Abstract
MicroRNA-155 (miRNA-155) is abundant in fibroblast-like synoviocytes (FLS) in rheumatoid arthritis (RA). Lysine-specific demethylase 1 (LSD1) has been found that it can ameliorate the severity of RA. Tumor necrosis factor-alpha, interleukin-1 beta, and interleukin-6 are key proinflammatory cytokines implicated in the pathogenesis of RA. In our study, we investigated whether miRNA-155 participates in the expression of LSD1 and proinflammatory cytokines in rheumatoid synovial cells. First of all, flow cytometry and cell counting kit-8 analysis were employed to explore the apoptosis and proliferation of FLS, respectively. Subsequently, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was applied to probe into the level of miRNA-155 in FLS when stimulated by miRNA-155 molecules. Moreover, RT-qPCR was used to explore the relative LSD1 miRNA expression in FLS when stimulated by miRNA-155 molecules, and Western blot and immunofluorescence assay were applied to probe into the expression level of LSD1. Finally, enzyme-linked immunosorbent assay was employed to analyze the secreting level of proinflammatory cytokines in FLS when stimulated by miRNA-155 molecules. RA-FLS showed a higher apoptosis rate than normal FLS. The cell proliferation of both HFLS and MH7A cells was promoted by miRNA-155 upregulation. Meanwhile, the expression of LSD1 and proinflammatory cytokines in the FLS of RA was also changed by miRNA-155 regulation. In conclusion, miRNA-155 participates in the expression of LSD1 and proinflammatory cytokines in rheumatoid synovial cells. These findings imply a potential function and interaction of miRNA-155 and LSD1.
1. Introduction
Rheumatoid arthritis (RA) is a kind of chronic autoimmune disorder featured by nonspecific inflammation of synovial membranes and joints. With the increased incidence rate and mortality percent, RA plagues approximately 1% of the world's population and brings enormous economic pressure and heavy social burden to the whole world [1, 2]. Earlier studies have shown that proinflammatory cytokines and inflammatory mediators are released in the pathological process of RA, which leads to chronic, symmetrical, and polysynovial arthritis. Moreover, these factors also cause recurrent and progressive joint pain and swelling, which eventually develop into joint damage, rigidity, and deformity, and even involve extraarticular organs [3, 4]. However, the mechanism of the pathological immune response of RA has not been elucidated.
Fibrosis exists in the pathological process of many diseases. There are large numbers of fibroblast-like synoviocytes (FLS) in human joints, which play considerable roles in the occurrence and persistence of RA. The synovial fluid produced by proliferative FLS increases the release of proinflammatory cytokines, which are crucial for both the joint destruction and the spread of inflammation in RA [5]. Tumor necrosis factor-alpha (TNF-α), interleukin-1 beta (IL-1β), and interleukin-6 (IL-6) are key proinflammatory cytokines implicated in the pathogenesis of RA [6, 7]. Targeted blocking of TNF-α and IL-6 is effective in the treatment of refractory RA patients [8, 9]. Chinese medicinal therapy is also a potential strategy to reduce the IL-1β-induced inflammation of RA [10].
Histone methylation is a dynamic process that influences the pathogenesis of RA. A considerable amount of studies have found that DNA methylation and histone modification may affect the progress of RA by coordinating the differentiation and function of immune cells [11, 12]. Therefore, regulation of histone methylation becomes a valid way to improve the RA treatment. Lysine-specific demethylase 1 (LSD1), a well-known lysine-specific demethylase, was initially found in 2004 [13]. Its function mainly depends on H3K4 (2 m) demethylase activity that brings an important physiological effect to many diseases [14]. Our previous research showed that LSD1-knockout mice effectively reduced RA development, especially decreased the joint injury and inflammatory response by abolishing LSD1 expression [15].
MicroRNAs (miRNAs), a class of small noncoding RNAs, are only 21-25 nucleotides. Many researchers focus on the implication of miRNAs in diseases. miRNA-155, as a member of the miRNAs family, regulates various diseases such as neoplastic diseases, cardiovascular disorders, and viral infection [16]. Moreover, miRNA-155 is crucial to the production of fibrosis; meanwhile, the dysfunction of miRNA-155 is closely related to the development of RA osteoclast differentiation [17–21]. miRNA-155−/− mice showed reduced bone destruction and osteoclast production. Besides, miRNA-155 can regulate gene expression with the modification of histones in some regions [22], which implied a significant function of miRNA-155 to histone modification. Furthermore, it has been reported that KDM1A (LSD1) was a target of miRNA-155 in prostate cancer [23].
In view of the above, we aimed at proving whether miRNA-155 can also regulate the expression of LSD1 in RA and thus affect the secretion of proinflammatory cytokines. We used miRNA-155 mimics and inhibitor to stimulate HFLS and MH7A cells to explore the effects of miRNA-155 on cell proliferation, and the expression of LSD1 and proinflammatory cytokines.
