 32P-γATP-labeling experiment showing that, among the citrus AGC1 kinases tested, only CsPK3 phosphorylates CsMAF1 in in vitro reactions. The CsPK3-mediated phosphorylation of CsMAF1 is significantly reduced by the S45D mutation. A protein loading control gel (bottom) shows the expressed kinases (asterisks) and CsMAF1 proteins (arrowheads).
(B) Subcellular localization of GFP-CsPK3 in N. benthamiana epidermal cells, showing that GFP-CsPK3 accumulates in the cytoplasm and nucleus of transformed cells (left panel). Cells were stained with Hoechst for nuclei visualization (middle panel) and the GFP and Hoechst images taken at 200× magnification were merged (right panel). The subcellular localization of CsPK3 was observed in at least ten randomly infiltrated leaf sectors.
(C) 32P-γATP-labeling assay showing CsPK3 autophosphorylation and that CsC34 significantly reduces CsPK3-mediated CsMAF1 phosphorylation, suggesting that CsC34 and CsPK3 compete for CsMAF1 binding. A protein loading control gel (bottom) shows the corresponding bands for CsPK3, CsC34 and CsMAF1 (arrowheads).
CsPK3 is a 67 kD protein encoded by a single citrus gene that is close related to the Arabidopsis (Arabidopsis thaliana) D6PKL2 kinase involved in auxin transport and phototropic responses (Supplemental Figure 7; Zourelidou et al., 2009; Willige et al., 2013; Zourelidou et al., 2014). In the C. sinensis genome, CsPK2 is the closest relative of CsPK3, sharing 74% amino acid sequence identity with it (Supplemental Figure 7). Nevertheless, CsPK2 did not phosphorylate CsMAF1 in vitro (Figure 7A), suggesting that CsPK3 specifically phosphorylates CsMAF1. In line with this, we found that the CsMAF1_S45D mutant was significantly less phosphorylated by CsPK3 than the wild-type CsMAF1, indicating that S45 is the major phosphosite targeted by this kinase (Figure 7A). Even though CsMAF1_S45D was weakly phosphorylated by CsPK3 in in vitro reactions, which could indicate a second CsPK3 site in CsMAF1, S45 was the only CsPK3 phosphosite detected by mass spectrometry analysis (Supplemental Figure 8).
CsPK3 labeled with GFP was found predominantly in the nucleus of Nicotiana benthamiana epidermal cells (Figure 7B), suggesting that it could phosphorylate CsMAF1 in vivo. Moreover, the data presented in Figures 4D and 5D indicate that CsPK3 and CsC34 recognize the same CsMAF1 region, that is, the α-helix 2. In agreement with this idea, we found that CsC34 significantly reduced CsMAF1 phosphorylation by CsPK3 in in vitro reactions (Figure 7C), thus confirming that CsC34 binds the S45 region of CsMAF1. Additionally, we noticed in these assays that CsPK3 undergoes autophosphorylation (Figures 7C, 8, and 9).
Figure 8.
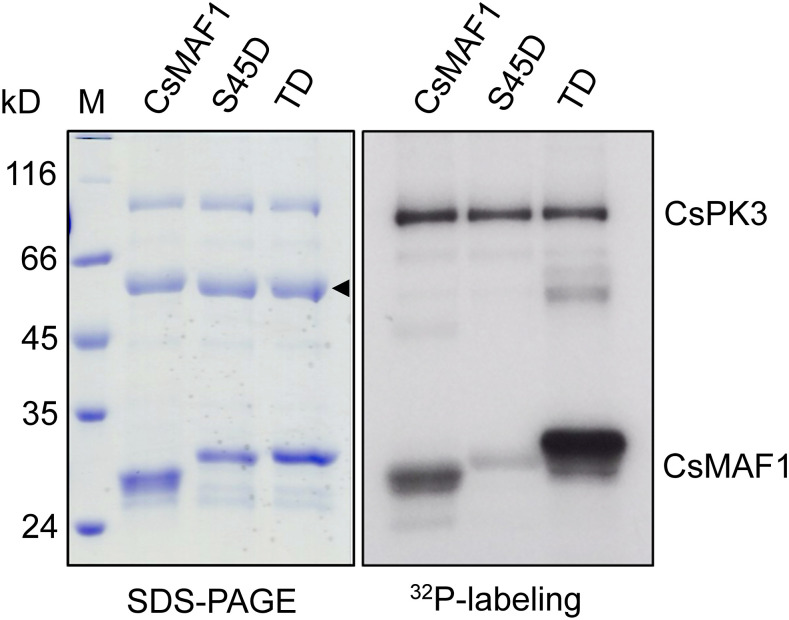
CsPK3-Mediated Phosphorylation of CsMAF1 Is Favored in the CsMAF1_TD Mutant (TD).
32P-γATP-labeling experiment showing that CsMAF1 phosphorylation by CsPK3 is significantly enhanced in the CsMAF1_TD mutant, relative to wild-type CsMAF1 or the CsMAF1_S45D mutant. Phosphorylated proteins separated on an SDS-PAGE gel (left panel) were detected by autoradiography (right panel). Bands corresponding to the autophosphorylated CsPK3 and phosphorylated CsMAF1 proteins are indicated on the right panel, whereas the CsMAF1 bands that migrate anomalously on SDS-PAGE gels (Soprano et al., 2017) are indicated by an arrowhead on the left panel.
Figure 9.
Two Protein Kinases Involved in CsMAF1 Regulation in Citrus.
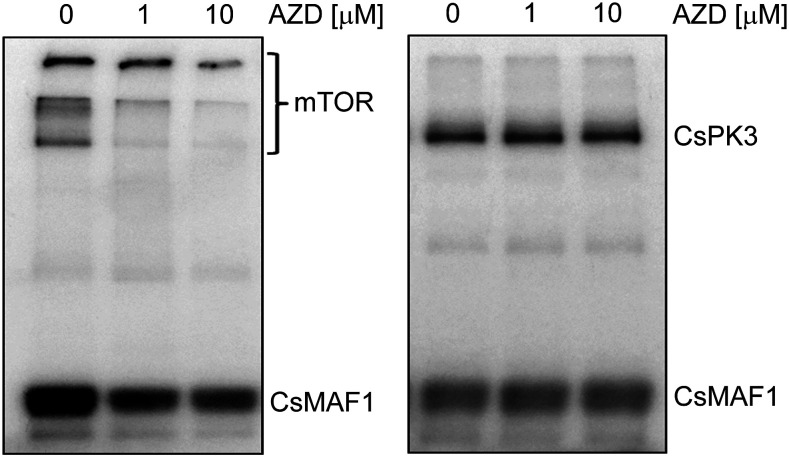
32P-γATP-labeling experiments showing that AZD inhibited the mTOR-mediated CsMAF1 phosphorylation, as well as the mTOR autophosphorylation (left panel), but not the CsPK3-mediated CsMAF1 phosphorylation or the CsPK3 autophosphorylation (right panel).
CsMAF1 Phosphorylation by CsPK3 Is Favored in the CsMAF1_TD Mutant
We showed previously that the mTOR-mediated phosphorylation of CsMAF1 was reduced in the CsMAF1_TD, but not in the CsMAF1_S45D mutant, as expected (Soprano et al., 2017). Here, however, we found that CsMAF1 phosphorylation by CsPK3 was significantly enhanced in the CsMAF1_TD mutant, relative to the wild-type CsMAF1 or the CsMAF1_S45D mutant (Figure 8), suggesting that phosphorylation of the phospholoop facilitates S45 phosphorylation by CsPK3. This idea is supported by CsMAF1 structural models that predict that the mTOR sites in the phospholoop are more solvent-exposed than S45 located in α-helix 2 (Figure 4A; Soprano et al., 2017).
To confirm that CsMAF1_TD was not phosphorylated by CsPK3 at another site, except S45, CsPK3-phosphorylated CsMAF1_TD samples were examined by mass spectrometry. The data show that S45 was the only phosphoresidue mapped in the CsMAF1_TD mutant (Supplemental Figure 8), thus supporting the notion that phosphorylation of the phospholoop favors S45 phosphorylation.
