Significance
Gene regulatory logic reflects the occupancy of cis elements by transcription factors and the configuration of promoters and enhancers. As the majority of genome-wide analyses have focused on adult cells, scant attention has been paid to embryonic cells, other than embryonic stem cells. Focusing on genome-wide comparative analyses of two stages of erythroblasts, we discovered that regulation of embryonic-specific genes is promoter-centric through Gata1, whereas adult-specific control is combinatorial enhancer-driven and requires Myb, which is confirmed by increased enhancer–promoter interactions of adult specific genes. Extending genome-wide comparative analyses more broadly to available datasets of diverse mouse and human cells and tissues, we conclude that the progressively increased enhancer dependence of cell-type–specific genes with developmental age is conserved during development.
Keywords: enhancer, erythropoiesis, HiChIP, GATA1, Myb
Abstract
How overall principles of cell-type–specific gene regulation (the “logic”) may change during ontogeny is largely unexplored. We compared transcriptomic, epigenomic, and three-dimensional (3D) genomic profiles in embryonic (EryP) and adult (EryD) erythroblasts. Despite reduced chromatin accessibility compared to EryP, distal chromatin of EryD is enriched in H3K27ac, Gata1, and Myb occupancy. EryP-/EryD-shared enhancers are highly correlated with red blood cell identity genes, whereas cell-type–specific regulation employs different cis elements in EryP and EryD cells. In contrast to EryP-specific genes, which exhibit promoter-centric regulation through Gata1, EryD-specific genes rely more on distal enhancers for regulation involving Myb-mediated enhancer activation. Gata1 HiChIP demonstrated an overall increased enhancer–promoter interactions at EryD-specific genes, whereas genome editing in selected loci confirmed distal enhancers are required for gene expression in EryD but not in EryP. Applying a metric for enhancer dependence of transcription, we observed a progressive reliance on cell-specific enhancers with increasing ontogenetic age among diverse tissues of mouse and human origin. Our findings highlight fundamental and conserved differences at distinct developmental stages, characterized by simpler promoter-centric regulation of cell-type–specific genes in embryonic cells and increased combinatorial enhancer-driven control in adult cells.
Interactions between chromatin and nuclear regulatory factors establish gene expression programs during development (1). Whereas chromatin landscapes have been elucidated by genome-wide chromatin profiling methods in numerous adult cell types (2, 3), scant attention has been paid to embryonic cell types in human and mouse systems, other than embryonic stem cells (ESCs). Despite well-characterized gene regulatory networks (GRNs) in embryos of some model animals (including Drosophila melanogaster, sea urchin, and chick) (4–6), whether the organization, or “logic,” of gene regulation differs between embryonic and adult cells remains to be explored. We began by examining these issues in the context of functionally analogous blood cells of two different stages of ontogeny, and then extended our findings more broadly to other cell types.
Primitive (EryP; also referred to as “embryonic”) and definitive (EryD; also referred to as “adult”) erythroid cells constitute distinct, temporally overlapping lineages with similar in vivo function and provide a unique opportunity to explore chromatin state at two different stages of ontogeny. Prior molecular studies have identified master erythroid-lineage transcription factors (TFs)—Gata1, Tal1, and Klf1—and several adult-specific factors, namely Bcl11a, Sox6, and Myb (7–10). Whereas transcriptional mechanisms have been studied extensively in EryD cells, we are unaware of analyses in EryP cells, which arise in the yolk sac as a distinct lineage.
EryP cells emerge in blood islands and mature as a semisynchronous cohort in the circulation. EryD cells, which are adult type, are generated within the fetal liver and later in the postnatal bone marrow (9). Gata1 plays a central role in the regulation of erythroid-specific genes in both EryP and EryD lineages and is required for their differentiation (11, 12). Gata1 collaborates with other critical TFs, including Scl/Tal1, Ldb1, Fog1, and Lmo2 (7, 9, 13). Knockout of each of these genes leads to defects in EryP and EryD cells (7, 9, 13). In contrast, the loss of Myb, which is expressed selectively in definitive type cells, impairs proliferation and differentiation of EryD, sparing EryP cells (8). Given the different reliance of EryP and EryD cells on TFs for their development, we have asked whether these related, but distinct, cell lineages in ontogeny differ in their fundamental regulatory organization and logic.
To address this question, we isolated mouse EryP and EryD erythroblasts and characterized transcriptomes, chromatin accessibility, histone modifications, transcription factor (TF) occupancies, and three-dimensional (3D) chromatin interactions. We observed that distal enhancer-dependent, cell-type–specific gene regulation is overall greater in EryD than in EryP, while cell-type–specific gene regulation in EryP is relatively promoter-centric, using Gata1. We hypothesized that these features reflect inherent differences between embryonic cells and more diverse, long-lived adult cells. Analyses of available datasets of diverse mouse and human cells and tissues provided further support for the unexpected finding that cell-type–specific regulatory logic changes with ontogeny.
Results
EryD-Specific Distal Accessible Chromatin Is Enriched for Active Enhancers.
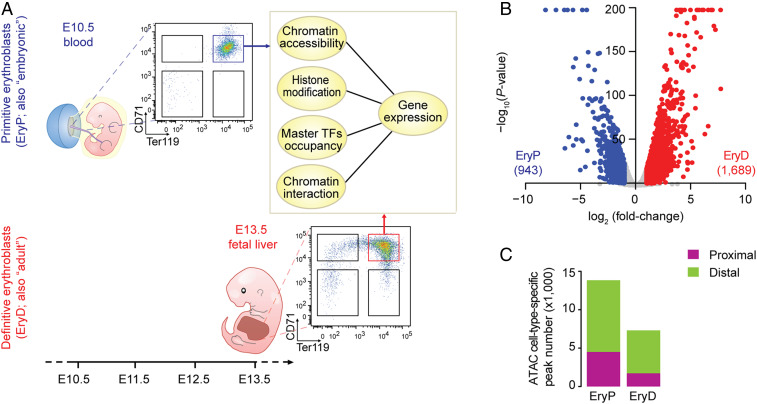
CD71+/Ter119+ EryP and EryD cells were isolated by FACS from embryonic day (E)10.5 embryonic peripheral blood and E13.5 fetal liver, respectively (14). We profiled transcriptomes, chromatin accessibility, histone modifications, and TF occupancies (Fig. 1A). Identity of the respective cell populations was assessed by the expression of globin and TFs characteristic of each lineage (SI Appendix, Figs. S1 A–C). Transcriptome analyses revealed 943 EryP-specific and 1,689 EryD-specific genes (log2(fold-change) > 1, P < 0.01) (Fig. 1B and Dataset S1). EryP-specific genes were modestly enriched with Gene Ontology (GO) terms associated with “metabolic process” (P = 3.2e-5), whereas EryD-specific expressed genes were significantly enriched in “cell cycle genes” (P = 2.5e-12) and “erythrocyte development” (P = 3.2e-8) (SI Appendix, Fig. S1D).
