Significance
We developed a strategy to avoid human subjectivity by assessing the quality of red blood cells using imaging flow cytometry and deep learning. We successfully automated traditional expert assessment by training a computer with example images of healthy and unhealthy morphologies. However, we noticed that experts disagree on ∼18% of cells, so instead of relying on experts’ visual assessment, we taught a deep-learning network the degradation phenotypes objectively from images of red blood cells sampled over time. Although training with diverse samples is needed to create and validate a clinical-grade model, doing so would eliminate subjective assessment and facilitate research. The time-based deep-learning strategy may also prove useful for other biological progressions, such as development and disease progression.
Keywords: stored blood quality, cell morphology, deep learning, weakly supervised learning
Abstract
Stored red blood cells (RBCs) are needed for life-saving blood transfusions, but they undergo continuous degradation. RBC storage lesions are often assessed by microscopic examination or biochemical and biophysical assays, which are complex, time-consuming, and destructive to fragile cells. Here we demonstrate the use of label-free imaging flow cytometry and deep learning to characterize RBC lesions. Using brightfield images, a trained neural network achieved 76.7% agreement with experts in classifying seven clinically relevant RBC morphologies associated with storage lesions, comparable to 82.5% agreement between different experts. Given that human observation and classification may not optimally discern RBC quality, we went further and eliminated subjective human annotation in the training step by training a weakly supervised neural network using only storage duration times. The feature space extracted by this network revealed a chronological progression of morphological changes that better predicted blood quality, as measured by physiological hemolytic assay readouts, than the conventional expert-assessed morphology classification system. With further training and clinical testing across multiple sites, protocols, and instruments, deep learning and label-free imaging flow cytometry might be used to routinely and objectively assess RBC storage lesions. This would automate a complex protocol, minimize laboratory sample handling and preparation, and reduce the impact of procedural errors and discrepancies between facilities and blood donors. The chronology-based machine-learning approach may also improve upon humans’ assessment of morphological changes in other biomedically important progressions, such as differentiation and metastasis.
Many clinically important assays involve expert assessment of images and the determination of the quality of red blood cells (RBCs) is no exception. RBCs are needed for life-saving blood transfusions and there is a worldwide shortage. RBCs are degraded by continued storage, yielding oxidative damage, decreased oxygen release capability, and membrane deformation, which can affect the in vivo circulation of transfused RBCs (1–6). Technological progress in the preservation and storage of cells has enabled blood banks to store RBCs at 1 to 6 °C for up to 8 wk in some countries (7–10). During storage, however, the loss of membrane integrity causes the red cells to morph reversibly from regular biconcave discocytes (smooth/crenated discs) into echinocytes (crenated discoid and spheroid), characterized by membrane protrusions or spicula. Eventually, these echinocytes further degrade irreversibly into spheroechinocytes (crenated spheres and smooth spheres) (11, 12). An increased presence of spheroechinocytes is associated with increased viscosity and disturbed capillary blood flow and oxygen delivery (2, 13), leading to decreased safety and efficacy of the transfusion. In addition to these degradation events during storage, each blood sample already contains a mixture of morphologies due to the cells’ varying biological ages. While prospective clinical trials have failed to show a clear relationship between the duration of RBC storage and patient outcomes, there continues to be a strong interest in understanding how the physiological changes that occur to RBCs during ex vivo storage are captured by their morphology, and in turn how this impacts RBC quality (14–18).
The quality of a stored blood unit is often assessed using microscopic examination or biochemical and biophysical assays, which are complex, time-consuming, and destructive to fragile cells (3–5, 12). In the microscopic approach, which is tedious and requires expertise, a sample is spread (smeared) on microscopic slides and the relative fractions of the six subclasses of RBCs (smooth disc, crenated disc, crenated discoid, crenated spheroid, crenated sphere, and smooth sphere) are estimated by manual cell counting (12, 19). These fractions are then multiplied by corresponding shape factors (1.0, 0.8, 0.6, 0.4, 0.2, and 0.0, respectively) and summed to yield the Morphology Index (MI), a quality metric for quantifying the morphological profile of RBCs during storage (20). This technique is labor-intensive, prone to subjective bias, and limited by small sample sizes. The smearing itself may affect the state of the fragile RBCs, causing adverse alterations to the sample’s true morphological profile. To avoid this adverse effect, wet preparations of RBC samples can sometimes be used to assess morphology to avoid the artifact created by the standard blood-smearing technique. However, this method requires even more expertise, does not remove subjective bias, and does not shorten the time required to perform the microscopic evaluation. Improved methods to objectively assess degradation events would thus improve quality assessment of manufactured blood products for transfusion and help identify donor factors and manufacturing methods that would produce higher-quality RBC products, potentially leading to better patient outcomes and helping meet the dramatically growing worldwide demand for stored blood (21).
Deep learning has shown great promise to detect biomedically important cell states in images (22). We hypothesized that a deep-learning–based morphological assessment approach might provide a reliable proxy for RBC quality (although we emphasize that RBC quality cannot be absolutely measured without treating patients and measuring outcomes). We therefore tested recent deep-learning methods on RBC images from three completely independent cohorts in two different countries using imaging flow cytometry (IFC) to assess whether: 1) A neural network might be trained to replicate an expert’s judgment in classifying the stages of RBC degradation in cell images and, going further, 2) whether a neural network might extract subtle degradation-related features of RBCs more objectively than humans. Success in the latter case would lend evidence that deep learning can extract features representing clinically important biological progressions from images that are not detectable to the human eye.
Results
Expert-Supervised Deep-Learning–Based Automation of the Standard RBC MI.
We aimed to devise improved methods to assess RBC blood quality by training deep convolutional neural networks to characterize the morphology of unstained RBCs at different time points during blood storage (Fig. 1 and SI Appendix, Fig. S1). We used an imaging flow cytometer to capture images of single RBCs as they flow through the instrument (23). The instrument naturally favors cells in suspension, such as blood cells, capturing images at a rate of hundreds to thousands of cells per second. This yields a large number of isolated, single-cell images well-suited to deep-learning algorithms, which learn from raw pixel information and benefit from a large pool of images to extract meaningful features. Through a hierarchical architecture of feature layers, a deep neural network identifies patterns in the input image relevant to discriminating morphologies of interest while suppressing irrelevant variations (24).
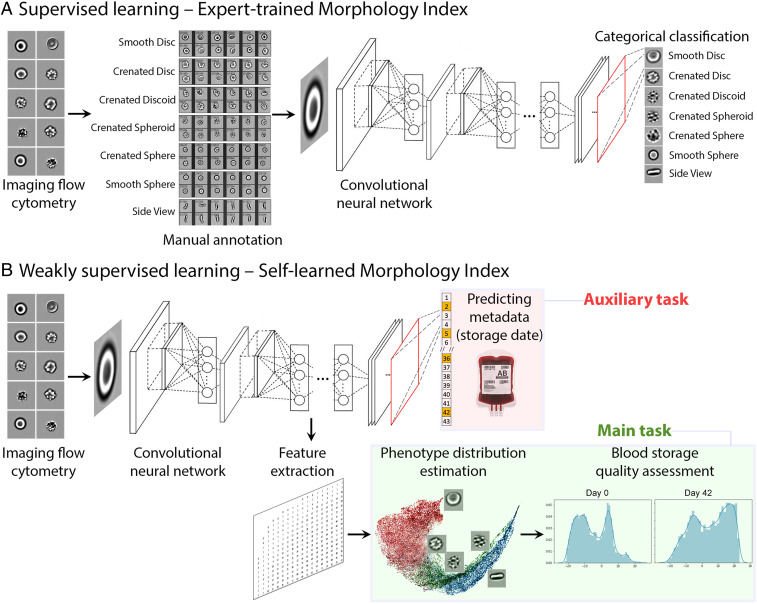
Fig. 1.
