Abstract
Drosha-dependent canonical microRNAs (miRNAs) play a crucial role in the biological functions and development of cancer. However, the effects of Drosha-independent non-canonical miRNAs remain poorly understood. In our previous work, we found a set of aberrant miRNAs, including some upregulated miRNAs, called Drosha-independent noncanonical miRNAs, in Drosha-knockdown gastric cancer (GC) cells. Surprisingly, Drosha-silenced GC cells still retained strong malignant properties (e.g., proliferation ability and cancer stem cell (CSC) characteristics), indicating that aberrantly upregulated non-canonical miRNAs may play an important role in the maintenance of the malignant properties in GC cells that express low Drosha levels. Here, we report that miR-6778–5p, a noncanonical miRNA, acts as a crucial regulator for maintenance of CSC stemness in Drosha-silenced GC cells. MiR-6778–5p belongs to the 5’-tail mirtron type of non-canonical miRNAs and is transcript splice-derived from intron 5 of SHMT1 (coding cytoplasmic serine hydroxymethyltransferase). It positively regulates expression of its host gene, SHMT1, via targeting YWHAE in Drosha-knockdown GC cells. Similar to its family member, SHMT2, SHMT1 plays a crucial role in folate-dependent serine/glycine inter-conversion in one-carbon metabolism. In Drosha wild type GC cells, SHMT2 mediates a mitochondrial-carbon metabolic pathway, which is a major pathway of one-carbon metabolism in normal cells and most cancer cells. However, in Drosha-silenced or Drosha low-expressing GC cells, miR-6778–5p positively regulates SHMT1, instead of SHMT2, thus mediating a compensatory activation of cytoplasmic carbon metabolism that plays an essential role in the maintenance of CSCs in gastric cancer (GCSCs). Drosha wild type GCSCs with SHMT2 are sensitive to 5-fluorouracil; however, Drosha low-expressing GCSCs with SHMT1 are 5-FU-resistant. The loss of miR-6778–5p or SHMT1 notably mitigates GCSC sphere formation and increases sensitivity to 5-fluorouracil in Drosha-knockdown gastric cancer cells. Thus, our study reveals a novel function of Drosha-independent noncanonical miRNAs in maintaining the stemness of GCSCs.
Keywords: Drosha-independent miRNA, miR-6778–5p, gastric CSC, cytoplasmic serine hydroxyl methyltransferase, one-carbon metabolism
Introduction
Gastric cancer (GC) is the fifth most common cancer and is a major cause of cancer mortality worldwide. In China, gastric cancer is the second leading cause of cancer deaths [1]. Although there are many treatments for gastric cancer in clinical practice, tumour metastasis and recurrence remain the most challenging problems in the treatment of gastric cancer. Thus, thoroughly exploring the pathological and molecular mechanisms of gastric cancer is at all imminent.
Recent studies indicate that cancer stem cells (CSCs) could play crucial roles in cancer progression and treatment resistance in gastric cancer [2, 3]. CSCs, a small subset of cells within the tumour bulk, have a dual capacity for self-renewal and pluripotent differentiation. They are considered to be the source of tumour initiation, development, metastasis, resistance to tumour treatment and recurrence [4]. Recently, researchers have found that CSCs in gastric cancer (GCSCs) express high levels of the surface marker CD44 and the stemness associated genes c-MYC and KLF4 [5, 6]. Compared to CSC studies in other tumours, however, there are few studies on the formation and maintenance of GCSCs, and their regulatory mechanisms are still unclear.
Drosha is critical for canonical microRNA (miRNA) biogenesis. Aberrant expression of Drosha has been implicated in the growth of cancer. Our previous study showed that aberrant Drosha expression may be associated with tumour malignancy in GC [7]. The invasion of gastric cancer cells was reduced by Drosha knockdown (Drosha KD) [8]. However, we also observed that downregulation of Drosha does not completely reverse the malignancy of gastric cancer, indicating that some unknown molecular mechanisms still maintain the malignant characteristics of gastric cancer cells. We found that some of miRNAs are abnormally upregulated in Drosha-silenced gastric cancer cells.
Aberrant expression of Drosha can cause processing of abnormal versions of microRNAs, leading to the occurrence and progression of cancer. Drosha is essential for canonical miRNA maturation; however, non-canonical microRNAs processed by alternative miRNA biogenesis pathways that bypass Drosha have been documented [9–15]. The first confirmed Drosha-independent miRNA biogenesis pathway was the mirtron pathway. Mirtrons, which serve as pre-miRNA mimics, are spliced and debranched from introns via spliceosome and debranching enzymes [11]. In addition, some mirtrons, so-called “tailed” mirtrons, contain an unstructured region 5’ or 3’ to a splicing-derived terminal intron hairpin[16]. Recently, another Drosha-independent miRNA has been discovered, that is derived from introns and produces pre-miRNA via RNA pol I [14, 15]. However, the function of these Drosha-independent miRNAs remains unclear. Only a few studies have found that mirtrons, similar to common miRNAs, can exert oncogenes or tumour suppressor genes in cancer [17, 18].
To investigate whether non-canonical miRNAs participate in the maintenance of malignancy of Drosha-low expressing or -silenced gastric cancer cells, we carefully analysed the abnormally upregulated miRNAs in Drosha-silenced gastric cancer cells. We found that most of these abnormal miRNAs belong to the noncanonical miRNA type, mirtrons (5’-tail mirtrons and 3’-tail mirtrons). Bioinformatics analysis revealed that these Drosha-independent miRNAs may be involved in cancer cell proliferation and CSC, among which atypical miRNA6778–5p (miR-6778–5p) is one of most enhanced 5’-tail mirtrons. Furthermore, miR-6778–5p is spliced from an intron of its host gene cytoplasmic, serine hydroxymethyltransferase (SHMT1), a key enzyme for cytoplasmic one-carbon metabolism. Metabolism changes in cancer have been reported to drive cell proliferation, survival and invasion [19–21], and metabolic reprogramming is crucial for tumourigenesis. Recent evidence indicates that metabolic pathways are involved in regulating the self-renewal and differentiation of SCs [22–27]. Compared to cancer cells, the metabolism of CSCs is more evident than glycolysis; while other CSCs can produce energy through classical mitochondrial oxidative phosphorylation or non-classical mitochondrial oxidative phosphorylation.
The SHMT gene encodes serine hydroxymethyltransferase (SHMT), which catalyses folate-dependent serine/glycine inter-conversion. There are two SHMT genes in the human genome: SHMT1 encodes a cytoplasmic isozyme in the cytosolic folate-dependent one-carbon metabolism process, and SHMT2 encodes the mitochondrial protein for maintenance of mitochondrial folate-dependent one-carbon metabolism. Generally, the SHMT2-mediated mitochondrial-carbon metabolic pathway is the major route through which one-carbon units are produced in normal cells and most cancer cells. Dysfunction of mitochondrial-carbon metabolic enzymes leads to impairment of mitochondrial carbon metabolism, and a compensatory activation of cytoplasmic carbon metabolism may play an essential role in maintenance of cell survival [28]. Here, we found that SHMT2 was downregulated in Drosha-knockdown or Drosha-decreased GC cells; however, SHMT1 was upregulated in these GC cells. Furthermore, SHMT1 has been shown to be a crucial factor in the folate-mediated one-carbon metabolism of CSCs. Zhang et al. observed that SHMT1 is overexpressed in tumour-initiating lung cancer cells (TICs) in non-small cell lung cancer (NSCLC) [29], suggesting that SHMT1 may be involved in the regulation of cancer stem cell activity by regulating cell metabolism; however, the underlying mechanisms remain to be clarified.
Furthermore, 14-3-3 protein epsilon (14-3-3ε), also called YWHAE, belongs to the 14-3-3 family of proteins. In general, 14-3-3 proteins interact with hundreds of proteins, affecting target protein activity, modification, and intracellular localization, and play a key role in signalling regulation in eukaryotes [30–32]. Study has shown that YWHAE is decreased in GC [33], suggesting that YWHAE may act as a tumour suppressor in GC.
In this study, we demonstrate that miR-6778–5p can maintain GCSC self-renewal by targeting YWHAE. YWHAE was found to inhibit the expression of SHMT1 by downregulating c-MYC, suggesting a positive feedback loop of SHMT1/miR-6778–5p/YWHAE/c-MYC/SHMT1 in gastric cancer cells. A high level of the cytosolic enzyme SHMT1 could replace the mitochondrial enzyme SHMT2 to promote CSC formation via cytoplasmic one-carbon metabolism, rather than mitochondrial-dependent one-carbon metabolism, in Drosha-knockdown gastric cancer cells. Therefore, our study revealed a novel mechanism of GCSC renewal and maintenance. Drosha-independent miR-6778–5p contributes to GCSCs and the malignant properties of gastric cancer via positive regulation of SHMT1 by targeting YWHAE, which participates in a folate-mediated cytoplasmic carbon metabolism pathway rather than mitochondrial carbon metabolism. Our study indicates a potential novel therapeutic strategy for gastric cancer.
Materials and Methods
Clinical samples
The gastric tumour tissues and their adjacent normal tissues used in this study were obtained from patients with gastric cancer without previous radiotherapy or chemotherapy at the First Affiliated Hospital of Chongqing Medical University. The investigation was approved by the ethics committee of Chongqing Medical University.
Cell culture, GCSC culture and sphere formation assays
The human gastric cancer cell lines MGC-803, SGC-7901, BGC-823 and NUGC-3 were kindly donated by Prof. Yang Ke in the Beijing Institute of Cancer Research (Beijing Shi, China). The Drosha-knockdown gastric cancer cells, including MGC-803 and SGC-7901 cells (called MGC-803 Drosha KD and SGC-7901 Drosha KD) were established as described previously[8]. MGC-803, BGC-823 and NUGC-3 parent cells and their corresponding Drosha KD cells were maintained in DMEM medium (Gibco, USA) supplemented with 10% FBS (Gibco, USA) at 37 °C in a humidified atmosphere containing 5% CO2; SGC-7901 and SGC-7901 Drosha KD cells were cultured in 1640 medium (Gibco, USA) supplemented with 10% FBS (Gibco, USA) at 37 °C in a humidified atmosphere containing 5% CO2.
Gastric cancer stem cells (GCSCs) were cultured as described previously [34]. GCSCs were enriched from MGC-803 or SGC-7901 cells using serum-free medium, which was composed of Dulbecco’s modified Eagle’s medium/F12 medium, 0.4% BSA (Sigma, USA), 20 μl/ml B27 supplement (Invitrogen, USA), 20 ng/ml basic fibroblast growth factor (bFGF, Gibco, USA), 10 ng/ml epidermal growth factor (EGF, Gibco, USA) and 2 μg/ml heparin. All GCSCs were kept in culture and passaged every 7 days. The percentage (%) of GCSC sphere formation was calculated as previously described[35]. The average volume of GCSC spheres (N=30 spheres) was calculated.
Plasmid constructs
The shRNAs specifically targeting miR-6778–5p and the YWHAE gene were separately reconstructed by sub-cloning into the pLVX-puro vector at the BamHI and EcoRI sites using PCR. The expression vectors of miR-6778–5p and SHMT1 were constructed by inserting the sequences of miR-6778–5p and SHMT1 cDNA into the pBABE-puro vector at the BamHI and EcoRI sites, respectively. The shRNA against SHMT1 was purchased from Hanheng Biotech (Shanghai, China). The pcDNA3.1-YWHAE was acquired from Addgene.
The splicing analysis of miR-6778/SHMT1 was performed as described previously[36]. Briefly, minigene constructs of miR-6778/SHMT1 (exons 5–6) was amplified from human genomic DNA by RT-PCR using the Phusion High-Fidelity DNA Polymerase (ThermoFisher Scientific). PCR products were digested with BamHI and HindIII and inserted into a pcDNA3.1 plasmid. The 5′- and 3′-splice site mutations were constructed using Phusion High-Fidelity DNA Polymerase with primers containing splicing site mutations. To generate a luciferase reporter that directly targeted YWHAE via miR-6778–5p, synthetic oligonucleotides(Invitrogen, USA) of the wild type or mutated binding sites of miR-6778–5p in the 3’-UTR of YWHAE was inserted into the pMIR-Reporter vector (Ambion, USA) at the Spe I and Hind III sites. The sequences described above are listed in Table S1 and Table S2.
miRNA array assay
The MGC-803/Drosha-shRNA (MGC-803/Drosha KD) and MGC-803/NC-shRNA (MGC-803/Drosha WT) control cells were used in the Agilent miRNA array (Agilent, Santa Clara, CA, USA). The raw data were normalized by Quantile algorithm using the Gene Spring Software12.6 (Agilent, Santa Clara, CA, USA). The different miRNAs were then grouped using hierarchical clustering with ‘complete’ using Heatmap.2 function (gplots package v.2.9.0).
