Significance
Recent experiments have suggested that the energy released by a chemical reaction can propel its enzyme catalyst (for example, alkaline phosphatase). However, this topic remains controversial, partially due to the indirect and ensemble nature of existing measurements. Here, we used recently developed single-molecule approaches to monitor directly the motions of individual proteins in aqueous solution and find that single alkaline phosphatase enzymes do not diffuse faster under catalysis. Instead, we demonstrate that interactions between the fluorescent dye and the enzyme’s substrate can produce the signature of apparent diffusion enhancement in fluorescence correlation spectroscopy, the standard ensemble assay currently used to study enzyme diffusion and indicate that single-molecule approaches provide a more robust means to investigate diffusion at the nanoscale.
Keywords: enzyme diffusion, fluorescence correlation spectroscopy (FCS), anti-Brownian electrokinetic (ABEL) trap, single-molecule diffusometry, single-particle tracking
Abstract
Theoretical and experimental observations that catalysis enhances the diffusion of enzymes have generated exciting implications about nanoscale energy flow, molecular chemotaxis, and self-powered nanomachines. However, contradictory claims on the origin, magnitude, and consequence of this phenomenon continue to arise. To date, experimental observations of catalysis-enhanced enzyme diffusion have relied almost exclusively on fluorescence correlation spectroscopy (FCS), a technique that provides only indirect, ensemble-averaged measurements of diffusion behavior. Here, using an anti-Brownian electrokinetic (ABEL) trap and in-solution single-particle tracking, we show that catalysis does not increase the diffusion of alkaline phosphatase (ALP) at the single-molecule level, in sharp contrast to the ∼20% enhancement seen in parallel FCS experiments using p-nitrophenyl phosphate (pNPP) as substrate. Combining comprehensive FCS controls, ABEL trap, surface-based single-molecule fluorescence, and Monte Carlo simulations, we establish that pNPP-induced dye blinking at the ∼10-ms timescale is responsible for the apparent diffusion enhancement seen in FCS. Our observations urge a crucial revisit of various experimental findings and theoretical models––including those of our own––in the field, and indicate that in-solution single-particle tracking and ABEL trap are more reliable means to investigate diffusion phenomena at the nanoscale.
At the nanoscale, passive Brownian diffusion dominates the mobility of molecules. Whether freely diffusing enzymes can harness chemical energy to generate additional mobility on top of Brownian motion is not well understood (1–3). Such a possibility seems to be supported by recent fluorescence correlation spectroscopy (FCS) measurements, which have shown that a number of nonmotor enzymes including F1-ATPase (4), urease (5–7), catalase (6, 8), alkaline phosphatase (ALP) (6), fructose bisphosphate aldolase (9), acetylcholinesterase (7), and hexokinase (10), enhance their diffusivities in the presence of substrates. Accordingly, various mechanisms, including fluctuations in pH (5), global temperature increase of the solution (11), force and charged product generation (5), oligomeric enzyme dissociation (12), and enzyme chemotaxis toward substrates (8), have been proposed to account for this phenomenon. Using FCS, we previously proposed a mechanistic link between the enhanced diffusion of catalase, urease, and ALP, and the heat released during their exothermic reactions (6). Within the framework of a stochastic theory, we proposed a “chemoacoustic effect” in which the heat released during catalytic turnover generates an asymmetric pressure wave that displaces the center of mass of the enzyme, manifesting as catalysis-enhanced enzyme diffusion. Arguing against this hypothesis, Illien et al. (9) reported that aldolase, an enzyme that catalyzes a slow and endothermic reaction, also exhibits enhanced diffusion in the presence of its substrate or a competitive inhibitor, which they show to be independent of the overall turnover rate of the reaction. These authors propose that the enhanced diffusion is due to conformational fluctuations that alter the enzyme’s hydrodynamic radius. More surprisingly, a catalytically inert tracer has also been reported to diffuse faster in the presence of active enzymes (13), leading to the suggestion that the energy released during enzyme catalysis can be transferred to and harnessed by its environment. Directly contradicting the FCS results by Illien et al. (9), Zhang et al. (14) and Günther et al. (15) recently reported no diffusion enhancement of aldolase using dynamic light scattering (14) and pulsed field gradient NMR (15), respectively. On the other hand, theory suggests that the energy required to account for the experimentally observed diffusion enhancement far exceeds the chemical power released in enzymatic reactions (11, 16). In addition, the change in the hydrodynamic radius of the enzyme needed to rationalize the observed diffusion enhancement is unlikely (2). To date, no unified theory has been proposed to rationalize these experimental observations, and publications in the field, either experimental or theoretical, rely almost exclusively on the validity of diffusion measurements made using FCS (1).
In FCS, time traces of light emitted while fluorescently labeled enzymes traverse a diffraction-limited confocal volume are recorded and analyzed in terms of the intensity autocorrelation function (17, 18). Because this autocorrelation is calculated over many molecules diffusing in and out of the focal volume, FCS only yields ensemble and time-averaged information. To extract the diffusion coefficient (D, µm2/s), an accurate fitting model is required but not always available (19). Many factors other than diffusion contribute to the shape of the autocorrelation function (20): For example, dye photophysics (18), sample heterogeneity (e.g., mixture of species with different D values), geometry of the confocal volume (21), and optical aberrations (22). Failure to account for these factors could lead to erroneous interpretations of FCS data, as highlighted in a recent publication (1).
These confounding effects prompted us to reexamine catalysis-enhanced enzyme diffusion using single-molecule techniques and performing additional control experiments. Here, in addition to FCS, we use in-solution single-particle tracking (SPT) and anti-Brownian electrokinetic (ABEL) trap-based diffusometry to cross-examine the diffusion of ALP. Our results reveal that the enzyme substrate p-nitrophenyl phosphate (pNPP) affects the photophysics of the dye and introduces artifacts in the FCS measurements. This finding sets the stage necessary for future investigation of enzyme diffusion at the nanoscale.
Results
Catalysis Is Neither Sufficient nor Necessary for the Apparent Diffusion Enhancement of ALP in FCS.
