Abstract
Chest X-ray is the first imaging technique that plays an important role in the diagnosis of COVID-19 disease. Due to the high availability of large-scale annotated image datasets, great success has been achieved using convolutional neural networks (CNN s) for image recognition and classification. However, due to the limited availability of annotated medical images, the classification of medical images remains the biggest challenge in medical diagnosis. Thanks to transfer learning, an effective mechanism that can provide a promising solution by transferring knowledge from generic object recognition tasks to domain-specific tasks. In this paper, we validate and a deep CNN, called Decompose, Transfer, and Compose (DeTraC), for the classification of COVID-19 chest X-ray images. DeTraC can deal with any irregularities in the image dataset by investigating its class boundaries using a class decomposition mechanism. The experimental results showed the capability of DeTraC in the detection of COVID-19 cases from a comprehensive image dataset collected from several hospitals around the world. High accuracy of 93.1% (with a sensitivity of 100%) was achieved by DeTraC in the detection of COVID-19 X-ray images from normal, and severe acute respiratory syndrome cases.
Keywords: DeTraC, Covolutional neural networks, COVID-19 detection, Chest X-ray images, Data irregularities
Introduction
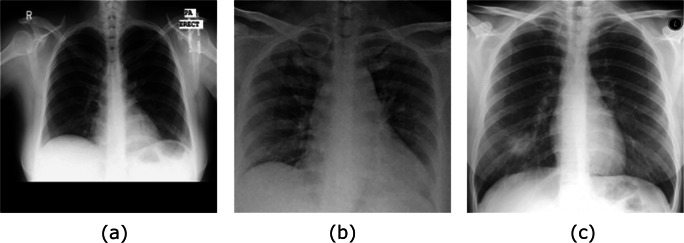
Diagnosis of COVID-19 is typically associated with both the symptoms of pneumonia and Chest X-ray tests [25]. Chest X-ray is the first imaging technique that plays an important role in the diagnosis of COVID-19 disease. Figure 1 shows a negative example of a normal chest X-ray, a positive one with COVID-19, and a positive one with the severe acute respiratory syndrome (SARS).
Fig. 1.
Examples of a) normal, b) COVID-19, and c) SARS chest X-ray images
Several classical machine learning approaches have been previously used for automatic classification of digitised chest images [7, 13]. For example, in [17], three statistical features were calculated from lung texture to discriminate between malignant and benign lung nodules using a Support Vector Machine SVM classifier. A grey-level co-occurrence matrix method was used with Backpropagation Network [22] to classify images from being normal or cancerous. With the availability of enough annotated images, deep learning approaches [1, 3, 30] have demonstrated their superiority over the classical machine learning approaches. CNN architecture is one of the most popular deep learning approaches with superior achievements in the medical imaging domain [14]. The primary success of CNN is due to its ability to learn features automatically from domain-specific images, unlike the classical machine learning methods. The popular strategy for training CNN architecture is to transfer learned knowledge from a pre-trained network that fulfilled one task into a new task [19]. This method is faster and easy to apply without the need for a huge annotated dataset for training; therefore many researchers tend to apply this strategy especially with medical imaging. Transfer learning can be accomplished with three major scenarios [16]: a) “shallow tuning”, which adapts only the last classification layer to cope with the new task, and freezes the parameters of the remaining layers without training; b) “deep tuning” which aims to retrain all the parameters of the pre-trained network from end-to-end manner; and (c) “fine-tuning” that aims to gradually train more layers by tuning the learning parameters until a significant performance boost is achieved. Transfer knowledge via fine-tuning mechanism showed outstanding performance in chest X-ray image classification [3, 9, 26].
Class decomposition [33] has been proposed with the aim of enhancing low variance classifiers facilitating more flexibility to their decision boundaries. It aims to the simplification of the local structure of a dataset in a way to cope with any irregularities in the data distribution. Class decomposition has been previously used in various automatic learning workbooks as a pre-processing step to improve the performance of different classification models. In the medical diagnostic domain, class decomposition has been applied to significantly enhance the classification performance of models such as Random Forests, Naive Bayes, C4.5, and SV M [20, 21, 37].
In this paper, we adapt our previously proposed convolutional neural network architecture based on class decomposition, which we term Decompose, Transfer, and Compose (DeTraC) model, to improve the performance of pre-trained models on the detection of COVID-19 cases from chest X-ray images.1 This is by adding a class decomposition layer to the pre-trained models. The class decomposition layer aims to partition each class within the image dataset into several sub-classes and then assign new labels to the new set, where each subset is treated as an independent class, then those subsets are assembled back to produce the final predictions. For the classification performance evaluation, we used images of chest X-ray collected from several hospitals and institutions. The dataset provides complicated computer vision challenging problems due to the intensity inhomogeneity in the images and irregularities in the data distribution.
The paper is organised as follow. In Section 2, we review the state-of-the-art methods for COVID-19 detection. Section 3 discusses the main components of DeTraC and its adaptation to the detection of COVID-19 cases. Section 4 describes our experiments on several chest X-ray images collected from different hospitals. In Section 5, we discuss our findings. Finally, Section 6 concludes the work.
Related work
In the last few months, World Health Organization (WHO) has declared that a new virus called COVID-19 has been spread aggressively in several countries around the world [18]. Diagnosis of COVID-19 is typically associated with the symptoms of pneumonia, which can be revealed by genetic and imaging tests. Fast detection of the COVID-19 can be contributed to control the spread of the disease.
Image tests can provide a fast detection of COVID-19, and consequently contribute to control the spread of the disease. Chest X-ray (CXR) and Computed Tomography (CT) are the imaging techniques that play an important role in the diagnosis of COVID-19 disease. The historical conception of image diagnostic systems has been comprehensively explored through several approaches ranging from feature engineering to feature learning.
