Significance
Half of eukaryotic proteins reside in organelles to which they are directed by dedicated targeting pathways, each recognizing unique targeting signals. Multiple proteins compete for any targeting pathway and might have different priority of reaching an organelle. However, the proteins with targeting priority, and the mechanisms underlying it, have not been explored. We developed a systematic tool to study targeting priority. We expressed a competitor protein and examined how it affects the localization of all other proteins targeted by the same pathway. We found several proteins with high targeting priority, dissected the mechanism of priority, and suggest that priority is governed by different parameters. This approach can be modified to study targeting priority in various organelles, cell types, and organisms.
Keywords: yeast, peroxisomes, protein targeting, systems cell biology
Abstract
Approximately half of eukaryotic proteins reside in organelles. To reach their correct destination, such proteins harbor targeting signals recognized by dedicated targeting pathways. It has been shown that differences in targeting signals alter the efficiency in which proteins are recognized and targeted. Since multiple proteins compete for any single pathway, such differences can affect the priority for which a protein is catered. However, to date the entire repertoire of proteins with targeting priority, and the mechanisms underlying it, have not been explored for any pathway. Here we developed a systematic tool to study targeting priority and used the Pex5-mediated targeting to yeast peroxisomes as a model. We titrated Pex5 out by expressing high levels of a Pex5-cargo protein and examined how the localization of each peroxisomal protein is affected. We found that while most known Pex5 cargo proteins were outcompeted, several cargo proteins were not affected, implying that they have high targeting priority. This priority group was dependent on metabolic conditions. We dissected the mechanism of priority for these proteins and suggest that targeting priority is governed by different parameters, including binding affinity of the targeting signal to the cargo factor, the number of binding interfaces to the cargo factor, and more. This approach can be modified to study targeting priority in various organelles, cell types, and organisms.
In eukaryotic cells, most proteins are encoded in the nuclear genome and are synthesized on cytosolic ribosomes. Approximately half of these proteins require targeting to specific organelles to execute their function (1). Organellar proteins are recognized and targeted to their correct cellular destination by various targeting pathways (2–4). Defects in protein targeting can lead to diseases that can be caused either by loss of protein function at its destination organelle, mistargeting to another organelle, or by accumulation of proteins in the cytosol, where they can misfunction or aggregate (5, 6).
Most organelle proteins have a targeting signal that is recognized by one or more cytosolic targeting factors. The complex of cargo protein with its targeting factor is directionally recruited to the correct organelle by binding receptor proteins on the destination membrane (4, 7, 8). Many cargo proteins carry the same type of targeting signal. For example, tens of proteins carry a peroxisomal targeting signal type I (PTS1), and hundreds of proteins carry a signal peptide to travel to the endoplasmic reticulum or a mitochondrial targeting sequence to travel to the inner mitochondrial compartments. Often proteins with a similar targeting signal also utilize the same targeting pathway (3). As many proteins are targeted via the same pathway, and the abundance of the cargo factor may become limiting, it has been shown that certain cargo proteins have evolved to have stronger targeting signals than others (9). Moreover, it is clear that some cargo can utilize multiple pathways to ensure optimal targeting, whereas others are restricted to a single pathway (4, 10, 11). Despite these anecdotal demonstrations, it is not yet clear for any pathway what all of the proteins that have evolved to be optimally targeted by it are, what the various mechanisms by which they do so are, and how this is rewired under the changing metabolic conditions of the cell.
To study targeting priority for entire pathways and in living cells, we developed a systematic tool that is based on high content imaging in the yeast Saccharomyces cerevisiae (hereafter called yeast). As a proof-of-concept, we used the well-described Pex5-mediated pathway that recognizes the PTS1 and targets most matrix (lumen) peroxisomal proteins (12–14). We expressed to very high levels a synthetic cargo protein with a PTS1 targeting signal and systematically examined how the competition over Pex5 affects the localization of each of the known peroxisomal proteins. We found that, as expected, the vast majority of PTS1-containing proteins were affected by the competition over Pex5. However, a few PTS1 proteins had targeting priority in a manner dependent on the carbon source consumed. By following up on priority cargoes, we suggest that they use various strategies to gain their priority, such as additional binding interface on Pex5 or targeting signals with strong affinity.
Results
Developing a Tool to Study Targeting Priority.
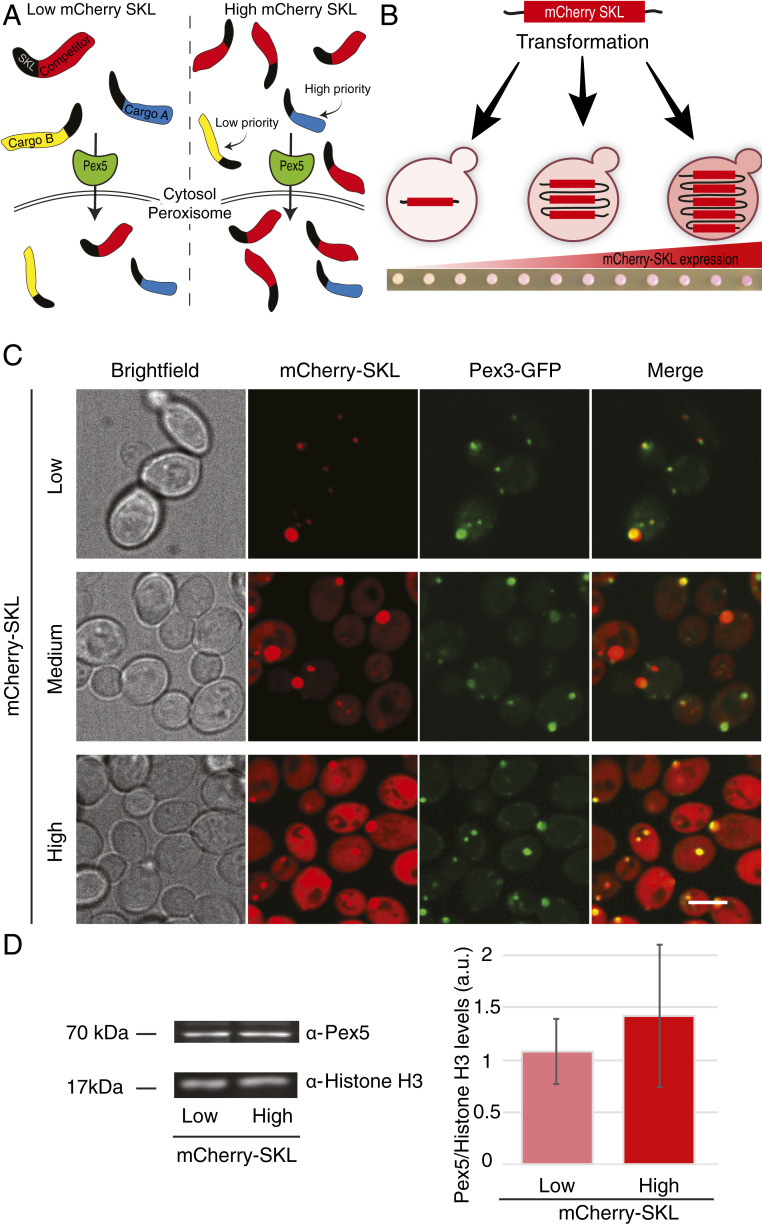
As a test case for our method of dissecting targeting priority of proteins to organelles, we used the Pex5-mediated targeting machinery of peroxisomal proteins in yeast. Pex5 mediates the targeting of most peroxisomal matrix proteins, mainly through binding to a PTS1 targeting signal (12, 14). This pathway is a good model as it is well characterized and has a well-known cargo repertoire. To induce conditions in which cargo proteins compete over a limited amount of Pex5, we expressed different levels of the mCherry protein tagged on its carboxy (C′) terminus with a well-studied PTS1 targeting signal (15) composed of the last three amino acids serine-lysine-leucine (mCherry-SKL) that are considered as a canonical targeting signal. Our rationale was that when the mCherry-SKL is expressed in low amounts, it will not affect targeting of peroxisomal matrix proteins since it will be one of many other cargoes requiring this pathway. However, when mCherry-SKL is expressed in high levels, it will compete with other Pex5 cargo proteins. Hence, only proteins that have high targeting priority would still be localized to peroxisomes (Fig. 1A).
