Cyclobutane pyrimidine dimers (CPDs) are predominant ultraviolet (UV) light-induced DNA lesions that can result in mutations and lead to skin cancers (1). CPD lesions are primarily repaired by nucleotide excision repair (NER), a highly conserved repair pathway (2). NER consists of two subpathways: transcription-coupled NER (TC-NER) and global genome NER (GG-NER) (2). GG-NER can occur anywhere in the genome, whereas TC-NER is dedicated for repair of bulky lesions at transcribed strands of active genes (2). Eukaryotic TC-NER is initiated by stalled RNA polymerase II (Pol II) at DNA damage sites, whereas the damage recognition in GG-NER DNA is mainly achieved by repair factor XPC (2). Many factors can modulate the repair efficiency and outcome in vivo, including the chromatin organization and genomic location of lesions, accessibility of repair factors, and occupancy of repair facilitators and suppressors. In PNAS, Duan et al. (3) take a genomic approach to investigate how the genome-wide contribution of several key protein factors would impact the TC-NER process of CPDs in yeast, providing important insights into how TC-NER is modulated in yeast chromatin.
In particular, Duan et al. (3) focus on three factors, Rad26, transcription factor IIH (TFIIH), and elongation factor Spt4/Spt5, that have dual roles in both TC-NER and transcription processes. During TC-NER, human Cockayne syndrome group B (CSB) protein and its yeast ortholog Rad26 play important roles in TC-NER. CSB/Rad26 is the first protein to be recruited to DNA damage-stalled Pol II (2). Human CSB, CSA, and UVSSA act cooperatively to recruit TFIIH to damage-stalled Pol II (4). TFIIH is involved in the lesion verification step as well as recruitment of repair factors for dual incision (2, 5). In contrast, yeast Spt4/Spt5 functions as a TC-NER suppressor and Rad26 is required to antagonize Spt4/Spt5’s suppression (6). Recent structure of the Pol II–Rad26 complex provided mechanistic insights into the roles of Rad26 in evicting Spt4/Spt5 and TC-NER initiation (7).
As an essential general transcription factor for transcription initiation, TFIIH unwinds the double-stranded DNA to facilitate the formation of the open complex with a transcription bubble that allows de novo RNA synthesis. When Pol II transits from initiation to the productive elongation stage, TFIIH is released from Pol II and yeast Spt4/Spt5 is recruited to Pol II to promote transcription elongation (6). Rad26 is also involved in transcription regulation of a subset of genes and stimulates transcription elongation in vitro (7, 8). Previous studies revealed significant occupancy heterogeneity of transcription/repair factors in different chromatin regions (9, 10). How does the occupancy heterogeneity of these factors affect TC-NER of DNA lesions in chromatin? A genome-wide mapping of DNA damage formation and repair would be important to answer this question.
Next-generation sequencing technologies have greatly facilitated our understanding of DNA damage formation and repair across the whole genome (9–12). Recently, quite a few high-resolution genome-wide DNA damage-mapping methods were developed (12–16), which can be categorized into three classes based on the mapping strategies. First, antibody-based methods (such as damage sequencing [Damage-seq] and excision repair sequencing [XR-seq]) take advantage of the specificity of antibodies against DNA damage and/or damage-binding proteins (such as TFIIH) (12–14). The antibody-based methods were employed to investigate the formation and dissect GG-NER and TC-NER excision dynamics of a variety of DNA lesions from bacteria to human (12–14). Second, chemical-based labeling methods (such as 8-oxo-7,8-dihydroguanine sequencing [OG-seq]) utilize specific chemical probes to label damage sites for mapping genome-wide distribution of DNA lesions (16). Third, repair enzyme-based methods (such as CPD sequencing [CPD-seq] and excision sequencing [Excision-seq]) take advantage of the specificity of repair enzymes that recognize DNA lesions (15, 17). Mao et al. (15) previously developed CPD-seq that employs T4 endonuclease V (T4 endoV) and apurinic/apyrimidinic endonuclease (APE1) to generate a free 3′-OH group immediately upstream of the CPD lesion, which is then ligated to an adaptor DNA for next-generation sequencing. In PNAS, Duan et al. (3) employed CPD-seq to investigate the genome-wide roles of Rad26 in TC-NER in yeast chromatin context.
