Significance
The fission yeast silent mating-type region provides an excellent system to ask how chromatic domains with opposite effects on gene expression coexist side by side along chromosomes and to investigate roles played by DNA elements and architectural proteins in the phenomenon. By showing that the IR-L and IR-R chromatin boundaries favor heterochromatin formation in the domain that separates them, dependent on each other and on binding sites for the architectural factor TFIIIC, our work brings to light an important function of these elements and supports the notion that similar types of interactions between boundaries might in other organisms as well stimulate heterochromatin formation in intervening chromosomal loops to actively shape gene expression landscapes.
Keywords: chromatin boundaries, gene silencing, single-cell studies, heterochromatin, fission yeast
Abstract
In fission yeast, the inverted repeats IR-L and IR-R function as boundary elements at the edges of a 20-kb silent heterochromatic domain where nucleosomes are methylated at histone H3K9. Each repeat contains a series of B-box motifs physically associated with the architectural TFIIIC complex and with other factors including the replication regulator Sap1 and the Rix1 complex (RIXC). We demonstrate here the activity of these repeats in heterochromatin formation and maintenance. Deletion of the entire IR-R repeat or, to a lesser degree, deletion of just the B boxes impaired the de novo establishment of the heterochromatic domain. Nucleation proceeded normally at the RNA interference (RNAi)-dependent element cenH but subsequent propagation to the rest of the region occurred at reduced rates in the mutants. Once established, heterochromatin was unstable in the mutants. These defects resulted in bistable populations of cells occupying alternate “on” and “off” epigenetic states. Deleting IR-L in combination with IR-R synergistically tipped the balance toward the derepressed state, revealing a concerted action of the two boundaries at a distance. The nuclear rim protein Amo1 has been proposed to tether the mating-type region and its boundaries to the nuclear envelope, where Amo1 mutants displayed milder phenotypes than boundary mutants. Thus, the boundaries might facilitate heterochromatin propagation and maintenance in ways other than just through Amo1, perhaps by constraining a looped domain through pairing.
Euchromatic and heterochromatic regions alternate along the chromosomes of eukaryotes, often separated by DNA elements with distinctive features and properties. In the fission yeast Schizosaccharomyces pombe, the 2-kb inverted repeats IR-L and IR-R mark the edges of a 20-kb heterochromatic domain that keeps mating-type information silent (SI Appendix, Fig. S1). Sharp transitions occur at these repeats in terms of gene expression (1) and histone modifications, particularly methylation of histone H3K9 precisely found between the two repeats (2).
In other organisms, similarly located DNA elements together with architectural proteins bound to them have been proposed to function as anchors for loop domains to participate in the physical and functional organization of genomes (3, 4). Whether IR-L and IR-R function in such a manner in fission yeast remains unknown. No evidence has been presented for physical or functional interactions between the two repeats, and whether they act as tethers to nuclear structures is unclear as deletion of both repeats has a very minor effect on the localization of the mating-type region at the nuclear periphery, if any (5). Not just the mode of action, but even the biological function of the repeats, is still not well-understood.
Structurally, IR-L and IR-R each contains five B-box motifs bound by the TFIIIC complex. TFIIIC is a multifunctional complex that exerts various effects on gene expression in addition to acting as a transcription factor for RNA polymerase III, including heterochromatin barrier effects and enhancer-blocking effects (6–11). At IR-L and IR-R, TFIIIC binds in the absence of RNA polymerase III (12, 13), similar to extra-TFIIIC binding sites (ETCs) in other organisms (14–16). By analogy with the S. pombe COC3 locus whose localization at the nuclear periphery depends on ETC sites, it has been speculated that TFIIIC might anchor the mating-type region to the nuclear periphery and perhaps in this way contribute to the formation of chromatin boundaries (13); however, direct evidence is lacking.
The nuclear rim protein Amo1 is another proposed anchor of the mating-type region to the nuclear periphery and in this case it has been reported that the peripheral localization of the mating-type region and its tight silencing depends on Amo1 (17). The interaction between Amo1 and the mating-type region is bridged by a protein complex called RIXC that associates along the entire silent region in a heterochromatin-dependent manner, and with the B boxes of the inverted repeats independent of heterochromatin (17). This redundant mode of association opens up the possibility that IR-L and IR-R have an Amo1-dependent tethering function even though deletion of both repeats does not detectably delocalize the mating-type region from the nuclear periphery (5). In the absence of the repeats, Amo1 interactions with heterochromatin maintain the domain at the nuclear envelope. To date, the nature of the interaction between RIXC and the B boxes, and a potential connection with TFIIIC, remains uncharacterized.
The position of the inverted repeats precisely at the edges of the heterochromatic domain is strongly suggestive of boundary activity. Yet, studies aimed at detecting heterochromatin expansion in mutants lacking the repeats or B boxes have identified only weak phenotypes unless conditions that foster heterochromatin formation are employed such as overexpressing the chromodomain protein Swi6, homolog of the major heterochromatin protein HP1 in other eukaryotes (1, 2, 13), or mutating the antisilencing factor Epe1 (18). However, early studies had suggested that the inverted repeats might have an additional function, in preventing encroachment of the heterochromatic domain by euchromatin (1, 19). In the mating-type region, RNA interference (RNAi) initiates heterochromatin formation at the cenH element located centrally (20). Heterochromatin formation catalyzed by the histone H3K9 methyltransferase Clr4 occurs rapidly within cenH and, in a subsequent step, the heterochromatic state propagates to the rest of the domain with slower kinetics (21). Once established, heterochromatin is maintained in an RNAi-independent manner (20) to control mating-type switching and to silence the mating-type donor loci (22). The aspect of heterochromatin formation or stability that would be affected by IR-L and IR-R is not known.
To better our understanding of boundary function, we created a series of IR-R mutants with which we performed multiple phenotypic characterizations. We present evidence that the boundary elements, not just their B boxes but other parts as well, participate in de novo heterochromatin establishment within the silenced domain and subsequently stabilize established heterochromatin in the following generations. Our experiments further uncover functional interactions between IR-L and IR-R that are essential to the robustness of the heterochromatic domain and that occur at least in part independent of tethering to the nuclear envelope by Amo1.
Results
Suppression of Gene Expression by the B Boxes and Other Parts of the IR-R Element.
