Significance
Primary stem cells are difficult to transfect, and their viability and function are impaired by traditional transfection methods. We show that nanostraws can be used to deliver RNA to primary human hematopoietic stem cells without any detectable negative effects. Nanostraw-treated cells show no alterations in gene expression and fully retain their proliferative capacity and their potential to regenerate blood cells of different lineages in vivo. This nonperturbative transfection method can benefit functional studies and clinical applications where minimal impact on stem cell function and viability is required.
Keywords: nanostraws, transfection, hematopoietic stem cell, electroporation, mRNA delivery
Abstract
Introduction of exogenous genetic material into primary stem cells is essential for studying biological function and for clinical applications. Traditional delivery methods for nucleic acids, such as electroporation, have advanced the field, but have negative effects on stem cell function and viability. We introduce nanostraw-assisted transfection as an alternative method for RNA delivery to human hematopoietic stem and progenitor cells (HSPCs). Nanostraws are hollow alumina nanotubes that can be used to deliver biomolecules to living cells. We use nanostraws to target human primary HSPCs and show efficient delivery of mRNA, short interfering RNAs (siRNAs), DNA oligonucleotides, and dextrans of sizes ranging from 6 kDa to 2,000 kDa. Nanostraw-treated cells were fully functional and viable, with no impairment in their proliferative or colony-forming capacity, and showed similar long-term engraftment potential in vivo as untreated cells. Additionally, we found that gene expression of the cells was not perturbed by nanostraw treatment, while conventional electroporation changed the expression of more than 2,000 genes. Our results show that nanostraw-mediated transfection is a gentle alternative to established gene delivery methods, and uniquely suited for nonperturbative treatment of sensitive primary stem cells.
Hematopoietic stem cells (HSCs) give rise to all different blood cells throughout life (1). They are used therapeutically in bone marrow transplantations for hematological malignancies (2) and congenital diseases that affect the blood system (3, 4). Just as other primary cell types, human hematopoietic stem and progenitor cells (HSPCs) are a scarce resource, and the number of cells that can be obtained is a limiting factor for clinical applications, such as transplantations (5), and functional assays. HSCs are targeted with nucleic acids for gene therapy purposes (6) and for functional studies to elucidate regulatory mechanisms of HSCs, such as self-renewal, differentiation, and malignant transformation (7). In such studies, foreign genetic material is conventionally introduced by viral transduction (6, 8) or electroporation (9). Integrating viruses such as retro- and lentivirus have low transduction efficiency, especially when aiming for a single integration event per cell (6). They also carry the risk of insertional mutagenesis, which can turn the treated cells into malignant, leukemia-causing cells (10, 11). Electroporation is very efficient, but it has been reported to cause severe damage and functional impairment in HSPCs (12, 13). Since low cell numbers are already a limiting factor for clinical use and in research applications, a nonperturbative gene delivery method, with less impact on the cell viability and function, is acutely needed.
Nanostraws have been used to efficiently deliver molecular cargos to cells with minimal effects on cell viability (14–16). Nanostraws are hollow aluminum oxide tubes (common diameter of 100 to 200 nm, length 1 to 3 μm) that are embedded in a cell culture-compatible polymer membrane. The nanostraws form a direct fluidic pathway from a cargo-containing compartment beneath the nanostraw membrane into the cytoplasm of cells cultured on top of the membrane. Adherent cells cultured on top of nanostraws pull themselves down onto the nanostraws, resulting in the nanostraws piercing through the cell membrane. The cargo is delivered into the cells by passive diffusion or electrokinetically driven by a weak, pulsed electric field (14, 17). Nanostraws have been shown to target small molecules, RNA, DNA, and proteins to different adherent cells in a tunable, nontoxic, and highly efficient manner (17). However, until now, nanostraws have not been shown to be able to gain intracellular access to primary cells in suspension.
Here, we establish a nanostraw-based transfection system for nonadherent primary cell types: centrifugation-enhanced nanostraw transfection (CeNT). We show that CeNT can be used as an efficient tool for small molecule and RNA delivery to human cord blood-derived CD34+ HSPCs. We show that CeNT is exceptionally gentle on this sensitive primary cell type, with no detectable effects on HSPC functionality and gene expression, making it an attractive alternative to other, more disruptive transfection methods.
Results and Discussion
Nanostraws Provide Intracellular Access to Primary Human Hematopoietic Stem and Progenitor Cells.