2. Materials and Methods
2.1. Preparation of Cells
Human fibroblast-like synoviocytes (HFLS) and human rheumatoid FLS (MH7A) were purchased from BNCC (Beijing, China). Cells were cultured with Dulbecco's modified eagle medium (Gibco, Carlsbad, CA, USA) including 10% fetal bovine serum (Gibco, Carlsbad, CA, USA), penicillin, and streptomycin (Thermo Fisher Scientific, Waltham, MA, USA. Final concentration: streptomycin, 100 μg/ml; penicillin, 100 U/ml). The cells were incubated in an incubator at 37°C and used at the 4th generation.
2.2. Cell Apoptosis Detection
Apoptosis was measured using a flow cytometer (Guava Technologies, San Francisco, CA, USA) according to a previous study with some modification [24]. In brief, when the cell coverage rate reaches up to 75%, the drug induces apoptosis; the drug action time and its concentration have been explored in advance. The cell suspension was obtained from cells by D-hanks, 1× binding buffer washing, trypsin digestion, centrifugation, and resuspension. 5 μl annexin V-FITC (Thermo Fisher Scientific, Waltham, MA, USA) was added to every 100 μl cell suspension (1 × 105 to 1 × 106 cells). 5 μl propidium iodide (PI, US Everbright® Inc., Suzhou, Jiangsu, China) staining (40 × PI dye solution diluted 10 times) was used for dyeing, and 1 × cell staining buffer was used to supplement the upper system to 300 μl. All experiments in our study were performed independently at least three times for each point described.
2.3. Cell Transfection
miRNA-155 mimics and inhibitor (5 nmol/100 μl), including their negative control (NC), were purchased (GenePharma Ltd., Shanghai, China). HFLS and MH7A cells in 6-well plates (2 × 105 cells/well) were transfected using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol.
2.4. Cell Proliferation Assay
Following transfection, cells were seeded into 96-well plates at a density of 2 × 103 cells/well in a humidified atmosphere of 5% CO2 at 37°C for 24 hours. The viability of cells was determined using cell counting kit-8 (CCK-8, Dojindo, Kyushu, Japan) according to the manufacturer's protocol at 24, 48, 72, and 96 hours of culture. For detecting the proliferation ability, the CCK-8 reagents were added in each well (10 μl/well). After incubating at 37°C with 5% CO2 for 1.5 hours, the optical density was measured at a wavelength of 450 nm. The proliferation curve was plotted with GraphPad Prism 8.0.
2.5. Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA extraction and RT-PCR were performed according to a previous study with some modification [25]. Total RNA was extracted from the passage 4 cells according to the instructions of TRIzol (Sigma, Shanghai, China). Briefly, cells were collected by centrifugation at 2000 rpm for 5 minutes and then mixed with 1 ml of TRIzol at room temperature for 5 minutes. Subsequently, the lysates were shaken for 10 seconds and stood at room temperature for 10 minutes. After centrifuging, the lysates at 12800 rpm at 4°C for 10 minutes, the supernatant was mixed with isochoric precooling isopropanol for 10 minutes and centrifuged at 12800 rpm and 4°C for another 10 minutes. 75% ethanol was used to wash the precipitate twice. When the precipitate was dry, it was dissolved in 30 μl RNase-free water (Thermo Fisher Scientific, Waltham, MA, USA) and quantified by a NanoDrop 100 spectrophotometer (ALLSHENG, Hangzhou, Zhejiang, China).
cDNA was synthesized according to the instructions of HiScript® Q RT SuperMix for qPCR (Vazyme, Nanjing, Jiangsu, China). Briefly, 4 × gDNA wiper Mix and 1 μg of total RNA were added to the PCR tube, then RNase-free H2O was complemented to 8 μl. After mixing, centrifugation was performed, and bathing at 42°C for 2 minutes. Subsequently, 4 μl 5 × qRT SuperMix II was added to the PCR tube. The following thermocycling conditions were applied for the cDNA synthesis: 55°C for 15 minutes and 85°C for 2 minutes. The obtained cDNA was stored at -80°C for future use.
SYBR Green mastermixes and a ViiA™ 7 real-time PCR system with 384-well block (Thermo Fisher Scientific, Waltham, MA, USA) were utilized to perform the PCR analysis. The information of forward and reverse primers is listed in Table 1. The following thermocycling conditions were applied for the PCR: Initial denaturation for 5 minutes at 95°C; 40 cycles of 95°C for 10 seconds, 60°C for 30 seconds; 1 cycle of dissociation for 95°C for 15 seconds, 60°C for 60 seconds and 95°C for 15 seconds. The expression level was normalized using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) small nuclear RNA and quantified using the 2-ΔΔCq.
Table 1.