Convolutional neural network (CNN) is one of the most popular and effective approaches in the diagnosis of COVD-19 from digitised images. Several reviews have been carried out to highlight recent contributions to COVID-19 detection [8, 15, 24]. For example, in [35], a CNN was applied based on Inception network to detect COVID-19 disease within CT. In [29], a modified version of ResNet-50 pre-trained network has been provided to classify CT images into three classes: healthy, COVID-19 and bacterial pneumonia. CXR were used in [23] by a CNN constructed based on various ImageNet pre-trained models to extract the high level features. Those features were fed into SVM as a machine learning classifier in order to detect the COVID-19 cases. Moreover, in [34], a CNN architecture called COVID-Net based on transfer learning was applied to classify the CXR images into four classes: normal, bacterial infection, non-COVID and COVID-19 viral infection. In [4], a dataset of CXR images from patients with pneumonia, confirmed COVID-19 disease, and normal incidents, was used to evaluate the performance of state-of-the-art convolutional neural network architectures proposed previously for medical image classification. The study suggested that transfer learning can extract significant features related to the COVID-19 disease.
Having reviewed the related work, it is evident that despite the success of deep learning in the detection of COVID-19 from CXR and CT images, data irregularities have not been explored. It is common in medical imaging in particular that datasets exhibit different types of irregularities (e.g. overlapping classes) that affect the resulting accuracy of machine learning models. Thus, this work focuses on dealing with data irregularities, as presented in the following section.
DeTraC method
This section describes in sufficient details the proposed method for detecting COVID-19 from CXR images. Starting with an overview of the architecture through to the different components of the method, the section discusses the workflow and formalises the method.
DeTraC architecture overview
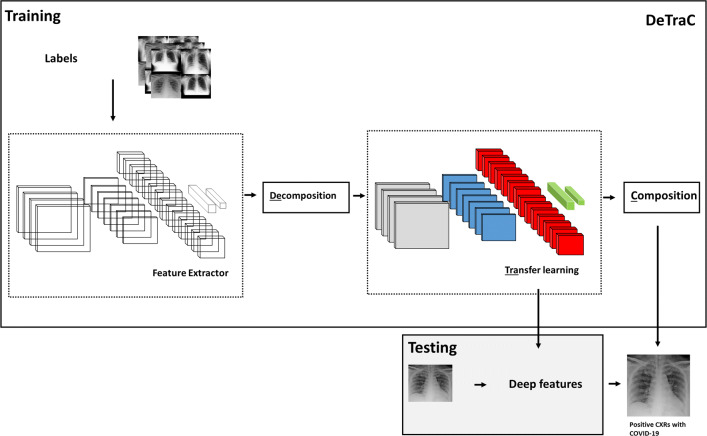
DeTraC model consists of three phases. In the first phase, we train the backbone pre-trained CNN model of DeTraC to extract deep local features from each image. Then we apply the class-decomposition layer of DeTraC to simplify the local structure of the data distribution. In the second phase, the training is accomplished using a sophisticated gradient descent optimisation method. Finally, we use a class composition layer to refine the final classification of the images. As illustrated in Fig. 2, class decomposition and composition components are added respectively before and after knowledge transformation from an ImageNet pre-trained CNN model. The class decomposition layer aiming at partitioning each class within the image dataset into k sub-classes, where each subclass is treated independently. Then those sub-classes are assembled back using the class-composition component to produce the final classification of the original image dataset.
Fig. 2.
De compose, Tra nsfer, and C ompose (DeTraC) model for the detection of COVID-19 from CXR images
Deep feature extraction
A shallow-tuning mode was used during the adaptation and training of an ImageNet pre-trained CNN model using the collected CXR image dataset. We used the off-the-shelf CNN features of pre-trained models on ImageNet (where the training is accomplished only on the final classification layer) to construct the image feature space. However, due to the high dimensionality associated with the images, we applied PCA [36] to project the high-dimension feature space into a lower-dimension, where highly correlated features were ignored. This step is important for the class decomposition to produce more homogeneous classes, reduce the memory requirements, and improve the efficiency of the framework.
Class decomposition layer
Now assume that our feature space (PCA’s output) is represented by a 2-D matrix (denoted as dataset A), and L is a class category. A and L can be rewritten as
| 1 |
where n is the number of images, m is the number of features, and k is the number of classes. For class decomposition, we used k-means clustering [38] to further divide each class into homogeneous sub-classes (or clusters), where each pattern in the original class L is assigned to a class label associated with the nearest centroid based on the squared euclidean distance (SED):
| 2 |
where centroids are denoted as cj.
Once the clustering is accomplished, each class in L will further divided into k subclasses, resulting in a new dataset (denoted as dataset B).
Accordingly, the relationship between dataset A and B can be mathematically described as:
| 3 |
where the number of instances in A is equal to B while C encodes the new labels of the subclasses (e.g. ). Consequently A and B can be rewritten as:
| 4 |
Transfer learning
With the high availability of large-scale annotated image datasets, the chance for the different classes to be well-represented is high. Therefore, the learned in-between class-boundaries are most likely to be generic enough to new samples. On the other hand, with the limited availability of annotated medical image data, especially when some classes are suffering more compared to others in terms of the size and representation, the generalisation error might increase. This is because there might be a miscalibration between the minority and majority classes. Large-scale annotated image datasets (such as ImageNet) provide effective solutions to such a challenge via transfer learning where tens of millions parameters (of CNN architectures) are required to be trained.
For transfer learning, we used, tested, and compared several ImageNet pre-trained models in both shallow- and deep- tuning modes, as will be investigated in the experimental study section. With the limited availability of training data, stochastic gradient descent (SGD) can heavily be fluctuating the objective/loss function and hence overfitting can occur. To improve convergence and overcome overfitting, the mini-batch (MB) of stochastic gradient descent (mSGD) was used to minimise the objective function, E(⋅), with cross-entropy loss
| 5 |
where xj is the set of input images in the training, yj is the ground truth labels while z(⋅) is the predicted output from a softmax function.