Fig. 1.
An in vivo tool to systematically study protein targeting priority. (A) Schematic representation of targeting competition created by the increasing levels of mCherry-SKL, indicated as “competitor” (red). (Left) When low levels of mCherry-SKL are expressed, peroxisomal matrix proteins are imported into the organelle. (Right) High levels of mCherry-SKL saturate the import machinery and create a competition for the limiting Pex5 targeting factor. Hence, proteins with low targeting priority (“cargo B,” yellow) will not be targeted to peroxisomes while proteins with high priority (“cargo A,” blue) will retain their capacity to target efficiently. (B) Schematic representation of a methodology to create targeting competition. An inert, peroxisome-destined, substrate with a peroxisomal targeting signal (mCherry-SKL) is integrated into the yeast genome in multiple copies, creating strains with increasing levels of mCherry-SKL. The increased mCherry expression is detected by the color of the colony and later verified by flow cytometry. (C) mCherry-SKL is localized to peroxisomes (colocalization with the peroxisomal membrane protein Pex3-GFP in diploid cells). When mCherry-SKL was expressed in high levels the mCherry signal was also detected in the cytosol implying that the targeting machinery is saturated. (Scale bar, 5 µm.) (D) Measurement of Pex5 levels in low and high mCherry-SKL strains grown in glucose. (Left) Western blot analysis with anti-Pex5 antibody. (Right) Quantification of Pex5 levels in the two different mCherry-expressing strains. Protein levels were normalized to the loading control, Histone H3. Values represent the mean ± SD of two biological repeats and four technical repeats.
To express high levels of the mCherry-SKL protein, we integrated into the yeast genome multiple copies of the mCherry-SKL–encoding gene driven by a strong promoter (15). This has the advantage of reaching very high expression levels that are not reachable using increasing strengths of promoters alone (16). Yeast colonies that highly expressed the competitor protein became intensely red; therefore, we were able to select colonies based on the intensity of their color and establish a collection of strains, each expressing different levels of mCherry-SKL (Fig. 1B).
To ensure that we indeed created a competition situation, we verified that when mCherry-SKL is expressed in low amounts, it is localized predominantly to peroxisomes, while when it is expressed at high amounts it is also localized to the cytosol (Fig. 1C). While cytosolic accumulation could also result from a feedback loop causing decreased degradation, the most likely explanation for this is that under these conditions the Pex5-targeting machinery becomes saturated. Indeed, we found that Pex5 was not up-regulated in the presence of elevated amounts of the mCherry-SKL competitor (Fig. 1D), supporting the hypothesis that Pex5 amounts are limiting. Taking these data together, we established an in vivo system that can be applied to study targeting priority.
Systematically Investigating Peroxisomal Proteins during Competition Uncovers a Subgroup of Cargo Proteins with High Targeting Priority.
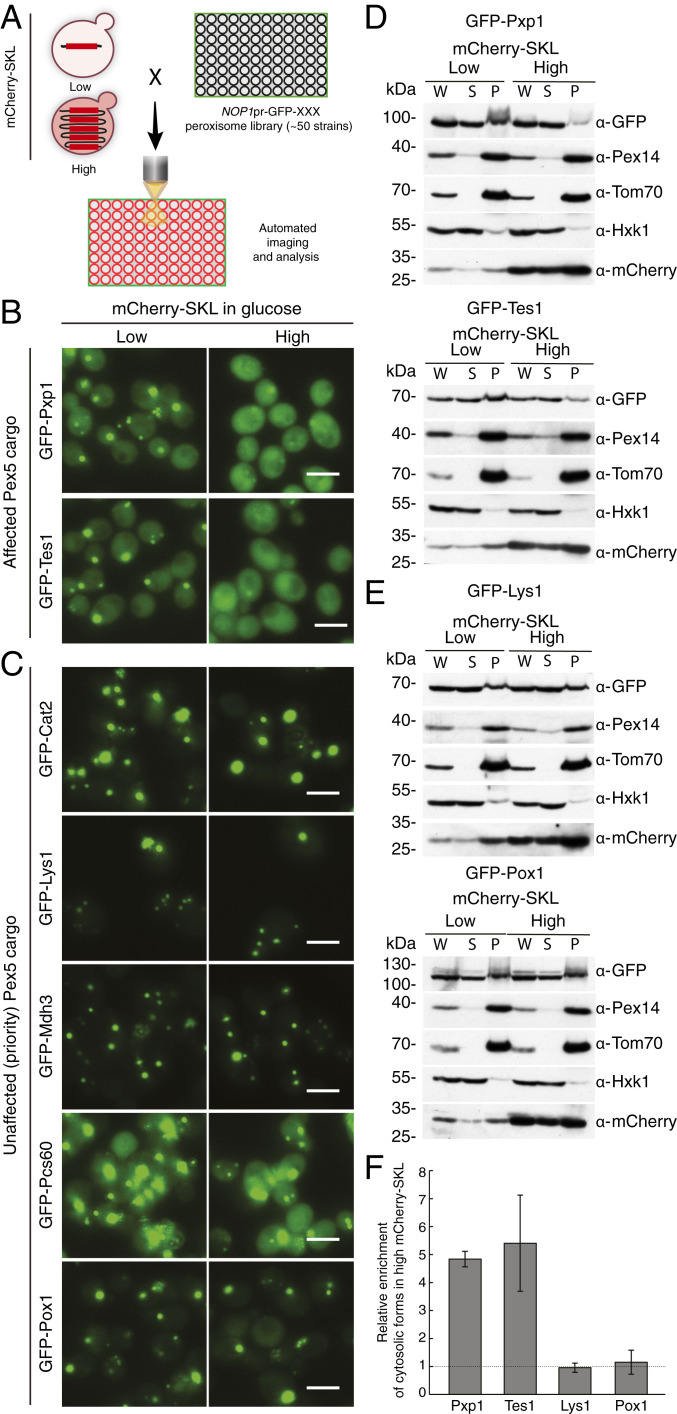
We chose two strains that had either low or high levels of the competitor (mCherry-SKL) that we verified had a homogenous level of expression. Using an automated procedure we used these to create two collections of strains: One expressing low (no competition) and one expressing high (competition) levels of mCherry-SKL, and each expressing all known peroxisomal proteins tagged with GFP at their amino (N′) terminus and expressed under a generic NOP1 promoter (17–19) (Fig. 2A). The N′ tagging ensured that the C′ PTS1 sequences were not obstructed. The strains were imaged on a high-content screening platform (20) and the abundance and localization of each GFP-tagged protein was compared between the low- and high-competition conditions. While abundance was evaluated computationally, localization was manually annotated using a binary measure: Unaffected (no visible difference in the GFP signal between low or high levels of mCherry-SKL) or affected (less/weaker GFP puncta and a stronger cytosolic GFP signal in the presence of high mCherry-SKL) (Dataset S1). We first verified that while the expression of the mCherry-SKL was increased in the high mCherry-SKL strains, it did not affect the expression level of the GFP-tagged peroxisomal proteins (SI Appendix, Fig. S1, glucose). Focusing on localization, we found that peroxisomal membrane proteins and matrix proteins that contain a PTS2 signal (these rely on Pex7 for targeting) (21) were not affected by the mCherry-SKL competitor (SI Appendix, Fig.S2). This gave us an indication that high levels of mCherry-SKL do not unspecifically alter peroxisomal physiology nor block the import machinery of matrix proteins. Unexpectedly, Pex18, a coreceptor of Pex7 (22), had reduced localization to peroxisomes when high levels of the competitor were expressed (Dataset S1). This hints that, in addition to the contribution of Pex18 to Pex7-mediated targeting, Pex18 may also have some interplay with the Pex5 targeting machinery in a way that should be further studied. Conversely, the localization to peroxisomes of the recently identified PTS1 targeting factor, Pex9 (9, 10), and one of its cargoes Gto1 (glutathione transferase omega-like 1 protein 1) was enhanced during competitive conditions rather than being reduced (Dataset S1), suggesting that it may function as a back-up pathway under these conditions.
