Significance
Cytometry technologies are able to profile immune cells at single-cell resolution. They are widely used for both clinical diagnosis and biological research. We developed a deep learning model for analyzing cytometry data. We demonstrated that the deep learning model accurately diagnoses the latent cytomegalovirus (CMV) in healthy individuals. In addition, we developed a method for interpreting the deep learning model, allowing us to identify biomarkers associated with latent CMV infection. The deep learning model is widely applicable to other cytometry data related to human diseases.
Keywords: CyTOF, flow cytometry, deep learning, cytomegalovirus, model interpretation
Abstract
Cytometry technologies are essential tools for immunology research, providing high-throughput measurements of the immune cells at the single-cell level. Existing approaches in interpreting and using cytometry measurements include manual or automated gating to identify cell subsets from the cytometry data, providing highly intuitive results but may lead to significant information loss, in that additional details in measured or correlated cell signals might be missed. In this study, we propose and test a deep convolutional neural network for analyzing cytometry data in an end-to-end fashion, allowing a direct association between raw cytometry data and the clinical outcome of interest. Using nine large cytometry by time-of-flight mass spectrometry or mass cytometry (CyTOF) studies from the open-access ImmPort database, we demonstrated that the deep convolutional neural network model can accurately diagnose the latent cytomegalovirus (CMV) in healthy individuals, even when using highly heterogeneous data from different studies. In addition, we developed a permutation-based method for interpreting the deep convolutional neural network model. We were able to identify a CD27- CD94+ CD8+ T cell population significantly associated with latent CMV infection, confirming the findings in previous studies. Finally, we provide a tutorial for creating, training, and interpreting the tailored deep learning model for cytometry data using Keras and TensorFlow (https://github.com/hzc363/DeepLearningCyTOF).
Modern cytometry technologies, including cytometry by time-of-flight mass spectrometry or mass cytometry (CyTOF), are able to characterize cell mixtures at the single-cell resolution with over 40 markers (1). Multidimensional cytometry data contains rich information that can be used to identify key cellular changes induced by diseases or other perturbations, such as viral infections, cancer immunotherapies, and vaccinations (2–4). In addition, cytometry measurements have been utilized for decades to diagnose a variety of conditions, such as leukemia, allergies, and infectious diseases (5–7).
The analysis of cytometry data typically starts with identifying cell populations by manual gating or by automated clustering using computational methods, including FLOCK, MetaCyto, flowSOM, and others (8–10). The subsequent analysis then uses summary statistics of the identified cell populations, including abundance and mean marker expression levels, to identify disease-associated cells or to predict clinical outcomes (11, 12). This approach is an intuitive way to analyze cytometry data and has yielded highly interpretable results. However, the approach has several disadvantages. First, in the cell gating step, the original cytometry data are reduced to summary statistics of cell subsets, potentially leading to the loss of important information such as the correlation between cell markers and the distribution of marker expression within each cell subset. Second, the commonly used approach requires all samples to be clustered in the same way, making it sensitive to batch effects and the choice of clustering methods. Finally, the approach may fail to detect cellular changes that do not lead to distinct cell populations, such as the continuous up-regulation of CTLA-4 in T cells in response to varying degrees of stimulation (13).
Several recent studies have explored alternative approaches to analyze cytometry data, bypassing the requirement for cell gating or cell clustering. We previously developed CytoDx, which fits the cytometry data using a two-stage linear model (14). Another study developed CellCNN to model the cytometry data using convolutional neural networks (15). Both of these methods utilize the full cytometry data, rather than the summary statistics from cell gating steps, therefore are more advantageous for disease diagnosis and identification of disease-associated cells (14, 15). However, these existing methods still use relatively simple models (linear regression and neural networks with a single convolutional layer). Both are only capable of combining cell markers linearly at the single-cell level, thus preventing them from capturing more complex combinatorial cellular phenotypes in cytometry measurement data.
The interpretation of the CytoDx and CellCNN models also remained a challenge. The methods developed in previous studies can only interpret parts of the models. To identify cell populations that are associated with outcomes of interest, both methods leverage the one-to-one correspondence between cells and the intermediate output of the model (the output of the cell-level model in CytoDx and convolutional layers in CellCNN). New methods are required to extract biological insights from the full models.
In this study, we developed and tested a framework for modeling cytometry data using a deep convolutional neural network (CNN), in which multiple hidden layers are used to model the high-dimensional cytometry data. Leveraging multiple large publicly available CyTOF datasets (472 samples from 9 studies) available in ImmPort (16–19), we demonstrate that the deep CNN model is able to diagnose asymptomatic cytomegalovirus infection with high accuracy, even in the presence of strong heterogeneity between datasets. In addition, we developed a permutation-based method to interpret the full deep CNN model. We identified a CD27- CD94+ CD8+ T cell population that is significantly increased in subjects with latent cytomegalovirus (CMV) infections across nine studies, confirming the findings in previous studies (20, 21). Interestingly, the CD27- CD94+ subset is increased in all four compartments (naive, effector, effector memory, and central memory) of CD8+ T cells, suggesting that CMV infection induces the CD94+ CD27- phenotype through a mechanism that is distinct from T cell activation and memory.
Results
A Deep CNN for Cytometry Data.
