Significance
Here we use the expression and accessibility data from a diverse set of cell types to learn a model for the dependence of the accessibility of a regulatory element on its DNA sequence and TF expression. Using GTEx samples with WGS data, we show that the noncoding variants predicted to affect accessibility are more strongly associated with the expression of nearby genes. To interpret a personal genome, we combine the sequence information with context-specific TF expression to prioritize variants and regulatory elements in any genomic region of interest. This approach should be helpful in the study of risk loci previously identified by GWAS. Results from analysis of height and WGS data from the GTEx project support this hypothesis.
Keywords: noncoding variants, regression model, personal genome, GWAS, fine-mapping analysis
Abstract
A person’s genome typically contains millions of variants which represent the differences between this personal genome and the reference human genome. The interpretation of these variants, i.e., the assessment of their potential impact on a person’s phenotype, is currently of great interest in human genetics and medicine. We have developed a prioritization tool called OpenCausal which takes as inputs 1) a personal genome and 2) a reference context-specific TF expression profile and returns a list of noncoding variants prioritized according to their impact on chromatin accessibility for any given genomic region of interest. We applied OpenCausal to 6,430 samples across 18 tissues derived from the GTEx project and found that the variants prioritized by OpenCausal are highly enriched for eQTLs and caQTLs. We further propose a strategy to integrate the predicted open scores with genome-wide association studies (GWAS) data to prioritize putative causal variants and regulatory elements for a given risk locus (i.e., fine-mapping analysis). As an initial example, we applied this method to a GWAS dataset of human height and found that the prioritized putative variants and elements are correlated with the phenotype (i.e., heights of individuals) better than others.
A person’s genome typically contains millions of variants which represent the differences between this personal genome and the reference human genome. The interpretation of these variants, i.e., the assessment of their potential impact on a person’s phenotype, is currently of great interest in human genetics and medicine. There have been many tools developed for interpreting coding variants through the prediction of their impact on the function of the gene products (1–5). For example, SIFT (sorting intolerant from tolerant) predicts the effects of all possible substitutions for each coding variant in the protein sequence using sequence homology (2); PolyPhen-2 predicts the possible impact of amino acid substitutions on the stability and function of human proteins using structural and comparative evolutionary considerations (4). These tools provide an integrated view to analyze the probable impact of particular coding variants on genomic functions. Noncoding variants, however, have been noticeably understudied due to the fact that one variant may have different effects in different tissues, at different developmental stages, and even in different individuals. Recently, several computational approaches have been developed for interpreting noncoding variants by utilizing various genomic and epigenomic annotations (6–10). Ritchie et al. developed genome-wide annotation of variants to prioritize noncoding variants by using a wide range of variant-specific annotations of different classes and at a wide range of genomic scales (6). Ward et al. designed HaploReg to expand genome-wide association studies (GWAS) tag variants into haplotype blocks and overlap the blocks with chromatin state annotations and eQTL (i.e. expression quantitative trait loci) to identify specific regulatory loci (7). Amlie-Wolf et al. proposed INFERNO (INFERring the molecular mechanisms of NOncoding genetic variants) as a tool for inferring the molecular mechanisms of noncoding genetic variants by integrating hundreds of functional genomics datasets spanning enhancer activity, transcription factor (TF) binding sites, and expression quantitative trait loci with GWAS summary statistics (8). However, all of these methods require a large amount of genomic data as input, directly limiting their application to a large scale of contexts. Importantly, most of the previous methods were designed using the summary data derived from single nucleotide polymorphisms (SNP) arrays of thousands of individuals, which fail to capture the individual-specific effect of genome-wide variants.
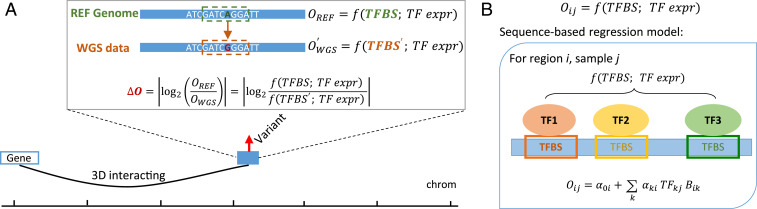
In this study, we focus on the interpretation of noncoding variants by using personal genomic sequences and reference context-specific expression profiles. Specifically, given a person’s noncoding variants within a genomic region of interest, we want to prioritize these variants according to their likely impacts on gene regulation in a tissue-specific manner. Fig. 1A illustrates a causal model for how a variant may impact gene regulation. In this model, a change (relative to the reference genome [REF]) in the sequence of a regulatory element (RE) alters the degree of chromatin accessibility of RE (i.e., openness). The change in the RE’s degree of chromatin accessibility then leads to more changes in the RE’s activity, ultimately affecting the expression of the target genes of that RE. Furthermore, a variant’s impact on the openness depends on the cellular context: if it affects the binding site of a TF that regulates accessibility, its impact will be larger for those cellular contexts where the TF is more highly expressed. These considerations led us to develop a prioritization tool called OpenCausal (Fig. 1) which takes as inputs 1) a personal genome and 2) a reference context-specific TF expression profile and subsequently outputs a list of noncoding variants prioritized according to their impact on openness for any given genomic region of interest. An important component of OpenCausal is Ropen, a tool designed for predicting the openness of an RE based on its sequence and its cellular context represented as a vector of TF expressions.
Fig. 1.
Model design. (A) Schematic overview of the OpenCausal approach. OpenCausal captures the change of chromatin accessibility caused by a variant, where the variation is derived from WGS data. (B) Schematic overview of the Ropen model. Ropen is a sequence-based regression model that predicts chromatin accessibility score for a RE using the expression of TFs binding on this region.