Fig. 2.
A high-content screen reveals a subgroup of Pex5 cargo proteins that have targeting priority. (A) A schematic representation of the high-content microscopy screen. Yeast strains with low or high levels of mCherry-SKL were mated with a collection of strains each expressing one peroxisomal protein tagged with GFP at the N′ terminus under the regulation of a constitutive NOP1 promoter. The mating was followed by sporulation and selection for haploids containing both the mCherry-SKL protein and a GFP-peroxisomal protein. Cells were imaged using a fluorescent microscope in glucose-containing media and the localization of each GFP peroxisomal protein was annotated in the presence of low or high mCherry levels. (B) Representative images of two Pex5 cargo proteins on the background of either low or high levels of mCherry-SKL. The GFP-tagged proteins were mainly localized to peroxisomes when low levels of mCherry-SKL were expressed but were mostly observed in the cytosol when high levels of mCherry-SKL were expressed. We dub such substrates as having low targeting priority. (Scale bars, 5 µm.) (C) Representative images of five Pex5 cargo proteins whose cellular localization was not affected by the mCherry-SKL protein expressed at very high levels. These proteins seem to have high targeting priority compared to mCherry-SKL. (Scale bars, 5 µm.) (D) Subcellular fractionation of cells expressing the indicated peroxisomal protein fused to GFP in the presence of low or high levels of mCherry-SKL. Fractions corresponding to whole-cell lysate (W), supernatant (S) that represents the cytosol, and pellet (P) that contains most organelles including peroxisomes, were obtained by differential centrifugation. The fractions were analyzed by SDS/PAGE and Western blot with antibodies against GFP, the peroxisomal protein Pex14, the mitochondrial protein Tom70, the cytosolic protein Hexokinase1 (Hxk1), and mCherry. Representative blots of GFP-Pxp1 and GFP-Tes1 indicate that the organellar localization of Pxp1 and Tes1is reduced in the presence of high levels of mCherry-SKL. (E) Representative blots showing that the localization of Lys1 and Pox1 is not altered when high levels of mCherry-SKL are expressed. (F) The intensities of bands representing the GFP-tagged Pxp1, Tes1, Lys1, and Pox1, were quantified from three independent experiments and adjusted to the intensity of the Ponceau staining in the corresponding lane. The supernatant/pellet ratio was calculated for each GFP-tagged protein. To examine if there is a difference in the supernatant/pellet ratio when different amounts of mCherry-SKL were expressed, we divided the ratio obtained in the presence of high mCherry-SKL by the ratio obtained in the presence of low mCherry-SKL. The graph represents the average of three independent experiments ±SD.
Focusing on the Pex5 cargo proteins [19 were correctly localized to peroxisomes in our collection (18, 23)], we observed that the localization of most (14 cargo proteins) was affected by the competition and that they accumulated in the cytosol (Dataset S1; examples shown in Fig. 2B). In support of the effect on Pex5 cargo being a direct result of saturating Pex5 amounts, we found that overexpressing Pex5 could rescue the targeting of such a PTS1 cargo, Pxp1 (SI Appendix, Fig. S3).
Interestingly, the localization of five Pex5 cargo proteins (carnitine acetyl-CoA transferase, Cat2; saccharopine dehydrogenase, Lys1; malate dehydrogenase 3, Mdh3; oxalyl-CoA synthetase, Pcs60; and fatty-acyl CoA oxidase, Pox1/Fox1) was not affected by the high levels of mCherry-SKL (Fig. 2C). Since targeting of these proteins seems completely unaffected by competition, we dub these proteins as having a high targeting priority. We verified the targeting priority observed by microscopy by performing subcellular fractionation experiments. Indeed, tracking two “low-priority” (Pxp1 and Tes1) and two “high-priority” (Lys1 and Pox1) proteins during low- and high-competition states uncovered that the low-priority proteins become mainly soluble (Fig. 2 D and F), while the high priority ones remain in the organelle pellet (Fig. 2 E and F). In summary, by systematically imaging all peroxisomal proteins in the presence of competition, we found that Pex5 cargoes have different targeting priorities.
Targeting Priority to Peroxisomes Is Dependent on Metabolic Conditions.
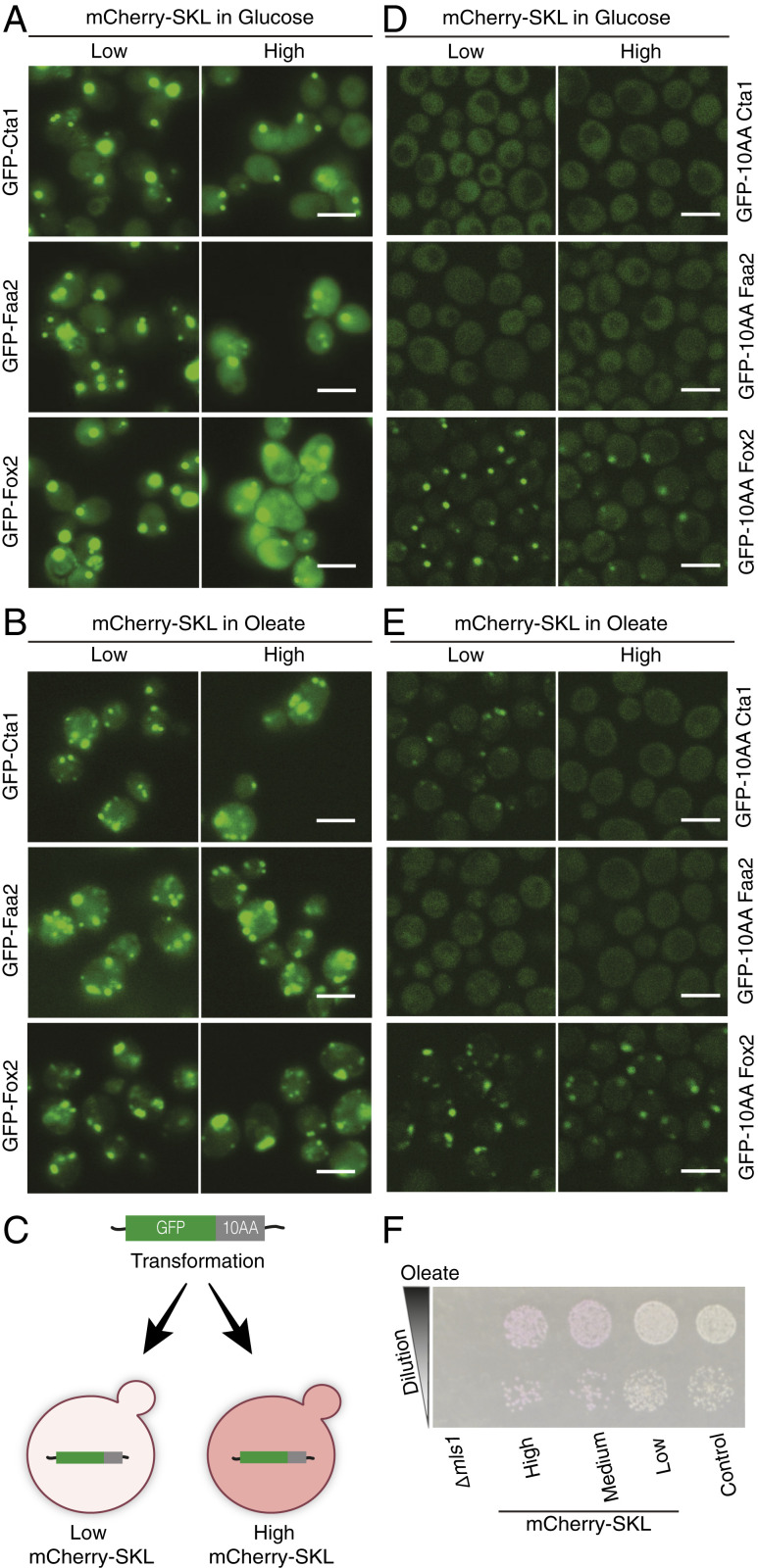
To examine if targeting priority of Pex5 cargo proteins is affected by the metabolic state of the cells, we repeated the high-content screen using the fatty acid oleate as a carbon source instead of glucose (Fig. 2A and Dataset S1). In S. cerevisiae the peroxisome is the sole organelle in which β-oxidation of fatty acids occurs, and hence peroxisomes are essential when cells rely on oleate (24). We observed that all proteins that had high-targeting priority in glucose maintained it in oleate (SI Appendix, Fig. S4A). Interestingly, three Pex5 cargo proteins [Cta1, peroxisomal catalase A; Faa2, fatty acyl-CoA synthetase; and Fox2, 3-hydroxyacyl-CoA dehydrogenase and enoyl-CoA hydratase (25–27)], which did not have priority in glucose (Fig. 3A), gained priority in oleate (Fig. 3B). As we saw in glucose, the localization of Pex9 and one of its cargo proteins Mls2 (malate synthase 2) was enhanced in oleate during competitive conditions (Dataset S1), suggesting that Pex9 may function as a back-up pathway under these conditions as well.