Two Protein Kinases Involved in CsMAF1 Regulation in Citrus
As observed in ScMAF1, CsMAF1 is regulated by at least two protein kinases. In addition to CsPK3, our data strongly indicate that a citrus mTOR homologue is involved in CsMAF1 regulation in citrus. This concept is supported by the observation that the specific mTOR inhibitor AZD8055 (AZD) completely blocked canker development induced by X. citri in citrus leaves, and that this inhibition was dependent on CsMAF1 (Soprano et al., 2017). Therefore, to rule out that the effect of AZD on canker development observed previously was due to a TOR kinase rather than CsPK3 inhibition, we tested whether AZD could inhibit the CsPK3-mediated CsMAF1 phosphorylation in vitro. As shown in Figure 9, AZD significantly inhibited the mTOR-mediated CsMAF1 phosphorylation, as well as mTOR autophosphorylation, but it did not inhibit the CsPK3-mediated CsMAF1 phosphorylation or CsPK3 autophosphorylation. These results strongly support the hypothesis that CsMAF1 is regulated by at least two protein kinases in citrus and that the effect of AZD on canker formation observed previously is likely due to the inhibition of a yet uncharacterized citrus TOR kinase. This idea is supported by the report that, in Arabidopsis, silencing of TOR was insufficient to have MAF1 fully dephosphorylated (Ahn et al., 2019).
The C-terminal Region of CsBRF1 Competes with CsMAF1 for CsC34 Interaction
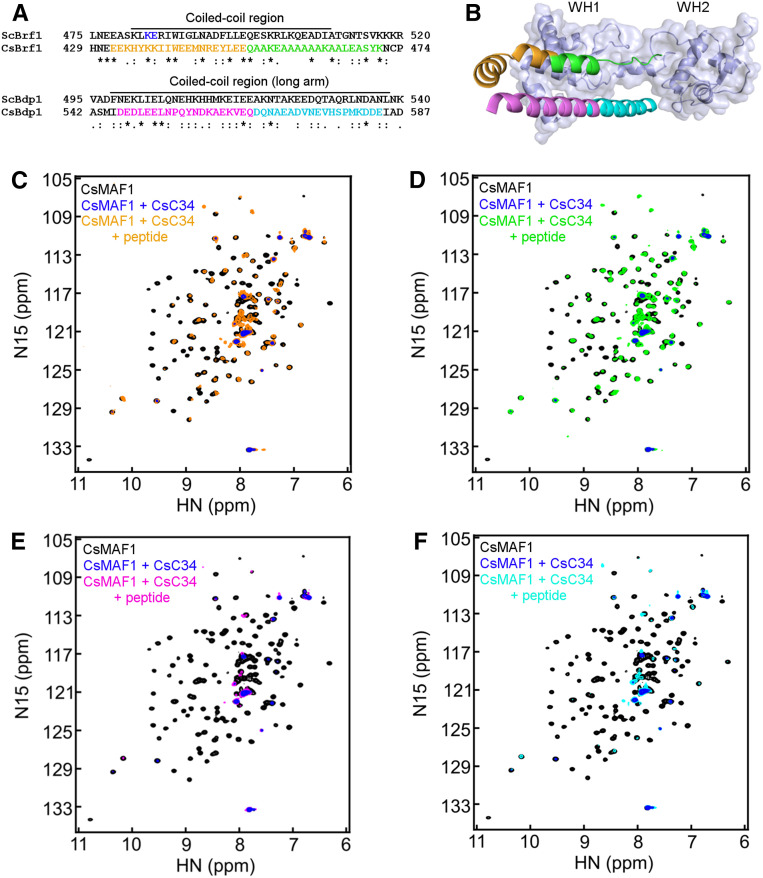
Recent structural data of the yeast RNA Pol III-TFIIIB complex revealed that the C-terminal regions of the TFIIIB components BRF1 (TFIIB-related factor 1) and BDP1 (B Double Prime 1) form a coiled-coil structure that stabilizes the WH2 domain of C34 over the Pol III cleft during preinitiation complex formation (Abascal-Palacios et al., 2018; Han et al., 2018; Vorländer et al., 2018). Since the yeast and human MAF1 bind BRF1 (Desai et al., 2005; Reina et al., 2006; Goodfellow et al., 2008) and CsMAF1 interacted predominantly with the corresponding WH1 domain of CsC34 (Figures 3C and 3D), we hypothesized that CsMAF1 could compete with BRF1 for CsC34 interaction. To test this idea, we identified the C. sinensis CsBRF1 and CsBDP1 homologues and mapped their C-terminal regions corresponding to the Brf1/Bdp1 coiled-coil structure that approaches the yeast C34 WH2 domain, based on sequence alignment (Figure 10A) and homology modeling (Figure 10B). Next, we probed by HSQC measurements the interaction of CsMAF1 with CsC34 in the presence and absence of CsBRF1 or CsBDP1 peptides covering the yeast Brf1/Bdp1 coiled-coil region (Figure 10A).
Figure 10.
The C-terminal Region of CsBRF1 Competes with CsMAF1 for CsC34 Binding.
(A) Protein sequence alignment of the C. sinensis CsBRF1 and CsBDP1 homologues (Genbank accessions XP_006479195.1 and XP_006487248.1, respectively) with the respective S. cerevisiae ScBrf1 and ScBdp1 proteins. Only the C-termini of the proteins are shown. The Brf1 and Bdp1 regions covering the coiled-coil structure found in the yeast RNA Pol III-TFIII B complex (Abascal-Palacios et al., 2018; Han et al., 2018; Vorländer et al., 2018) are indicated above the alignment. The peptides used in the HSQC experiments are colored in orange and green in CsBRF1, and magenta and cyan in CsBDP1, whereas the blue residues in ScBrf1 are those that when mutated to alanines decreased the Brf1 interaction with C34 (Andrau et al., 1999).
(B) Structural model of the CsC34 WH1 and WH2 domains (light blue) near the coiled-coil structure formed by the C-terminal helices of CsBRF1 and CsBDP1, based on the cryo-EM yeast RNA Pol III-TFIIIB complex (PDB ID 6F41; Vorländer et al., 2018). Colored elements represent the peptide sequences depicted in (A).
(C) and (D) NMR HSQC spectra of 15N-labeled-CsMAF1 (black signals) in the presence of CsC34 (blue signals), compared with CsMAF1 + CsC34 + the CsBRF1 peptides EEKHYKKIIWEEMNREYLEE (orange signals; [C]) or QAAKEAAAAAAKAALEASYK (green signals; [D]), showing that both CsBRF1 peptides restored most of the CsMAF1 signals lost upon CsC34 interaction.
(E) and (F) NMR HSQC spectra of 15N-labeled-CsMAF1 (black signals) in the presence of CsC34 (blue signals), compared with CsMAF1 + CsC34 + the CsBDP1 peptides DEDLEELNPQYNDKAEKVEQ (magenta signals; [E]) or DQNAEADVNEVHSPMKDDE (cyan signals; [F]) showing that none of the CsBDP1 peptides restored the CsMAF1 signals lost upon CsC34 binding.
We found that while the CsBRF1 C-terminal peptides EEKHYKKIIWEEMNREYLEE and QAAKEAAAAAAKAALEASYK restored most of the CsMAF1 NMR signals lost upon CsC34 interaction (Figures 10C and 10D, respectively), none of the CsBDP1 C-terminal peptides matching the yeast Bdp1 long arm (Vorländer et al., 2018), DEDLEELNPQYNDKAEKVEQ and DQNAEADVNEVHSPMKDDE (Figure 10A), restored the CsMAF1 signals under the same binding conditions (Figures 10E and 10F, respectively). Importantly, the CsBRF1 peptide QAAKEAAAAAAKAALEASYK disrupted the CsMAF1-CsC34 interaction more effectively than peptide EEKHYKKIIWEEMNREYLEE (Figures 10C and 10D), a result that is in line with our structural model that predicts that peptide QAAKEAAAAAAKAALEASYK has a larger interaction area with the CsC34 WH1 domain than peptide EEKHYKKIIWEEMNREYLEE (Figure 10B). Moreover, our results agree with genetic studies that show that mutations in this region of the yeast Brf1 protein (Figure 10A) significantly decreased Brf1 interaction with C34 (Andrau et al., 1999).