Specifically, we used assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) data on 42 Encyclopedia of DNA Elements (ENCODE) samples across 18 cellular contexts to train the prediction model, Ropen. The effectiveness of Ropen was validated on 6,430 samples across 18 tissues from the Genotype-Tissue Expression (GTEx) project as a target gene’s expression is well correlated with the predicted openness of its associated REs (Fig. 2). Based on Ropen, we developed OpenCausal by computing causal scores (denoted by ΔO) to quantify the impact of variants on RE openness. We validated the usefulness of the scores by demonstrating that variants with high causal scores are highly enriched for eQTLs and caQTLs (caQTL: chromatin accessibility quantitative trait locus; Fig. 3). To provide an initial example of this model, we applied it to prioritize variants in GWAS loci for height. First, using the reference TF expression profile of muscle, we calculated the open scores of variants located in muscle-specific open regions for those donors with available whole-genome sequencing (WGS) data. Then, we defined a statistics variant causality score (VCS) by simultaneously considering the influence of variants on the chromatin accessibility of REs and the relationship between REs and the given trait. For a given risk locus derived from GWAS summary data, we conducted a fine-mapping analysis to prioritize the WGS-based variants in this loci based on their VCSs. We assessed the prioritized variants by checking the correlation between the genotypes of variants and the phenotype of donors and found that the prioritized variants indeed show higher correlations than other variants. We further extended this approach to prioritize REs within GWAS risk loci and validated that the prioritized REs are more correlated with the phenotype of donors. These results provided an initial validation of the usefulness of OpenCausal for the prioritization of variants and REs in GWAS risk loci.
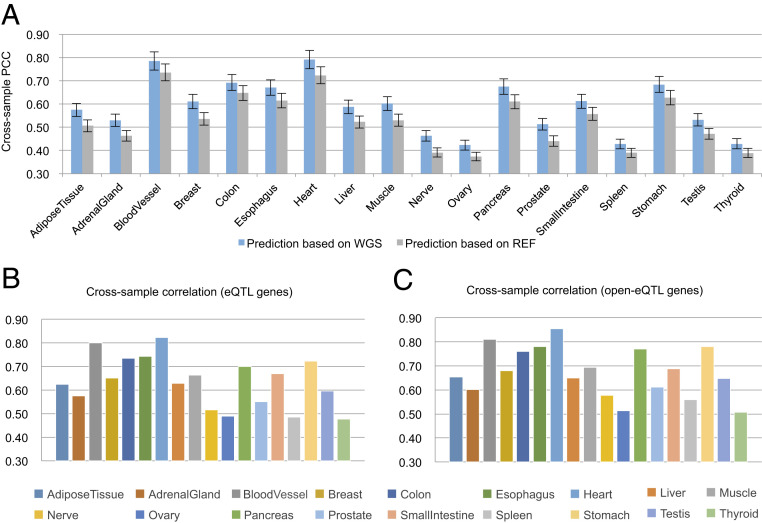
Fig. 2.
Performance of gene expression prediction for GTEx samples. (A) Comparison between prediction using WGS-based chromatin accessibility scores with that using REF-based chromatin accessibility scores in terms of cross-sample correlation. (B) Performance of expression prediction on genes involved in eQTL interactions. (C) Performance of expression prediction on genes involved in eQTL interactions whose variants are located in REs.
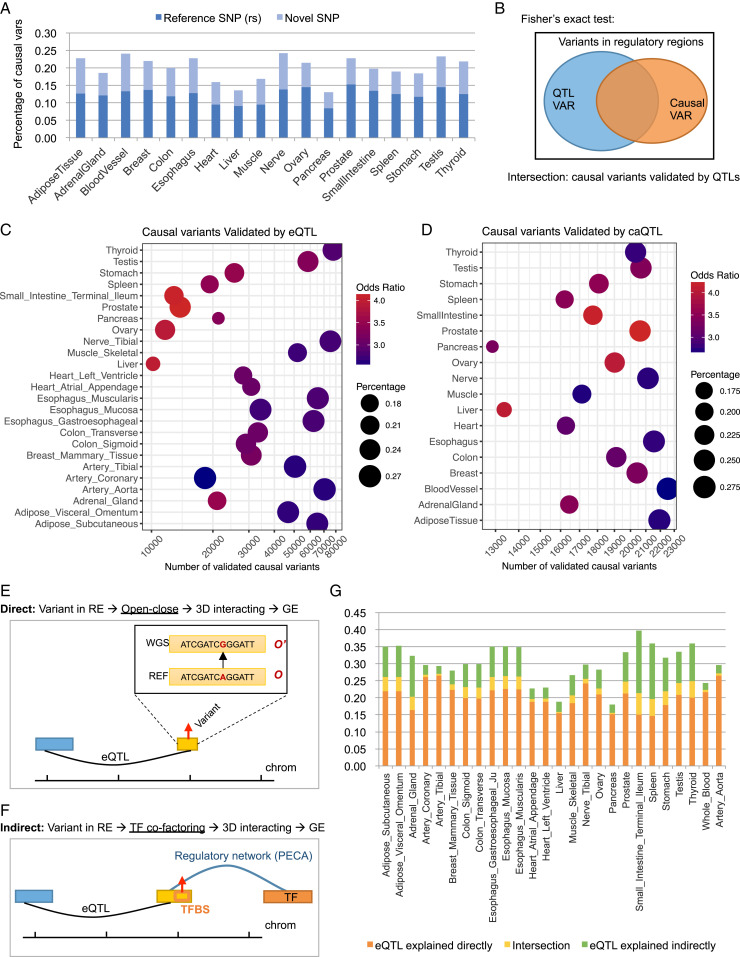
Fig. 3.