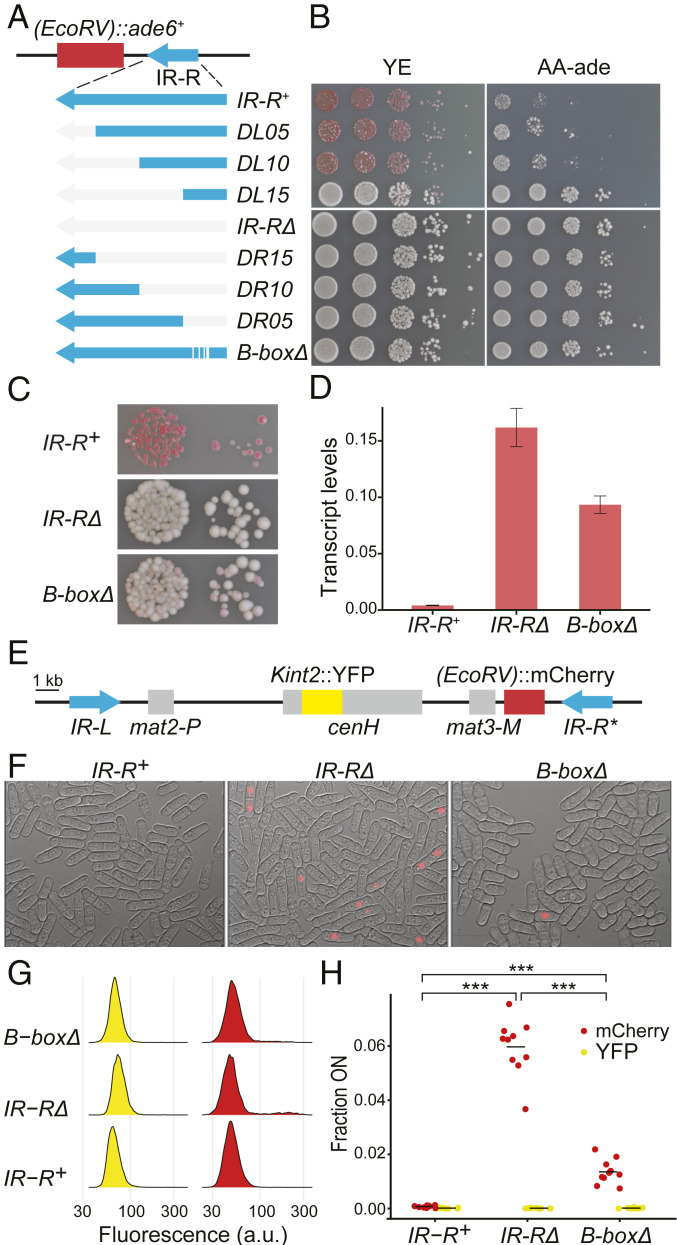
To evaluate the function of the inverted repeats and B boxes in heterochromatic gene silencing, we first examined expression of the S. pombe ade6+ gene inserted near the mat3-M mating-type cassette in strains lacking IR-R or portions of IR-R (SI Appendix, Fig. S1). Seven partial deletions were generated by removing incrementally ∼500 bp of the 2.1-kb element starting from either end or by precisely excising the B boxes (Fig. 1A and SI Appendix, Fig. S1).
Fig. 1.
Alleviated gene silencing in mutants of the IR-R boundary. (A) Deletion series and position of the (EcoRV)::ade6+ reporter gene. (B) Tenfold serial dilutions of cell suspensions were spotted onto the indicated media. Poor growth on AA-ade and red color on YE indicate (EcoRV)::ade6+ repression. (C) Zoomed-in view of YE plates. (D) (EcoRV)::ade6+ transcript analysis by qRT-PCR, normalized to act1+. Three independent cultures were processed for each strain. Error bars indicate SD. (E) Representation of the entire heterochromatic domain showing the location of the two fluorescent reporter genes Kint2::YFP within the heterochromatin nucleation center cenH and (EcoRV)::mCherry. (F) Representative images of cells with the indicated boundary mutations, overlaying red fluorescence and bright-field channels. More images are shown in SI Appendix, Fig. S4. (G) Representative histograms of fluorescence intensities for Kint2::YFP (yellow) and EcoRV)::mCherry (red) in IR-R+ and mutant strains. More histograms are shown in SI Appendix, Fig. S5. a.u., arbitrary units. (H) Fraction of the cell population expressing Kint2::YFP or (EcoRV)::mCherry for IR-R+ and mutant strains, each represented by 10 independent cultures propagated in parallel [ANOVA for (EcoRV)::mCherry, F = 223, P = 2.0 × 10−16]. Horizontal bars indicate mean values (***P < 0.001). The proportion of cells expressing (EcoRV)::mCherry was significantly higher in the IR-RΔ (P = 4.2 × 10−15) and B-boxΔ (P = 0.0005) mutants compared with the IR-R+ strain. In addition, the proportion was significantly higher in the IR-RΔ mutant than in the B-boxΔ strain (P = 1.7 × 10−14).
The reporter used, (EcoRV)::ade6+, is normally silenced by heterochromatin (23). This results in adenine auxotrophy and in the accumulation of a red pigment in colonies grown on medium with a low concentration of adenine (Fig. 1 B and C). Here, all strains lacking the IR-R B boxes (B-boxΔ, DR05, DR10, DR15, and IR-RΔ) produced light pink or white colonies on low-adenine medium and grew well in the absence of adenine, indicative of (EcoRV)::ade6+ derepression. The smallest deletions (B-boxΔ and DR05) had less of an effect than the larger deletions. In addition, nested deletions from the side of IR-R adjacent to the heterochromatic domain (DL05, DL10, and DL15) gradually derepressed (EcoRV)::ade6+, in a variegated manner, even when the five B boxes remained (Fig. 1 B and C). In particular, DL15 removing 1,622 bp of IR-R+ but not the B boxes caused a pronounced derepression. The greatest loss of silencing was observed when the whole of IR-R was deleted (IR-RΔ). These observations suggested that the B boxes are essential for tight silencing, but also that the whole of IR-R is required for optimal silencing.
As the deleted DNA was not replaced by exogenous fragments, one effect of the deletions is to reduce the distance between the (EcoRV)::ade6+ reporter and euchromatin. This raises the possibility that the difference between B-boxΔ and IR-RΔ cells stems from the reduced distance. However, in a slightly different setting in a previous study, ura4+ placed on the heterochromatic side of IR-R was derepressed to the same extent by deletion of IR-R or by replacement of IR-R with 2 kb of λ DNA (1). The fact that a level of silencing comparable to what remains here in the B-boxΔ mutant was not attained with just λ DNA argues for a B box-independent sequence-specific effect by IR-R.
For a more quantitative evaluation of (EcoRV)::ade6+ expression, qRT-PCR was performed to measure transcript levels in some of the strains (Fig. 1D). As expected from the plating assays, expression of (EcoRV)::ade6+ was greatly enhanced by deleting IR-R, resulting in a >15-fold increase in messenger RNA. (EcoRV)::ade6+ expression was enhanced to a lower degree by deleting just the B boxes (Fig. 1D). This suggests that molecular differences at the mating-type region affecting transcriptional activity exist between these two strains, but that the Ade6 protein concentration in the strains lacking the B boxes is sufficient for some degree of adenine prototrophy.
The growth assays and transcript analysis of the (EcoRV)::ade6+ reporter thus reveal a role for the B boxes in heterochromatic silencing at a population level and furthermore indicate that the B boxes operate synergistically or in parallel with the rest of IR-R to achieve full silencing.
Single-Cell Measurements Detect Two Subpopulations with Distinct Expression States in IR-R Mutants.
We turned to fluorescent reporters, a YFP (yellow fluorescent protein) gene at the Kint2 site within cenH and mCherry at the (EcoRV) site near mat3-M (Fig. 1E), to visualize silencing defects in IR-R mutants at a single-cell level. Similar to previous experiments (21), expression of both YFP and mCherry was controlled by the S. pombe ura4+ promoter and terminator of transcription, and the fluorescent proteins were targeted to the nucleus by the SV40 nuclear localization sequence. Quantitative fluorescence microscopy was carried out with the IR-R+, B-box∆, and IR-R∆ strains (Fig. 1 F–H and SI Appendix, Figs. S2–S4). Images were acquired for 10 independent cultures of each strain and fluorescence intensities were measured for 1,500 to 15,000 cells in each culture (Table 1 and SI Appendix, Fig. S1). clr4∆ mutants served as reference for the expressed state (SI Appendix, Fig. S2).
Table 1.