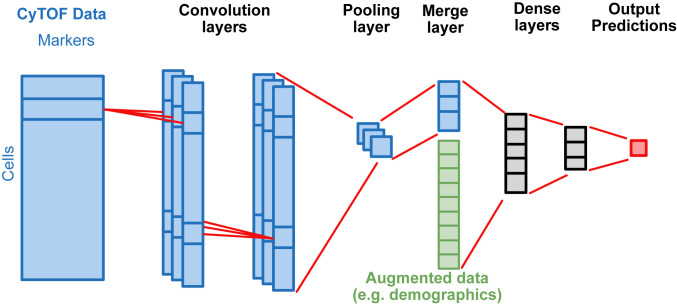
We designed a deep CNN architecture tailored to the cytometry data. The input into the model is the raw cytometry data, which are matrices with rows representing cells and columns representing markers. The outputs of the model are sample-level information of interest, such as disease diagnosis, drug responsiveness, or the presence of a genetic deficiency. The internal layers of the deep CNN model include multiple convolutional layers to extract cell-level features, a pooling layer to aggregate the cell-level features into sample level features, and dense layers to capture the interaction between the sample-level features (Fig. 1).
Fig. 1.
Schematic diagram showing the structure of the deep CNN model. The model takes arcsinh-transformed cytometry data (dimension equals no. of cells × no. of markers) as input, extracts cellular features using convolution layers (filter size equals to 1 × no. of markers) and aggregates cellular features using max or average pooling. The aggregated features can be augmented with other non-cytometry data. The dense layers combine the augmented data and predict outcomes of interest, which can be either continuous or categorical variables.
A key characteristic of cytometry data is that it represents an unordered collection of cells. The data representation is similar to the point cloud in computer vision (22). In order to model this type of data in an efficient way, the neural network needs to be invariant to the permutation of rows in the data (23). We achieved this by 1) designing “one-cell” filters in convolutional layers, which combines all marker information within the same row, but not across rows and 2) applying either max or mean function over all cells in the pooling layer, both of which are invariant to the permutation of data.
In addition to cytometry data, the deep CNN model allows the incorporation of external information, such as demographics (age, gender, and race) and results from other experiments. Specifically, the output of the pooling layer can be combined with other sample-level information to improve model performance and to adjust for control variables (Fig. 1).
The Deep CNN Model Accurately Predicts Asymptomatic CMV Infection.
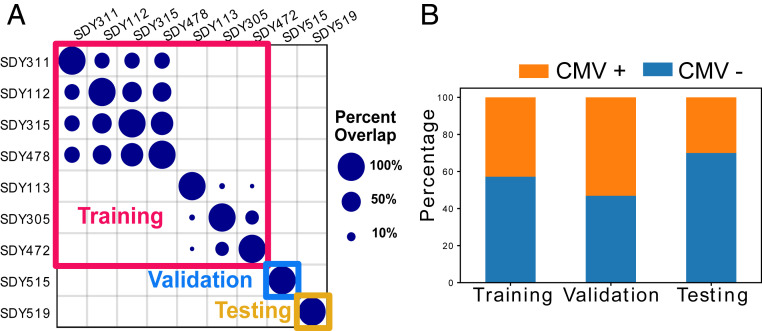
To test the performance of the deep CNN model, we applied it to nine CyTOF datasets to train it to diagnose asymptomatic CMV infection. The dataset spans nine human immunology studies and contains 596 peripheral blood mononuclear cell (PBMC) samples from 313 subjects (16–19). We split the nine studies into training, validation, and testing datasets. To ensure an unbiased performance evaluation, we selected SDY515 and SDY519 as validation and testing datasets, which do not share subjects with other studies (Fig. 2).
Fig. 2.
Overview of the CyTOF datasets. (A) Overlap of subjects between nine studies and the split of the studies into training, validation, and testing datasets. The dot size represents the percentage of overlapping subjects between studies. (B) Percentages of CMV-positive and CMV-negative individuals in training, validation, and testing datasets.
We trained and optimized the deep CNN model using training and validation datasets. The final model is evaluated using the test dataset. The deep CNN model is able to diagnose the CMV infection with high accuracy (area under the receiver operating curve [AUROC] = 0.94, area under the precision-recall curve [AUPRC] = 0.91). To benchmark the performance of the deep CNN model, we trained and tested several existing methods, including CytoDx, CellCNN, and FlowSOM (10, 14, 15). The 1,000-fold bootstrap analysis shows that the deep CNN model outperforms the existing methods (Fig. 3).
Fig. 3.
The performance of deep CNN and other methods. We used Deep CNN, CytoDx, CellCNN, and FlowSOM to diagnose latent CMV in the test dataset. (A) The performances of the models measured by the receiver-operator characteristics curves. (B) The areas under the receiver-operator characteristics curves. The error bars represent the SD. The SD and P values are measured by 1,000-fold bootstrapping. (C) The performances of the models measured by the precision-recall curves. (D) The areas under the precision-recall curves. The error bars represent the SD. The SD and P values are measured by 1,000-fold bootstrapping. *P < 0.05; **P < 0.01.
We tested the robustness of the model against the choice of training, validation, and testing dataset. In each iteration, we randomly assigned one study as the validation dataset, one study as the testing dataset, and the rest of the studies as the training dataset to train and evaluate the deep CNN model. We repeated the process 10 times and found that the model is able to diagnose CMV accurately in all iterations (AUROC ranges from 0.93 to 0.97, see SI Appendix, Table S1).