OpenCausal detects causal variants for REs. (A) Percentage of detected causal variants in different tissues. Reference SNP (rs) represents variants overlapped with the reported reference SNPs. Novel SNP (ns) represents variants that have not been reported as reference SNPs. (B) Schematic overview of Fisher’s exact test. (C) Validation of detected causal variants using tissue-specific eQTL data. (D) Validation of detected causal variants using caQTL data. Odds ratio represents the odds ratios calculated from Fisher’s exact test. Percentage means the percentage of eQTL/caQTL variants covered by the detected causal variants. (E and F) Schematic overviews of the direct regulatory mechanism (E) and the indirect regulatory mechanism (F) for the interpretation of eQTL interactions. (G) Percentage of interpreted tissue-specific eQTL interactions.
Results
Design of OpenCausal.
Taking a personal genome and a TF expression profile as inputs, OpenCausal aims to detect causal variants on REs according to their impact on openness. Fig. 1A illustrates a causal model for how a variant may impact gene regulation. In this model, a change (relative to the REF) in the sequence of an RE alters the degree of chromatin accessibility of that RE (i.e., openness). This leads to changes in the activity of that RE and subsequently affects the expression of the target genes of that RE. To develop OpenCausal according to this model, we first design a tool named Ropen to predict the openness of an RE based on its sequence and the cellular context represented as a vector of TF expressions (Fig. 1B).
Ropen is a regression model for RE’s openness scores and the expression of the selected TFs—those with TF binding sites (TFBSs) in the RE—as predictors. For each RE, the TFBS is derived from scanning along that sequence of RE and matching the motifs of each TF to that sequence using a motif scanning tool, Hypergeometric Optimization of Motif EnRichment (HOMER) (11). Since only those sequence patterns (typically 8 to 12 bp in length) consistent with the motif profile of a TF can be recognized as TFBSs, variants located in regulatory regions in a personal genome may cause the appearances or disappearances of TFBSs. Hence, our Ropen model leverages this information to predict the differences between changes in the RE’s accessibility in one’s personal genome and those in the REF.
In order to train Ropen, we fit millions of region-specific regression models, each capturing the relation between the openness of a given genomic region and the expression of the TFs with binding affinity to this region. We trained these models based on paired ribonucleic acid sequencing (RNA-seq) and ATAC-seq data from the ENCODE project (12) (42 pairs across 18 tissues; Dataset S1). To derive a list of regulatory genomic regions, we merged ATAC-seq peaks across all samples, yielding a total of 2,965,129 REs. We then calculated the chromatin accessibility scores of these REs and quantified the TF binding affinity on each RE using HOMER. Next, for each RE, we fit a regression model (the Ropen model) to predict its chromatin accessibility score in a given context based on the TF expression profile in this context and the TF binding affinities on this region (Fig. 1B).
To assess model performance, we used leave-one-out cross-validation. Briefly, we predicted the chromatin accessibility of one tissue using the model trained on the other tissues. Each tissue is considered as a testing set once. Ropen achieved Pearson correlation coefficients (PCCs) of 0.628 to 0.833 (SI Appendix, Fig. S1A) and area under the receiver operating characteristic (AUROCs) of 0.939 to 0.996 (SI Appendix, Fig. S1B) in 18 testing sets (details of the evaluation process are shown in SI Appendix, Text S1). We also compared the performance of Ropen that takes motif occurring frequency as inputs with the performance of Ropen that takes motif scores as inputs. We found that Ropen achieved identical performance in both scenarios (SI Appendix, Text S2 and Figs. S2 and S3). Next, we compared Ropen model with two baseline models: one predicts chromatin accessibility scores by regressing on the expression of all TFs regardless of the TF binding information, and the other predicts chromatin accessibility by averaging open signals across all training samples. The results showed that our Ropen model consistently outperformed the baseline models (SI Appendix, Fig. S1 A and B). We further categorized the REs into four groups based on their cross-sample variation (i.e., SD across training samples, std) and examined the performance of Ropen in each group. The results showed that Ropen achieves better performance in the regions with higher cross-sample variation than those with lower variation (SI Appendix, Fig. S1C). Moreover, we observed that the difference in the performance between Ropen and the mean-based baseline model is much larger in the most dynamic group than that in all of the REs (0.081 vs. 0.030; SI Appendix, Fig. S1D). This shows that Ropen has made effective use of the tissue-specific TF expression data that capture the variation across different samples.
Validation of Ropen on GTEx Samples.
We collected paired RNA-seq and WGS data on 6,430 samples across 18 tissues (Dataset S1) from the GTEx Project and used Ropen to compute the chromatin accessibility scores based on the paired WGS and TF expression for these samples.
We used the proportion of gene expression variation explained by these openness scores to assess the performance of Ropen. First, the GTEx samples of each tissue were divided uniformly into five subsets: one for testing and the others for training. Then, for each tissue, we fit a regression model to predict gene expression using openness signals of the REs located in a neighborhood of the target gene (Methods). We used two statistics to evaluate the model performance: cross-gene Pearson’s correlation and cross-sample Pearson’s correlation (SI Appendix, Fig. S4). We filtered out the genes whose expressions are constant across all tissues (say, the SD of expression across samples < 0.2), leaving an average of 25,298 genes to be predicted in different tissues. The results showed that the prediction of gene expression achieved cross-gene PCCs of 0.98 to 0.99 (SI Appendix, Fig. S5) and cross-sample PCCs of 0.42 to 0.79 (Fig. 2A) across tissues. This indicates that the chromatin accessibility predicted by Ropen based on the expression of a small number of genes (i.e., 688 TFs) can indeed explain a substantial portion of the variance in expression genome-wide and thus suggests that the predictions by Ropen are informative.