Primer sequences and their amplified bands' size range. Sequences are listed 5′→3′.
| Gene | Primer (forward) | Primer (reverse) | Length (bp) |
|---|---|---|---|
| GAPDH | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT | 96 |
| miRNA-155 | GCGCCTCCTACATATTAGCA | GTGCAGGGTCCGAGGT | 77 |
2.6. Western Blot (WB)
Protein extraction and WB were performed as previously described, with some modification [26]. Protein was extracted from the passage 4 cells when the cell coverage rate reaches up to 90% according to the instructions of SDS lysis buffer (Beyotime, Shanghai, China). The cells taken from the incubator were washed twice with phosphate buffer saline (Beyotime, Shanghai, China) after discarding the culture solution, then added to the cell lysate and placed on ice at 4°C. Blew with the tip of the pipette until the cells are fully lysed, transferred the lysed samples into the centrifuge tube, and continued to lyse on the ice for 10 minutes. After cracking, the supernatant was transferred to a new centrifuge tube and centrifuged at 4°C, 12000 rpm for 10 minutes, adding loading buffer at 100°C, 20 minutes; after water bath, the centrifuge tube at 4°C, 12000 rpm, centrifuged for 1 minute, and stored at -20°C for standby. Subsequently, a total of 12 μg of protein was loaded into SDS-PAGE according to the instructions of the SDS-PAGE gel preparation kit (Beyotime, Shanghai, China), and transferred to polyvinylidene fluoride membranes (Thermo Fisher Scientific, Waltham, MA, USA). The membranes were blocked with Tris-buffered saline Tween-20 (TBST, Beyotime, Shanghai, China) containing 5% skimmed milk (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan) dissolved in for 1 hour at room temperature. Subsequently, the membranes were rinsed with TBST three times and incubated with primary antibody dissolved in antibody diluted by blocking solution at room temperature for 2 hours. Membranes were then incubated with the corresponding secondary antibodies (Rabbit. no. Ab129195; 1 : 10,000, Abcam, Shanghai, China) at room temperature for 1 hour. Protein bands were visualized using the Image J (1.51j8 version, Bethesda, MD, USA). The expression level of the targeted protein was normalized to that of GAPDH.
2.7. Immunofluorescence Assay (IFA)
To investigate the effect of miRNA-155 on the expression and distribution of LSD1 in cells, the fixed cell slides were blocked with 3% H2O2 for 5 minutes, permeabilized with 0.5% Triton X-100 (Solarbio, Beijing, China) for 30 minutes, and then blocked with 5% rabbit serum albumin for 15 minutes. Subsequently, the cells were incubated with mouse-derived antibody diluted in 0.1% Triton at 37°C for 1 hour, followed by a further incubation at 37°C for 30 minutes with a rabbit secondary antibody to Mouse IgG (Anti-KDM1A/LSD1, ab129195, Abcam, Shanghai, China) at 1 μg/ml (shown in green). Nuclear DNA was labeled in blue with DAPI. The slides were photographed with blue light, and the expression of antibodies in tissues was determined according to the distribution of nuclei under blue light.
2.8. Enzyme-Linked Immunosorbent Assay (ELISA)
To determine the effect of miRNA-155 and LSD1 on the secretion of proimmunology cytokines by FLS, HFLS and MH7A cells were transfected with miRNA-155 mimics and inhibitor. A total of 48 hours following transfection, the concentration of TNF-α, IL-1β, and IL-6 in the cell culture supernatant (obtained by centrifugation at 13500 rpm for 5 minutes at room temperature) was performed using ELISA kits (mIbio, Shanghai, China) according to the instructions.
2.9. Statistics Analysis
All data are presented as mean ± standard deviation. The comparison between every two groups was evaluated using an unpaired two-tailed t-test. It was considered to be statistically significant that the P value was less than 0.05. All statistical analyses were performed using GraphPad Prism 8.0 for Windows (San Diego, CA, USA).
3. Results
3.1. RA Induces the Cell Apoptosis of FLS
To investigate whether RA causes the cell apoptosis, we set out to identify the cell apoptosis ratio between normal FLS and rheumatoid FLS. HFLS and MH7A cells were cultured until the 4th generation (Figure 1(a)) and analyzed by flow cytometry (Figure 1(b)). The histogram analysis indicated that the apoptotic ratio of MH7A cells is significantly higher than that of HFLS cells (Figure 1(c)). The results showed that RA can induce cell apoptosis easily.
Figure 1.

The growth and apoptosis of normal FLS and RA-FLS. (a) HFLS and MH7A cells were cultured and observed until the 4th generation. Scale bar, 1 cm. (b, c) The apoptosis of HFLS and MH7A cells was evaluated by flow cytometry. ∗P < 0.05.
3.2. Upregulation of miRNA-155 Promotes the Proliferation of HFLS and MH7A Cells
Unlike the cancer cell proliferation [24, 27], the mechanism of RA cell proliferation remained unknown. We next used miRNA-155 to identify its regulation on the proliferation of HFLS and MH7A cells. We firstly checked the expression of miRNA-155 in these two groups of cells and after being interfered by miRNA-155 mimics and inhibitor. RT-qPCR results indicated that the miRNA-155 level of MH7A cells was significantly higher than that of HFLS cells, and it can be upregulated or downregulated individually after miRNA-155 mimics or inhibitor treatment in HFLS and MH7A cells (Figures 2(a) and 2(b)). Following transfection with miRNA-155 mimics and inhibitor, CCK-8 assay was used to determine the proliferation of HFLS and MH7A cells (Figures 2(c) and 2(d)). The results displayed that miRNA-155 mimics can promote the proliferation of both HFLS and MH7A cells. Consistently, miRNA-155 inhibitor also inhibits the proliferation of both HFLS and MH7A cells.