Together, the results suggest that the C-terminal region of CsBRF1, but not CsBDP1, competes with CsMAF1 for CsC34 binding.
DISCUSSION
Despite intensive research over the last two decades, our knowledge of precisely how MAF1 represses Pol III transcription and how phosphorylation affects this process is incomplete. Here we show that CsMAF1 binds preferentially to the WH1 domain of CsC34 and that its phosphoregulatory region, comprising the phospholoop and adjacent α-helix-2, not only contributes to this binding but also regulates it. We present evidence that phosphorylation of the TOR sites in the phospholoop is sufficient to decrease CsMAF1 affinity to CsC34 and derepress Pol III transcription in yeast cells. These data are thus in line with the observation that the Cryo-EM density of ScMAF1 overlaps with the second WH domain of C34 (Vannini et al., 2010). Indeed, a recently published high-resolution Cryo-EM structure of yeast Pol III in complex with MAF1 reveals that ScMAF1 binds to the WH2 domain of C34 through polar interactions involving the conserved PDH/YDFS motif (Vorländer et al., 2020). This agrees with our data showing that the WH1 domain of CsC4 is structurally equivalent to the WH2 domain of yeast C34 (Figure 3C).
The ScMAF1 structure in this new Cryo-EM Pol III-MAF1 complex (PDB ID 6TUT) lacks, however, the phosphoregulatory region (Vorländer et al., 2020). Although this region is not required for C34 interaction in yeast (Vorländer et al., 2020), when the crystal structure of CsMAF1 (PDB ID 5U50) with an extended α-helix-2 (chain D) is aligned to the yeast Pol III-MAF1 complex, we notice that α-helix-2 also approaches the WH2 domain of C34 or the WH1 domain of the CsC34 model (Supplemental Figure 9). Despite its proximity to the WH1 domain of CsC34, however, the TOR sites S59 and T62, which are visible only in CsMAF1 protomer D, are exposed to the solvent, whereas S47 faces the CsC34 WH1 domain, as opposed to S45 (Supplemental Figure 9). Moreover, according to this structural alignment, the α-helix-2 segment where S45 and S47 sit is also close to the region where the C-terminal domain of Brf1 docks into the WH2 domain of yeast C34 (Supplemental Figure 9). This is noteworthy because the corresponding peptides of CsBRF1 competed with CsMAF1 for CsC34 interaction (Figure 10B). In addition, we noticed that the side chains of the two aspartates of the CsMAF1 PDYDFS loop also protrude toward the CsC34 model as observed in the yeast Pol III-MAF1 complex (Supplemental Figure 9; Vorländer et al., 2020), suggesting that this conserved loop acts as an anchor for MAF1-C34 binding in all organisms.
By comparing the structural elements of CsMAF1 implicated in CsC34 interaction in monocot and dicot plants, we noticed that, besides the PDYDFS loop, the α-helix-2 segment is also conserved among plant MAF1 proteins. Notably, S45 and S47 are fully conserved in monocots and dicots, and although the phospholoop region is less conserved than the α-helix-2 region, the mTOR sites S59 and S66 are also invariable in plant MAF1 (Supplemental Figure 10). This suggests that the phospholoop and α-helix-2 play a critical role in the regulation of MAF1 binding to C34 in plants.
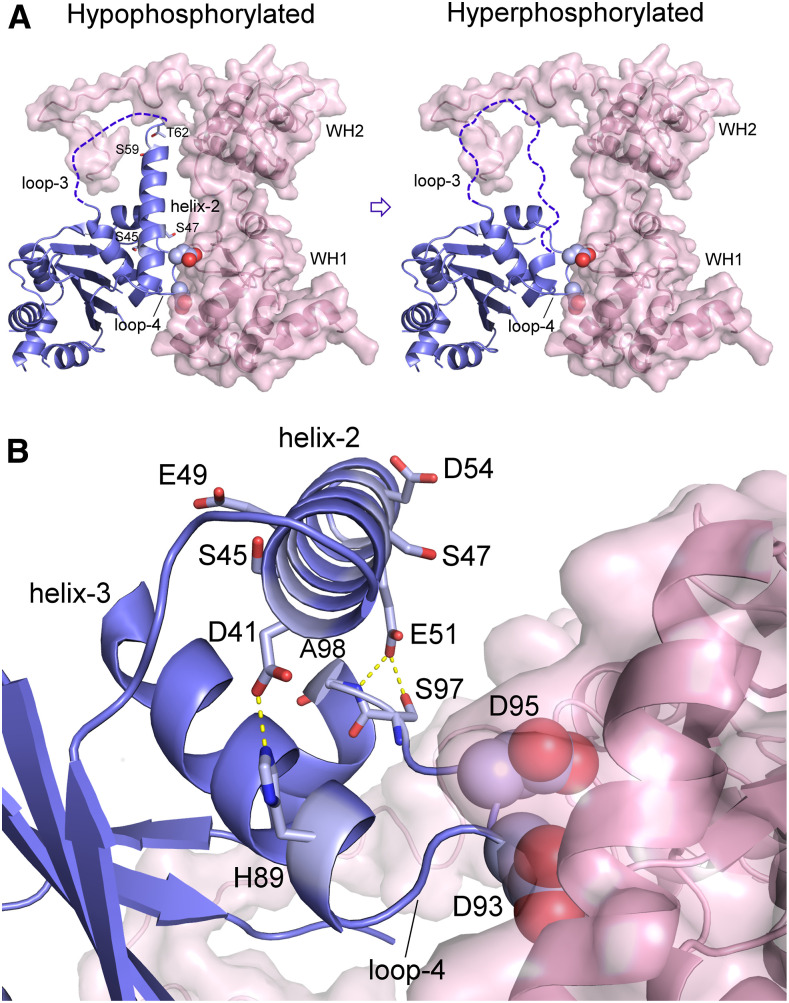
A close inspection of the different protomers in the CsMAF1 crystal (PDB ID 5U50) reveals that the GKSLDT segment that connects the α-helix-2 to the phospholoop can be structured (chain D) or unstructured (chain A; Soprano et al., 2017). This change in the secondary structure of this region offered us a clue as to how phosphorylation of the phospholoop and α-helix-2 might regulate the binding of CsMAF1 to CsC34. Thus, we propose a model to explain how progressive phosphorylation of CsMAF1 affects the interaction with CsC34 (Figure 11A). This model considers that phosphorylation starts with the more solvent-exposed Ser residues of the phospholoop, including the conserved S66 (Supplemental Figure 10). As additional residues of the phospholoop and α-helix-2 are phosphorylated, particularly T62 and S59, the α-helix-2 becomes unstable and progressively unfolds, facilitating phosphorylation of S47 and S45. This is thought to cause further conformational changes in this region that would eventually disrupt local interactions with the WH1 domain and/or destabilize the PDYDFS anchor (Figure 11A). This model of CsMAF1 regulation by hyperphosphorylation is supported by the fact that phosphorylation of S45 by CsPK3 was favored in the CsMAF1_TD mutant (Figure 8) and that both CsMAF1_TD and CsMAF1_S45D proteins assume a similar conformation that differs from that of wild-type CsMAF1 (Supplemental Figure 6). In addition, it is interesting to note that α-helix-2 carries two conserved glutamate (E49 and E51) and two aspartate (D41 and D54) residues (Supplemental Figure 10) that, together with S45 and S47, are evenly distributed along the helix and which confer a strong acidic character to the helix (Figure 11B). Thus, the addition of phosphate groups in this region is thought to destabilize the helix through charge repulsion. Remarkably, a close inspection of the CsMAF1 structure reveals that the side chain of E51 makes a polar contact with the main chain of S97 and A98 from one side of the PDYDFS loop, whereas D41 holds the H89 side chain from the other side of the PDYDFS loop (Figure 11B). Therefore, it is clear from the CsMAF1 structure that the α-helix-2 helps to hold the PDYDFS anchor in place to interact with the WH1 domain of CsC34 (Figure 11B).