Evaluation and composition
In the class decomposition layer of DeTraC, we divide each class within the image dataset into several sub-classes, where each subclass is treated as a new independent class. In the composition phase, those sub-classes are assembled back to produce the final prediction based on the original image dataset. For performance evaluation, we adopted Accuracy (ACC), Specificity (SP) and Sensitivity (SN) metrics. They are defined as:
| 6 |
| 7 |
| 8 |
where TP is the true positive in case of COVID-19 case and TN is the true negative in case of normal or other disease, while FP and FN are the incorrect model predictions for COVID-19 and other cases.
More precisely, in this work we are coping with a multi-classification problem. Consequently, our model has been evaluated using a multi-class confusion matrix of [28]. Before error correction, the input image can be classified into one of (c) non-overlapping classes. As a consequence, the confusion matrix would be a (Nc × Nc) matrix, and TP, TN, FP and FN for a specific class i are defined as:
| 9 |
| 10 |
| 11 |
| 12 |
where xii is an element in the diagonal of the matrix.
Procedural steps of DeTraC model
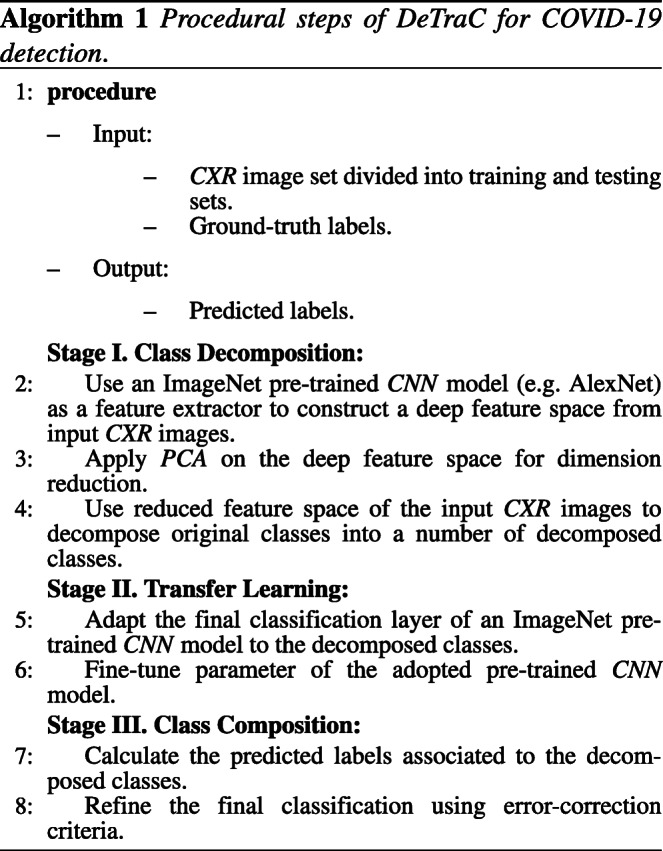
Having discussed the mathematical formulations of DeTraC model, in the following, the procedural steps of DeTraC model is shown and summarised in Algorithm 1.
DeTraC establishes the effectiveness of class decomposition in detecting COVID-19 from CXR images. The main contribution of DeTraC is its ability to deal with data irregularities, which is one of the most challenging problems in the detection of COVID-19 cases. The class decomposition layer of DeTraC can simplify the local structure of a dataset with a class imbalance. This is achieved by investigating the class boundaries of the dataset and adapt the transfer learning accordingly. In the following section, we experimentally validate DeTraC with real CXR images for the detection of COVID-19 cases from normal and SARS cases.
Experimental study
This section presents the dataset used in evaluating the proposed method, and discusses the experimental results.
Dataset
In this work we used a combination of two datasets:
Parameter settings
All the experiments in our work have been carried out in MATLAB 2019a on a PC with the following configuration: 3.70 GHz Intel(R) Core(TM) i3-6100 Duo, NVIDIA Corporation with the donation of the Quadra P5000GPU, and 8.00 GB RAM.
We applied different data augmentation techniques to generate more samples including flipping up/down and right/left, translation and rotation using random five different angles. This process resulted in a total of 1764 samples. Also, a histogram modification technique was applied to enhance the contrast of each image. The dataset was then divided into two groups: 70% for training the model and 30% for evaluation of the classification performance.
For the class decomposition layer, we used AlexNet [12] pre-trained network based on shallow learning mode to extract discriminative features of the three original classes. AlexNet is composed of 5 convolutional layers to represent learned features, 3 fully connected layers for the classification task. AlexNet uses 3 × 3 max-pooling layers with ReLU activation functions and three different kernel filters. We adopted the last fully connected layer into three classes and initialised the weight parameters for our specific classification task. For the class decomposition process, we used k-means clustering [38]. In this step, as pointed out in [2], we selected k = 2 and hence each class in L is further divided into two clusters (or subclasses), resulting in a new dataset (denoted as dataset B) with six classes (norm_1, norm_2, COVID-19_1,COVID-19_2, SARS_1, and SARS_2), see Table 1.
Table 1.
Sample distribution in each class of the CXR dataset before and after class decomposition
| Original labels | norm | COVID-19 | SARS | |||
|---|---|---|---|---|---|---|
| # instances | 80 | 105 | 11 | |||
| Decomposed labels | norm_1 | norm_2 | COVID-19_1 | COVID-19_2 | SARS_1 | SARS_2 |
| # instances | 441 | 279 | 666 | 283 | 63 | 36 |
In the transfer learning stage of DeTraC, we used different ImageNet pre-trained CNN networks such as AlexNet [12], VGG19 [27], ResNet [31], GoogleNet [32], and SqueezeNet [10]. The parameter settings for each pre-trained model during the training process are reported in Table 2.
Table 2.