Fig. 3.
Targeting priority to peroxisomes is dependent on the metabolic conditions. (A) The localization of GFP-Cta1 (peroxisome catalase), GFP-Faa2 (fatty acyl-CoA synthetase), and GFP-Fox2 (fatty acid oxidase) all under regulation of the NOP1 promoter, was affected by the high mCherry-SKL levels when cells were grown in glucose. (B) The localization was not affected in oleate making Cta1, Faa2, and Fox2 condition-specific priority cargo. (C) Schematic showing how GFP was fused at its C′ to the last 10 aa (10AA) of different cargo proteins and expressed from a genomic copy in the presence of low or high levels of mCherry-SKL, allowing examination of whether targeting priority is encoded in the PTS1 of proteins. (D) GFP was fused at its C′ to the last 10 aa of Cta1, Faa2, or Fox2. The GFP fusions had low targeting priority in glucose. (E) GFP fused to the last 10 aa of Fox2 had a targeting priority in oleate, suggesting that in the case of Fox2 the oleate-specific targeting priority is embedded in the last 10 aa. (Scale bars in A, B, D, and E, 5 µm.) (F) Dilution assay of strains expressing three levels of mCherry-SKL on oleate. No significant growth defect was observed in the presence of high levels of mCherry-SKL. A strain lacking Mls1 (malate synthetase 1), which is essential for growth on oleate, was used as a control for the oleate media, and a control yMS721, strain was used for growth baseline.
Since all GFP-tagged proteins were expressed under a generic promoter, this reduced the possibility that oleate-specific targeting priority is determined by an increased level of transcription in our experimental setting. However, to verify this, we compared protein intensity of all peroxisomal proteins between the two conditions (using GFP levels as a proxy) (SI Appendix, Fig. S4B) and by specifically measuring protein levels of cargo proteins that had priority in oleate but not in glucose (SI Appendix, Fig. S4C). We found that, indeed, the protein levels were not elevated, ruling out elevation by posttranscriptional or translational regulation. Since mCherry levels were more variable in oleate than in glucose (SI Appendix, Fig. S1), we ensured that our results were not simply due to lower levels of competition by checking that the mCherry levels in the strains that had oleate-specific priority are not the lowest in the distribution. Ruling out these potential sources of false positives, we suggest that targeting priority can be modulated depending on the metabolic requirements of the cell. This could occur through posttranslational modifications of the targeting factor Pex5 or of the specific cargo, such that binding to certain cargo is enhanced.
To examine if the PTS1 is sufficient to enable oleate-specific targeting priority, we fused the last 10 aa of Cta1, Faa2, or Fox2 to the C′ of GFP and integrated the different GFP fusions into the yeast genome of strains expressing low or high levels of mCherry-SKL (Fig. 3C). We then grew the cells in glucose or in oleate and examined if the last 10 aa were sufficient to bestow targeting priority to the GFP fusions in glucose or in oleate. The GFPs with the last 10 aa of Cta1 or Faa2 did not have high targeting priority, neither in glucose nor in oleate (Fig. 3 D and E), suggesting that for these proteins the oleate-specific targeting priority information was encoded elsewhere. In contrast, the GFP last 10 aa of Fox2 behaved exactly like full-length Fox2; it was affected by the competition in glucose (Fig. 3D) but was not affected by the competitor when grown in oleate (Fig. 3E). This demonstrates that in the case of Fox2, the last 10 aa are sufficient to enable oleate-specific targeting priority. While it is not clear how the same 10 aa provide condition-specific priority, we hypothesize that they may become posttranslationally modified in oleate. In support of this, while protein levels were not altered for GFP-Fox2, we detected a difference in the pattern in which GFP-Fox2 (full protein) ran in an SDS gel in oleate compared to glucose (SI Appendix, Fig. S4C).
A central function of peroxisomes is the breakdown of fatty acids by β-oxidation. We noticed that the proteins that gained priority in oleate are enzymes involved in β-oxidation and in removal of H2O2, a product of β-oxidation. This suggests that part of the reason to employ targeting priority is to ensure that sufficient amounts of the peroxisomal proteins required for growth in oleate are localized to peroxisomes, even under suboptimal growth conditions (simulated in our experiment by high mCherry-SKL levels). Indeed, the cells expressing high levels of mCherry-SKL did not have a severe growth defect when grown on oleate, despite the fact that many peroxisomal proteins were not targeted correctly (Fig. 3F). These results indicate that protein targeting to peroxisomes can be adjusted upon environmental changes and cellular requirements.
Strong Binding Affinity of the PTS1 to Pex5 Provides One Way of Obtaining Targeting Priority.
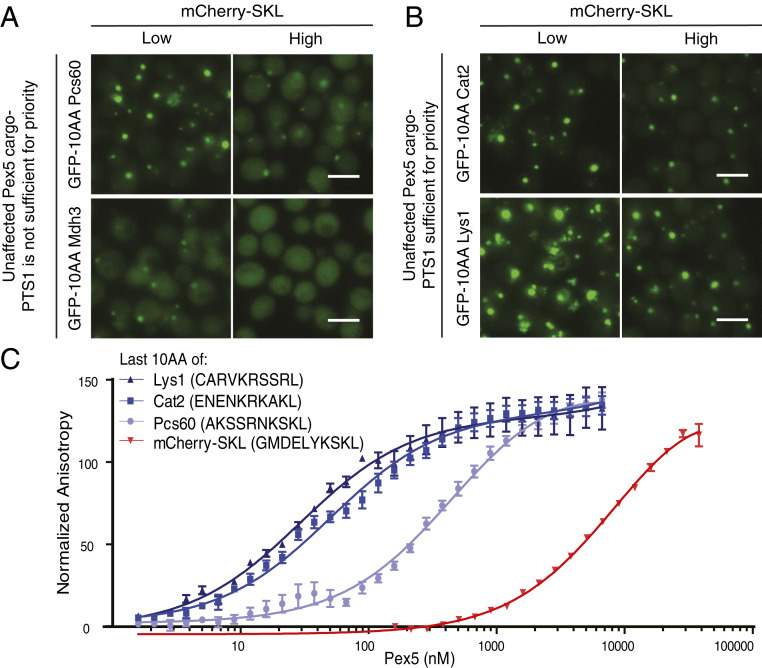
We decided to focus on the five Pex5 cargo proteins (Cat2, Lys1, Mdh3, Pcs60, and Pox1) that had targeting priority in both glucose and oleate to identify the molecular mechanisms by which their targeting priority is gained. It was previously shown that Pox1 does not contain a PTS1 signal and that it binds to Pex5 by a unique mechanism (28). Hence, we suggest that the way by which Pox1 gains priority is by avoiding competition on the same binding site. It is not yet clear if Pex5 can bind two cargo proteins simultaneously (a PTS1 and a non-PTS1 cargo, for example) or simply that the alternate binding interface provides stronger affinity.
