Significance
Here we report a synthetic lethal interaction between the epitranscriptomic modifier FTO and the tumor suppressor VHL. Notably, FTO inhibition reduced the growth of both HIF wild type and HIF-deficient tumors. These findings reveal an epitranscriptomic vulnerability of VHL-deficient cells and identify a potential HIF-independent therapeutic target for ccRCC tumors.
Keywords: synthetic lethality, FTO, kidney cancer, von Hippel–Lindau, SLC1A5
Abstract
Loss of the von Hippel–Lindau (VHL) tumor suppressor is a hallmark feature of renal clear cell carcinoma. VHL inactivation results in the constitutive activation of the hypoxia-inducible factors (HIFs) HIF-1 and HIF-2 and their downstream targets, including the proangiogenic factors VEGF and PDGF. However, antiangiogenic agents and HIF-2 inhibitors have limited efficacy in cancer therapy due to the development of resistance. Here we employed an innovative computational platform, Mining of Synthetic Lethals (MiSL), to identify synthetic lethal interactions with the loss of VHL through analysis of primary tumor genomic and transcriptomic data. Using this approach, we identified a synthetic lethal interaction between VHL and the m6A RNA demethylase FTO in renal cell carcinoma. MiSL identified FTO as a synthetic lethal partner of VHL because deletions of FTO are mutually exclusive with VHL loss in pan cancer datasets. Moreover, FTO expression is increased in VHL-deficient ccRCC tumors compared to normal adjacent tissue. Genetic inactivation of FTO using multiple orthogonal approaches revealed that FTO inhibition selectively reduces the growth and survival of VHL-deficient cells in vitro and in vivo. Notably, FTO inhibition reduced the survival of both HIF wild type and HIF-deficient tumors, identifying FTO as an HIF-independent vulnerability of VHL-deficient cancers. Integrated analysis of transcriptome-wide m6A-seq and mRNA-seq analysis identified the glutamine transporter SLC1A5 as an FTO target that promotes metabolic reprogramming and survival of VHL-deficient ccRCC cells. These findings identify FTO as a potential HIF-independent therapeutic target for the treatment of VHL-deficient renal cell carcinoma.
Kidney cancer remains a leading cause of cancer-related deaths worldwide. Clear cell renal cell carcinoma (ccRCC) is the most common form of kidney cancer, and it is estimated that only 11% of patients with a metastatic form of ccRCC will survive 5 years (1). Elucidating the molecular drivers of ccRCC progression and survival is critical for the development of effective therapies to target advanced-stage kidney cancer.
The von Hippel–Lindau (VHL) tumor suppressor is inactivated in a majority of ccRCC tumors (2, 3). VHL is the substrate recognition component of an E3 ubiquitin ligase complex containing the elongins B and C (4–6), Cullin-2 (6), and Rbx1 (7) that targets the hydroxylated, oxygen-sensitive α-subunits of HIFs (HIF-1, -2, and -3) for ubiquitination and degradation by the 26S proteasome (8, 9). VHL loss results in the constitutive activation of HIF targets, including the proangiogenic factors VEGF and PDGF (10). As a result, ccRCC tumors are highly vascularized and respond to antiangiogenic therapies. While antiangiogenic therapy has significantly increased progression-free survival in patients with metastatic renal cancer, the majority of patients treated with these agents eventually have disease progression (11, 12). New agents targeting HIF-2 have entered clinical trials for kidney cancer, and many of these patients ultimately have disease progression due to the development of resistance or bypass pathways (13–15). These findings underscore the need for additional therapeutic strategies for the treatment of ccRCC, particularly those that impair the growth and survival of renal cell carcinoma cells directly.
In the era of personalized medicine, synthetic lethality is an attractive strategy to identify vulnerabilities and drug targets for cancers with known genetic drivers that are difficult to directly target, such as tumor suppressor genes (16, 17). Synthetic lethality occurs when the combined mutation and/or inhibition of two genes results in cell death (18). Traditionally, candidate synthetic lethal (SL) partners have been identified through large-scale functional screens in cell lines using shRNA, CRISPR, or small molecule libraries. Using these approaches, a number of VHL SL interactions have been identified (19–24). More recently, computational methods have been utilized to predict SL partners of somatic mutations in human cancer using genomic data sets (25, 26). These approaches have advantages over cell line-based screens in that they enable genomewide analysis of SL partners for cancer-associated mutations directly within human tissue specimens.
In this report, we have utilized an innovative computational method, mining synthetic lethals (MiSL), to identify SL partners of VHL inactivation in ccRCC based on human ccRCC datasets. We identify fat mass- and obesity-associated protein (FTO), an N6-methyladenosine (m6A) RNA demethylase, as an HIF-independent SL partner of VHL in vitro and in vivo. Moreover, we identify the glutamine transporter SLC1A5 as an important functional FTO target in VHL-deficient ccRCC cells. These findings reveal mechanisms driving the growth and survival of VHL-deficient ccRCC and broaden the field for the development of a new class of inhibitors in kidney cancer therapy.
Results
MiSL Identifies FTO as an SL Partner of VHL Loss in Human ccRCC Specimens.
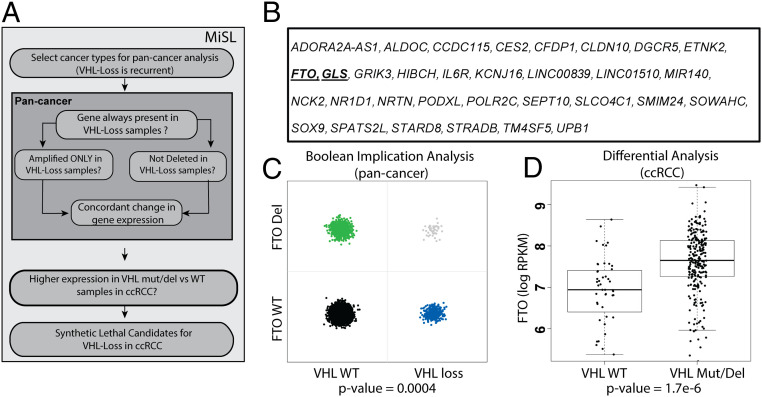
To identify molecular targets that exploit inherent vulnerabilities of VHL-deficient ccRCC, we utilized MiSL, a computational algorithm that analyzes large pan-cancer genomic and transcriptomic patient datasets to identify SL partners of cancer-specific genomic alterations (26). The underlying assumption of MiSL is that, across multiple cancers, SL partners of a genomic alteration will be amplified more frequently or deleted less frequently in primary tumor samples harboring the alteration (26). Here we applied MiSL to pan-cancer TCGA data to identify SL partners of VHL loss in multiple datasets (Fig. 1A). For our analysis, the VHL-loss feature was defined using VHL mutation and deletion data (more details provided in Methods). From the entire set of available TCGA tumor types, the algorithm identified 26 tumor types where VHL loss was recurrent. Samples from these tumor types were analyzed to identify genes that were either rarely deleted in the presence of VHL loss or amplified only in VHL-loss samples. Boolean implications (statistical “if/then” relationships) were used to efficiently extract the required relationships from genomic data. Specifically, we searched for two types of Boolean implications: (i) if VHL loss is present, then gene B is not deleted, which is a mutual exclusion relationship (a HILO Boolean implication); or (ii) if gene B is amplified, then VHL loss is present (a HIHI Boolean implication [27]). The Boolean implication analysis was restricted to gene deletions and amplifications that were unlikely to be passenger alterations. To maximize the discovery of SL genes for VHL-deficient ccRCC, the preliminary SL candidates were further filtered down to only include genes significantly differentially overexpressed in the presence of VHL mutations and/or deletions versus the wild type (WT) in ccRCC. This step eliminated genes unlikely to be essential in the context of VHL loss in ccRCC. Genes satisfying all of the above-mentioned filters became the candidate list of SL partners for VHL loss in ccRCC (Fig. 1B and Dataset S1).