Previous studies have demonstrated that the CMV prevalence is significantly different between age, sex, and race groups (24–26). Therefore, augmenting the CyTOF data with demographic data can potentially improve the performance of the deep CNN model. We tested the augmented model and found that its performance is similar to the nonaugmented model, suggesting that demographics data do not provide additional information to the model in this particular case (Fig. 3).
Next, we characterized the performance of the deep CNN model when the number of samples and the number of markers are reduced. The original training data contains 333 samples. We downsampled the sample number to 266, 200, 133, and 67 (80%, 60%, 40%, and 20% of the original sample size) and used the reduced datasets to train new deep CNN models. In order to fairly compare the performance, we used the original testing data for all models. We found that a sample size of 200 is required to maintain the performance (SI Appendix, Fig. S1A). We then tested the deep CNN model with reduced numbers of markers. We first permuted each marker and tested how the permutation affects the performance of the deep CNN model. This allows us to rank the importance of each marker (SI Appendix, Fig. S1B). We iteratively delete the markers, from least important to the most important. At each iteration, we train and test a new deep CNN model. We found that the model maintains performance when the number of markers is greater than 8 (SI Appendix, Fig. S1C).
The Deep CNN Model Mitigates Batch Effects across Studies.
Visual inspection reveals an obvious heterogeneity between CyTOF data from different studies (Fig. 4A). Despite the heterogeneity, the deep CNN model is able to accurately diagnose CMV infection in all nine datasets, suggesting that the model is able to extract CMV-related signals from noises caused by batch effects and other non-CMV related differences in the immune system. We measured the cross-study heterogeneity using a Kruskal–Wallis test in each layer of the deep CNN model. We found that the heterogeneity is gradually mitigated across the layers of the deep CNN model (Fig. 4 B–G). The heterogeneity is the strongest at the input layer (P = 4.7 × 10−74) but became insignificant in the output layer (P = 0.16). Notably, the heterogeneity is not only reduced among the studies within the training dataset but also mitigated across the training, validation, and test dataset. The results suggest that the deep CNN model is robust and is generalizable to data outside the training dataset.
Fig. 4.
The deep CNN model mitigates the heterogeneity in cytometry data. (A) A principal component analysis plot visualizing the heterogeneity between cytometry data from SDY311 and SDY315. (B–F) Dot plots showing the mean values in different layers of the deep CNN model, including input (B), first convolutional layer (C), second convolutional layer (D), dense layer (E), and output layer (F). Each dot represents a sample in the studies. (G) Bar plot showing the batch effects in different layers in the deep CNN model, measured by the negative logarithm of P values. (H) Percent of variations in different layers that can be attributed to age, study, gender, race, and CMV status. ANOVA is used to decompose the variations. P values reported in B–G are from Kruskal–Wallis tests.
To examine how the deep learning model processes different types of heterogeneities, we decomposed into three categories: batch effect, non-CMV biological differences, and biological differences due to CMV infection (Fig. 4H). Due to the limited data available, it is challenging to characterize all non-CMV biological differences. However, we were able to attribute part of the variations to a few documented factors, including age, race, and gender. We decomposed the variation using a linear model (Y ∼ age + gender + race + study + CMV). The analysis shows that the majority of variations in the input came from technical variations. The layers in the deep learning model reduce the technical variations while amplifying the CMV-related variations. The variation from age, gender, and race are small in the input data but are not completely eliminated by the deep learning model. Because the CMV infection correlates with age (P = 0.0016) and race (P = 0.02), it is difficult to fully separate their effect in an observational study.
The Deep CNN Model Identifies Associations between Immune Cell Subsets and CMV Infection.
Leveraging the one-to-one correspondence between cells and internal nodes in the convolution layers, we first use the activation values of the convolution layers to identify cells associated with CMV infection. Using the cell definitions from the Human Immunology Project Consortium (27), we identified 24 well-characterized cell populations from the CyTOF data. For each cell population, we quantified the mean activation value in the convolution layers. In the first convolution layer, memory B cells, CD8+ T effector memory (T-EM) cells and CD4+ T central memory (T-CM) cells have the highest mean activation value from the three filters, respectively. In the second convolution layer, effector CD8+ T cells, plasmablasts, and CD8+ T-EM cells have the highest mean activation value from the three filters, respectively. The natural killer T (NKT) cells are also highly activated in the first filter of the second convolution layer (Fig. 5A). To test if the highly activated cells are associated with CMV infection, we quantified their percentage within PBMC from CMV-positive and CMV-negative subjects from all nine studies. We found that two of the three cell populations (memory B cells and CD4+ T-CM cells) activated by the first convolution layer are associated with CMV infection. The cell subsets activated in the second convolutional layers (effector CD8+ T cells, plasmablasts, and CD8+ T-EM cells) are all significantly associated with CMV infection (SI Appendix, Fig. S2).
Fig. 5.