Figure 11.
Mechanistic Model of How Phosphorylation of CsMAF1 Affects the Binding of CsMAF1 to CsC34.
(A) CsMAF1 in its unphosphorylated or hypophosphorylated form shows a flexible phospholoop (loop-3) with an extended α-helix-2. Phosphorylation would start with the phospholoop (loop-3) residues that are more solvent exposed, including S66. As additional sites are phosphorylated, particularly T62 and S59, the α-helix-2 becomes unstable and progressively unfolds, facilitating phosphorylation of S47 and S45. This would cause further conformational changes in this region, which would eventually disrupt local interactions with the WH1 domain and/or destabilize the PDYDFS anchor (the two aspartates shown as spheres).
(B) Close view of the region where the α-helix-2 approaches the WH1 domain of CsC34, showing that the side chain of E51 makes a polar contact with the main chain of S97 and A98 from one side of the PDYDFS loop, whereas D41 holds the H89 side chain from the other side of the PDYDFS loop. The addition of phosphate groups at S45 and S47 is thought to destabilize the helix through charge repulsion.
Moreover, our model of CsMAF1 regulation by phosphorylation is reminiscent of the mechanism by which human c-Src kinase is phospho-regulated. In this kinase, the ordered activation loop forms an α-helix that stabilizes the inactive conformation of the kinase, but which upon phosphorylation becomes unfolded, thus restoring kinase activity (Xu et al., 1999).
Prior studies showed that S45 is phosphorylated by murine PKA and its phosphomimetic mutation S45D retarded the CsMAF1 transit from the nucleoplasm to the nucleolus (Soprano et al., 2017; Soprano et al., 2018). Here we show that S45 is specifically phosphorylated by the citrus CsCPK3 kinase, which carries the typical extended T-loop and PIF motif found in plant AGC kinases (Bögre et al., 2003; Rademacher and Offringa, 2012). In this group of kinases, the T-loop needs to be phosphorylated for kinase activation (Bögre et al., 2003; Zegzouti et al., 2006a; Rademacher and Offringa, 2012). Thus, although CsPK3 can be a substrate of other citrus kinases, our labeling experiments show that CsPK3 autoactivates itself and this explains its in vitro activity upon CsMAF1. Not surprisingly, many CsPK3-related kinases, including the Arabidopsis PID (PINOID), WAG1 and WAG2, and tomato Adi3 (AvrPto-dependent Pto-interacting protein 3), also display autoactivation properties (Zegzouti et al., 2006a; Zegzouti et al., 2006b; Ek-Ramos et al., 2010).
Like all plant AGC kinases, CsPK3 has a specific insertion segment that separates the Mg+2 binding site from the T-loop (Supplemental Figure 7). At least for some of these kinases, this insertion segment dictates the subcellular localization of the protein (Zegzouti et al., 2006b; Ek-Ramos et al., 2010; Rademacher and Offringa, 2012). This is the case of tomato Adi3, which has an NLS within the insertion segment that is responsible not only for its nuclear localization, but also for its activity as a cell death suppressor (Ek-Ramos et al., 2010). Interestingly, CsPK3, which accumulates in the nucleus of N. benthamiana cells, also carries an NLS motif that overlaps with that of Adi3 (Supplemental Figure 7).
CsPK3 is closely related to the Arabidopsis D6PKL2 kinase that influences cell growth and morphogenesis through the activation of efflux pumps required for polar auxin transport (Zourelidou et al., 2009; Willige et al., 2013; Zourelidou et al., 2014). Auxin, a key component of the TOR signaling pathway in plants (Soprano et al., 2018; Ryabova et al., 2019), promotes citrus canker, and prevents CsMAF1 accumulation in the nucleolus, whereas inhibitors of polar auxin transport block canker formation induced by X. citri (Cernadas and Benedetti, 2009; Soprano et al., 2017). Thus, besides CsMAF1, CsPK3 might target additional proteins of the auxin-TOR signaling pathway in citrus involved in cell growth and morphogenesis. Moreover, because the auxin-TOR signaling pathway in plants also controls nutrient and energy availability and is linked to defense mechanisms, it is normally targeted by pathogen effector molecules whose function is to evade defenses and exploit host cell resources to favor pathogen growth (Soprano et al., 2018). In the case of the citrus–X. citri interaction, the effector molecule PthA4 is critical to induce the cell hypertrophy and hyperplasia needed for epidermal rupture and bacterial dispersion (Brunings and Gabriel, 2003). Thus, by targeting CsMAF1, PthA4 is thought to sequester CsMAF1 to enhance the tRNA and 5S rRNA synthesis required for cell growth and division.
METHODS
Bacterial Strains, Plasmids, and Growth Conditions
Escherichia coli cells were grown at 37°C in Luria-Bertani (LB) medium (10 g/L tryptone, 5 g/L yeast (Saccharomyces cerevisiae) extract, 10 g/L sodium chloride, pH 7.5). For plasmid selection, the following antibiotics were used at the specified final concentrations: kanamycin, 50 mg/L; ampicillin, 100 mg/L; spectinomycin, 50 mg/L; and gentamicin, 10 mg/L. The bacterial strains and plasmids used in this study are listed in Supplemental Table 1. Nicotiana benthamiana seeds were germinated on nutrient soil, transplanted individually to pots, and grown at 24°C under T8 light-emitting diode tubes (18W) with a 16-h-day/8-h-night photoperiod.
Cloning Procedures
The oligonucleotide sequences used for the cloning procedures are listed in Supplemental Table 2. Synthetic DNA fragments encoding the C. sinensis RNA Pol III subunits CsC160, CsC82, and CsC34 were subcloned in pET28a (Novagen) for E. coli expression (Supplemental Table 1). The CsC34 coding region was amplified by PCR from the pET28a construct and fused to EGFP or NusA. The corresponding fragments were subcloned into the pBbE1a and pET28a vectors, respectively, to obtain the 6xHis-EGFP-CsC34 and 6xHis-NusA-3xFLAG-CsC34 constructs (Supplemental Table 1). The DNA fragments encoding the WH domains of CsC34 were also amplified from the pET28a construct, cloned in pENTR-D-Topo (Thermo Fisher Scientific), and moved into pDEST17 (Thermo Fisher Scientific), whereas those corresponding to the CsCPK1, CsCPK2, CsCPK3, and CsCPK4 genes were amplified from a C. sinensis cDNA library (Domingues et al., 2010), cloned in pENTR-D-Topo, and transferred to pDEST15 (Thermo Fisher Scientific) for bacterial expression (Supplemental Table 1). The cDNA encoding the kinase CsPK3 was also transferred to the p*7WGF2 binary vector for plant expression. The 6xHis-CsMAF1_dCT and 6xHis-CsMAF1_S45D_dCT constructs were subcloned into pET28a and pYGEX-4T1 for bacterial and yeast expression, respectively (Supplemental Table 1). All DNA constructs were confirmed by DNA sequencing.