Fig. 1.
Mining of synthetic lethals (MiSL) algorithm identifies FTO as a VHL synthetic lethal partner in human cancer tissues. (A) Overview of the MiSL analysis to identify VHL synthetic lethal partners in pan-cancer datasets (adapted from Sinha et al. [26]). (B) List of the top 32 hits for VHL synthetic lethal partners in ccRCC. (C) Boolean implication analysis for FTO deletions and VHL loss in patient samples across multiple TCGA tumor types. Each point is a tumor sample. The x-axis indicates if a sample has VHL loss or not, and the y-axis indicates if a sample has FTO deletions or not. A small amount of noise has been added to the points to enable better visualization. The sparseness of the top right quadrant indicates much fewer samples with combined loss than expected by chance (P value of mutual exclusion = 0.0004). (D) FTO mRNA expression in the TCGA ccRCC patient samples with VHL mutations and/or deletions versus VHL wild type samples (P value of differential expression = 1.7e-06).
Among the top VHL SL candidates, MiSL identified a previously validated VHL synthetic lethal pathway such as the glutaminase (GLS) pathway. Our computational analysis found that GLS is selectively amplified only in the presence of the VHL mutation (Fig. 1B and Dataset S1). Importantly, GLS inhibitors are currently in clinical trials for the treatment of kidney cancer, demonstrating that MiSL can identify established and clinically relevant targets for ccRCC (28). We searched for novel druggable targets among the top candidates and identified the N6-methyladenosine (m6A) demethylase fat-mass and obesity-associated protein (FTO) as a potential druggable SL candidate for VHL loss in ccRCC (Fig. 1B and Dataset S1). MiSL identified FTO as a potential SL partner of VHL because FTO deletions are mutually exclusive with VHL loss in pan-cancer analysis across all applicable TCGA tumors (HILO Boolean implication; P = 0.0004; Fig. 1C). Additionally, analysis of the ccRCC TCGA patient samples revealed that tumor specimens with VHL deletion or mutation expressed increased levels of FTO mRNA compared to VHL wild type tumors (Fig. 1D and SI Appendix, Fig. S1). It should be noted that, since FTO deletions are rare in ccRCC samples, this potential SL relationship would have been missed in a ccRCC-only analysis. FTO belongs to the AlkB family of Fe(II)- and 2-oxoglutarate–dependent N6-methyladenosine RNA demethylases to regulate pre-mRNA splicing and mRNA translation, degradation, and nuclear export (29, 30). FTO has recently been shown to play an oncogenic role in leukemia, melanoma, and breast cancers (31–34). However, the role of FTO in kidney cancer remains unknown.
FTO Is Highly Expressed in ccRCC Compared to Normal Kidney Tissue.
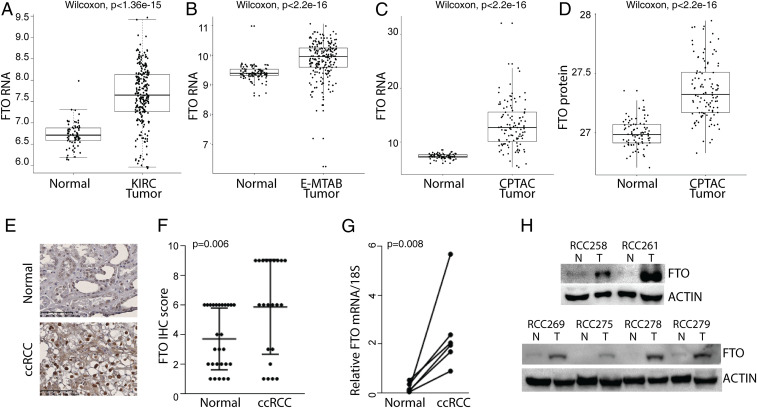
Having identified FTO as a potential SL partner of VHL-deficient ccRCC using MiSL, we sought to further examine FTO expression in ccRCC and normal adjacent tissue. Analysis of the 538 ccRCC samples and 72 normal kidney samples from TCGA revealed that FTO expression is significantly increased in ccRCC tumors with VHL deletions or mutations compared to normal kidney tissue (Fig. 2A and SI Appendix, Fig. S2A). Analysis of the E-MTAB-6692 dataset (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-6692/), a metadataset comprising a total of 347 samples including both primary tumors and tumor-free renal tissues from six independent GEO datasets, further confirmed FTO is overexpressed in ccRCC compared to normal renal tissues (P < 2.2e-16; Fig. 2B). Additionally, analysis of transcriptomic and proteomic data of treatment-naive ccRCC and paired normal tissues in the Clinical Proteomic Tumor Analysis Consortium (CPTAC) containing 110 treatment-naive ccRCC tumors and 84 matched normal samples revealed that FTO is overexpressed at both the transcriptomic and protein levels in tumor tissues relative to normal tissues (Fig. 2 C and D and ref. 35). Immunohistochemical analysis of FTO expression in a ccRCC tissue microarray containing 30 pairs of ccRCC and adjacent tissue further confirmed that FTO protein levels were increased in ccRCC tumor tissue compared to normal adjacent tumor tissue (Fig. 2 E and F). Finally, we obtained six pairs of ccRCC tumors and adjacent tissue to analyze FTO expression at the mRNA and protein level. In all six pairs, FTO expression was increased in ccRCC tumors compared to normal adjacent tissue (Fig. 2 G and H). These data confirm that FTO expression is increased in ccRCC relative to normal kidney tissue. It should be noted that FTO mRNA expression levels are not significantly altered across VHL-deficient ccRCC tumor stages I–IV (SI Appendix, Fig. S2B). Taken together, these data demonstrate that FTO is highly expressed in ccRCC tumors.
Fig. 2.