Figure 2.

The effects of miRNA-155 on cell viability of normal FLS and RA-FLS. (a, b) Both HFLS and MH7A cells (2 × 105 cells/well) were transfected with 5 μl miRNA-155 mimics and inhibitor for 48 hours. Total RNA was extracted to detect the expression of miRNA-155 by RT-qPCR. Results were normalized by GAPDH miRNA. ∗P < 0.05. (c) HFLS cells were transfected with miRNA-155 mimics and inhibitor for 48 hours, followed by the assessment of cell viability by CCK-8 assay. ∗P < 0.05. (d) MH7A cells were transfected with miRNA-155 mimics and inhibitor for 48 hours, followed by the assessment of cell viability by CCK-8 assay. ∗P < 0.05.
3.3. Upregulation of miRNA-155 Decreases the Expression of LSD1 in RA
LSD1 has been known as a target of miRNA-155. To further clarify the effect of miRNA-155 on LSD1, we detected the expression level of the relative LSD1 miRNA in HFLS and MH7A cells after miRNA-155 mimics and inhibitor interference. We treated cells with miRNA-155 mimics and inhibitor to identify the relationship between LSD1 and miRNA-155. RT-qPCR results showed that the miRNA level of LSD1 in the inhibitor group increased. Correspondingly, the miRNA level of LSD1 decreased in the mimics group (Figures 3(a) and 3(b)). Moreover, LSD1 protein expression was also regulated by miRNA-155 mimics and inhibitor, which was consistent with the RT-qPCR data (Figures 3(c) and 3(d)).
Figure 3.
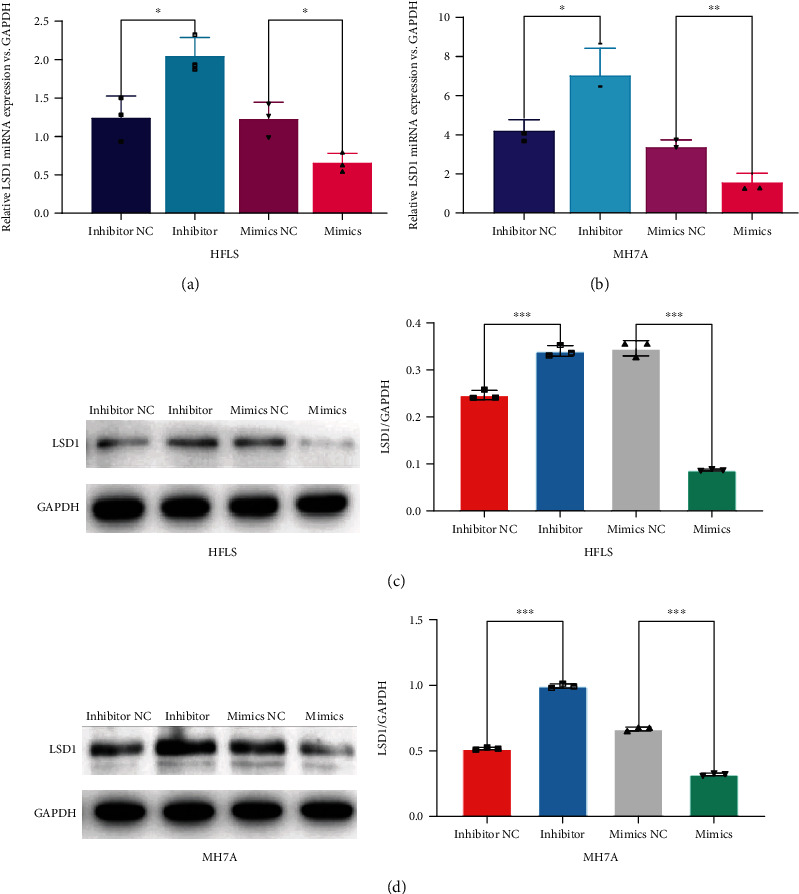
The regulation of miRNA-155 on LSD1 expression in normal FLS and RA-FLS. (a, b) Both HFLS and MH7A cells were transfected with miRNA-155 mimics and inhibitor for 48 hours. Total RNA was extracted to detect the expression of LSD1 miRNA by RT-qPCR. Results were normalized by GAPDH miRNA. ∗P < 0.05. (c, d) The protein levels of LSD1 in both HFLS and MH7A cells were detected by Western blot. Results were normalized by GAPDH protein. ∗P < 0.05.
In order to further verify the effect of miRNA-155 on the expression of LSD1, we used IFA to observe whether the distribution of LSD1 changed in cells stimulated by treating miRNA-155 mimics and inhibitor. We observed that the distribution of LSD1 was in the nucleus, and its expression in both two cells was stimulated significantly by miRNA-155 inhibitor treatment. On the contrary, LSD1 expression decreased in the miRNA-155 mimics group (Figures 4(a) and 4(b)). These results indicated that miRNA-155 can target and negatively regulate LSD1 in both HFLS and MH7A cells.