We further focused on the remaining four PTS1 proteins (Cat2, Lys1, Mdh3, and Pcs60) and examined if their priority is determined by their PTS1 signal. To test this, we fused the last 10 aa of these four PTS1 proteins as well as low-priority controls, to the C′ of GFP and integrated the different GFP fusions into the yeast genome of strains expressing low or high levels of mCherry-SKL (Fig. 3C). For two of the proteins, Mdh3 and Pcs60, targeting priority information was not encoded in the PTS1 since GFP fused to their last 10 aa could not provide priority in the face of high competition (Fig. 4A). When GFP was fused to the last 10 aa of Cat2 or Lys1, it was targeted efficiently even when high levels of mCherry-SKL were expressed (Fig. 4B); hence, for these two proteins targeting priority is encoded in the last 10 aa.
Fig. 4.
Strong binding affinity of the PTS1 to Pex5 provides one way of obtaining targeting priority. (A) GFP was fused to the last 10 aa of the priority targets Mdh3 or Pcs60 and localization was measured under low or high competition in glucose. Priority was not encoded in the PTS1 context of these two proteins. (Scale bars, 5 µm.) (B) GFP was fused to the last 10 aa of the priority targets Cat2 or Lys1 and localization was measured under low or high competition in glucose. For these two constructs, the last 10 aa were enough to endow priority supporting the notion that the information that governs priority is embedded in the last 10 aa of these cargo proteins. (Scale bars, 5 µm.) (C) Binding affinity of various PTS1 peptides was quantified using fluorescence anisotropy. Both Cat2 and Lys1 have strong binding affinities for Pex5, which explain their targeting priority. Interestingly, Pcs60 PTS1 is a weaker binder to Pex5, which correlates with its inability to confer priority when coupled to GFP. The last 10 aa of the mCherry-SKL competitor has a lower affinity, despite ending with an SKL. Calculated Kds from the anisotropy experiment are: Lys1 peptide 29.3 ± 3.65 nM, Cat2 peptide 49.94 ± 4.1 nM, Pcs60 peptide 454.7 ± 43.81 nM, mCherry-SKL peptide 9,844 ± 1,177 nM.
To verify that the PTS1 of Cat2 and Lys1 indeed confers priority through higher association with Pex5, we directly measured binding affinity to Pex5 by fluorescence anisotropy (29). In vitro binding of the full-length yeast Pex5 to fluorescent peptides containing the last 10 aa of Cat2, Lys1, and Pcs60, as well as to mCherry-SKL, showed that the binding affinity of Pex5 to the last 10 aa of Cat2 and Lys1 is ∼10-fold higher than the binding of Pex5 to the last 10 aa of Pcs60. This strongly supports the theory that high-affinity interaction with Pex5 is sufficient to confer priority and that both Cat2 and Lys1 employ this strategy to secure targeting (Fig. 4C). Surprisingly, both the mCherry competitor and Pcs60 end with SKL, which is considered to be the strongest PTS1 signal, but still they have a different binding affinity to Pex5. This supports the notion that the PTS1 strength is not simply determined by the last 3 aa but rather by a wider context (13, 30).
Targeting Priority Is Governed by Various Molecular Mechanisms.
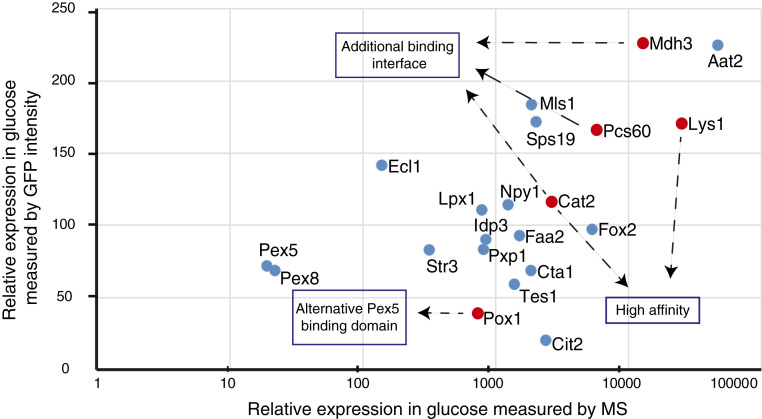
While high affinity explains the priority targeting of Cat2 and Lys1, and we hypothesize that binding to an alternate interface of Pex5 explains the targeting priority of Pox1, the priority mechanism for Pcs60 and Mdh3 remained unclear. The ability of a cargo protein to bind to its targeting factor is a combination of both affinity and abundance; therefore, we hypothesized that another possibility to gain targeting priority is simply by having very high protein amounts. Hence, we examined the protein abundance, previously measured by mass spectrometry (MS) (31) of all native PTS1 proteins and plotted them against the GFP intensity measured in our strains. While Pcs60, Lys1, and Mdh3 are indeed very highly expressed, some proteins that demonstrated similar or higher expression, in both the MS data and the GFP intensity data (such as Aat2), did not have a targeting priority in our assay (Fig. 5). Hence, high concentrations may support targeting for Pcs60 and Mdh3, but this precludes the option that high concentrations of the protein in the cytosol are enough to confer targeting priority. Additionally, it was previously suggested that Cat2, Pcs60, and Mdh3 bind to Pex5 not only through the PTS1 motif but also through other motifs (25, 32, 33). Having multiple binding sites of a single protein to the targeting factor might therefore also increase binding affinity and determine targeting priority. One additional option for obtaining priority lies in the unique feature of the peroxisomal translocon that allows translocation of entire complexes (34). For example, malate dehydrogenases form homo-complexes (35) that may be imported in an assembled state forming a complex that contains several targeting signals, creating avidity.
Fig. 5.
Different parameters govern targeting priority. Relative expression of native Pex5 and its cargo proteins measured by MS in cells grown in glucose (taken from ref. 36) plotted versus the GFP intensity measured in the NOP1 N′ tagged strains, which correlates to the protein levels. Cargo proteins with high targeting priority are marked in red. The graph implies that targeting priority is guided by different parameters and is not simply governed by protein abundance.
In summary, herein we established a tool to systematically study targeting priority of proteins to organelles. To test our set-up, we chose to use the well-characterized Pex5-mediated targeting of peroxisomal proteins. Using systematic imaging, we identified a subgroup of Pex5 cargo proteins that has targeting priority in both glucose and oleate and others that are oleate-specific. The discovery that targeting priority is dependent on the metabolic state of the cell suggests that priority could be regulated by posttranslational modifications (36).
We suggest that targeting priority can be obtained by different mechanisms, including harboring nonstandard targeting signals, encoding a strong binding affinity to the targeting factor, or maintaining multiple binding sites.
The tool that we developed here enabled us to supply a holistic overview on targeting priority to organelles and demonstrate some of the parameters that can govern targeting priority, as well as their physiological significance. The competition tool that we developed can be easily modified to unravel targeting priority to other organelles and is not limited to yeast. Understanding the rules that govern targeting of proteins to different cellular compartments will not only shed light on how the basic unit of life—a cell—functions, but also has great potential for therapeutic and biotechnological uses.