FTO expression is increased in ccRCC compared to normal tissue. (A) FTO mRNA expression in 538 ccRCC patient samples as compared to 72 normal kidney samples from TCGA data (P = 1.36e-15). (B) FTO mRNA expression in ccRCC tumors versus normal tissues in the E-MTAB-6692 dataset (P < 2.2e-16). E-MTAB-6692 is a metadataset comprising 347 samples including both primary tumors and tumor-free renal tissues from six independent GEO datasets. To minimize interplatform variation, only datasets generated from the GPL570 platform (Affymetrix Human Genome U133 Plus 2.0 Array) were processed to develop the metadataset. Each dataset was preprocessed with robust multichip average (RMA) normalization, merged, and batch effect-corrected via Combat method. (C and D) FTO mRNA (C) and protein (D) expression in ccRCC patient samples versus matched normal tissues in CTPAC dataset (P < 2.2e-16). (E) Immunohistochemical analysis of FTO in a ccRCC tissue microarray (TMA) containing 30 ccRCC tumor and adjacent normal tissue pairs. (F) Quantification of FTO expression by immunohistochemical analysis in ccRCC TMA samples as compared to adjacent normal tissue. For scoring the percentage of cells with positive FTO staining, the percentage of cells with the core were evaluated: no positive staining = 0, 1–25% = 1, 25–50% = 2, 50–100% = 3. The staining intensity of cells with positive FTO staining were evaluated: no staining = 0, weak staining = 1, moderate staining = 2, strong intense staining = 3. The two scores were multiplied to result in an overall score ranging from 0 to 9. (G and H) Real-time PCR (G) and Western blot (H) analysis of FTO expression in ccRCC tumor and adjacent normal tissues.
FTO Inhibition Selectively Reduces the Growth and Survival of VHL-Deficient ccRCC Cells.
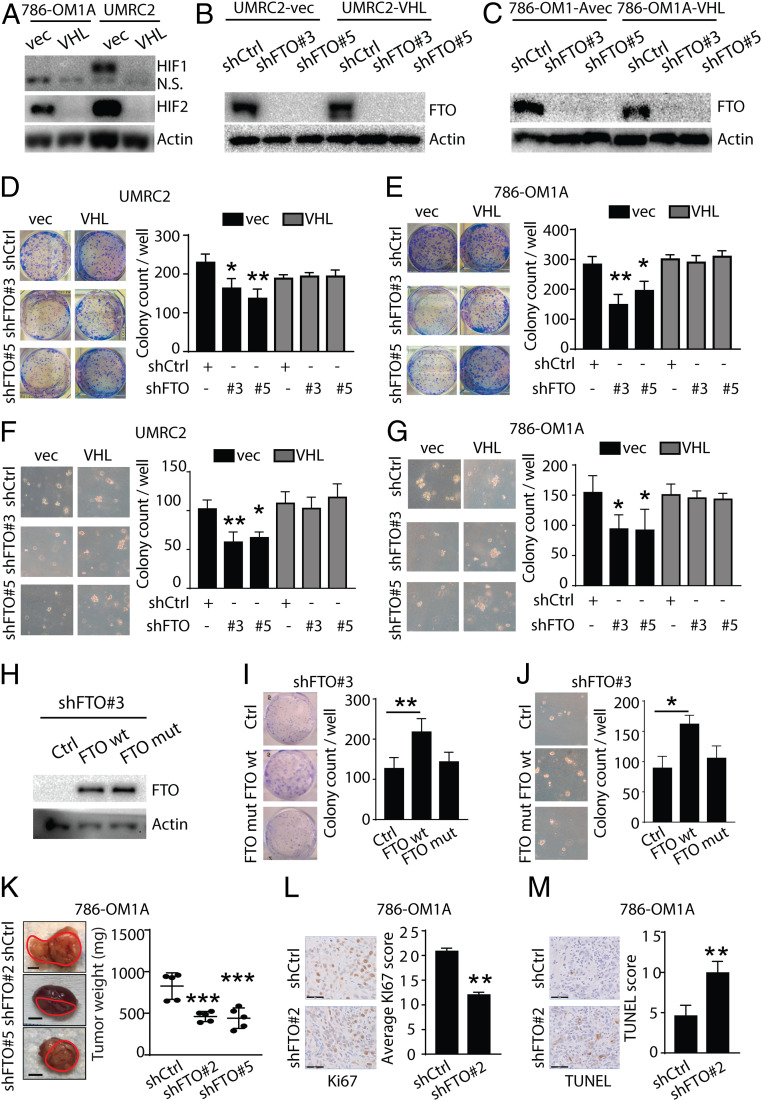
To investigate the functional role of FTO in VHL wild type and VHL-deficient ccRCC tumor growth and survival, we performed loss-of-function studies with a genetic approach to knock down FTO expression in ccRCC isogenic cell lines that are VHL-deficient (786-OM1A-vec and UMRC2-vec) or VHL-reconstituted (786-OM1A-VHL, UMRC2-VHL). Consistent with the role of VHL in targeting HIF-1 and HIF-2 for proteasomal degradation, VHL expression resulted in loss of HIF-1 and HIF-2 expression in UMRC2-VHL cells and loss of HIF-2 in 786-OM1A-VHL cells (Fig. 3A and refs. 8 and 9). FTO protein levels did not change with VHL reconstitution (Fig. 3 B and C). Knockdown of FTO expression in the UMRC2 and 786-OM1A isogenic cell lines using two independent shRNA hairpins was verified at the protein level (Fig. 3 B and C). Because it was difficult to maintain efficient knockdown of FTO expression after selection, we determined the growth and survival of VHL-deficient ccRCC cells at early time points after viral infection with the FTO shRNA hairpins. Using cell viability and 2D clonogenic assays, we observed that the growth and survival of VHL-deficient UMRC2-vec and 786-OM1A-vec cells was reduced in FTO knockdown cells compared to the control cells (Fig. 3 D and E and SI Appendix, Fig. S3 A–C). In addition, 3D soft agar assays confirmed that FTO knockdown results in decreased anchorage-independent growth of VHL-deficient cells (Fig. 3 F and G). In contrast, FTO knockdown did not affect the growth and survival of UMRC2-VHL or 786-OM1A-VHL reconstituted cells (Fig. 3 D–G and SI Appendix, Fig. S3 A–C). To confirm that the reduction in VHL-deficient ccRCC growth and survival was due to on-target effects of FTO shRNA, we restored FTO expression in shFTO#3 (located within the 3′UTR region of the gene) UMRC2-vec cells and found that FTO expression enhanced the growth and survival of these cells, demonstrating on-target effects of the shFTO#3 construct (Fig. 3 H–J). Moreover, to determine if VHL-deficient ccRCC growth and survival is dependent upon its enzymatic activity, we expressed a FTO demethylase mutant (carrying two point mutations, H231A and D233A, which disrupt enzymatic activity [36]) in shFTO#3 cells (Fig. 3H). In contrast to the FTO-wild type construct, the FTO enzymatic mutant construct did not enhance the growth and survival of FTO-knockdown ccRCC cells (Fig. 3 I and J). In addition, treatment with meclofenamic acid, a small molecule that inhibits FTO demethylase activity (37), reduced the growth and survival of 786-OM1A-vec cells compared to 786-OM1A-VHL cells (SI Appendix, Fig. S3 D and E). Meclofenamic acid also reduced the growth and survival of 786-0M1A-VHL cells compared to vehicle-treated cells, suggesting that there are off-target effects of this drug in these cells (SI Appendix, Fig. S3 D and E). Together, these data indicate that FTO enzymatic activity is important for FTO-mediated growth and survival of VHL-deficient ccRCC cells.