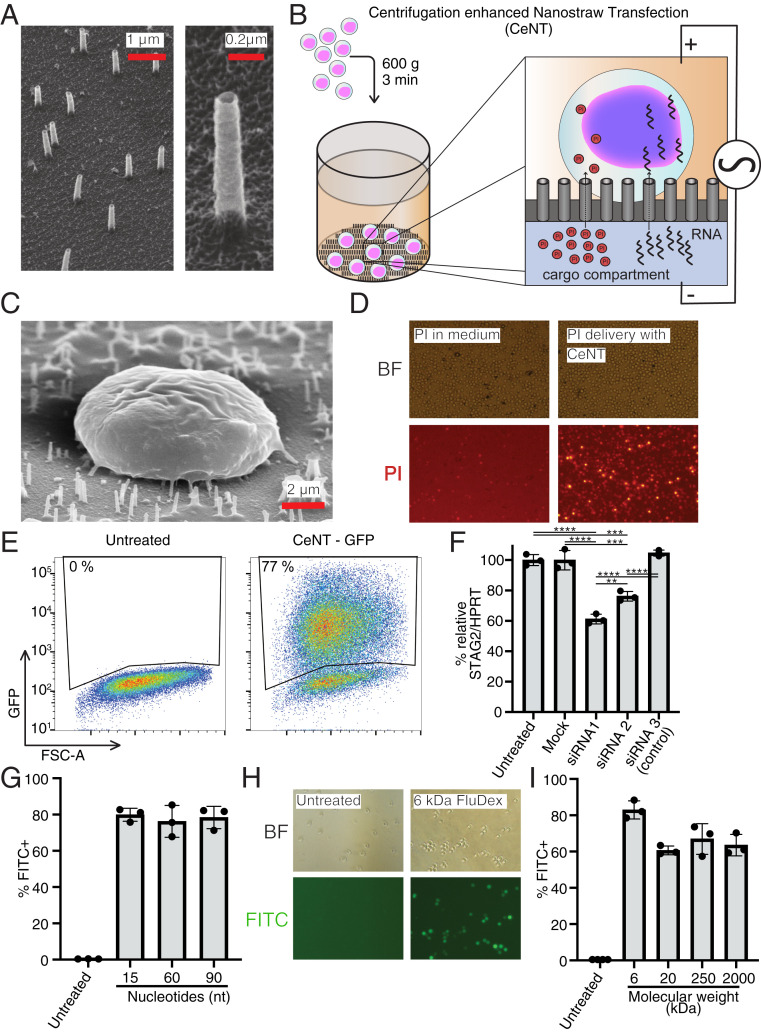
Human primary stem cells, including HSPCs, are challenging to transfect and sensitive to stress that can be caused by the chosen transfection method (9). We attempted to overcome the challenge of transfecting HSPCs by using nanostraws, which have previously been shown to have low impact on cell function and viability. In order to make nanostraws applicable to nonadherent HSPCs, we produced nanostraws as previously described (15, 16, 18) with modifications to the geometry and starting material. The nanostraws were fabricated to have 100- to 130-nm diameter, ∼1-μm length, and an areal density of 3 × 107 cm−2, which corresponds to about 15 nanostraws per cell with 8-μm diameter (Fig. 1A). The nanostraw membrane was fixed to the bottom of a 5-mm plastic tube with the nanostraws pointing into the tube, forming a nanostraw cell culture container. To mimic the tight attachment to the surface that is necessary for successful transfection, we applied an external force by centrifugation to press the cells directly onto the nanostraw membrane (Fig. 1B). To prevent damage to the nanostraws during centrifugation, we adjusted the geometry of the nanostraw membrane by making shorter (1 μm) nanostraws in a mechanically more stable polymer. To that end, we exchanged the track-etched membrane from polycarbonate (PC) to polyethylene terephthalate (PET), which was found to have a higher mechanical strength at fixed film thickness. We applied a low-power, pulsed square electric field across the cells to locally destabilize the cell membrane just above the nanostraw tips and to electrokinetically drive cargo into the cells (14, 15). To account for the less close interface between HSPCs and the nanostraws compared to adherent cells, we applied 40 V instead of the 20-V pulses which are commonly used for adherent cells (14, 16). Thereby, we developed a relatively quick transfection method, taking ∼10 min, including the centrifugation step, and termed it centrifugation-enhanced nanostraw transfection (CeNT).
Fig. 1.
Small molecule and RNA delivery to HSPCs by CeNT. (A) Thirty-degree tilted view SEM pictures of nanostraw membrane. (B) Schematic overview of CeNT procedure. Cargo is delivered into CD34+ HSPCs through nanostraws by application of a gentle, pulsed electric field. (C) A 45° tilted view SEM picture of CD34+ cell on nanostraw membrane. (D) Fluorescent microscopy images of PI delivery to CD34+ cells with CeNT. (Left) Control condition with PI in the medium with the cells. (Right) PI in cargo compartment. (E) Representative FACS plot of CD34+ cells that were exposed to GFP mRNA through CeNT, 6 h post treatment. (F) Relative STAG2 expression levels 2 d after siRNA treatment with CeNT. siRNA1 and siRNA2 target STAG2, and siRNA3 is a control with no specific target (n = 3, **P < 0.005, ***P < 0.0005, ****P < 0.00005). (G) Efficiency of CeNT-mediated DNA oligo delivery to HSPCs (n = 3). (H) Bright-field and fluorescent microscopy images of HSPCs that were targeted with FITC-labeled dextran (6 kDa) using CeNT. (I) Delivery efficiency of FITC-labeled dextrans of different sizes to HSPCs using CeNT (n = 3).
To qualitatively assess whether CeNT can provide intracellular access to the cytosol of human CD34+ HSPCs (Fig. 1C), we delivered propidium iodide (PI), which is a small molecule that does not usually cross the membrane of live cells and only stains dead cells and debris (19). Upon delivering PI to HSPCs using CeNT, ∼70% of the cells took up the dye (Fig. 1D). As a negative control, the voltage pulses were applied with PI added directly to the cell culture medium instead of the cargo compartment, which resulted in practically no uptake of the dye (Fig. 1D). The cytosolic injection of PI shows that CeNT can be used to deliver small nonpermeable molecules to CD34+ cells and encouraged us to attempt delivery of larger molecules which can be used to modify and regulate gene expression.
Efficient RNA Delivery to Human HSPCs by CeNT.
Next, we investigated whether more complex cargoes can be delivered to CD34+ HSPCs using nanostraws by delivering an mRNA encoding for the fluorescent reporter protein GFP. We chose to deliver mRNA because it is generally well-tolerated by human CD34+ cells, while plasmid DNA delivery has cytotoxic effects on this cell type (20). A further benefit of using RNA over plasmid DNA is that it does not have to translocate into the nucleus, reducing the biological complexity and allowing faster detection of the expressed protein. Six hours after treatment, we observed efficient GFP expression in more than 75% of treated CD34+ cells (Fig. 1E). The GFP fluorescence intensity in CeNT-treated cells appeared to be uniformly distributed within the treated cell population (Fig. 1E).