Two alternate machine-learning pipelines to assess RBC quality by morphology. The input of the pipeline is single-cell RBC images from imaging flow cytometry. (A) Supervised learning automates the classification of cells into expert-defined categories (SI Appendix, Figs. S1 and S3). The neural network ResNet50 was trained to classify each individual cell into one of the seven morphology classes (smooth disc, crenated disc, crenated discoid, crenated spheroid, crenated sphere, smooth sphere, and side view), as guided by expert annotations of those classes. (B) Weakly supervised learning, by contrast, learns a new quality metric independent of human input, the SMI. The network ResNet50 was trained to identify the storage date of the blood unit a given RBC could belong to, as an auxiliary task. The morphological features extracted by a layer of the network during the training phase can be then used to assign each cell to a point along a continuum from healthy to degraded.
We first developed a supervised classifier (Fig. 1A), where the machine-learning model is supervised to “learn” to categorize cells into the six morphological classes of RBCs mentioned above, plus an additional side-view class where the true class was indiscernible. We collected blood samples from healthy volunteers at two sites on different continents (Canadian Blood Services, hereafter “Canadian,” and the University Hospital of Geneva, hereafter “Swiss”) and processed red cell units using standard blood bank protocols (25), followed by IFC analysis every 3 to 7 d until expiration at 6 wk (SI Appendix, Figs. S1A and S2) (25–27). Five researchers annotated ∼52,700 RBCs spanning across the blood units (SI Appendix, Fig. S3), creating the largest freely available public dataset of its kind (see Data Availability; ∼67,400 cells including the undecidable class and held-out dataset, described later). The brightfield images of ∼40,900 annotated RBCs were then used as ground truth to train a ResNet50 model (28), a well-known neural network for image classification (29).
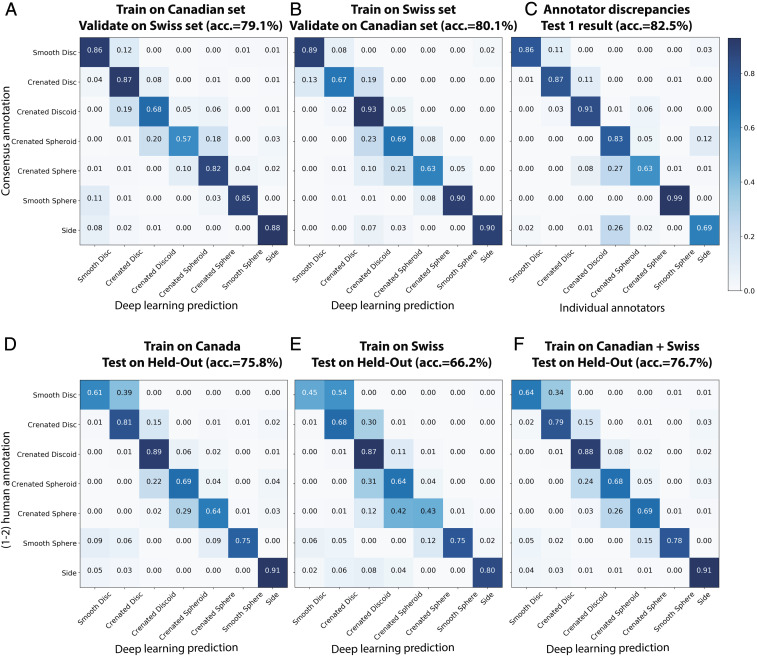
This fully supervised model was able to approximate human annotators in categorizing cells into one of the seven expert-defined morphological classes. Taking great care not to “contaminate” any test sets with cell images from samples used for training (SI Appendix, Fig. S4), we assessed the performance and robustness of this fully supervised model in several tests. First (Validation in SI Appendix, Fig. S4), we observed strong accuracy (79.1 to 80.1% agreement with experts) of the models; as a baseline, a random classifier yields only 14.3% accuracy for seven classes. These values were obtained for the optimized network trained on images solely from one site and tested on the other, even though the samples were prepared by different facilities across continents without any prior knowledge of each other (Fig. 2 A and B). We hypothesize the simplicity of label-free IFC contributes to this success across cohorts. Training the network on combined Canadian and Swiss training data (Test 2 in SI Appendix, Fig. S4) achieved an average accuracy of 76.7% on a held-out dataset, which was only tested a single time prior to submission of this report (Fig. 2 D–F); this approaches the 80.3% accuracy (average recall of 0.80, precision of 0.81, and F1-score 0.80) seen on the nonheld-out data that was used in optimizing the network (Test 1 in SI Appendix, Fig. S4), indicating the model is not overfit. To further assess the robustness and the variability of the classification model when different subsamples of the training data are selected, a 10-fold leave-one-out cross-validation approach has also been conducted. We iterated the training-validation partitions in which 9 of 10 bags (green blocks in SI Appendix, Fig. S5 A–J) are used to train a model that is then evaluated on the remaining bag (red block in SI Appendix, Fig. S5 A–J). This procedure is then repeated for 10 possible choices for the left-out bag, and the predictive performance scores from the 10 runs are then reported as an average classification accuracy of 86.7% ± 3.5% (mean ± SD) as well as receiver operating characteristic (which plots sensitivity as a function of one minus specificity) curves and its associated area under the curve (SI Appendix, Fig. S5 A–L). Because all of these accuracy values are comparable to the accuracy between different experts (82.5%) (Fig. 2C), we conclude the trained deep-learning model is roughly as effective as an expert. With proper clinical validation and ideally additional training images from other facilities, this strategy could be implemented for routine automated assessment of RBCs by IFC. We freely provide the trained model for training and testing on data from other blood bank facilities (Code Availability).
Fig. 2.
Supervised deep learning (Automated Morphology Index) approaches human-level performance for assessing RBC morphology. (A and B) Validation of the supervised deep-learning classifier across two facilities, which include distinct instruments, operators, sample preparation procedures, and donors. Samples were collected independently, with no effort to standardize across the two sites. Most “errors” are in chronologically adjacent categories. Confusion matrices show the prediction of seven categories of RBC morphologies performed by a ResNet50 model (A) trained on the Canadian dataset (n = ∼15,500 cells) and tested on Swiss data (n = ∼25,400 cells) and (B) vice versa; comparable accuracy is seen in both cases. (C) Discrepancies between five human annotators when assigning the exact same cells (n = 1,500) into RBC morphology classes. Detailed analysis of human discrepancies is shown in SI Appendix, Figs. S6 and S10; the average is shown here. (D–F) Validation of the trained supervised models on held-out datasets (Test 2 in SI Appendix, Fig. S4). The held-out datasets were not used in training and were only tested once, immediately before the submission of the work. As is the case for the supervised deep learning model in A and B, most of the errors are in adjacent classes, pointing to inconsistency in human-defined categories (SI Appendix, Fig. S11). Because the accuracy shown in F, 76.7%, is comparable to that between experts (in C, 82.5%), we conclude the trained deep-learning model is roughly as effective as an expert.
Despite this successful result, we questioned whether visual inspection by experts best captures RBC storage lesions. As mentioned above, each individual expert only agrees with the experts’ consensus around 82.5% of the time (Fig. 2C). This means that an automated method trained to replicate an expert cannot do better than 82.5%. We noted that most of the experts’ discrepancies, as is also the case for the supervised deep-learning model, occurred between adjacent RBC subclasses (SI Appendix, Figs. S6 and S10), indicating that classification of RBCs into discrete “bins,” whether human-annotated or automated, may be a poor fit to this relatively continuous biological process (visualized in SI Appendix, Fig. S7).