Fig. 3.
FTO inhibition reduces VHL-deficient ccRCC growth and survival. (A) Western blot analysis of HIF1 and HIF2 expression in UMRC2 and 786-OM1A cells transfected with empty vector and VHL. Actin was used as loading control. (B and C) Western blot analysis of FTO expression in UMRC2-vec or UMRC2-VHL (B) and 786-OM1A-vec or 786-OM1A-VHL (C) cells transfected with shControl, shFTO#3, and shFTO#5. Actin was used as loading control. (D–G) Picture (Left) and quantification (Right) of 2D colonies (D and E) and 3D soft agar colonies (F and G) in UMRC2-vec or UMRC2-VHL (D and F) and 786-OM1A-vec or 786-OM1A-VHL (E and G) cells transfected with shControl, shFTO#3, and shFTO#5. Data represent the average ± SD (*P < 0.05 and **P < 0.01 for indicated group vs. shCtrl). (H) Western blot analysis of FTO expression in UMRC2-vec shFTO#3 cells transfected with control, FTO wild type (FTO wt), or FTO demethylase mutant (FTO mut) expression constructs. (I and J) Picture (Left) and quantification (Right) of 2D colonies (I) and 3D soft agar colonies (J) in UMRC2-vec-shFTO#3 cells transfected with control, wild type FTO (FTO wt), or mutant FTO (FTO mut). Data represent the average ± SD (*P < 0.05 and **P < 0.01 for indicated group vs. control). (K) Macroscopic picture (Left) and total tumor weight (Right; n = 5 for shCtrl and n = 6 for shFTO#2 and shFTO#5) of primary tumors taken from mouse orthotopic subrenal capsule injection of shCtrl, shFTO#2, and shFTO#5 786-OM1A cells. Data represent the average ± SD (***P < 0.001 for indicated group vs. shCtrl). (L and M) IHC (Left) staining of Ki67 (L) and TUNEL (M) and quantification (Right) of primary tumors taken from mouse orthotopic subrenal capsule injection of shCtrl or shFTO#2 786-OM1A cells (three mice, five fields per tumor). Data represent the average ± SD (**P < 0.01).
To further confirm the role of FTO inhibition in VHL-deficient ccRCC growth and survival, we utilized an siRNA approach to knock down FTO expression. We observed a similar reduction in the growth and survival of VHL-deficient, but not VHL-reconstituted, UMRC2 and 786-OM1A cells treated with siFTO smart pools compared to cells treated with nontargeting siRNA smart pools (SI Appendix, Fig. S3 F–K). Finally, we utilized a CRISPR-Cas9 approach to deplete FTO in UMRC2 ccRCC cells with two independent sgRNAs (SI Appendix, Fig. S3 L and M). CRISPR-Cas9–mediated FTO depletion resulted in a significant reduction in colony formation in VHL-deficient UMRC2 cells (SI Appendix, Fig. S3M).
We next investigated the functional role of FTO in ccRCC tumor growth in vivo. We performed orthotopic ccRCC tumor xenograft studies in FTO control and knockdown 786-OM1A cells (SI Appendix, Fig. S3N). Primary ccRCC tumor growth under the renal capsule was reduced in mice injected with FTO-knockdown tumor cells compared to mice injected with FTO wild type shControl tumor cells (Fig. 3K). We investigated FTO knockdown efficiency in the tumors that did grow from the shFTO cells lines by immunohistochemistry and observed that FTO expression was present in some of the tumor cells, suggesting that, in a polyclonal population, cells that expressed FTO were selected to grow in vivo (SI Appendix, Fig. S3O). The reduction in tumor growth of FTO-knockdown cells was associated with reduced proliferation (Ki67) and increased apoptosis (TUNEL) compared to control tumors (Fig. 3 L and M). Together these data demonstrate that FTO knockdown selectively reduces the growth and survival of VHL-deficient ccRCC cells.
FTO Synthetic Lethality in VHL-Deficient ccRCC Cells Is Independent of HIF-1 and HIF-2.
VHL plays a central role in the regulation of the hypoxic signaling pathway. VHL loss results in the constitutive activation of the hypoxia-inducible factors HIF-1 and HIF-2, where HIF functions as an oncogenic driver of tumor progression (14, 38). However, in addition to regulating HIF, VHL has been reported to regulate additional substrates including Jade-1, p53, and ZHX2 (39–42). Therefore, FTO synthetic lethality in VHL-deficient cells may occur in an HIF-dependent or HIF-independent manner.
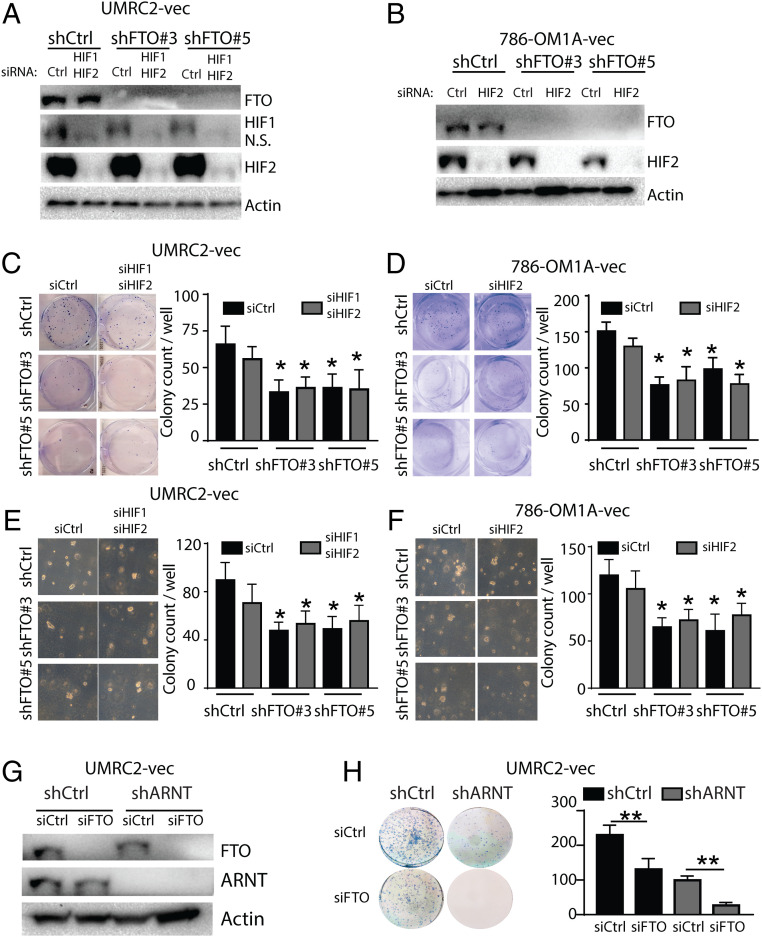
To determine whether the synthetic lethal interaction between FTO inhibition and VHL inactivation in VHL-deficient ccRCC cells is HIF-dependent, we first utilized an siRNA approach to knock down HIF-1 and HIF-2 in control or FTO-knockdown UMRC-2-vec or 786-OM1A-vec cells (Fig. 4A). Small hairpin RNA (shRNA)-mediated FTO knockdown reduced 2D colony survival and anchorage-independent growth in both HIF-1 and HIF-2 wild type and knockdown cells (Fig. 4 C–F). We observed a similar reduction in UMRC2-vec and 786-OM1A-vec growth and survival when FTO was knocked down by FTO small interfering RNAs (siRNAs; SI Appendix, Fig. S4 A–J). As previously reported, we found that short-term knockdown of HIF in VHL-deficient ccRCC cells did not significantly reduce cell growth or survival under high serum conditions (Fig. 4 C–F and refs. 3 and 38). To investigate whether FTO inhibition reduces the cell growth and survival of ccRCC cells with stable HIF knockdown, we generated UMRC2-vec cells with shControl or shARNT hairpins. ARNT is the common binding partner for HIF-1, and HIF-2 and ARNT knockdown reduces HIF-1 and HIF-2 transcriptional activity (43, 44). Knockdown of ARNT did not change FTO expression (Fig. 4G and SI Appendix, Fig. S4K). In these stable cultures, ARNT knockdown resulted in reduced colony growth and survival compared to shControl cells (Fig. 4H and SI Appendix, Fig. S4L). Importantly, FTO knockdown reduced the growth and survival of both shControl and shARNT UMRC2-vec cells (Fig. 4H and SI Appendix, Fig. S4L). Together, these data suggest that FTO inhibits the growth and survival of VHL-deficient cells in an HIF-independent manner.