To confirm that other functional RNA species can also be delivered using CeNT, we targeted small interfering RNAs (siRNAs) into HSPCs. Small interfering RNAs can be used to temporarily regulate gene expression by degrading a targeted mRNA in the cell. We used siRNAs against STAG2, a subunit of the cohesin complex, which has previously been successfully knocked down with lentivirally delivered shRNAs (7). Two days after the treatment, qPCR analysis was performed to determine the efficiency of the knockdown. Treatment with two different siRNAs significantly reduced the expression of STAG2 relative to the commonly used reference gene HPRT, while mock treatment or CeNT of a nontargeting siRNA did not affect STAG2 levels (Fig. 1F). Taken together, our results show successful delivery and translation of exogenous mRNA, and also targeted down-regulation of endogenous genes with siRNA by CeNT.
Impact of Nanostraw Length and Cargo Size on Delivery Efficiency.
After establishing nanostraw-mediated intracellular access to HSPCs, we investigated whether nanostraw length and cargo sizes have an impact on delivery efficiency and cell viability. To identify the appropriate length range, we manufactured nanostraws of different lengths, ranging from 200 nm to 3,200 nm (SI Appendix, Fig. S1A), and used them to deliver GFP mRNA to CD34+ cells. One day after the delivery, we analyzed GFP fluorescence using flow cytometry and found that lengths up to 1,100 nm resulted in satisfactory delivery efficiencies of 50% or higher (SI Appendix, Fig. S1B). Longer nanostraws performed progressively worse, with 3,200 nm length being completely unsuitable for delivery. Nanostraws are manufactured by removing layers of polymer to expose the alumina nanochannels that are already embedded in the membrane. The length of the nanostraws can be increased by removing more polymer, but the total length of the alumina channels is constant. Therefore, we do not expect increased clogging with longer nanostraws because the cargo has to travel the same distance through the alumina nanochannels regardless of how long the nanostraws are. It is possible that the reduced rigidity and stiffness of longer nanostraws might prevent successful cell piercing and cause the observed lower delivery efficiencies. Treated cells maintained their viability, as determined by 7AAD and Annexin V staining, compared to untreated cells, regardless of nanostraw length (SI Appendix, Fig. S1C).
A transfection method is most useful if it can be used to deliver diverse cargos of different sizes. Therefore, we investigated if the efficiency of nanostraw-mediated delivery is affected by the molecular size of the cargo. To this end, we used nanostraws to target HSPCs with FITC-tagged DNA oligonucleotides of different lengths and fluorescein-labeled dextran molecules of molecular weights ranging from 6 kDa to 2,000 kDa. The delivery efficiency was determined by flow cytometry directly after the cargo delivery and a washing step. DNA oligos could be delivered with average efficiencies ranging from 76% to 80% (Fig. 1G). Delivery of the shortest, 6-kDa dextran was delivered to 83% of treated cells on average. Longer dextrans (20, 250, and 2,000 kDa) were delivered with average efficiencies ranging from 61% to 67% (Fig. 1 H and I). We also stained treated cells with the dead-cell stain 7AAD and found that there is no immediate toxicity resulting from the delivery of differentially sized molecules (SI Appendix, Fig. S2 A and B). Taken together, these results indicate that delivery of smaller molecules is slightly more efficient than delivery of larger molecules. Despite this, even extremely large molecules of up to 2,000 kDa can successfully be delivered using nanostraws, which confirms that molecular weight is not a limiting factor for CeNT within the tested size range.
HSPCs Are Fully Viable and Functional after CeNT Treatment.
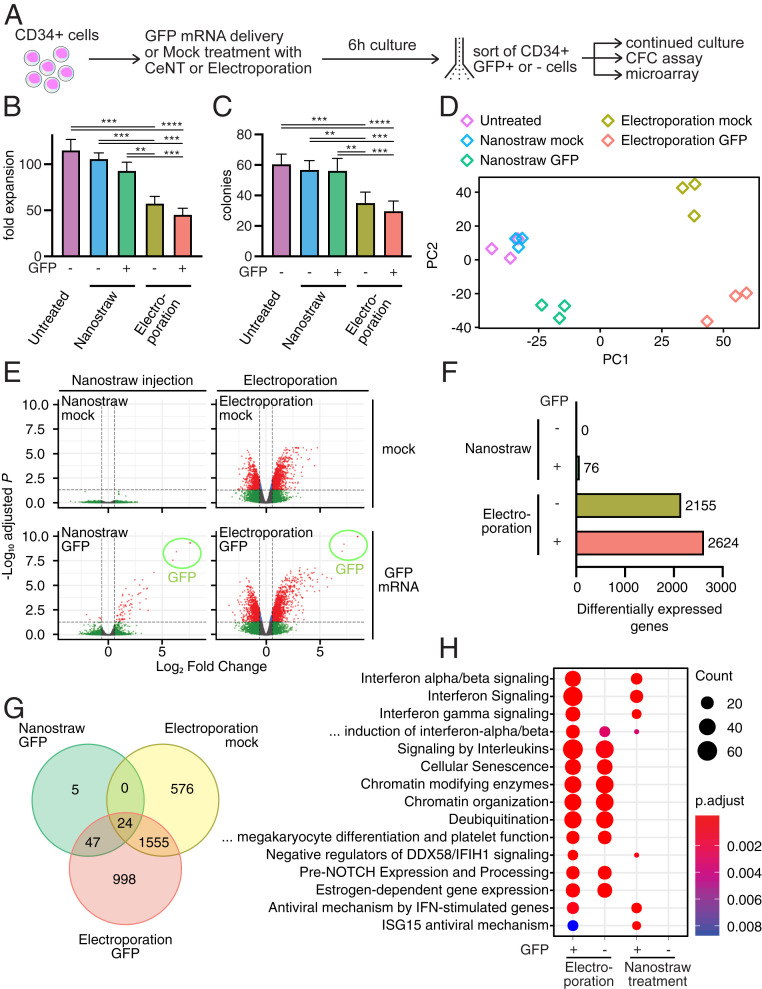