Among enzymes that have been reported to exhibit catalysis-enhanced diffusion, ALP shows the highest diffusion enhancement with its substrate pNPP (6) (Fig. 1A), the magnitude of which is difficult to reconcile with theoretical predictions based on energetic coupling between the enzyme and its environment (2). We therefore sought to investigate the enhanced diffusion of ALP by performing additional control experiments in FCS (Fig. 1 B and C), and by using alternative methods to measure diffusion at the single-molecule level (Fig. 1D). We purified commercial bovine intestinal ALP by size-exclusion chromatography (SI Appendix, Fig. S1A) and fluorescently labeled the enzyme with JF646 or Atto647N (SI Appendix, Fig. S1C). JF646 was chosen for its superior brightness and photostability (23), as shown in recent SPT and localization microscopy experiments (24). Using FCS, we recorded fluorescence transients of JF646-labeled ALP for 300 s, calculated the autocorrelation function G(τ) every 10 s, and fitted G(τ) with a simple model to estimate the diffusion coefficient (D) for each 10-s window (Fig. 2 A–C). The average D value obtained over the 300-s window is 45.6 ± 2.9 µm2/s (Fig. 2C, data from 0 to 300 s). We then added 2 mM pNPP substrate to the same solution during data acquisition.
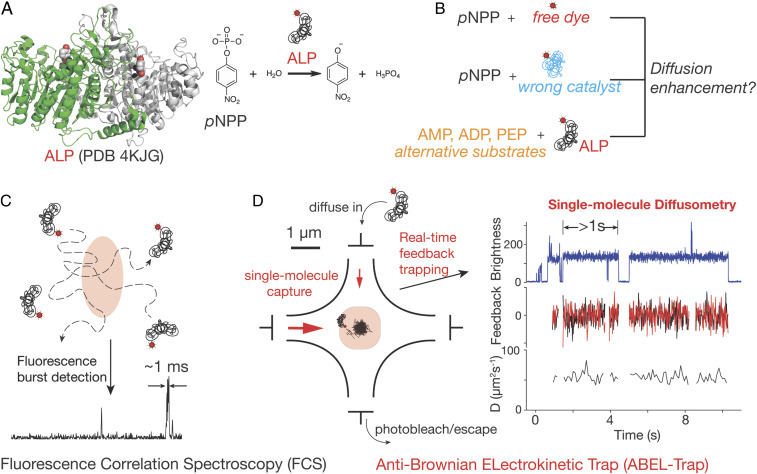
Fig. 1.
Revisiting catalysis enhanced diffusion of ALP. (A) Structure of ALP (Left) and the reaction it catalyzes (Right). The two protomers of ALP are colored in green and gray. pNPP (red and white) and zinc (black) are shown as spheres. During a reaction, ALP removes the phosphate group from pNPP and it has been shown previously that catalysis enhances the diffusivity of ALP. (B) Control experiments including pNPP with free dye, pNPP with wrong catalyst, and ALP reacting with alternative substrates are carried out to examine the role of catalysis on ALP diffusivity. (C) FCS estimates ensemble-averaged diffusion coefficients from autocorrelation analysis of fluorescence bursts in a confocal volume. (D) Principles of ABEL trap-based single-molecule diffusometry (see text for details). The diffusion coefficient of individual molecules can be measured for several seconds.
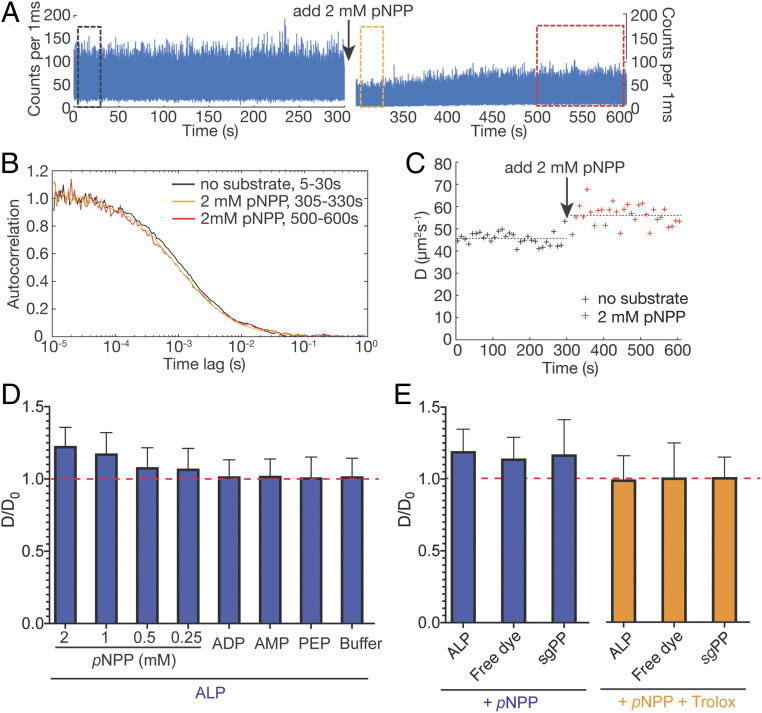
Fig. 2.
Catalysis is neither sufficient nor necessary for enhanced diffusion of ALP in FCS. (A) FCS time trace of ALP-JF646 before (Left) and after (Right) adding 2 mM pNPP. The slow equilibration of the quenching following initial addition of pNPP might contribute to the partial fluorescence recovery. (B) Normalized autocorrelation curves of data in black, orange, and red rectangle boxes in A. (C) D values extracted from fitting every 10 s of data in A, with the mean of 30 D values plotted as dashed lines. (D) pNPP, but not other substrates (saturating concentrations of ADP, AMP, PEP) or buffer control cause apparent diffusion enhancement of ALP-JF646 (plotted as ratio of D/D0, where D0 and D are the mean diffusion coefficients before and after substrate addition, respectively). D and D0 are measured and averaged as in C. The red horizontal dashed line indicates where D/D0 = 1. (E) pNPP (blue bars) not only causes apparent diffusion enhancement of ALP-JF646, but also that of free dye (JF646) and of the wrong catalyst (sgPP-JF646). The apparent diffusion enhancement can be abrogated with a dye triplet state quencher, Trolox (orange bars). The data are processed and represented similar to D.