Fig. 4.
FTO regulates VHL-deficient ccRCC cell growth and survival in an HIF-independent manner. (A and B) Western blot analysis of FTO, HIF1, and HIF2 expression in UMRC2-vec (A) and 786-OM1A-vec (B) cells transfected with indicated shRNA and siRNA. Actin was used as loading control. (C–F) Picture (Left) and quantification (Right) of 2D colonies (C and D) and 3D soft agar colonies (E and F) in UMRC2-vec (C and E) and 786-OM1A-vec (D and F) cells transfected with indicated shRNA and siRNA. (G) Western blot analysis of FTO and ARNT expression in shCtrl and shARNT UMRC2 cells transfected with siControl and siFTO. Actin was used as loading control. (H) Picture (Left) and quantification (Right) of 2D colonies in shCtrl and shARNT UMRC2 cells transfected with siControl and siFTO. Data represent the average ± SD. Student t test against corresponding control conditions (P < 0.05 and **P < 0.01).
Transcriptome-Wide m6A-Seq and mRNA-Seq Analyses Identify Potential FTO Targets in ccRCC.
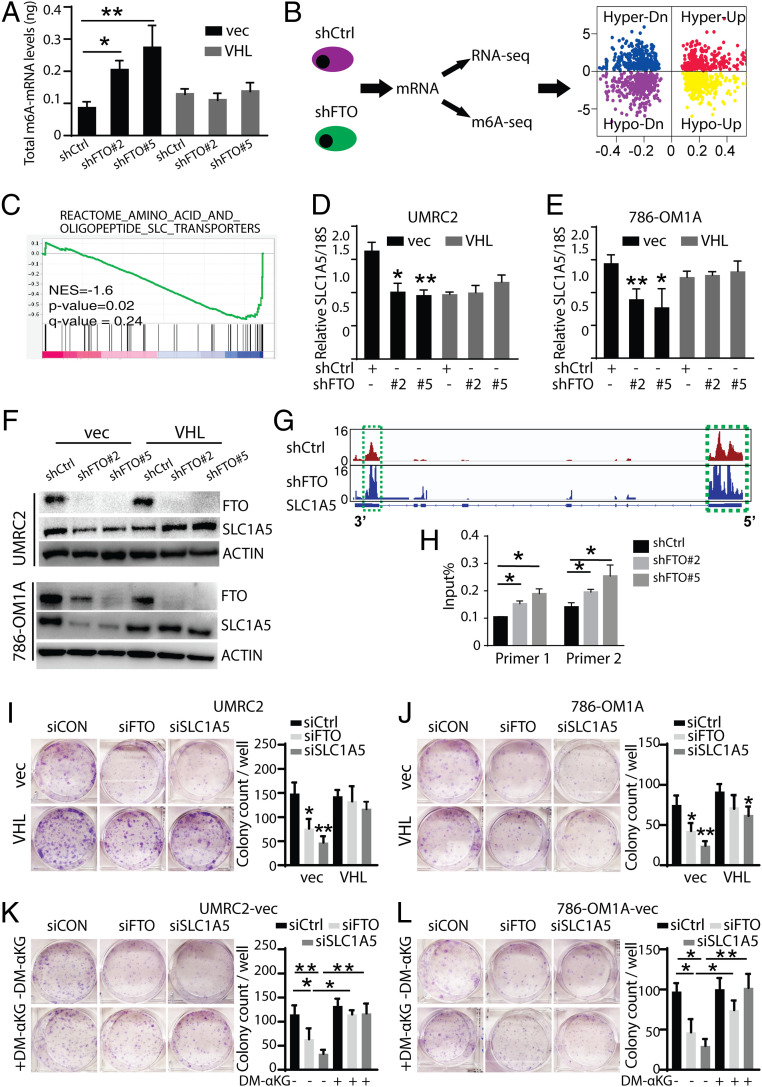
To identify functional FTO targets in VHL-deficient ccRCC cells, we performed transcriptome-wide m6A sequencing and RNA sequencing analysis of two independent FTO-knockdown and control ccRCC cell lines. A complete list of differentially expressed genes based on RNA-seq analysis of shFTO and shControl UMRC2-vec cells is provided in Dataset S2 (45). FTO knockdown in VHL-deficient ccRCC cells resulted in a global increase in m6A methylation levels (Fig. 5A and SI Appendix, Fig. S5 A and B). Interestingly, global m6A methylation levels were not changed in FTO-deficient VHL-wild type cells (Fig. 5A and SI Appendix, Fig. S5C). Analysis using exomePeak (46) identified a total of 2,680 and 4,049 m6A peaks showing a significant (P < 0.05) increase and decrease in abundance (normalized to input) in shFTO cells relative to shControl cells, and they were termed hyper- and hypomethylated m6A peaks, respectively (Fig. 5B and SI Appendix, Fig. S5D; differential analysis for the complete set of peaks identified by exomePeak are in Dataset S3 (45). Since FTO is a demethylase, we further focused our analysis on hypermethylated genes. Integrative analysis of the m6A-seq and RNA-seq data identified 652 hypermethylated m6A peaks where the RNA transcripts were significantly down-regulated (n = 340; hyper-down) or up-regulated (n = 312; hyper-up) in shFTO cells relative to shControl cells (complete list in Dataset S4, ref. 45). Gene set enrichment analysis (GSEA) using the molecular signature database (MSigDB) revealed that genes down-regulated in shFTO vs. shControl cells were significantly enriched in pathways related to amino acid and peptide SLC transport and glucose transport (Fig. 5C and SI Appendix, Fig. S5E). It is known that VHL-deficient ccRCC cells undergo metabolic reprogramming that involves the activation of glycolysis and lactate production, making them dependent upon exogenous glutamine and glutamine metabolism for the synthesis of fatty acids, amino acids, and nucleosides, as well as for the generation of glutathione and redox balance (28, 47). Interestingly, among the genes identified in the amino acid and peptide SLC transport pathway, the glutamine transporter SLC1A5 is hypermethylated and down-regulated in FTO-knockdown ccRCC cells (Dataset S4). High SLC1A5 expression is associated with tumor progression and reduced overall survival in ccRCC patients (48). We further confirmed that SLC1A5 mRNA and protein levels were decreased upon FTO knockdown in VHL-deficient UMRC2-vec and 786-OM1A-vec cells (Fig. 5 D–F). The m6A-sequencing data indicated that FTO targets the 5′ UTR and 3′ UTR of the SLC1A5 transcript, as FTO inhibition results in an increase in m6A methylation within these regions (Fig. 5G). We validated site-specific increases in SLC1A5 m6A methylation within the 3′ UTR region by m6A-qRT-PCR analysis (Fig. 5H). We next investigated SLC1A5 function in ccRCC cells. SLC1A5 knockdown recapitulated the selective decrease in colony survival caused by FTO knockdown in UMRC2-vec and 786-OM1A-vec cells compared to UMRC2-VHL and 786-OM1A-VHL cells (Fig. 5 I and J). Previous studies have shown that exogenous dimethyl-αKG (DM-αKG) treatment, which converts