In the next experiment, we performed a more detailed investigation into the effect of CeNT on cell viability and function, and compared it directly with conventional electroporation using a commercially available kit specifically designed for CD34+ cells (Fig. 2A). We subjected the same batch of HSPCs to CeNT-based delivery of GFP mRNA or mock treatment, as well as conventional electroporation with GFP mRNA or mock (SI Appendix, Fig. S3A). The same electric pulse was applied in all mock conditions, but without mRNA present in the delivery buffers. All conditions were performed in parallel and in triplicates, and the cells were cultured under the same conditions (cell concentration, medium) in parallel to untreated cells. After 6 h, live CD34+GFP+ or CD34+GFP− cells were sorted for continued culture, colony-forming cell (CFC) assays, and gene expression analysis. We also cultured unsorted cells of each condition and measured cell viability with 7AAD and Annexin V staining after 1 and 2 d in culture. We found that nanostraw-treated cells were equally viable as untreated cells at both time points, whereas electroporation led to a strong reduction of viability (SI Appendix, Fig. S3 B and C).
Fig. 2.
CD34+ HSPCs are viable and fully functional after CeNT. (A) Schematic outline of experiment. (B) Fold expansion of cells treated with different conditions after 2 wk in culture (n = 9; n = 3 biological and n = 3 technical replicates; **P < 0.005, ***P < 0.0005, ****P < 0.00005). Below the x-axis, “GFP -” denotes mock treatment without mRNA. (C) Total number of colonies derived from sorted, live CD34+GFP+ or CD34+GFP− cells after 2 wk in culture (n = 8 to 9; n = 3 biological and n = 2 to 3 technical replicates; **P < 0.005, ***P < 0.0005, ****P < 0.00005). (D) Principal component analysis of sorted, live CD34+GFP+/− cells (n = 3). (E) Volcano plots of CeNT and electroporation conditions compared to untreated cells 6 h after treatment (n = 3). (F) Number of differentially expressed genes (fold change > 1.5, adj. P < 0.05). (G) Venn diagram visualizing differentially expressed genes that are overlapping in different treatment conditions. (H) Pathway enrichment analysis of up-regulated genes.
To assess the proliferative capacity of treated cells, we plated equal numbers of sorted live cells and determined the total number of live cells after 2 wk in culture (Fig. 2B). Completely untreated cells expanded 115 ± 21-fold during a 2-wk period, and nanostraw mock- and nanostraw GFP-treated cells expanded 106 ± 12- and 93 ± 14-fold, respectively. Electroporated cells (mock and GFP) expanded substantially less, 57 ± 15- and 45 ± 11-fold, respectively. Overall, this shows that the proliferative capacity of CeNT-treated cells, in contrast to electroporated cells, is not significantly affected by nanostraw treatment.
To investigate whether CeNT treatment affects the functional capacity of HSPCs to form colonies, we performed a colony-forming cell (CFC) assay. Live CD34+ cells were plated on semisolid, methylcellulose MethoCult medium with differentiation-promoting cytokines. After 14 d, the number and type of colonies were determined. Untreated, nanostraw mock-treated, and nanostraw GFP-treated cells gave rise to similar numbers of colonies, 60 ± 8, 57 ± 6, and 56 ± 8, respectively. Electroporated cells, both with and without GFP, showed greatly reduced colony forming capability (35 ± 8, 30 ± 7), even though the same number of live cells was plated (Fig. 2C). The colony-forming capacity was thus significantly lower in both electroporation settings compared to untreated and nanostraw-treated cells. Additionally, nanostraw-treated cells and untreated cells gave rise to similar sized colonies, whereas electroporated cells resulted in smaller and less dense colonies. This is in line with previously published data that show more than 75% reduction of colony-forming capacity in human CD34+ cells upon electroporation (13). In summary, the ability of CD34+ progenitor cells to mature and form colonies is not affected by nanostraw treatment, which is key for clinical applications and functional studies.
CeNT Treatment Does Not Perturb Gene Expression.
To detect more subtle effects of CeNT on the cells, we performed global gene expression analysis. Six hours after treatment, we sorted CD34+GFP+ or CD34+GFP− cells and performed microarray analysis. We used principal component analysis (PCA) to visualize how the gene expression compared between the different treatments. Both mock and GFP CeNT-treated cells, unlike electroporated cells, clustered together closely with untreated cells in the PCA (Fig. 2D). This indicates that the gene expression profiles of nanostraw-treated HSPCs are very similar to untreated HSPCs.