Weakly Supervised, Deep-Learning–Based Self-Learned MI.
We thus investigated an alternative training strategy based on weakly supervised learning (30–33), in which the neural network learns the morphological properties of RBCs independently from visual categories defined by experts. The fundamental strategy is to train the network to predict an auxiliary but biologically meaningful property: The storage duration of the blood unit from which each cell was sampled (Fig. 1B). Although storage duration correlates with RBC health, predicting storage age is not the goal for two reasons. First, the storage age of blood bags is typically already known, thus there is no need to predict it. Second, storage duration correlates imperfectly with RBC health, in the same way that individual humans’ age and health are correlated but not completely predictive. For RBCs of identical storage duration, there are dramatic biological age variations and cell heterogeneity that are more medically relevant than storage age. Nevertheless, weak supervision in this context means that the model is trained on a variable (RBC storage duration) that causes the network to pay attention to features in images that correspond to this variable. Once a network is trained, storage duration predictions themselves are ignored, and an intermediate layer of the network is used to compute thousands of features from the input images; these features should capture morphological changes that occur in response to storage. A dimensionality reduction method is then applied to map cells onto a linear continuum that captures this biological phenomenon.
Following this strategy, we trained the ResNet50 network to estimate (regress) the storage duration of RBC images. Because no human annotation is required with this strategy, we could use more than one million RBCs pooled from the entire joint dataset (Canadian and Swiss, not mapping to any abovementioned datasets). Not surprisingly, given biological age variations and cell heterogeneity, the model was not particularly accurate in predicting the age of a blood unit from a single-cell image in the held-out test sets (average error was 18.87 ± 6.96 d in a prediction range spanning 48 d), nor did it show strong ability to predict the known morphological classes, based on the ∼40,900 annotated cells used in the previous supervised learning framework (SI Appendix, Fig. S8A).
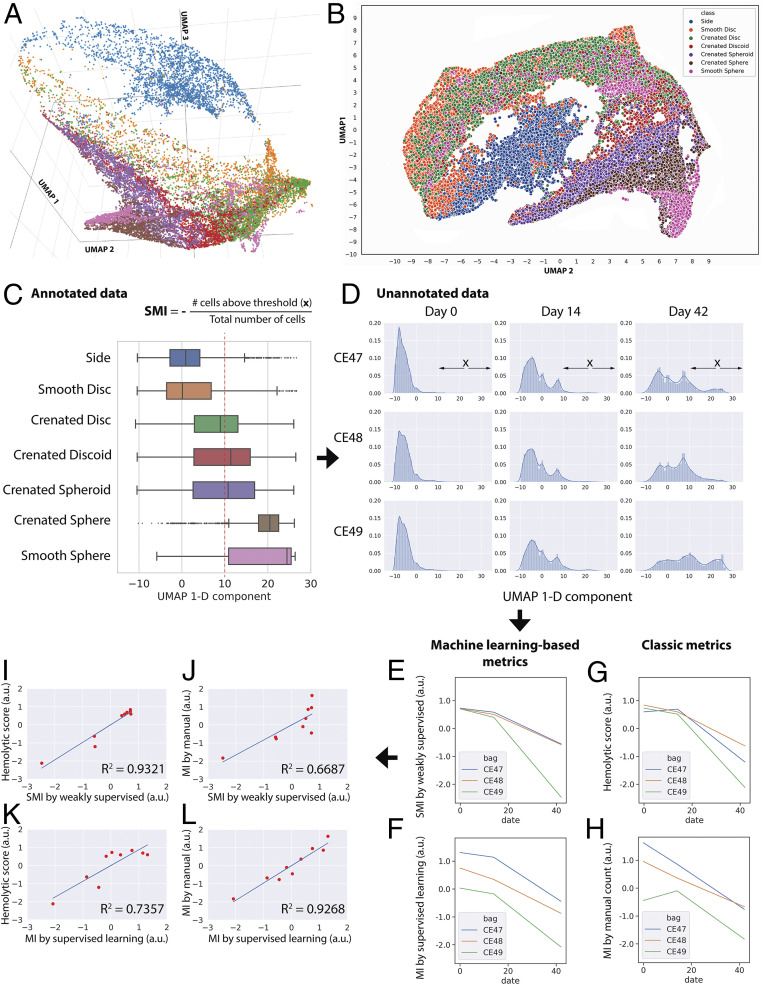
Nevertheless, an intermediate layer of this trained network (Materials and Methods) learned to extract single-cell features that revealed a meaningful order of morphological progression. Visually inspecting an embedding space obtained with Uniform Manifold Approximation and Projection (UMAP) (34, 35) suggests that the single-cell features could be approximately aligned on a low-dimensional manifold (Fig. 3 A and B) (see ref. 36). This progression proceeds correctly from healthy to unhealthy cell phenotypes: Discocytes (smooth discs and crenated discs) to echinocytes (crenated discoids and crenated spheroids) to spherocytes (crenated spheres and smooth spheres). The progression is confirmed by the annotated cells, but the linear pattern is detectable even in their absence. The trajectory also positioned side-view cells in proximity to disc-like cell classes, which is sensible because only disc-shaped objects could present in flank angles, while spheres are spherical regardless of the view. Other trajectory recovery methods, such as diffusion map (37) and diffusion pseudotime (38, 39), did not provide as clear a resolution of the progression; they are well-suited to trajectories that branch (40) (Materials and Methods). In contrast, the same analysis using classic image features extracted by CellProfiler (41) organized cells into discrete clusters (SI Appendix, Fig. S9) rather than a continuous progression of morphologies.
Fig. 3.
Data-driven ordering of RBC morphologies by weakly supervised learning allows robust blood quality assessment. (A–C) We discovered a relatively linear progression for major morphological classes of RBCs using features extracted from an intermediate layer (Res4a_ReLU) of the trained weakly supervised model projected into low-dimensional space using a UMAP algorithm. This continuum is observed in 3D (A), 2D (B), or 1D (C) embedding space and interactive 3D-PCA, t-SNE, and UMAP projections of 7,000 representative cells can be explored in a public browser-based tool (ref. 36), select colors and labels for better visualization). Color coding in A, B, and C is consistent, showing that the extracted weakly supervised features place cells along their correct biological progression, from discocytes (smooth discs, crenated discs) to echinocytes (crenated discoids, crenated spheroids, crenated spheres) to spherocytes (smooth sphere). The boxes in A overlap due to continuous transitions between morphology categories, which could not be further resolved by the chosen ResNet50 architecture. The red dotted line in C indicates the threshold in the 1D UMAP, above which RBCs were categorized as unhealthy; this includes most spheroechinocytes (crenated spheroid, crenated spheres, and smooth spheres). The increasing fraction of unhealthy cells (x) over the total number of cells is termed SMI. (D) Distribution of unannotated cells according to the 1D UMAP of weakly supervised features. For each blood unit, deep-learning features were extracted from label-free images of 20,000 cells by a trained weakly supervised neural network. The extracted features were then projected in 1D UMAP space (x axis of each histogram). The shift of distributions from the left to the right as time progresses is clearly visible (more healthy biconcave RBCs are toward the negative end of the x axis; spheroechinocytes are toward the positive end). (E–H) Results from different approaches for evaluating the quality of blood units, with the y axes unified to the same scale. (E) Blood quality according to the proposed SMI. (F) Blood quality according to our automated MI morphology analysis using a fully supervised classifier. (G) Blood quality as assessed by a physiological assay for hemolysis. (H) Blood quality as assessed by expert manual MI morphology analysis (∼4,000 cells per blood unit per time point). (I–L) Pairwise comparisons between proposed machine learning approaches and classic methods for evaluating the quality of red cell units. (I) There is a stronger correlation between the proposed weakly supervised-based quality assessment and hemolytic readouts (coefficient of determination, R2 = 0.93) than that of (J) human manual annotations of morphology (R2 = 0.67). (K and L) In contrast, the proposed supervised learning-based method shows the opposite trend. The x and y axes of plots I–L were unified to the same scale.