RNA extraction and qRT-PCR
Total RNA was extracted using Trizol (Invitrogen, USA) according to the manufacturer’s instructions. The purified RNAs were used for reverse transcription using the PrimeScript RT Reagent Kit (TaKaRa, Dalian, China). qRT-PCR was carried out using a SYBR Premix Ex TaqTM II kit (TaKaRa, Dalian, China). Relative gene expression was determined using the 2Ct(internal control)−Ct(gene) method. The primers used are listed in Table S3.
Chromatin immunoprecipitation (ChIP) assay and Luciferase reporter assay
ChIP assays were conducted with a ChIP Kit (Thermo, USA) using c-MYC and IgG antibodies. The primers used for amplifying potential SHMT1 binding sites were as follows: forward: 5’- ACATAGTTAGACCCCCATCTC-3’; reverse: 5’- CGCGTGTGTGATAGCATCTCG-3’. SHMT1 promoters with a c-MYC binding motif (WT) or SHMT1 promoters without a c-MYC binding motif (MUT) were constructed using the pGL3 Basic system (Promega, USA). pGL3-SHMT1-promoter-WT or pGL3-SHMT1-promoter-Mut was transfected into MGC-803 cells together with the pCMV-Renilla control. At 48h after transfection, the luciferase activity was detected. Transient transfection and luciferase assays were performed as described previously [36]. HEK-293, MGC-803 or SGC-7901 cells were co-transfected with miR-6778–5p mimics (GenePharma, Shanghai) and pMIR-YWHAE 3’-UTR (wild-type or mutant) and pRL-TK control vector using lipofectamine 2000 (Invitrogen, USA). Renilla and firefly luciferase activity were measured with a Dual-Luciferase Reporter System (Promega, USA) after 48 h in culture.
Western blot analysis (WB analysis)
Cells were collected and lysed with RIPA buffer (containing protease inhibitors). Cell lysates were subsequently dissolved in 10% SDS-PAGE and subjected to western blotting. The following primary antibodies were used: anti-Drosha (1:1000; Abcam UK), anti-SHMT1 (1:1000; Abcam, UK), anti-YWHAE (1:1000; Abcam, UK), anti-SHMT2(1:1000; Abcam, UK), anti-MTHFD2 (1:1000, Abcam, UK), anti-CD44 (1:500; Santa Cruz, USA), anti-c-MYC (1:500; Santa Cruz, USA), anti-KLF4 (1:500; Santa Cruz, USA), and anti-β-Actin (1:1000; Bioshop, Canada). The appropriate horseradish peroxidase-conjugated secondary antibodies were subsequently applied, and images were captured using Scion image software.
Immunohistochemistry (IHC)
IHC analysis was performed as described previously [36]. Briefly, the tumour tissues were fixed with 4% paraformaldehyde. Paraffin-embedded specimens were sliced into 4 μm sections. The sections were treated with 3% hydrogen peroxide in methanol for 10 min to block endogenous peroxidase activity and then incubated with rabbit primary antibodies against CD44 (1:100), KLF4 (1:100) and c-MYC (1:100) overnight at 4 °C. After incubating with a polymer helper solution, polyperoxidase-anti-rabbit IgG, the sections were stained with diaminobenzidine (DAB). The CSC biomarker staining intensities were scored as follows: 0, no staining; 1+, 1–25%; 2+, 26–50%; 3+, >50% staining.
Metabolite Analysis
Gastric cancer cells (1.5×105) were plated in 6-well plates and cultured in DMEM for 24 h. After washing the cells with PBS, “starved media” including MEM (Invitrogen, USA), MEM vitamin solution (Sigma, USA) and D-Glucose solution (Sigma, USA) was added to the plates, and the cells were maintained in culture for 3 h. Then, the starved media was replaced with “analytic media” (including MEM (Invitrogen, USA), MEM vitamin solution (Sigma, USA), D-Glucose solution (Sigma, USA) and 2,3,3-2H-serine (Cambridge Isotope Laboratories)), and the cells were cultured for another 16 h and used for CSC enrichment.
GCSCs were cultured in serum-free medium as described above. GCSC spheres were carefully collected from the 6-well plate in a tube and centrifuged, and the supernatant was discarded. After washing with 1 ml of ice-cold PBS, 0.4 ml of pre-cooled methanol was added to the cell mixture, and the cells were incubated at −80 °C for 30 min. Then, 0.4 ml of ddH2O was added. Metabolite analysis by LC-MS/MS metabolomics was performed by Profleader (Shanghai, China).
LC-MS/MS analyses were performed using a Vanquish UHPLC system (Thermo Fisher) coupled with an Orbitrap Q Exactive HF-X mass spectrometer (Thermo Fisher). Samples were injected into a Hyperil Gold column (100×2.1 mm, 1.9 μm) using a 16-min linear gradient at a flow rate of 0.2 mL/min. The eluents for the positive polarity mode included eluent A (0.1% FA in water) and eluent B (methanol). The eluents for the negative polarity mode were eluent A (5 mM ammonium acetate, pH 9.0) and eluent B (methanol). The solvent gradient was set as follows: 2% B, 1.5 min; 2–100% B, 12.0 min; 100% B, 14.0 min; 100–2% B, 14.1 min; 2% B, 16 min. A Q Exactive HF-X mass spectrometer was operated in positive/negative polarity mode with a spray voltage of 3.2 kV, capillary temperature of 320°C, sheath gas flow rate of 35 arb and aux gas flow rate of 10 arb.
Xenograft experiments
Gastric cancer stem cells (1×105) were mixed with 100 μL of PBS and subcutaneously injected into 4-week-old female nude mice. Tumour volumes were measured using two calliper measurements every three days (volume = ½ [L × W2]). After four weeks, animals were euthanized. Tumour tissues were fixed in 4% paraformaldehyde and subjected to immunohistochemistry (IHC) analysis. The animal experiments and animal care were in accordance with institutional guidelines.
Statistical analysis
Statistical significance was determined with SPSS 17.0 software. All results are presented as the means ± SD of at least three independent determinations. Continuous variables were compared between two groups analysed using an independent Student’s t-test. A value of P<0.05 was considered statistically significant.
Results
1. Knockdown of Drosha leads to atypical novel microRNAs in gastric cancer cells
The biogenesis process of canonical microRNAs (miRNAs) requires Drosha and Dicer. A set of upregulated non-canonical miRNAs were identified in Drosha-knockdown (Drosha KD) gastric cancer (GC) MGC-803 cells by miRNA array analysis (Figure 1A). Non-canonical microRNAs processed by alternative miRNA biogenesis pathways that bypass Drosha have been documented in Caenorhabditis elegans and plants [11]. Two types of major atypical miRNAs exist (mirtrons and 5-capped miRNAs). Mirtrons are products derived from short hairpin introns by alternative splicing; 5’-capped miRNAs and microprocessor-independent 7-methylguanosine (m7G)-capped miRNAs are products derived from introns and transcribed by RNA pol I (Figure 1B). To classify the non-canonical miRNAs, we analysed the structure of these upregulated miRNAs using RNA structure 5.7 software and the UCSC (http://genome.ucsc.edu/) database. Some of the upregulated non-canonical miRNAs in Drosha-silenced GC cells are mirtrons (5’-tail mirtrons and 3’-tail mirtrons) (Figure 1C). Among them, miR-6760–5p is a known human 5’-tailed mirtron, and its expression was notably increased in the Drosha-knockdown cells (Figure 1A), consistent with previous findings [16]. In addition to miR-6760–5p, several other non-canonical miRNAs were identified. For example, hsa-mir-6778–5p is a human 5’-tailed mirtron located the in SHMT1 gene. The pre-miRNA structures of miR-6760 and miR-6778 have some typical characteristics, such as a typical pyrimidine rich region in the terminal AG splice acceptor, the 3’ end of their pre-miRNA matches the splice acceptor in their introns, and the end of the miR-6778/miR-6778* duplex and miR-6760/miR-6760* duplex exhibits a 3′-overhang (Figure 1D).
Figure 1. Knockdown of Drosha leads to novel non-canonical microRNAs in gastric cancer cells.
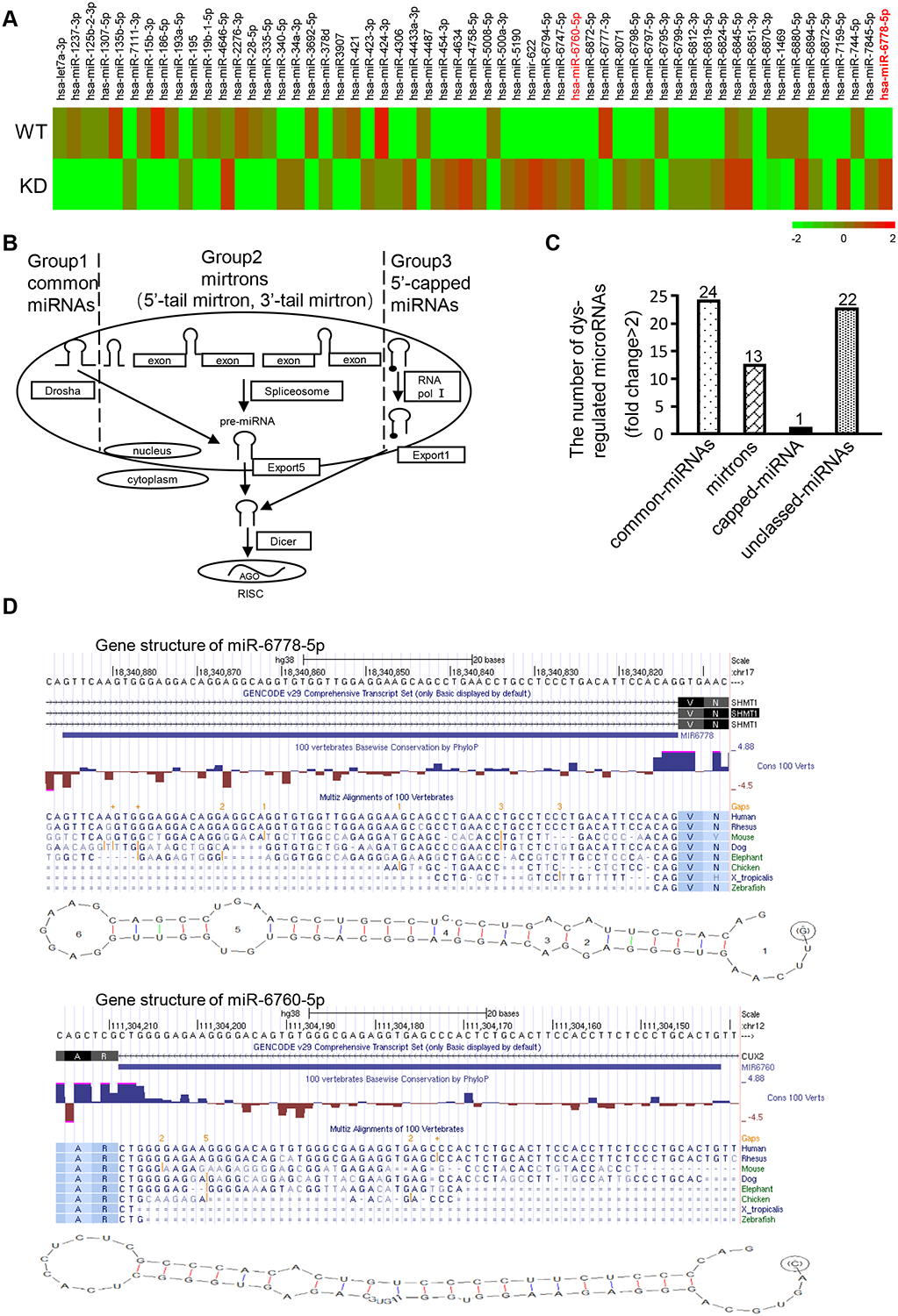
(A) A heatmap of aberrant miRNAs identified by a miRNA array of Drosha-knockdown cells. (B) Schematic diagram of canonical and splicing-mediated microRNA (miRNA) biogenesis. (C) Classification of the aberrant miRNAs in Drosha-knocked down gastric cancer cells using bioinformatics analysis. (D) Structure of a 5’ tailed mirtron. The predicted secondary structures of the investigated pre-miR-6778–5p and a known 5’ tailed mirtron (pre-miR-6760–5p) are shown. The sequences of 5’ tailed mirtrons were compared with those of their host genes, SHMT1 and CUX2, using RNA structure 5.7 software and the UCSC database (http://genome.ucsc.edu/). Human SHMT1 (miR-6778–5p) and and human CUX2 (miR-6760–5p) have the typical pyrimidine-rich region in the terminal AG splice acceptor area; the 3’ end of their pre-miRNA matches the splice acceptor in their introns.