Fig. 1.
Distal chromatin accessibility differs in EryP and EryD erythroblasts. (A) Isolation of EryP and EryD and experiment outline. (B) Transcriptomic analysis of EryP and EryD erythroblasts. (C) Comparisons of EryP-/EryD-specific ATAC peak numbers and their genomic distribution.
Nucleosome eviction is regarded as a primary step in transcriptional activation (15, 16). To explore chromatin structure at the genome-wide level, we examined chromatin accessibility in EryP and EryD by Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) (SI Appendix, Fig. S1 E and F). We assessed genome-wide differential accessibility between EryP and EryD using MAnorm (17) and observed 13,838 EryP-specific (4,506 at proximal and 9,332 at distal regions) and 7,315 EryD-specific (1,726 at proximal and 5,589 at distal regions) accessible regions (Fig. 1C and SI Appendix, Fig. S1G), indicative of greater overall accessibility of EryP chromatin at both proximal and distal regions.
To evaluate whether the different accessibility at distal regions in EryP and EryD relates to putative enhancer activity, we performed chromatin immunoprecipitation sequencing (ChIP-seq) for H3K27ac. As expected, H3K27ac distributed across both proximal and distal regions (SI Appendix, Fig. S1 E and F). In contrast to the greater overall accessibility in EryP, we identified 5,137 EryP-specific H3K27ac peaks and 7,164 EryD-specific peaks (Fig. 2A and SI Appendix, Fig. S2A). While the numbers of cell-type–specific H3K27ac peaks at proximal regions were comparable between EryD and EryP (1,652 vs. 1,841), we observed more cell-type–specific distal H3K27ac peaks in EryD than in EryP (5,512 vs. 3,296) (Fig. 2A). We categorized the distal accessible regions based on H3K27ac peaks into two subgroups, active (with H3K27ac) or open only (without H3K27ac). A small fraction (22%) of EryP-specific distal accessible regions were active, which was similar to that in shared distal accessible regions (25%) (Fig. 2B). In contrast, the percentage was much greater (65%) in EryD-specific distal accessible regions (Fig. 2B and SI Appendix, Fig. S2B), suggesting that EryD-specific distal accessible regions are relatively enriched for more active chromatin, presumably distal active enhancers. Accordingly, we indeed identified 2,087 EryP-specific, 4,986 EryD-specific, and 3,258 shared active enhancers (Fig. 2C), which confirmed the greater overall number of H3K27ac peaks in EryD-specific distal regions. This relative difference became more pronounced upon enumeration of “super-enhancers” (SEs). As shown in Fig. 2D, the number of EryD-specific SEs greatly exceeded that in EryP. Thus, the increased number of cell-type–specific active enhancers characterizes EryD distal accessible chromatin.
Fig. 2.
EryD-specific gene regulation is distal enhancer-driven. (A) Comparisons of EryP-/EryD-specific H3K27ac ChIP-seq peaks and their genomic distribution. (B) Categorization of distal accessible regions with H3K27ac. Heatmaps of ATAC-seq and H3K27ac ChIP-seq signal, centered at the ATAC-seq peak summits. Chromatin distal accessible regions were grouped into three groups: EryP-specific, EryD-specific, and shared peaks. Pie charts show the percentage of “active” (ATAC + H3K27ac) and “open” (ATAC-only) of each group, based on the presence of H3K27ac peaks. (C and D) Venn diagrams of EryP-/EryD-specific active enhancers (C) and super-enhancers (D). (E) Association study of EryP-/EryD-specific active enhancers and gene expression using C-score analysis. Rows are EryP-/EryD-specific genes. Columns are EryP-/EryD-specific enhancers. Also see SI Appendix, Fig. S2A and Table S3 and Materials and Methods.
EryD-Specific Transcription Is Distal Enhancer-Driven.