Figure 4.

The cellular distribution of LSD1 in normal FLS and RA-FLS when transfected with miRNA-155 molecules. (a) Representative imaging of LSD1 distribution in both HFLS and MH7A cells. Immunofluorescence staining used LSD1 antibody (green) and DAPI (blue). Scale bar, 500 μm. (b) Semiquantitative analysis of (a). ∗P < 0.05.
3.4. Upregulation of miRNA-155 Decreases the Secretion of Proinflammatory Cytokines in RA
It is known that multiple proinflammatory cytokines were secreted during RA, including TNF-α, IL-1β, and IL-6 [6, 7]. Here, we attempt to find out whether miRNA-155 participates in regulating the secretion of TNF-α, IL-1β, and IL-6 in HFLS and MH7A cells. The secretion of TNF-α, IL-1β, and IL-6 in cell supernatant was examined by ELISA, respectively. Data analysis showed that these factors were all markedly raised in miRNA-155 inhibitor-treated cells compared with the miRNA-155 inhibitor NC group. Similarly, their secretion also decreased by the treatment of miRNA-155 mimics (Figure 5). Thus, these results revealed that miRNA-155 is associated with the expression of some proinflammatory cytokines in RA.
Figure 5.
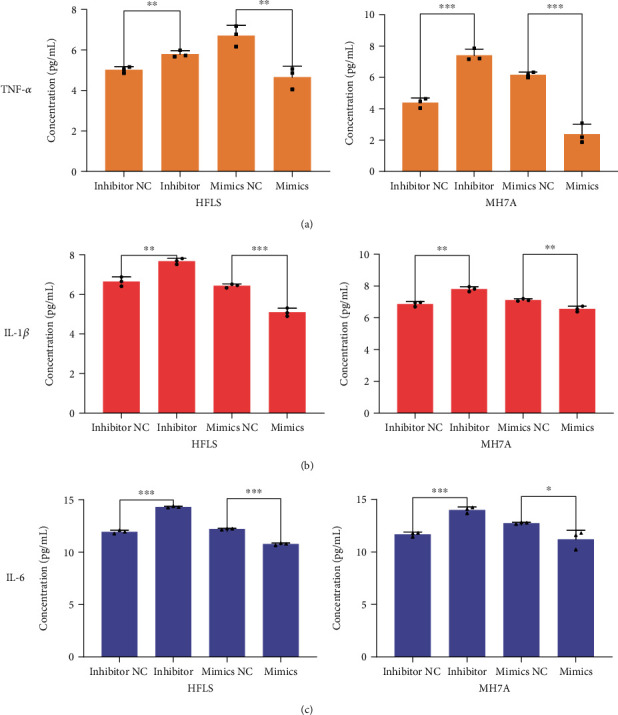
The effects of miRNA-155 on the production of proinflammatory cytokines in normal FLS and RA-FLS. (a) The levels of TNF-α in the supernatant of both HFLS and MH7A cells were measured by ELISA assay. ∗P < 0.05. (b) The levels of IL-1β in the supernatant of both HFLS and MH7A cells were measured by ELISA assay. ∗P < 0.05. (c) The levels of IL-6 in the supernatant of both HFLS and MH7A cells were measured by ELISA assay. ∗P < 0.05.
4. Discussion
RA is characterized by the improved proliferation of FLS and aggregation of proinflammatory cytokines in the joints. Previous studies have shown that RA is mainly due to the improved proliferation, migration, and invasion capacity of FLS [28]. Some evidence showed that miRNAs play huge roles in immune system diseases [29–32]. In addition, some miRNAs modulated by drug treatment also have important effects on osteogenic differentiation. For example, miR-21-5p can affect MC3T3-E1 cell proliferation, apoptosis, osteogenic differentiation, and matrix mineralization of osteoblasts by Icariin [33], which is known as an antitumor drug [25, 26]. However, the specific role of miRNA-155 in RA has not been elucidated.
In our study, we firstly cultured the obtained HFLS and MH7A cells and detected their apoptosis percent at the 4th generation, the results showed that the apoptosis percent of MH7A cells was significantly higher than that of HFLS cells. It may be due to that MH7A cells contain more inflammation factors and possess stronger migration and invasion abilities than HFLS cells.
Subsequently, we used miRNA-155 mimics and miRNA-155 inhibitor to stimulate HFLS and MH7A cells, we randomly divided the samples into two groups; one group was used to detect the expression of miRNA-155 in cells by RT-qPCR, and the other group was used to detect the cell proliferation by CCK-8 analysis. The results suggest that miRNA-155 mimics can promote the expression of miRNA-155 and the proliferation of FLS cells (not only normal FLS but also RA-FLS). During the occurrence of some autoimmune diseases, miRNA-155 has been proved to promote cell proliferation [34, 35]. We also found the same phenomenon in RA, which may further explain the function of miRNA-155 on cell proliferation in autoimmune diseases. In addition, some studies have shown that miRNA-155 abounds in B cells, monocytes, and macrophages from synovial tissues of RA patients [36, 37]. On this basis, our results showed a similar conclusion in inflammatory cells but more clearly elucidated a direct function of miRNA-155 on inflammatory cell proliferation.