We therefore defined the recovered 1D UMAP manifold from healthy to unhealthy as a new metric of blood unit quality, self-learned MI (SMI), where cells that possess higher values in the 1D manifold of deep-learning features are associated with older storage duration and lower quality for blood transfusion (Fig. 3C). Validating this metric is challenging, given that there is no perfect ground truth. Expert morphological annotation cannot be deemed as correct, given intraexpert discrepancies as mentioned above. The current regulatory gold standard for RBC quality requires radio-labeling (or biotinylating) RBCs, transfusing them into volunteers, and measuring the percentage that circulates after 24 h, with 75% being the threshold. This was not feasible for our study and furthermore is a methodology that many in the field seek to replace, as it does not capture the ultimate endpoint of interest, oxygen delivery (42).
We therefore subjected three blood units to two parallel quality assessments at weeks 0, 2, and 6 of the storage period. The quality assessments were 1) a biochemical assay for hemolysis, which focuses on red cell stability, and 2) IFC with the standard expert morphology classification, MI. These two assessments correlate, but not strongly (R2 = 0.65) (SI Appendix, Fig. S8D). Two blood units were analyzed during the validation of the weakly supervised framework (Test 3 in SI Appendix, Fig. S4), and one was held out and tested a single time prior to the submission of this manuscript (Test 4 in SI Appendix, Fig. S4). A morphological ordering of single cells shows the expected degradation events over time (Fig. 3 D–H).
We found that the SMI of the three blood units corresponded better to the physiological–biochemical readout, the hemolytic score (coefficient of determination, R2 = 0.93) (Fig. 3I) than to the classic inspection-based MI (R2 = 0.67) (Fig. 3J). This suggests that the SMI can produce measures of blood quality that are more consistent with biochemical readouts, and less consistent with a subjective morphological inspection. The automated MI by fully supervised learning showed the opposite trend (R2 = 0.74 compared to the hemolytic score and R2 = 0.93 compared to classic MI) (Fig. 3 K and L), indicating that fully supervised models carry over subjective biases and are less consistent with more objective biochemical readouts. Applying a healthy/unhealthy threshold (as in SMI) instead of indexing (0, 0.2, 0.4, 0.6, 0.8, 1, as in MI) using manually annotated images is also less correlated to the hemolytic score (R2 = 0.85) (SI Appendix, Fig. S8E), indicating that the improvement is due to the weakly supervised approach rather than a change in thresholding versus indexing.
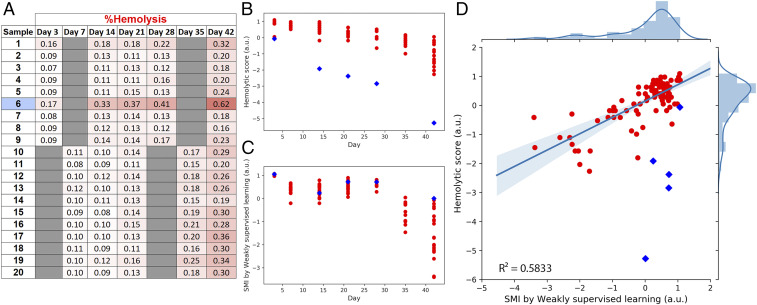
As a final test of generalizability and robustness, we combined the Swiss and Canadian training data and tested the SMI scoring system on an additional 20 red cell units sampled at five storage durations acquired by a third facility, the Blood for Research Facility (netCAD, Vancouver, BC, Canada) (Fig. 4). Again, we observed the low-dimensional manifold progression of cells from healthy to degenerated. Furthermore, with the caveat of one sample particularly prone to hemolyze, likely due to unknown donor factors (Materials and Methods), we observed the expected correlation between SMI and hemolytic scores (Fig. 4D). The R2 of 0.58 is lower than that observed in the tests in Fig. 3, but still indicates the ability of the SMI strategy to be relatively robust to samples collected by different operators at different clinical locations.
Fig. 4.
Generalizability of SMI to blood samples from a third facility. (A) Additional data for comparison of SMI (as developed in this report) and conventional hemolysis scores of 20 red cell units sampled at five storage durations were analyzed at Canadian Blood Services in Edmonton, Alberta. (B) Hemolytic scores based on the standard physiological hemolysis tests for the collected red cell units. Sample 6 showed an elevated level of hemolysis from day 3 to day 42 (deeper red shades in the table, blue diamonds in B–D), which is likely due to donor factors (Materials and Methods). This data point is therefore marked as blue in the data plots but excluded from statistics. (C) SMI scores by weakly supervised learning of the corresponding red cell units. (D) The correlation between hemolysis and SMI scoring systems. Coefficient of determination R2 = 0.5833. Shaded bands around the regression line display the 95% confidence interval for the regression estimate. With the inclusion of elevated hemolyzed sample (sample 6, shown as blue diamonds), the coefficient of determination R2 is 0.2520, likely because the current neural network was not trained to tolerate certain confounding factors such as donor factors that lead to unusually high hemolysis levels.
Discussion
Methods and metrics for the assessment of RBC quality are rapidly developing and uncertain, given the lack of sufficient clinical data to conclusively determine ideal proxies (whether morphological or biochemical) for in vivo circulation or for the clinical outcomes of interest (42, 43). Our work does not aim to resolve this controversy nor claim the superiority of any one assessment method over others. Rather, in this study we present two strategies that are capable of providing more reliable and convenient quantitative data in future studies of RBC quality that aim to resolve some of these controversies and identify useful donor factors.
The first strategy used supervised deep learning to automate and standardize the current standard blood-quality scoring procedure, which is based on expert visual classification of RBCs into morphological classes and computation of the MI; this work automates and standardizes a tedious and subjective assay, providing near expert-level results. The second strategy derived an SMI to measure blood quality using weakly supervised deep learning trained on storage age; this approach went beyond human vision and matched physiologically relevant physical tests of RBC quality better than expert manual morphology assessment, while avoiding assessment subjectivity. It is important to note that the SMI failed to recognize the unusually high hemolysis levels of one blood sample (Fig. 4). The discrepancy between morphology and hemolysis in this instance, and as observed in prior studies (44, 45), is precisely the phenomenon that the field wishes to scrutinize in order to determine the underlying causative factors of this discrepancy; our methodology makes this easier to study. If the field conclusively determines that hemolysis, as measured here, is an ideal target metric for patient outcomes, then rare samples like this one would need to be collected and included in the training of SMI models.
We tested for overfitting, a common machine-learning problem that yields success on one set of data but failure on data from other facilities: Here, we obtained similar accuracy when the model was trained and tested across entirely different patient cohorts (Swiss vs. Canadian, whose samples were prepared completely independently on different continents and without knowledge of the others’ protocol and set-up). Robustness was further confirmed using samples from a third independent site. This generalizability is presumably because sample preparation and imaging for brightfield IFC have few variables and parameters. We anticipate that the system would likely benefit from retraining on a broader, consortium-scale collection of data, including multiple donor demographics, preparation procedures, and manufacturing facilities, as well as inclusion of samples that are hemolysis-sensitive. This would allow testing the power and limitations of the two new strategies, especially with respect to actual clinical transfusion outcomes or proxies agreed upon by the field as being relevant to clinical transfusion outcomes (42).