Remarkably, we did not detect any significantly up- or down-regulated genes in response to mock nanostraw treatment (Fig. 2 E and F). This shows that subjecting CD34+ cells to nanostraw treatment does not cause a strong change at the transcriptional level, further corroborating the previous results, which show that cell viability and function are not perturbed. When delivering GFP mRNA using nanostraws, 76 genes were up- or down-regulated. Electroporation had a strong impact on global gene expression, with over 2,000 genes affected, regardless of whether GFP mRNA was present or not (Fig. 2F). Of note, 71 of 76 differentially regulated genes in the nanostraw GFP delivery were also differentially regulated in the electroporation GFP condition (Fig. 2G), indicating that these genes are specifically associated with the presence of GFP mRNA.
Pathway enrichment analysis showed that differentially regulated genes in the nanostraw GFP condition were mostly related to response mechanisms to foreign RNAs, such as IFN signaling (Fig. 2H) (21). These pathways were also enriched in the electroporation GFP condition, but not the electroporation mock condition, supporting the notion that these pathways are related to the delivery of exogenous GFP mRNA. In both electroporation conditions, several pathways related to cellular stress were up-regulated. Importantly, we did not see any up-regulation of these stress-related pathways in nanostraw-treated cells. This may be one of the reasons for the better cell function of nanostraw-treated cells over electroporated cells. Moreover, the absence of differentially expressed genes from CeNT is of great benefit for studying the immediate effect of introduced biomolecules without any interfering signals from the delivery method.
CeNT-Treated HSPCs Engraft in Immunocompromised Mice.
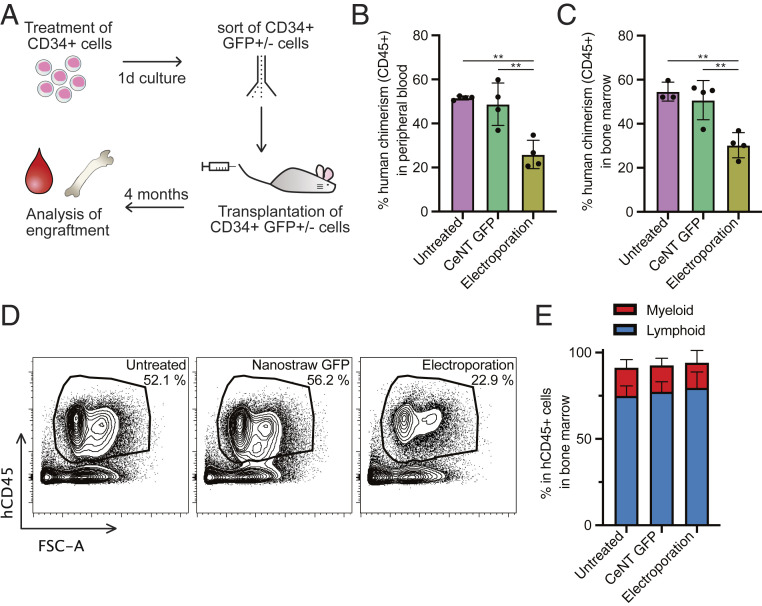
Our previous in vitro experiments confirmed that hematopoietic progenitor cells are not negatively affected by CeNT. Next, we wanted to investigate the impact of CeNT on functional, long-term repopulating HSCs and their multilineage engraftment potential in vivo. CD34+ cells were treated with nanostraw injection and mock electroporation. After 1 d in culture, live CD34+GFP+ or CD34+GFP− cells were sorted and injected in equal numbers into sublethally irradiated, immunocompromised mice (Fig. 3A). For the nanostraw-treated cells, only GFP+ cells were sorted to ensure that only cells that were successfully targeted with the nanostraws were transplanted (SI Appendix, Fig. S4). After 4 mo, the percentage of human cells in the peripheral blood was 52 ± 1% in the untreated and 49 ± 10% in the nanostraw-treated condition, indicating that the engraftment potential of human HSPCs is not impaired by nanostraw treatment. Cells treated with conventional electroporation had significantly lower engraftment potential, with 26 ± 6% (Fig. 3B). Similar results were observed in the bone marrow (Fig. 3 C and D). Nanostraw-treated cells gave rise to both myeloid and lymphoid cells, and we observed a similar lineage distribution in all conditions (Fig. 3E). This further confirms that the functional properties of human HSPCs are maintained upon CeNT.
Fig. 3.
CeNT-treated hematopoietic stem cells successfully engraft in mice. In the CeNT GFP condition, only GFP+ cells were sorted and transplanted. (A) Outline of experiment. Percentage of human CD45+ cells 4 mo posttransplantation in (B) peripheral blood and (C) bone marrow (n = 3 to 4; **P < 0.005). (D) Representative FACS plots showing the frequency of human CD45+ cells in mouse bone marrow 4 mo posttransplantation. (E) Percentage of lymphoid (CD19+ B cells and CD3+ T cells) and myeloid (CD33+) cells within the human CD45+ population in the mouse bone marrow.
Conclusion
Human HSPCs are recalcitrant to transfection (22). Currently existing methods for nucleic acid delivery, such as electroporation, have greatly expanded our knowledge of HSC biology but have limitations with regard to safety, toxicity, or preservation of regular cell function. We show that CeNT is an efficient alternative method in the transfection toolbox that can be used for delivering nonpermeable small molecules, siRNA, and mRNA to primary human HSPCs. CeNT-delivered RNA species execute their expected biological function, and the treatment is exceptionally gentle and does not cause any functional impairment. Human HSPCs that were treated with CeNT completely retained their proliferative capacity and colony-forming potential. They also engraft in immunocompromised mice with similar efficiency as untreated cells and result in long-term and multilineage blood production. This confirms that even naïve and undifferentiated HSCs, which are particularly difficult to target, can be targeted using CeNT.