Materials and Methods
Yeast Media.
SD media used in this study contains 6.7 g/L yeast nitrogen base and 2% glucose, with complete amino acid mix, unless written otherwise; 1 g/L of monosodium glutamic acid replaces ammonium sulfate in the SD if antibiotics are used. When mentioned, 300 mg/L Hygromycin B (CAS#: 31282-04-9, Roche) was used. S oleate media was made with synthetic media, 0.2% oleate (Sigma) +0.1% Tween 80 and a complete amino acids mix.
Yeast Strains, Plasmids, and Primers.
S. cerevisiae strains used in this study are described in Dataset S2, excluding strains that were included in the yeast libraries and were frozen as a library and not as single strains. Strains yMS5243 to yMS5252 are shown in Fig. 3 D and E and the strains yMS4346, yMS4347, yMS4350, yMS4351, yMS4353, yMS4355, yMS4356, yMS4357, yMS4359, and yMS4360 are shown in Fig. 4.
Primers are described in Dataset S3 and plasmids are described in Dataset S4.
Cells were genetically manipulated using a transformation method that includes the usage of lithium-acetate, polyethylene glycol, and single-stranded DNA (37). Primers for manipulations and validation were designed using Primers-4-Yeast (38).
Plasmid pMS934 (p69_TDH3) (39) was modified using a restriction-free method (40) with the primers described in Dataset S3 creating plasmid pMS816 (mCherry-SKL).
The pYM-based pMS555 plasmid that was originally used for N-terminal GFP tagging (17) was modified to contain the last 10 aa of Cat2, Cta1, Faa2, Fox2, Lys1, Mdh3, and Pcs60 at the C′ of the GFP sequence. Using these plasmids, GFP-PTS1 was genomically integrated into the HO locus in strains expressing low and high levels of mCherry-SKL (Figs. 3 D and E and 4 A and B).
The mCherry-SKL strains were based on the yMS721 (41). pMS816 plasmid was integrated into the yeast genome after linearization by a unique Mun1 restriction enzyme (ThermoFisher Scientific) in the TDH3 promoter (39). After Hygromycin B (300 mg/L) antibiotics selection, single colonies were hand-picked to create several hundred candidates. The candidates grew overnight in synthetic media [SD(MSG) with Hygromycin B selection], and then their mCherry fluorescence levels were measured by flow cytometry to determine the expression level of the mCherry-SKL protein.
Flow cytometer measurements and analysis were done using an LSRII system (BD Biosciences). Flow cytometry was conducted with excitation at 488 nm and emission at 525 ± 25 nm for GFP samples. For mCherry, excitation was conducted at 594 nm and emission at 610 ± 10 nm. The average number of cells analyzed was 30,000.
Yeast Library Preparation.
To create collections of haploid strains containing both the mCherry-SKL and the peroxisomal proteins tagged with a GFP, automated mating, sporulation, and haploid selection steps were taken (42). Strains expressing low or high levels of mCherry-SKL were crossed with a collection of ∼90 strains of controls and known peroxisomal proteins tagged with GFP at their N′ terminus and expressed under the constitutive NOP1 promoter (17–19). A RoToR bench-top colony arrayer (Singer Instruments) was used to handle libraries (42, 43). In brief, mating was performed on rich medium plates. Diploid cell selection was performed on SD(MSG)-URA-Hygromycin B. The cells were then transferred for 7 d to nitrogen starvation plates to induce sporulation. Selection of haploid cells with the desired mutations was performed by transferring cells to SD(MSG)-URA-LYS-ARG+Hygromycin B plates. Spores with α-mating type were selected in the absence of leucine. To select against remaining diploid cells the plates contained the toxic amino acid derivatives Canavanine and Thialysine (Sigma-Aldrich).
Yeast Culturing for High-Content Screening.
To visualize the strains containing low/high levels of mCherry-SKL and a GFP-tagged peroxisomal protein, we used an automated microscopy set-up. In short, for growth in liquid media we used a RoToR arrayer to transfer the cells from agar plates into 384-well polystyrene plates (Greiner Bio-One). We then grew the liquid cultures overnight at 30 °C in SD(MSG)-URA+Hygromycin B media in a LiCONiC incubator. We used a JANUS liquid handler (PerkinElmer) connected to the incubator to dilute the strains to an OD600 of ∼0.2 into plates containing SD medium. Plates were incubated at 30 °C for 4 h in SD medium. For the screen performed in oleate, cells were transferred with OD600 of ∼0.2 into plates containing S oleate medium. Plates were incubated at 30 °C for 20 h. The cultures were then transferred by the liquid handler into glass-bottom 384-well microscope plates (Matrical Bioscience) coated with Con A (Sigma-Aldrich). After 20 min, wells were washed four times and then left with SD-Riboflavin complete medium in case of the glucose screen or DDW in case of oleate screen, to obtain a cell monolayer and remove nonadherent cells. We then transferred the plates to a ScanR automated inverted fluorescent microscope system (Olympus) using a robotic swap arm (Hamilton). Images were recorded using a 60× air lens (NA 0.9) and with an ORCA-ER charge-coupled device camera (Hamamatsu) at room temperature. We acquired the images in a GFP channel (excitation filter 490/20 nm, emission filter 535/50 nm) and an mCherry channel (excitation filter 572/35 nm, emission filter 632/60 nm). We manually reviewed the images using the ImageJ analysis program (https://imagej.net/Downloads).
Manual Microscopy.
Manual microscopy was performed (Figs. 1C, 3 D and E, and 4 A and B) using the VisiScope Confocal Cell Explorer system. The system is composed of a Yokogawa spinning disk scanning unit (CSU-W1) coupled with an inverted Olympus IX83 microscope. We acquired the images using a 60× oil lens and a connected PCO-Edge sCMOS camera, controlled by VisView software, with a wavelength of 561 nm (mCherry) and 488 nm (GFP). For slight linear adjustments to brightness and contrast, we transferred the images to ImageJ. For image visualization of the yeast cells we used the brightfield channel to segment the cells. All images taken are of haploid cells, except Fig. 1C, which shows images of diploids.
Protein Extraction and Western Blot Analysis.
Proteins from cells expressing low or high levels of mCherry-SKL were extracted by Urea protein extraction. In short, 10 mL of 0.5 OD600 yeast cells were harvested by centrifugation. Next, 200 µL of lysis buffer containing 8 M Urea, 50 mM Tris, protease inhibitor (Merck) pH = 7 were added to the pellet and the cell wall was broken down by vortexing at high speed with ready-to-use glass beads (Scientific Industries) at 4 °C for 10 min; 25 µL of 20% SDS were added and the extracts were incubated at 95 °C for 5 min. Protein extracts were transferred into new tubes and stored at −20 °C. Samples in Fig. 1D were analyzed by SDS/PAGE and Western blotting using an anti-Pex5 antibody 1:10,000 (44) (kindly provided by Ralf Erdmann, Ruhr-Universität Bochum, Bochum, Germany), and anti-Histone H3 (Abcam, # ab1791; 1:10,000) was used as the loading control. The anti-Histone H3 antibody was diluted in a mix of 3% BSA, 0.01% sodium azide and Phenol red in PBS (Biological Industries) solution. IRDye 680LT goat anti-rabbit IgG secondary antibody (LI-COR Biosciences) was used at 1:10,000 dilution, followed by scanning using the Odyssey Imaging System (LI-COR Biosciences).
Subcellular Fractionation.
Cells were grown in 100 mL of synthetic medium containing 2% glucose (SD) for 5 to 6 h until OD600 of 1 to 2. Cells were then harvested by centrifugation (4,000 × g, 5 min, room temperature) and the pellet was resuspended with 25 mL water. Next, cells were pelleted again as above and kept at −20 °C till further processing.