Such an effort would be worthwhile: Our proposed assay offers simple, label-free sample preparation, enabling nonexperts to assess the quality of stored blood. This is in contrast to microscopic examination (which requires experts and whose smearing step may damage the sample), conventional biochemical/biophysical assays (which require complex laboratory procedures), or IFC followed by manual gating (11, 12, 25) (which adds a step and is subjective). Although substantial engineering and testing would be needed, in principle the presented strategy could be adapted to an inexpensive laser-free imaging flow cytometer for resource-poor situations. Improved techniques to monitor blood product quality would revolutionize efforts to personalize allocation of blood products based on factors thought to impact RBC quality, including donor characteristics (age, sex, ethnicity, frequency of donation) (44, 46–50). Like many artificial-intelligence–driven analysis systems introduced in recent years, the goal need not be to entirely eliminate expert interaction but instead to screen samples or cell images to identify the most readily classifiable, so that the expert’s time is used on samples or cells that are more borderline.
More broadly, in this work we found that machine learning can surpass humans’ visual assessment of biomedically important morphological changes that occur over time. The weakly supervised approach discovered the natural progression of RBC deterioration without relying on human observations. In several applications, machine-learning–based systems have proven superior to humans but these have been straightforward supervised tasks (classification), including natural image classification (51), radiology (52), dermatology (53, 54), and pathology (55). Conversely, here machine learning itself reveals a clinically important chronological progression of cells based on their morphology, as has been previously done using other data types, most commonly mRNA levels (56–59), and also biomarker staining (60). Our weakly supervised strategy based on chronology might be applied to the morphological analysis of a variety of other noisy biological processes that occur over time, such as differentiation and metastasis.
Materials and Methods
Sample Preparation.
For the initial rounds of training, 18 red cell concentrate units were collected; 10 (bags A to J) at the Blood for Research Facility, Centre for Innovation, Canadian Blood Services, and 8 (bags CE44 to CE52) at the Transfusion Center of the University Hospital of Geneva, Switzerland. The Canadian Blood Services Research Ethics Board approved (Protocol #nc0058) the collection of the blood products used in this study that were obtained from volunteer, healthy blood donors, who provided written, informed consent. The utilization of blood samples from healthy donors for research was approved by the Ethical Committee of the University Hospital of Geneva. As this was general approval for the use of blood samples for nondiagnostic anonymized research signed by all donors, there is no specific ethical committee approval number. Written informed consent was received from participants, and samples were anonymized prior to inclusion in the study. Further details about sample protocols have been described in Pinto et al. (25) and in SI Appendix, Fig. S1.
An additional (third) dataset comprised of hemolytic and IFC measurements of 20 red cell units sampled at 5 storage durations (total 100 data points) were collected at the Blood for Research Facility (netCAD, Vancouver, Canada) and shipped to Canadian Blood Services in Edmonton, Alberta for testing (Fig. 4). The sample preparation protocol for IFC was similar to that of the other Canadian samples. Samples were then analyzed at the University of Alberta Faculty of Medicine and Dentistry Flow Cytometry Facility. One sample in this batch showed an elevated hemolysis levels compared to the others (Fig. 4A). It is unlikely that this was due to bacterial contamination as no common visual indicators of bacterial contamination were present and the hemolysis levels, although higher than the other samples, are still acceptable at day 42 (<0.8%). Additionally, a review of the IFC images themselves at two time points did not reveal any significant presence of bacteria. This outlier is therefore more likely caused by donor factors that make this unit more susceptible to hemolysis; this could not be confirmed as the sample was not available for further investigation.
IFC Data Acquisition and IDEAS Analysis.
For each sample, 5 μL of red cell concentrate were suspended in 200 μL of PBS (magnesium and calcium-free) in a 1.5-mL low-retention microfuge tube (Sigma T4816-250A). Samples were placed on an Amnis ImageStreamX Mark II (Amnis, EMD Millipore), five laser two-camera system (ASSIST calibrated) with a brightfield area lower limit of 50 m2 used to eliminate debris and speed beads. Channels 1, 9 (brightfield), and 12 (dark-field) were used to capture 100,000 brightfield/darkfield RBC images per sample using the low-speed/high-sensitivity settings at 60× magnification (0.9 numerical aperture, 0.33 per square pixel resolution, 40-μm field-of-view, 2.5-μm depth-of-field). The IFC measurements were repeated for each scheduled time point throughout the blood storage.
The instrument-associated analysis software IDEAS v6.2 was used to preliminarily process the acquired IFC data to remove out-of-focus cells, artifacts, debris, and clumped objects, as previously described (25–27). Images of in-focus single cells were then used for manual annotation and downstream deep-learning analysis. Brightfield and darkfield images were exported in .CIF or .TIF formats. Darkfield images were ignored for the final results shown in this study.
Ground Truth Annotation.
For the supervised machine learning procedure, each RBC was manually annotated by assigned human annotators, in consultation with an RBC morphology expert. Five annotators with different backgrounds (biologists, engineers, and a hematologist) were tasked to manually label allocated RBCs (see next section) as smooth disc, crenated disc, crenated discoid, crenated spheroid, crenated sphere, smooth sphere, side-view, and undecidable class. The undecidable category includes debris or cells that are blurry, blebbed, or folded, and typically represent artifacts of the testing process (SI Appendix, Fig. S3, bottom row; see also description in figure legend). Brightfield and darkfield images of annotated cells were then exported as .TIF.
Data Splitting and Validation Strategy.
The overall strategy is schematized in SI Appendix, Fig. S4.
Training.
Image data from replicate samples of bags A, B, D, E, F, H, CE47, CE49, CE50, and CE52 were pooled together. About 17,000 cells of that pooled dataset were annotated by three different annotators. Two annotators were tasked to annotate images from the same blood bags, but different individual cells from them; one annotated cells with an even object index and the other, cells with an odd object index. Finally, one additional annotator reviewed every cell individually and flagged dubious annotation mistakes for correction or removal.
Test 1.
A class-balanced set of ∼1,500 cells pooled from bags C, G, and I (SI Appendix, Fig. S3) were selected to test interobserver variation and labeling replicability between the five annotators; that is, each individual was tasked to label the exact same cells using an in-house web application (SI Appendix, Fig. S12).
Test 3 (morphology).
Image sets randomly sampled from (unpooled) bags CE47 and CE49 were used to test the robustness of the trained neural network on imbalanced data. During and after Tests 1 and 2, if suboptimal settings were detected, retraining of the supervised and weakly supervised models were allowed and optimization with improved parameters was implemented until the models were satisfactorily considered final. Once finalized, no further changes to the model weights were allowed and only a single inference was done on the hold-out test sets.
Tests 3 and 4 (physiology).
In particular, bags CE47, CE48, and CE49 have parallel data for both morphological (assayed by an IFC) and physiological (assayed by hemolysis test) assessments. Physiological readouts were used as a means to validate conclusions drawn by morphological findings.
Tests 2 and 4.
More than 20,000 annotated cells of bags C, G, I, J, CE44, CE45, CE48, and CE51 were kept held-out during the development and optimization of the machine learning algorithms. These data were unlocked only when all machine learning models were final. The prediction on this held-out data were computed a single time, immediately before the submission of the report for the final validation of the trained models.
Supervised Deep Learning.