Interestingly, fluorescence intensity was quenched by ∼50% immediately after substrate addition (even though the volume added is negligible) but recovered to 70% of the initial intensity within 150 s, remaining relatively stable thereafter (Fig. 2 A, Right). Such pNPP-induced fluorescence quenching effect was also observed in bulk experiments with the free dye (SI Appendix, Fig. S2A). Analysis of 300 s of data obtained after pNPP addition gave a mean D value for the enzyme of 56.1 ± 4.7 µm2/s (Fig. 2C, data after 300 s), corresponding to a 23% apparent diffusion enhancement of ALP. Notably, the dispersion in extracted D values increases after pNPP addition (Fig. 2C, red crosses). Consistent with previous findings (6), the diffusion enhancement decreases as pNPP concentration is lowered (Fig. 2D, first four groups with pNPP). Control experiments adding buffer did not give rise to any diffusion enhancement, indicating that the effect observed with pNPP was not due to perturbation of the experimental setup during sample addition (Fig. 2D, buffer group). Labeling ALP with the previously used Atto647N dye yielded similar results (SI Appendix, Fig. S2B). In summary, these experiments confirmed previous FCS experiments reporting ALP-enhanced diffusion in the presence of pNPP.
To test whether the diffusion enhancement of ALP observed in the presence of 2 mM pNPP originated from enzyme catalysis, we performed several control experiments. First, we measured free JF646 dye before and after addition of 2 mM pNPP. As shown in Fig. 2E, addition of pNPP induced 15% apparent diffusion enhancement of free JF646 dye and a significant increase in the dispersion of D values (as characterized by the size of the error bars in Fig. 2E; see free dye group), similar in magnitude to those seen in experiments with ALP-JF646. Second, we measured the diffusion coefficient of Atto647N-labeled Streptococcus gordonii (sg) inorganic pyrophosphatase (25) (sgPP-Atto647N) (see SI Appendix, Materials and Methods for details) before and after addition of 2 mM pNPP. Even though pNPP is not a substrate of sgPP, we again observed a 17% apparent D enhancement and increase in the dispersion of D (Fig. 2E, sgPP group). Third, we measured ALP-JF646 in the presence of other ALP substrates, including 2 mM AMP (adenosine monophosphate), 4 mM ADP (adenosine diphosphate), and 2 mM PEP (phosphoenolpyruvate). ALP remains active with these substrates (SI Appendix, Fig. S1B) but no apparent D enhancement was observed with any of them (Fig. 2D, ADP, AMP, PEP groups, and SI Appendix, Fig. S2B). Unlike pNPP, these compounds do not quench the fluorescence of JF646 dye in bulk (SI Appendix, Fig. S2A). Similar results were obtained when ALP was labeled with Atto647N dye. Collectively, these data indicate that catalysis is neither sufficient nor necessary for the apparent D enhancement of ALP in the presence of pNPP in FCS (Fig. 2 D and E).
ABEL Trap Experiments Reveal that pNPP Induces Dye Quenching and Blinking at the Millisecond Timescale.
To further investigate the physical origin of the apparent diffusion enhancement observed in FCS, we turned to ABEL trap-based single-molecule diffusometry (26), a recently developed technique capable of measuring the diffusion coefficient of individual molecules in solution. The ABEL trap senses the molecular position in real time via fluorescence detection and applies an electric voltage to exert an electrokinetic feedback control that counteracts the molecule’s Brownian motion (27). This procedure allows ∼1- to 10-s continuous trapping of individual molecules, an observation time window three to four orders of magnitude longer than that in FCS (26, 28–30). The feedback voltages can then be used to reconstruct the diffusion trajectory and to estimate the diffusion coefficient of the individual molecules. Unlike FCS, in which the emitted light intensity fluctuates due to both the number of molecules transiently traversing the confocal volume and the photophysics of the dye (e.g., quenching, blinking, and so forth), in the ABEL trap the signal arises from a single molecule and variations in dye emission can be directly observed. Accordingly, the diffusion coefficient values obtained in ABEL trap experiments are not subject to artifacts arising from dye photophysics.
We first trapped single ALP-JF646 molecules without substrate. A typical dataset is shown in Fig. 3A. Here, single molecules of ALP-JF646 diffuse into an ∼3-µm × 3-µm trapping area and are captured for multiple seconds before escaping (usually due to photobleaching of the dye). After one molecule leaves, another one stochastically enters the area and becomes trapped, giving rise to a brightness trace (Fig. 3 A, Top). Only one molecule is captured at a time and feedback voltages (Fig. 3 A, Middle) are applied every time an emitted photon arrives at the detector to keep the molecule trapped. The feedback voltages are used to determine the average diffusion coefficient of the molecule (Fig. 3 A, Bottom). Enzymes labeled with one or two JF646 dyes can be easily differentiated by their initial brightness and the number of discrete photobleaching steps (Fig. 3 A, Top Left). Due to the nonspecific labeling scheme used here, we cannot draw any conclusion regarding the oligomerization state of the enzyme (i.e., monomer or dimer) from molecule brightness. On the other hand, the diffusion coefficient can be used to infer the oligomerization state of the protein (30). D values extracted from many single molecules are narrowly distributed around 54.8 µm2/s (Fig. 3 A, Right histogram). This value matches well the diffusion coefficient (55.8 µm2/s) predicted from a hydrodynamic model (31) based on the enzyme’s crystal structure (PDB ID code 4KJG), suggesting that the enzyme exists as a stable homodimer within our experimental time window (1 to 2 h) and enzyme concentration (20 pM).