The deep CNN model identifies associations between immune cell subsets and CMV infection. (A) The mean activation value in the convolutional layers in each cell population. The activation values are normalized by dividing the activation values by the highest value in the filter. (B) The workflow for interpreting the full deep CNN model. (C) A decision tree identifies the cells that lead to the largest changes in model output (ΔY) when up-sampled. Each node represents a cell subset. The rules by which the populations split are indicated inside the nodes. The values in each node represent the percent of the subset in the total population and the average change of model output (ΔY) when cells are up-sampled. The red box highlights the node with the highest mean ΔY. (D) Scatter plots showing the gating of the CD8+ CD3+ CD27- CD94+ cells and the composition of naive (T-N), effector (T-E), effector memory (T-EM), and central memory (T-CM) compartment in bulk CD8+ T cells and in CD8+ CD3+ CD27- CD94+ T cells. (E) The percentage of CD8+ CD3+ CD27- CD94+ cells in CMV+ and CMV- subjects across nine studies. P values are from two-way ANOVA models, with CMV infection and study as two factors. The P values of the CMV infection variable are reported.
Next, we inspected beyond the convolutional layers and hope to identify the key immune differences by interpreting the full deep CNN model. We developed a permutation-based interpretation procedure (Methods), which is inspired by the Local Interpretable Model-agnostic Explanations (LIME) approach (28). Briefly, we iteratively up-sampled each cell by copying it to replace other randomly chosen cells within the sample. We then applied the deep CNN model on both the original data and the permuted data. The difference in the model output (ΔY) quantifies the impact of each cell on the output of the deep learning model.
We then built a decision tree to identify cell subsets that have a high impact on the deep CNN model (Fig. 5B). We choose to use decision tree models because of their high interpretability and the structural similarity between decision trees and the hieratical cell gating. The decision tree identifies a CD8+ CD3+ CD27- CD94+ population that induces the highest ΔY (Fig. 5C). We manually identified the population based on the rules specified by the decision tree model (Fig. 5D). We noticed that the decision tree bisects the markers into positive and negative regions in a way that is consistent with manual gating. We previously have developed a computational tool named MetaCyto that can identify cell subsets based on their definitions (9). We used MetaCyto to identify the CD8+ CD3+ CD27- CD94+ population across all nine studies and found that the population is consistently increased in all studies (Fig. 5E and Methods). The result is consistent with previous studies that demonstrated the up-regulation of CD94 and down-regulation of CD27 in CMV-positive individuals (20, 21).
We further inspected the composition of the CD8+ CD3+ CD27- CD94+ population. We found that the CD8+ CD3+ CD27- CD94+ population does not correspond to any of the four well-characterized subsets of CD8+ T cells (naive, effector, central memory, and effector memory CD8+ T cells). Rather, all four subsets are present in the CD8+ CD3+ CD27- CD94+ population (Fig. 5D). Among the four subsets, the effector and effector memory cells are enriched in CD8+ CD3+ CD27- CD94+ cells compared to the bulk CD8+ T cells population. We then quantified the proportion of CD27- CD94+ cell subsets within the naive, effector, central memory, and effector memory CD8+ T cells. We found that CD27- CD94+ cells are increased in all four T cell compartments (SI Appendix, Fig. S5), suggesting that CMV infection induces the CD94+ CD27- phenotype through a mechanism that is distinct from T cell activation and memory.
To test the stability of the data interpretation procedure, we randomly chose studies to be training, validation, and testing datasets (same as SI Appendix, Table S1). In each iteration, we trained the deep CNN models and applied the data interpretation procedure to the models. We found that the ΔY values from different models are consistent with each other (SI Appendix, Fig. S6, median correlation = 0.92). The decision trees all identified the association between CD94+ CD27- T cells and CMV infection (SI Appendix, Fig. S7). While CD8 is not explicitly used in the decision trees, manual inspection showed that most of the CD3+ CD94+ CD27- cells are CD8 positive.
In addition to the decision tree, we visualized the ΔY and marker profiles using t-distributed Stochastic Neighbor Embedding (t-SNE) plots. Manual inspection of the plots revealed the CD8+ CD3+ CD27- CD94+ subsets to be CD4- and CD56+ (SI Appendix, Fig. S8).
Discussion
A key advantage of deep learning has been its ability to jointly optimize the feature extraction and classification steps to maximize the prediction accuracy, leading to its success in tasks involving unstructured data, such as image recognition and natural language processing (29, 30). This advantage makes the deep learning model a natural choice for analyzing cytometry data. The traditional cell-gating methods can be viewed as a way to extract features from the cytometry data. Because the cell gating step is disconnected from the later classification process, the cell gating results are often not optimized for identifying cell populations that are most associated with the outcome of interest. In the deep CNN model, the back-propagation algorithm iteratively updates the convolution layers based on classification accuracy, therefore achieving higher sensitivity in detecting cell subsets that are associated with the output of interest.
A previous study described a novel method called CellCNN (15), which uses a single layer convolutional neural network to analyze cytometry data. While this work was innovative, a limitation of the CellCNN model is that the single convolutional layer is only able to extract cellular features by combining cell markers linearly. We extend the CellCNN model by introducing multiple convolution layers and dense layers, allowing the extraction of cellular features using complex nonlinear combinations of markers. Our results show that the deep CNN model is able to identify cell populations that require multilevel hierarchical gatings, such as plasmablast, effector memory CD8+ T cells, and NKT cells. In addition, the multiple layers of the deep CNN model are able to mitigate the batch effects, making the model more generalizable across studies.
Our analysis (SI Appendix, Fig. S1) shows that while the deep CNN model requires a large number of samples (≥200), it performs well with a relatively small number of markers (≥8). Therefore, the deep CNN model can be applied to flow cytometry data in clinical settings, where panels with 8–12 markers are routinely used. In addition, the deep CNN model can be applied to flow cytometry data from large-scale studies, such as several datasets shared in the ImmPort database (SDY702, SDY887, and SDY998) (31–33).