In PNAS, Duan et al. take a genomic approach to investigate how the genome-wide contribution of several key protein factors would impact the TC-NER process of CPDs in yeast, providing important insights into how TC-NER is modulated in yeast chromatin.
Li and Smerdon previously reported Rad26-independent TC-NER in a RAD26 full-deletion yeast strain (i.e., rad26Δ) (18). They further studied the contribution of Rad26-dependent and Rad26-independent TC-NER in a few individual genes. TC-NER is largely Rad26-independent on some highly transcribed genes (such as GAL1), whereas TC-NER is Rad26-dependent on slowly and moderately transcribed genes (such as URA3 and RPB2 genes) (6). However, the genome-wide distribution pattern of Rad26-dependent and Rad26-independent pathways is not fully understood. To address this question, Duan et al. (3) analyzed the TC-NER of CPDs across ∼5,200 Pol II-transcribed yeast genes in wild-type cells, a rad26Δ strain, and a Rad26 ATPase-dead strain. Their data indicate that Rad26 is generally required for TC-NER in most yeast genes, and Rad26-independent TC-NER mainly occurs in a few highly transcribed genes.
Li and Smerdon also reported TC-NER activities at a short region immediately downstream of transcription start site (TSS) at GAL1 and RPB2 genes (6, 18). It was proposed the association of TFIIH with Pol II at TSS could directly contribute to TC-NER activities (18, 19). An earlier study using genome-wide XR-seq observed TC-NER repair activities near TSS and transcription termination site (TTS) regions; however, it is not clear whether these TC-NER activities at these regions require Rad26 or not (14). To address this question, the authors performed in-depth genome-wide analysis of TC-NER in regions nearby TSS (200 bp upstream to 650 bp downstream of the TSS) and TTS (i.e., from −300 bp to +200 bp relative to the polyadenylation site). The authors found that Rad26 is critical for TC-NER downstream of the first (+1) nucleosome in transcribed regions as well as around TTS regions. In contrast, the TC-NER within ∼30 bp downstream of the TSS is Rad26-independent. Duan et al. (3) confirm that this Rad26-independent TC-NER near TSS is widespread for many genes.
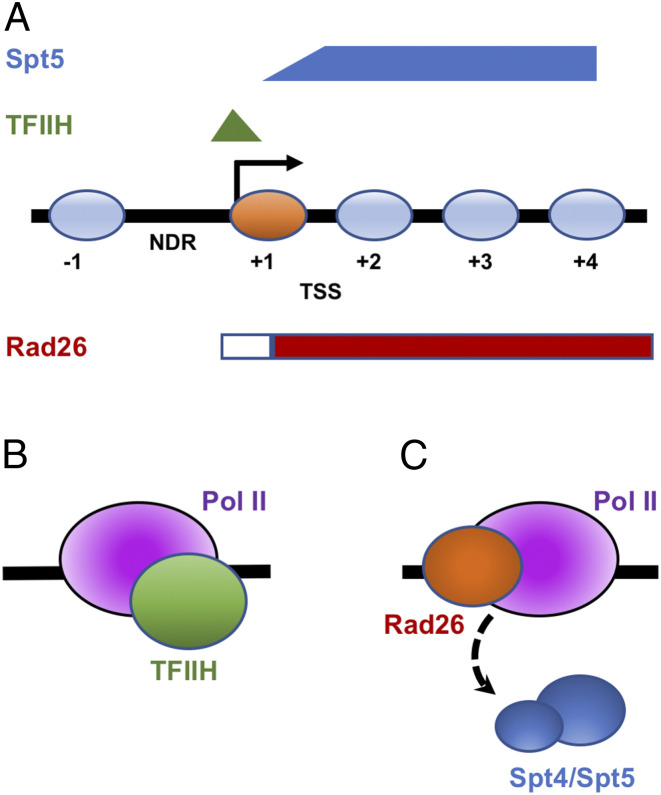
Why is Rad26 specifically required for TC-NER downstream, but not upstream, of the +1 nucleosome? The authors hypothesize that occupancy fluctuation of Pol II and/or transcription/repair factors may correlate with this differential Rad26 requirement in TC-NER at TSS regions. Duan et al. (3) first checked the Pol II density at the +1 nucleosome and found that the occupancy pattern of Pol II is not fully correlated with the Rad26 requirement in TC-NER. The authors then analyzed the occupancy patterns of TFIIH and Spt5 (9, 10) and found combined distribution of TFIIH and Spt4/Spt5 has a nice correlation with the pattern of differential Rad26 requirement in TC-NER at TSS regions (Fig. 1A). The authors suggest that, at the TSS-proximal side of the +1 nucleosome, the high occupancy of Pol II-associated TFIIH and low Spt4/5 occupancy allows cells to directly activate TC-NER without the requirement of Rad26 (Fig. 1B). In contrast, Pol II releases TFIIH and binds Spt4/Spt5 for productive elongation downstream of +1 nucleosome. Rad26 is required to remove highly occupied Spt5, the TC-NER suppressor, in order to activate TC-NER at downstream nucleosomes (Fig. 1C). Indeed, compared to the rad26Δ strain, TC-NER activity is significantly restored in the rad26Δspt4Δ strain across the genome, particularly in the downstream nucleosomes, confirming the genome-wide suppression role of Spt4/Spt5 at downstream nucleosomes in rad26Δ.