Protein Expression and Purification
The recombinant 6xHis-PthA2-4, 6xHis-CsMAF1, and derived mutants were expressed and purified essentially as previously described by Domingues et al. (2010) and Soprano et al. (2017). The recombinant proteins 6xHis-EGFP-Cs160, 6xHis-EGFP-CsC82, 6xHis-EGFP-CsC34, 6xHis-NusA-3xFLAG-CsC34, 6xHis-WH1, 6xHis-WH2, 6xHis-EGFP, and 6xHis-NusA were expressed in E. coli BL21 Star (DE3; Novagen) with the addition of 0.4 mM isopropyl-β-D-thiogalactopyranoside (IPTG) to cells grown to mid-exponential phase at 20°C or 37°C under agitation (200 rpm). Bacterial cells were harvested by centrifugation, and cell pellets were resuspended in 50 mM Tris-HCl (pH 7.5) containing 300 mM NaCl, 10 mM imidazole, 1 mg/mL lysozyme, and 10 µg/mL DNase I. Cells were ruptured by sonication and the suspension was centrifuged at 14,000 × g at 4°C for 50 min and filtered through a 0.45 µm filter (Merck Millipore). Recombinant proteins were purified by Ni-NTA affinity chromatography (GE Healthcare) using an imidazole gradient for protein elution. Protein fractions were concentrated and resolved on a HiLoad Superdex 200 (16/600) column (GE Healthcare) and eluted in 50 mM Tris-HCl buffer (pH 7.5) containing 300 mM NaCl and 1 mM DTT.
The recombinant protein kinases GST-CsPK1-4 were purified using a GST-affinity column (GE Healthcare) equilibrated with 1× PBS buffer, pH 7.4. After elution in PBS containing 7 mg/mL glutathione, protein fractions were dialyzed against PBS buffer containing 1 mM DTT.
Purified proteins were concentrated using Amicon Ultra centrifugal devices (Merck Millipore) and quantified on a NanoDrop spectrophotometer (Thermo Scientific). Protein purity was evaluated by SDS-PAGE and purified proteins were stored at −80°C in 5% (v/v) glycerol.
Far-Protein Gel Blot Assays
Far-protein gel blot assays were performed as described by Hall (2004). Briefly, lysates of E. coli cultures expressing the recombinant proteins as targets were resolved on 12% (w/v) acrylamide-SDS gels. The proteins were transferred to nitrocellulose membranes, which were blocked with 1× PBS‐TT‐milk (10 mM Na2HPO4, 2 mM KH2PO4, 2.7 mM KCl, 137 mM NaCl, 0.1% [v/v] Triton X‐100, 0.1% [v/v] Tween‐20, and 5% [v/v] powdered milk, pH 7.4) for 1 h at 4°C. Membranes were probed with 50 μg/mL GST-CsMAF1, GST-CsMAF1_dCT, or 3xFLAG-CsC34 in the same buffer for 12 h at 4°C under slow agitation. Membranes were washed four times with PBS‐TT and incubated with the goat anti‐GST (1:5000) serum (GE Healthcare, 274577-01) or the rabbit anti‐FLAG (1:5000) serum (Sigma-Aldrich, F7425) for 2 h at 4°C. After being washed, the membranes were incubated with the respective goat (Sigma-Aldrich, A5420) or rabbit (GE Healthcare, NA934) peroxidase-conjugated secondary antibodies at 1:10000 dilution. A mouse anti-6xHis serum (1:5000; GE Healthcare, 274710-01) and an anti-mouse secondary antibody (1:10000; GE Healthcare, NA931V) were used for control assays. Peroxidase activity was assayed using the ECL Advanced system (Thermo Fischer Scientific), and protein bands were detected by chemiluminescence.
Microscale Thermophoresis
For MST analysis, 6xHis-EGFP-CsC34 was purified using a nickel-affinity column, according to manufacturer’s instructions (GE Healthcare). The protein was diluted to a working concentration of 50 nM in titration buffer (1× PBS, pH 7.4), and then mixed with increasing amounts (1.5 nM to 200 μM) of the ligands (CsMAF1, CsMAF1_TD, or CsMAF1_S45D). Protein mixtures were loaded on Monolith™ NT.115 premium-coated capillaries (Nano Temper Technologies) and analyzed on a NanoTemper Monolith NT115 device (NanoTemper Technologies) using the excitation/emission wavelengths (557/574 nm) for capturing EGFP fluorescence. MST analysis was conducted in 60% light-emitting diode and 20% MST power for n = 3. The affinity of the 6xHis-PthA4 protein to GST-CsMAF1 or GST-CsMAF1_dCT was determined as described above, except that purified 6xHis-PthA4 was fluorescently labeled with the NHS–ester chemistry (MonolithTM NT Protein Labeling Kit RED-NHS, NanoTemper Technologies). Labeled 6xHis-PthA4 was further purified on a Sephadex G25 column, according to the manufacturer’s instructions (GE Healthcare), and the MST measurements were performed using the excitation/emission wavelengths (650/670 nm) for capturing the NHS fluorescence. Dissociation constants were calculated using the Hill equation Y=Bmax*X^h/(Kd^h + X^h), where Bmax is the maximum specific binding in the same units as Y; Kd is the ligand concentration needed for a half-maximum binding at equilibrium, expressed in the same units as X; and h is the Hill slope. Values presented are the means ± standard deviations of three independent measurements.
In Vitro Phosphorylation and Phosphosite Mapping
CsMAF1 was phosphorylated in vitro using purified GST-CsPK3 or the human mTOR/RAPTOR/MLST8 complex (Sigma-Aldrich) in the presence of 500 mCi/mmol γ32P-ATP (PerkinElmer). The proteins were separated by SDS-PAGE and detected by autoradiography, as previously described by Soprano et al. (2017). For mass spectrometry determination of CsPK3 and mTOR phosphosites, ∼25 µg of phosphorylated CsMAF1 in 50 mM NH4HCO3 was mixed with 25 μL 0.2% (v/v) RapiGest SF (Waters) and incubated at 80°C for 15 min. Disulfide bonds were reduced with 100 mM DTT and incubated at 60°C for 15 min. The reduced Cys side chains were alkylated by the addition of 300 mM iodoacetamide and further incubated in the dark at room temperature for 30 min. Proteolytical digestion was done by the addition of 0.25 µg of proteomics grade chymotrypsin or trypsin in 50 mM NH4HCO3, and the mixture was incubated at 37°C overnight. The removal of the acid-labile mass spectrometry (MS) compatible detergent was done by the addition of 10 μL of 5% (v/v) TFA, and the mixture was incubated at 37°C for 90 min. After digestion, the phosphopeptide mixtures were centrifuged at 18,000 g, 6°C for 30 min, and analyzed on the microUPLC-ESI-qTOF-MS and XEVO G2 systems (Waters). Before injection, columns were equilibrated with 93% (v/v) mobile phase A (water with 0.1% formic acid) and 7% (v/v) mobile phase B (acetonitrile containing 0.1% formic acid) at 40°C. Peptides were trapped on a ACQUITY UPLC Symmetry C18 Trap column (Waters) at 15 µL/min flow rate for 4 min. Phosphopeptides were separated from the trap column by gradient elution to an analytical column ACQUITY UPLC M-Class HSS T3 column (Waters) at 5 µL/min flow rate with a 7 to 40% (v/v) acetonitrile gradient over 28 min, followed by a 4.5-min rinse with 85% (v/v) acetonitrile. The column was re-equilibrated at initial conditions for 21 min. Data independent acquisition mode (MSE) was performed by operating the instrument at positive ion V mode, applying the MS and tandem mass spectrometry functions over 0.5 s intervals with 6 V low energy and 15 to 45 V high energy collision to collect the peptide mass to charge ratio (m/z) and the product ion information to deduce the amino acid sequence. Capillary voltage and source temperature were set to 3.0 kV and 80°C, respectively. Phosphopeptides were then identified with a Proteome ID (UP000027120) specific database (UniProt). After an assessment query, the software automatically sets the peptide and fragment mass tolerances. Tolerated modifications were Cys carbamidomethylation, phosphorylation (STY), and oxidation of Met. Raw data processing and protein identification were performed using the ProteinLynx Global Server 3.0.3 (Waters) with the following parameters: minimum of 3 fragment ions matched per peptide, minimum of 5 fragment ions matched per protein, minimum of 1 unique peptide matched per protein, 1 possible chymotrypsin or trypsin missed cleavage, carbamidomethylation of Cys as fixed modification, and oxidation of Met as variable modification. The data were filtered using high peptide confidence with less than 1% false discovery rate, and potential phosphorylated sites were manually validated.