Parameters settings for each pre-trained model used in our experiments
| Pre-trained model | Learning rate | MB-Size | Weight decay | Learning rate-decay |
|---|---|---|---|---|
| AlexNet | 0.001 | 256 | 0.001 | 0.9 every 3 epochs |
| VGG19 | 0.001 | 32 | 0.0001 | 0.9 every 2 epochs |
| GoogleNet | 0.001 | 128 | 0.001 | 0.95 every 3 epochs |
| ResNet | 0.0001 | 128 | 0.0001 | 0.95 every 5 epochs |
| SqueezeNet | 0.001 | 256 | 0.0001 | 0.9 every 2 epochs |
Validation and comparisons
To demonstrate the robustness of DeTraC, we used different ImageNet pre-trained CNN models (such as AlexNet, VGG19, ResNet, GoogleNet, and SqueezeNet) for the transfer learning stage in DeTraC. For a fair comparison, we also compared the performance of the different versions of DeTraC directly with the pre-trained models in both shallow- and deep- tuning modes. The results are summarised in Table 3, confirming the robustness and effectiveness of the class decomposition layer when used with pre-trained ImageNet CNN models.
Table 3.
COVID-19 Classification performance (on both original and augmented test cases) before and after applying DeTraC, obtained by AlexNet, VGG19, ResNet, GoogleNet, and SqueezeNet in case of shallow- and deep- tuning modes
| Pre-trained Model | Tuning Mode | without class decomposition | with class decomposition (DeTraC) | ||||
|---|---|---|---|---|---|---|---|
| Acc | SN | SP | Acc | SN | SP | ||
| (%) | (%) | (%) | (%) | (%) | (%) | ||
| AlexNet | shallow | 88.78 | 83.75 | 90.01 | 92.63 | 89.43 | 90.18 |
| VGG19 | shallow | 90.35 | 90.8 | 89.8 | 93.42 | 89.71 | 95.7 |
| ResNet | shallow | 91.14 | 78.17 | 90.65 | 92.12 | 64.13 | 94.2 |
| GoogleNet | shallow | 90.16 | 86.73 | 89.83 | 91.01 | 76.03 | 82.6 |
| SqueezeNet | shallow | 81.66 | 73.49 | 91.05 | 91.68 | 90.43 | 89.83 |
| AlexNet | deep | 93.84 | 91.73 | 90.30 | 95.66 | 97.53 | 93.49 |
| VGG19 | deep | 94.59 | 91.64 | 93.08 | 97.35 | 98.23 | 96.34 |
| ResNet | deep | 92.5 | 65.01 | 94.3 | 95.12 | 97.91 | 91.87 |
| GoogleNet | deep | 93.68 | 92.59 | 91.52 | 94.71 | 97.88 | 95.76 |
| SqueezeNet | deep | 92.24 | 95.04 | 88.61 | 94.90 | 95.70 | 94.71 |
As shown by Fig. 3, DeTraC with VGG19 has achieved the highest accuracy of 97.35%, sensitivity of 98.23%, and specificity of 96.34%. Moreover, Fig 3 shows the learning curve accuracy and loss between training and test obtained by DeTraC. Also, the Area Under the receiver Curve (AUC) was produced as shown in Fig 4.
Fig. 3.
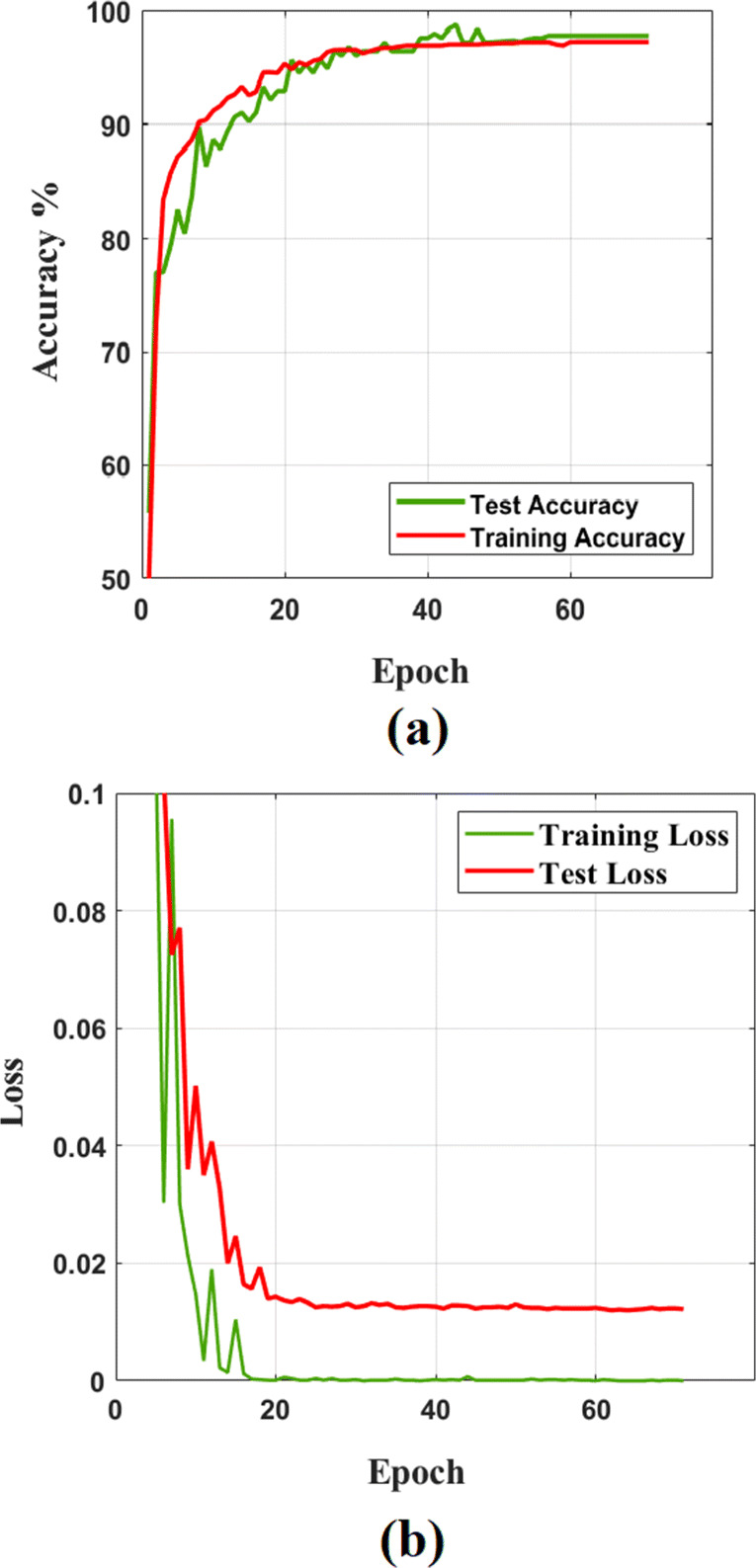
The learning curve accuracy (a) and error (b) obtained by DeTraC model when VGG19 is used as a backbone pre-trained model
Fig. 4.