Fraction of the cell population expressing the kint2::YFP or (EcoRV)::mCherry reporter in wild-type and mutant cell cultures
| Genotype | YFP-ON cells, mean | mCherry-ON cells, mean | n |
| IR-R+ | 0.1 × 10−3 | 0.1 × 10−2 | 16 |
| IR-RΔ | 0.2 × 10−3 | 7.1 × 10−2 | 19 |
| B-boxΔ | 0.2 × 10−3 | 1.8 × 10−2 | 16 |
| IR-LΔ IR-R+ | 0.2 × 10−3 | 0.4 × 10−3 | 12 |
| IR-LΔ IR-RΔ | 0.4 × 10−3 | 14.7 × 10−2 | 15 |
| IR-LΔ B-boxΔ | 0.3 × 10−3 | 1.7 × 10−2 | 12 |
| amo1Δ IR-R+ | 0.5 × 10−3 | 0.1 × 10−2 | 6 |
| amo1Δ IR-RΔ | 0.5 × 10−3 | 28.5 × 10−2 | 6 |
| amo1Δ B-boxΔ | 0.1 × 10−3 | 9.3 × 10−2 | 6 |
| amo1Δ IR-LΔ IR-R+ | 0.3 × 10−3 | 0.1 × 10−2 | 6 |
| amo1Δ IR-LΔ IR-RΔ | 0.9 × 10−3 | 52.8 × 10−2 | 6 |
| amo1Δ IR-LΔ B-boxΔ | 0.2 × 10−3 | 7.9 × 10−2 | 6 |
n represents the number of independent cultures examined in each case.
Silencing of Kint2::YFP was very stringent in the three clr4+ strains examined, independent of changes at the IR-R+ element (Fig. 1 G and H). Silencing of (EcoRV)::mCherry, on the other hand, was stringent in the wild type but lost in a fraction of the cell populations for the B-box∆ and IR-R∆ mutants (Fig. 1 F–H and SI Appendix, Figs. S3 and S4). At least for the IR-RΔ mutant, where more cells were derepressed and the pattern thus more easily seen, the distribution of fluorescence intensities was bimodal with a subpopulation expressing (EcoRV)::mCherry to a level similar to the clr4Δ mutant and the other subpopulation tightly repressing the reporter similar to the IR-R+ strain (Fig. 1G and SI Appendix, Figs. S2 and S3).
The proportions of ON and OFF cells were determined in each culture by setting arbitrary intensity thresholds of ∼150 for YFP and ∼100 for mCherry based on clr4∆ and clr4+ histograms accompanied by visual examination of all micrographs to verify the calls (Fig. 1H, Table 1, and SI Appendix, Fig. S3 and Table S1). Approximately 6% of IR-RΔ cells and 1% of B-boxΔ cells showed derepression in this experiment and similar values were obtained in subsequent experiments (SI Appendix, Table S1). The more widespread derepression by IR-RΔ than by B-boxΔ reflects what had been observed with (EcoRV)::ade6+ strains (Fig. 1 A–D). In the case of (EcoRV)::ade6+, inheritance of the Ade6 protein [estimated at ∼80,000 molecules per cell when expressed from the endogenous locus (24) with a half-life of 18 h (25)] is likely to contribute to adenine prototrophy during colony growth, masking the cell-to-cell heterogeneity seen with (EcoRV)::mCherry.
The pattern of (EcoRV)::mCherry expression in IR-R mutants raised the questions of whether and how the populations of OFF and ON cells interconvert. Bimodal patterns of gene expression have been observed for other heterochromatin mutants such as for the Saccharomyces cerevisiae sir1 mutants, where defects in heterochromatin establishment and maintenance result in mixed populations of OFF and ON cells (26–28). Here we considered that heterochromatin might be labile in the absence of boundary, in which case ON cells would arise from heterochromatin loss in the OFF cells. It also appeared possible that the absence of boundary would reduce the efficiency of heterochromatin formation; reduced heterochromatin establishment would increase the proportion of ON cells in the populations. These possibilities were investigated in several ways.
Participation of the IR-R Boundary in De Novo Heterochromatin Establishment.
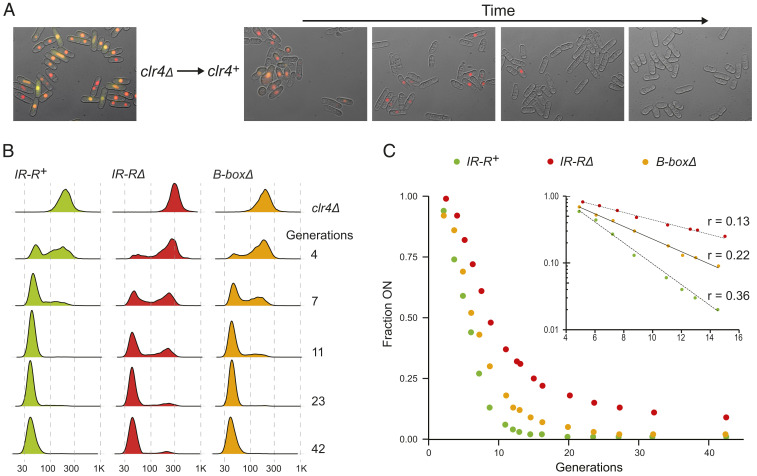
Heterochromatin establishment was monitored by reintroducing clr4+ by genetic crosses into clr4∆ strains harboring the Kint2::YFP and (EcoRV)::mCherry fluorescent reporters, using prototrophies to select for the desired recombinant progeny at spore germination as described in Materials and Methods, similar to ref. 21. Kint2::YFP (EcoRV)::mCherry cells could thus be examined from the first divisions after they acquired the clr4+ gene and subsequently at regular time intervals by sampling liquid cultures maintained in exponential phase. The rate of silencing of the fluorescent reporters was then used as a proxy for the rate of heterochromatin establishment. Rates were measured in the IR-R mutants to dissect the effect these have on heterochromatin establishment (Fig. 2 and SI Appendix, Fig. S5). In this assay, cells with no YFP signal have nucleated heterochromatin at cenH, while cells with no mCherry signal have established heterochromatin near mat3-M.
Fig. 2.
Slow de novo heterochromatin establishment caused by mutations at the IR-R boundary. (A) Outline of the experiment where clr4Δ is replaced with clr4+ through a cross, followed by imaging of liquid cultures maintained in exponential growth phase for several days. Images are from the IR-R+ strain. (B) Histograms of (EcoRV)::mCherry cell fluorescence for IR-R+, IR-RΔ, and B-boxΔ cell cultures at the indicated generations following reintroduction of clr4+. Fluorescence is indicated on the x axis in arbitrary units. (C) Fraction of cell populations expressing (EcoRV)::mCherry over time following the reintroduction of clr4+. The values used to produce the curves are in SI Appendix, Table S2. (C, Inset) Semilog plots of the same curves and the rates of exponential decay of the ON state for the three mating-type regions examined, IR-R+, IR-RΔ, and B-boxΔ.
Heterochromatin establishment at cenH was fast and not affected by IR-R mutations (SI Appendix, Fig. S5 and Table S2). (Kint2)::YFP was silenced in the vast majority of cells within 10 generations, reflecting what had been observed previously for the wild type (21) and consistent with our steady-state measurements where no expression of (Kint2)::YFP was detected (Fig. 1 G and H and SI Appendix, Fig. S4).
In contrast, silencing of (EcoRV)::mCherry occurred at different rates in the three strains (Fig. 2 B and C and SI Appendix, Table S2). For cells with wild-type IR-R, silencing was almost fully established at the population level 20 generations after spore germination. In the IR-RΔ strain, even after ∼40 generations, 10% of cells still expressed the normally silent region (Fig. 2C). In a strain lacking the B boxes, heterochromatin establishment occurred at a rate intermediate between the wild type and IR-RΔ (Fig. 2C). The mean lifetimes of the ON state (2.8 generations for IR-R+; 7.7 for IR-RΔ; 4.5 for B-boxΔ) again reflect the differences previously observed between the B-box∆ and IR-R∆ mutants and further support the notion that molecular differences exist between the two mutants, in this case resulting in different kinetics of heterochromatin establishment.
Requirement for the IR-R Boundary in Heterochromatin Maintenance.