Functional Complementation in Yeast
The yeast wild-type strain BY4742 and the Maf1-deletion mutant YI3945 (Kwapisz et al., 2002) were transformed with the citrus CsMAF1 gene and its derivative mutant CsMAF1_TD cloned into pYEX-4T1 (Clontech) as previously described by Soprano et al. (2013). The Maf1-deletion mutant strain was also transformed with empty vector pYEX-4T1. Transformants were grown on SC-Ura (SC medium lacking uracil) plates at 30°C for 4 d. Cells were incubated in 1× PBS buffer, pH 7.4, containing 20 mg/mL lyticase for 30 min at room temperature. Total RNA was isolated using Dicrect-Zol RNA miniprep plus (Zymo Research) and quantified by spectrophotometry. Extracted RNA was used to prepare cDNA, using a cDNA synthesis kit (Applied Biosystems) with random oligos. The expression of tRNA genes was quantified using RT-qPCR and the oligo sequences described in the Supplemental Table 2. The relative fold change in target gene expression was calculated using the equation 2ΔΔCt (Livak and Schmittgen, 2001), using UB6 as the reference gene. Two technical replicates and three biological replicates were used for each gene. Statistical analysis was conducted using Student's t test (P < 0.05; Supplemental Data Set). For immunoblot analysis, yeast cells were harvested and resuspended in 50 mM Tris-HCl (pH 8.0) containing 100 mM NaCl, 0.1% (v/v) Tween-20, and 1 mg/mL lysozyme. Cells were ruptured by sonication and after centrifugation (15,000 × g) at 4°C for 45 min, protein extracts were quantified by the Pierce BCA assay (Thermal Fisher Scientific). Equal amounts of proteins were loaded on 10% (w/v) acrylamide-SDS gels and transferred onto polyvinylidene fluoride membranes (GE Healthcare). Protein bands were detected by immunoblotting using the goat anti‐GST or mouse anti-GAPDH (Proteintech, 60004-1-IG) serum (1:5000) and secondary peroxidase-conjugated antibodies (1:10000). Peroxidase activity was assayed using the ECL Advanced system (Thermo Fischer Scientific), and protein bands were detected by chemiluminescence.
NMR Spectroscopy
Uniformly 15N-labeled 6xHis-CsMAF1 and derivative mutants 6xHis-CsMAF1_dCT, 6xHis-CsMAF1_dCT_TD, and 6xHis-CsMAF1_dCT_S45D were produced in E. coli BL21(DE3) cells grown at 37°C in M9 minimal medium supplemented with 15N-ammonium chloride (Sigma-Aldrich) to OD600nm = 0.6, followed by induction with 0.4 mM IPTG at 20°C for 16 h. Labeled proteins were purified by affinity and size-exclusion chromatography as described above and dialyzed against 50 mM sodium phosphate buffer (pH 7.4).
For titration experiments, labeled proteins were diluted to a final concentration of 150 µM in phosphate buffer, pH 7.4, containing 10% (v/v) of deuterated water (Cambridge Isotopes, Inc.). NMR experiments were performed at 40°C using a Varian/Agilent Inova 600 MHz spectrometer equipped with a triple-resonance (1H/13C/15N) and Z-axis pulsed-field gradient cryogenic probe, operating at a 1H frequency of 599.881 MHz, at the Brazilian Biosciences National Laboratory. Titration experiments were performed by measuring a series of two-dimensional 1H-15N HSQC spectra, acquired with 2048 complex data points in the acquisition domain and 256 increments in the time domain. Labeled MAF1 proteins were titrated against purified 6xHis-PthA4, NusA-CsC34, or NusA alone at a 1:5 to a 1:20 ligand:CsMAF1 molar ratio. For titrations with the test peptides, CsC34 was incubated for 5 min with the test peptide at a 5:1 peptide:CsMAF1 molar ratio before the addition of CsMAF1. All spectra were processed using NMRPipe and NMRView software (Johnson and Blevins, 1994; Delaglio et al., 1995).
CsPK3 Subcellular Localization
The cDNA encoding CsPK3 was amplified and subcloned into the p*7WGF2 vector (Karimi et al., 2002). The constructs were inserted into the Agrobacterium tumefaciens EHA105 cells, which were grown in yeast extract peptone medium supplemented with 50 mg/L rifampicin and 100 mg/L spectinomycin for 20 h at 28°C and 200 rpm. The cells were pelleted and washed in 10 mM MES, pH 5.6, containing 10 mM MgCl2 and 0.2 mM acetosyringone, as previously described by Soprano et al. (2017). The cells were suspended in the same buffer and incubated for 3 h at room temperature in the dark. The bacterial suspension was diluted in the same buffer to an OD600nm = 0.2 and used to infiltrate young leaves of 3-week-old N. benthamiana plants. Forty-eight hours after bacterial inoculation, a solution of 5 µg/mL Hoechst 34,580 (Sigma-Aldrich) in 1× PBS buffer, pH 7.4, containing 0.03% (v/v) Triton X-100, was infiltrated into the same preinfiltrated leaf sectors. Leaf discs from the inoculated leaf sectors were immediately excised and analyzed on a Leica DM6 FS fluorescence microscope equipped with LAS X software.
Molecular Modeling and Structural Alignments
Homology models of CsC34, CsBRF1, and CsBDP1 were built using SWISS-MODEL in the automated mode. CsC34 was modeled against the yeast Pol III structure 5FJ8 (Hoffmann et al., 2015), whereas CsBRF1 and CsBDP1 were modeled using the yeast Pol III structure 6F41 (Vorländer et al., 2018) as a template. Protein structures were aligned and analyzed using the software PyMOL v1.8.2 (https://pymol.org/2/).
Statistical Analysis
For the qPCR analyses, three biological samples with two technical replicates were used for measuring the expression levels of each target gene (tRNAHis, tRNATyr, and 5S rRNA). A paired (two tailed) Student's t test (P < 0.05) was performed to evaluate the differences between the means of control (empty vector) and CsMAF1-expressing cells (CsMAF1 and CsMAF1_TD; Supplemental Data Set).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers CsC34 (XP_006475959); CsPK3 (XP_006467259); CsBRF1 (XP_006479195.1); CsBDP1 (XP_006487248.1).
Supplemental Data
Supplemental Figure 1. Polymorphic residues found in the C-terminal regions of PthAs.
Supplemental Figure 2. CsMAF1_dCT shows better HSQC signal dispersion compared with full-length CsMAF1.
Supplemental Figure 3. CsC34 is homologous to the yeast RNA Pol III C34 subunit and to several uncharacterized plant proteins annotated as ‘DNA-directed RNA Pol III subunit Rpc6’.
Supplemental Figure 4. Triple-labeled CsMAF1 shows a depletion of NMR peaks compared with single-labeled CsMAF1.
Supplemental Figure 5. Mass spectrometry analysis of mTOR-phosphorylated CsMAF1.
Supplemental Figure 6. The phosphomimetic CsMAF1_S45D and CsMAF1_TD mutants show similar NMR HSQC spectra relative to CsMAF1.
Supplemental Figure 7. CsPK3 is a member of the AGC1 subfamily of plant protein kinases.
Supplemental Figure 8. Mass spectrometry analysis of CsPk3-phosphorylated CsMAF1.
Supplemental Figure 9. The α-helix-2 of CsMAF1 approaches the WH1 domain of CsC34.
Supplemental Figure 10. CsMAF1 phosphosites found in α-helix-2 and loop-3 are conserved among MAF1 from monocots and dicots.
Supplemental Table 1. Strains and plasmids used in this study.
Supplemental Table 2. Oligonucleotides used in this study.
Supplemental Data Set. Statistical analysis of RT-qPCR data.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
AUTHOR CONTRIBUTIONS
M.O.A. conducted most of the experiments including cloning, protein expression and purification, sample preparation for mass spectrometry, FWB, and yeast and plant assays; M.O.A. and M.L.S. carried out the NMR experiments; M.O.A., F.A.H.B., and A.C.M.F. conducted the MST assays; M.O.A. and C.E.B. wrote the article; C.E.B. acquired funding and supervised the project.