We further checked the performance of gene expression prediction on genes that are involved in eQTL interactions (referred as eQTL genes). We collected tissue-specific eQTL interactions from the GTEx project (9,348 genes per tissue on average) and predicted the expression of eQTL genes for each tissue. The results showed that the prediction on eQTL genes achieved a higher average cross-sample PCC of 0.63 compared with an average cross-sample PCC of 0.59 of the prediction on all genes (Fig. 2B). This implies that genes related to eQTL interactions are better explained by open signals. Then, we further restricted genes to those involved in eQTL interactions whose SNPs are located in REs (8,099 genes per tissue on average, referred as open-eQTL genes). On this subset of genes, the gene expression prediction achieved an even higher average cross-sample PCC of 0.67, which explained 46.21% of the expression level of open-eQTL genes (; Fig. 2C).
Next, we wondered whether chromatin accessibility prediction based on TFBS information derived from WGS data is better than that derived from the REF. Since we did not have the true chromatin accessibility of GTEx samples as ground truth, we instead compared the performance of gene expression prediction based on the openness scores derived from WGS versus that from REF. We observed that the predictions based on WGS consistently outperformed predictions based on REF (Fig. 2A), indicating that WGS data provides more reliable TFBS information for the prediction of chromatin accessibility than REF. As expected, the improvement of prediction performance based on WGS data, though clear and consistent across all 18 tissues, was modest compared with the baseline. This is mainly because the predictor variables in both methods already include the context-specific TF expression profiles, which can explain a large portion of the variability in chromatin accessibility. Thus, given TF expression, the variability explainable by adding WGS sequence information is limited. On the other hand, we stress that a WGS-based model is necessary if our aim is to understand the impact of variants in personal genomics studies in which expression data are typically not available for the subjects. Therefore, instead of comparing the predictive power of WGS data with context-specific TF expression data, a more important question to ask is whether it is possible to leverage the predictive power of the former to obtain useful interpretation of the variants found in a personal genome. In the following sections, we will address this question by developing the variant prioritization tool OpenCausal. We will also show that the variants identified as having high impact on chromatin accessibility do indeed have a stronger correlation with phenotype than the variants identified as having low impact (see Fig. 5).
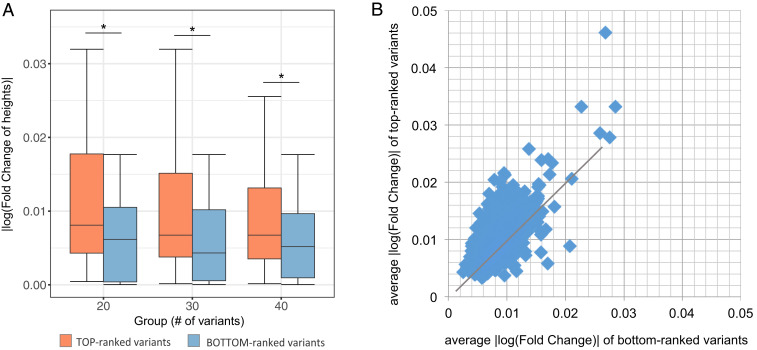
Fig. 5.
Validation of prioritized putative causal variants for height GWAS. (A) Comparison between K top-ranked putative causal variants and K bottom-ranked variants (K = 20, 30, and 40) in the IGF1-related risk locus. y axis is the absolute log value of fold change between the average height of donors with the minor allele and that of donors with the major allele (i.e., |log FC|). *P < 0.05. (B) Comparison between top-ranked variants and bottom-ranked variants for 2,953 risk loci. Each dot represents a risk locus. The y-axis value of each dot is the average |log FC| of 40 top-ranked variants, and x axis is that of 40 bottom-ranked variants in this locus.
Identification of Causal Variants for REs.
On the basis of the sequence-based Ropen model, we designed OpenCausal, a method to detect causal variants for REs in a personal genome (Fig. 1A). First, for any variant in a given RE of a personal genome, we used Ropen to compute the openness score of the RE based on 1) the sequence of the RE in the personal genome and 2) the tissue-specific TF expression profile for the individual (or a corresponding reference profile if the personal one is not available). Next, substituting the personal genome with the REF, we can compute another openness score for this RE in the same tissue. The log fold change ∆O between these two openness scores then gives us an estimate of the impact of this variant on the accessibility of the RE in that specific sample. We call ∆O the causal score of this variant and use it to quantify the impact of this variant on the accessibility in both sample-specific and tissue-specific manner. If ∆O > 0.1, this variant is regarded as a causal variant in this tissue for that person.
To identify causal variants from WGS data for the GTEx samples, we first filtered out REs whose accessibility scores are constant across all ENCODE samples, leaving a list of 1,712,731 REs. Then, we applied the OpenCausal approach to 6,430 GTEx samples of 18 tissues for which both RNA-seq data and WGS data are available, to detect sample-specific causal variants for 1,712,731 REs. For each sample of a given tissue, we calculated the causal scores and identified sample-specific causal variants using the above definition. Then, we obtained tissue-specific causal variants by merging sample-specific causal variants across samples of the same tissue. As a result, OpenCausal identified an average of 1,400,041 causal variants for REs in different tissues (Dataset S2). The percentage of causal variants among all variants in open regions of each tissue is shown in Fig. 3A (ranging from 13.07 to 24.25%). We further checked the overlap between the detected causal variants and reference SNPs and found that 61.80% of detected causal variants could be found as reference SNPs while the remaining 38.20% were SNPs that had not been reported as reference SNP before.