The fact that a proportion of (EcoRV)::mCherry clr4+ cells expressed mCherry in cultures started from single cells (Fig. 1 F–H), together with the adenine prototrophy of (EcoRV)::ade6+ boundary mutants (Fig. 1 B–D), suggested that heterochromatin might be unstable in boundary mutants, in addition to being established at a slower rate than for wild-type cells. To measure stability, clr4+ cells with the Kint2::YFP and (EcoRV)::mCherry reporters were placed under small agar patches in minimal medium and imaged by time-lapse fluorescence microscopy at each generation for six or seven generations. The formation of 22 microcolonies for each strain was observed in this way. Both gains and losses of the silenced state were observed as well as clonal inheritance of each state for multiple divisions (Movies S1–S9).
Derepression of YFP was never observed under these conditions, confirming that heterochromatin at cenH is unaffected by IR-R. As expected, in wild-type cells, heterochromatin was also stable at EcoRV. No case of (EcoRV)::mCherry derepression was observed among the imaged cells (n = 685 divisions). In contrast, derepression of (EcoRV)::mCherry occurred readily in the boundary mutants. Each event was scored by visual inspection, allowing an estimate of the rates of silencing loss. This showed that the derepression rate was increased to a similar extent in the B-box∆ and IR-R∆ mutants, respectively, 3.5 × 10−2 (n = 482 divisions) and 3.1 × 10−2 (n = 655 divisions) loss of silencing events per cell division.
Role of IR-L in Setting Up the Right Domain Boundary.
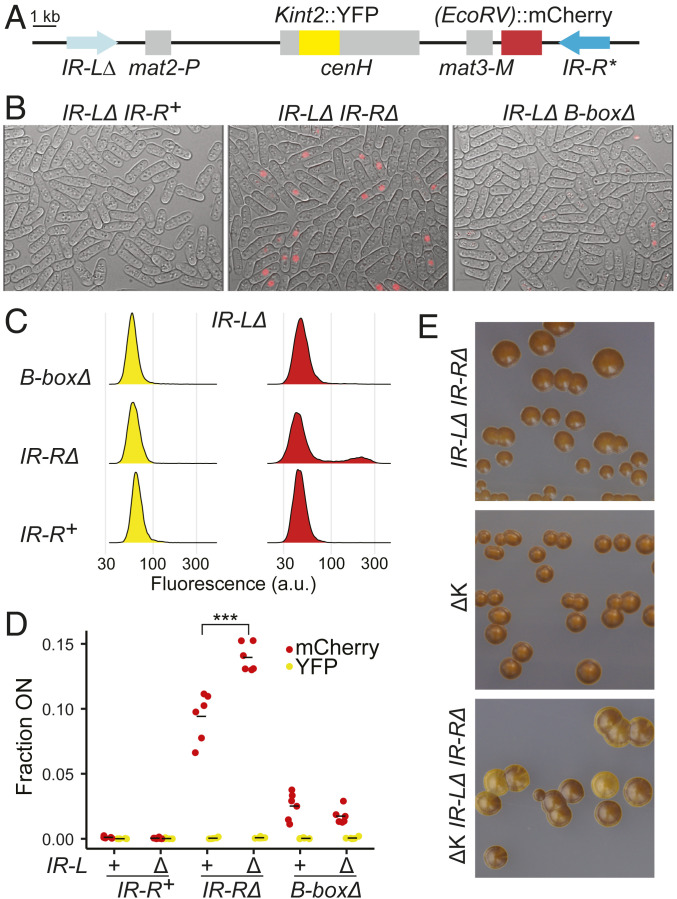
The distribution profiles of (EcoRV)::mCherry expression gave us the opportunity to investigate a potential interplay between the left and right boundaries by examining the expression of (EcoRV)::mCherry in strains lacking IR-L (Fig. 3 A–D, Table 1, and SI Appendix, Figs. S6 and S7 and Table S1). It had previously been observed that deletion of IR-L derepressed a reporter gene near the mat2-P mating-type cassette [(XbaI)::ura4+] but did not detectably affect (EcoRV)::ura4+ expression near mat3-M (1). Consistently, here (EcoRV)::mCherry remained silent when IR-L but not IR-R was deleted. When both IR-L and IR-R were deleted, a synergistic derepression of (EcoRV)::mCherry occurred. Fluorescence intensity did not increase in individual mCherry-positive cells, but higher proportions of mCherry-positive cells were present in the double mutant compared with the mutant lacking only IR-R. The effect was seen in all cultures propagated in parallel for this specific experiment and corroborated by subsequent measurements, for a total of 16 cultures of IR-R∆ cells and 12 cultures of IR-L∆ IR-R∆ cells (SI Appendix, Table S1). On average, compared with ∼7% derepressed cells in the IR-RΔ mutant, ∼15% were derepressed by the double-boundary deletion. In contrast, no synergistic effect was detected when only the B boxes were deleted together with IR-L (Fig. 3 B–D, Table 1, and SI Appendix, Figs. S3–S7 and Table S1). Thus, IR-L participates in the repression of (EcoRV)::mCherry, located 20 kb away, close to the other edge of the heterochromatic domain.
Fig. 3.
Synergistic effects of the IR-L and IR-R boundaries on heterochromatic silencing at (EcoRV)::mCherry. (A) Representation of the mating-type region in this experiment showing the deletion of IR-L in light blue. (B) Images of cells combining IR-LΔ with mutations at IR-R, overlaying red fluorescence and bright-field channels. Individual channels are shown in SI Appendix, Fig. S6. (C) Representative histograms of fluorescence intensities for mutants combining IR-LΔ with mutations at IR-R. More histograms are shown in SI Appendix, Fig. S5. (D) Same as Fig. 1A for strains with the indicated genotypes, examining six independent cultures for each genotype (ANOVA, F = 198.01, P = 2.22 × 10−16, ***P < 0.001). The proportion of cells expressing (EcoRV)::mCherry was significantly higher in the IR-LΔ IR-RΔ mutant compared with the IR-RΔ mutant (P = 1.2 × 10−7). (E) Iodine staining of colonies showing the dark ΔK mutant epitype in the presence or absence of boundaries. Light staining reflects poor mating-type switching due to heterochromatin defects.
The Boundaries Are Required for RNAi-Independent Heterochromatin in cenH Mutants.
Mutants lacking the cenH element (ΔK mutants) display a bistable state where cells are either similar to wild type or lack heterochromatin over the mat2-P mat3-M region (29, 30). Each state has a mean half-life greater than 1,000 generations (30), and thus the two phenotypes are clearly distinguishable not only at the colony level but also upon restreaking of colonies. The bistability is believed to reflect a de novo heterochromatin establishment defect in ΔK cells due to loss of the RNAi-dependent heterochromatin nucleation center cenH (20); establishment would still occur, but inefficiently. Here, we wondered whether deletion of the domain boundaries would affect the bistable phenotype of ΔK mutants. We created a strain identical to one of the original ΔK mutants (30) but lacking both boundaries (Fig. 3E).
The two phenotypes of the ΔK mutant are easily observed on sporulation plates by iodine staining of colonies. In this procedure, spores, but not vegetative cells, are stained dark by iodine vapors (31). Because heterochromatin is required for efficient mating-type switching and thus for homothallic mating and sporulation (22, 32, 33), the ΔK mutant epitype with established heterochromatin forms brown colonies similar to wild-type (h90) strains, while the ΔK mutant epitype lacking heterochromatin forms light mottled colonies like clr4Δ strains (29, 30). We found that in a ΔK mutant lacking both boundaries the dark epitype could not be readily propagated (Fig. 3E). Colonies were not as uniformly stained as when the boundaries were present and, upon restreaking, light colonies and sectors appeared at high frequency, which was not the case when the boundaries were present (Fig. 3E). Thus, a stable heterochromatic state could not be attained in the absence of boundaries. Similarly, Wang and Moazed found that the boundaries stabilized heterochromatin induced by artificially tethering Clr4 (34).