Acknowledgments
We acknowledge the use of the LNBio facilities ‘LEC’, ‘MAS,’ ‘LIB,’ and ‘NMR’ and thank Jaqueline Rodrigues da Silva and Jackeline de Lima Zanella for technical help. We also thank Magdalena Boguta for providing the yeast maf1 mutant. This work was supported by São Paulo Research Foundation (FAPESP; grants 2018/08535-4 and 2014/50880-0, and a fellowship 2017/18570-9 to M.O.A.); and the Ministry of Science, Technology and Innovation | Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq; fellowships 303238/2016-0 to C.E.B. and 164832/2017-3 to M.O.A.).
References
- Abascal-Palacios G., Ramsay E.P., Beuron F., Morris E., Vannini A.(2018). Structural basis of RNA polymerase III transcription initiation. Nature 553: 301–306. [DOI] [PubMed] [Google Scholar]
- Abe V.Y., Benedetti C.E.(2016). Additive roles of PthAs in bacterial growth and pathogenicity associated with nucleotide polymorphisms in effector-binding elements of citrus canker susceptibility genes. Mol. Plant Pathol. 17: 1223–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn C.S., Lee D.H., Pai H.S.(2019). Characterization of Maf1 in Arabidopsis: Function under stress conditions and regulation by the TOR signaling pathway. Planta 249: 527–542. [DOI] [PubMed] [Google Scholar]
- Andrau J.C., Sentenac A., Werner M.(1999). Mutagenesis of yeast TFIIIB70 reveals C-terminal residues critical for interaction with TBP and C34. J. Mol. Biol. 288: 511–520. [DOI] [PubMed] [Google Scholar]
- Blombach F., Makarova K.S., Marrero J., Siebers B., Koonin E.V., van der Oost J.(2009). Identification of an ortholog of the eukaryotic RNA polymerase III subunit RPC34 in Crenarchaeota and Thaumarchaeota suggests specialization of RNA polymerases for coding and non-coding RNAs in Archaea. Biol. Direct 4: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bögre L., Okrész L., Henriques R., Anthony R.G.(2003). Growth signalling pathways in Arabidopsis and the AGC protein kinases. Trends Plant Sci. 8: 424–431. [DOI] [PubMed] [Google Scholar]
- Boguta M., Czerska K., Zoładek T.(1997). Mutation in a new gene MAF1 affects tRNA suppressor efficiency in Saccharomyces cerevisiae. Gene 185: 291–296. [DOI] [PubMed] [Google Scholar]
- Bonhoure N., et al. (2015). Loss of the RNA polymerase III repressor MAF1 confers obesity resistance. Genes Dev. 29: 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunings A.M., Gabriel D.W.(2003). Xanthomonas citri: Breaking the surface. Mol. Plant Pathol. 4: 141–157. [DOI] [PubMed] [Google Scholar]
- Cai Y., Wei Y.H.(2016). Stress resistance and lifespan are increased in C. elegans but decreased in S. cerevisiae by mafr-1/maf1 deletion. Oncotarget 7: 10812–10826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernadas R.A., Benedetti C.E.(2009). Role of auxin and gibberellin in citrus canker development and in the transcriptional control of cell-wall remodeling genes modulated by Xanthomonas axonopodis pv. citri. Plant Sci. 177: 190–195. [Google Scholar]
- Chen C.Y., Lanz R.B., Walkey C.J., Chang W.H., Lu W., Johnson D.L.(2018). Maf1 and repression of RNA polymerase III-mediated transcription drive adipocyte differentiation. Cell Rep. 24: 1852–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieśla M., Towpik J., Graczyk D., Oficjalska-Pham D., Harismendy O., Suleau A., Balicki K., Conesa C., Lefebvre O., Boguta M.(2007). Maf1 is involved in coupling carbon metabolism to RNA polymerase III transcription. Mol. Cell. Biol. 27: 7693–7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A.(1995). NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6: 277–293. [DOI] [PubMed] [Google Scholar]
- Desai N., Lee J., Upadhya R., Chu Y., Moir R.D., Willis I.M.(2005). Two steps in Maf1-dependent repression of transcription by RNA polymerase III. J. Biol. Chem. 280: 6455–6462. [DOI] [PubMed] [Google Scholar]
- Domingues M.N., De Souza T.A., Cernadas R.A., de Oliveira M.L., Docena C., Farah C.S., Benedetti C.E.(2010). The Xanthomonas citri effector protein PthA interacts with citrus proteins involved in nuclear transport, protein folding and ubiquitination associated with DNA repair. Mol. Plant Pathol. 11: 663–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ek-Ramos M.J., Avila J., Cheng C., Martin G.B., Devarenne T.P.(2010). The T-loop extension of the tomato protein kinase AvrPto-dependent Pto-interacting protein 3 (Adi3) directs nuclear localization for suppression of plant cell death. J. Biol. Chem. 285: 17584–17594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow S.J., Graham E.L., Kantidakis T., Marshall L., Coppins B.A., Oficjalska-Pham D., Gérard M., Lefebvre O., White R.J.(2008). Regulation of RNA polymerase III transcription by Maf1 in mammalian cells. J. Mol. Biol. 378: 481–491. [DOI] [PubMed] [Google Scholar]
- Hall R.A.(2004). Studying protein-protein interactions via blot overlay or Far Western blot. Methods Mol. Biol. 261: 167–174. [DOI] [PubMed] [Google Scholar]
- Han Y., Yan C., Fishbain S., Ivanov I., He Y.(2018). Structural visualization of RNA polymerase III transcription machineries. Cell Discov. 4: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann N.A., Jakobi A.J., Moreno-Morcillo M., Glatt S., Kosinski J., Hagen W.J., Sachse C., Müller C.W.(2015). Molecular structures of unbound and transcribing RNA polymerase III. Nature 528: 231–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B.A., Blevins R.A.(1994). NMR View: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4: 603–614. [DOI] [PubMed] [Google Scholar]
- Kantidakis T., Ramsbottom B.A., Birch J.L., Dowding S.N., White R.J.(2010). mTOR associates with TFIIIC, is found at tRNA and 5S rRNA genes, and targets their repressor Maf1. Proc. Natl. Acad. Sci. USA 107: 11823–11828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A.(2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Khanna A., Pradhan A., Curran S.P.(2015). Emerging roles for Maf1 beyond the regulation of RNA polymerase III activity. J. Mol. Biol. 427: 2577–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwapisz M., Smagowicz W.J., Oficjalska D., Hatin I., Rousset J.P., Zoładek T., Boguta M.(2002). Up-regulation of tRNA biosynthesis affects translational readthrough in maf1-delta mutant of Saccharomyces cerevisiae. Curr. Genet. 42: 147–152. [DOI] [PubMed] [Google Scholar]
- Lee Y.L., Li Y.C., Su C.H., Chiao C.H., Lin I.H., Hsu M.T.(2015). MAF1 represses CDKN1A through a Pol III-dependent mechanism. eLife 4: e06283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leśniewska E., Boguta M.(2017). Novel layers of RNA polymerase III control affecting tRNA gene transcription in eukaryotes. Open Biol. 7: 170001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Tsang C.K., Wang S., Li X.X., Yang Y., Fu L., Huang W., Li M., Wang H.Y., Zheng X.F.(2016). MAF1 suppresses AKT-mTOR signaling and liver cancer through activation of PTEN transcription. Hepatology 63: 1928–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D.(2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Michels A.A.(2011). MAF1: A new target of mTORC1. Biochem. Soc. Trans. 39: 487–491. [DOI] [PubMed] [Google Scholar]