Next, we validated the identified causal variants using tissue-specific eQTL data collected from the GTEx project, based on the assumption that if a variant can cause a significant change in the accessibility of the RE where it locates, it should be more likely to affect distal gene expression. We checked in each tissue the overlap between causal variants identified by OpenCausal and variants involved in eQTL interactions, yielding an average of 37,794 validated causal variants across different tissues. We conducted Fisher’s exact tests regarding all variants located in REs of a specific tissue as background (Fig. 3B). The results showed that the number of causal variants validated by eQTL data were significantly higher than expected, with odds ratios ranging from 2.52 to 4.17 (all P values < 2.2 × 10−16, Fisher’s exact tests; Fig. 3C). In addition, we collected 297,308 lead caQTL (chromatin accessibility QTL) variants from (13) as another validation set in which the caQTL variants are defined as putative lead variants for open peaks. Then, we checked the enrichment of our detected causal variants in these lead caQTL variants. The results showed that an average of 22.77% of lead caQTL variants were detected as causal variants for REs by our approach, with odds ratios ranging from 2.63 to 4.29 across tissues (all P values < 2.2 × 10−16, Fisher’s exact tests; Fig. 3D).
From the above analysis, we noticed that 23.12% of eQTL variants were identified as causal variants by OpenCausal (Fig. 3C) on average. A possible regulatory mechanism behind this can be described as follows: A variant located in an RE causes the change of TFBS for pioneer TFs, which may influence the open signal of this region and changes its state of being open to closed. Then, this change can influence the expression level of a distal gene through three-dimensional chromatin interacting. Eventually, the relationship between variations in REs and the change of distal gene expression is captured by eQTL interactions (the direct regulatory process; Fig. 3E). We can explain the regulatory mechanism of 23.12% of eQTL interactions using our approach with this understanding in mind. In addition, there is another situation in which the variation in an RE does not necessarily change the accessibility of this region but still can influence the distal gene expression by disturbing the binding affinity of cofactors or nonpioneer TFs (the indirect regulatory process; Fig. 3F). We detected this kind of variant by checking whether the variants are located at the TFBS of its regulating TFs, where the TF-RE interactions are derived from the regulatory networks constructed with the chromatin accessibility scores predicted by Ropen model using our previously developed gene regulatory network inference model PECA (a statistical approach based on paired expression and chromatin accessibility) (14) (SI Appendix, Text S3 and Dataset S3). In this way, we identified an average of 9.78% of eQTL interactions that can be explained by the indirect regulatory process. Altogether, our model directly and indirectly explained the regulatory mechanism of an average of 30.23% of eQTL interactions across different tissues (Fig. 3G).
Prioritization of Putative Causal Variants for GWAS Trait.
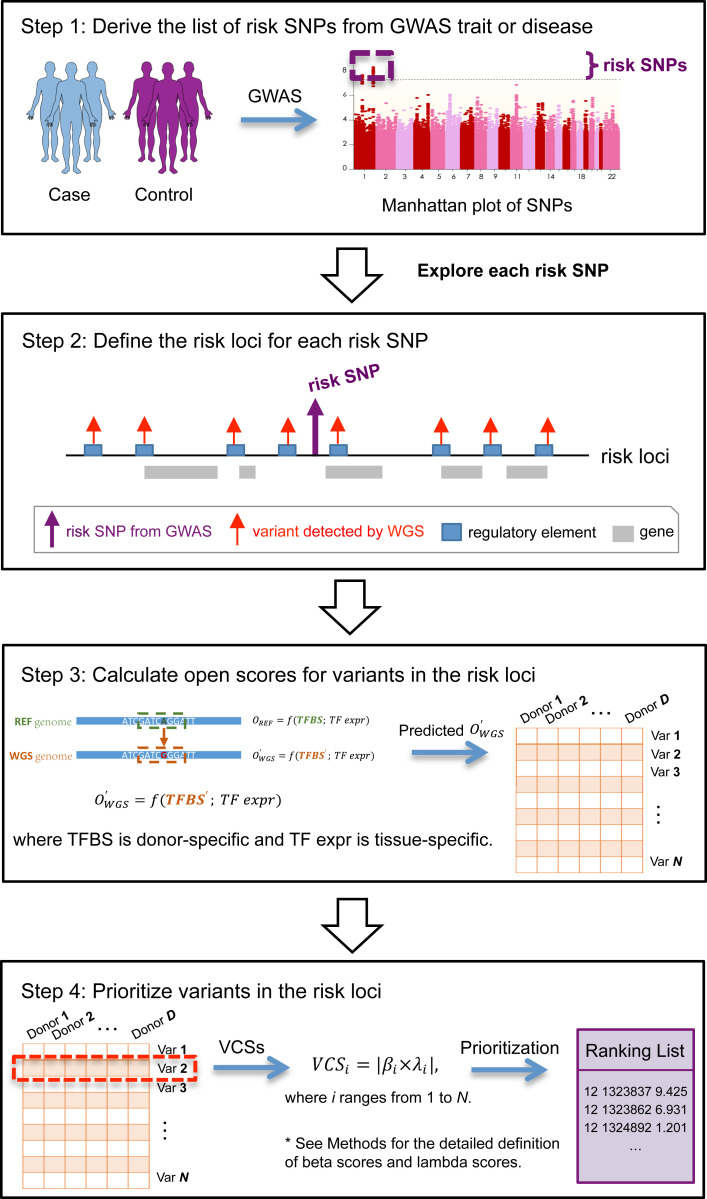
GWAS have been widely used to identify the genomic regions on chromosomes that harbor genetic determinants of complex traits (15–17). However, SNPs detected by the GWAS study—tag SNPs—typically do not have a direct causal relation to the trait (18, 19). The association between a tag SNP and a trait can be indirect due to the association between a tag SNP and a causal SNP, which in turn is associated with a trait. However, since patterns of linkage disequilibrium among SNPs can be complex, it is challenging to determine the underlying causal variants (18). We show below a useful statistical measure that assesses the impact of a given variant on the openness of a regulatory region, which helps with detecting functional noncoding variants that are most likely to be causally related to a trait.