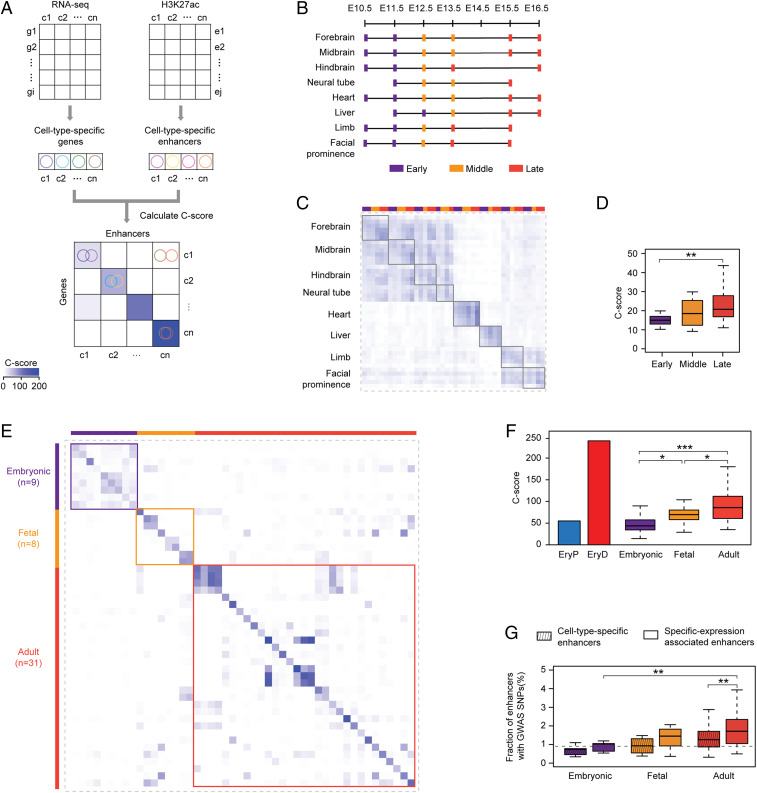
To systematically assess the association between cell-type–specific active enhancers and differential gene expression, we introduced a computational metric, designated C-score (SI Appendix, Fig. S3A). The calculation strategy is similar to Genomic Regions Enrichment of Annotations Tool (GREAT) (18), which is widely used by the genomics community to study functional enrichment associated with a subset of genomic regions. Similar to GREAT, the C-score analysis starts with assigning EryP-/EryD-specific enhancers to target genes (using the “nearest neighbor gene” approach) (17). Next, the statistical significance of association between assigned target genes and cell-type–specific genes (Dataset S1) is quantified by a P value inferred by using the Fisher’s exact test. Finally, the C-score is defined as the −log10(P value) (SI Appendix, Fig. S3 A, Left and SI Appendix, Table S3). Of note, C-score analysis evaluates the association between a set of specific chromatin features (e.g., H3K27ac peaks and ATAC-seq peaks) and cell-type–specific transcriptional activities (SI Appendix, Fig. S3 A, Left). By using C-score analysis (Fig. 2E), we found that activities of cell-type–specific enhancers were more strongly associated with cell-type–specific genes in EryD cells (C-score = 248) than in EryP cells (C-score = 54). The divergence in the observed association might suggest that EryP- and EryD-specific gene expression rely on different regulatory elements, whereas EryD-specific gene expression is relatively enhancer-driven.
To exclude bias in our calculation, we tested other mapping approaches and alternative scoring metrics for the C-score (SI Appendix, Fig. S3A and Materials and Methods). As shown in SI Appendix, Fig. S3 B–D, approaches of “multiple genes mapping” and “various mapping distance” achieved C-score patterns comparable to Fig. 2E with original setting. To assess the effect of calculating P values using an alternative statistical test, we chose the binomial test, which was used in GREAT analysis to assess functional significance of cis-regulatory elements across the entire genome (18), and referred to the corresponding results as the G-score (SI Appendix, Fig. S3 A, Right). In all these analyses, we arrived at the same inference that the association between cell-type–specific enhancers and cell-type–specific genes is greater in EryD than in EryP cells (SI Appendix, Fig. S3 B–E), indicating this conclusion is unlikely due to a technical artifact of the C-score analysis procedure.
We next performed control analyses with EryP-/EryD-specific ATAC-seq peaks and observed strong correlations with EryP-/EryD-specific genes, respectively (SI Appendix, Fig. S1 H, Left and SI Appendix, Table S3), consistent with the established association between chromatin accessibility and transcriptional activities (15, 16), suggesting C-score analysis indeed reflects the association between a set of specific chromatin features and cell-type–specific transcription. C-score can be affected by sample size. We thereby performed C-score analysis with distal EryP-/EryD-specific ATAC-seq peaks, as we observed fewer distal ATAC-seq peaks in EryD as compared to EryP (5,589 vs. 9,332) (Fig. 1C). In agreement with C-score analysis with EryP-/EryD-specific enhancers (Fig. 2E), we observed that the C-score of distal ATAC-seq peaks in EryD was also substantially greater than that in EryP (261 vs. 88) (SI Appendix, Fig. S1 H, Right and SI Appendix, Table S3). These data indicate that conclusions derived from C-score analysis of EryP-/EryD-specific enhancers are not biased by sample size and also demonstrate that both EryD-specific enhancers and EryD-specific distal accessible regions are more strongly associated with cell-type–specific genes in EryD cells than in EryP cells.
Distal enhancers are critical for gene expression, but the weak correlation between EryP-specific enhancers and EryP-specific genes seemed at first incompatible. To interrogate the overall relevance of distal enhancers in EryP, we performed GREAT analysis of EryP-/EryD-shared enhancers, which revealed that most GO terms were associated with red blood cell functions, such as “erythrocyte homeostasis” (P = 2.0e-13) (SI Appendix, Fig. S2C), suggesting that shared enhancers correlate highly with red blood cell identity genes in both EryP and EryD (9). The locus control region (LCR) of β-globin genes, which is required for transcription of embryonic and adult β-like globin genes (19, 20), was also identified as an EryP-shared and EryD-shared enhancer (SI Appendix, Fig. S2E), indicating essential functions of shared enhancers in both EryP and EryD cells. Motif analysis in shared enhancers showed that the most enriched motifs corresponded to master TFs (TAL1 and GATA1) (SI Appendix, Fig. S2D), consistent with established roles of Tal1 and Gata1 at both stages (9, 11, 12). Thus, EryP-/EryD-shared enhancers correlate highly with red blood cell identity, whereas EryP-specific enhancers exhibit weak correlation with EryP-specific genes. Taken together, we speculate that transcription of EryD-specific genes is largely distal enhancer-driven, whereas EryP-specific genes rely more heavily on other regulatory elements.
Gata1 Controls EryP-Specific Gene Transcription through Proximal Elements.
TFs and distinctive chromatin structures coordinately regulate transcription. To explore the regulation of EryP-specific genes, we performed ChIP-seq of Gata1 and Tal1 in EryP and EryD cells (SI Appendix, Fig. S1F). The total number of cell-type–specific Tal1 peaks and their genomic distribution patterns were similar in EryP and EryD (Fig. 3A and SI Appendix, Fig. S4A), and the majority of cell-type–specific Tal1 peaks (>90%) resided at distal regions (Fig. 3A). The C-score of EryD-specific Tal1 peaks in EryD was greater than that in EryP (Fig. 3B), suggesting a role of Tal1 in enhancer-dependent regulation of EryD-specific genes, but less so for EryP-specific genes.