Fig. 1.
TFIIH and Spt5 distribution modulates the requirement of Rad26 in TC-NER. (A) Plot of occupancies of TFIIH (green) and Spt5 (blue) at TSS region and the pattern of Rad26 requirement in TC-NER. Nucleosomes are shown in light blue ovals, except +1 nucleosome (orange). Regions for Rad26-independent and Rad26-dependent TC-NER are shown in white and red bars, respectively. (B) Model for a Rad26-independent TC-NER at TSS-proximal side of +1 nucleosome. TFIIH (green) interacts with Pol II (purple) to activate TC-NER without Rad26. (C) Model for Rad26-dependent TC-NER at downstream nucleosomes. Rad26 (orange) is required to evict Spt4/Spt5 (blue) for TC-NER.
In summary, Duan et al. (3) report genome-wide analysis of distribution of Rad26-dependent and Rad26-independent TC-NER in chromatin. The authors reveal that the differential requirement of Rad26 in TC-NER has a nice correlation with the distribution of TFIIH and Spt4/Spt5 at TSS regions across the genome. A model of how TFIIH and Spt4/Spt5 control the Rad26 requirement is proposed to explain this correlation. These results greatly expand our understanding of the genome-wide roles of Rad26 in TC-NER and its interplay with TFIIH and Spt4/Spt5 at the whole-genome level in chromatin.
Interestingly, CSB-independent TC-NER was also reported in the TSS-proximal coding region (−40 to +20 nt) in a human gene (JUN) (20). This raises the important question of whether human cells have a mechanism similar to that reported in PNAS. While Rad26 (CSB), TFIIH, and Spt4/Spt5 (DRB-sensitivity-inducing factor [DSIF], Spt4/Spt5 homolog) are highly conserved from yeast to humans, there are some important variations between yeast and humans. First, promoter-proximal pausing and its release during early elongation (immediate downstream of TSS) function as a widespread and major regulatory mechanism in higher eukaryotes (such as metazoans) (21). This pausing is mediated by NELF (negative elongation factor) in collaboration with DSIF. In sharp contrast, such pausing has not been observed in budding yeast, which lacks homologs of all NELF subunits (21). Second, while the role of yeast Spt4/Spt5 as suppressor in TC-NER and the role of Rad26 in antagonizing this suppression is well documented in yeast (6), it is not clear whether human DSIF and CSB have similar roles. Third, in humans the recruitment of TFIIH to stalled Pol II is mediated cooperatively by CSB, CSA, and UVSSA. In contrast, it is not clear how TFIIH is recruited in yeast, which lacks a homolog of UVSSA, and whether Rad26 has a similar role (as CSB) in TFIIH recruitment. Given these variations, it would also be very interesting to expand this study in other species, particularly in human cells. Finally, a rapid growing list of new TC-NER factors has been identified in the past a few years, and next-generation damage-mapping methods would offer great opportunities to dissect genome-wide roles of these new factors on TC-NER in chromatin.