In our study, RT-qPCR and WB results showed that miRNA-155 mimics and inhibitor treatment regulated the expression of LSD1, especially in MH7A cells. These results implied that miRNA-155 can affect cell proliferation through LSD1, and its effect was obvious in the inflammatory cells. Although LSD1 has been reported to promote the cell proliferation in some tumor diseases [38, 39], its function in autoimmune diseases, especially in RA, still needs to be studied. Our results provided new evidence to elucidate that miRNA-155 related cell proliferation may through LSD1 regulation. Therefore, the molecular mechanism of LSD1 in RA will be our future research direction to explore the possible targeted therapy.
In order to explore whether miRNA-155 can affect joint inflammation by regulating LSD1, we used miRNA-155 mimics and inhibitor to interfere with HFLS and MH7A cells. The results showed that miRNA-155 mimics could inhibit the expression of LSD1 and proinflammatory cytokines in both HFLS and MH7A cells. On the contrary, miRNA-155 inhibitor could promote the expression of LSD1 and proinflammatory cytokines in both HFLS and MH7A cells. However, several studies found that miRNA-155−/− mice showed reduced bone destruction and osteoclast production [22]. Moreover, overexpression of miRNA-155 in monocytes leads to an increase of TNFα, IL-1β, and IL-6 [40]. Our previous study showed that the joint injury and inflammatory response of RA model mice were improved to a certain extent after the LSD1 gene knockdown [15]. The complexity of RA and the different cell types may be the main reasons for the different conclusions. However, the interaction of miRNA-155 and LSD1, which closely related to the inflammation, could identify the potential roles in FLS cells. It will guide us to further investigate the function of LSD1 during inflammation, and to confirm that miRNA-155 can regulate proteins to affect the expression of proinflammatory cytokines [29].
Although our research cannot cover the whole process of RA, we try to explain the development of inflammation. The internal environment of the body is quite complicated. A certain gene, protein, or factor alone cannot determine the occurrence and outcome of the disease. Due to the change of culture conditions and the adaptability of the cell itself, the in vitro environment will affect cell proliferation, apoptosis, invasion, secretion, etc. We hope that our research will help to provide more evidence and new strategies for drugs and the fundamental prevention and treatment of RA.
Acknowledgments
The study was supported by grants from the National Natural Science Foundation of China (81501866), the Science & Technology Bureau of Nantong (MA2019005), the Health Commission of Nantong (JC2019075), the Jiangsu Provincial Young Medical Talent Foundation (QNRC2016411), the Jiangsu Six-One Project (LGY2018035), the Jiangsu 333 Talent Peak Program (To W.L.and J.F.), and the Nantong 226 High-level Talents Project (To J. F.).
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
Authors' Contributions
Ziliang Yu, Hao Liu, and Jianbo Fan contributed equally to this work.
References
- 1.Fazal S. A., Khan M., Nishi S. E., et al. A clinical update and global economic burden of rheumatoid arthritis. Endocrine, Metabolic & Immune Disorders Drug Targets. 2018;18(2):98–109. doi: 10.2174/1871530317666171114122417. [DOI] [PubMed] [Google Scholar]
- 2.van der Woude D., van der Helm-van Mil A. H. M. Update on the epidemiology, risk factors, and disease outcomes of rheumatoid arthritis. Best Practice & Research. Clinical Rheumatology. 2018;32(2):174–187. doi: 10.1016/j.berh.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Smolen J. S., Aletaha D., McInnes I. B. Rheumatoid arthritis. Lancet. 2016;388(10055):2023–2038. doi: 10.1016/S0140-6736(16)30173-8. [DOI] [PubMed] [Google Scholar]
- 4.McInnes I. B., Schett G. The pathogenesis of rheumatoid arthritis. The New England Journal of Medicine. 2011;365(23):2205–2219. doi: 10.1056/NEJMra1004965. [DOI] [PubMed] [Google Scholar]
- 5.Noss E. H., Brenner M. B. The role and therapeutic implications of fibroblast-like synoviocytes in inflammation and cartilage erosion in rheumatoid arthritis. Immunological Reviews. 2008;223(1):252–270. doi: 10.1111/j.1600-065X.2008.00648.x. [DOI] [PubMed] [Google Scholar]
- 6.McInnes I. B., Buckley C. D., Isaacs J. D. Cytokines in rheumatoid arthritis - shaping the immunological landscape. Nature Reviews Rheumatology. 2016;12(1):63–68. doi: 10.1038/nrrheum.2015.171. [DOI] [PubMed] [Google Scholar]