2. The 5’-tail mirtron miR-6778–5p is a transcript splice derived from intron 5 of SHMT1
To validate the microarray data, the efficiency of Drosha knockdown and the related microRNA expression was verified in the gastric cancer cell lines MGC-803, SGC-7901, BGC-823 and NUGC-3. Indeed, the upregulation of microRNAs was verified (Figure 2A) by qRT-PCR in the Drosha-knockdown gastric cancer cells (Figure S1A). Among them, miR-6778–5p was significantly increased in Drosha KD MGC-803 cells, which was confirmed in other Drosha KD gastric cancer cells (Figure 2B). In addition, a high level of miR-6778–5p was detected in gastric tumours expressing low Drosha levels compared to gastric tumours expressing high Drosha levels (Figure 2C). To further understand that miR-6778–5p is a Drosha-independent miRNA, we compared the expression levels of miR-6778–5p and a known canonical miRNA, miR-21, in gastric cancer cells and tissues. We found that miR-6778–5p levels were significantly lower than miR-21 levels in Drosha wild type (Drosha WT) cells. Loss of Drosha obviously decreased the expression levels of miR-21 in MGC-803 cells, while miR-6778–5p levels were notably increased under knockdown of Drosha (Figure S1B). Moreover, we found that miR-6778–5p levels were significantly lower than miR-21 levels in Drosha-higher gastric tumour tissues, and conversely, the miR-6778–5p levels were distinctly higher than the miR-21 levels in Drosha-lower gastric tumour tissues (Figure S1C).
Figure 2. The 5’ tail mirtron miR-6778–5p is a transcript splice derived from the intron 5 of SHMT1.
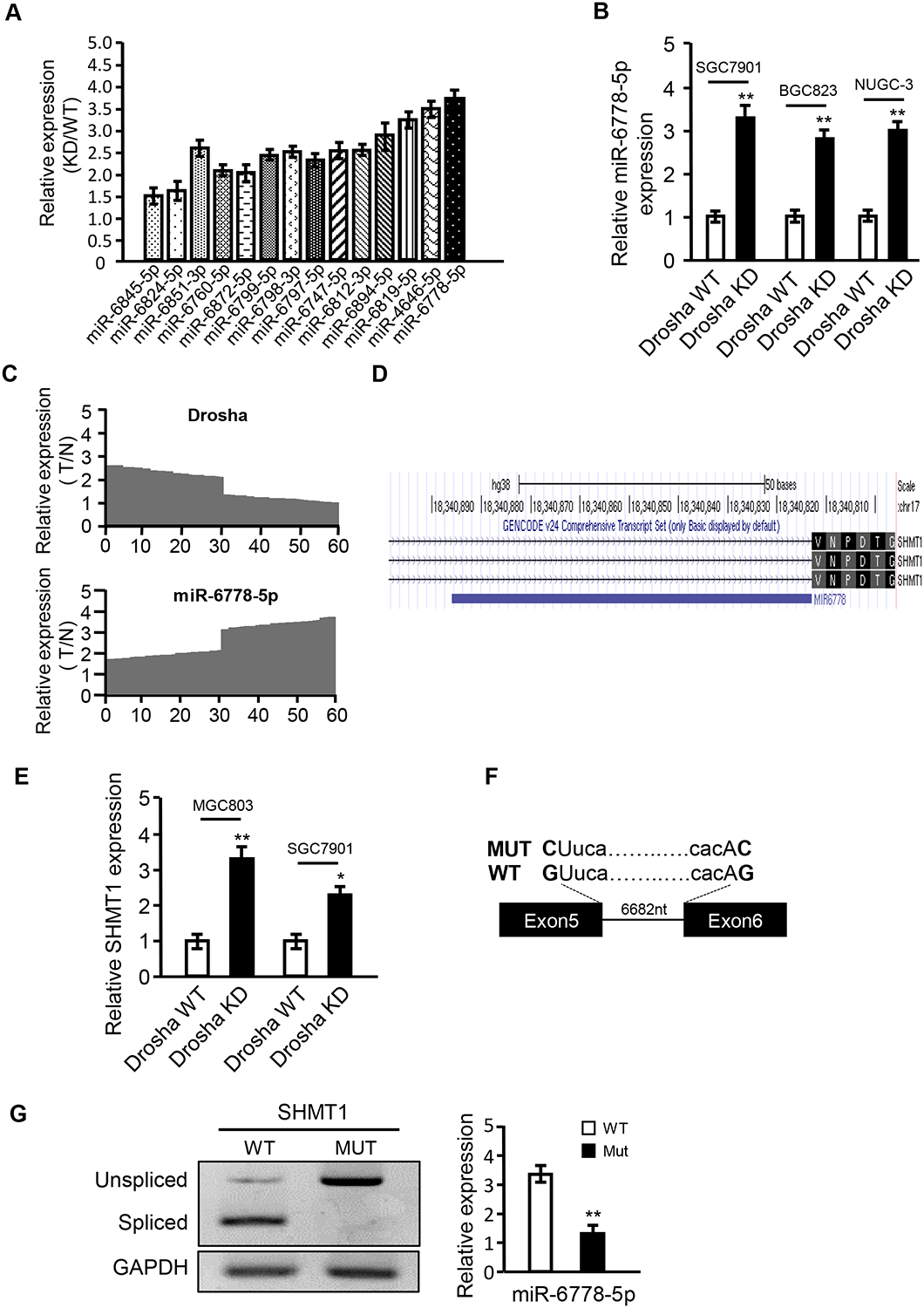
(A) qRT-PCR was used to confirm the upregulated miRNAs identified by microarray analysis. KD: MGC-803/Drosha knockdown gastric cancer cells; WT: MGC-803/Drosha wild type gastric cancer cells. U6 was used as an internal control. (B) qRT-PCR analysis of miR-6778–5p expression in Drosha wild type (Drosha WT) and Drosha-knockdown (Drosha KD) gastric cancer cells. U6 was used as an internal control. (C) qRT-PCR was used to detect the expression levels of Drosha and miR-6778–5p in gastric cancer tissues and their paired gastric normal tissues (n=60). T: gastric tumour tissues; N: normal gastric tissues. U6 was used as an internal control. (D) The sequences are compared between pre-miR-6778–5p and its host gene, SHMT1, using the UCSC database (http://genome.ucsc.edu/). (E) qRT-PCR was used to analyse the endogenous mRNA expression of SHMT1 in MGC-803/Drosha KD and SGC-7901/Drosha KD gastric cancer cells. (F, G) Identification of the Drosha-independent 5’-tail mirtron miR-6778–5p. (F) Minigene structure of miR-6778–5p. Boxes and lines indicate exons and introns of SHMT1, respectively. The splice site mutation is shown in bold. (G) Splicing analysis of the SHMT1 transcript. Wild type and splicing-deficient minigene transcripts were transfected into MGC-803 cells, and mRNA products were analysed by RT-PCR (left panels). MiR-6778–5p expression in wild type and splicing-deficient cells was analysed by qRT-PCR (right panels)
To understand the origin of mirtron miR-6778–5p, we searched for its host gene using the UCSC (http://genome.ucsc.edu/) database and found that miR-6778–5p may be derived from intron 5 of the SHMT1 gene (Figure 2D). The mRNA levels of SHMT1 were also upregulated in Drosha KD MGC-803 and other Drosha KD GC cells (e.g. SGC7901) (Figure 2E). To determine whether miR-6778–5p is spliced from its host gene, SHMT1, we constructed a plasmid harbouring minigenes of intron 5 and spanning the coding exons of SHMT1, in which the wild-type (WT) minigene encompasses the natural sequence, while the mutant (MUT) minigene contains variants affecting the G residues at the 5’ splice donor site (GU changed to CU) and 3′ splice acceptor sites (AG changed to AC) (Figure 2F). As expected, introns were effectively spliced in the bulk of the analysed mRNA in WT minigene-transfected GC cells, while mRNAs in the MUT variants retained the un-spliced exon–intron–exon structure (Figure 2G left panel). Furthermore, the expression of mature miR-6778–5p was downregulated in MUT minigene-transfected GC cells compared to that in WT minigene-transfected GC cells (Figure 2G right panel). To explore why SHMT1 expression was increased in Drosha KD GC cells, we screened transcription factors regulate SHMT1 using a bioinformatics analysis (Promoter 2.0, JASPAR). Compared the predicted transcription factor with the up-regulated genes in our previous mRNA expression profiles, c-MYC was found to be a potential transcription factor of SHMT1 (Figure S1D). Using chromatin immunoprecipitation and dual-luciferase assays, we verified that c-MYC could directly bind to the SHMT1 promoter (Figure S1E) and regulate SHMT1 transcriptional activity (Figure S1F). Furthermore, the mRNA and protein levels of c-MYC were increased in Drosha KD MGC803 cells (Figure S1G–S1H).
3. Effects of miR-6778–5p on gastric cancer stem cells (GCSC)
Our previous studies confirm that loss of nuclear Drosha is related to cell invasion in GC [8]. However, progressive cell growth and gastric cancer stem cell self-renewal showed little change in all cases (Figure S2A–2B). CD44+/high cells can be regarded as CSCs in gastric cancer [37]. According to a bioinformatics analysis, higher levels of CD44 are present in gastric tumours than in normal tissue samples (Figure S2C). However, there were no notable changes in CD44 between Drosha low- and high-expressing gastric tumours (Figure S2D), and CSC-related markers were not differently detected in GCSCs derived from Drosha-knockdown cells (Drosha KD CSCs) and Drosha wild type cells (Drosha WT CSCs) (Figure S2E). We aimed to determine whether the aberrant upregulation of non-canonical miR-6778–5p is involved in the maintenance of malignancy in Drosha-knockdown GC cells. Indeed, higher miR-6778–5p and CD44 levels were detected in GC tumour tissues than in their matched non-tumour tissues (Figure 3A). Higher levels of miR-6778–5p were found in Drosha low-expressing gastric tumours than in Drosha high-expressing gastric tumours; however, the CD44 mRNA levels were not different in these GC tissues (data not shown). Consistent with these findings, similar results were obtained in the Drosha-knockdown gastric cancer cells (Figure S2F), indicating that miR-6778–5p may be a participant in the maintenance of GCSC stemness. To verify our hypothesis, stably interfering miR-6778–5p was transfected into Drosha KD MGC-803 and SGC-7901 GC cells using a specific shRNA, and CSC formation was examined using miR-6778–5p wild type and knock down GC cells. Stably interfering miR-6778–5p expression in Drosha-knockdown GC cells (Drosha KD) (Figure 3B) significantly decreased CSC formation efficiencies and CSC sphere volume compared to that in Drosha wild type GC cells (Figure 3C–3D and Figure S2G). Accordingly, the levels of stem the cell markers CD44, KLF4, and c-MYC were also notably reduced in the Drosha and miR-6778–5p double-silenced GCSC spheres compared to those in Drosha WT GCSCs (Figure 3E). Similarly, the GCSC formation efficiency was also decreased in Drosha KD cells, but not in Drosha WT cells, when miR-6778–5p inhibitors were used (Figure S2H). After efficient transfection of ectopic miR-6778–5p into Drosha WT gastric cancer cells (MGC-803/miR-6778–5p and SGC-7901/miR-6778–5p) using lentivirus (Figure 3F), the CSC sphere formation efficiency and sphere size were increased compared to their controls (Figure 3G–3I). Correspondingly, the expression levels of the CSC-related proteins CD44, KLF4 and c-MYC were enhanced (Figure 3J). Collectively, these data confirm that miR-6778–5p plays a facilitative role in gastric cancer stem cells.