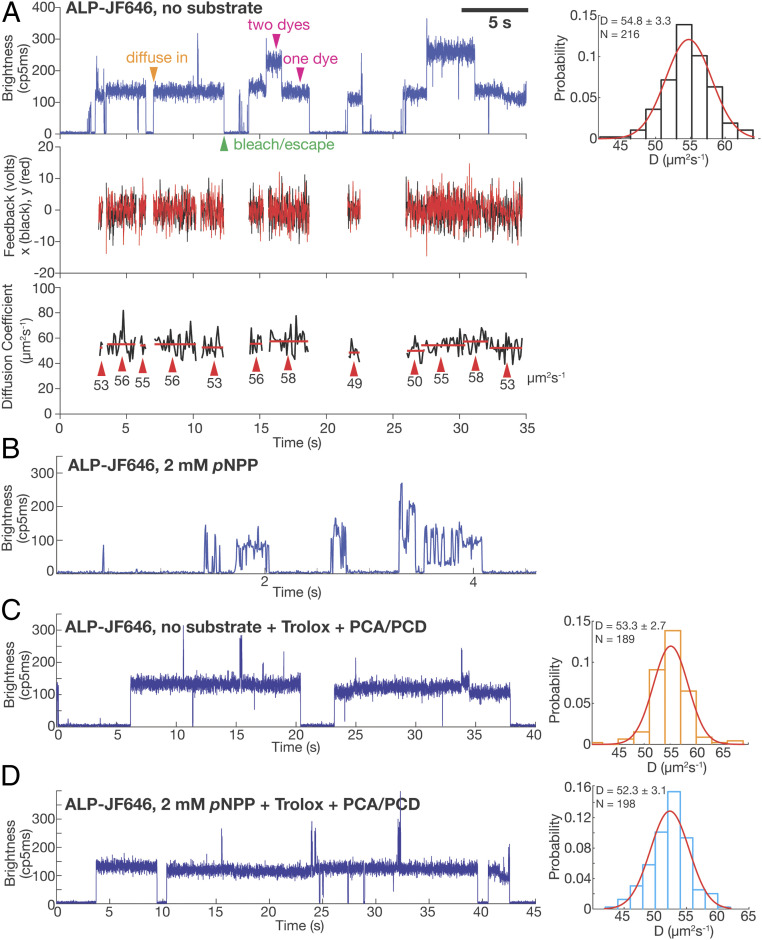
Fig. 3.
ABEL trap-based single-molecule diffusometry reveals no catalysis-induced diffusion enhancement of ALP. (A) A representative ABEL trap trace of ALP-JF646 with no substrate. (Top Left) Brightness plot (fluorescence intensity, photon counts per 5 ms) of the detected fluorescence signal. A single fluorophore corresponds to a brightness level of ∼130 (counts per 5 ms), whereas dual-fluorophore corresponds to ∼260 (counts per 5 ms). Transient spikes in the brightness trace are caused by brief co-occupancy of two molecules in the trapping region. The orange arrowhead (diffuse in) denotes the start of a successful trapping event. Magenta arrowheads denote the regions of the data that correspond to one or two dyes per protein molecule. The green arrowhead denotes the end of a trapping event, due to either photobleaching of the dye or escape of the trapped molecule. (Middle) The corresponding feedback voltages (x in black, y in red) applied in order to counteract Brownian motion and keep the molecule in trap. The feedback is only plotted when there is a molecule in the trap. (Bottom) D of each trapped molecule calculated every 100 ms (black trace). The red lines indicate the mean of each identified molecule (see SI Appendix for details of molecule identification procedure), with the value written under the red lines and denoted with red arrows. The mean D of each molecule is used to build up the histogram on the Top Right. (Top Right) D histogram of ALP-JF646 without substrate and a Gaussian fit to the data. Mean ± Std of D and number of single molecules trapped (N) are displayed on the top left corner. (B) A representative ABEL trap intensity trace of ALP-JF646 with 2mM pNPP. (C) A representative ABEL trap intensity trace (Left) and D histogram (Right) of ALP-JF646 with no substrate in the Trolox + PCA/PCD buffer. (D) A representative ABEL trap intensity trace (Left) and D histogram (Right) of ALP-JF646 with 2 mM pNPP in the Trolox + PCA/PCD buffer.
Next, we attempted to trap ALP-JF646 under catalysis by adding 2 mM pNPP. Under this condition we could no longer trap stably individual ALP-JF646 molecules. Instead, only brief transients with large brightness fluctuations and lasting from milliseconds to hundreds of milliseconds were observed (Fig. 3B). Because the ABEL trap relies on photon detection to counteract diffusion, trapping is not stable when the dye enters frequently (or for a prolonged period of time) a dark state. Therefore, to characterize these photophysical effects induced by pNPP, we measured emission from single JF646 dyes attached to surface immobilized DNA duplexes (SI Appendix, Fig. S3A). Without pNPP, stable dye emission lasting tens of seconds was observed (SI Appendix, Fig. S3B). Addition of 2 mM pNPP resulted in rapid on–off switching (blinking) and a dramatic reduction of the on-time of the fluorophore (SI Appendix, Fig. S3C), confirming the quenching and blinking effects induced by pNPP seen in the ABEL trap. These observations are also consistent with the quenching observed in FCS and bulk experiments (Fig. 2A and SI Appendix, Fig. S2A).
Single-molecule fluorophore quenching and blinking have been studied extensively (32), and several additives are known to rescue the molecules from the dark state. Indeed, when we included a commonly used antiblinking reagent (Trolox with oxygen removal by protocatechuic acid, PCA, and protocatechuic deoxygenase, PCD) (33, 34) in our trapping buffer, the pNPP-induced dye blinking effect was completely suppressed, allowing us to trap single ALP-JF646 molecules in the presence of pNPP (Fig. 3 D, Left) for multiple seconds in the ABEL trap. Thus, by suppressing pNPP-induced dye quenching and blinking, we identified the experimental conditions that allowed us to study the diffusion behavior of individual enzyme molecules under catalysis.
ABEL Trap Reveals No Catalysis-Enhanced Diffusion of ALP.
Using this buffer system, we compared the distributions of D values obtained for ALP-JF646 with and without pNPP. In the presence of the antiblinking reagent and in the absence of substrate, ALP molecules showed a narrow distribution (D = 53.3 ± 2.7 µm2/s, n = 189) (Fig. 3C), similar to the values obtained in the absence of the reagent (Fig. 3A). With 2 mM pNPP, we saw no difference in the distribution of the diffusion coefficient (D = 52.3 ± 3.1 µm2/s, n = 198) (Fig. 3D). Notably, the enzyme remains active in all buffer conditions (SI Appendix, Fig. S1D). Thus, the ABEL trap data do not agree with the FCS results and instead indicate that there is no catalysis-induced diffusion enhancement of ALP.
SPT Reveals No Catalysis-Enhanced Diffusion of ALP.