Fig. 3.
Myb is required for distal enhancer activation and Gata1 binding in EryD. (A and B) Comparison of EryP-/EryD-specific Tal1 peaks and their genomic distribution (A). Corresponding association study of EryP-/EryD-specific Tal1 peaks and gene expression is shown in B. (C–E) Comparison of EryP-/EryD-specific Gata1 peaks and their genomic distribution (C). Corresponding association studies of EryP-/EryD-specific Gata1 peaks and gene expression are shown at genome-wide (D), proximal (E, Upper) and distal (E, Lower) regions. (F) Scatter plot of the difference of motif enrichment scores (y axis) and gene expression (x axis) reveals Myb (red spot) as an EryD-specific transcription factor. y axis represents the log2 fold change of the percentage of EryP-/EryD-specific enhancers with motifs, while x axis represents the log2 fold change of gene expression of the cognate TFs. Broken lines indicate threshold at fold change of 1.5 on motif enrichment (y axis) and threshold at fold change of 4 of gene expression (x axis). (G) Heatmaps of the normalized ChIP-seq reads of Gata1, H3K27ac ChIP-seq, p300, and Myb, centered around the Gata1 peak summits in EryP and EryD. EryP-/EryD-specific Gata1 peaks were separated into proximal (purple) and distal (green) regions. Myb and p300 ChIP-seq were performed in E13.5 fetal liver. (H and I) ChIP-seq signal density plots of Gata1 (H) and H3K27ac (I) in MEL cells treated with control shRNA (Ctrl shRNA) or Myb shRNA. Signals included in plots were restricted to EryD-specific distal regions as shown in Fig. 3G. y axis is normalized ChIP-seq reads, log2(RPKM + 1).
Gata1 ChIP-seq revealed that cell-type–specific Gata1 occupancy at distal regions was significantly greater in EryD than EryP (89% versus 57%), despite a comparable overall number of Gata1 peaks (Fig. 3C and SI Appendix, Fig. S4B). In marked contrast, 43% EryP-specific Gata1 peaks were located in proximal regions in EryP (Fig. 3C), indicative of greater proximal Gata1 binding. We also observed strong association between EryP-specific Gata1 peaks and EryP-specific expressed genes (Fig. 3D). In particular, EryP-specific genes were more enriched in EryP-specific Gata1 proximal peaks, whereas EryD-specific genes were more enriched in EryD-specific Gata1 distal peaks (Fig. 3E and SI Appendix, Fig. S3 F–I). These data suggest that Gata1 regulates EryP-specific transcription predominantly through proximal regions. Upon comparison of Gata1 with H3K27ac profiles, we observed that Gata1 occupancy indeed colocalized with H3K27ac in both EryP and EryD (Fig. 3G). Taken together, Gata1 distinct genomic distribution and C-score analyses of Gata1 cell-type–specific peaks demonstrate that EryP-specific genes are regulated principally through Gata1 occupancy at proximal regions, whereas EryD-specific genes are controlled to a larger extent by distal enhancers, involving distal Gata1 occupancy.
Myb Mediates Enhancer Activation and Gata1 Distal Occupancy in EryD.
We next sought to identify TFs that mediate EryD-specific enhancer activity and the distal Gata1 distribution. Motif analysis in EryP-specific and EryD-specific enhancers suggested putative EryD-specific regulators (SI Appendix, Fig. S2D). We ranked TF motifs according to their relative enrichment, as well as expression of the cognate TFs in EryP and EryD (Fig. 3F). The Myb motif was enriched in EryD-specific enhancers, and Myb was specifically expressed in EryD (Fig. 3F). Previous work demonstrated that fetal liver erythropoiesis is defective in Myb null mice (8), and the transcriptional coactivator CBP/p300 (SI Appendix, Fig. S4C), acetylating H3K27 (21), interacts with Myb through its KIX domain (22). Therefore, we hypothesized that interaction of Myb and p300 may mediate activation of EryD-specific enhancers and promote Gata1 distal occupancy. To this end, we performed ChIP-seq of Myb in E13.5 fetal liver cells and performed bioChip-seq of p300 in cells that were harvested from p300+/fb;Rosa26+/BriA mice (23) (SI Appendix, Fig. S4D). Consistent with this hypothesis, Myb bound exclusively to distal regions and cooccupied with Gata1, and the occupancy of p300 and Myb was highly colocalized with EryD-specific Gata1-bound enhancers (Fig. 3G) (P < 2.2e-16, Fisher’s exact test). To examine effects of Myb loss, we attenuated Myb expression in mouse erythroleukemia (MEL) cells with Doxycycline (Dox)-inducible shRNA directed to Myb or a control shRNA. Of note, MEL cells showed comparable chromatin landscape profiles as EryD (Fig. 3G and SI Appendix, Fig. S4E). Myb shRNA decreased overall Gata1 binding and H3K27ac at EryD-specific Gata1 occupied distal regions (Fig. 3 H and I and SI Appendix, Fig. S4E). Taken together, these findings provide evidence that Myb is essential for EryD-specific enhancer activation and Gata1 distal occupancy.
Gata1 HiChIP Confirms Increased Enhancer–Promoter Interactions in EryD.