- Michels A.A., Robitaille A.M., Buczynski-Ruchonnet D., Hodroj W., Reina J.H., Hall M.N., Hernandez N.(2010). mTORC1 directly phosphorylates and regulates human MAF1. Mol. Cell. Biol. 30: 3749–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir R.D., Lee J., Haeusler R.A., Desai N., Engelke D.R., Willis I.M.(2006). Protein kinase A regulates RNA polymerase III transcription through the nuclear localization of Maf1. Proc. Natl. Acad. Sci. USA 103: 15044–15049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oficjalska-Pham D., Harismendy O., Smagowicz W.J., Gonzalez de Peredo A., Boguta M., Sentenac A., Lefebvre O.(2006). General repression of RNA polymerase III transcription is triggered by protein phosphatase type 2A-mediated dephosphorylation of Maf1. Mol. Cell 22: 623–632. [DOI] [PubMed] [Google Scholar]
- Oler A.J., Cairns B.R.(2012). PP4 dephosphorylates Maf1 to couple multiple stress conditions to RNA polymerase III repression. EMBO J. 31: 1440–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palian B.M., Rohira A.D., Johnson S.A., He L., Zheng N., Dubeau L., Stiles B.L., Johnson D.L.(2014). Maf1 is a novel target of PTEN and PI3K signaling that negatively regulates oncogenesis and lipid metabolism. PLoS Genet. 10: e1004789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta K., Lefebvre O., Martin N.C., Smagowicz W.J., Stanford D.R., Ellis S.R., Hopper A.K., Sentenac A., Boguta M.(2001). Maf1p, a negative effector of RNA polymerase III in Saccharomyces cerevisiae. Mol. Cell. Biol. 21: 5031–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan A., Hammerquist A.M., Khanna A., Curran S.P.(2017). The C-box region of MAF1 regulates transcriptional activity and protein stability. J. Mol. Biol. 429: 192–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher E.H., Offringa R.(2012). Evolutionary adaptations of plant AGC kinases: From light signaling to cell polarity regulation. Front Plant Sci 3: 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina J.H., Azzouz T.N., Hernandez N.(2006). Maf1, a new player in the regulation of human RNA polymerase III transcription. PLoS One 1: e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverendo M., Argüello R.J., Polte C., Valečka J., Camosseto V., Auphan-Anezin N., Ignatova Z., Gatti E., Pierre P.(2019). Polymerase III transcription is necessary for T cell priming by dendritic cells. Proc. Natl. Acad. Sci. USA 116: 22721–22729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideout E.J., Marshall L., Grewal S.S.(2012). Drosophila RNA polymerase III repressor Maf1 controls body size and developmental timing by modulating tRNAiMet synthesis and systemic insulin signaling. Proc. Natl. Acad. Sci. USA 109: 1139–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D.N., Wilson B., Huff J.T., Stewart A.J., Cairns B.R.(2006). Dephosphorylation and genome-wide association of Maf1 with Pol III-transcribed genes during repression. Mol. Cell 22: 633–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins J., Veras I., Cabarcas S., Willis I., Schramm L.(2007). Human Maf1 negatively regulates RNA polymerase III transcription via the TFIIB family members Brf1 and Brf2. Int. J. Biol. Sci. 3: 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryabova L.A., Robaglia C., Meyer C.(2019). Target of Rapamycin kinase: Central regulatory hub for plant growth and metabolism. J. Exp. Bot. 70: 2211–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty M., Noguchi C., Wilson S., Martinez E., Shiozaki K., Sell C., Mell J.C., Noguchi E.(2020). Maf1-dependent transcriptional regulation of tRNAs prevents genomic instability and is associated with extended lifespan. Aging Cell 19: e13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shor B., Wu J., Shakey Q., Toral-Barza L., Shi C., Follettie M., Yu K.(2010). Requirement of the mTOR kinase for the regulation of Maf1 phosphorylation and control of RNA polymerase III-dependent transcription in cancer cells. J. Biol. Chem. 285: 15380–15392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soprano A.S., Abe V.Y., Smetana J.H., Benedetti C.E.(2013). Citrus MAF1, a repressor of RNA polymerase III, binds the Xanthomonas citri canker elicitor PthA4 and suppresses citrus canker development. Plant Physiol. 163: 232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soprano A.S., et al. (2017). Crystal structure and regulation of the citrus Pol III repressor MAF1 by auxin and phosphorylation. Structure 25: 1360–1370.e4. [DOI] [PubMed] [Google Scholar]
- Soprano A.S., Smetana J.H.C., Benedetti C.E.(2018). Regulation of tRNA biogenesis in plants and its link to plant growth and response to pathogens. Biochim. Biophys. Acta. Gene Regul. Mech. 1861: 344–353. [DOI] [PubMed] [Google Scholar]
- Sun Y., et al. (2019). Maf1 ameliorates cardiac hypertrophy by inhibiting RNA polymerase III through ERK1/2. Theranostics 9: 7268–7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towpik J., Graczyk D., Gajda A., Lefebvre O., Boguta M.(2008). Derepression of RNA polymerase III transcription by phosphorylation and nuclear export of its negative regulator, Maf1. J. Biol. Chem. 283: 17168–17174. [DOI] [PubMed] [Google Scholar]
- Vannini A., Ringel R., Kusser A.G., Berninghausen O., Kassavetis G.A., Cramer P.(2010). Molecular basis of RNA polymerase III transcription repression by Maf1. Cell 143: 59–70. [DOI] [PubMed] [Google Scholar]
- Vorländer M.K., Baudin F., Moir R.D., Wetzel R., Hagen W.J.H., Willis I.M., Müller C.W.(2020). Structural basis for RNA polymerase III transcription repression by Maf1. Nat. Struct. Mol. Biol. 27: 229–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorländer M.K., Khatter H., Wetzel R., Hagen W.J.H., Müller C.W.(2018). Molecular mechanism of promoter opening by RNA polymerase III. Nature 553: 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige B.C., Ahlers S., Zourelidou M., Barbosa I.C., Demarsy E., Trevisan M., Davis P.A., Roelfsema M.R., Hangarter R., Fankhauser C., Schwechheimer C.(2013). D6PK AGCVIII kinases are required for auxin transport and phototropic hypocotyl bending in Arabidopsis. Plant Cell 25: 1674–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis I.M.(2018). Maf1 phenotypes and cell physiology. Biochim. Biophys. Acta. Gene Regul. Mech. 1861: 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis I.M., Moir R.D.(2007). Integration of nutritional and stress signaling pathways by Maf1. Trends Biochem. Sci. 32: 51–53. [DOI] [PubMed] [Google Scholar]
- Xu W., Doshi A., Lei M., Eck M.J., Harrison S.C.(1999). Crystal structures of c-Src reveal features of its autoinhibitory mechanism. Mol. Cell 3: 629–638. [DOI] [PubMed] [Google Scholar]
- Zegzouti H., Anthony R.G., Jahchan N., Bögre L., Christensen S.K.(2006a). Phosphorylation and activation of PINOID by the phospholipid signaling kinase 3-phosphoinositide-dependent protein kinase 1 (PDK1) in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 6404–6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegzouti H., Li W., Lorenz T.C., Xie M., Payne C.T., Smith K., Glenny S., Payne G.S., Christensen S.K.(2006b). Structural and functional insights into the regulation of Arabidopsis AGC VIIIa kinases. J. Biol. Chem. 281: 35520–35530. [DOI] [PubMed] [Google Scholar]
- Zhang S., Li X., Wang H.Y., Steven Zheng X.F.(2018). Beyond regulation of pol III: Role of MAF1 in growth, metabolism, aging and cancer. Biochim. Biophys. Acta. Gene Regul. Mech. 1861: 338–343. [DOI] [PubMed] [Google Scholar]
- Zourelidou M., et al. (2014). Auxin efflux by PIN-FORMED proteins is activated by two different protein kinases, D6 PROTEIN KINASE and PINOID. eLife 3: e02860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zourelidou M., Müller I., Willige B.C., Nill C., Jikumaru Y., Li H., Schwechheimer C.(2009). The polarly localized D6 PROTEIN KINASE is required for efficient auxin transport in Arabidopsis thaliana. Development 136: 627–636. [DOI] [PubMed] [Google Scholar]