Figure 3. Effects of miR-6778–5p on GCSC enrichment.
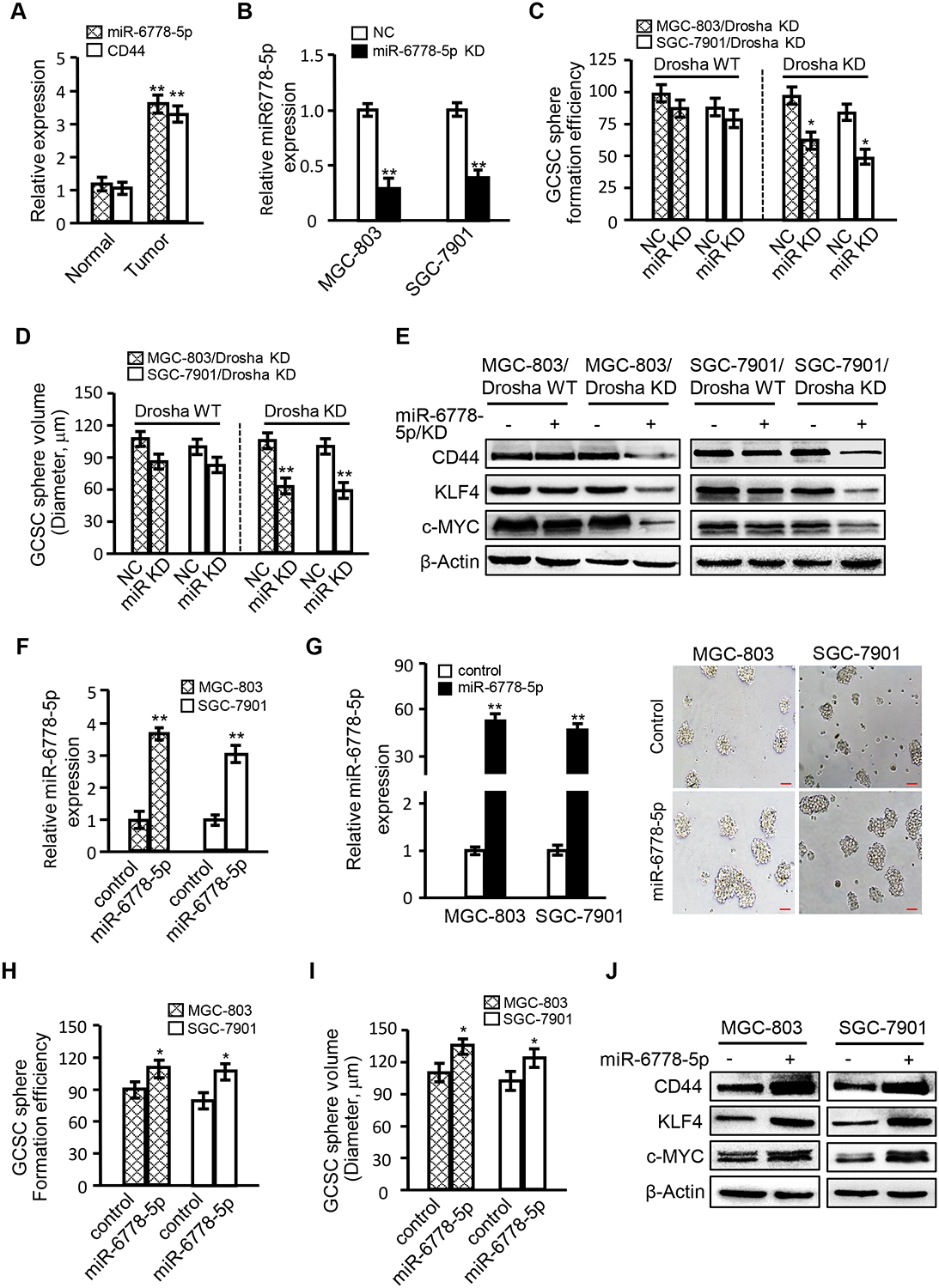
(A) qRT-PCR was used to assess miR-6778–5p and CD44 levels in GC tissues and para-cancerous tissues. (B) The interference efficiency of miR-6778–5p in MGC-803 and SGC-7901 GC cells was detected by qRT-PCR. U6 was used as an internal control. (C) Histograms show the CSC sphere formation efficiency. (D) Histograms show the CSC sphere sizes. (E) Western blotting was used to determine CSC-associated gene expression in the indicated gastric cancer cells. (F) qRT-PCR was used to evaluate the miR-6778–5p levels in gastric cancer cells transfected with ectopic miR-6778–5p or its control vector. U6 worded as an internal control. (G) The overexpression efficiency of miR-6778–5p in MGC-803 and SGC-7901 GC cells was detected by qRT-PCR (left panels). Images of GCSC spheres derived from MGC-803/miR-6778–5p, SGC-7901/miR-6778–5p and their controls (magnification: ×100; scale bar: 100 μm) (right panels). (H, I) Histograms show the CSC sphere formation efficiency and the sphere volume. (J) The expression levels of CSC-associated genes were determined by western blotting in the indicated gastric cancer cells.
4. YWHAE, a target of miR-6778–5p, regulates SHMT1 through downregulation of c-MYC
To identify the targets of miR-6778–5p, all of the potential target genes predicted by software programs (TargetScan7.2, miRanda and MirTarget2) were compared with the downregulated genes in our mRNA profiles. Three candidate genes were identified in MGC-803 Drosha KD cells (Figure 4A), and significant downregulation of YWHAE was demonstrated in Drosha-knockdown gastric cancer cells (Figure 4B). In the bioinformatics analysis, a binding site of miR-6778–5p was identified in the YWHAE 3′-UTR (Figure S3A). Indeed, miR-6778–5p could inhibit YWHAE 3′-UTR luciferase activity and mutation of the miR-6778–5p binding sites in the 3′-UTR of YWHAE diminished the responsiveness of the YWHAE 3′-UTR to miR-6778–5p (Figure 4C and Figure S3B). Furthermore, overexpression of miR-6778–5p in parental GC cells reduced the YWHAE protein level (Figure 4D, upper panel), and knockdown of miR-6778–5p in Drosha-silenced GC cells increased YWHAE protein (Figure 4D, down panel). Thus, YWHAE is the target gene of miR-6778–5p, which is consistent with previous studies in colon cancer cells [38].
Figure 4. YWHAE, a target of miR-6778–5p, regulates SHMT1through downregulation of c-MYC.
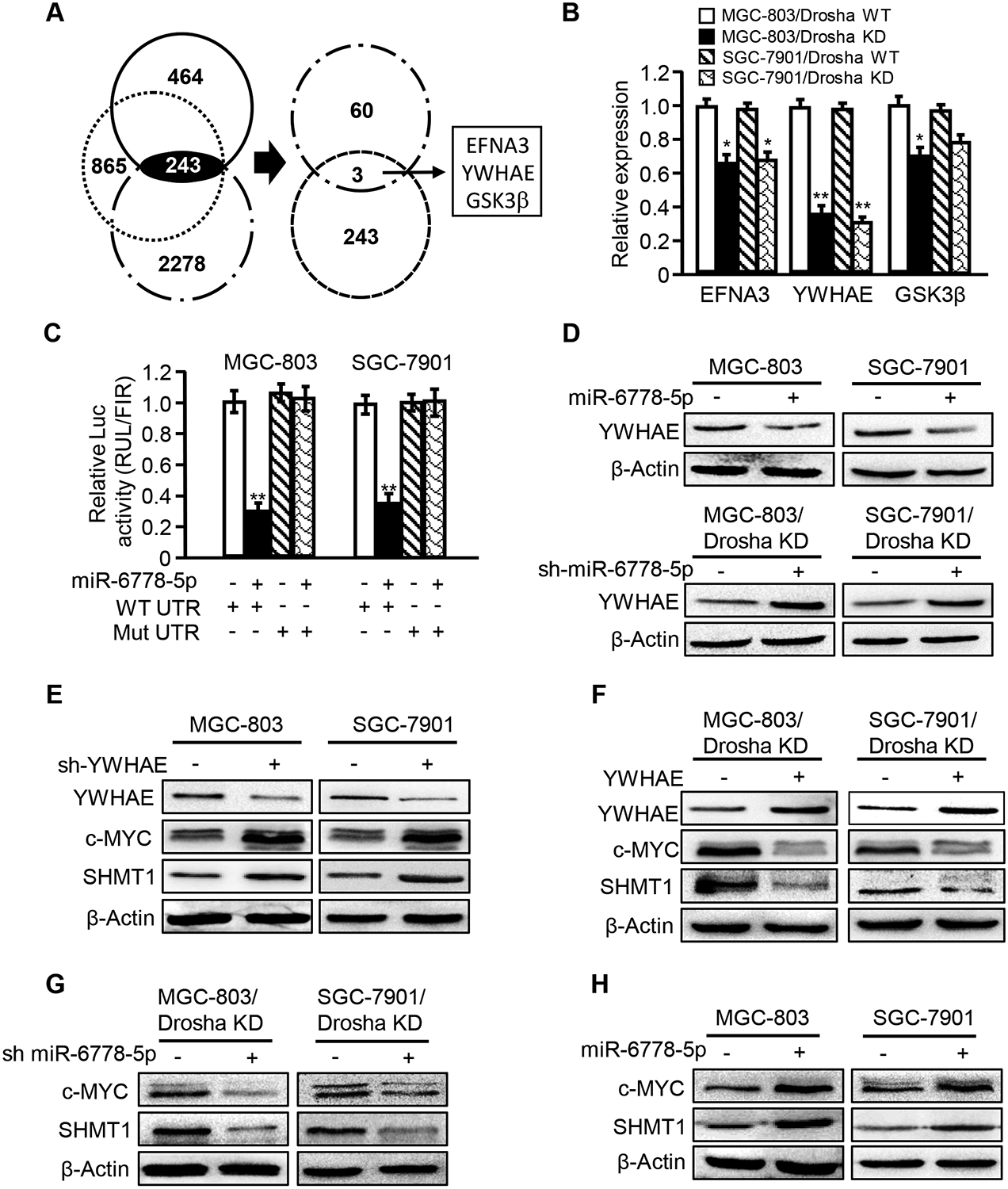
(A) A Venn diagram shows the putative targets of miR-6778–5p in Drosha-knockdown (Drosha KD) gastric cancer cells. (B) EFNA3, YWHAE and GSK3β expression levels in Drosha-knockdown cells were determined by qRT-PCR. (C) The wild-type YWHAE 3’-UTR reporter or the miR-6778–5p binding site mutant reporter was transfected into MGC-803 and SGC-7901 cells, and the inhibitory effect of miR-6778–5p on luciferase activity was detected. (D) YWHAE protein levels were determined by western blot analysis in MGC-803 and SGC-7901 cells transfected with ectopic miR-6778–5p or a control vector and in Drosha-knockdown MGC-803 and SGC-7901 or Drosha and miR-6778–5p double knockdown cells. (E-H) Western blot analysis was performed to determine the YWHAE, c-MYC and SHMT1 protein levels in the indicated gastric cancer cells.