Our analyses identified enhancer-dependent regulatory logic that distinguishes EryP-specific and EryD-specific gene activation. Distal enhancers regulate gene expression through long-range interactions to promoters of target genes. Long-range chromatin interactions are mediated via Gata1/Tal1/Lmo2/Ldb1 complexes in erythroid cells (9, 13). To determine if enhancer-dependent gene regulation increases in EryD cells, we performed Gata1 HiChIP (24, 25) to profile the enhancer–promoter (E-P) interactions. Interaction matrices at progressively higher resolution revealed that Gata1 HiChIP in EryD exhibited more evident chromatin interactions than in EryP at 5-kb resolution (Fig. 4A and SI Appendix, Fig. S5A), as well as more loops within representative 400-kb EryD-specific expressed loci (Mgll and Abtb1) (Fig. 4B and SI Appendix, Fig. S5 B and C). We next characterized E-P interactions of EryP-/EryD-specific genes by examining reads distribution within ±100-kb window from TSS. In contrast to sporadic E-P interactions of EryP-specific genes in both EryP and EryD cells (Fig. 4 C, Left), promoters of EryD-specific genes exhibited frequent interactions with surrounding enhancers in EryD cells, as compared with EryP cells (Fig. 4 C, Right). E-P loops of EryP-specific genes were similar in number between EryP and EryD (P = 0.47, permutation test), whereas E-P loops of EryD-specific genes were significantly greater in number in EryD cells (P < 0.01, permutation test) (Fig. 4D and SI Appendix, Fig. S5 D and E). These observations provide additional evidence that EryP-specific gene activation employ promoter-centric regulatory logic, in which long-range E-P interactions are less enriched, whereas EryD-specific gene expression depends to a greater extent on enhancer-driven logic, in which frequent E-P interactions are observed. To test if E-P interaction is overall greater in EryD cells, we counted E-P loops in EryP- and EryD-common expressed genes (Materials and Methods), and observed that E-P loops of common expressed genes in EryD cells were significantly greater in number in EryD than in EryP (P < 0.01, permutation test) (Fig. 4D and SI Appendix, Fig. S5 D and E). The EryD-specific expression of Lmo2 (5.7 ± 0.15 in EryP vs. 8.9 ± 0.11 in EryD) (SI Appendix, Fig. S1A), a bridging factor for DNA binding factors (Gata1 and Tal1) and Ldb1 (9, 13), provided additional evidence to support the finding that Gata1 contributes to a greater degree in mediating E-P interactions in EryD than in EryP. Taken together, these observations indicate that increased long-range interactions are correlated with EryD-specific genes in EryD cells.
Fig. 4.
Gata1 HiChIP confirms increased E-P interactions in EryD. (A) Interaction maps of Gata1 HiChIP in EryP and EryD at 500-kb and 5-kb resolution, normalized by square root of vanilla coverage (VC SQRT). Zoom-in views of highlighted regions are in B. (B) Gata1 HiChIP 3D signal enrichment at the Mgll and Abtb1 loci in EryP and EryD cells. (C) Average profiles of E-P interactions of EryP-/EryD-specific genes. Promoters of EryP-specific or EryD-specific genes were set as anchors, and enhancers were defined by Gata1 peaks ±100 kb away from TSS. The average of reads within E-P interactions was plotted. y axis is the average of normalized reads for E-P interactions per gene. Each enhancer bin in the x axis indicates the rank position of the enhancer based on its distance to promoter. Genomic baseline (gray bars) are E-P interactions of a set of randomly selected genes of matched size in both EryP and EryD cells. (D) Quantification of E-P loops in EryP and EryD cells at 25-kb resolution. P value represents permutation test in 1,000 random genes selection of matched size. (E–G) Validation of EryP-shared and EryD-shared enhancers (Enhancer 1, E1). v4C interaction profiles at the E1 in EryP and EryD cells were aligned in E. Quantitative transcript analysis upon E1 deletion in MEL cells and mESCs-derived CD71+/Ter119+ cells were presented in F and G, respectively. (H–J) Validation of EryD-specific enhancer (Enhancer 2, E2). v4C interaction profiles at the E2 in EryP and EryD cells were aligned in H. Quantitative transcript analysis upon E2 deletion in MEL cells and mESCs-derived CD71+/Ter119+ cells were presented in I and J, respectively. Virtual 4C interaction profiles were generated at 1-kb resolution (E and H). E1 and E2 are highlighted in yellow, and promoters of flanking genes are highlighted in gray (E and H). ChIP-seq and RNA-seq track of EryP (blue highlighted) and those of EryD cells (red highlighted) were aligned to the selected genomic region (B, E, and H). Experiments were replicated at least twice in F, G, I, and J. Error bars indicate the SEM; n = 3 (F, G, I, and J). *P < 0.05, **P < 0.01, ***P < 0.001, unpaired one-tailed Student’s t test (F, G, I, and J) and permutation test (D).
CRISPR/Cas9 Genomic Editing Confirms Putative Enhancer-Driven Logic in EryD.
The increased E-P interactions on EryD-specific genes may occur via EryD-specific enhancers or shared enhancers. We next disrupted specific enhancers and shared enhancers to interrogate their requirement for gene expression. To evaluate contribution of EryP-/EryD-shared enhancers in both EryP and EryD, we selected loci with surrounding EryD-specific and EryP-/EryD-common expressed genes. As a representative example, within the Trmo, Hemgn, Anp32b, and Nans locus (chr4: 46410632–46411247), enhancer 1 (yellow highlighted), or E1 for short, is occupied by Gata1 and H3K27ac in both EryP and EryD (Fig. 4E). Trmo and Nans are EryD-specific genes, whereas Hemgn and Anp32b are EryP-/EryD-common expressed genes (SI Appendix, Fig. S5F). We applied virtual 4C (v4C) analysis (25) of Gata1 HiChIP to examine differential E-P interactions at the locus. Here, we set E1 as an anchor point and visualized all interaction reads occurring with E1. As shown in the top two tracks (Fig. 4E), interaction reads between E1 and the Hemgn promoter (gray highlighted) were indeed greater in EryD than in EryP (27.2 in EryD vs. 19.7 in EryP at Hemgn promoter, and 22.2 in EryD vs. 8.7 in EryP at E1). This observation suggests that E-P interactions of the EryP-/EryD-shared enhancer (E1) correlate with Hemgn gene expression in EryD cells, but not in EryP cells.