Protocols for image preprocessing and deep-learning training of the supervised classification are similar to our previously established label-free imaging flow cytometry machine vision framework (61). In brief, the input images were contrast-stretched channel-wise and resized to 48 × 48 pixels by cropping or padding. To counter illumination variations in image inputs, the data were zero-centered using channel-wise mean subtraction and augmentation was implemented, such as random combinations of horizontal or vertical flips, horizontal, or vertical shifts (up to 50% of the image size), and rotations up to 180°. We implemented a ResNet50 architecture (62) (SI Appendix, Fig. S13), with categorical cross-entropy as the loss function and accuracy as the performance metric. The model was compiled using the Adam optimizer with a learning rate of 0.0001. The learning rate was reduced by a factor of 10 when the validation loss failed to improve for 10 consecutive epochs. The model was trained for a maximum of 512 epochs, although early stopping generally terminated training before 200 epochs when there is no improvement in the validation loss after 50 consecutive epochs, as detailed in Doan et al. (61). Training and validation data were randomly undersampled per blood unit across cell types to create a balanced dataset. Eighty percent of sampled data were assigned to the training dataset, with the remaining 20% assigned to internal validation of the model during its training. Prediction metrics included recall, precision, F1-score, and weighted accuracy.
Weakly Supervised Learning.
Regression model.
The architecture of the weakly supervised ResNet50 neural network is essentially similar to that of the supervised ResNet50, except for two modifications: 1) We removed the last seven-class (categorical) layer and replaced with a dense layer without activation function (for regression purpose instead of classification) and 2) we used “mean absolute error” as a loss function for the weakly supervised regression model, instead of “categorical cross-entropy” as in the supervised classification model.
The weakly supervised ResNet50 was trained to predict the age of storage time for each presented single-cell RBC image. In the last layer of this architecture, the duration of 49-d storage was regressed to a real number in a continuous range from −5 to 5. This range was adopted to introduce a contrast between short (negative) and long (positive) duration values, which facilitates learning-relevant morphology features. After the training phase, the intermediate and penultimate layers of the network, including Res4a_ReLU, Res5a_ReLU, and pool5 were benchmarked for the efficiency of feature extraction: the features extracted as each layer were used as inputs to train a support vector machine to classify 1,500 cells of bags C, G, and I into seven morphological categories; Res4a_ReLU was selected as the layer of choice given the best support vector machine classification reports. This layer was then used as a feature extractor to retrieve embeddings of cells from brightfield images. The direct outputs from the last layer (regression) were also tested for self-learned morphology trajectory recovery and MI (SI Appendix, Fig. S8 B and C).
Dimensionality reduction.
The set of features (1,024) extracted by the regression model was visualized using UMAP in three dimensions and two dimensions, which revealed that cells lay approximately on a 1D manifold. We explored methods to recover this manifold, including t-distributed stochastic neighbor embedding (t-SNE) (63), UMAP (34, 35), diffusion map (37), and diffusion pseudotime (38, 39) (SI Appendix, Fig. S14).
Ultimately, we used UMAP to map cell deep-learning embeddings onto a 1D distribution. The parameters used for generating the 1D UMAP (calculated independently of the 2D and 3D visualizations in Fig. 3) were as follows: 12 nearest neighbors were set to approximate the overall shape of the manifold using a Euclidean metric; effective minimum distance between embedded points was set at 0.1; a spectral embedding method was used to initialize UMAP embedding; 200 training epochs and a learning rate of 1.0 was used to optimize the embedding. The seed used by the random number generator was kept constant at 42 throughout the study. The distribution of cells along this unidirectional UMAP axis allowed the estimation of the cell degradation phenotype for the given blood unit. Based on the visual inspection of a subset of annotated data (merged bags A, B, D, E, F, H, CE50, and CE52), we categorized all RBCs below a manually selected threshold in the component space of the 1D UMAP as healthy, which when summed can exclude most spheroechinocytes (crenated spheroid, crenated spheres, and smooth spheres), thought to have negative attributes for blood transfusions (SI Appendix, Figs. S15 and S16; see legends for details about threshold selection). The fraction of unhealthy cells (x) over the total number of cells is termed SMI.
Physiological (Hemolysis) Assay.
For the data shown in Fig. 3 E–L, at time points day 0, day 14, and day 42, storage media was collected by performing double centrifugation at 2,000 × g for 10 min to remove RBCs. The supernatant was added to an equal volume of Drabkin’s solution (Sigma). Hemoglobin concentrations were determined spectrophotometrically at 540 nm. Hemolysis is determined as a percentage of lysed erythrocytes and was calculated based on an average total hemoglobin concentration of 181.6 g/L and an average hematocrit of 54% (n = 122 blood units) (SI Appendix, Table S1).
For the 20 units in the additional dataset (Fig. 4), hemolysis measurements were performed following the testing facility protocol as previously described (46), with the exception that the supernatant preparation (storage media collection) was performed as described above.
Conventional Image Analysis.
Images contained within .CIF files were stitched into montages by using a Python script. Cellular objects from the montages were identified (segmented) using CellProfiler 3.1.8 (41, 64). More than 600 object features were extracted by a series of built-in measurement modules, including measuring object intensity, size, shapes, textures, and correlations. Data cleaning and feature selection were performed by Cytominer (65) to remove features with near-zero variance and features that have poor correlation across replicates. Redundant features that are highly correlated were then identified and only one feature for each of these groups was retained. After pruning, 135 relevant cell features were retained, in which no pair of features had a correlation greater than the 95% cutoff threshold.
Code Availability.
The complete vignette of fully supervised and weakly supervised learning for red blood cell morphology analysis is disseminated in GitHub (73). The code for the web-based application for human annotation can be found in GitHub (74). We disseminated a more generalizable deep learning package, Deepometry (28). This open-source pipeline eases the analytic workflow for single-cell images, from handling raw images to operating the neural network ResNet50 architecture. This workflow was originally built for imaging flow cytometry data but can be readily adapted for microscopic images of isolated single objects. Unlike other deep-learning frameworks, which are limited to three-channel RGB images, our modification of ResNet50 allows researchers to use any number of stained or unstained channels. Deepometry embedding outputs can be viewed using public web-based visualization tools, such as Tensorflow projector (http://projector.tensorflow.org/) or Morpheus (https://clue.io/morpheus), for interactive inspection.
Supplementary Material
Acknowledgments
We thank the staff of the netCAD Blood for Research Facility, Centre for Innovation, Canadian Blood Services, Sophie Waldvogel, and all the staff at the Transfusion Center of the University Hospital of Geneva (Switzerland) for providing blood samples and quality-control data, and the generosity of the blood donors who made this research possible; T. C. Chang for consultations associated with validating the selection of images for the truth populations used for analysis, and for the development of the red blood cell gating and filtering template on the IDEAS software platform; The Lunenfeld Tanenbaum Research Institute flow cytometry facility for providing access for image flow cytometry experiments (supported through grants from the Canada Foundation For Innovation); M. H. Rohban for his expert consultations on developing fundamental concepts and critical elements of the machine-learning and deep-learning frameworks throughout the study; and Maren Buettner for critical feedback on the manuscript. Funding for this project was provided by US National Science Foundation/UK Biotechnology and Biological Sciences Research Council Joint Grant NSF DBI 1458626 and BB/N005163 (to A.E.C. and P.R.); Biotechnology and Biological Sciences Research Council Grant BB/P026818/1 (to P.R.); Natural Sciences and Engineering Research Council of Canada and the Canadian Institutes of Health Research, via a Collaborative Health Research Projects Grant 315271 “Characterization of blood storage lesions using photoacoustic technologies” (to M.C.K. and J.P.A.); and a grant administered by Carigest S.A. of Geneva, Switzerland (to A.R.). The Canadian Blood Services research program is funded by the federal (Health Canada), provincial, and territorial Ministries of Health. Experiments were performed at the University of Alberta Faculty of Medicine & Dentistry Flow Cytometry Facility, which receives financial support from the Faculty of Medicine & Dentistry and Canada Foundation for Innovation awards to contributing investigators. The views expressed herein do not represent the views of the Canadian federal government or any other funding agencies.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001227117/-/DCSupplemental.