As previously mentioned, an elevated miR-6778–5p level could facilitate GCSC enrichment. We determined whether YWHAE plays a role in GCSC enrichment. YWHAE rescue in Drosha-knockdown MGC-803 or SGC-7901 cells attenuated the GCSC sphere formation efficiency (Figure S3C), while silencing of YWHAE in parental MGC-803 and SGC-7901 cells promoted GCSC sphere formation (Figure S3D). YWHAE is a suppressor of c-MYC expression[33], and c-MYC serves as a transcriptional regulator for SHMT1 expression (Figure S1), indicating that YWHAE can affect SHMT1 expression through c-MYC. As expected, YWHAE silencing led to increased c-MYC and SHMT1 levels in parental GC cells (Figure 4E), and ectopic YWHAE significantly mitigated c-MYC and SHMT1 protein levels in Drosha-knockdown GC cell lines (Figure 4F). Administration of MG132 (a proteasome inhibitor) to cells resulted in increased c-MYC protein in YWHAE over-expressing MGC-803 cells (Figure S3E), suggesting that YWHAE may regulate the stability of c-MYC. Additionally, the expression levels of c-MYC and SHMT1 were significantly decreased in Drosha and miR-6778–5p double knockdown GC cells (Figure 4G) and were increased in ectopic miR-6778–5p-expressing parental GC cells (Figure 4H), indicating miR-6778–5p can regulate c-MYC and SHMT1 via YWHAE. Together, these data reveal that miR-6778–5p can positively regulate SHMT1 expression through YWHAE/c-MYC signalling.
5. SHMT1 regulates the self-renewal capacity of GCSCs
SHMT encodes serine hydroxymethyltransferase (SHMT), which catalyses folate-dependent serine/glycine inter-conversion. The human genome contains two SHMT genes: SHMT1, which encodes a cytoplasmic isozyme in the cytosolic folate-dependent one-carbon metabolism process, and SHMT2, which encodes the mitochondrial gene for maintenance of mitochondrial folate-dependent one-carbon metabolism. Deletion of SHMT2 or MTHFD2 (another enzyme in mitochondrial folate-dependent one-carbon metabolism) has been reported to lead to inactivation of mitochondrial folate-dependent one-carbon metabolism in tumour cells, which could be partly compensated by a compensatory increase in endogenic SHMT1, thus supporting the survival and growth of cancer cells. We observed that SHMT2 and MTHFD2 were downregulated in Drosha-knockdown GC cells (Figure S4A–S4B), suggesting an inactivation of mitochondrial folate-dependent one carbon metabolism in Drosha-knockdown GC cells.
Next, we investigated whether SHMT1 is essential to GCSC. A high level of SHMT1 was found in gastric tumour tissues by bioinformatics analysis (Figure S4C). In addition, relatively higher SHMT1 and CD44 mRNA levels were found in gastric tumour samples (Figure S4D). Furthermore, increased SHMT1 and decreased SHMT2 levels were found in Drosha-low-expressing gastric tumours compared to those in Drosha-high-expressing gastric tumours according to qRT-PCR analysis (Figure S4E). Expression of stably interfering SHMT1 in Drosha-knockdown GC cells significantly decreased their sphere formation potential (Figure 5A–5D); however, knockdown of SHMT1 in Drosha wild type GC cells had no observable effects on the sphere formation potential (Figure S4F–S4H). To further investigate this finding, a constructor encoding SHMT1 was stably transfected into parental MGC-803 and SGC-7901 cells. Compared to control cells, ectopic SHMT1 significantly promoted CSC sphere formation and increased the expression levels of stem cell markers (Figure 5E–5H).
Figure 5. SHMT1 is involved in the maintenance of GCSC stemness.
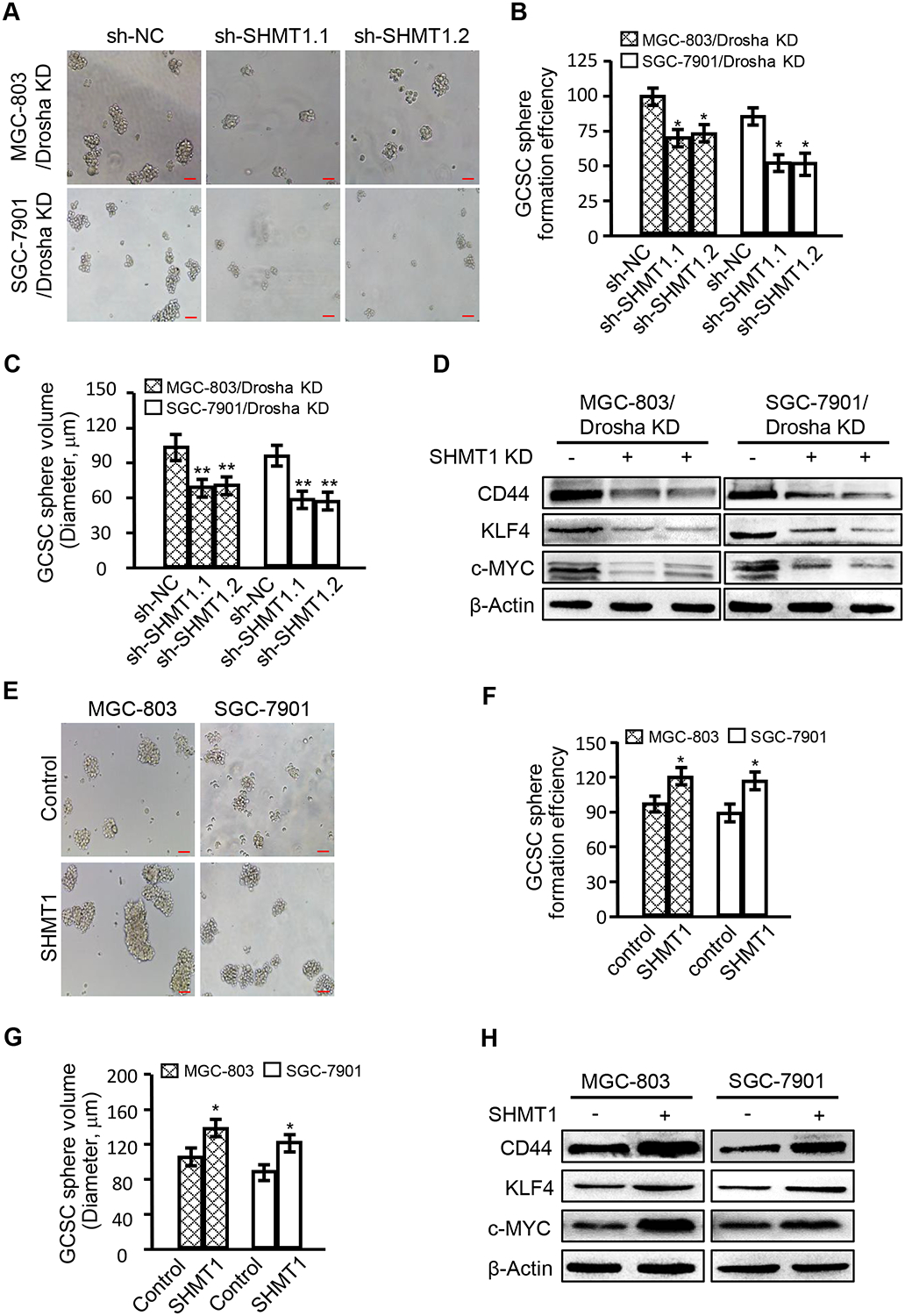
(A-D) Short hairpin RNAs (sh-SHMT1.1 and sh-SHMT1.2) that act against SHMT1 were stably transfected into GC MGC-803 and SGC7901 cells. Silencing of SHMT1, CSC sphere formation, sphere sizes, and CSC-related gene expression were evaluated in Drosha-knockdown MGC-803 and SGC-7901 cells. (A) Images of GCSC spheres (magnification: ×100; scale bar: 100 μm); (B) CSC sphere formation efficiency; (C) CSC sphere sizes. (D) CSC-associated gene expression by western blotting. (E-H). Ectopic SHMT1 was transfected into MGC-803 and SGC-7901 parent cells, and CSC sphere formation, sphere sizes, and CSC-related gene expression were evaluated. (E) Images of GCSC sphere formation in the indicated gastric cancer cells (magnification: ×100; scale bar: 100 μm). (F) CSC sphere formation efficiency. (G) CSC sphere sizes. (H) CSC-associated gene expression in the indicated GCSCs.
6. MiR-6778–5p contributes to GCSC maintenance via regulation of cytoplasmic one-carbon metabolism by feedback regulation of SHMT1
SHMT is involved in folate-dependent serine/glycine inter-conversion in one-carbon metabolism, which provides metabolic materials for nucleotide synthesis and participates in methionine recycling [39, 40]. As previously mentioned, the protein levels of SHMT2 and MTHFD2 associated with mitochondrial folate metabolism were obviously decreased in Drosha-knockdown GC cells (Figure S4A–S4B). However, SHMT1, a cytoplasmic folate metabolism-related metabolic enzyme, was markedly increased in Drosha knockdown GCSCs (Figure 2E). These findings suggested an intriguing scientific hypothesis: Drosha-independent miR-6778–5p regulation of the YWHAE/SHMT1 axis may govern cytoplasmic folate-dependent one-carbon metabolism rather than the canonical mitochondrial one-carbon metabolism in GCSCs.
To confirm our hypothesis, miR-6778–5p, SHMT1 and SHMT2 were knocked down in Drosha WT and Drosha KD gastric cancer cells, and GCSC stemness maintenance was observed. As shown in Figure 6A and Figure S5A, compared to Drosha-WT GC cells, silencing of miR-6778–5p (labelled as miR KD) or loss of SHMT1, rather than SHMT2, in Drosha-knockdown GC cells led to a significant decrease in sphere formation and sphere sizes in GCSCs; however, knockdown of SHMT2 rather than SHMT1 in Drosha-WT GC cells led to a reduction in sphere formation. Serine supplements could rescue the sphere formation potential, suggesting that carbon metabolism is important for GCSC formation, and the cytosolic carbon metabolism in Drosha-knockdown GCs replaces the mitochondrial carbon metabolism in Drosha-WT GCs and plays an essential role in GCSC maintenance. To support this finding, metabolites of serine in GCSCs were tracked using [2,3,3-2H] labelled serine and analysed by liquid chromatography-mass spectroscopy (LC-MS). SHMT1-catalysed cytosolic one-carbon metabolism can generate M+2 dTTP, whereas mitochondrial one-carbon metabolic pathway is catalysed by SHMT2, which results in products such as M+1 dTTP [28]. Consistent with the previous findings, increased M+2 dTTP content and decreased M+1 dTTP content were detected in Drosha-knockdown GCSCs, in contrast to Drosha-WT GCSCs (Figure 6B). Silencing of SHMT2, rather than miR-6778–5p or SHMT1, in Drosha-WT GCSCs attenuated M+1 dTTP content; however, knockdown of either miR-6778–5p, SHMT1 or SHMT2 in Drosha-knockdown GCSCs cells did not impact M+1 dTTP content (Figure 6C, left panel). In contrast, loss of miR-6778–5p, SHMT1 or SHMT2 had little effect on M+2 dTTP in Drosha-WT GCSCs; however, silencing of miR-6778–5p or SHMT1, rather than SHMT2, notably decreased M+2 dTTP in Drosha-knockdown GCSCs (Figure 6C, right panel). These data demonstrate that a robust miR-6778–5p-SHMT1 signalling axis-mediated cytosolic one-carbon metabolism is in Drosha-knockdown GC cells contributes to GCSC stemness maintenance.
Figure 6. miR-6778–5p-SHMT1 signalling contributes to GCSC maintenance via regulation of cytoplasmic one-carbon metabolism.