For subcellular fractionation, the cells pellet was thawed on ice, washed with 1 mL water, centrifuged as above, and the pellet was weighed. Then, cells were resuspended in 400 µL of resuspension buffer (100 mM Tris, 10 mM DTT, pH 7.2) per 0.2-g pellet and incubated for 10 min at 30 °C while shaking at 450 rpm. Cells were harvested as above and resuspended in 1 mL spheroblasting buffer (1.2 M sorbitol, 20 mM KPI, pH 7.2) followed by pelleting of the cells as above. The pellet was resuspended with 1 mL spheroblasting buffer supplemented with Zymolyase (6-mg/g pellet) and incubated for 1 h at 30 °C while shaking at 450 rpm. The efficiency of spheroblasting was tested by measuring the OD600 of spheroblasts diluted 1:100 in either water or 1.2 M sorbitol and, from this step on, cells were kept at 4 °C.
After spheroblasting was completed, cells were harvested (3,000 × g, 5 min, 4 °C). The pellet was resuspended with 3 mL homogenization buffer (0.6 M sorbitol, 10 mM Tris, 1 mM EDTA, 2 mM Phenylmethanesulfonyl fluoride [PMSF], pH 7.2), and cell lysate was obtained by douncing 20 times in an ice bath. Protein concentration was determined by Bradford assay and proteins from 200 µL of the lysate suspension were taken as whole-cell lysate fraction and proteins were precipitated by methanol-chloroform (see below). The rest of the lysate was centrifuged (600 × g 5 min, 4 °C) to remove unruptured cells and cell debris and the supernatant was centrifuged at high speed (25,000 × g, 10 min, 4 °C). Then, 200 µL of the supernatant were taken as the cytosolic fraction and proteins were precipitated using methanol-chloroform. The pellet was washed with SEM buffer (250 mM sucrose, 80 mM KCl, 10 mM Mops, 2 mM PMSF, 1 mM EDTA, pH 7.2) and harvested again (25,000 × g, 10 min, 4 °C) to obtain the peroxisomes and mitochondria fraction. All fractions were dissolved in Laemmli buffer at a concentration of 2 mg/mL and heated for 10 min at 95 °C before analysis by SDS/PAGE and Western blotting using an anti-GFP antibody 1:2,000 (Torrey Pines), anti-Tom70 1:4,000 (Pineda, custom-made), anti-Pex14 1:2,500 (kindly provided by Ralf Erdmann), anti-Hxk1 1:4,000 (Biotrend), and anti-mCherry 1:10,000 (Abcam). Horseradish peroxidase-conjugates were used as secondary antibodies. Band intensities were quantified with AIDA software (Raytest).
Chloroform/Methanol Protein Precipitation.
Chloroform/methanol precipitation was performed as follows: 4 vol of methanol were added to 200 µL aqueous sample and the mixture was vortexed for 5 s. Then, 1 vol of chloroform was added to the sample, vortexed for 5 s, and 3 vol of water were added to the sample and vortexed 20 s. The mixture was centrifuged (16,000 × g, 1 min, room temperature), the upper layer was removed, and 3 vol of methanol were added and vortexed thoroughly for 20 s. The mixture was centrifuged again (16,000 × g, 1 min, room temperature), the supernatant was discarded, and the pellet containing the proteins was dried at 40 °C before analysis.
Serial Dilutions on Oleate.
Serial 10-fold dilutions were created by growing cells overnight in 0.2% glucose and back dilution of ∼6 h to start with OD600 = 0.1 of all strains of interest in liquid media, and diluting them in 10-fold increments. Next, 2.5 µL of each dilution were plated using Finnpipette F1 Multichannel Pipettes (ThermoFisher Scientific) on oleate agar plates and imaged using Canon PC1591 digital camera after 2 to 8 d (as indicated) of growth at 30 °C.
Protein Purification.
Full-length S. cerevisiae Pex5 was cloned in a petM30 vector. Pex5 was expressed in autoinduction medium (45) with 5 h at 37 °C and 26 h at 20 °C. Cells were harvested, resuspended in lysis buffer (50 mM Hepes pH 7.5, 150 mM NaCl, 20 mM imidazole, protease inhibitor [Roche], DNase [Sigma], and lysozyme [Sigma]), homogenized 1 h at 4 °C, and lysed by sonication. Lysate was then cleared by centrifugation and the supernatant loaded onto Ni-NTA resin (Qiagen). Bound protein was washed with buffer (50 mM Hepes pH 7.5, 750 mM NaCl, 20 mM Imidazole) and the protein eluted with low salt buffer (50 mM Hepes pH 7.5, 150 mM NaCl, 250 mM Imidazole). The eluate was then dialyzed against Hepes pH 7.5, 150 mM NaCl, 0.5 mM TCEP and simultaneously digested with 1 mg of tobacco etch virus (TEV)-protease. Undigested protein and TEV protease were removed by a second Ni-NTA step and flow-through containing Pex5 were concentrated for gel filtration (Hiload 16/60 Superdex 200 pg, GE Healthcare). Relevant fractions were pooled together and the protein was concentrated, flash-frozen in liquid nitrogen, and stored at −80 °C.
Fluorescence Anisotropy.
FITC-labeled peptides corresponding to the carboxyl-terminal (9 or 10) amino acids of Lys1, Cat2, Pcs60, and mCherry-SKL (Lys1: FITC-YARVKRSSRL, Cat2: FITC-YNENKRKAKL, Pcs60: FITC-YKSSRNKSKL, mCherry-SKL: FITC-GMDELYKSKL; Genscript) were solubilized in water and used in the assay at a final concentration of 10 nM. A tyrosine was added at the N terminal of some peptides for concentration determination; in these cases only the last 9 aa of the PTS1 protein were used. Measurements of fluorescence anisotropy changes were performed in black 96-well plates (Greiner) with an Infinite M1000 plate reader (Tecan) with excitation/detection at 470/530 nm. The experiment was performed in 50 mM Hepes pH 7.5, 150 mM NaCl. A concentration range of 6.6 µM to 1.5 nM (for Lys1, Cat2, and Pcs60) or 38 µM to 160 nM for mCherry-SKL was obtained by serial dilution and each concentration measured in triplicate. Three independent experiments were performed and binding data were normalized and analyzed using Prism (GraphPad software). Kinetic information was obtained by least-square fitting of a Binding–Saturation model with one binding site.
Supplementary Material
Acknowledgments
We thank Ralf Erdmann for kindly sharing the anti-Pex5 and anti-Pex14 antibodies; Bettina Warscheid, Silke Oeljeklaus, Miriam Eisenstein, Noa Dahan, and Yuri Bykov for critical reading of the manuscript and for helpful suggestions; and Bettina Warscheid and Silke Oeljeklaus for sharing the data shown in Fig. 5 and for kindly sharing plasmid pMS742. The work in the M.S. laboratory is supported by the European Research Council Consolidator Grant Peroxisystem 64660, a European Union International Training Network grant (PERICO 812968), and by the Minerva Foundation with funding from the Federal German Ministry for Education and Research (712935). The robotic set-up in the M.S. laboratory was purchased through kind support of the Blythe Brenden-Mann Foundation. The joint work of the M.S and D.R. laboratories was supported by a collaborative grant from the German Israeli Foundation (GIF I-1458-412.13/2018). M.S. is an incumbent of the Dr. Gilbert Omenn and Martha Darling Professorial Chair in Molecular Genetics. J.B. is supported by the EI3POD programme. E.Y. is supported by the Ariane de Rothschild women doctoral program. The work in the M.W. laboratory is supported by the Deutsche Forschungsgemeinschaft (WI 1058/9-2) via the Pertrans network.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. W.A.P. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920078117/-/DCSupplemental.
Data Availability.
All study data are included in the main text and SI Appendix.