We performed CRISPR/Cas9 genome editing in definitive-stage (EryD) MEL cells (26, 27) using paired guide RNAs (sgRNAs) targeting flanking regions of E1. The deletion size was refined by Gata1 ChIP-seq peaks (Fig. 4E and SI Appendix, Fig. S5K). Transcripts of four genes in the E1 locus were examined in day 5 differentiated WT and ΔE1 MEL cells. Hemgn transcripts, as well as Trmo, were significantly reduced in ΔE1 MEL cells (Fig. 4F). To evaluate enhancer contribution in EryP cells, we utilized mouse embryonic stem cells (mESCs) (28, 29), as no primitive-stage erythroid cell lines are available. mESCs were differentiated into erythroblasts in vitro (SI Appendix, Fig. S5H), and CD71+/Ter119+ cells were isolated by FACS for qRT-PCR analysis (29) (SI Appendix, Fig. S5I). Gene expression of Hbb-b1, Hbb-bh1, and Hbb-by in mESC-derived CD71+/Ter119+ cells indicated their primitive-stage origin (SI Appendix, Fig. S5J) (28, 30). In contrast to MEL cells, Hemgn and Trmo expression was maintained in E1-deleted mESC-derived CD71+/Ter119+ cells (Fig. 4G). This example of editing EryP-/EryD-shared enhancers sheds light on the differential contribution of enhancers to gene expression—the shared enhancer is required for target gene expression in EryD, but not in EryP cells.
We validated the requirement of EryD-specific enhancer, enhancer 2, or E2 (chr12: 111517811–111518523), within the Tnfaip2 and Eif5 locus (Fig. 4H and SI Appendix, Fig. S5K). Tnfaip2 is an EryD-specific gene, whereas Eif5 is an EryP-/EryD-common expressed gene (SI Appendix, Fig. S5G). A similar strategy was applied to validate E2 in MEL cells and mESC-derived CD71+/Ter119+ cells. Expression of Tnfaip2 was markedly down-regulated in E2-deleted MEL cells (Fig. 4I), whereas Tnfaip2 expression was unaffected in E2-deleted, mESC-derived CD71+/Ter119+ cells (Fig. 4J), in agreement with the v4C prediction at Tnfaip2 promoter in EryD (Fig. 4H). Thus, enhancer deletions provide additional evidence that EryD-specific distal enhancers are essential for EryD-specific gene expression at the selected locus in EryD cells.
Taken together, genome-wide profiling of E-P interactions and enhancer inactivation at selected loci demonstrated long-range interactions are increased at EryD-specific genes and associated enhancers are essential for EryD-specific gene expression, supporting conclusions derived from correlative studies. Specifically, promoter-centric regulatory logic of EryP-specific genes is manifest through proximal occupancy by Gata1, whereas distal enhancer-driven activation in EryD involves Myb and extensive E-P interactions (SI Appendix, Fig. S5L).
Enhancer-Driven Regulatory Logic Correlates with Development in Mouse Tissues.
To explore whether our findings are broadly relevant to other cell contexts, we systematically evaluated the relative contribution of enhancer activities to gene regulation at different developmental stages by applying C-score analysis to mouse ENCODE consortium datasets (2) (Fig. 5A and Materials and Methods). Multiple embryonic tissues, including heart, limb, liver, and neural systems, were collected anatomically, and the age was annotated as the number of days post coitum from E10.5 to E16.5 (Fig. 5B). In 42 qualified samples, we defined cell-type–specific genes (500 per cell type), based on RNA-seq data, and defined cell-type–specific enhancers (5,000 per cell type), based on H3K27ac ChIP-seq signal (Materials and Methods). For each sample, the association between the sample-specific genes and sample-specific enhancers was estimated by C-score (Fig. 5A and SI Appendix, Fig. S3A). To provide a statistical analysis of C-scores across developmental ages and to have sufficient samples in each stage to perform statistical comparisons, we assigned the beginning 1–2 developmental ages and the last 1–2 developmental ages into “Early” and “Late,” respectively. The remaining developmental ages were considered as “Middle” (Fig. 5B). In general, C-scores progressively increased with developmental age (Fig. 5C and SI Appendix, Fig. S6C and Dataset S2), and C-scores in the “Late” group were significantly higher than those in the “Early” group (Fig. 5D). Thus, progressive reliance of cell-specific gene expression on distal enhancers with developmental age appears to be conserved during mouse development.
Fig. 5.
Enhancers contribute to cell-type–specific gene expression progressively with increasing age of ontogeny. (A) Schematic of unbiased evaluation of contribution of cell-type–specific enhancer to gene expression in ENCODE and Roadmap datasets. (B–D) Contribution of cell-type–specific enhancer to gene expression in ENCODE datasets. (B) The schematic diagram of tissues and developmental ages. C-score matrix (C) and statistical analysis of C-scores (D) are presented. (E and F) Contribution of cell-type–specific enhancer to gene expression in Roadmap datasets is represented by C-score matrix (E) and statistical analysis of C-scores (F). Samples were classified into three stages as highlighted. (G) Enrichment analysis of GWAS SNPs in cell-type–specific enhancers and specific-expression associated enhancers in Roadmap datasets. In box plot, the box represents the 25th and 75th percentiles, while the whiskers represent the 5th and 95th percentiles. *P < 0.05, **P < 0.01, ***P < 0.001, unpaired one-tailed Student’s t test (D, F, and G).
Enhancer-Driven Regulatory Logic Correlates with Ontogeny in Human Cells.
As a second, independent test of our hypothesis, we interrogated datasets of 48 diverse human cell types from the NIH Roadmap Epigenomics project (31). Similar analytical methods were applied to Roadmap datasets (Fig. 5A), and we observed that the C-scores within one cell type (diagonal values) were much higher than those across cell types (nondiagonal values) (Fig. 5E and SI Appendix, Fig. S6B), consistent with the concept of tissue-specific enhancers (32). Cell types were classified into three groups according to their developmental stage. We observed that the overall correlation scores for the “Fetal” and “Adult” groups were significantly higher than those in the “Embryonic” group, whereas correlation scores increased progressively with developmental age (Fig. 5F and SI Appendix, Figs. S3 J–M and S6D and Dataset S3). Recent studies suggest that disease-associated DNA sequence variation occurs largely in enhancers (2, 33). We next investigated the extent to which disease-associated SNPs occur in “cell-type–specific enhancers” and in “specific-expression associated enhancers,” a subset of cell-type–specific enhancers, of which nearby associated genes were also cell-type–specific expressed (Fig. 5A). We observed that adult “cell-type–specific enhancers” are highly enriched in SNPs from the Genome-wide Association Studies (GWAS) (Fig. 5G). Moreover, “specific-expression associated enhancers” were more strongly enriched with SNPs than “cell-type–specific enhancers” in the adult group (Fig. 5G), implying that these variants might be associated with gene regulation and diseases in adult. A possible explanation for the lack of enrichment of diseased associated SNPs in embryonic-specific enhancers is that such genetic variation may lead to gene expression changes that lead to prenatal or perinatal lethality. Taken together, our observations provide persuasive evidence that distinct contributions of combinatorial enhancer-driven regulation at embryonic and adult stages, respectively, represent a conserved theme through ontogeny.