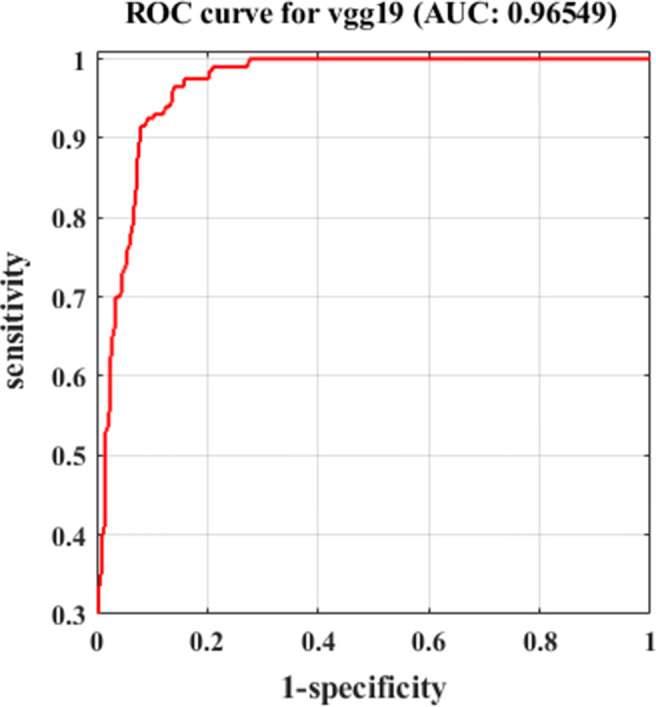
The ROC analysis curve by training DeTraC model based on VGG19 pre-trained model
Based on the above, deep-tuning mode provides better performance in all the cases. Thus, the last set of experiments is conducted to show the effectiveness of DeTraC on detecting original Covid-19 cases (i.e. after removing the augmented test images). Table 4 demonstrates the classification performance of all the models, used in this work, on the original test cases. As illustrated by Table 4, DeTraC outperformed all pre-trained models with a large margin in most cases. Note that the only case when a deep-tuned pre-trained model (VGG19) showed a higher sensitivity (100%) in detecting COVID-19 cases than DeTraC (87.09%) achieved at the cost of a very low specificity (53.44%) when compared to DeTraC (100%). This explains the notable accuracy boost when applying DeTraC on the deep-tuned VGG19 architecture (+ 25.36%).
Table 4.
COVID-19 Classification performance (on the original test cases) before and after applying DeTraC, obtained by AlexNet, VGG19, ResNet, GoogleNet, and SqueezeNet in case of deep-tuning mode
| Pre-trained Model | Tuning Mode | Without class decomposition | With class decomposition (DeTraC) | ||||
|---|---|---|---|---|---|---|---|
| Acc | SN | SP | Acc | SN | SP | ||
| (%) | (%) | (%) | (%) | (%) | (%) | ||
| AlexNet | deep | 58.62 | 67.41 | 48.14 | 89.10 | 82.1 | 84.30 |
| VGG19 | deep | 67.74 | 100 | 53.44 | 93.10 | 87.09 | 100 |
| ResNet | deep | 84.48 | 83.87 | 85.18 | 93.10 | 100 | 85.18 |
| GoogleNet | deep | 75.86 | 54.83 | 72.10 | 89.65 | 90.32 | 88.89 |
| SqueezeNet | deep | 70.68 | 54.81 | 88.88 | 82.75 | 83.87 | 81.48 |
Discussion
Training CNN s can be accomplished using two different strategies. They can be used as an end-to-end network, where an enormous number of annotated images must be provided (which is impractical in medical imaging). Alternatively, transfer learning usually provides an effective solution with the limited availability of annotated images by transferring knowledge from pre-trained CNN s (that have been learned from a bench-marked large-scale image dataset) to the specific medical imaging task. Transfer learning can be further accomplished by three main scenarios: shallow-tuning, fine-tuning, or deep-tuning. However, data irregularities, especially in medical imaging applications, remain a challenging problem that usually results in miscalibration between the different classes in the dataset. CNN s can provide an effective and robust solution for the detection of the COVID-19 cases from CXR images and this can be contributed to control the spread of the disease.