Data Availability.
Annotated data of ∼67,400 cells (including undecidable class and held-out dataset) can be found in Figshare (66). Unannotated data for weakly supervised learning can be found in Figshare (67). The 3D-PCA, t-SNE, and UMAP visualization of supervised learning embeddings (penultimate layer, pool5) for 7,000 annotated RBCs are available in ref. 68; extracted features are available in Figshare (69). The 3D-PCA, t-SNE, and UMAP visualization of weakly supervised learning embeddings (intermediate layer, Res4a_ReLU) for 7,000 annotated RBCs are available in ref. 36; extracted features are available in Figshare (70). The 3D-PCA, t-SNE, and UMAP visualization of classic image features (extracted by CellProfiler) for 5,000 cells randomly selected from the pooled annotated Swiss test sets (33,467 RBCs) are available in ref. 71; extracted features are available in GitHub (72).
References
- 1.Holme S., Current issues related to the quality of stored RBCs. Transfus. Apheresis Sci. 33, 55–61 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Hess J. R., Red cell changes during storage. Transfus. Apheresis Sci. 43, 51–59 (2010). [DOI] [PubMed] [Google Scholar]
- 3.D’Alessandro A., Liumbruno G., Grazzini G., Zolla L., Red blood cell storage: The story so far. Blood Transfus. 8, 82–88 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D’Alessandro A., Liumbruno G. M., Red blood cell storage and clinical outcomes: New insights. Blood Transfus. 15, 101–103 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D’Alessandro A. et al., An update on red blood cell storage lesions, as gleaned through biochemistry and omics technologies. Transfusion 55, 205–219 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Sowemimo-Coker S. et al., Development of a statistical model for predicting in vivo viability of red blood cells: Importance of red cell membrane changes: SP28. Transfusion 55, 56A–57A (2015). [Google Scholar]
- 7.D’Alessandro A., Righetti P. G., Zolla L., The red blood cell proteome and interactome: An update. J. Proteome Res. 9, 144–163 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Cancelas J. A. et al., Additive solution-7 reduces the red blood cell cold storage lesion. Transfusion 55, 491–498 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Shih A. W. et al.; QMiP Investigators on behalf of the Biomedical Excellence for Safer Transfusion (BEST) Collaborative , Not all red cell concentrate units are equivalent: International survey of processing and in vitro quality data. Vox Sang. 114, 783–794 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Council of Europe , Guide to the Preparation, Use and Quality Assurance of Blood Components: Recommendation No. R (95) 15, (Council of Europe, 2007). [Google Scholar]
- 11.Roussel C. et al., Spherocytic shift of red blood cells during storage provides a quantitative whole cell-based marker of the storage lesion. Transfusion 57, 1007–1018 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Longster G. H., Buckley T., Sikorski J., Derrick Tovey L. A., Scanning electron microscope studies of red cell morphology. Changes occurring in red cell shape during storage and post transfusion. Vox Sang. 22, 161–170 (1972). [DOI] [PubMed] [Google Scholar]
- 13.Hess J. R., Solheim B. G., “Red blood cell metabolism, preservation, and oxygen delivery” in Principles of Transfusion Medicine, Simon T. L., McCullough J., Snyder E. L., Solheim B. G., Strauss R. G., Eds. (Wiley Online, 2016), pp. 97–109. [Google Scholar]
- 14.Steiner M. E. et al., Effects of red-cell storage duration on patients undergoing cardiac surgery. N. Engl. J. Med. 372, 1419–1429 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhabangi A. et al., Effect of transfusion of red blood cells with longer vs shorter storage duration on elevated blood lactate levels in children with severe anemia: The TOTAL randomized clinical trial. JAMA 314, 2514–2523 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Fergusson D. A. et al., Effect of fresh red blood cell transfusions on clinical outcomes in premature, very low-birth-weight infants: The ARIPI randomized trial. JAMA 308, 1443–1451 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Heddle N. M. et al., Effect of short-term vs. long-term blood storage on mortality after transfusion. N. Engl. J. Med. 375, 1937–1945 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Lacroix J. et al.; ABLE Investigators; Canadian Critical Care Trials Group , Age of transfused blood in critically ill adults. N. Engl. J. Med. 372, 1410–1418 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Usry R. T., Moore G. L., Manalo F. W., Morphology of stored, rejuvenated human erythrocytes. Vox Sang. 28, 176–183 (1975). [DOI] [PubMed] [Google Scholar]
- 20.Tchir J. D. R., Acker J. P., Holovati J. L., Rejuvenation of ATP during storage does not reverse effects of the hypothermic storage lesion. Transfusion 53, 3184–3191 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Carson J. L. et al.; Clinical Transfusion Medicine Committee of the AABB , Red blood cell transfusion: A clinical practice guideline from the AABB. Ann. Intern. Med. 157, 49–58 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Ching T. et al., Opportunities and obstacles for deep learning in biology and medicine. J. R. Soc. Interface 15, 20170387 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zuba-Surma E. K., Ratajczak M. Z., Analytical capabilities of the ImageStream cytometer. Nethods Cell Biol 102, 207–230 (2011). [DOI] [PubMed] [Google Scholar]
- 24.LeCun Y., Bengio Y., Hinton G., Deep learning. Nature 521, 436–444 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Pinto R. N., “Application of image flow cytometry for the characterization of red blood cell morphology” in High-Speed Biomedical Imaging and Spectroscopy: Toward Big Data Instrumentation and Management II, Tsia K. K., Goda K., Eds. (Proceedings of the SPIE, 2017), Vol. 10076. [Google Scholar]
- 26.Pinto R. N., “Application of Image flow cytometry and photoacoustics for the characterization of red blood cell storage lesions” Master’s thesis, Ryerson University, Toronto, ON, Canada (2017).
- 27.Pinto R. N. et al., Label-free analysis of red blood cell storage lesions using imaging flow cytometry. Cytometry A 95, 976–984 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Doan M., McQuin C., Goodman A., Deepometry: Image classification for imaging (flow) cytometry. https://github.com/broadinstitute/deepometry. Accessed 30 June 2018.
- 29.Baltruschat I. M., Nickisch H., Grass M., Knopp T., Saalbach A., Comparison of deep learning approaches for multi-label chest X-ray classification. Sci. Rep. 9, 6381 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oquab M., Bottou L., Laptev I., Sivic J., “Is object localization for free?-weakly-supervised learning with convolutional neural networks” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, (IEEE, 2015), pp. 685–694. [Google Scholar]
- 31.Joulin A., van der Maaten L., Jabri A., Vasilache N., Learning Visual Features from Large Weakly Supervised Data in Computer Vision – ECCV 2016, (Lecture Notes in Computer Science, Springer, Cham, 2016). [Google Scholar]
- 32.Gross S., Ranzato M., Szlam A., “Hard mixtures of experts for large scale weakly supervised vision” in 2017 IEEE Conference on Computer Vision and Pattern Recognition, (IEEE, 2017), pp. 5085–5093. [Google Scholar]
- 33.Caicedo J. C., McQuin C., Goodman A., Singh S., Carpenter A. E., “Weakly supervised learning of single-cell feature embeddings” in 2018 IEEE Conference on Computer Vision and Pattern Recognition, (IEEE, 2018), pp. 9309–9318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McInnes L., Healy J., Saul N., Großberger L., UMAP: Uniform manifold approximation and projection. J. Open Source Softw. 3, 861 (2018). [Google Scholar]
- 35.Becht E. et al., Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Doan M., Visualization of red blood cell weakly-supervised learning embeddings. TensorFlow. http://projector.tensorflow.org/?config=https://raw.githubusercontent.com/carpenterlab/2019_doan_pnas/master/DL_WeaklySupervised/Data/Step3/Output/Annotated/projector_config.pbtxt. Deposited 10 July 2020.