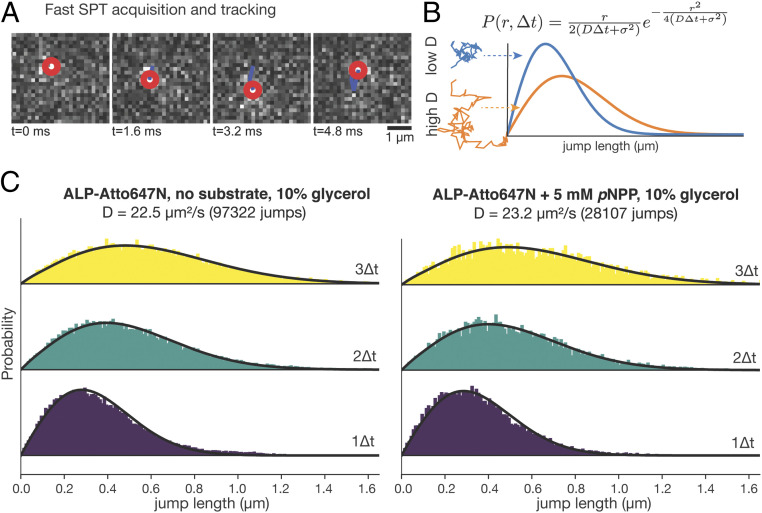
To cross-validate the new single-molecule observation that ALP does not diffuse faster under catalysis conditions, we performed high-speed, in-solution SPT experiments of the enzyme with and without pNPP. SPT has emerged as a powerful approach to track the movement of individual molecules (23, 35). By imaging fluorescent molecules at high rate, it is possible to localize their positions in successive frames (Fig. 4A). The jump-length distribution from these time courses can be used to derive their diffusion coefficients, D (Fig. 4B) (36, 37). Unlike FCS, this method is less sensitive to dye photophysics, as dark state or blinking fluorophores will not appear in successive images for reconstruction of diffusion time courses. Using high-speed SPT with stroboscopic illumination (37) (SI Appendix, Materials and Methods), we validated the robustness of this method (SI Appendix, Fig. S4) and imaged ALP-Atto647N at ∼600 Hz (1.7 ms per frame) in buffer containing 10% glycerol, to slow down the diffusion. The acquired traces were then analyzed using a population model (36) that expresses the distribution of jump lengths assuming a Brownian, free-diffusion model (Fig. 4B). Fitting both histograms of jump lengths to the model yielded similar diffusion coefficients with and without substrate (D = 22.5 µm2/s with no substrate, and D = 23.3 µm2/s with 5 mM pNPP) (Fig. 4C), confirming the ABEL trap finding that catalysis does not enhance the diffusivity of ALP at the single-molecule level. Consistent with the quenching and blinking effects of pNPP on the dye, we detected ∼10-fold fewer particles in the presence than in the absence of pNPP (SI Appendix, Fig. S5A) and the intensity of the detected spots was also lower (SI Appendix, Fig. S5B).
Fig. 4.
SPT reveals no catalysis-induced diffusion enhancement of ALP. (A) Representative images of high-speed SPT acquisition and tracking of single molecules. The red circle denotes the center of the detected molecule in each frame. The blue line denotes the molecule’s diffusion trajectory. (B) Modeling jump length distribution under free diffusion assumptions. Jump length distribution histograms of molecules with low and high Ds are plotted in blue and orange, respectively. (C) Jump length distribution histograms of ALP-Atto647N with no substrate (Left) and with 5 mM pNPP (Right). The yellow, green, and purple histograms are distributions of jump lengths between 3∆t (∆t =1.7 ms), 2∆t and 1∆t, respectively. The calculated D and the number of total jumps are written on the top of the histograms.
The Apparent Diffusion Enhancement of ALP in FCS Is Caused by pNPP-Induced Photophysics of the Dye.
The new single-molecule results prompted us to reexamine our previous FCS experiments. Because FCS relies on fitting the decay of the intensity correlation function in the 0.1- to 50-ms range to extract the diffusion coefficient, we hypothesize that pNPP-induced dye blinking––which was observed at the ∼10-ms timescale in the ABEL trap experiments––is responsible for the apparent diffusion enhancement of ALP in FCS. We note that fluorophore blinking has been proposed as a possible basis for enhanced enzyme diffusion (38). To test this hypothesis, we carried out FCS experiments in Trolox-based antiblinking buffer, which suppresses pNPP-induced dye blinking (Fig. 3D). Significantly, no pNPP-induced diffusion enhancement of ALP-JF646, free JF646 dye, and sgPP-Atto647N was observed (Fig. 2E, +Trolox groups), confirming the hypothesis that pNPP-induced dye photophysics is responsible for the apparent diffusion enhancement of ALP in FCS.