Here, we adapted and validated a deep convolutional neural network, called DeTraC, to deal with irregularities in a COVID-19 dataset by exploiting the advantages of class decomposition within the CNN s for image classification. DeTraC is a generic transfer learning model that aims to transfer knowledge from a generic large-scale image recognition task to a domain-specific task. In this work, we validated DeTraC with a COVID-19 dataset with imbalance classes (including 105 COVID-19, 80 normal, and 11 SARS cases). DeTraC has achieved high accuracy of 98.23% with VGG19 pre-trained ImageNet CNN model, confirming its effectiveness on real CXR images. To demonstrate the robustness and the contribution of the class decomposition layer of DeTraC during the knowledge transformation using transfer learning, we also used different pre-trained CNN models such as ALexNet, VGG, ResNet, GoogleNet, and SqueezeNet. Experimental results showed high accuracy in dealing with the COVID-19 cases used in this work with all the pre-trained models trained in a deep-tuning mode. More importantly, significant improvements have been demonstrated by DeTraC when compared to the pre-trained models without the class decomposition layer. Finally, this work suggests that the class decomposition layer with a pre-trained ImageNet CNN model (trained in a deep-tuning mode) is essential for transfer learning, see Figs. 5 and 6, especially when data irregularities are presented in the dataset.
Fig. 5.
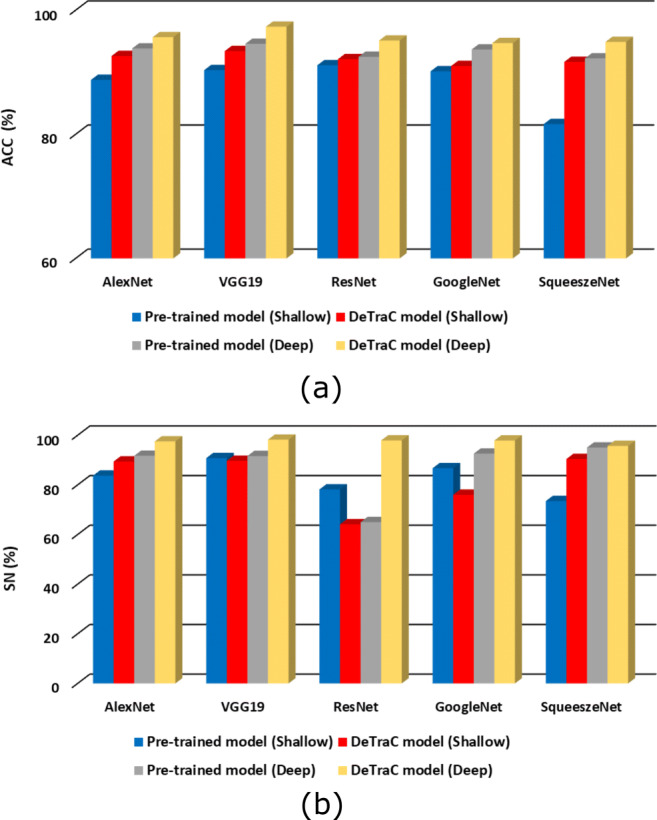
The accuracy (a) and sensitivity (b), on both original and augmented test cases, obtained by DeTraC model when compared to different pre-trained models, in shallow- and deep-tuning modes
Fig. 6.
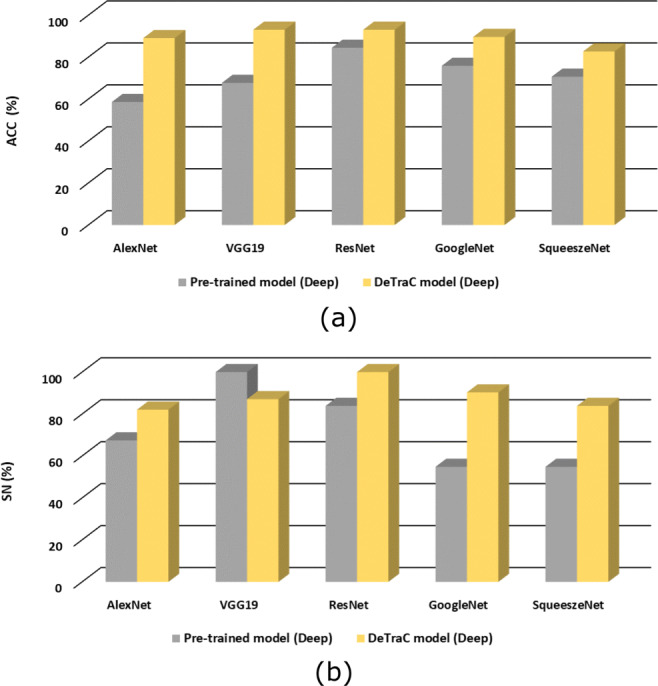
The accuracy (a) and sensitivity (b), on the original test cases only, obtained by DeTraC model when compared to different pre-trained models, in shallow- and deep-tuning modes
Conclusion and future work
Diagnosis of COVID-19 is typically associated with the symptoms of pneumonia, which can be revealed by genetic and imaging tests. Imagine test can provide a fast detection of the COVID-19 and consequently contribute to control the spread of the disease. CXR and CT are the imaging techniques that play an important role in the diagnosis of COVID-19 disease. Paramount progress has been made in deep CNN s for medical image classification, due to the availability of large-scale annotated image datasets. CNN s enable learning highly representative and hierarchical local image features directly from data. However, the irregularities in annotated data remains the biggest challenge in coping with real COVID-19 cases from CXR images.
In this paper, we adapted DeTraC, a deep CNN architecture, that relies on a class decomposition approach for the classification of COVID-19 images in a comprehensive dataset of CXR images. DeTraC showed effective and robust solutions for the classification of COVID-19 cases and its ability to cope with data irregularity and the limited number of training images too. We validated DeTraC with different pre-trained CNN models, where the highest accuracy has been obtained by VGG19 in DeTraC. With the continuous collection of data, we aim in the future to extend the experimental work validating the method with larger datasets. We also aim to add an explainability component to enhance the usability of the model. Finally, to increase the efficiency and allow deployment on handheld devices, model pruning, and quantisation will be utilised.