to glutamate and glutamine inside the cell, protects VHL-deficient ccRCC cells from glutaminase I (GLS1) inhibitor-mediated growth suppression (47). Therefore, we investigated whether DM-αKG treatment could protect VHL-deficient ccRCC cells from growth suppression mediated by SLC1A5 or FTO knockdown. Exogenous DM-αKG treatment restored the survival of UMRC2-vec and 786-OM1A-vec cells with SLC1A5 and FTO knockdown, indicating that FTO inhibition may reduce intracellular glutamine uptake needed for ccRCC growth and survival (Fig. 5 K and L). Therefore, we next examined whether FTO knockdown alters glutamine consumption in VHL-deficient ccRCC cells. FTO inhibition reduced glutamine consumption rates in UMRC2-vec cells compared to UMRC2-vec control cells (SI Appendix, Fig. S5G). These data indicate that SLC1A5 is a functionally important FTO target in VHL-deficient ccRCC cells and suggest that FTO may play a role in the metabolic reprogramming of VHL-deficient ccRCC cells.
Fig. 5.
Integration of transcriptome-wide m6A-seq and mRNA-seq assays identify SLC1A5 as an FTO target in ccRCC. (A) Global m6A changes of UMRC2-vec and UMRC2-VHL cell lines by m6A mRNA-ELISA. (B) Workflow for RNA sequencing and m6A sequencing of UMRC2-vec transfected with shControl, shFTO#2, and shFTO#5. The quadrant plot indicates m6A levels and mRNA transcript abundance levels. The x-axis refers to log2 fold change in mRNA transcript abundance levels between shFTO and control cells. The y-axis indicates log2 fold change of m6A levels between shFTO and control cells. The different quadrants represent (I) hypermethylated and up-regulated genes (red), (II) hypermethylated and down-regulated genes (blue), (III) hypomethylated and up-regulated genes (yellow), and (IV) hypomethylated and down-regulated genes (purple). (C) Gene set enrichment analysis (GSEA) for the SLC transporter gene set from MSigDB. Genes were ranked based on the degree of differential expression between shFTO and shControl cells. A negative enrichment score indicates that genes belonging to these gene sets were significantly down-regulated in shFTO compared to shControl cells. (D and E) Real-time PCR analysis of SLC1A5 expression in shControl or shFTO UMRC2 (D) and 786-OM1A (E) cells. Data represent the average ± SD (*P < 0.05 and **P < 0.01 for indicated group vs. shCtrl). (F) Western blot analysis of FTO and SLC1A5 expression in UMRC2 (Top) and 786-OM1A (Bottom) shControl or shFTO cells. Data represent the average ± SD (*P < 0.05 and **P < 0.01). (G) The m6A abundances in SLC1A5 transcripts in shFTO and shControl UMRC2 cells as determined by m6A-seq. (H) Gene-specific m6A qPCR analysis of m6A level in SLC1A5 mRNA in shFTO and shControl UMRC2 cells. Primers 1 and 2 are targeting the 3′ UTR region of SLC1A5. Data represent the average ± SD (*P < 0.05 for indicated group vs. shCtrl). (I and J) Picture (Left) and quantification (Right) of 2D colonies in UMRC2-vec and UMRC2-VHL (L) or 786-OM1A-vec and 786-OM1A-VHL (J) cells transfected with siCtrl, siFTO, and siSLC1A5. (K and L) Picture (Left) and quantification (Right) of 2D colonies in UMRC2-vec (K) and 786-OM1A-vec (L) cells transfected with siCtrl, siFTO, and siSLC1A5 and cultured with or without dimethyl α-ketoglutarate (DM-αKG). Data represent the average ± SD. Student t test against corresponding control conditions (**P < 0.05 and **P < 0.01).
Discussion
RNA methylation and its regulatory machinery are rapidly emerging as new therapeutic targets that influence gene expression and protein translation in cancer. N6-methyladenosine (m6A) is the most common internal mRNA modification. In 2011, FTO was discovered as the first m6A demethylase. It was further demonstrated that m6A is a reversible and dynamic RNA modification that can regulate gene expression and biological processes similar to DNA and histone modifications (36). Elucidating the roles of epitranscriptomic modifiers in development and disease is an active area of investigation. Using an innovative computational platform that mines primary tumor genomic and transcriptomic data, we discovered a synthetic lethal interaction between the m6A demethylase FTO and the tumor suppressor VHL in renal cell carcinoma cells.