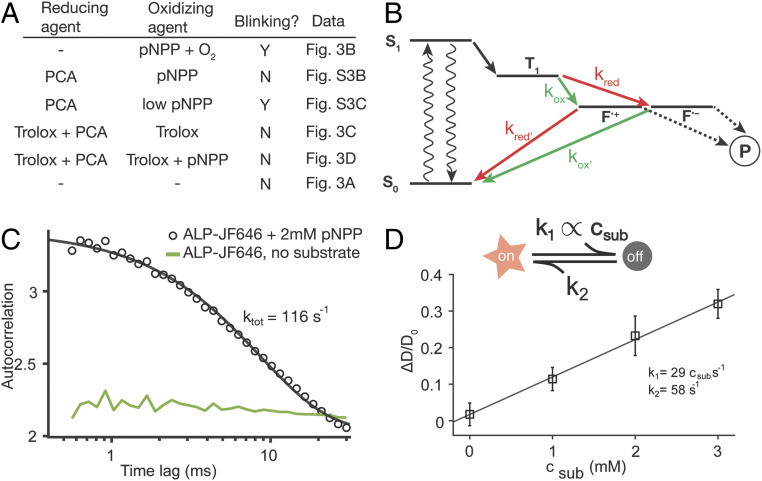
By performing ABEL trap experiments of ALP-JF646 under different buffer conditions, we were able to elucidate the mechanism of pNPP-induced dye blinking. First, we found that not all components in the Trolox-based antiblinking mixture are needed to suppress blinking in the presence of 2 mM pNPP. A single component, 2 mM PCA, is sufficient (SI Appendix, Fig. S6B). However, 2 mM PCA does not prevent blinking of the dye (SI Appendix, Fig. S6C) in the presence of substoichiometric quantities of pNPP (200 µM, O2 removed), suggesting that matching concentrations of pNPP and PCA are needed for stable emission. All of our observations regarding pNPP-induced JF646 photophysics (Fig. 5A) can be reconciled by the reducing and oxidizing mechanism of dye quenching proposed by Vogelsang et al. (39). To achieve a stable emission in the redox framework, both a reducing and an oxidizing agent are needed in the absence of oxygen to efficiently depopulate the triplet state (T1), as well as the radical cation (F•+) or anion states (F•−), which result from the oxidation and reduction of the triplet state, respectively. Here, pNPP contains a nitro group, making it a plausible oxidizing agent; PCA is a known reducing agent (40) and Trolox, when dissolved in solution, contains both reducing and oxidizing components (41). We thus propose that pNPP quenches JF646 fluorescence by random collision and subsequent oxidization of JF646 triplet state to create a long-lived, cation state (F•+) (Fig. 5B). If this cation state is not efficiently scavenged by a reductant (e.g., PCA and/or Trolox) in solution, the fluorophore stays dark for extended periods (>10 ms). This mechanism is quite general and suggests that pNPP-induced quenching is not limited to the JF646 dye. The same mechanism has been shown to operate with other red dyes of different structural families (39). Indeed, when we measured the emission of single immobilized DNA duplexes labeled with the Atto647 dye (SI Appendix, Fig. S7 A and B), addition of 2 mM pNPP (SI Appendix, Fig. S7C) induces rapid blinking of the dye on the timescale of ∼10 ms (SI Appendix, Fig. S7C). On the other hand, addition of 2.5 mM PEP (a redox-inactive substrate of ALP) does not induce blinking (SI Appendix, Fig. S7D). Recently, Günther et al. (1) reported quenching of Alexa 488 by pNPP, manifested as a reduction of fluorescent lifetime of the dye in the presence of 2 to 4 mM of this substrate.
Fig. 5.
pNPP-induced dye photophysics is responsible for the apparent diffusion enhancement of ALP in FCS. (A) Summary of all ABEL trap ALP-JF646 experiment buffer conditions with identified oxidizing and reducing agents and blinking outcome, showing consistency with the reducing and oxidizing framework as depicted in B. (B) A proposed model of pNPP-induced dye photophysics based on the reducing and oxidizing framework. The dye triple state (T1) can be oxidized (green arrows) or reduced (red arrows) into charge separated states (F•+ and F•−), which are not fluorescent and maybe prone to photobleaching (P). An oxidizing agent in solution needs to be balanced by the presence of a reducing agent (and vice versa) for blinking suppression, otherwise, the molecule is trapped in a dark radical state (F•+ or F•−) and blinks off. (C) Extracting pNPP-induced blinking kinetics from ABEL trap data. Intensity autocorrelation curve of ALP-JF646 with 2 mM pNPP (black circles, from the data represented by Fig. 3B) shows a pronounced decay between 1 ms and 30 ms and is fitted with a single exponential function (black line) to extract the total rate. The intensity autocorrelation of ALP-JF646 without substrate (from Fig. 3A) is shown in green for reference (amplitude multiplied by 10 for clarity). (D) Monte Carlo simulation of FCS experiments with a substrate-induced blinking model (Upper schematic, see text and SI Appendix for details), using the listed parameters constrained by the rate extracted in C. The extracted apparent diffusion coefficient (averaged over 10 independent simulation runs) is plotted against the substrate concentration and fit with a line. Error bars represent SD.
Finally, to evaluate quantitatively whether pNPP-induced dye photophysics can give rise to the magnitude (∼20%) of apparent D enhancement seen in the FCS experiments, we conducted Monte Carlo simulations of our FCS experiments (see SI Appendix, Materials and Methods and Fig. S8 for details), where the dye can switch from an “on” state to an “off” state with a rate (k1) that depends linearly on substrate concentration and a constant off-to-on rate (k2) that corresponds to spontaneous return of the dye to the ground state, S0 (Fig. 5D). The total switching rate (ktot = k1 + k2 = 116 s−1) at 2 mM pNPP was directly extracted from ABEL trap data (Fig. 3B) by fitting the observed intensity autocorrelation function in the range of 0.5 to 20 ms to a single exponential decay (Fig. 5C). Given that it is difficult to determine k1 and k2 uniquely in our experiments, we used k1 = k2 = ktot/2 as a rough estimate. The simulated intensity trace was subjected to autocorrelation analysis and fitted with the same one-species model without photophysics (SI Appendix, Fig. S8). The simulation results successfully recapitulate the magnitude (∼20%) of the apparent D enhancement seen in our FCS experiments (Fig. 5D).
Taken together, all of the results presented above indicate that pNPP-induced dye blinking is responsible for the apparent diffusion enhancement of JF646, Atto647N, ALP-Atto647N, sgPP-Atto647N, and ALP-JF646 observed in FCS.
Discussion
Catalysis-fueled propulsion of biomolecules at the nanoscale is no doubt a fascinating concept and has potential applications in the field of nanoscience and medicine (42–49). However, mounting experimental and theoretical evidence argue against the mechanism, scale, and even the existence of such phenomenon (1, 2, 14, 16, 38, 50). The vast majority of publications documenting enhanced enzyme diffusion upon catalysis were performed with FCS, which is prone to artifacts, such as free dye contamination, dissociation of enzyme quaternary structure, and dye photophysics that could result in apparent enhanced enzyme diffusion. A deeper experimental and theoretical investigation of this phenomenon calls for better tools, and direct single-molecule measurements of molecular diffusion at the nanoscale could offer the clearest picture.