- 37.Coifman R. R. et al., Geometric diffusions as a tool for harmonic analysis and structure definition of data: Diffusion maps. Proc. Natl. Acad. Sci. U.S.A. 102, 7426–7431 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haghverdi L., Büttner M., Wolf F. A., Buettner F., Theis F. J., Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods 13, 845–848 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Wolf A. F., Angerer P., Theis F. J., Scanpy for analysis of large-scale single-cell gene expression data. bioRxiv:10.1101/174029 (9 August 2017).
- 40.Saelens W., Cannoodt R., Todorov H., Saeys Y., A comparison of single-cell trajectory inference methods. Nat. Biotechnol. 37, 547–554 (2019). [DOI] [PubMed] [Google Scholar]
- 41.McQuin C. et al., CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol. 16, e2005970 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vostal J. G. et al., Proceedings of the Food and Drug Administration’s public workshop on new red blood cell product regulatory science 2016. Transfusion 58, 255–266 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hess J. R., Measures of stored red blood cell quality. Vox Sang. 107, 1–9 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Jordan A. et al., Assessing the influence of component processing and donor characteristics on quality of red cell concentrates using quality control data. Vox Sang. 111, 8–15 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Tzounakas V. L. et al., Donor-specific individuality of red blood cell performance during storage is partly a function of serum uric acid levels. Transfusion 58, 34–40 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Acker J. P. et al., A quality monitoring program for red blood cell components: In vitro quality indicators before and after implementation of semiautomated processing. Transfusion 54, 2534–2543 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Heddle N. M. et al., The association between blood donor sex and age and transfusion recipient mortality: An exploratory analysis. Transfusion 59, 482–491 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Zeller M. P. et al., Sex-mismatched red blood cell transfusions and mortality: A systematic review and meta-analysis. Vox Sang. 114, 505–516 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Roubinian N. H. et al.; NHLBI Recipient Epidemiology and Donor Evaluation Study-III (REDS-III) , Association of donor age, body mass index, hemoglobin, and smoking status with in-hospital mortality and length of stay among red blood cell-transfused recipients. Transfusion 59, 3362–3370 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roubinian N. H. et al., Effect of donor, component, and recipient characteristics on hemoglobin increments following red blood cell transfusion. Blood 134, 1003–1013 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He K., Zhang X., Ren S., Sun J., Delving deep into rectifiers: Surpassing human-level performance on ImageNet classification. arXiv [cs.CV] (2015).
- 52.Rajpurkar P. et al., Deep learning for chest radiograph diagnosis: A retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med. 15, e1002686 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Esteva A. et al., Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haenssle H. A. et al.; Reader study level-I and level-II Groups , Man against machine: Diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann. Oncol. 29, 1836–1842 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Coudray N. et al., Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 24, 1559–1567 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Galen P. et al., Single-cell RNA-seq reveals AML hierarchies relevant to disease progression and immunity. Cell 176, 1265–1281.e24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jang J. S. et al., Molecular signatures of multiple myeloma progression through single cell RNA-Seq. Blood Cancer J. 9, 2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Young M. D. et al., Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science 361, 594–599 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vieira Braga F. A. et al., A cellular census of human lungs identifies novel cell states in health and in asthma. Nat. Med. 25, 1153–1163 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Damond N. et al., A map of human type 1 diabetes progression by imaging mass cytometry. Cell Metab. 29, 755–768.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Doan M. et al., Label-free leukemia monitoring by computer vision. Cytometry A 97, 407–414 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.He K., Zhang X., Ren S., Sun J., “Deep residual learning for image recognition” in 2016 IEEE Conference on Computer Vision and Pattern Recognition, (IEEE, 2016). [Google Scholar]
- 63.van der Maaten L., Hinton G., Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008). [Google Scholar]
- 64.Carpenter A. E. et al., CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singh S., et al. , Cytominer. https://github.com/cytomining/cytominer/. Accessed 9 May 2020.
- 66.Doan M., et al. , Annotated images of different phenotypes of red blood cells. Figshare. https://figshare.com/articles/URL7_Annotated_Data/12432506. Deposited 6 May 2020.
- 67.Doan M., et al. , Unannotated images of red blood cells. Figshare. https://figshare.com/articles/URL8_Unnotated_Data/12432959. Deposited 6 May 2020.
- 68.Doan M., et al. , Visualization of red blood cell supervised learning embeddings. TensorFlow. http://projector.tensorflow.org/?config=https://raw.githubusercontent.com/carpenterlab/2019_doan_pnas/master/DL_Supervised/Data/Step4/Output/projector_config.pbtxt. Deposited 10 July 2020.
- 69.Doan M., et al. , Supervised learning embeddings (penultimate layer, pool5) for 7,000 annotated RBCs. Figshare. https://figshare.com/articles/URL10_Supervised_Visualization/12433181. Deposited 6 May 2020.
- 70.Doan M., et al. , Weakly supervised learning embeddings (intermediate layer, Res4a_ReLU) for 7,000 annotated RBCs. Figshare. https://figshare.com/articles/URL11_WeaklySupervised_Visualization/12433226. Deposited 6 May 2020.
- 71.Doan M., et al. , Visualization of red blood cell conventional image features (extracted by CellProfiler). TensorFlow. http://projector.tensorflow.org/?config=https://raw.githubusercontent.com/carpenterlab/2019_doan_pnas/master/CellProfiler_Feature_extraction/Data/Step4/Output/projector_config.pbtxt. Deposited 10 July 2020.
- 72.Doan M., et al. , Conventional image features (extracted by CellProfiler) for 5,000 cells. Github. https://github.com/carpenterlab/2019_doan_pnas/tree/master/CellProfiler_Feature_extraction/Data/Step4/Output. Deposited 6 May 2020.
- 73.Doan M., Caicedo J., Singh S., Supervised classification and weakly supervised regression for Label-free assessment of red blood cell storage lesions. Github. https://github.com/carpenterlab/2019_doan_pnas. Deposited 10 July 2020.
- 74.Doan M., Caicedo J., Annotation tool for single-cell classification. Github. https://github.com/broadinstitute/single-cell-annotation. Deposited 29 March 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Annotated data of ∼67,400 cells (including undecidable class and held-out dataset) can be found in Figshare (66). Unannotated data for weakly supervised learning can be found in Figshare (67). The 3D-PCA, t-SNE, and UMAP visualization of supervised learning embeddings (penultimate layer, pool5) for 7,000 annotated RBCs are available in ref. 68; extracted features are available in Figshare (69). The 3D-PCA, t-SNE, and UMAP visualization of weakly supervised learning embeddings (intermediate layer, Res4a_ReLU) for 7,000 annotated RBCs are available in ref. 36; extracted features are available in Figshare (70). The 3D-PCA, t-SNE, and UMAP visualization of classic image features (extracted by CellProfiler) for 5,000 cells randomly selected from the pooled annotated Swiss test sets (33,467 RBCs) are available in ref. 71; extracted features are available in GitHub (72).