The gentle nature of this delivery method is further underlined by the minimal impact on the gene expression of treated cells. While conventional bulk electroporation causes dysregulation of several thousand genes, global gene expression is virtually unaffected by CeNT. This allows for the detection of subtle changes in gene expression that are caused purely by the presence of the introduced RNA. Such changes are at risk for being masked by the profound effects on gene expression that other methods have. CeNT makes it possible to study the immediate effect of transient mRNA delivery on primary cells without having interfering signals from the transfection method. This is useful for elucidating subtle mechanisms and pathways that might remain hidden otherwise.
Limited cell numbers make it difficult to study the biology of primary cells. Any treatment of such cells should not reduce the already low number of available cells, and also not impair their function. This becomes especially important when working with patient-derived cells, which are even more sensitive to stress. CeNT is an efficient method for targeting sensitive primary stem cells, and we expect it to be useful for both clinical applications and functional studies, where maximal cell recovery, viability, and performance are essential.
Methods
Ethics Statement.
Work with primary human samples was approved by the regional ethical committee for Lund/Malmö (Regionala Etikprövningsnämnden i Lund/Malmö), approval no. 2010–696. Informed consent was obtained from mothers of the umbilical cord blood donors, and all samples were deidentified prior to use in the study. All animal experiments were approved by the regional animal experiment ethical review board in Malmö/Lund.
Production and Assembly of Nanostraws.
Nanostraws were developed in close collaboration with Navan Technologies. Track-etched polyethylene terephthalate (PET) membranes (GVS) with pore diameter of 100 nm, pore density of 3 × 107 cm−2, and membrane thickness of 12 μm were used as templates for nanostraw fabrication. The membranes were coated in a Savannah S100 Atomic Layer Deposition reactor (Cambridge Nanotech) using trimethylaluminum and water as precursors. About 10 nm aluminum oxide was deposited. The top-layer aluminum oxide was removed using inductively coupled plasma reactive ion etching (ICP-RIE) in an APEX SLR plasma etcher (Plasma Therm) using an Ar plasma at 100 W RF, 250 W ICP for 2 min. Oxygen-based RIE was used to form the nanostraws by removing some of the PET membrane using 25 W RF, 500 W ICP for 90 s. Successful processing was confirmed by cutting out a small piece of the nanostraw membrane, which was coated by 4 nm Pd:Pt and imaged by scanning electron microscopy (SEM) in a LEO 1560 SEM (Zeiss) using a thermal field emission gun operating at 15 kV. The processed nanostraw membranes were attached to the bottom of cell culture-compatible plastic tubes using double-sided tape in order to form a cell culture chamber. The tubes had an inner diameter of 5 mm and could hold up to 300 µL of liquid. The assembled cell culture chambers were sterilized in a custom-built UV/ozone box for 5 min. For the nanostraw length series, the following processing steps were performed: after ALD coating as described above, the membrane (same batch) was cut into smaller pieces for oxygen RIE. Oxygen RIE etching times ranging from 1 s to 360 s were used to form nanostraws of lengths from 200 nm to 3,200 nm. The nanostraw length was estimated from 30° tilted view SEM images of nanostraws standing up on their substrate (for lengths up to 1 μm). Due to charging effects, nanostraws longer than 1 μm had to be broken and measured while lying down on the substrate.
Cell Culture.
Umbilical cord blood was collected at Skåne University Hospital and Helsingborg Hospital. Mononuclear cells were extracted using Lymphoprep tubes (Alere Technologies, no. 1019818). CD34+ cells from mixed donors were isolated from mononuclear cells with a CD34 MicroBead Kit (Miltenyi Biotec, no. 130–046-703) according to the manufacturer’s instructions and cryopreserved in FBS supplemented with 10% DMSO. CD34+ cells were cultured in StemSpan SFEM (Stemcell Technologies, no. 09650) with human SCF (PeproTech, no. 300–07), human TPO (PeproTech, no. 300–18), and human Flt3L (PeproTech, no. 300–19) at 100 ng/mL each at 37 °C and 5% CO2.
CeNT Treatment.
For experiments in Fig. 2, CD34+ cells were thawed and cultured for 1 d. The cells were split and treated with the following conditions: nanostraw mock, nanostraw GFP, electroporation mock, electroporation GFP, and completely untreated. Each condition was performed in separate, independent triplicates. For each nanostraw treatment, 150,000 cells were centrifuged (600 × g, 3 min, 20 °C) onto the nanostraw membrane. A droplet containing the cargo was placed on top of a Au-coated glass slide. For the mock condition, the membrane was placed on top of a 10-μL 0.1× PBS droplet or, for the GFP condition, onto a 10-μL 0.1× PBS droplet containing 2 μg EGFP mRNA (L-7601; TriLink BioTechnologies). A pulsed square electric field (40 V, 40 Hz, 200 μs, 3 × 40 s) was applied in between the Au slide and a Pt electrode dipped into the cell culture container using a Grass S48 Stimulator (Astro-Med). The applied voltage at the Pt electrode and the current flowing through were monitored using an oscilloscope (PICOSCOPE 2206B; Pico Technology). The Pt electrode was kept at positive bias in order to help drive negatively charged mRNA through the nanostraws and into the cells. For each electroporation treatment, 1 × 106 cells were either electroporated with GFP mRNA or mock electroporated without any cargo, as described in more detail below. Cells were then cultured with exactly the same cell concentration in each condition. After 6 h, live 7AAD-CD34+GFP+, or GFP− for mock conditions, cells were sorted for a proliferation assay, a CFC assay, and microarray analysis. Each individual treatment condition was sorted before the next replicates were sorted to account for any time-dependent effects. During the sort, the efficiency of the GFP delivery was recorded.