Biographies
Asmaa Abbas
was born in Assiut city, Egypt. She finished her B.Sc. in Computer Science from the Department of Mathematics at Assiut University in 2004. Asmaa has got MSc in computer science from the Mathematics Department at the Assiut University in 2020. She is currently a researcher at Mathematics Department in Assiut University. Her research interests include deep learning, medical image analysis, and data mining.

Mohammed M. Abdelsamea
is currently a Senior Lecturer in Data and Information Science at the School of Compu- ting and Digital Technology, Birmingham City University. Before joining BCU, he worked for the School of Computer Science at Nottingham Univer- sity, Mechanochemical Cell Biology at Warwick University, Nottingham Molecular Pathology Node (NMPN), and Division of Cancer and Stem Cells both at Nottingham Medical School. In 2016, he was a Marie Curie Research Fellow at the School of Computer Science at Nottingham University. Before moving to the UK, he worked as an Assistant Professor of Computer Science for Assiut University, Egypt. He was also a Visiting Researcher at Robert Gordon University, Aberdeen, UK. Abdelsamea has received his Ph.D. degree (with Doctor Europaeus) in Computer Science and Engineering from IMT - Institute for Advanced Studies, Lucca, Italy. His main research interests are in computer vision including image processing, deep learning, data mining and machine learning, pattern recognition, and image analysis.

Mohamed Medhat Gaber
is a Professor in Data Analytics at the School of Computing and Digital Technology, Birmingham City University.Mohamed received his PhD from Monash University, Australia. He then held appointments with the University of Sydney, CSIRO, and Monash University, all in Australia. Prior to joining Birmingham City University, Mohamed worked for the Robert Gordon University as a Reader in Computer Science and at the University of Portsmouth as a Senior Lecturer in Computer Science, both in the UK. He has published over 200 papers, coauthored 3 monograph-style books, and edited/co-edited6 books on data mining and knowledge discovery. His work has attracted well over four thousand citations, with an h-index of 35. Mohamed has served in the program committees of major conferences related to data mining, including ICDM, PAKDD, ECML/PKDD and ICML. He has also co-chaired numerous scientific events on various data mining topics. Professor Gaber is recognised as a Fellow of the British Higher Education Academy (HEA). He is also a member of the International Panel of Expert Advisers for the Australasian Data Mining Conferences. In 2007, he was awarded the CSIRO teamwork award.

Footnotes
The developed code is available at https://github.com/asmaa4may/DeTraC_COVId19.
This article belongs to the Topical Collection: Artificial Intelligence Applications for COVID-19, Detection, Control, Prediction, and Diagnosis
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Asmaa Abbas, Email: asmaa.abbas@science.aun.edu.eg.
Mohammed M. Abdelsamea, Email: mohammed.abdelsamea@bcu.ac.uk
Mohamed Medhat Gaber, Email: mohamed.gaber@bcu.ac.uk.
References
- 1.Abbas A, Abdelsamea MM (2018) Learning transformations for automated classification of manifestation of tuberculosis using convolutional neural network. In: 2018 13Th international conference on computer engineering and systems (ICCES), IEEE, pp 122–126
- 2.Abbas A, Abdelsamea MM, Gaber MM. Detrac: Transfer learning of class decomposed medical images in convolutional neural networks. IEEE Access. 2020;8:74901–74913. doi: 10.1109/ACCESS.2020.2989273. [DOI] [Google Scholar]
- 3.Anthimopoulos M, Christodoulidis S, Ebner L, Christe A, Mougiakakou S. Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Trans Med Imaging. 2016;35(5):1207–1216. doi: 10.1109/TMI.2016.2535865. [DOI] [PubMed] [Google Scholar]
- 4.Apostolopoulos ID, Mpesiana TA (2020) Covid-19: Automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med 1 [DOI] [PMC free article] [PubMed]
- 5.Candemir S, Jaeger S, Palaniappan K, Musco JP, Singh RK, Xue Z, Karargyris A, Antani S, Thoma G, McDonald CJ. Lung segmentation in chest radiographs using anatomical atlases with nonrigid registration. IEEE Trans Med Imaging. 2014;33(2):577–590. doi: 10.1109/TMI.2013.2290491. [DOI] [PubMed] [Google Scholar]
- 6.Cohen JP, Morrison P, Dao L (2020) Covid-19 image data collection. arXiv:2003.11597
- 7.Dandıl E, Çakiroğlu M, Ekşi Z, Özkan M, Kurt ÖK, Canan A (2014) Artificial neural network-based classification system for lung nodules on computed tomography scans. In: 2014 6Th international conference of soft computing and pattern recognition (soCPar), IEEE, pp 382–386
- 8.Dong D, Tang Z, Wang S, Hui H, Gong L, Lu Y, Xue Z, Liao H, Chen F, Yang F, Jin R, Wang K, Liu Z, Wei J, Mu W, Zhang H, Jiang J, Tian J, Li H (2020) The role of imaging in the detection and management of covid-19: a review. IEEE Rev Biomed Eng 1–1 [DOI] [PubMed]
- 9.Gao M, Bagci U, Lu L, Wu A, Buty M, Shin HC, Roth H, Papadakis GZ, Depeursinge A, Summers RM, et al. Holistic classification of ct attenuation patterns for interstitial lung diseases via deep convolutional neural networks. Comput Meth Biomechan Biomed Eng Imaging Visual. 2018;6(1):1–6. doi: 10.1080/21681163.2015.1124249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iandola FN, Han S, Moskewicz MW, Ashraf K, Dally WJ, Keutzer K (2016) Squeezenet:, Alexnet-level accuracy with 50x fewer parameters and< 0.5 mb model size. arXiv:1602.07360