Here, we have taken the first steps toward this goal. By directly measuring the diffusion coefficient of individual enzyme molecules, we uncover an apparent discrepancy between the diffusion behavior of ALP in the presence of pNPP measured with FCS (∼20% enhancement) and with single-molecule techniques (no enhancement). On the basis of several crucial control FCS experiments, single-molecule diffusometry, dye photophysics measurements on the surface, and simulation, we conclude that the apparent diffusion enhancement of ALP observed in FCS is caused by pNPP-induced quenching and blinking of the dye. A transient quenching mechanism has previously been proposed by Bai and Wolynes (38), and indeed our results here agree well with that suggestion. We propose that pNPP-induces dye quenching and blinking through a redox-based electron transfer mechanism. Notably, dye photophysics, depending on the mechanism, can have timescales ranging from nanoseconds to seconds. However, substrate-induced redox blinking, as proposed here, is particularly detrimental to diffusion estimates in FCS because its timescale (∼10 ms) may coincide with the diffusion transit timescale of molecules through the confocal volume. For this reason, we urge caution in interpreting FCS measurements when adding compounds that have distinct redox properties (e.g., hydrogen peroxide).
Given the results presented here and similar concerns voiced by others (1), we believe that the experimental evidence for enhanced enzyme diffusion needs to be critically reevaluated. We advocate the use of single-molecule techniques such as in-solution SPT and ABEL trap-based diffusometry. These methods extend the observation time of molecular diffusion from ∼1 ms (as in FCS) to several seconds, thus enabling molecule-by-molecule analysis and more precise characterization of diffusion coefficient [compared to more modern, information-rich analysis of FCS data (51)] while decoupling photophysical artifacts from diffusion coefficient estimation. The ability to uncover the full distribution of diffusive populations from single-molecule analysis will allow us to test alternative hypotheses of enhanced diffusion, such as redistribution of enzyme oligomerization states associated with substrate binding or catalysis, which has been suggested for aldolase (14, 52). As examples of this new measurement capability, single-molecule diffusometry (by ABEL trapping) recently revealed the nucleotide-dependent shift of quaternary structure of rubisco activase between monomers, dimers, and hexamers (30). Here we have confirmed that ALP molecules remain as dimers under our experimental conditions.
While our experiments with ALP do not support enhanced enzyme diffusion, we do not preclude these phenomena to still hold true for other enzymes. For example, using a different SPT setup, Xu et al. (53) have recently shown that the diffusion coefficient of urease is enhanced by 300% in the presence of its substrate, although the exact cause of this large diffusion enhancement remains enigmatic.
Materials and Methods
A detailed description of the materials and methods is given in the SI Appendix, Materials and Methods. Briefly, ALP from bovine intestinal mucosa was purchased from Sigma-Aldrich (Cat # P7923), further purified in house, and fluorescently labeled using amine-NHS chemistry. ABEL trap-based single-molecule diffusometry was implemented as previously described (26) and high-speed SPT was carried out according to the recent protocol (36).
Supplementary Material
Acknowledgments
We thank Philip Tinnefeld for comments and suggestions of the manuscript; members in the S.M., C.B., X.D., and Zimmer laboratories for insightful comments and discussions; Ana Robles and Astou Tangara for microscope maintenance and set-up; and Ronen Gabizon for experimental assistance and discussions. Q.W. acknowledges Evangelos Gatzogiannis for the loan of a PI P-563 piezo stage. A.S. is supported by Swedish Research Council International Postdoc Grant 2017-00389. This work was funded by a grant from the US Department of Energy, Office of Basic Energy Sciences, Division of Materials Sciences and Engineering under contract DE-AC02-05CH11231 (to C.B.); National Institutes of Health Grant R01GM032543 (to C.B.), and UO1-497 EB021236 (to X.D.); National Science Foundation Grant MCB 1616591 (to S.M.); and a Lewis-Sigler Fellowship of Princeton University (to Q.W.). S.M. is a Chan Zuckerberg Biohub Investigator. C.B. is an investigator with the Howard Hughes Medical Institute.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. A.E.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2006900117/-/DCSupplemental.
Data Availability.
All study data are included in the main text and SI Appendix.
References
- 1.Günther J.-P., Börsch M., Fischer P., Diffusion measurements of swimming enzymes with fluorescence correlation spectroscopy. Acc. Chem. Res. 51, 1911–1920 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y., Hess H., Enhanced diffusion of catalytically active enzymes. ACS Cent. Sci. 5, 939–948 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kondrat S., Popescu M. N., Brownian dynamics assessment of enhanced diffusion exhibited by “fluctuating-dumbbell enzymes”. Phys. Chem. Chem. Phys. 21, 18811–18815 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Börsch M. et al., Conformational changes of the H+-ATPase from Escherichia coli upon nucleotide binding detected by single molecule fluorescence. FEBS Lett. 437, 251–254 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Muddana H. S., Sengupta S., Mallouk T. E., Sen A., Butler P. J., Substrate catalysis enhances single-enzyme diffusion. J. Am. Chem. Soc. 132, 2110–2111 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riedel C. et al., The heat released during catalytic turnover enhances the diffusion of an enzyme. Nature 517, 227–230 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jee A.-Y., Dutta S., Cho Y.-K., Tlusty T., Granick S., Enzyme leaps fuel antichemotaxis. Proc. Natl. Acad. Sci. U.S.A. 115, 14–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sengupta S. et al., Enzyme molecules as nanomotors. J. Am. Chem. Soc. 135, 1406–1414 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Illien P. et al., Exothermicity is not a necessary condition for enhanced diffusion of enzymes. Nano Lett. 17, 4415–4420 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Zhao X. et al., Substrate-driven chemotactic assembly in an enzyme cascade. Nat. Chem. 10, 311–317 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Golestanian R., Enhanced diffusion of enzymes that catalyze exothermic reactions. Phys. Rev. Lett. 115, 108102 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Jee A. Y., Chen K., Tlusty T., Zhao J., Granick S., Enhanced diffusion and oligomeric enzyme dissociation. J. Am. Chem. Soc. 141, 20062–20068 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Zhao X. et al., Enhanced diffusion of passive tracers in active enzyme solutions. Nano Lett. 17, 4807–4812 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y., Armstrong M. J., Bassir Kazeruni N. M., Hess H., Aldolase does not show enhanced diffusion in dynamic light scattering experiments. Nano Lett. 18, 8025–8029 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Günther J. P., Majer G., Fischer P., Absolute diffusion measurements of active enzyme solutions by NMR. J. Chem. Phys. 150, 124201 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Feng M., Gilson M. K., A thermodynamic limit on the role of self-propulsion in enhanced enzyme diffusion. Biophys. J. 116, 1898–1906 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su D., Hou Y., Dong C., Ren J., Fluctuation correlation spectroscopy and its applications in homogeneous analysis. Anal. Bioanal. Chem. 411, 4523–4540 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Rigler R., Elson E. S., Fluorescence Correlation Spectroscopy: Theory and Applications, (Springer Berlin Heidelberg, 2001). [Google Scholar]