In order to interpret the convolutional layers, we grouped the cells into previously defined cell subsets and quantified the mean activation value in each cell subset. We identified multiple cell subsets associated with CMV infection, including effector CD8+ T cells, plasmablasts, and CD8+ effector memory cells. Interestingly, not all of the cell subsets identified from the first convolution layer are associated with CMV infection. In contrast, all of the subsets identified from the second convolutional layer are significantly associated with CMV infection. The results suggest that the first convolution layer captures intermediate cellular features that do not directly correlate with CMV infection but are essential for identifying CMV-associated cell subsets in the later convolution layers.
To assess the marker importance, we iteratively permuted each marker and measured the decrease of model performance (SI Appendix, Fig. S1B). It should be noted that the procedure tends to underestimate the importance of markers that are correlated with each other. For example, while CD94+ CD27- CD8+ T cells are associated with CMV infection, the measured importance of CD94 is low. CD94 is correlated with several markers in CD8+ T cells, including CD56 (correlation = 0.58), CD16 (correlation = 0.37) and CD161 (correlation = 0.31), all of which have high importance. Due to the information redundancy between these markers, the performance of the model may not decrease when CD94 is permuted. Cautions should be taken when interpreting the marker importance, as markers with low importance may still be associated with CMV infection.
The current study has several limitations. First, our analysis of the CMV datasets is a retrospective study. Future studies are needed to prospectively validate the diagnostic model and test the causal relationship between immune cells and CMV infection. Second, the deep neural network requires a large dataset for training, limiting its use in small-scale studies. The limitation can be potentially solved by transfer learning (34). Publicly available cytometry data can be used to pretrain the network for extracting cellular features from the markers. The last dense layers of the pretrained model can then be trained using task-specific data for predicting the outcome of interest. Third, the current CNN model predicts the clinical outcome using cytometry data from a single time point. In many cases, the histories of the immune states are important for diagnosis or prediction. For example, the change of the immune system before and after vaccination is predictive of the vaccine responses (2, 35). In future studies, we will combine the CNN model with recurrent neural networks (RNN), such as a Long Short-Term Memory (LSTM) model, to model the change of the immune system over time.
Latent infection with CMV is asymptomatic and induces limited perturbation of the immune system, making it a challenging task to diagnose the latent CMV using CyTOF data of peripheral blood samples. Despite the subtlety of changes in the immune system, the deep CNN model is able to diagnose the latent CMV infection with high accuracy. The result suggests that the deep CNN model can potentially be used to diagnose more severe conditions, including autoimmune diseases, cancer, and symptomatic infections. We envision the use of CyTOF and deep CNN as a screening tool for diagnosing a wide range of conditions, whose results can be further confirmed by established disease-specific laboratory tests, such as the serological test for diagnosing CMV infection (36).
Methods
Data Preparation.
We first queried the ImmPort database to identify samples from healthy individuals with both CyTOF and CMV antibody titer data. The query identified 472 samples from nine studies, including SDY112, SDY113, SDY305, SDY311, SDY315, SDY472, SDY478, SDY515, and SDY519 as of March 2019 (16–19). We downloaded CyTOF data and transformed the raw cytometry signal using arcsinh transformation (y = arcsinh(x/5)). To combine CyTOF samples, we included 27 markers that are present in data from all nine studies and subsampled 10,000 cells from each sample. The final CyTOF data are organized into a three-dimensional matrix (472 samples × 27 markers × 10,000 cells).
Deep CNN Architecture.
The deep CNN model takes cytometry matrices as inputs. For each sample, the matrix profiles multiple markers (columns) for single cells (rows). Convolution layers are used after the input layer to extract cellular features from the cytometry data. The filter size in the first convolution layer is 1 × m × 1, where m is the number of markers in cytometry data. The filter size used in the subsequent convolutional layers is 1 × 1 × f, where f is the number of filters in the previous convolution layer. The cellular features of the last convolution layer are pooled into sample-level features using either max or mean pooling. The pooling layer is followed by dense layers, which combine the features extracted by the convolutional layers and summarized by the pooling layers. In the output layer, a logistic regression combines the output of the last dense layer to predict binary outcomes. For continuous outcomes, linear regression is used. For each layer, batch normalization is used for regularization and to facilitate model training. We used Rectified Linear Unit (ReLU) as the activation function for all internal layers.
Training, Optimization, and Testing of the Deep CNN Model.
We used the Adam algorithm, a variant of the gradient descent, to identify the best parameters in the neural network (37), with binary cross-entropy as the loss function. To prevent overfitting, the performance of the model is tested at each epoch using the validation data. The parameters that give rise to the best validation result are used in the final model.
The hyperparameters of the deep learning model include the number of convolution layers, the number of filters in the convolution layers, the type of pooling layer (max or mean pooling), the number and size of the dense layers and the learning rate. We performed a grid search to optimize hyperparameters using the training and validation datasets. The optimized model for diagnosing CMV contains two convolution layers with three filters in each layer, a mean pooling layer, and a three-node dense layer. The model is trained with a learning rate of 0.0001, batch size of 60 and total epochs of 500. The performance of the optimized model is tested using the test dataset (SDY519), which has not been used during the training and optimization processes.