Protein Factors: Amo1, Bqt4, Lem2, and Man1.
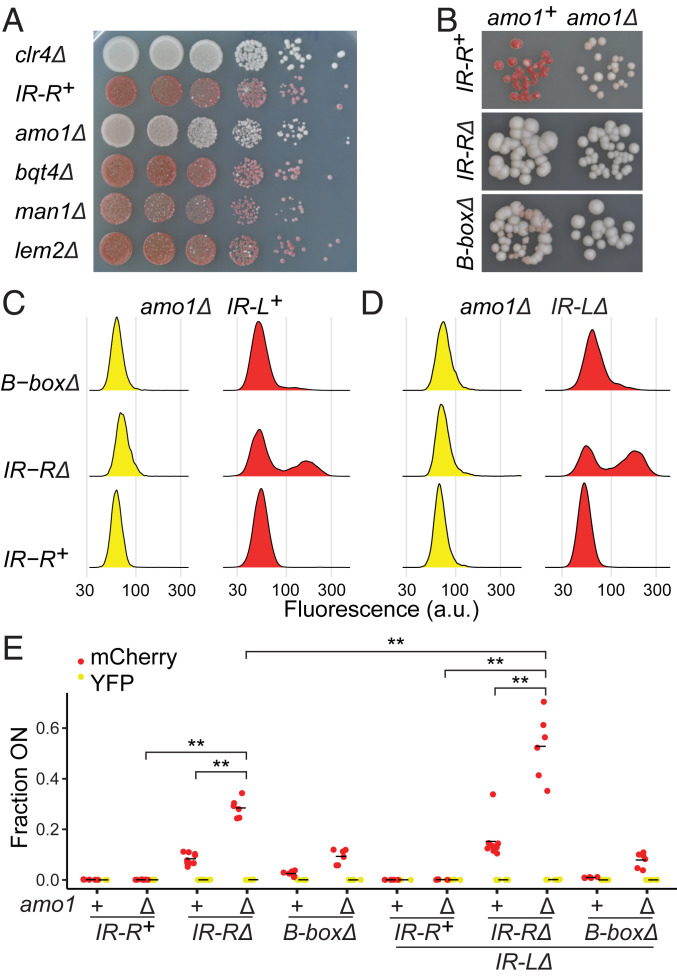
Recently, two studies have suggested that the localization of the mating-type region at the nuclear periphery might be part of its silencing mechanism. First, it was reported that H3K9me is considerably reduced in the mating-type region in mutants lacking the nuclear membrane protein Bqt4 (35). Second, mutants in the nuclear rim protein Amo1 were also reported to reduce H3K9me in the mat2-mat3 region, in particular outside cenH, attributed to defective maintenance (17). In the case of Amo1, reduced H3K9me was accompanied by partial delocalization from the nuclear periphery (17). In the case of Bqt4, reduced H3K9me was accompanied by partial delocalization from the nuclear periphery specifically during replication (35). Because anchoring at the nuclear periphery by the boundary elements could be part of their mechanism of action, we conducted an epistasis analysis to investigate the effects of combining mutations at IR-R with mutations in potential anchors at the nuclear envelope. We included a lem2Δ mutant (36), for which Barrales et al. previously detected a 5- to 25-fold increase of (EcoRV)::ura4+ transcript (37), a reporter at the same location as (EcoRV)::ade6+, as well as a man1Δ mutant for which no effect on heterochromatin has been reported, to our knowledge.
Silencing of the sensitive (EcoRV)::ade6+ reporter appeared unaltered in the bqt4Δ, lem2Δ, and man1Δ mutants (Fig. 4A). While somewhat surprising in the cases of bqt4Δ and lem2Δ, silencing near mat2-P is also maintained in these mutants (17). We thus focused on amo1Δ, which showed reduced silencing of (EcoRV)::ade6+ (Fig. 4 A and B), consistent with its effect near mat2-P (17). (EcoRV)::ade6+ derepression by amo1Δ was not as pronounced as (EcoRV)::ade6+ derepression by IR-RΔ according to the light pink colony color of amo1Δ colonies, and there appeared to be cumulative defects in all cases when combining partial deletions of IR-R and amo1Δ, as shown for B-boxΔ in Fig. 4B. Quantitative analysis with (EcoRV)::mCherry confirmed these conclusions as fluorescent cells were more abundant in the amo1Δ IR-RΔ and amo1Δ B-boxΔ mutants than in each single mutant (Fig. 4E, Table 1, and SI Appendix, Figs. S3 and S4 and Table S1), with amo1Δ having a very small effect on its own [on the order of 0.1% of amo1Δ cells showed (EcoRV)::mCherry expression by fluorescence microscopy]. Furthermore, (EcoRV)::mCherry expression was also detected in a greater proportion of cells in the amo1Δ IR-LΔ IR-RΔ mutant than in the IR-LΔ IR-RΔ mutant (Fig. 4E, Table 1, and SI Appendix, Figs. S6 and S7 and Table S1). Altogether, these phenotypes indicate Amo1-independent functions of the boundaries in heterochromatic silencing, without ruling out some form of combined action. In addition, the great proportion of derepressed cells in the amo1Δ IR-LΔ IR-RΔ mutant (53% on average), greater than in the amo1Δ IR-RΔ mutant (29% on average), underscores the effect of IR-L at a distance.
Fig. 4.
Independent effects of the nuclear envelope protein Amo1 and boundary elements on heterochromatin integrity. (A) Four nuclear envelope proteins were tested for their requirement in the silencing of (EcoRV)::ade6+ by plating assay. As in Fig. 1, red colony color denotes repression, and pink or white denotes expression. A clr4Δ mutant is shown for comparison. (B) Epistasis analysis of amo1Δ and mutations at IR-R conducted with (EcoRV)::ade6+. (C–E) Epistasis analysis of amo1Δ and mutations at IR-R conducted with (EcoRV)::mCherry as in Fig. 1 G and H but examining six independent cultures for each genotype (nine for IR-RΔ and IR-LΔ IR-RΔ) [Kruskal–Wallis for (EcoRV)::mCherry, H = 73.13, P = 3.1 × 10−11, **P < 0.01]. The proportion of cells expressing (EcoRV)::mCherry was significantly higher in the IR-RΔ amo1Δ mutant than in the IR-R+ amo1Δ (P = 0.003) and IR-RΔ (P = 0.002) mutants. It was also significantly higher in the IR-RΔ IR-LΔ amo1Δ mutant than in the IR-LΔ IR-R+ amo1Δ (P = 0.003) and IR-LΔ IR-RΔ (P = 0.002) strains. Finally, the proportion was significantly higher in the IR-LΔ IR-RΔ amo1Δ mutant than the IR-RΔ amo1Δ strain (P = 0.003).
Discussion
Our experiments provide evidence that the IR-L and IR-R boundary elements in the fission yeast mating-type region facilitate heterochromatin propagation and maintenance over the silenced domain. They also reveal subelements that can be functionally separated, indicating B box-dependent and -independent mechanisms of boundary activity. How might boundaries facilitate the propagation of heterochromatin? The tethering of IR-L and IR-R to the nuclear envelope, physical interactions between them, or the binding of protein complexes might affect chromatin dynamics in various ways that we discuss below. A mechanistic clue comes from the fact that the left and right repeats functionally interact to form the heterochromatic domain.
All our experiments point to gene silencing being alleviated in boundary mutants. Not all cells were equally affected in populations; quite the contrary, single-cell measurements uncovered the coexistence of repressed and derepressed subpopulations capable of interconverting. Cells expressing the (EcoRV)::mCherry reporter appeared in the descendants of repressed cells and clonal expression was observed (Movies S5 and S8), indicating that heterochromatin maintenance occasionally fails in boundary mutants and that the resulting expressed chromatic state is epigenetically inherited. Consistently, the heterochromatic state of the ΔK mutant was also destabilized by the absence of boundaries (Fig. 3E).