- 7.Alam J., Jantan I., Bukhari S. N. A. Rheumatoid arthritis: recent advances on its etiology, role of cytokines and pharmacotherapy. Biomedicine & Pharmacotherapy. 2017;92:615–633. doi: 10.1016/j.biopha.2017.05.055. [DOI] [PubMed] [Google Scholar]
- 8.Bek S., Bojesen A. B., Nielsen J. V., et al. Systematic review and meta-analysis: pharmacogenetics of anti-TNF treatment response in rheumatoid arthritis. The Pharmacogenomics Journal. 2017;17(5):403–411. doi: 10.1038/tpj.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Narazaki M., Tanaka T., Kishimoto T. The role and therapeutic targeting of IL-6 in rheumatoid arthritis. Expert Review of Clinical Immunology. 2017;13(6):535–551. doi: 10.1080/1744666X.2017.1295850. [DOI] [PubMed] [Google Scholar]
- 10.Zhai K. F., Duan H., Cui C. Y., et al. Liquiritin from Glycyrrhiza uralensis attenuating rheumatoid arthritis via reducing inflammation, suppressing angiogenesis, and inhibiting MAPK signaling pathway. Journal of Agricultural and Food Chemistry. 2019;67(10):2856–2864. doi: 10.1021/acs.jafc.9b00185. [DOI] [PubMed] [Google Scholar]
- 11.Qiu H., Wu H., Chan V., Lau C. S., Lu Q. Transcriptional and epigenetic regulation of follicular T-helper cells and their role in autoimmunity. Autoimmunity. 2017;50(2):71–81. doi: 10.1080/08916934.2017.1284821. [DOI] [PubMed] [Google Scholar]
- 12.Meng M., Liu H., Chen S., et al. Methylation of H3K27 and H3K4 in key gene promoter regions of thymus in RA mice is involved in the abnormal development and differentiation of iNKT cells. Immunogenetics. 2019;71(7):489–499. doi: 10.1007/s00251-019-01124-x. [DOI] [PubMed] [Google Scholar]
- 13.Shi Y., Lan F., Matson C., et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 14.Huang Y., Vasilatos S. N., Boric L., Shaw P. G., Davidson N. E. Inhibitors of histone demethylation and histone deacetylation cooperate in regulating gene expression and inhibiting growth in human breast cancer cells. Breast Cancer Research and Treatment. 2012;131(3):777–789. doi: 10.1007/s10549-011-1480-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu W., Fan J.-B., Xu D.-W., et al. Knockdown of LSD1 ameliorates the severity of rheumatoid arthritis and decreases the function of CD4 T cells in mouse models. International Journal of Clinical and Experimental Pathology. 2018;11(1):333–341. [PMC free article] [PubMed] [Google Scholar]
- 16.Faraoni I., Antonetti F. R., Cardone J., Bonmassar E. miR-155 gene: a typical multifunctional microRNA. Biochimica et Biophysica Acta. 2009;1792(6):497–505. doi: 10.1016/j.bbadis.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 17.Yan Q., Chen J., Li W., Bao C., Fu Q. Targeting miR-155 to treat experimental scleroderma. Scientific Reports. 2016;6(1) doi: 10.1038/srep20314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Artlett C. M., Sassi-Gaha S., Hope J. L., Feghali-Bostwick C. A., Katsikis P. D. Mir-155 is overexpressed in systemic sclerosis fibroblasts and is required for NLRP3 inflammasome-mediated collagen synthesis during fibrosis. Arthritis Research & Therapy. 2017;19(1) doi: 10.1186/s13075-017-1331-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayashi T., Kaneda T., Toyama Y., Kumegawa M., Hakeda Y. Regulation of receptor activator of NF-kappa B ligand-induced osteoclastogenesis by endogenous interferon-beta (INF-beta ) and suppressors of cytokine signaling (SOCS). The possible counteracting role of SOCSs- in IFN-beta-inhibited osteoclast formation. The Journal of Biological Chemistry. 2002;277(31):27880–27886. doi: 10.1074/jbc.M203836200. [DOI] [PubMed] [Google Scholar]
- 20.Mann M., Barad O., Agami R., Geiger B., Hornstein E. miRNA-based mechanism for the commitment of multipotent progenitors to a single cellular fate. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(36):15804–15809. doi: 10.1073/pnas.0915022107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J., Zhao H., Chen J., et al. Interferon-β-induced miR-155 inhibits osteoclast differentiation by targeting SOCS1 and MITF. FEBS Letters. 2012;586(19):3255–3262. doi: 10.1016/j.febslet.2012.06.047. [DOI] [PubMed] [Google Scholar]
- 22.Blüml S., Bonelli M., Niederreiter B., et al. Essential role of microRNA-155 in the pathogenesis of autoimmune arthritis in mice. Arthritis and Rheumatism. 2011;63(5):1281–1288. doi: 10.1002/art.30281. [DOI] [PubMed] [Google Scholar]
- 23.Daniunaite K., Dubikaityte M., Gibas P., et al. Clinical significance of miRNA host gene promoter methylation in prostate cancer. Human Molecular Genetics. 2017;26(13):2451–2461. doi: 10.1093/hmg/ddx138. [DOI] [PubMed] [Google Scholar]