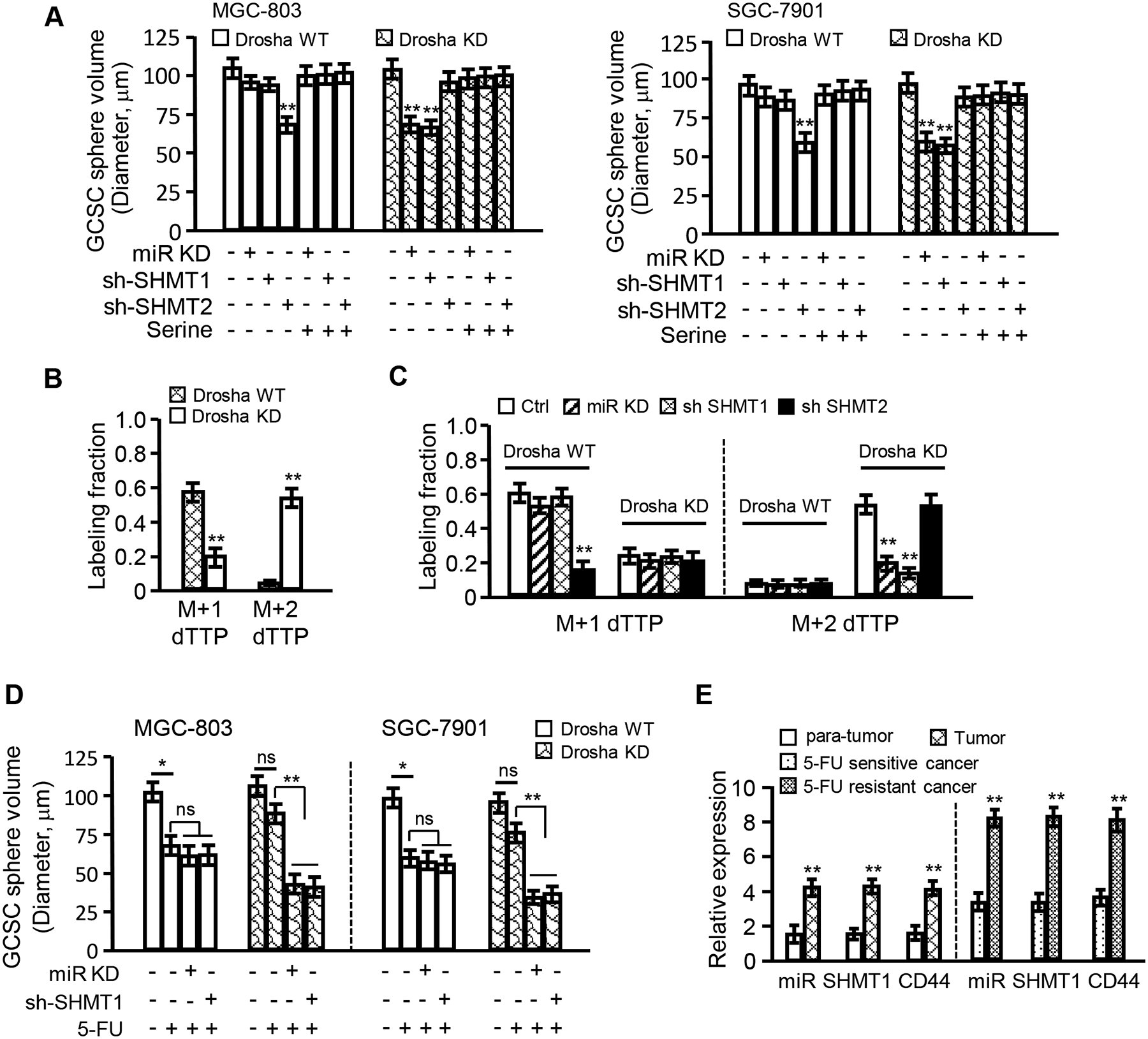
(A) Histograms show the sphere volumes of GCSCs derived from the indicated gastric cancer cells. Addition of serine (0.8 mM) could party rescue CSC formation in miR-6778–5p or SHMT1-knockdown gastric cancer cells and could especially rescue CSC formation in Drosha-knockdown gastric cancer cells. (B) Isotopic tracing analysis of relative M+1 and M+2 dTTP in GCSCs derived from Drosha WT and Drosha KD MGC-803 cells cultured in medium with [2,3,3-2H]-serine. (C) Isotopic tracing analysis of M+1 and M+2 dTTP in GCSCs derived from the indicated gastric cancer cells cultured in medium with [2,3,3-2H]-serine. (D) Histograms show the sphere volumes of GCSCs derived from the indicated Drosha WT or Drosha KD gastric cancer cells treated with or without 30 μg/ml of 5-FU (a chemotherapeutic drug for gastric cancer). (E) The miR-6778–5p, SHMT1 and CD44 levels in gastric tumour tissues and their control normal tissues and 5-FU sensitive or resistant gastric tumour tissues were evaluated by qRT-PCR.
It has been indicated that dTTP, the metabolite of one-carbon metabolism, is involved in CSC enrichment of colorectal cancer and chemotherapy resistance [41, 42]. To understand whether miR-6778–5p-SHMT1 signalling axis-mediated cytosolic one-carbon metabolism affects the sensitivity of GCSCs to chemotherapy, 5-fluorouracil (5-FU; a metabolic disruptor, that impedes dTTP synthesis) was used to examine CSC formation. As shown in Figure 6D and Figure S5B, 5-FU could decrease GCSC formation of Drosha WT gastric cancer cells compared to Drosha-knocked down GCs. Knockdown of miR-6778–5p or SHMT1 in Drosha WT gastric cancer cells caused no significant reduction of CSC formation in contrast to the control cells under 5-FU treatment; however, loss of miR-6778–5p or SHMT1 in Drosha-silenced gastric cancer cells notably reduced GCSC sphere formation under 5-FU treatment. In addition, higher levels of SHMT1 and CD44 were detected in gastric tumour tissues than in para-cancerous tissues. Higher miR-6778–5p, SHMT1 and CD44 levels were found in 5-FU-resistant gastric tumours than in 5-FU-sensitive tumours (Figure 6E), indicating that miR-6778–5p-SHMT1 signalling axis-mediated cytosolic carbon metabolism has a potential clinical chemotherapeutic value.
7. Effect of miR-6778–5p and SHMT1 on gastric cancer tumourigenesis in vivo
To investigate the effects of miR-6778–5p/SHMT1 signalling on gastric cancer tumourigenesis in vivo, MGC-803/Drosha WT, MGC-803/Drosha WT/miR-6778–5p KD, MGC-803/Drosha WT/SHMT1 KD, MGC-803/Drosha KD, MGC-803/Drosha KD/miR-6778–5p KD and MGC-803/Drosha KD/SHMT1 KD GCSCs were subcutaneously injected into nude mice. As shown, knockdown of either miR-6778–5p or SHMT1 inhibited the tumourigenic potential of GCSCs and tumour growth (Figure 7A–7B). However, the tumourigenic abilities of MGC-803/Drosha WT/miR-6778–5p KD or MGC-803/Drosha WT/SHMT1 KD GCSCs were similar to those of MGC-803/Drosha WT GCSCs (Figure S6A–S6B). Compared to the control mice, exogenous serine could restore tumour growth in tumour-burden mice injected with MGC-803/Drosha KD/miR-6778–5p KD GCSCs and MGC-803/Drosha KD/SHMT1 KD GCSCs, and 5-FU obviously suppressed the tumourigenesis and tumour growth (Figure 7A–7B). Similarly, exogenous serine could promote tumour growth in tumour-burden mice injected with MGC-803/Drosha WT/miR-6778–5p KD or MGC-803/Drosha WT/SHMT1 KD GCSCs, and 5-FU obviously suppressed tumourigenesis and tumour growth (Figure S6A–S6B). Consistently, high levels of CD44, KLF4, and c-MYC were detected in Drosha-knockdown alone tumours. Knockdown of miR-6778–5p or SHMT1 led to reduced CD44, KLF4, and c-MYC protein levels, while administration of serine to these mice restored the expression of CD44, KLF4, and c-MYC. The lowest CD44, KLF4, and c-MYC levels were found in 5-FU-treated mice (Figure 7C). These data support that the miR-6778–5p/SHMT1 signalling axis is critical to maintain stemness in Drosha downregulated gastric tumours, and interference in miR-6778–5p/SHMT1 signalling may enhance their sensitivity to 5-FU chemotherapy drugs.
Figure 7. Effects of miR-6778–5p and SHMT1 on gastric cancer tumourigenesis in vivo.
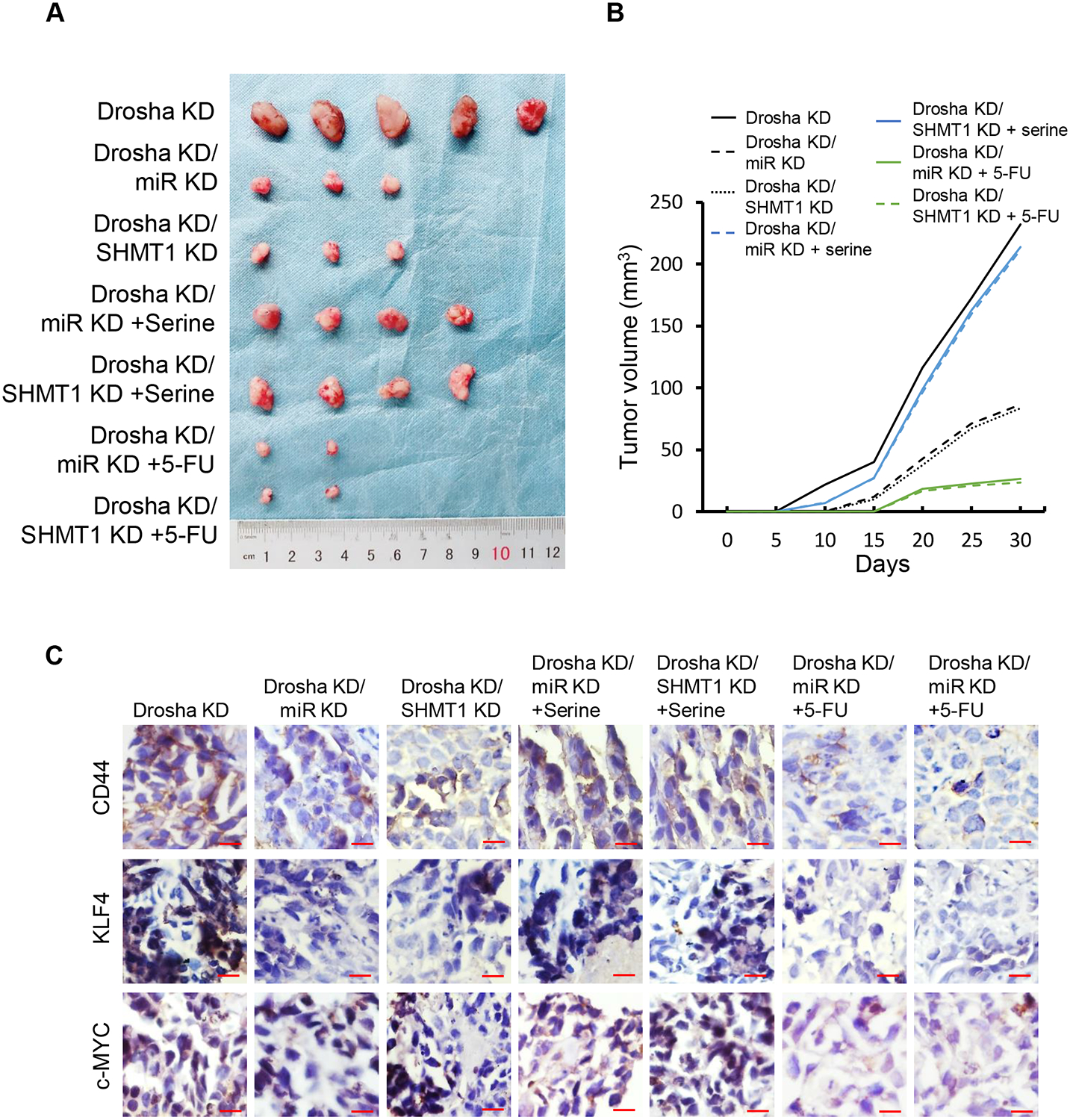
The indicated GCSCs (1×105) were subcutaneously injected into nude mice. Images of tumour sizes and their corresponding tumour growth curve are shown. The mice were injected with MGC-803/Drosha KD/miR-6778–5p KD, MGC-803/Drosha KD/SHMT1 KD, and the control cells MGC-803/Drosha KD. The mice were given serine (130 mg/kg) and 5-FU (30 mg/kg) at 5 days after cell injection. (A) Representative images of tumours in the indicated nude mice xenografts. (B) Tumour growth curve corresponding to (A). (C) Representative images of CSC-associated gene expression by IHC staining in the indicated xenografts (magnification: ×400; scale bar: 50 μm).