Training and Optimization of CytoDx, CellCNN, and FlowSOM.
To test the performance of CytoDx, CellCNN, and FlowSOM, we used the same training, validation and testing datasets that had been applied to the deep CNN model (Fig. 2A). We trained two CytoDx models using the CytoDx R package. The first model uses the arcsinh transformed cytometry data as input. The second model uses the rank-transformed cytometry data and the two-way interactions between each pair of markers. We used the validation dataset to evaluate the two models and found the second model to be superior. We benchmarked its performance using the test dataset.
We performed a grid search for the CellCNN model (number of filters ranging from 2 to 10, drop out rate ranging from 0.1 to 0.9). Using the validation dataset, we chose an optimal set of hyper-parameters (number of filters equals 5, drop out rate equals 0.2). Adam algorithm is used for training the model with a learning rate of 0.0001. The trained model is evaluated using the testing dataset.
Using FlowSOM, we clustered the cells data using a 10-by-10 self-organizing map (SOM) and identified 20 metaclusters from the SOM result. We derived summary statistics of the identified cell subsets, including percentage in PBMC and mean fluorescence intensity (MFI) of cell markers. We then trained a Random Forest model (number of trees = 100) to predict the latent CMV infection in the subjects using results from FlowSOM as input. The optimized models were evaluated using the testing dataset.
Measurement of the Heterogeneity between Datasets.
We calculated the average marker intensities of each sample as a surrogate to measure heterogeneity between studies. For internal layers of the deep CNN model, we calculated the average activation value of each sample in each layer. We then use the Kruskal–Wallis test (also known as the one-way ANOVA on ranks) to test if the average marker or activation values are significantly different between studies. We used the nonparametric Kruskal-Wallis test because the activation values are not normally distributed due to the use of ReLU and logistic activation functions.
We used a linear regression model (Y∼ age + gender + race + study + CMV) to decompose the variation in deep CNN layers. Y is the average linear activation value (before applying relu or logistic functions) at each layer of the deep CNN model.
Quantifying Activation Value in Cell Populations.
We extracted the activation values of the internal nodes in each filter of the convolutional layers. Using definitions from the Human Immunology Project Consortium, We identified 24 immune cell subsets from the CyTOF data. We mapped the activation values to the 24 cell populations and calculated the mean activation value for each population. We normalized the mean activation value to the maximum activation value in each convolutional layer.
Measuring Marker Importance.
For each marker, we permuted the values across all cells and patients. We then fed the modified input data into the trained deep CNN model to obtain a new AUROC. The feature importance is measured by the difference between the new AUROC and the original AUROC. The process is performed 100 times. We then calculated the mean and SD of AUROC decrease.
Permutation Based Interpretation of Deep CNN Model.
For each cell in cytometry data, we up-sampled the cell by copying it to replace other randomly chosen cells within the sample. We then applied the deep CNN model on both the original data and the permuted data. The difference in the model output (ΔY) quantifies the impact of the cell on the output of the deep learning model. We repeated the process until ΔY is calculated for all cells in SDY519.
We choose to up-sample the cells, rather than delete the cell, to evaluate its impact. This is because cytometry data contain a large number of cells, so deleting a single cell has limited impact on the model output. However, we can up-sample the cell to replace a significant proportion of cells in the sample, therefore inducing a significant change to the model output. We up-sampled every cell to 1% or 5% of the total population and found that the ΔY are highly correlated between the two scenarios, suggesting that the ΔY is robust to the level of upsampling (SI Appendix, Fig. S3). We chose to up-sample each cell to 5% of the sample in this study.
Decision trees were trained using the CyTOF data as inputs and ΔY as outputs. The DecisionTreeClassifier function in the scikit-learn package is used to construct the decision tree. To determine the depth of the decision tree, we constructed decision trees with maximum depth from 2 to 10. We measured the performance of the decision trees using the correlation between observed ΔY and fitted ΔY. We used the “elbow” method and determined an optimal depth of 4 (SI Appendix, Fig. S4). Specifically, we measured the performance of the decision trees using the correlation between the ΔY and the fitted values. We found that the performance of the decision tree increased rapidly from depth of 1–4 but slowed down afterward.
Quantify Cell Populations Using MetaCyto.
To identify the cell subset with the highest ΔY, we inspected the decision tree model and identified the hierarchical decision rule that leads to the leaf with the highest mean ΔY (CD8 > 2.4, CD27 < 1.38, CD3 > 2.14, CD94 > 0.82). We noticed that the decision tree bisects the markers into positive and negative regions in a way that is consistent with manual gating. We, therefore, specified the cell definition to be CD8+ CD27- CD3+ CD94+, which can be used as input in our previously developed MetaCyto R package. Using the “searchCluster” function in MetaCyto (9), we quantified the proportion of CD8+ CD27- CD3+ CD94+ subset across nine studies. Briefly, MetaCyto determines an optimal cutoff value that bisects each marker into positive and negative regions. MetaCyto chooses the optimal cutoff value using a silhouette scanning approach, which scans the cutoff across the range of a marker and evaluates the quality of the bisections using the silhouette statistics. The bisection that leads to the highest silhouette statistics is selected. The procedure is performed independently for each study. Using the same procedure, we quantified the proportion of CD27- CD94+ cells within naive, effector, effector memory, and central memory CD8+ T cells.
tSNE Visualization of CyTOF Data.
tSNE algorithm was used to visualize the 27-dimensional CyTOF data in a 2-dimensional plot. TSNE function in the sklearn package was used to generate the plots (perplexity = 30).