Acknowledgments
D.W.’s research is supported by NIH Grant GM 102362.
Footnotes
The author declares no competing interest.
See companion article, “Genome-wide role of Rad26 in promoting transcription-coupled nucleotide excision repair in yeast chromatin,” 10.1073/pnas.2003868117.
References
- 1.Friedberg E. C., et al. , DNA Repair and Mutagenesis (ASM Press, Washington, DC, 2006). [Google Scholar]
- 2.Hanawalt P. C., Spivak G., Transcription-coupled DNA repair: Two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 9, 958–970 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Duan M., Selvam K., Wyrick J. J., Mao P., Genome-wide role of Rad26 in promoting transcription-coupled nucleotide excision repair in yeast chromatin. Proc. Natl. Acad. Sci. U.S.A. 117, 18608–18616 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Weegen Y., et al. , The cooperative action of CSB, CSA, and UVSSA target TFIIH to DNA damage-stalled RNA polymerase II. Nat. Commun. 11, 2104 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Compe E., Egly J. M., TFIIH: When transcription met DNA repair. Nat. Rev. Mol. Cell Biol. 13, 343–354 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Li S., Transcription coupled nucleotide excision repair in the yeast Saccharomyces cerevisiae: The ambiguous role of Rad26. DNA Repair (Amst.) 36, 43–48 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Xu J., et al. , Structural basis for the initiation of eukaryotic transcription-coupled DNA repair. Nature 551, 653–657 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vélez-Cruz R., Egly J. M., Cockayne syndrome group B (CSB) protein: At the crossroads of transcriptional networks. Mech. Ageing Dev. 134, 234–242 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Rhee H. S., Pugh B. F., Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483, 295–301 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baejen C., et al. , Genome-wide analysis of RNA polymerase II termination at protein-coding genes. Mol. Cell 66, 38–49.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Wyrick J. J., Roberts S. A., Genomic approaches to DNA repair and mutagenesis. DNA Repair (Amst.) 36, 146–155 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu J., Selby C. P., Adar S., Adebali O., Sancar A., Molecular mechanisms and genomic maps of DNA excision repair in Escherichia coli and humans. J. Biol. Chem. 292, 15588–15597 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu J., Adar S., Selby C. P., Lieb J. D., Sancar A., Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. Genes Dev. 29, 948–960 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W., Adebali O., Yang Y., Selby C. P., Sancar A., Single-nucleotide resolution dynamic repair maps of UV damage in Saccharomyces cerevisiae genome. Proc. Natl. Acad. Sci. U.S.A. 115, E3408–E3415 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mao P., Smerdon M. J., Roberts S. A., Wyrick J. J., Chromosomal landscape of UV damage formation and repair at single-nucleotide resolution. Proc. Natl. Acad. Sci. U.S.A. 113, 9057–9062 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding Y., Fleming A. M., Burrows C. J., Sequencing the mouse genome for the oxidatively modified base 8-oxo-7,8-dihydroguanine by OG-Seq. J. Am. Chem. Soc. 139, 2569–2572 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ransom M., Bryan D. S., Hesselberth J. R., High-resolution mapping of modified DNA nucleobases using excision repair enzymes. Methods Mol. Biol. 1672, 63–76 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Li S., Smerdon M. J., Rpb4 and Rpb9 mediate subpathways of transcription-coupled DNA repair in Saccharomyces cerevisiae. EMBO J. 21, 5921–5929 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Svejstrup J. Q., Mechanisms of transcription-coupled DNA repair. Nat. Rev. Mol. Cell Biol. 3, 21–29 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Tu Y., Bates S., Pfeifer G. P., Sequence-specific and domain-specific DNA repair in xeroderma pigmentosum and Cockayne syndrome cells. J. Biol. Chem. 272, 20747–20755 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Adelman K., Lis J. T., Promoter-proximal pausing of RNA polymerase II: Emerging roles in metazoans. Nat. Rev. Genet. 13, 720–731 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]