Scanning Electron Microscopy (SEM).
For SEM of HSPCs on nanostraws, additional sample preparation was needed. HSPCs were centrifuged onto the nanostraw membrane, followed by three rinses in PBS to remove cell media. The cells were then fixed in 2.5% glutaraldehyde. The next day, the cells were dehydrated in an EtOH drying series of 10 min each in 30%, 50%, 75%, 90%, 95%, and 99.5% EtOH. Critical point drying (CPD) was used to completely remove the EtOH from the cells with minimal disruption of the cellular shape in a Quorum K850 CPD (Quorum Technologies). The CP dried samples were coated with 4 nm Pd:Pt and imaged with a LEO 1560 SEM (Zeiss).
Propidium Iodide Delivery.
Propidium iodide (PI) was diluted to 200 μg/mL in 0.1× PBS, and 10 μL was placed on the bottom electrode. CD34+ cells were centrifuged (600 × g, 3 min) onto the nanostraw membrane and placed onto the PI-containing droplet. A pulsed electric field (20 to 30 V, 40 Hz, 200 μs, 2× 40 s) was applied to drive the PI delivery into the cytoplasm of the cells. After the treatment, the bottom of the nanostraw membrane was rinsed and the cells were directly imaged with an Olympus IX70 fluorescence microscope. The delivery efficiency was estimated by dividing the number of PI-positive cells (red) by the total number of cells that were counted in an image taken at 40× magnification. As a negative control, the experiment was performed with a 10-μL 0.1× PBS droplet without any PI placed below on the bottom electrode and the same absolute amount of PI that was used in the previous delivery added directly into the cell containing medium. After application of the pulsed electric field, cells were imaged as above.
Dextran and DNA Oligo Delivery.
FITC-labeled DNA oligos (15 nt, 60 nt, and 90 nt) with the following random sequences were ordered from Integrated DNA Technologies: 15 nt, ACTGGTCAAC/iFluorT/GGTC; 60 nt, ACTGGTCAAC/iFluorT/GGTCATCCTGAAGT AAATGCTATGCGACTGATTGGGCTACGCTCCGCTA; and 90 nt, ACGCGGCTAGGG ACTGGTCAAC/iFluorT/GGTCATCCTGAAGTAAATGCTATGCGACTGATTGGGCTACGCTCCGCTAAAAAAGTGCTAAAGGCAG. They were delivered by placing a 10-μL droplet of 0.1× PBS containing 1 nmol DNA oligo under the nanostraw membrane and applying a pulsed electric field (40 V, 40 Hz, 200 μs, 3 × 40 s). Fluorescein–dextran (6 kDa, 20 kDa, 250 kDa, and 2,000 kDa) was purchased from Fina Biosolutions and delivered by placing a 10-μL H2O droplet containing 1 μg/μL fluorescein–dextran under the nanostraw membrane and applying a pulsed electric field (40 V, 40 Hz, 200 μs, 3 × 40 s).
Electroporation.
Electroporation was performed using the Nucleofector 2b Device (Lonza) and the Human CD34+ Cell Nucleofector Kit (VAPA-1003; Lonza) according to the manufacturer’s instructions. For each electroporation, 1 × 106 cells were treated. In conditions where GFP was delivered, 10 μg Cleancap EGFP mRNA (L-7601; TriLink BioTechnologies) was added to the electroporation mix.
siRNA Delivery.
We designed an siRNA against STAG2 with the target sequence GCAGUUCUUACAGCUUUGUUU (siRNA1). It was synthesized together with two predesigned siRNAs (siRNA2-STAG2, SASI_Hs02_00311139; siRNA3 nontargeting control, SIC001) by Sigma Aldrich/Merck. A total of 30,000 CD34+ cells were centrifuged (600 × g, 3 min) onto a nanostraw membrane, placed onto a 10-μL 0.1× PBS droplet containing 20 pmol siRNA, and exposed to a pulsed electric field (40 V, 40 Hz, 200 μs, 3 × 40 s). Each siRNA delivery was performed in triplicates. Two days after the treatment, RNA was extracted from the cells using the RNeasy Micro Kit (Qiagen) and reverse-transcribed using the SuperScript IV First-Strand Synthesis System (Thermo Fisher Scientific). qPCR was performed with TaqMan assays for STAG2 (Hs00198227_m1) and HPRT (Hs99999909_m1) using the 7900HT Fast Real-Time PCR System (Applied Biosystems).
Flow Cytometry/FACS.
The following anti-human antibodies were used for staining: sort for microarray, long-term culture, and CFC assay, CD34-A700 (BD Pharmingen no. 561440, clone 581); bone marrow and peripheral blood engraftment analysis, CD45-APC (Biolegend no. 304037, clone HI30), CD33-PE (Biolegend no. 303404, clone WM53), CD19-BV605 (BD Horizon no. 562653 clone SJ25-C1), and CD3-PE-Cy7 (Invitrogen no. 25–0038-42, clone UCHT1). 7AAD (Sigma-Aldrich, no. A9400) was used to discriminate dead cells. Stained cells were analyzed with a BD LSRFortessa or BD FACSCanto or sorted with a BD FACSAria IIu or BD FACSAria III. Data were analyzed with FlowJo 10.6.1 (BD).
Cell Viability Assay.
Cell viability was determined using the 7AAD/PE Annexin V Apoptosis Detection KIT I (BD Biosciences, no. 559763) according to the manufacturer’s instructions.
Proliferation Assay.
For each sample, 300 live human CD34+GFP+/− cells (n = 3) were seeded in a 96-well U-bottom plate. After 2 wk in culture, total live cell numbers were determined with flow cytometry.