Boundary mutations also impaired the de novo establishment of heterochromatin, in a step subsequent to heterochromatin nucleation at cenH by RNAi (Fig. 2 and SI Appendix, Fig. S5). Shared mechanisms are expected to establish and maintain chromatic states; for example, the read–write activity of chromatin-modifying enzymes, or nucleosome turnover, will affect both. Here, while silencing was lost at similar rates in mutants lacking the whole of IR-R or just the B boxes, the de novo establishment was differentially affected by the two mutations, more profoundly by the absence of IR-R. Thus, IR-R appears to have a B box-dependent action affecting both establishment and maintenance, and a B box-independent action more specific for de novo establishment, altogether resulting in different distributions of ON and OFF cells in IR-RΔ and B-boxΔ strains. The incremental loss of boundary activity caused by the nested deletions of IR-R in the part that does not contain the B boxes is reminiscent of the progressive loss of activity of ars elements subjected to similar deletions (38). This might be relevant to the mode of action of IR-R as IR-R possesses ars activity on plasmids (1) and exerts a control on DNA replication in the chromosome (39), suggesting the boundaries might couple DNA and chromatin replication. This is also suggested by the spectrum of mutations affecting heterochromatic silencing at (EcoRV)::ade6+ that define many replication factors (40).
In the context of the literature, we considered that a B box-dependent action of the boundaries might occur through Amo1. Amo1 and its interacting partner RIXC have been proposed to tether the mating-type region to the nuclear periphery in a subnuclear compartment where reduced histone turnover stabilizes heterochromatin (17). The tethering would rely on the association of RIXC with heterochromatin throughout the silent domain and with the B boxes independent of heterochromatin (17). In both RIXC and Amo1 mutants the mating-type region is displaced from the nuclear periphery and silencing defects are observed (17). Here, we observed a small derepression of (EcoRV)::ade6+ in the amo1Δ mutant (Fig. 4), consistent with the originally detected defect for amo1Δ in a sensitized background lacking the REII silencer near mat2-P (17). Derepression by amo1Δ was less pronounced than for the B-boxΔ mutant and cumulative effects were observed when combining the amo1Δ and B-boxΔ mutations (Fig. 4 and Table 1). Thus, while confirming a requirement for Amo1 for tight heterochromatic silencing, these phenotypes also suggest different mechanisms of action for Amo1 and the B-box elements that would account for the B box-dependent silencing of (EcoRV)::ade6+ and (EcoRV)::mCherry in amo1Δ cells. One possibility is that other factors such as TFIIIC or Sap1 still exert a repression through the B boxes in the absence of Amo1.
Even larger phenotypic differences were observed between amo1Δ and full boundary-deletion mutants as well as increased derepression when combining amo1Δ with IR-RΔ or IR-LΔ IR-RΔ (Figs. 4 and 5 and Table 1). Thus, not just the B boxes but the whole boundaries as well retain their functionality in the absence of Amo1. Conversely, repression by Amo1 can occur in the absence of boundaries, this last effect plausibly due to tethering of the mating-type region to the nuclear periphery by RIXC and Amo1 through the rest of the heterochromatic domain (Fig. 5).
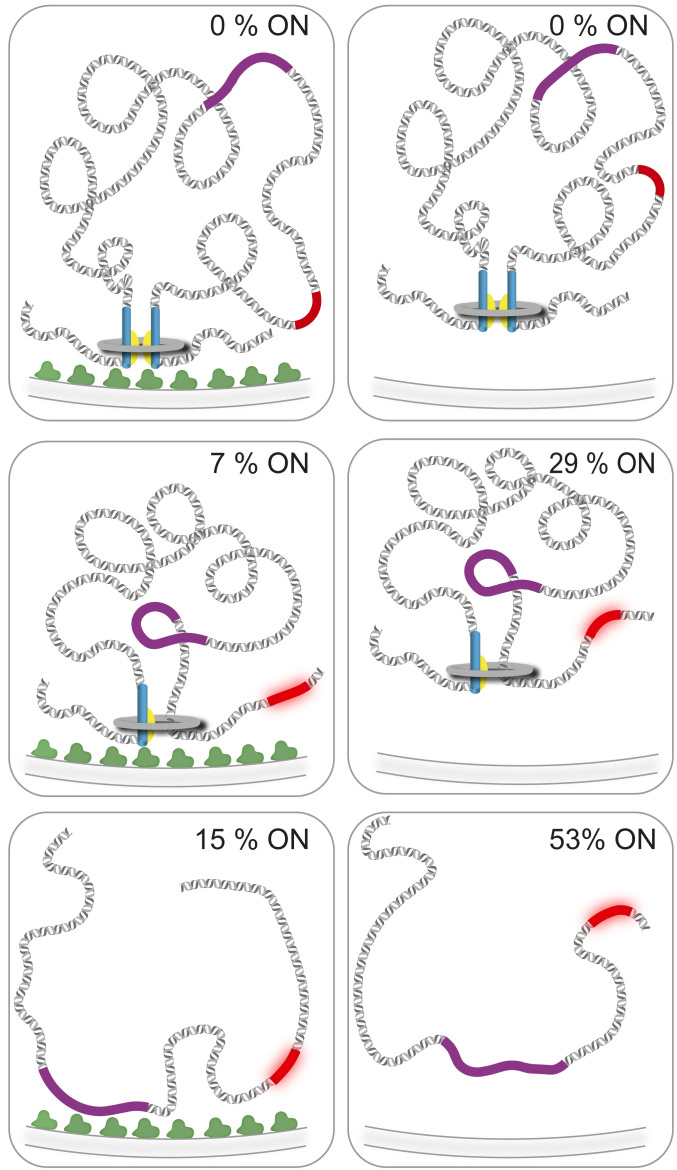
Fig. 5.
Loop domain model for the mating-type region. The mating-type region is anchored at the nuclear periphery by the Amo1 protein (in green, Left; Right, amo1Δ strains). (Top) The two boundaries (in blue, with TFIIIC binding sites in yellow) pair to define a heterochromatic domain in which the mCherry reporter (in red) is silenced independent of Amo1. A hypothetical condensin ring is represented in gray. (Middle) In the absence of the IR-R boundary, the mCherry gene occasionally slides out of the loop domain, resulting in heterochromatin loss and mCherry expression in some cells. Heterochromatin loss is compounded by delocalization from the nuclear envelope and higher histone turnover in the absence of Amo1. (Bottom) When both boundaries are deleted, the loop domain falls apart. Heterochromatin nucleated at cenH (purple) maintains nuclear envelope association through Amo1 but heterochromatin propagation and maintenance at mCherry are inefficient. Loss of silencing is again compounded by the absence of Amo1.