Statistical Analysis.
We performed 1,000-fold bootstrapping to test if the performances of two machine learning models are equal. In each iteration, we sampled from the testing dataset with replacement and evaluated the performance of the two models using AUC. We calculated the P value as the percentage of interactions in which a model outperforms the other. We measured the batch effect in each layer using the Kruskal–Wallis test. We used a two-way ANOVA model to test the association between a cell subset and CMV infection, in which the proportion of the cell subset is regressed on CMV infection and study.
Supplementary Material
Acknowledgments
We thank Mark Davis and Harry Greenberg for openly sharing their CyTOF measurement data on the ImmPort database, facilitating research like ours. We thank Patrick Dunn, Elizabeth Thomson, Henry Schaefer, Daniel Wong, Dmytro Lituiev, Benjamin Glicksberg, and Matthew Elliott for helpful discussions. We thank Boris Oskotsky for server support. The research reported in this publication was supported by National Institute of Allergy and Infectious Diseases Bioinformatics Support Contract HHSN316201200036W. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. T.M. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2003026117/-/DCSupplemental.
Availability of Data and Code.
The CyTOF and anti-CMV antibody titer data are publically available on ImmPort (38–46). We provided a tutorial demonstrating how to create, train, and interpret the deep CNN model (https://github.com/hzc363/DeepLearningCyTOF). All of the codes used in the study are available on GitHub (https://github.com/hzc363/Deep_learning_CyTOF_Code).
References
- 1.Spitzer M. H., Nolan G. P., Mass cytometry: Single cells, many features. Cell 165, 780–791 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakaya H. I. et al., Systems biology of vaccination for seasonal influenza in humans. Nat. Immunol. 12, 786–795 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michlmayr D. et al., Comprehensive innate immune profiling of chikungunya virus infection in pediatric cases. Mol. Syst. Biol. 14, e7862 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spitzer M. H. et al., Systemic immunity is required for effective cancer immunotherapy. Cell 168, 487–502.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rawstron A. C. et al., Reproducible diagnosis of chronic lymphocytic leukemia by flow cytometry: An European research initiative on CLL (ERIC) & European society for clinical cell analysis (ESCCA) harmonisation project. Cytometry B Clin. Cytom. 94, 121–128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ocmant A. et al., Flow cytometry for basophil activation markers: The measurement of CD203c up-regulation is as reliable as CD63 expression in the diagnosis of cat allergy. J. Immunol. Methods 320, 40–48 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Farias M. G., de Lucena N. P., Dal Bó S., de Castro S. M., Neutrophil CD64 expression as an important diagnostic marker of infection and sepsis in hospital patients. J. Immunol. Methods 414, 65–68 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Qian Y. et al., Elucidation of seventeen human peripheral blood B-cell subsets and quantification of the tetanus response using a density-based method for the automated identification of cell populations in multidimensional flow cytometry data. Cytometry B Clin. Cytom. 78 (suppl. 1), S69–S82 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu Z. et al., MetaCyto: A tool for automated meta-analysis of mass and flow cytometry data. Cell Rep. 24, 1377–1388 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Gassen S. et al., FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A 87, 636–645 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Bruggner R. V., Bodenmiller B., Dill D. L., Tibshirani R. J., Nolan G. P., Automated identification of stratifying signatures in cellular subpopulations. Proc. Natl. Acad. Sci. U.S.A. 111, E2770–E2777 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Gassen S., Vens C., Dhaene T., Lambrecht B. N., Saeys Y., FloReMi: Flow density survival regression using minimal feature redundancy. Cytometry A 89, 22–29 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Ni Choileain S. et al., The dynamic processing of CD46 intracellular domains provides a molecular rheostat for T cell activation. PLoS One 6, e16287 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu Z., Glicksberg B. S., Butte A. J., Robust prediction of clinical outcomes using cytometry data. Bioinformatics 35, 1197–1203 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arvaniti E., Claassen M., Sensitive detection of rare disease-associated cell subsets via representation learning. Nat. Commun. 8, 14825 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhattacharya S. et al., ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Sci. Data 5, 180015 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kronstad L. M., Seiler C., Vergara R., Holmes S. P., Blish C. A., Differential induction of IFN-α and modulation of CD112 and CD54 expression govern the magnitude of NK cell IFN-γ response to influenza A viruses. J. Immunol. 201, 2117–2131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miron M. et al., Human lymph nodes maintain TCF-1hi memory T cells with high functional potential and clonal diversity throughout life. J. Immunol. 201, 2132–2140 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alpert A. et al., A clinically meaningful metric of immune age derived from high-dimensional longitudinal monitoring. Nat. Med. 25, 487–495 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Stijn A. et al., Human Cytomegalovirus Infection Induces a Rapid and Sustained Change in the Expression of NK Cell Receptors on CD8+ T Cells. J. Immunol. 180, 4550–4560 (2008). [DOI] [PubMed] [Google Scholar]
- 21.van Baarle D., Kostense S., van Oers M. H. J., Hamann D., Miedema F., Failing immune control as a result of impaired CD8+ T-cell maturation: CD27 might provide a clue. Trends Immunol. 23, 586–591 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Rusu R. B., Marton Z. C., Blodow N., Dolha M., Beetz M., Towards 3D Point cloud based object maps for household environments. Robot. Auton. Syst. 56, 927–941 (2008). [Google Scholar]
- 23.Qi C. R., Su H., Mo K., Guibas L. J., PointNet: Deep learning on point sets for 3D classification and segmentation. arXiv:161200593 (2 December 2016).