Once we have the WGS data of an individual, we can apply OpenCausal to detect individual-specific causal variants for REs in different tissues for this individual, where the TF expression input is based on a reference profile (say, obtained by averaging the TF-expression profiles in the GTEx expression data in a given tissue). We propose a method to integrate these variants and their associated chromatin accessibility scores with GWAS summary data to prioritize variants in GWAS loci for a trait (Fig. 4). Briefly, for a given risk SNP identified from GWAS summary data, we define the 200-kb region centering at this SNP as a risk locus. Then, we define VCSs for variants in the risk locus by simultaneously considering the influence of variants on the chromatin accessibility of REs and the relationship between REs and the given trait (Methods). Finally, by ranking variants according to their VCS scores, we can prioritize the variants based on their potential to affect chromatin accessibility in tissue contexts relevant to the GWAS trait. We note that in the above method, in order to interpret a personal genome without associated expression or accessibility data, the open scores are calculated using a reference profile (of tissue-specific TF expression) representing the population average of profiles, rather than using the profile from that particular donor.
Fig. 4.
Workflow of the prioritization of genetic variants for GWAS trait.
Next, we illustrate this method using an initial example of the GWAS trait of height. Since we had the phenotype information (i.e., heights) from 635 GTEx donors, which can be used for validation, we chose height for demonstrative purposes. We wanted to see how well our method prioritizes variants falling within a risk locus. In this work, the risk loci for height were obtained based on the summary statistics of a GWAS metaanalysis of about 700,000 individuals of height (19).
As a sanity check, we first examined a risk locus around the known height-related gene IGF1 (20). This is a 200-kb region centering at a highly significant risk SNP rs12579077 (P = 3.4 × 10−32) 2 kb upstream of IGF1. For each of 635 GTEx donors with WGS data, we applied OpenCausal to predict individual-specific open scores in the tissue context of muscle, which is known to be relevant for height (21–24). We calculated the VCS scores for all variants detected in the risk locus and ranked these variants according to the scores. Next, we assessed whether the phenotype is more strongly correlated with genotype stratification by top-ranked variants or that by bottom-ranked variants. To ensure that the number of donors with minor alleles is large enough for evaluation, we filtered out variants whose frequency of minor allele is less than 10. This yielded a list of 157 variants in the IGF1-related risk locus. For each of these variants, we calculated the absolute value of the log fold change between the average height of donors with the minor alleles and that with the major alleles (i.e., log |FC|). We then compared the log |FC| of K top-ranked variants with that of K bottom-ranked variants. The result shows that variants prioritized to the top of the list are more related to the phenotype than variants ranked at the bottom of the list (K = 20, 30, and 40; Fig. 5A). The strategy of deciding the number of causal variants is discussed in SI Appendix, Text S4 and Fig. S6.
To draw a more general conclusion, we collected 3,290 height-associated risk SNPs, distributed across the genome, from the combined GWAS metaanalysis of ∼700,000 individuals (19). After merging SNPs that are close to each other (say, the distance between two risk SNPs closer than 10 kb), we obtained 2,953 risk SNPs. For each risk SNP, we again defined the 200-kb region centering at this SNP as a risk locus. Using the method described above, we prioritized the variants in each risk locus in the tissue context of muscle. Then, for each risk locus, we compared the average log |FC| of 40 top-ranked variants with that of 40 bottom-ranked variants. Results show that the genotypes of top-ranked variants can better stratify the donors than that of the bottom-ranked variants in a high proportion (75.62%) of the risk loci (Fig. 5B). Furthermore, this proportion increases to 85.89% if we exclude essentially tied cases (i.e., cases where the difference between the average log |FC| of top-ranked variants and that of bottom-ranked variants is less than 5 × 10−3). To sum up, by checking the correlation between the genotypes of variants and the phenotype of donors, we have illustrated that our method is effective in the prioritization of putative causal variants in GWAS risk loci.
Finally, we extended this approach to prioritize REs within GWAS risk loci (SI Appendix, Text S5). This extension is important as it allows the interpretation of rare variants including those unique to a person’s genome. We applied it to analyze 2,953 GWAS risk loci for human height. Since each locus has already passed a very stringent threshold for association, our goal is to further assess the relative importance and the tissue-specific role of the REs within each locus. For each of 19 tissues (those in Dataset S1, plus blood tissue), our method provided a prioritization (i.e., ranking) of the REs within each locus. If in at least one tissue, the height prediction based on the top 40 REs is significantly more accurate [i.e., P < 0.00263 = (0.05)/19] than that based on the bottom 40 REs, we regard the prioritization as significantly correct for this locus. Conversely, if the prediction based on the bottom 40 REs is significantly more accurate, the prioritization is regarded as significantly incorrect for this locus. We use fivefold cross-validation to evaluate the prioritizations, where the ranking is done based on four subgroups of donors and the evaluation of prediction accuracy is done on the left-out subgroup, and all modeling was sex-specific. Among all of the loci, the prioritizations were significantly correct in 1,562 loci (Dataset S4), and many comparisons reached a very high level of significance (73 P values were in the range of 2.5 × 10−9 to 9.5 × 10−6). In contrast, the prioritizations were significantly incorrect in only 24 loci (Dataset S5). This result provided a strong validation for our approach to the prioritization of REs. Furthermore, by examining the tissues associated with the significant results for a given locus in Dataset S4, one can identify the relevant tissue contexts for that locus–trait association.
Methods
Data Collection and Preprocessing.
We collected paired RNA-seq and ATAC-seq data for 42 samples of 18 tissues from the ENCODE project (12). Chromatin accessibility peaks were called from binary alignment map (BAM) files of ATAC-seq samples using MACS2 (i.e. model-based analysis of ChIP-Seq v2) (25) with default settings. We then merged peaks of all samples and extended the length of each peak to 500 bp surrounding its middle site, yielding a list of 2,965,129 REs. Processed fragments per kilobase million (FPKMs) of RNA-seq samples were downloaded from the ENCODE project. The detailed information of ENCODE samples is described in Dataset S1. WGS data and RNA-seq data of GTEx samples were obtained from the dbGaP accession number phs000424.v7.p2 on 30 May 2019. Tissue-specific eQTL interactions were downloaded from the GTEx Portal on 30 May 2019.