Discussion
Through comparative genome-wide analyses of embryonic and adult erythroblasts, we made the unanticipated observation that the dependence of cell-specific gene expression on distal enhancers increases with developmental stage. In contrast to embryonic-specific erythroid genes, adult cell-specific gene expression is more highly dependent on distal enhancer regulation. As this conclusion is primarily inferred from the integration of cell-specific gene expression, transcription factor binding, and active histone marks, we employed HiChIP of the master erythroid transcription factor GATA1 and CRISPR/Cas9-mediated deletion of selected enhancers to test initial conclusions. Indeed, we observed increased E-P interactions of cell-type–specific genes in adult erythroid cells, as compared with embryonic erythroid cells, and demonstrated the requirements for distal enhancers at selected loci in adult, but not embryonic, cells. Analyses of available datasets of diverse mouse and human cells and tissues further broadened the scope of the finding that enhancer-dependent, cell-type–specific regulatory logic increases with ontogeny. Taken together, our findings reveal that the extent to which cell-specific gene expression relies on distal enhancers is not constant in development. As the vast majority of genome-wide analyses have focused on adult type cells, in which distal enhancers dominate transcriptional programs, our observations were unexpected.
Gene regulatory logic reflects the occupancy of cis elements by transcription factors and the chromatin configuration. Tissue-specific enhancers may be brought in close proximity to promoters by looping (34–36), which has been revealed by ChIA-PET, HI-C, superresolution microscopy, and quantitative live-imaging (34, 35, 37–39). Recent studies have shown that chromatin organization is reconfigured during development, stem cell differentiation, and somatic cell reprogramming (38, 40–42), suggesting stage-specific configuration instructs regulatory logic. Embryonic red cells represent a transient lineage, which is replaced by definitive red cells (adult lineage) that sustain the individual throughout life (9). Consistent with these notions, Gata1 HiChIP revealed a greater number of E-P interactions of in EryD, as compared to embryonic EryP cells (Fig. 4 C and D). The greater involvement of enhancers in long-range regulation in EryD requires activation of distal enhancers. We identified Myb as an EryD-specific regulator, which exclusively bound to EryD-specific distal enhancers (Fig. 3G). Loss of Myb decreased overall H3K27ac deposition and Gata1 occupancy within distal enhancer regions (Figs. 3 H and I), suggesting that Myb is essential for EryD-specific distal enhancer activation. Disruption, inversion, or insertion of enhancers can perturb tissue-specific chromatin architecture and lead to inappropriate expression of target gene (43–47). Here, CRISPR/Cas9-mediated deletion of selected enhancers resulted in a striking decrease in target gene expression in adult stage MEL cells, but not in mESC-derived erythroblasts (Fig. 4 E–J). These observations imply that E-P configuration is developmental age-specific, involving specific transcription factors and enhancer activation.
Distal enhancers maintain cell identities (32, 36). Functional enrichment using GREAT analysis shows that EryP-/EryD-shared enhancers, including the LCR region of β-globin genes (SI Appendix, Fig. S2E), are largely enriched with red blood cell functions (SI Appendix, Fig. S2C). In contrast, the dominant role of adult-specific distal enhancers may reflect a greater need to respond to complex and changing environmental cues, including cell–cell interactions, soluble cytokines, and mechanic forces that trigger signal transduction to the nucleus (34, 35). The shift of regulatory logic from promoter-centric in embryonic cells to enhancer-dependent in adult cells for the subset of genes that define each cell type mirrors increased complexity of pathways and extracellular niches in the adult stage (48). The greater involvement of distal enhancers in adult is relevant to human diseases, as the vast majority of disease-associated variations occur within noncoding genomic sequences (2, 33). Promoter-centric regulation in embryonic cells and greater enhancer-dependent control in adult cells for cell-specific genes constitute a previously unrecognized theme in network organization. Further in-depth genome-wide studies in different species and cell types, including programmable 3D genome rewiring, may provide additional insights into the regulatory logic employed at different stages of ontogeny.
Materials and Methods
Details of materials and methods of primary cell isolation, cell culture, mESCs erythroid differentiation, library preparation, and data analysis of RNA-seq, ChIP-seq, ATAC-seq and HiChIP, data visualization, C-score analysis, processing of Roadmap and Encode datasets, CRISPR/Cas9 deletion, and statistical analysis can be found in SI Appendix.
Supplementary Material
Acknowledgments
We thank Nicole Flanagan, John Daley, and Xiaoji Wu, Partha Pratim Das, Sidinh Luc, Hye Ji Cha, and Yan Kai for technical help and helpful discussions. Research was supported by funding from NIH Cooperative Centers of Excellence in Hematology Award 5U54DK11805 (to S.H.O.), NIH Grants R01HL119099 and R01HG009663 (to G.-C.Y. and S.H.O.), and National Natural Science Foundation of China Grant 31871317 (to J.H.). S.H.O. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008672117/-/DCSupplemental.
Data Availability.
The RNA-seq, ChIP-seq, ATAC-seq, and HiChIP data generated in this study have been deposited in the Gene Expression Omnibus (GEO) database under accession no. GSE112717. All other study data are included in the article and SI Appendix.