- 24.Zhang F., Zhang Y.-Y., Sun Y.-S., et al. Asparanin A fromAsparagus officinalisL. induces G0/G1 cell cycle arrest and apoptosis in human endometrial carcinoma Ishikawa cells via mitochondrial and PI3K/AKT signaling pathways. Journal of Agricultural and Food Chemistry. 2019;68(1):213–224. doi: 10.1021/acs.jafc.9b07103. [DOI] [PubMed] [Google Scholar]
- 25.Sun Y. S., Thakur K., Hu F., Zhang J. G., Wei Z. J. Icariside II inhibits tumorigenesis via inhibiting AKT/Cyclin E/ CDK 2 pathway and activating mitochondria-dependent pathway. Pharmacological Research. 2020;152:p. 104616. doi: 10.1016/j.phrs.2019.104616. [DOI] [PubMed] [Google Scholar]
- 26.Sun Y. S., Thakur K., Hu F., Cespedes-Acuña C. L., Zhang J. G., Wei Z. J. Icariside II suppresses cervical cancer cell migration through JNK modulated matrix metalloproteinase-2/9 inhibition in vitro and in vivo. Biomedicine & Pharmacotherapy. 2020;125:p. 110013. doi: 10.1016/j.biopha.2020.110013. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y. Y., Zhang F., Zhang Y. S., et al. Mechanism of juglone-induced cell cycle arrest and apoptosis in Ishikawa human endometrial cancer cells. Journal of Agricultural and Food Chemistry. 2019;67(26):7378–7389. doi: 10.1021/acs.jafc.9b02759. [DOI] [PubMed] [Google Scholar]
- 28.Huber L. C., Distler O., Tarner I., Gay R. E., Gay S., Pap T. Synovial fibroblasts: key players in rheumatoid arthritis. Rheumatology (Oxford) 2006;45(6):669–675. doi: 10.1093/rheumatology/kel065. [DOI] [PubMed] [Google Scholar]
- 29.Miao C. G., Yang Y. Y., He X., et al. New advances of microRNAs in the pathogenesis of rheumatoid arthritis, with a focus on the crosstalk between DNA methylation and the microRNA machinery. Cellular Signalling. 2013;25(5):1118–1125. doi: 10.1016/j.cellsig.2013.01.024. [DOI] [PubMed] [Google Scholar]
- 30.Shi D. L., Shi G. R., Xie J., Du X. Z., Yang H. MicroRNA-27a inhibits cell migration and invasion of fibroblast-like synoviocytes by targeting follistatin-like protein 1 in rheumatoid arthritis. Molecules and Cells. 2016;39(8):611–618. doi: 10.14348/molcells.2016.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miao C. G., Yang Y. Y., He X., et al. MicroRNA-152 modulates the canonical Wnt pathway activation by targeting DNA methyltransferase 1 in arthritic rat model. Biochimie. 2014;106:149–156. doi: 10.1016/j.biochi.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 32.Hong B. K., You S., Yoo S. A., et al. MicroRNA-143 and -145 modulate the phenotype of synovial fibroblasts in rheumatoid arthritis. Experimental & Molecular Medicine. 2017;49(8):p. e363. doi: 10.1038/emm.2017.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lian F., Zhao C., Qu J., et al. Icariin attenuates titanium particle-induced inhibition of osteogenic differentiation and matrix mineralization via miR-21-5p. Cell Biology International. 2018;42(8):931–939. doi: 10.1002/cbin.10957. [DOI] [PubMed] [Google Scholar]
- 34.Xu L., Leng H., Shi X., Ji J., Fu J., Leng H. MiR-155 promotes cell proliferation and inhibits apoptosis by PTEN signaling pathway in the psoriasis. Biomedicine & Pharmacotherapy. 2017;90:524–530. doi: 10.1016/j.biopha.2017.03.105. [DOI] [PubMed] [Google Scholar]
- 35.Ren L., Zhao Y., Huo X., Wu X. MiR-155-5p promotes fibroblast cell proliferation and inhibits FOXO signaling pathway in vulvar lichen sclerosis by targeting FOXO3 and CDKN1B. Gene. 2018;653:43–50. doi: 10.1016/j.gene.2018.01.049. [DOI] [PubMed] [Google Scholar]
- 36.Kurowska-Stolarska M., Alivernini S., Ballantine L. E., et al. MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(27):11193–11198. doi: 10.1073/pnas.1019536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alivernini S., Kurowska-Stolarska M., Tolusso B., et al. MicroRNA-155 influences B-cell function through PU.1 in rheumatoid arthritis. Nature Communications. 2016;7(1) doi: 10.1038/ncomms12970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao C., Vasilatos S. N., Bhargava R., et al. Functional interaction of histone deacetylase 5 (HDAC5) and lysine-specific demethylase 1 (LSD1) promotes breast cancer progression. Oncogene. 2017;36(1):133–145. doi: 10.1038/onc.2016.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lv T., Yuan D., Miao X., et al. Over-expression of LSD1 promotes proliferation, migration and invasion in non-small cell lung cancer. PLoS One. 2012;7(4) doi: 10.1371/journal.pone.0035065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elmesmari A., Fraser A. R., Wood C., et al. MicroRNA-155 regulates monocyte chemokine and chemokine receptor expression in rheumatoid arthritis. Rheumatology (Oxford) 2016;55(11):2056–2065. doi: 10.1093/rheumatology/kew272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.