A clue for how the boundaries might function is provided by the synergy between IR-L and IR-R (Figs. 3 and 4, Table 1, and SI Appendix, Figs. S3, S4, S6, and S7 and Table S1). Derepression of (EcoRV)::mCherry at a distance by IR-LΔ, in a situation where heterochromatic silencing remains at Kint2::YFP in the intervening region, argues against the boundaries functioning as individual roadblocks against a linear spread of chromatic modifications, here euchromatic modifications spreading inward in the absence of boundaries. The synergy rather suggests the existence of a loop domain between the two boundaries, similar to models of boundary activity in Drosophila and other organisms (3). The mating-type region is delocalized from the nuclear periphery in Amo1 mutants (17), yet we find that silencing is not affected nearly to the same extent by amo1Δ as by boundary deletions, indicating attachment to the nuclear envelope is not how the domain would be constrained. Instead, homologous pairing between the two elements together with TFIIIC binding might define the bottom of the loop (Fig. 5). In S. pombe, as in other organisms, condensin localizes to TFIIIC binding sites (12, 41–43). A loop constrained at its base by a condensin ring, or possibly by the long coil-coiled protein Sap1 detected at that location (21, 44), could create a domain of increased interactions, boosting positive feedbacks for nucleosomal modifications and heterochromatin propagation from the cenH nucleation center present in the loop (44) (Fig. 5). As it forms, heterochromatin would be further compacted by cohesin (45, 46) and altered nucleosomal contacts (47). Deletion of a single boundary would not preclude formation of the proposed looped domain, as a loop could be initiated and reeled in from the intact side (47) but the edge of the domain on the side lacking the boundary would not be precisely positioned, resulting in the observed variegation of the silencing phenotype on that side only, as observed (1). Only when both boundaries are deleted would silencing be affected at a distance due to loop disruption. Even then, silencing would remain in a proportion of cells as heterochromatin formation would still be initiated at cenH which, among other effects, would maintain the mating-type region at the nuclear envelope (Fig. 5).
In the budding yeast S. cerevisiae, a transfer RNA (tRNA) gene near the HMR silent mating-type cassette at one end of chromosome III facilitates long-range interactions with the HML locus at the other end of chromosome III and stabilizes gene silencing at both locations (48). Moreover, nucleoporins recruit HMR to the nuclear periphery through this tRNA gene and thereby contribute to HMR silencing (49). While the proposed loop domain for the S. pombe mating-type region (Fig. 5) helps understand observations specific to the fission yeast system, it thus appears possible that the two organisms rely on partially shared mechanisms to facilitate heterochromatin formation.
Materials and Methods
Growth Conditions and Genetic and Molecular Manipulations of S. pombe.
Media (YES, EMM2, MSA) and genetic manipulations including transformations, genetic crosses, and chromosomal DNA preparations were according to published fission yeast protocols (50–53). YE (5 g/L yeast extract and 30 g/L glucose, a rich medium with low adenine concentration) and AA-ade [dropout medium lacking adenine (54)] were used to assay (EcoRV)::ade6+ expression. Plasmid and strain constructions are detailed in SI Appendix. Strain genotypes are presented in SI Appendix, Table S3 and oligonucleotides are listed in SI Appendix, Table S4.
Heterochromatin Establishment Experiments.
Reintroduction of the clr4+ gene was conducted essentially as in ref. 21. Strains with the mating-type regions of interest in a clr4∆ arg12∆ background, SC32, SC36, and SC37, were crossed with the clr4+ arg12+ strain PA90. In the progeny, arg12+ was used to select for the tightly linked clr4+ gene and (XmnI)::ade6+ on the euchromatic side of IR-L+ was used to select for the mating-type region originating from SC32, SC36, or SC37. Practically, isolated colonies of all strains were patched onto rich medium (YES) at the beginning of the experiment and the patches were used the following day to prepare mating mixes on sporulation plates (MSA supplemented with adenine, leucine, and uracil; MSA uses arginine as nitrogen source). Two to three days later, free spores were prepared by resuspending each mating mix in 1 mL β-glucuronidase solution (Roche Diagnostics) diluted 1:100 in H2O and filter-sterilized, and incubating at 30 °C overnight to lyse vegetative cells and degrade ascus walls. The free spores were pelleted, washed with H2O, resuspended in 1 mL EMM2 minimal medium supplemented with uridine but lacking arginine and adenine, and placed at 30 °C to induce germination of Ade+ Arg+ spores. Liquid cultures inoculated with the germinated spores were kept in a shaking incubator at 30 °C and maintained in exponential phase for several days by diluting them in EMM2 containing uridine, keeping a culture volume of 1 mL throughout the experiment. Cell counting and imaging were performed at regular intervals to produce growth curves and to measure fluorescence. Crosses were monitored by plating the original random spore preparations on nonselective YES plates and by scoring all markers for 100 spore colonies, and subsequently by plating and genotyping cells from the growing cultures.
Acquisition and Analysis of Fluorescence Images.
Liquid cultures were set up in 1 to 2 mL EMM2 minimum medium supplemented as needed and incubated with vigorous agitation at 30 °C. The cultures were inoculated with free spore preparations for heterochromatin establishment experiments or with single colonies to measure the proportion of cells expressing Kint2::YFP or (EcoRV)::mCherry in strains of interest, in which case on the order of 6 to 10 cultures were inoculated in parallel for each strain. Fluorescence images were acquired with an Xcyto 10 Quantitative Cell Imager run with XcytoView 1.0.109.0 (ChemoMetec) by placing 50 µL culture onto a disposable two-chamber glass slide (ChemoMetec) and imaging 10,000 cells or 50 fields. Exposure times of 1 s were used for both YFP and mCherry. To exclude spores present at the early time points of heterochromatin establishment experiments and occasional cell aggregates, gating was imposed based on cell perimeter. Images were analyzed with XcytoView 1.0.109.0 software and visually inspected to introduce corrections when necessary. Files retrieved in .fcs format were further analyzed and plotted in R. A Nikon Ti Eclipse microscope was used for time-lapse microscopy and other fluorescence images were acquired with a Zeiss Axioplan microscope, in both cases using 100× objectives.
Heterochromatin Maintenance Experiments.
Time-lapse microscopy was performed by imaging isolated cells and their descendants every 3 h. For each strain, SC50, SC51, and SC52, ∼2 µL cell suspension was spotted onto a small agarose patch in supplemented EMM2 medium, left to dry a few minutes, and inverted onto a 24 × 50-mm no. 1.5 microscope coverslip that could accommodate the three patches. The coverslip was used to close a small homemade imaging chamber and positioned on the stage of the Nikon Ti Eclipse microscope enclosed in a temperature-controlled chamber kept at 30 °C. Approximately 10 positions were chosen under each patch using NIS-Elements software and images were collected every 3 h for 20 h in the bright-field, YFP, and mCherry channels, using a combination of autofocus and the Perfect Focus System to maintain the focus. The resulting time-lapse series were hand-counted to determine the number of total cell divisions and the number of cell divisions in which the fluorescent reporters had been turned on.
Supplementary Material
Acknowledgments
We thank current and former laboratory members for discussions and funding agencies, the Novo Nordisk Foundation (Grant NNF19OC0058686 to G.T. and Scholarship to S.J.C.) and Carlsberg Foundation (Grants CF15-0853 and CF19-0200 to G.T.), for supporting our work. We are also grateful to Søren Kjærulff and Rune Troelsgaard Pedersen at ChemoMetec for help in using an Xcyto 10 Quantitative Cell Imager. Strains were provided by Shelley Sazer (SS2343 for the lem2::natMX4 allele) and National BioResource Project Yeast (FY33658 for the bqt4::natMX6 allele).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010062117/-/DCSupplemental.
Data Availability.
All study data are included in the article, SI Appendix, and movies.