References
- 1.Huh W.-K. et al., Global analysis of protein localization in budding yeast. Nature 425, 686–691 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Blobel G., Sabatini D. D., . “Ribosome-membrane interaction in eukaryotic cells” in Biomembranes, Manson L. A., Ed. (Springer US, 1971), pp. 193–195. [Google Scholar]
- 3.Kunze M., Berger J., The similarity between N-terminal targeting signals for protein import into different organelles and its evolutionary relevance. Front. Physiol. 6, 259 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aviram N., Schuldiner M., Targeting and translocation of proteins to the endoplasmic reticulum at a glance. J. Cell Sci. 130, 4079–4085 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Slawecki M. L. et al., Identification of three distinct peroxisomal protein import defects in patients with peroxisome biogenesis disorders. J. Cell Sci. 108, 1817–1829 (1995). [DOI] [PubMed] [Google Scholar]
- 6.Carapito R. et al., Mutations in signal recognition particle SRP54 cause syndromic neutropenia with Shwachman-Diamond-like features. J. Clin. Invest. 127, 4090–4103 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurzchalia T. V. et al., The signal sequence of nascent preprolactin interacts with the 54 K polypeptide of the signal recognition particle. Nature 320, 634–636 (1986). [DOI] [PubMed] [Google Scholar]
- 8.Schatz G., Dobberstein B., Common principles of protein translocation across membranes. Science 271, 1519–1526 (1996). [DOI] [PubMed] [Google Scholar]
- 9.Ghosh D., Berg J. M., A proteome-wide perspective on peroxisome targeting signal 1(PTS1)-Pex5p affinities. J. Am. Chem. Soc. 132, 3973–3979 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Yifrach E. et al., Characterization of proteome dynamics in oleate reveals a novel peroxisome targeting receptor. J. Cell Sci. 129, 4067–4075 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Effelsberg D., Cruz-Zaragoza L. D., Schliebs W., Erdmann R., Pex9p is a novel yeast peroxisomal import receptor for PTS1-proteins. J. Cell Sci. 129, 4057–4066 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Walter T., Erdmann R., Current advances in protein import into peroxisomes. Protein J. 38, 351–362 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Brocard C., Hartig A., Peroxisome targeting signal 1: Is it really a simple tripeptide? Biochim. Biophys. Acta 1763, 1565–1573 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Gould S. J., Keller G. A., Hosken N., Wilkinson J., Subramani S., A conserved tripeptide sorts proteins to peroxisomes. J. Cell Biol. 108, 1657–1664 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nötzel C., Lingner T., Klingenberg H., Thoms S., Identification of new fungal peroxisomal matrix proteins and revision of the PTS1 consensus. Traffic 17, 1110–1124 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Kintaka R., Makanae K., Moriya H., Cellular growth defects triggered by an overload of protein localization processes OPEN. Sci. Rep. 6, 31774 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yofe I. et al., One library to make them all: Streamlining the creation of yeast libraries via a SWAp-tag strategy. Nat. Methods 13, 371–378 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dahan N., Schuldiner M., Zalckvar E., “Peroxisome mini-libraries: Systematic approaches to study peroxisomes made easy” in Peroxisomes, Schrader M., Ed. (Methods Molecular Biology, vol. 1595, Humana Press, New York, 2017) pp. 305–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weill U. et al., Genome-wide SWAp-tag yeast libraries for proteome exploration. Nat. Methods 15, 617–622 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Breker M., Gymrek M., Schuldiner M., A novel single-cell screening platform reveals proteome plasticity during yeast stress responses. Cell Biol. 200, 839–850 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grunau S. et al., Peroxisomal targeting of PTS2 pre-import complexes in the yeast Saccharomyces cerevisiae. Traffic 10, 451–460 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Stein K., Schell-Steven A., Erdmann R., Rottensteiner H., Interactions of Pex7p and Pex18p/Pex21p with the peroxisomal docking machinery: Implications for the first steps in PTS2 protein import. Mol. Cell. Biol. 22, 6056–6069 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schäfer A., Kerssen D., Veenhuis M., Kunau W. H., Schliebs W., Functional similarity between the peroxisomal PTS2 receptor binding protein Pex18p and the N-terminal half of the PTS1 receptor Pex5p. Mol. Cell. Biol. 24, 8895–8906 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hiltunen J. K. et al., The biochemistry of peroxisomal β-oxidation in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 27, 35–64 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Rymer Ł., Kempiński B., Chełstowska A., Skoneczny M., The budding yeast Pex5p receptor directs Fox2p and Cta1p into peroxisomes via its N-terminal part near the FxxxW domain. J. Cell Sci. 131, jcs216986 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Hiltunen J. K. et al., Peroxisomal multifunctional beta-oxidation protein of Saccharomyces cerevisiae. Molecular analysis of the fox2 gene and gene product. J. Biol. Chem. 267, 6646–6653 (1992). [PubMed] [Google Scholar]
- 27.Hettema E. H. et al., The ABC transporter proteins Pat1 and Pat2 are required for import of long-chain fatty acids into peroxisomes of Saccharomyces cerevisiae. EMBO J. 15, 3813–3822 (1996). [PMC free article] [PubMed] [Google Scholar]
- 28.Klein A. T. J., van den Berg M., Bottger G., Tabak H. F., Distel B., Saccharomyces cerevisiae acyl-CoA oxidase follows a novel, non-PTS1, import pathway into peroxisomes that is dependent on Pex5p. J. Biol. Chem. 277, 25011–25019 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Jameson D. M., Ross J. A., Fluorescence polarization/anisotropy in diagnostics and imaging. Chem. Rev. 110, 2685–2708 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeLoache W. C., Russ Z. N., Dueber J. E., Towards repurposing the yeast peroxisome for compartmentalizing heterologous metabolic pathways. Nat. Commun. 7, 11152 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morgenstern M. et al., Definition of a high-confidence mitochondrial proteome at quantitative scale. Cell Rep. 19, 2836–2852 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elgersma Y. et al., Analysis of the carboxyl-terminal peroxisomal targeting signal 1 in a homologous context in Saccharomyces cerevisiae. J. Biol. Chem. 271, 26375–26382 (1996). [DOI] [PubMed] [Google Scholar]
- 33.Hagen S. et al., Structural insights into cargo recognition by the yeast PTS1 receptor. J. Biol. Chem. 290, 26610–26626 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walton P. A., Hill P. E., Subramani S., Import of stably folded proteins into peroxisomes. Mol. Biol. Cell 6, 675–683 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minárik P., Tomášková N., Kollárová M., Antalík M., Malate dehydrogenases-structure and function. Gen. Physiol. Biophys. 21, 257–265 (2002). [PubMed] [Google Scholar]
- 36.Schummer A., Fischer S., Oeljeklaus S., Warscheid B., Study of peroxisomal protein phosphorylation by functional proteomics. Methods Mol. Biol. 1595, 267–289 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Gietz R. D., Woods R. A., Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350, 87–96 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Yofe I., Schuldiner M., Primers-4-Yeast: A comprehensive web tool for planning primers for Saccharomyces cerevisiae. Yeast 31, 77–80 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Kafri M., Metzl-Raz E., Jona G., Barkai N., The cost of protein production. Cell Rep. 14, 22–31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Unger T., Jacobovitch Y., Dantes A., Bernheim R., Peleg Y., Applications of the restriction free (RF) cloning procedure for molecular manipulations and protein expression. J. Struct. Biol. 172, 34–44 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Breslow D. K. et al., A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat. Methods 5, 711–718 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cohen Y., Schuldiner M., Advanced methods for high-throughput microscopy screening of genetically modified yeast libraries. Methods Mol. Biol. 781, 127–159 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Tong A. H. Y., Boone C., Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol. Biol. 313, 171–192 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Albertini M. et al., Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell 89, 83–92 (1997). [DOI] [PubMed] [Google Scholar]
- 45.Studier F. W., Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the main text and SI Appendix.