- 11.Jaeger S, Karargyris A, Candemir S, Folio L, Siegelman J, Callaghan F, Xue Z, Palaniappan K, Singh RK, Antani S, Thoma G, Wang Y, Lu P, McDonald CJ. Automatic tuberculosis screening using chest radiographs. IEEE Trans Med Imaging. 2014;33(2):233–245. doi: 10.1109/TMI.2013.2284099. [DOI] [PubMed] [Google Scholar]
- 12.Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. In: Advances in neural information processing systems, pp 1097–1105
- 13.Kuruvilla J, Gunavathi K. Lung cancer classification using neural networks for ct images. Comput Meth Prog Biomed. 2014;113(1):202–209. doi: 10.1016/j.cmpb.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 14.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 15.Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, Bai J, Lu Y, Fang Z, Song Q, et al. (2020) Artificial intelligence distinguishes covid-19 from community acquired pneumonia on chest ct. Radiology p 200905 [DOI] [PMC free article] [PubMed]
- 16.Li Q, Cai W, Wang X, Zhou Y, Feng DD, Chen M (2014) Medical image classification with convolutional neural network. In: 2014 13Th international conference on control automation robotics & vision (ICARCV), IEEE, pp 844–848
- 17.Manikandan T, Bharathi N. Lung cancer detection using fuzzy auto-seed cluster means morphological segmentation and svm classifier. J Med Syst. 2016;40(7):181. doi: 10.1007/s10916-016-0539-9. [DOI] [PubMed] [Google Scholar]
- 18.Organization WH, et al. (2020) Coronavirus disease 2019 (covid-19); situation report 51
- 19.Pan SJ, Yang Q. A survey on transfer learning. IEEE Trans Knowl Data Eng. 2009;22(10):1345–1359. doi: 10.1109/TKDE.2009.191. [DOI] [Google Scholar]
- 20.Polaka I, Borisov A. Clustering-based decision tree classifier construction. Technol Econ Dev Econ. 2010;16(4):765–781. doi: 10.3846/tede.2010.47. [DOI] [Google Scholar]
- 21.Polaka I et al (2013) Clustering algorithm specifics in class decomposition no: Applied Information and Communication Technology
- 22.Sangamithraa P, Govindaraju S (2016) Lung tumour detection and classification using ek-mean clustering. In: 2016 International conference on wireless communications, signal processing and networking (wiSPNET), IEEE, pp 2201–2206
- 23.Sethy PK, Behera SK (2020) Detection of coronavirus disease (covid-19) based on deep features
- 24.Shi F, Wang J, Shi J, Wu Z, Wang Q, Tang Z, He K, Shi Y, Shen D (2020) Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for covid-19 IEEE Reviews in Biomedical Engineering [DOI] [PubMed]
- 25.Shi H, Han X, Jiang N, Cao Y, Alwalid O, Gu J, Fan Y, Zheng C (2020) Radiological findings from 81 patients with covid-19 pneumonia in wuhan, china: a descriptive study The Lancet Infectious Diseases [DOI] [PMC free article] [PubMed]
- 26.Shin HC, Roth HR, Gao M, Lu L, Xu Z, Nogues I, Yao J, Mollura D, Summers RM. Deep convolutional neural networks for computer-aided detection: Cnn architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging. 2016;35(5):1285–1298. doi: 10.1109/TMI.2016.2528162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simonyan K, Zisserman A (2014) Very Deep Convolutional Networks for Large-Scale Image Recognition, International Conference on Learning Representations
- 28.Sokolova M, Lapalme G. A systematic analysis of performance measures for classification tasks. Inform Process Manage. 2009;45(4):427–437. doi: 10.1016/j.ipm.2009.03.002. [DOI] [Google Scholar]
- 29.Song Y, Zheng S, Li L, Zhang X, Zhang X, Huang Z, Chen J, Zhao H, Jie Y, Wang R, et al. (2020) Deep learning enables accurate diagnosis of novel coronavirus (covid-19) with ct images medRxiv [DOI] [PMC free article] [PubMed]
- 30.Sun W, Zheng B, Qian W (2016) Computer aided lung cancer diagnosis with deep learning algorithms. In: Medical imaging 2016: computer-aided diagnosis, vol. 9785, p. 97850z. International society for optics and photonics
- 31.Szegedy C, Ioffe S, Vanhoucke V, Alemi AA (2017) Inception-v4, inception-resnet and the impact of residual connections on learning. In: Thirty-first AAAI conference on artificial intelligence
- 32.Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A (2015) Going deeper with convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 1–9
- 33.Vilalta R, Achari MK, Eick CF (2003) Class decomposition via clustering: a new framework for low-variance classifiers Third IEEE international conference on data mining, IEEE, pp 673–676
- 34.Wang L, Wong A (2020) Covid-net: A tailored deep convolutional neural network design for detection of covid-19 cases from chest radiography images [DOI] [PMC free article] [PubMed]
- 35.Wang S, Kang B, Ma J, Zeng X, Xiao M, Guo J, Cai M, Yang J, Li Y, Meng X, et al. (2020) A deep learning algorithm using ct images to screen for corona virus disease (covid-19) medRxiv [DOI] [PMC free article] [PubMed]
- 36.Wold S, Esbensen K, Geladi P. Principal component analysis. Chemometr Intell Lab Syst. 1987;2(1-3):37–52. doi: 10.1016/0169-7439(87)80084-9. [DOI] [Google Scholar]
- 37.Wu J, Xiong H, Chen J. Cog: local decomposition for rare class analysis. Data Min Knowl Disc. 2010;20(2):191–220. doi: 10.1007/s10618-009-0146-1. [DOI] [Google Scholar]
- 38.Wu X, Kumar V, Quinlan JR, Ghosh J, Yang Q, Motoda H, McLachlan GJ, Ng A, Liu B, Philip SY, et al. Top 10 algorithms in data mining. Knowl Inform Syst. 2008;14(1):1–37. doi: 10.1007/s10115-007-0114-2. [DOI] [Google Scholar]