We demonstrate that genetic inhibition of FTO selectively reduces the growth and survival of VHL-deficient renal cell carcinoma cells. Integrated analysis of m6A methylation and RNA sequencing revealed that FTO knockdown increases m6A methylation and decreases the expression of the glutamine transporter SLC1A5. SLC1A5 is the primary glutamine transporter in cancer cells and plays an important role in maintaining the growth and survival of glutamine-dependent cancers (49–51). Indeed, SLC1A5 is emerging as an important therapeutic target for glutamine-dependent cancers (52). Previous studies have shown that VHL-deficient ccRCC cells are dependent upon glutamine to support tumor growth and survival (28, 47). As a result, GLS1 inhibitors that target mitochondrial glutaminase and the conversion of glutamine to glutamate are currently being evaluated in early-phase clinical trials in ccRCC (CB-839; Calithera Biosciences). Our study indicates that high SLC1A5 expression, mediated at least in part by FTO, contributes to ccRCC growth and survival. Future studies are needed to determine the oncogenic role and therapeutic potential of targeting SLC1A5 in ccRCC.
We define a mechanism by which FTO inhibition reduces VHL-deficient renal cell carcinoma viability, growth, and survival in an HIF-independent manner. It is well established that VHL plays a central role in the regulation of the adaptive hypoxic signaling pathway through the regulation of the hypoxia-inducible transcription factors HIF-1 and HIF-2 (53). This is supported by a key oncogenic role of HIF-2 in renal cell carcinoma (14, 38). However, there are a growing number of HIF-independent VHL substrates and activities. For example, the zinc fingers and homeoboxes 2 protein (ZHX2) was recently identified as a novel VHL substrate that promotes VHL-deficient tumor growth and survival associated with the activation of NF-κB signaling (42). The TANK binding kinase I (TBK1) was also recently identified as a novel VHL substrate that promotes VHL-deficient ccRCC tumorigenesis by enhancing p62 stability and ccRCC proliferation (54). Additionally, CDK4/6 inhibitors have recently been identified as HIF-independent synthetic lethal partners in VHL-deficient renal cell carcinoma cells (24). Our studies identify an epitranscriptomic dependency of VHL-deficient cells mediated by FTO-independent of HIF activity. Future studies are needed to further explore how FTO inhibition regulates HIF-independent activities in VHL-deficient cells.
The preclinical studies described in this study indicate an important role for FTO in ccRCC tumor growth and survival. These findings raise the intriguing possibility that therapeutic targeting of FTO may be an effective anticancer strategy in ccRCC. As an Fe2+ and 2-oxoglutarate (2OG)-dependent AlkB dioxygenase that targets m6A RNA substrates, there are several classes of FTO inhibitors that have been identified, including those that compete for FTO binding to m6A substrates or its substrate 2OG (32, 37, 55–58). In this study, we investigated the efficacy of meclofenamic acid, an FTO inhibitor that competes with FTO binding to m6A substrates (37), in VHL-deficient and reconstituted ccRCC cells. While meclofenamic acid reduced the growth and survival of 786-OM1A-vec cells compared to 786-OM1A-VHL cells, it also reduced the growth and survival of 786-0M1A-VHL cells compared to vehicle-treated cells, suggesting that there are off-target effects of meclofenamic acid in ccRCC cells. While current inhibitors serve as important tool compounds to study FTO activity, the activity of these drugs is still limited due to off-target effects and poor pharmacodynamics. Next-generation drugs are needed to optimize FTO inhibition for clinical translation. Our studies support future preclinical studies investigating the combination FTO inhibitors with current standard of care for ccRCC, which is an antiangiogenic agent (axitinib) and a T cell checkpoint inhibitor (pembrolizumab). We hypothesize that targeting multiple distinct pathways that contribute to VHL-associated progression may enhance the efficacy and or reduce recurrent disease in ccRCC patients.
In summary, we have identified FTO as a critical factor to maintain the growth and survival of VHL-deficient renal cell carcinoma cells in an HIF-independent manner. Our findings suggest that FTO may provide an epitranscriptomic regulation of metabolic reprogramming of VHL-deficient renal cell carcinoma cells and identify therapeutic targets for the treatment of renal cell carcinoma.
Materials and Methods
Detailed analysis of application of MiSL algorithm for identification of SL partners of VHL-deficient ccRCC, FTO transcriptomic and proteomic analysis, cell lines and cell culture, patient samples, CRISPR knockdown, shRNA and siRNA, cell viability assay, cell growth assay (crystal violet), 2D colony formation assay, 3D soft agar assay, apoptosis assay, immunohistochemical staining, TUNEL staining, RNA isolation and quantitative real-time PCR (qRT-PCR), Western blot and antibodies, m6A ELISA, RNA-seq and m6A-seq assays, gene-specific m6A qPCR, glutamine consumption assay by liquid chromatography/mass spectrometry, and animal studies are provided in SI Appendix, Materials and Methods. Statistical analysis was performed using Prism software (GraphPad). In all cases, ANOVA followed by two-tailed, unpaired Student t tests were performed to analyze statistical differences between groups. P values < 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by NIH Grant CA-198291 (A.J.G. and E.B.R.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. M.C.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2000516117/-/DCSupplemental.
Data Availability.
The raw data files of sequencing experiments have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus. The accession number is GSE139123.
References
- 1.Escudier B., Szczylik C., Porta C., Gore M., Treatment selection in metastatic renal cell carcinoma: Expert consensus. Nat. Rev. Clin. Oncol. 9, 327–337 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Latif F. et al., Identification of the von Hippel-Lindau disease tumor suppressor gene. Science 260, 1317–1320 (1993). [DOI] [PubMed] [Google Scholar]
- 3.Iliopoulos O., Kibel A., Gray S., Kaelin W. G. Jr., Tumour suppression by the human von Hippel-Lindau gene product. Nat. Med. 1, 822–826 (1995). [DOI] [PubMed] [Google Scholar]
- 4.Duan D. R. et al., Inhibition of transcription elongation by the VHL tumor suppressor protein. Science 269, 1402–1406 (1995). [DOI] [PubMed] [Google Scholar]
- 5.Kibel A., Iliopoulos O., DeCaprio J. A., Kaelin W. G. Jr., Binding of the von Hippel-Lindau tumor suppressor protein to Elongin B and C. Science 269, 1444–1446 (1995). [DOI] [PubMed] [Google Scholar]
- 6.Pause A. et al., The von Hippel-Lindau tumor-suppressor gene product forms a stable complex with human CUL-2, a member of the Cdc53 family of proteins. Proc. Natl. Acad. Sci. U.S.A. 94, 2156–2161 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamura T. et al., Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284, 657–661 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Jaakkola P. et al., Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Ivan M. et al., HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science 292, 464–468 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Rankin E. B., Giaccia A. J., The role of hypoxia-inducible factors in tumorigenesis. Cell Death Differ. 15, 678–685 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li L., Kaelin W. G. Jr., New insights into the biology of renal cell carcinoma. Hematol. Oncol. Clin. North Am. 25, 667–686 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rini B. I., Metastatic renal cell carcinoma: Many treatment options, one patient. J. Clin. Oncol. 27, 3225–3234 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Cho H. et al., On-target efficacy of a HIF-2α antagonist in preclinical kidney cancer models. Nature 539, 107–111 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen W. et al., Targeting renal cell carcinoma with a HIF-2 antagonist. Nature 539, 112–117 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Courtney K. D. et al., Phase I dose-escalation trial of PT2385, a first-in-class hypoxia-inducible factor-2α antagonist in patients with previously treated advanced clear cell renal cell carcinoma. J. Clin. Oncol. 36, 867–874 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan D. A., Giaccia A. J., Harnessing synthetic lethal interactions in anticancer drug discovery. Nat. Rev. Drug Discov. 10, 351–364 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang A., Garraway L. A., Ashworth A., Weber B., Synthetic lethality as an engine for cancer drug target discovery. Nat. Rev. Drug Discov. 19, 23–38 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Hartwell L. H., Szankasi P., Roberts C. J., Murray A. W., Friend S. H., Integrating genetic approaches into the discovery of anticancer drugs. Science 278, 1064–1068 (1997). [DOI] [PubMed] [Google Scholar]