References
- 1.Thon G., Bjerling P., Bünner C. M., Verhein-Hansen J., Expression-state boundaries in the mating-type region of fission yeast. Genetics 161, 611–622 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Noma K., Allis C. D., Grewal S. I., Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293, 1150–1155 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Chetverina D. et al., Boundaries of loop domains (insulators): Determinants of chromosome form and function in multicellular eukaryotes. BioEssays 39, 1600233 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rao S. S. et al., A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alfredsson-Timmins J., Henningson F., Bjerling P., The Clr4 methyltransferase determines the subnuclear localization of the mating-type region in fission yeast. J. Cell Sci. 120, 1935–1943 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Donze D., Adams C. R., Rine J., Kamakaka R. T., The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 13, 698–708 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simms T. A. et al., TFIIIC binding sites function as both heterochromatin barriers and chromatin insulators in Saccharomyces cerevisiae. Eukaryot. Cell 7, 2078–2086 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raab J. R. et al., Human tRNA genes function as chromatin insulators. EMBO J. 31, 330–350 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ebersole T. et al., tRNA genes protect a reporter gene from epigenetic silencing in mouse cells. Cell Cycle 10, 2779–2791 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott K. C., Merrett S. L., Willard H. F., A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr. Biol. 16, 119–129 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Scott K. C., White C. V., Willard H. F., An RNA polymerase III-dependent heterochromatin barrier at fission yeast centromere 1. PLoS One 2, e1099 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwasaki O., Tanaka A., Tanizawa H., Grewal S. I., Noma K., Centromeric localization of dispersed Pol III genes in fission yeast. Mol. Biol. Cell 21, 254–265 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noma K., Cam H. P., Maraia R. J., Grewal S. I., A role for TFIIIC transcription factor complex in genome organization. Cell 125, 859–872 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Kirkland J. G., Raab J. R., Kamakaka R. T., TFIIIC bound DNA elements in nuclear organization and insulation. Biochim. Biophys. Acta 1829, 418–424 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donze D., Extra-transcriptional functions of RNA polymerase III complexes: TFIIIC as a potential global chromatin bookmark. Gene 493, 169–175 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Stutzman A. V., Liang A. S., Beilinson V., Ikegami K., Transcription-independent TFIIIC-bound sites cluster near heterochromatin boundaries within lamina-associated domains in C. elegans. Epigenetics Chromatin 13, 1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holla S. et al., Positioning heterochromatin at the nuclear periphery suppresses histone turnover to promote epigenetic inheritance. Cell 180, 150–164.e15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia J. F., Al-Sady B., Madhani H. D., Intrinsic toxicity of unchecked heterochromatin spread is suppressed by redundant chromatin boundary functions in Schizosacchromyces pombe. G3 (Bethesda) 5, 1453–1461 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh G., Klar A. J., The 2.1-kb inverted repeat DNA sequences flank the mat2,3 silent region in two species of Schizosaccharomyces and are involved in epigenetic silencing in Schizosaccharomyces pombe. Genetics 162, 591–602 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall I. M. et al., Establishment and maintenance of a heterochromatin domain. Science 297, 2232–2237 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Obersriebnig M. J., Pallesen E. M., Sneppen K., Trusina A., Thon G., Nucleation and spreading of a heterochromatic domain in fission yeast. Nat. Commun. 7, 11518 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thon G., Maki T., Haber J. E., Iwasaki H., Mating-type switching by homology-directed recombinational repair: A matter of choice. Curr. Genet. 65, 351–362 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thon G., Bjerling K. P., Nielsen I. S., Localization and properties of a silencing element near the mat3-M mating-type cassette of Schizosaccharomyces pombe. Genetics 151, 945–963 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marguerat S. et al., Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell 151, 671–683 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christiano R., Nagaraj N., Fröhlich F., Walther T. C., Global proteome turnover analyses of the yeasts S. cerevisiae and S. pombe. Cell Rep. 9, 1959–1965 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pillus L., Rine J., Epigenetic inheritance of transcriptional states in S. cerevisiae. Cell 59, 637–647 (1989). [DOI] [PubMed] [Google Scholar]
- 27.Xu E. Y., Zawadzki K. A., Broach J. R., Single-cell observations reveal intermediate transcriptional silencing states. Mol. Cell 23, 219–229 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Dodson A. E., Rine J., Heritable capture of heterochromatin dynamics in Saccharomyces cerevisiae. eLife 4, e05007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grewal S. I., Klar A. J., Chromosomal inheritance of epigenetic states in fission yeast during mitosis and meiosis. Cell 86, 95–101 (1996). [DOI] [PubMed] [Google Scholar]
- 30.Thon G., Friis T., Epigenetic inheritance of transcriptional silencing and switching competence in fission yeast. Genetics 145, 685–696 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bresch C., Müller G., Egel R., Genes involved in meiosis and sporulation of a yeast. Mol. Gen. Genet. 102, 301–306 (1968). [DOI] [PubMed] [Google Scholar]
- 32.Jakočiūnas T., Holm L. R., Verhein-Hansen J., Trusina A., Thon G., Two portable recombination enhancers direct donor choice in fission yeast heterochromatin. PLoS Genet. 9, e1003762 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maki T., Ogura N., Haber J. E., Iwasaki H., Thon G., New insights into donor directionality of mating-type switching in Schizosaccharomyces pombe. PLoS Genet. 14, e1007424 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X., Moazed D., DNA sequence-dependent epigenetic inheritance of gene silencing and histone H3K9 methylation. Science 356, 88–91 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ebrahimi H., Masuda H., Jain D., Cooper J. P., Distinct “safe zones” at the nuclear envelope ensure robust replication of heterochromatic chromosome regions. eLife 7, e32911 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gonzalez Y., Saito A., Sazer S., Fission yeast Lem2 and Man1 perform fundamental functions of the animal cell nuclear lamina. Nucleus 3, 60–76 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barrales R. R., Forn M., Georgescu P. R., Sarkadi Z., Braun S., Control of heterochromatin localization and silencing by the nuclear membrane protein Lem2. Genes Dev. 30, 133–148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maundrell K., Hutchison A., Shall S., Sequence analysis of ARS elements in fission yeast. EMBO J. 7, 2203–2209 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toteva T. et al., Establishment of expression-state boundaries by Rif1 and Taz1 in fission yeast. Proc. Natl. Acad. Sci. U.S.A. 114, 1093–1098 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jahn L. J. et al., Dependency of heterochromatin domains on replication factors. G3 (Bethesda) 8, 477–489 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuen K. C., Slaughter B. D., Gerton J. L., Condensin II is anchored by TFIIIC and H3K4me3 in the mammalian genome and supports the expression of active dense gene clusters. Sci. Adv. 3, e1700191 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haeusler R. A., Pratt-Hyatt M., Good P. D., Gipson T. A., Engelke D. R., Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev. 22, 2204–2214 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bada M., Walther D., Arcangioli B., Doniach S., Delarue M., Solution structural studies and low-resolution model of the Schizosaccharomyces pombe sap1 protein. J. Mol. Biol. 300, 563–574 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Dodd I. B., Micheelsen M. A., Sneppen K., Thon G., Theoretical analysis of epigenetic cell memory by nucleosome modification. Cell 129, 813–822 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Sanulli S. et al., HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature 575, 390–394 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nonaka N. et al., Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol. 4, 89–93 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Ganji M. et al., Real-time imaging of DNA loop extrusion by condensin. Science 360, 102–105 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamdani O. et al., tRNA genes affect chromosome structure and function via local effects. Mol. Cell. Biol. 39, e00432-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruben G. J. et al., Nucleoporin mediated nuclear positioning and silencing of HMR. PLoS One 6, e21923 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petersen J., Russell P., Growth and the environment of Schizosaccharomyces pombe. Cold Spring Harb. Protoc. 2016, pdb.top079764 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murray J. M., Watson A. T., Carr A. M., Transformation of Schizosaccharomyces pombe: Lithium acetate/dimethyl sulfoxide procedure. Cold Spring Harb. Protoc. 2016, pdb.prot090969 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Ekwall K., Thon G., Genetic analysis of Schizosaccharomyces pombe. Cold Spring Harb Protoc. 2017, pdb.top079772 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Ekwall K., Thon G., Spore analysis and tetrad dissection of Schizosaccharomyces pombe. Cold Spring Harb. Protoc. 2017, pdb.prot091710 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Rose M., Winston F., Hieter P., Methods in Yeast Genetics: A Laboratory Course Manual, (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1990). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article, SI Appendix, and movies.