Definition of Chromatin Accessibility Scores.
For each ATAC-seq sample, we denote the number of reads falling into each RE of length L (i.e., a 500-bp peak) as N. To remove the effect of sequencing depth, we choose a background window of length W surrounding this peak and denote the number of reads falling into this window as M. The chromatin accessibility openness score is formally defined as the fold change of the read counts per base pair and can be simply calculated as
where the length of REs L is set to be 500 bp and the length of a background window W is 1 Mb, according to the suggestion from refs. 26 and 27.
Ropen Model.
Ropen regresses the chromatin accessibility scores of REs using the expression of TFs as predictors. For each RE, we train a locus-level regression model. Specifically, for a biological sample j, we calculate the chromatin accessibility score of RE i based on ATAC-seq data. The TFBSs are obtained by scanning each RE for motifs of different TFs using HOMER (11) with default settings. The regression process can be formulated as the following optimization problem:
where is the log format of expression of TF k in sample j and represents the occurring frequency that region i contains the binding site of TF k; . Here N is the total number of REs, and M is the number of TFs. is selected using the cross-validation strategy based on the mean squared error. We solve the optimization problem of each RE using the elastic net (28).
Gene Expression Prediction Using Chromatin Accessibility Scores.
For each gene, we train a regression model using chromatin accessibility scores of the REs neighboring this gene. Specifically, we detect REs of each tissue from ATAC-seq data. Then, we rank the tissue-specific REs based on their distances to a given gene. To predict the expression of a gene, we train a regression model using the chromatin accessibility scores of K REs nearest to this gene as predictors. This process can be formulated as the following optimization problem:
where is the log form of the expression of gene g in sample j, is the set of K neighboring REs for gene g, and is the chromatin accessibility score of kth neighbor region in sample j; . Here T is the total number of genes, and Q is the number of samples in each tissue. K is set as 30 in our case. We solve the optimization problem of each gene using the elastic net (28).
Design of OpenCausal.
Based on the Ropen model, we can predict the chromatin accessibility score of a given region using the TF expression and genomic sequence information as input, which can be denoted as
where the TFBS can be derived from the sequences of the REF, or alternatively, it can be learned from WGS data. We use the change of chromatin accessibility scores before and after SNP mutation to measure the influence of a variant on an RE. To quantify this influence, we define the absolute value of log fold change between chromatin accessibility scores calculated based on the REF and that calculated based on WGS data as the causal score for the given region, formulated as
Definition of VCS.
First, we assess the marginal influence of REs on the phenotype of a given trait (say, heights) using a regression model,
where is the heights of donors, is the predicted open scores of kth RE, D is the number of donors used in this regression model, and K is the number of REs located on the given risk locus. SEX is a Boolean vector that indicates the sex of donors. In this way, beta scores can be used to reflect the correlation between REs and heights. Variants located on the same RE share the same beta score.
Then, for each variant, we defined a lambda score to access the influence of mutated variants,
where is the predicted open score of a variant in donor i. is the number of donors with the minor allele, and is the number of donors with the major allele. For each variant, is the difference between the average open scores of donors with allele 1 and those with allele 2. Thus, lambda scores reflect the influence of variants on the chromatin accessibility of REs.
Finally, we define the VCS as the absolute value of the product of beta scores and lambda scores, to reflect the importance of variants for a specific trait, denoted as
To avoid the possible information leakage, the VCSs for the GTEx donors were computed using fivefold cross-validation; i.e., we use a subset of donors (say, one of five fold) to train the above regression model for beta scores and use the other donors (say, the other four of five fold) to implement the prioritization and evaluation analysis.
Discussion and Conclusions
In this paper, we propose the method of OpenCausal to prioritize genetic variants based on their impact on chromatin accessibility of REs. The core component of this method is a sequence-based regression model designed to predict the chromatin accessibility scores of REs using genomic sequences and TF expression. By capturing the change of chromatin accessibility scores after involving personal genetic variants on the sequences of REs, we quantify the impact of variants on the accessibility of regulatory regions. The Ropen model has been trained on 42 samples of 18 tissues from the ENCODE project. We statistically evaluated the performance of the Ropen model using a cross-tissue validation strategy. On the basis of Ropen, we have developed the OpenCausal method and applied it to 6,430 samples of 18 tissues derived from the GTEx project. First, we validated the effectiveness of Ropen on GTEx samples by showing that a target gene’s expression is well correlated with the predicted openness of its associated REs. Then, we applied OpenCausal to calculate causal scores for REs in different tissues. The quality of causal scores was validated since variants with high causal scores are highly enriched for eQTLs and caQTLs. As an initial application of OpenCausal, we applied it to prioritize variants in a GWAS data set on human height. For a given risk locus derived from GWAS summary data, we conducted a fine-mapping analysis to prioritize the WGS-based variants in this loci based on VCS scores. We validated the putative causal variants by checking the correlation between genotypes of variants and phenotypes of donors. The results showed that heights stratified by variants with high VCSs are more differentially distributed than those stratified by other variants. This indicates that our method can effectively prioritize variants that are related to the phenotype of a given trait, which provides a point of view for the fine-mapping analysis.