- 19.Sutphin P. D. et al., Targeting the loss of the von Hippel-Lindau tumor suppressor gene in renal cell carcinoma cells. Cancer Res. 67, 5896–5905 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Turcotte S. et al., A molecule targeting VHL-deficient renal cell carcinoma that induces autophagy. Cancer Cell 14, 90–102 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolff N. C. et al., High-throughput simultaneous screen and counterscreen identifies homoharringtonine as synthetic lethal with von Hippel-Lindau loss in renal cell carcinoma. Oncotarget 6, 16951–16962 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chakraborty A. A. et al., HIF activation causes synthetic lethality between the VHL tumor suppressor and the EZH1 histone methyltransferase. Sci. Transl. Med. 9, eaal5272 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson J. M. et al., Rho-associated kinase 1 inhibition is synthetically lethal with von Hippel-Lindau deficiency in clear cell renal cell carcinoma. Oncogene 36, 1080–1089 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicholson H. E. et al., HIF-independent synthetic lethality between CDK4/6 inhibition and VHL loss across species. Sci. Signal. 12, eaay0482 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jerby-Arnon L. et al., Predicting cancer-specific vulnerability via data-driven detection of synthetic lethality. Cell 158, 1199–1209 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Sinha S. et al., Systematic discovery of mutation-specific synthetic lethals by mining pan-cancer human primary tumor data. Nat. Commun. 8, 15580 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sahoo D., Dill D. L., Gentles A. J., Tibshirani R., Plevritis S. K., Boolean implication networks derived from large scale, whole genome microarray datasets. Genome Biol. 9, R157 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gameiro P. A. et al., In vivo HIF-mediated reductive carboxylation is regulated by citrate levels and sensitizes VHL-deficient cells to glutamine deprivation. Cell Metab. 17, 372–385 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lan Q. et al., The critical role of RNA m6A methylation in cancer. Cancer Res. 79, 1285–1292 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Gerken T. et al., The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 318, 1469–1472 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang S. et al., m6A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat. Commun. 10, 2782 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su R. et al., R-2HG exhibits anti-tumor activity by targeting FTO/m(6)A/MYC/CEBPA signaling. Cell 172, 90–105.e23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z. et al., FTO plays an oncogenic role in acute myeloid leukemia as a N6-methyladenosine RNA demethylase. Cancer Cell 31, 127–141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niu Y. et al., RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol. Cancer 18, 46 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clark D. J. et al., Integrated proteogenomic characterization of clear cell renal cell carcinoma. Cell 179, 964–983.e31 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jia G. et al., N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang Y. et al., Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 43, 373–384 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kondo K., Kim W. Y., Lechpammer M., Kaelin W. G. Jr., Inhibition of HIF2alpha is sufficient to suppress pVHL-defective tumor growth. PLoS Biol. 1, E83 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mikhaylova O. et al., The von Hippel-Lindau tumor suppressor protein and Egl-9-Type proline hydroxylases regulate the large subunit of RNA polymerase II in response to oxidative stress. Mol. Cell. Biol. 28, 2701–2717 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roe J. S. et al., p53 stabilization and transactivation by a von Hippel-Lindau protein. Mol. Cell 22, 395–405 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Zhou M. I., Wang H., Foy R. L., Ross J. J., Cohen H. T., Tumor suppressor von Hippel-Lindau (VHL) stabilization of Jade-1 protein occurs through plant homeodomains and is VHL mutation dependent. Cancer Res. 64, 1278–1286 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Zhang J. et al., VHL substrate transcription factor ZHX2 as an oncogenic driver in clear cell renal cell carcinoma. Science 361, 290–295 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang G. L., Jiang B. H., Rue E. A., Semenza G. L., Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. U.S.A. 92, 5510–5514 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wood S. M., Gleadle J. M., Pugh C. W., Hankinson O., Ratcliffe P. J., The role of the aryl hydrocarbon receptor nuclear translocator (ARNT) in hypoxic induction of gene expression. Studies in ARNT-deficient cells. J. Biol. Chem. 271, 15117–15123 (1996). [DOI] [PubMed] [Google Scholar]
- 45.Xiao Y., Sinha S., Rankin E. B., Next generation sequencing and m6A sequencing facilitates quantitative analysis of wild type and genetic FTO knockdown ccRCC cells transcriptomes. NCBI Gene Expression Omnibus. http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE139123. Deposited 20 October 2019.
- 46.Meng J., Cui X., Rao M. K., Chen Y., Huang Y., Exome-based analysis for RNA epigenome sequencing data. Bioinformatics 29, 1565–1567 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okazaki A. et al., Glutaminase and poly(ADP-ribose) polymerase inhibitors suppress pyrimidine synthesis and VHL-deficient renal cancers. J. Clin. Invest. 127, 1631–1645 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y. et al., High expression of Solute Carrier Family 1, member 5 (SLC1A5) is associated with poor prognosis in clear-cell renal cell carcinoma. Sci. Rep. 5, 16954 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Romero R. et al., Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat. Med. 23, 1362–1368 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hassanein M. et al., SLC1A5 mediates glutamine transport required for lung cancer cell growth and survival. Clin. Cancer Res. 19, 560–570 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nicklin P. et al., Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 136, 521–534 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schulte M. L. et al., Pharmacological blockade of ASCT2-dependent glutamine transport leads to antitumor efficacy in preclinical models. Nat. Med. 24, 194–202 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaelin W. G., Jr., The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat. Rev. Cancer 8, 865–873 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Hu L. et al., TBK1 is a synthetic lethal target in cancer with VHL loss. Cancer Discov. 10, 460–475 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peng S. et al., Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1. Sci. Transl. Med. 11, eaau7116 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Qiao Y. et al., A novel inhibitor of the obesity-related protein FTO. Biochemistry 55, 1516–1522 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Huang Y. et al., Small-molecule targeting of oncogenic FTO demethylase in acute myeloid leukemia. Cancer Cell 35, 677–691.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen B. et al., Development of cell-active N6-methyladenosine RNA demethylase FTO inhibitor. J. Am. Chem. Soc. 134, 17963–17971 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data files of sequencing experiments have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus. The accession number is GSE139123.