Discussion
Drosha is essential for canonical microRNA (miRNA) biogenesis. However, non-canonical miRNAs, including mirtron (3’-tail mirtron and 5’-tail mirtron) and 5’-capped microRNA, exist that are independent of Drosha processing. The canonical miRNAs are known to be involved in regulation of various biological processes in tumours, such as tumourigenesis, invasion, metastasis, and drug resistance. However, the biological functions of non-canonical miRNAs remain unclear. Our previous studies revealed that Drosha silencing cannot efficiently abolish the malignant factors of gastric cancer, including tumour cell proliferation and CSC characteristics. We found that a non-canonical miRNA, 5’-tail mirtron miR-6778–5p, can maintain the stemness of gastric cancer stem cell (GCSC) via positive feedback regulation of expression of its host gene, SHMT1, and cytoplasmic one-carbon metabolism. Our findings may provide novel insight into understanding the complicated characteristics of gastric cancer and potential new treatment strategies.
Aberrant Drosha-independent miRNAs, including the so-called non-canonical miRNAs, are present in Drosha downregulated gastric cancer cells. Recently, several studies have identified non-canonical miRNAs in higher organisms and human tissues, such as miRNA3547 and miRNA6927 in mice and miRNA6807 in humans [16]. All of these non-canonical miRNAs are mirtrons (3’-tail mirtrons and 5’-tail mirtrons) and 5’-capped microRNAs. In this study, we discovered a set of aberrant non-canonical miRNAs, including miR-6778–5p and miR-6760–5p (one of the known 5’-tail mirtrons), in Drosha-knockdown gastric cancer cells. MiR-6778–5p, similar to miR-6760–5p, is an abnormal transcript splice of the intron of the host gene (here, miR-6778–5p is a transcript splice of intron 5 of the SHMT1 gene; miR-6760–5p is a transcript splice of the CUX2 gene). However, the molecular mechanism by which non-canonical miRNAs are increased under knockdown/suppression of Drosha remains unclear. One possibility is a competing relationship between small RNAs, as reported by Young-Kook Kima [15]. This study showed that the processing ability of pre-miRNA of noncanonical miRNAs by subsequent enzymes (such as DICER) is increased in the absence of pre-miRNA of canonical miRNAs. Another possibility is positive feedback regulation between non-canonical miRNAs and their host genes, as reported in the current study.
Non-canonical miR-6778–5p contributes gastric tumour malignancy via targeting YWHAE and upregulating c-MYC to promote expression of the host gene, SHMT1. Consistent with a previous study [38], we confirmed that YWHAE is the target of miR-6778–5p. A previous study indicated that a low YWHAE level can promote gastric cancer development, which closely relies on c-MYC [33]. YWHAE is a transcriptional inhibitor of c-MYC. Silencing of miR-6778–5p in Drosha-knockdown gastric cancer cells significantly decreased c-MYC levels, and ectopic miR-6778–5p could upregulate c-MYC expression in Drosha wild type gastric cancer cells. c-MYC acted as a direct regulator of SHMT1 expression by binding to a specific c-MYC binding site in the promoter of the host gene, SHMT1. Therefore, non-canonical miR-6778–5p was experimentally indicated to regulate its host gene through the YWHAE-c-MYC axis via a feedback loop. Our study indicated that aberrant non-canonical miRNAs (e.g., miR-6778–5p) in Drosha-knockdown gastric cancer cells may contribute to tumour development.
Metabolic reprogramming is crucial for tumourigenesis and tumour development [43]. In the current study, we confirmed that SHMT1 is a key regulator of cytoplasmic folate-related one-carbon metabolism, which catalyses the reversible conversion of serine to glycine and the conversion of tetrahydrofolate to methylenetetrahydrofolate [44–46]. Generally, the SHMT2-mediated mitochondrial carbon metabolic pathway is the major pathway by which one-carbon units are produced in normal cells and most cancer cells [28, 44]. Dysfunction of mitochondrial carbon metabolic enzymes (SHMT2 and MTHDF2) leads to impairment of mitochondrial carbon metabolism, and compensatory activation of cytoplasmic carbon metabolism may play an essential role in the maintenance of cell survival [28]. High levels of SHMT1 have been detected in non-small cell lung cancer [29]. Here, our study provides strong evidence to support an altered carbon metabolic pathway in gastric cancer cell; that is, cytoplasmic one-carbon metabolism rather than canonical mitochondrial one-carbon metabolism plays a pivotal role in the maintenance of CSC stemness in Drosha-knockdown or Drosha low-expressing gastric cancer.
Different metabolites were observed between cytoplasmic one-carbon metabolism and mitochondrial one-carbon metabolism. Ducker et al. found that cytosolic SHMT1 is helpful for production of M+2 dTTP, while SHMT2-mediated mitochondrial one-carbon metabolism tends to produce M+1 dTTP[28]. 5-Fluorouracil (5-FU), is a chemotherapeutic drug for clinical GC treatment, which impedes dTTP synthesis[47]. We observed a high level of SHMT1 and the CSC biomarker CD44 in 5-FU-resistant gastric cancer patients. Interference of the miR-6778–5p/YWHAE/SHMT1 axis in gastric cancer provided GCSCs derived from Drosha-knockdown gastric cancer with an enhanced sensitivity to 5-FU treatment, and obviously reduced CSC stemness and tumourigenesis. Our findings may provide a potential new therapeutic approach for future gastric cancer treatment.
In conclusion, miR-6778–5p is a Drosha-independent miRNA. MiR-6778–5p can promote expression of its host gene, SHMT1, through targeting of YWHAE, which relieves inhibition of c-MYC, thus upregulating the transcriptional activity of SHMT1. A high level of SHMT1 mediates cytoplasmic one-carbon metabolism to produce M+2 dTTP, which promotes CSC enrichment and stemness maintenance in gastric cancer. Impeding the miR-6778–5p/YWHAE/SHMT1 signalling axis can enhance the sensitivity of gastric cancer to 5-FU chemotherapy.
Supplementary Material
Figure 8. A proposed working model.
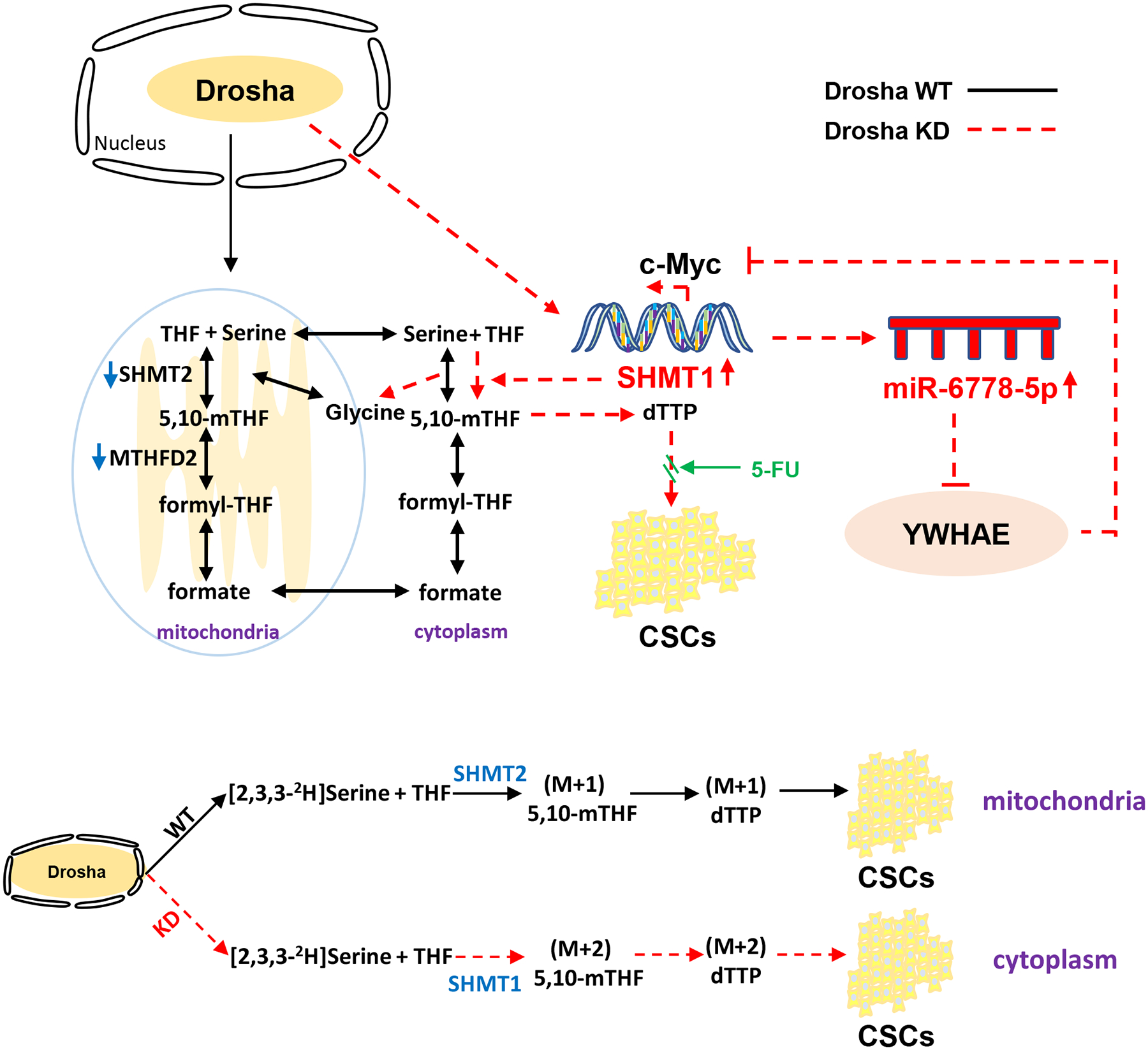
A schematic representation depicts the role of the miR-6778–5p/YWHAE/SHMT1 axis in the maintenance of gastric cancer stem cell stemness through regulating cytoplasmic one-carbon metabolism. In Drosha wild type (Drosha WT) gastric cancer cells, SHMT2-catalysed mitochondrial one-carbon metabolism generates (M+1) 5,10-mTHF and M+1 dTTP to promote CSC enrichment and stemness maintenance. However, loss of Drosha leads an enhanced miRNA6778–5p, which targets YWHAE, an inhibitor of c-MYC, to positively up-regulate the expression of its host gene, SHMT1. SHMT1 catalyses cytosolic one-carbon metabolism instead of mitochondrial-carbon metabolism to generate (M+2) 5,10-mTHF and M+2 dTTP, thus fuels CSC enrichment and stemness maintenance in gastric cancer Drosha–decreased or knocked down gastric cancers. Impeding the miR-6778–5p/YWHAE/SHMT1 signalling axis can enhance the sensitivity of gastric cancer to 5-FU chemotherapy.
Acknowledgements
This work was supported in part by National Natural Science Foundation of China (NSFC 31671481, NSFC 81472476 and NSFC 81172296); and National Natural Science Foundation of China (NSFC 81802447) for Yan-e Du; And also supported in part by the outstanding Postgraduate Fund of Chongqing Medical University (2017) and Chongqing Graduate Research and Innovation Project of the Chongqing Education Committee (2017) (CYS17162) for Maojia Zhao.
Footnotes
Conflict of Interest
The authors declare no competing financial interests.