The encouraging performance of VCS scores in prioritizing noncoding variants is attributed to the ability of our prediction model in integrating sequence information with expression data to capture functional variants. As the most important component of OpenCausal, the Ropen model can be independently applied to predict chromatin accessibility scores. This kind of model is currently urgently in demand because RNA-seq data are usually easier to be obtained than ATAC-seq data, but in some cases we may need paired RNA-seq and ATAC-seq data. Furthermore, according to previous studies, the frequency of variants has a large effect on the success of fine mapping by statistical methods (29). WGS can be informative for fine-mapping rare variants. Our method provides insight into detecting functional noncoding rare variants by quantifying the influence of variants on the chromatin state of open regions. For an individual, if the WGS data and context-specific expression data are available, OpenCausal can be applied to detect individual-specific causal variants for REs in different tissues for this individual. The VCS scores designed by our method can be regarded as an annotation for the variants to help WGS data to prioritize common variants and rare variants. In this way, OpenCausal provides a valuable resource for the detection of functional noncoding variants and the interpretation of how genetic variants are involved in the regulatory process and further related to diseases. With the accumulation of available WGS data, we believe there will be more and more methods developed to detect and fine-map rare variants.
Supplementary Material
Acknowledgments
The Genotype-Tissue Expression project was supported by the Common Fund of the Office of the Director of the National Institutes of Health. All GTEx Data were downloaded from The database of Genotypes and Phenotypes (dbGaP). Z.D. and W.H.W. were supported by National Institutes of Health Grants P50HG007735 and R01HG010359. W.L. and R.J. were supported by National Key Research and Development Program of China Grant 2018YFC0910404; the National Natural Science Foundation of China Grants 61873141, 61721003, and 61573207; and the Tsinghua-Fuzhou Institute for Data Technology. W.L. was supported by a scholarship from the Chinese Scholarship Council that supported her work as a visiting student at Stanford University. We thank Shining Ma and Qiao Liu for their helpful suggestions.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1922703117/-/DCSupplemental.
Data Availability.
Source code and data are freely available at https://github.com/liwenran/OpenCausal.
References
- 1.Shameer K., Tripathi L. P., Kalari K. R., Dudley J. T., Sowdhamini R., Interpreting functional effects of coding variants: Challenges in proteome-scale prediction, annotation and assessment. Brief. Bioinform. 17, 841–862 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar P., Henikoff S., Ng P. C., Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4, 1073–1081 (2009). [DOI] [PubMed] [Google Scholar]
- 3.McLaren W. et al., The ensembl variant effect predictor. Genome Biol. 17, 122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adzhubei I. A. et al., A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pollard K. S., Hubisz M. J., Rosenbloom K. R., Siepel A., Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20, 110–121 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ritchie G. R., Dunham I., Zeggini E., Flicek P., Functional annotation of noncoding sequence variants. Nat. Methods 11, 294–296 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ward L. D., Kellis M., HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–D934 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amlie-Wolf A. et al., INFERNO: Inferring the molecular mechanisms of noncoding genetic variants. Nucleic Acids Res. 46, 8740–8753 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kircher M. et al., A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu Q. et al., Systematic tissue-specific functional annotation of the human genome highlights immune-related DNA elements for late-onset Alzheimer’s disease. PLoS Genet. 13, e1006933 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heinz S. et al., Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.ENCODE Project Consortium , The ENCODE (ENCyclopedia of DNA elements) project. Science 306, 636–640 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Kumasaka N., Knights A. J., Gaffney D. J., High-resolution genetic mapping of putative causal interactions between regions of open chromatin. Nat. Genet. 51, 128–137 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duren Z., Chen X., Jiang R., Wang Y., Wong W. H., Modeling gene regulation from paired expression and chromatin accessibility data. Proc. Natl. Acad. Sci. U.S.A. 114, E4914–E4923 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nikpay M. et al., A comprehensive 1,000 genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 47, 1121–1130 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuchsberger C. et al., The genetic architecture of type 2 diabetes. Nature 536, 41–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ripke S. et al.; Schizophrenia Working Group of the Psychiatric Genomics Consortium , Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaid D. J., Chen W., Larson N. B., From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat. Rev. Genet. 19, 491–504 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yengo L. et al.; GIANT Consortium , Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of european ancestry. Hum. Mol. Genet. 27, 3641–3649 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okada Y. et al., A genome-wide association study in 19 633 Japanese subjects identified LHX3-QSOX2 and IGF1 as adult height loci. Hum. Mol. Genet. 19, 2303–2312 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Rauch F., Bailey D. A., Baxter-Jones A., Mirwald R., Faulkner R., The “muscle-bone unit” during the pubertal growth spurt. Bone 34, 771–775 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Klover P., Hennighausen L., Postnatal body growth is dependent on the transcription factors signal transducers and activators of transcription 5a/b in muscle: A role for autocrine/paracrine insulin-like growth factor I. Endocrinology 148, 1489–1497 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Han D.-S. et al., Skeletal muscle mass adjusted by height correlated better with muscular functions than that adjusted by body weight in defining sarcopenia. Sci. Rep. 6, 19457 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barton E. R. et al., Deletion of muscle GRP94 impairs both muscle and body growth by inhibiting local IGF production. FASEB J. 26, 3691–3702 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y. et al., Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li W., Wong W. H., Jiang R., DeepTACT: Predicting 3D chromatin contacts via bootstrapping deep learning. Nucleic Acids Res. 47, e60 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li W., Wang M., Sun J., Wang Y., Jiang R., Gene co-opening network deciphers gene functional relationships. Mol. Biosyst. 13, 2428–2439 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Zou H., Hastie T., Regularization and variable selection via the elastic net. J. R. Stat. Soc. B 67, 301–320 (2005). [Google Scholar]
- 29.Wu Y., Zheng Z., Visscher P. M., Yang J., Quantifying the mapping precision of genome-wide association studies using whole-genome sequencing data. Genome Biol. 18, 86 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source code and data are freely available at https://github.com/liwenran/OpenCausal.