- 24.van Boven M. et al., Infectious reactivation of cytomegalovirus explaining age- and sex-specific patterns of seroprevalence. PLOS Comput. Biol. 13, e1005719 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colugnati F. A., Staras S. A., Dollard S. C., Cannon M. J., Incidence of cytomegalovirus infection among the general population and pregnant women in the United States. BMC Infect. Dis. 7, 71 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fowler K. B. et al., Racial and ethnic differences in the prevalence of congenital cytomegalovirus infection. J. Pediatr. 200, 196–201.e1 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Finak G. et al., Standardizing flow cytometry immunophenotyping analysis from the human immunophenotyping consortium. Sci. Rep. 6, 20686 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ribeiro M. T., Singh S., Guestrin C., Why Should I Trust You?, (Explaining the Predictions of Any Classifier, 2016). [Google Scholar]
- 29.Lai S., Xu L., Liu K., Zhao J., “Recurrent convolutional neural networks for text classification” in Proceedings of the Twenty-Ninth AAAI Conference on Artificial Intelligence, (The AAAI Press, Palo Alto, CA, 2015), pp. 2267–2273. [Google Scholar]
- 30.He K., Xhang X., Ren S., Sun J., “Deep residual learning for image recognition” in 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), (Institute of Electrical and Electronics Engineers, Piscataway, NJ, 2016), pp. 770–778, https://ieeexplore.ieee.org/stamp/stamp.jsp?arnumber=7780459. [Google Scholar]
- 31.Zhang F. et al.; Accelerating Medicines Partnership Rheumatoid Arthritis and Systemic Lupus Erythematosus (AMP RA/SLE) Consortium , Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol. 20, 928–942 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomic A., Tomic I., Dekker C. L., Maecker H. T., Davis M. M., The FluPRINT dataset, a multidimensional analysis of the influenza vaccine imprint on the immune system. Sci. Data 6, 214 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thome J. J. C. et al., Spatial map of human T cell compartmentalization and maintenance over decades of life. Cell 159, 814–828 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin H. C. et al., Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imaging 35, 1285–1298 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vahey M. T. et al., Expression of genes associated with immunoproteasome processing of major histocompatibility complex peptides is indicative of protection with adjuvanted RTS,S malaria vaccine. J. Infect. Dis. 201, 580–589 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Ross S. A., Novak Z., Pati S., Boppana S. B., Overview of the diagnosis of cytomegalovirus infection. Infect. Disord. Drug Targets 11, 466–474 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kingma D. P., Ba J., Adam: A method for stochastic optimization. arXiv:14126980 (22 December 2014).
- 38.Davis M., T cell responses to H1N1v and a longitudinal study of seasonal influenza vaccination (TIV) SLVP015 2011. ImmPort. https://www.immport.org/shared/study/SDY112. Accessed 4 March 2019.
- 39.Greenberg H., Plasmablast response to inactivated and live attenuated influenza vaccines (TIV3/TIV3 ID/LAIV) SLVP021 2011. ImmPort. https://www.immport.org/shared/study/SDY113. Accessed 4 March 2019.
- 40.Greenberg H., Plasmablast response to inactivated and live attenuated influenza vaccines (TIV3/TIV3 ID) SLVP021 2012. ImmPort. https://www.immport.org/shared/study/SDY311. Accessed 4 March 2019.
- 41.Davis M., T cell responses to H1N1v and a longitudinal study of seasonal influenza vaccination (TIV) SLVP015 2010. ImmPort. https://www.immport.org/shared/study/SDY311. Accessed 4 March 2019.
- 42.Davis M., T cell responses to H1N1v and a longitudinal study of seasonal influenza vaccination (TIV) SLVP015 2012. ImmPort. https://www.immport.org/shared/study/SDY315. Accessed 4 March 2019.
- 43.Greenberg H., Plasmablast response to inactivated and live attenuated influenza vaccines (TIV3/TIV3 ID) in SLVP021 2013. ImmPort. https://www.immport.org/shared/study/SDY472. Accessed 4 March 2019.
- 44.Davis M., T cell responses to H1N1v and a longitudinal study of seasonal influenza vaccination SLVP015 2013. ImmPort. https://www.immport.org/shared/study/SDY478. Accessed 4 March 2019.
- 45.Davis M., Monozygotic and Dizygotic Twin Pair T-Cell Responses to Influenza Vaccination SLVP018 2010. ImmPort. https://www.immport.org/shared/study/SDY515. Accessed 4 March 2019.
- 46.Davis M., Monozygotic and Dizygotic Twin Pair T-Cell Responses to Influenza Vaccination SLVP018 2011. ImmPort. https://www.immport.org/shared/study/SDY519. Accessed 4 March 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The CyTOF and anti-CMV antibody titer data are publically available on ImmPort (38–46). We provided a tutorial demonstrating how to create, train, and interpret the deep CNN model (https://github.com/hzc363/DeepLearningCyTOF). All of the codes used in the study are available on GitHub (https://github.com/hzc363/Deep_learning_CyTOF_Code).