References
- [1].Allemani C, Matsuda T, Di Carlo V, Harewood R, Matz M, Niksic M, Bonaventure A, Valkov M, Johnson CJ, Esteve J, Ogunbiyi OJ, Azevedo ESG, Chen WQ, Eser S, Engholm G, Stiller CA, Monnereau A, Woods RR, Visser O, Lim GH, Aitken J, Weir HK, Coleman MP, Group CW, Global surveillance of trends in cancer survival 2000–14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries, Lancet, 391 (2018) 1023–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ji C, Yang L, Yi W, Xiang D, Wang Y, Zhou Z, Qian F, Ren Y, Cui W, Zhang X, Zhang P, Wang JM, Cui Y, Bian X, Capillary morphogenesis gene 2 maintains gastric cancer stem-like cell phenotype by activating a Wnt/beta-catenin pathway, Oncogene, 37 (2018) 3953–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mao J, Liang Z, Zhang B, Yang H, Li X, Fu H, Zhang X, Yan Y, Xu W, Qian H, UBR2 Enriched in p53 Deficient Mouse Bone Marrow Mesenchymal Stem Cell-Exosome Promoted Gastric Cancer Progression via Wnt/beta-Catenin Pathway, Stem cells, 35 (2017) 2267–2279. [DOI] [PubMed] [Google Scholar]
- [4].Moharil RB, Dive A, Khandekar S, Bodhade A, Cancer stem cells: An insight, Journal of oral and maxillofacial pathology : JOMFP, 21 (2017) 463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhang X, Hua R, Wang X, Huang M, Gan L, Wu Z, Zhang J, Wang H, Cheng Y, Li J, Guo W, Identification of stem-like cells and clinical significance of candidate stem cell markers in gastric cancer, Oncotarget, 7 (2016) 9815–9831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kim YS, Lee HJ, Park JM, Han YM, Kangwan N, Oh JY, Lee DY, Hahm KB, Targeted molecular ablation of cancer stem cells for curing gastrointestinal cancers, Expert review of gastroenterology & hepatology, 11 (2017) 1059–1070. [DOI] [PubMed] [Google Scholar]
- [7].Zhang H, Hou Y, Xu L, Zeng Z, Wen S, Du YE, Sun K, Yin J, Lang L, Tang X, Liu M, Cytoplasmic Drosha Is Aberrant in Precancerous Lesions of Gastric Carcinoma and Its Loss Predicts Worse Outcome for Gastric Cancer Patients, Digestive diseases and sciences, 61 (2016) 1080–1090. [DOI] [PubMed] [Google Scholar]
- [8].Xu L, Hou Y, Tu G, Chen Y, Du YE, Zhang H, Wen S, Tang X, Yin J, Lang L, Sun K, Yang G, Tang X, Liu M, Nuclear Drosha enhances cell invasion via an EGFR-ERK1/2-MMP7 signaling pathway induced by dysregulated miRNA-622/197 and their targets LAMC2 and CD82 in gastric cancer, Cell death & disease, 8 (2017) e2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Rasschaert P, Figueroa T, Dambrine G, Rasschaert D, Laurent S, Alternative splicing of a viral mirtron differentially affects the expression of other microRNAs from its cluster and of the host transcript, RNA biology, 13 (2016) 1310–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Havens MA, Reich AA, Duelli DM, Hastings ML, Biogenesis of mammalian microRNAs by a non-canonical processing pathway, Nucleic acids research, 40 (2012) 4626–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ruby JG, Jan CH, Bartel DP, Intronic microRNA precursors that bypass Drosha processing, Nature, 448 (2007) 83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC, Mammalian mirtron genes, Molecular cell, 28 (2007) 328–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Castellano L, Stebbing J, Deep sequencing of small RNAs identifies canonical and non-canonical miRNA and endogenous siRNAs in mammalian somatic tissues, Nucleic acids research, 41 (2013) 3339–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Xie M, Li M, Vilborg A, Lee N, Shu MD, Yartseva V, Sestan N, Steitz JA, Mammalian 5’-capped microRNA precursors that generate a single microRNA, Cell, 155 (2013) 1568–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kim YK, Kim B, Kim VN, Re-evaluation of the roles of DROSHA, Export in 5, and DICER in microRNA biogenesis, Proceedings of the National Academy of Sciences of the United States of America, 113 (2016) E1881–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ladewig E, Okamura K, Flynt AS, Westholm JO, Lai EC, Discovery of hundreds of mirtrons in mouse and human small RNA data, Genome research, 22 (2012) 1634–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sakai E, Miura Y, Suzuki-Kouyama E, Oka K, Tachibana M, Kawabata K, Sakurai F, Mizuguchi H, A mammalian mirtron miR-1224 promotes tube-formation of human primary endothelial cells by targeting anti-angiogenic factor epsin2, Scientific reports, 7 (2017) 5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Jones D, Li Y, He Y, Xu Z, Chen H, Min W, Mirtron microRNA-1236 inhibits VEGFR-3 signaling during inflammatory lymphangiogenesis, Arteriosclerosis, thrombosis, and vascular biology, 32 (2012) 633–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Marchetti P, Trinh A, Khamari R, Kluza J, Melanoma metabolism contributes to the cellular responses to MAPK/ERK pathway inhibitors, Biochimica et biophysica acta. General subjects, 1862 (2018) 999–1005. [DOI] [PubMed] [Google Scholar]
- [20].Kinnaird A, Dromparis P, Saleme B, Gurtu V, Watson K, Paulin R, Zervopoulos S, Stenson T, Sutendra G, Pink DB, Carmine-Simmen K, Moore R, Lewis JD, Michelakis ED, Metabolic Modulation of Clear-cell Renal Cell Carcinoma with Dichloroacetate, an Inhibitor of Pyruvate Dehydrogenase Kinase, European urology, 69 (2016) 734–744. [DOI] [PubMed] [Google Scholar]
- [21].Kory N, Wyant GA, Prakash G, Uit de Bos J, Bottanelli F, Pacold ME, Chan SH, Lewis CA, Wang T, Keys HR, Guo YE, Sabatini DM, SFXN1 is a mitochondrial serine transporter required for one-carbon metabolism, Science, 362 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Nakada D, Saunders TL, Morrison SJ, Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells, Nature, 468 (2010) 653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Steinhauser ML, Bailey AP, Senyo SE, Guillermier C, Perlstein TS, Gould AP, Lee RT, Lechene CP, Multi-isotope imaging mass spectrometry quantifies stem cell division and metabolism, Nature, 481 (2012) 516–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Snyder V, Reed-Newman TC, Arnold L, Thomas SM, Anant S, Cancer Stem Cell Metabolism and Potential Therapeutic Targets, Frontiers in oncology, 8 (2018) 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fawal MA, Jungas T, Kischel A, Audouard C, Iacovoni JS, Davy A, Cross Talk between One-Carbon Metabolism, Eph Signaling, and Histone Methylation Promotes Neural Stem Cell Differentiation, Cell reports, 23 (2018) 2864–2873 e2867. [DOI] [PubMed] [Google Scholar]
- [26].Deshmukh A, Deshpande K, Arfuso F, Newsholme P, Dharmarajan A, Cancer stem cell metabolism: a potential target for cancer therapy, Molecular cancer, 15 (2016) 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Luo M, Wicha MS, Targeting Cancer Stem Cell Redox Metabolism to Enhance Therapy Responses, Seminars in radiation oncology, 29 (2019) 42–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ducker GS, Chen L, Morscher RJ, Ghergurovich JM, Esposito M, Teng X, Kang Y, Rabinowitz JD, Reversal of Cytosolic One-Carbon Flux Compensates for Loss of the Mitochondrial Folate Pathway, Cell metabolism, 23 (2016) 1140–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhang WC, Shyh-Chang N, Yang H, Rai A, Umashankar S, Ma S, Soh BS, Sun LL, Tai BC, Nga ME, Bhakoo KK, Jayapal SR, Nichane M, Yu Q, Ahmed DA, Tan C, Sing WP, Tam J, Thirugananam A, Noghabi MS, Pang YH, Ang HS, Mitchell W, Robson P, Kaldis P, Soo RA, Swarup S, Lim EH, Lim B, Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis, Cell, 148 (2012) 259–272. [DOI] [PubMed] [Google Scholar]
- [30].Aitken A, Collinge DB, van Heusden BP, Isobe T, Roseboom PH, Rosenfeld G, Soll J, 14-3-3 proteins: a highly conserved, widespread family of eukaryotic proteins, Trends in biochemical sciences, 17 (1992) 498–501. [DOI] [PubMed] [Google Scholar]
- [31].Quayle SN, Sadar MD, 14-3-3 sigma increases the transcriptional activity of the androgen receptor in the absence of androgens, Cancer letters, 254 (2007) 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Qiu Y, Zhou Z, Li Z, Lu L, Li L, Li X, Wang X, Zhang M, Pretreatment 14-3-3 epsilon level is predictive for advanced extranodal NK/T cell lymphoma therapeutic response to asparaginase-based chemotherapy, Proteomics. Clinical applications, 11 (2017). [DOI] [PubMed] [Google Scholar]
- [33].Leal MF, Ribeiro HF, Rey JA, Pinto GR, Smith MC, Moreira-Nunes CA, Assumpcao PP, Lamarao LM, Calcagno DQ, Montenegro RC, Burbano RR, YWHAE silencing induces cell proliferation, invasion and migration through the up-regulation of CDC25B and MYC in gastric cancer cells: new insights about YWHAE role in the tumor development and metastasis process, Oncotarget, 7 (2016) 85393–85410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ma Y, Fu HL, Wang Z, Huang H, Ni J, Song J, Xia Y, Jin WL, Cui DX, USP22 maintains gastric cancer stem cell stemness and promotes gastric cancer progression by stabilizing BMI1 protein, Oncotarget, 8 (2017) 33329–33342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhou M, Hou Y, Yang G, Zhang H, Tu G, Du YE, Wen S, Xu L, Tang X, Tang S, Yang L, Cui X, Liu M, LncRNA-Hh Strengthen Cancer Stem Cells Generation in Twist-Positive Breast Cancer via Activation of Hedgehog Signaling Pathway, Stem cells, 34 (2016) 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Du YE, Tu G, Yang G, Li G, Yang D, Lang L, Xi L, Sun K, Chen Y, Shu K, Liao H, Liu M, Hou Y, MiR-205/YAP1 in Activated Fibroblasts of Breast Tumor Promotes VEGF-independent Angiogenesis through STAT3 Signaling, Theranostics, 7 (2017) 3972–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Takaishi S, Okumura T, Tu S, Wang SS, Shibata W, Vigneshwaran R, Gordon SA, Shimada Y, Wang TC, Identification of gastric cancer stem cells using the cell surface marker CD44, Stem cells, 27 (2009) 1006–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Li H, Jin X, Liu B, Zhang P, Chen W, Li Q, CircRNA CBL.11 suppresses cell proliferation by sponging miR-6778–5p in colorectal cancer, BMC cancer, 19 (2019) 826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Yang M, Vousden KH, Serine and one-carbon metabolism in cancer, Nature reviews. Cancer, 16 (2016) 650–662. [DOI] [PubMed] [Google Scholar]
- [40].Locasale JW, Serine, glycine and one-carbon units: cancer metabolism in full circle, Nature reviews. Cancer, 13 (2013) 572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Tsubouchi T, Soza-Ried J, Brown K, Piccolo FM, Cantone I, Landeira D, Bagci H, Hochegger H, Merkenschlager M, Fisher AG, DNA synthesis is required for reprogramming mediated by stem cell fusion, Cell, 152 (2013) 873–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Shen Y, Tong M, Liang Q, Guo Y, Sun HQ, Zheng W, Ao L, Guo Z, She F, Epigenomics alternations and dynamic transcriptional changes in responses to 5-fluorouracil stimulation reveal mechanisms of acquired drug resistance of colorectal cancer cells, The pharmacogenomics journal, 18 (2018) 23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Venkatanarayan A, Raulji P, Norton W, Chakravarti D, Coarfa C, Su X, Sandur SK, Ramirez MS, Lee J, Kingsley CV, Sananikone EF, Rajapakshe K, Naff K, Parker-Thornburg J, Bankson JA, Tsai KY, Gunaratne PH, Flores ER, IAPP-driven metabolic reprogramming induces regression of p53-deficient tumours in vivo, Nature, 517 (2015) 626–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Anderson DD, Stover PJ, SHMT1 and SHMT2 are functionally redundant in nuclear de novo thymidylate biosynthesis, PloS one, 4 (2009) e5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Macfarlane AJ, Perry CA, McEntee MF, Lin DM, Stover PJ, Shmt1 heterozygosity impairs folate-dependent thymidylate synthesis capacity and modifies risk of Apc(min)-mediated intestinal cancer risk, Cancer research, 71 (2011) 2098–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Paone A, Marani M, Fiascarelli A, Rinaldo S, Giardina G, Contestabile R, Paiardini A, Cutruzzola F, SHMT1 knockdown induces apoptosis in lung cancer cells by causing uracil misincorporation, Cell death & disease, 5 (2014) e1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wang C, Li X, Zhang J, Ge Z, Chen H, Hu J, EZH2 contributes to 5-FU resistance in gastric cancer by epigenetically suppressing FBXO32 expression, OncoTargets and therapy, 11 (2018) 7853–7864. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