References
- 1.Long H. K., Prescott S. L., Wysocka J., Ever-changing landscapes: Transcriptional enhancers in development and evolution. Cell 167, 1170–1187 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.ENCODE Project Consortium , An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim T. H. et al., A high-resolution map of active promoters in the human genome. Nature 436, 876–880 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonn S. et al., Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat. Genet. 44, 148–156 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Oliveri P., Tu Q., Davidson E. H., Global regulatory logic for specification of an embryonic cell lineage. Proc. Natl. Acad. Sci. U.S.A. 105, 5955–5962 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stathopoulos A., Levine M., Genomic regulatory networks and animal development. Dev. Cell 9, 449–462 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Cantor A. B., Orkin S. H., Transcriptional regulation of erythropoiesis: An affair involving multiple partners. Oncogene 21, 3368–3376 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Mucenski M. L. et al., A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell 65, 677–689 (1991). [DOI] [PubMed] [Google Scholar]
- 9.Palis J., Primitive and definitive erythropoiesis in mammals. Front. Physiol. 5, 3 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sankaran V. G. et al., Developmental and species-divergent globin switching are driven by BCL11A. Nature 460, 1093–1097 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujiwara Y., Browne C. P., Cunniff K., Goff S. C., Orkin S. H., Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc. Natl. Acad. Sci. U.S.A. 93, 12355–12358 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pevny L. et al., Development of hematopoietic cells lacking transcription factor GATA-1. Development 121, 163–172 (1995). [DOI] [PubMed] [Google Scholar]
- 13.Cantor A. B., Katz S. G., Orkin S. H., Distinct domains of the GATA-1 cofactor FOG-1 differentially influence erythroid versus megakaryocytic maturation. Mol. Cell. Biol. 22, 4268–4279 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koulnis M. et al., Identification and analysis of mouse erythroid progenitors using the CD71/TER119 flow-cytometric assay. J. Vis. Exp., 2809 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thurman R. E. et al., The accessible chromatin landscape of the human genome. Nature 489, 75–82 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neph S. et al., Circuitry and dynamics of human transcription factor regulatory networks. Cell 150, 1274–1286 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang J. et al., Dynamic control of enhancer repertoires drives lineage and stage-specific transcription during hematopoiesis. Dev. Cell 36, 9–23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLean C. Y. et al., GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bender M. A., Bulger M., Close J., Groudine M., Beta-globin gene switching and DNase I sensitivity of the endogenous beta-globin locus in mice do not require the locus control region. Mol. Cell 5, 387–393 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Bender M. A. et al., Independent formation of DnaseI hypersensitive sites in the murine beta-globin locus control region. Blood 95, 3600–3604 (2000). [PubMed] [Google Scholar]
- 21.Tie F. et al., CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 136, 3131–3141 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai P. et al., CBP as a transcriptional coactivator of c-Myb. Genes Dev. 10, 528–540 (1996). [DOI] [PubMed] [Google Scholar]
- 23.Zhou P. et al., Mapping cell type-specific transcriptional enhancers using high affinity, lineage-specific Ep300 bioChIP-seq. eLife 6, e22039 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mumbach M. R. et al., HiChIP: Efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods 13, 919–922 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mumbach M. R. et al., Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat. Genet. 49, 1602–1612 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ganguly S., Skoultchi A. I., Absolute rates of globin gene transcription and mRNA formation during differentiation of cultured mouse erythroleukemia cells. J. Biol. Chem. 260, 12167–12173 (1985). [PubMed] [Google Scholar]
- 27.Sheffery M., Marks P. A., Rifkind R. A., Gene expression in murine erythroleukemia cells. Transcriptional control and chromatin structure of the alpha 1-globin gene. J. Mol. Biol. 172, 417–436 (1984). [DOI] [PubMed] [Google Scholar]
- 28.Choi K., Kennedy M., Kazarov A., Papadimitriou J. C., Keller G., A common precursor for hematopoietic and endothelial cells. Development 125, 725–732 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Fraser S. T., Isern J., Baron M. H., Maturation and enucleation of primitive erythroblasts during mouse embryogenesis is accompanied by changes in cell-surface antigen expression. Blood 109, 343–352 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kingsley P. D. et al., Ontogeny of erythroid gene expression. Blood 121, e5–e13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernstein B. E. et al., The NIH Roadmap epigenomics mapping consortium. Nat. Biotechnol. 28, 1045–1048 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andersson R. et al., An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hnisz D. et al., Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schoenfelder S., Fraser P., Long-range enhancer-promoter contacts in gene expression control. Nat. Rev. Genet. 20, 437–455 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Stadhouders R., Filion G. J., Graf T., Transcription factors and 3D genome conformation in cell-fate decisions. Nature 569, 345–354 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Furlong E. E. M., Levine M., Developmental enhancers and chromosome topology. Science 361, 1341–1345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li G. et al., Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148, 84–98 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krijger P. H. et al., Cell-of-origin-specific 3D genome structure acquired during somatic cell reprogramming. Cell Stem Cell 18, 597–610 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fukaya T., Lim B., Levine M., Enhancer control of transcriptional bursting. Cell 166, 358–368 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dixon J. R. et al., Chromatin architecture reorganization during stem cell differentiation. Nature 518, 331–336 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Laat W., Duboule D., Topology of mammalian developmental enhancers and their regulatory landscapes. Nature 502, 499–506 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y. et al., Transcriptionally active HERV-H retrotransposons demarcate topologically associating domains in human pluripotent stem cells. Nat. Genet. 51, 1380–1388 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tickle C., Towers M., Sonic Hedgehog signaling in limb development. Front. Cell Dev. Biol. 5, 14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lovén J. et al., Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuijers J. et al., Transcriptional dysregulation of MYC reveals common enhancer-docking mechanism. Cell Rep. 23, 349–360 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kragesteen B. K. et al., Dynamic 3D chromatin architecture contributes to enhancer specificity and limb morphogenesis. Nat. Genet. 50, 1463–1473 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kragesteen B. K., Brancati F., Digilio M. C., Mundlos S., Spielmann M., H2AFY promoter deletion causes PITX1 endoactivation and Liebenberg syndrome. J. Med. Genet. 56, 246–251 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Heinz S., Romanoski C. E., Benner C., Glass C. K., The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol. 16, 144–154 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq, ChIP-seq, ATAC-seq, and HiChIP data generated in this study have been deposited in the Gene Expression Omnibus (GEO) database under accession no. GSE112717. All other study data are included in the article and SI Appendix.