- 19.Krichevsky O., Bonnet G., Fluorescence correlation spectroscopy: The technique and its applications. Rep. Prog. Phys. 65, 251–297 (2002). [Google Scholar]
- 20.Kandula H. N., Jee A. Y., Granick S., Robustness of FCS (fluorescence correlation spectroscopy) with quenchers present. J. Phys. Chem. A 123, 10184–10189 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Hess S. T., Webb W. W., Focal volume optics and experimental artifacts in confocal fluorescence correlation spectroscopy. Biophys. J. 83, 2300–2317 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banachowicz E., Patkowski A., Meier G., Klamecka K., Gapiński J., Successful FCS experiment in nonstandard conditions. Langmuir 30, 8945–8955 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Grimm J. B. et al., Bright photoactivatable fluorophores for single-molecule imaging. Nat. Methods 13, 985–988 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Basu S. et al., FRET-enhanced photostability allows improved single-molecule tracking of proteins and protein complexes in live mammalian cells. Nat. Commun. 9, 2520 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ilias M., Young T. W., Streptococcus gordonii soluble inorganic pyrophosphatase: An important role for the interdomain region in enzyme activity. Biochim. Biophys. Acta 1764, 1299–1306 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Wang Q., Moerner W. E., Single-molecule motions enable direct visualization of biomolecular interactions in solution. Nat. Methods 11, 555–558 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Cohen A. E., Moerner W. E., Method for trapping and manipulating nanoscale objects in solution. Appl. Phys. Lett. 86, 093109 (2005). [Google Scholar]
- 28.Wang Q., Moerner W. E., An adaptive anti-Brownian electrokinetic trap with real-time information on single-molecule diffusivity and mobility. ACS Nano 5, 5792–5799 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Wang Q., Goldsmith R. H., Jiang Y., Bockenhauer S. D., Moerner W. E., Probing single biomolecules in solution using the anti-Brownian electrokinetic (ABEL) trap. Acc. Chem. Res. 45, 1955–1964 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q., Serban A. J., Wachter R. M., Moerner W. E., Single-molecule diffusometry reveals the nucleotide-dependent oligomerization pathways of Nicotiana tabacum Rubisco activase. J. Chem. Phys. 148, 123319 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Ortega A., Amorós D., García de la Torre J., Prediction of hydrodynamic and other solution properties of rigid proteins from atomic- and residue-level models. Biophys. J. 101, 892–898 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ha T., Tinnefeld P., Photophysics of fluorescent probes for single-molecule biophysics and super-resolution imaging. Annu. Rev. Phys. Chem. 63, 595–617 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rasnik I., McKinney S. A., Ha T., Nonblinking and long-lasting single-molecule fluorescence imaging. Nat. Methods 3, 891–893 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Aitken C. E., Marshall R. A., Puglisi J. D., An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J. 94, 1826–1835 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen H. et al., Single particle tracking: From theory to biophysical applications. Chem. Rev. 117, 7331–7376 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Hansen A. S. et al., Robust model-based analysis of single-particle tracking experiments with Spot-On. eLife 7, e33125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hansen A. S., Pustova I., Cattoglio C., Tjian R., Darzacq X., CTCF and cohesin regulate chromatin loop stability with distinct dynamics. eLife 6, e25776 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bai X., Wolynes P. G., On the hydrodynamics of swimming enzymes. J. Chem. Phys. 143, 165101 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Vogelsang J. et al., A reducing and oxidizing system minimizes photobleaching and blinking of fluorescent dyes. Angew. Chem. Int. Ed. Engl. 47, 5465–5469 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Jovanovic S. V., Steenken S., Tosic M., Marjanovic B., Simic M. G., Flavonoids as antioxidants. J. Am. Chem. Soc. 116, 4846–4851 (1994). [Google Scholar]
- 41.Cordes T., Vogelsang J., Tinnefeld P., On the mechanism of Trolox as antiblinking and antibleaching reagent. J. Am. Chem. Soc. 131, 5018–5019 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Dey K. K., Sen A., Chemically propelled molecules and machines. J. Am. Chem. Soc. 139, 7666–7676 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Jee A.-Y., Cho Y.-K., Granick S., Tlusty T., Catalytic enzymes are active matter. Proc. Natl. Acad. Sci. U.S.A. 115, E10812–E10821 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arqué X. et al., Intrinsic enzymatic properties modulate the self-propulsion of micromotors. Nat. Commun. 10, 2826 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghosh S. et al., Motility of enzyme-powered vesicles. Nano Lett. 19, 6019–6026 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Saper G., Hess H., Synthetic systems powered by biological molecular motors. Chem. Rev. 120, 288–309 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Niebel B., Leupold S., Heinemann M., An upper limit on Gibbs energy dissipation governs cellular metabolism. Nat. Metab. 1, 125–132 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Elenewski J. E., Velizhanin K. A., Zwolak M., Topology, landscapes, and biomolecular energy transport. Nat. Commun. 10, 4662 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keya J. J., Kabir A. M. R., Kakugo A., Synchronous operation of biomolecular engines. Biophys. Rev. 12, 401–409 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown A. I., Sivak D. A., Theory of nonequilibrium free energy transduction by molecular machines. Chem. Rev. 120, 434–459 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Jazani S. et al., An alternative framework for fluorescence correlation spectroscopy. Nat. Commun. 10, 3662 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tolan D. R., Schuler B., Beernink P. T., Jaenicke R., Thermodynamic analysis of the dissociation of the aldolase tetramer substituted at one or both of the subunit interfaces. Biol. Chem. 384, 1463–1471 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Xu M., Ross J. L., Valdez L., Sen A., Direct single molecule imaging of enhanced enzyme diffusion. Phys. Rev. Lett. 123, 128101 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the main text and SI Appendix.