Colony-Forming Assay.
A total of 167 live human CD34+GFP+/− cells were sorted into 1 mL MethoCult H4230 (Stemcell Technologies, no. 04230) with 20% IMDM, 25 ng/mL hSCF, 50 ng/mL GM-CSF, 25 ng/mL IL3, and 2 U/mL EPO. The cells were cultured in six-well plates for 2 wk at 37 °C and 5% CO2. Colonies were counted and scored by an experienced person who was blinded to the sample identity.
Microarray.
A total of 20,000 live, human CD34+GFP+/− cells were sorted into lysis buffer and immediately snap-frozen at −80 °C. Total RNA was extracted with the RNeasy Micro Kit (Qiagen) according to the manufacturer’s protocol. Purity and integrity of the RNA was assessed with the Agilent 2100 Bioanalyzer with the RNA 6000 Pico LabChip reagent set (Agilent). Sample preparation and processing with Affymetrix Human Gene 2.0 arrays were performed at an Affymetrix service provider and core facility, “KFB–Center of Excellence for Fluorescent Bioanalytics” (Regensburg, Germany).
Microarray Data Analysis.
Microarray data were normalized with the robust multiarray average (rma) function of the R (23) package oligo, version 1.49.0 (24), and annotated with ENTREZIDs from the Bioconductor (25) annotation data package org.Hs.eg.db, version 3.8.2, using the AnnotationDbi package, version 1.47.0. Principal component analysis was performed with the prcomp function of the stats package, version 3.6.0, and plotted with the ggplot2 package, version 3.1.1 (26). Differentially expressed genes (FC > 1.5, adjusted P < 0.05) were detected using the limma package, version 3.41.2 (27), and volcano plots were created using the EnhancedVolcano package, version 1.3.0. Pathway enrichment analysis of up-regulated genes was performed using clusterProfiler, version 3.13.0 (28).
Animal Experiments.
Cord blood-derived human CD34+ cells were thawed and cultured for 1 d before they were divided for the following treatment conditions: untreated, electroporated with Human CD34+ Cell Nucleofector Kit (VAPA-1003; Lonza), and CeNT with GFP mRNA. The treated cells were cultured for 1 d. Live (7AAD-)CD34+ and, if GFP was part of the treatment, GFP+ cells were sorted for the following transplantation. For each treatment condition, sublethally irradiated (300 cGy) NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ. (NSG) mice were injected with 59,000 cells in 500 μL PBS/2% FBS into the tail vein. Bone marrow and peripheral blood were analyzed for engraftment and lineage distribution of human cells with flow cytometry after 4 mo.
Statistics.
When comparing multiple groups, GraphPad Prism 8 was used to perform one-way ANOVA with Tukey’s multiple comparison test. Data in figures are shown as mean ± SD, and significance is indicated with asterisks (*P < 0.05, **P < 0.005, ***P < 0.0005, ****P < 0.00005).
Supplementary Material
Acknowledgments
We thank the Lund University BMC animal house facility and FACS core for support. Nanostraws were produced and characterized at the Lund Nano Laboratory. J.L. acknowledges support from the Swedish Research Council, the Swedish Cancer Foundation, the Swedish Pediatric Cancer Foundation, the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (Grant 648894), and the StemTherapy program at Lund University. A.S. was supported by the Swedish Cancer Foundation and Lady TATA memorial trust. C.N.P. acknowledges funding from ERC-CoG Grant NanoPokers (662206), the Swedish Foundation for Strategic Research (ITM17 Grant), the Swedish Research Council, The Crafoord Foundation, and NanoLund. M.H. acknowledges funding from the Knut and Alice Wallenberg Foundation under the Stanford Postdoctoral Fellowship Program.
Footnotes
Competing interest statement: M.H. is Chief Technology Officer at Navan Technologies, Inc., a startup commercializing nanostraws.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001367117/-/DCSupplemental.
Data Availability.
Microarray data were deposited in the Gene Expression Omnibus (GEO) database of the National Center for Biotechnology Information (NCBI) under the GEO accession number GSE151027 (29).
References
- 1.Shizuru J. A., Negrin R. S., Weissman I. L., Hematopoietic stem and progenitor cells: Clinical and preclinical regeneration of the hematolymphoid system. Annu. Rev. Med. 56, 509–538 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Vogel W., Scheding S., Kanz L., Brugger W., Clinical applications of CD34(+) peripheral blood progenitor cells (PBPC). Stem Cells 18, 87–92 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Mahmoud H. K. et al., Allogeneic hematopoietic stem cell transplantation for non-malignant hematological disorders. J. Adv. Res. 6, 449–458 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faraci M. et al., Allogeneic hematopoietic stem cell transplantation in congenital disorders: A single-center experience. Pediatr. Transplant. 21 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Mayani H., Wagner J. E., Broxmeyer H. E., Cord blood research, banking, and transplantation: Achievements, challenges, and perspectives. Bone Marrow Transplant. 55, 48–61 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Naldini L., Genetic engineering of hematopoiesis: Current stage of clinical translation and future perspectives. EMBO Mol. Med. 11, e9958 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galeev R. et al., Genome-wide RNAi screen identifies cohesin genes as modifiers of renewal and differentiation in human HSCs. Cell Rep. 14, 2988–3000 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Genovese P. et al., Targeted genome editing in human repopulating haematopoietic stem cells. Nature 510, 235–240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diener Y., Bosio A., Bissels U., Delivery of RNA-based molecules to human hematopoietic stem and progenitor cells for modulation of gene expression. Exp. Hematol. 44, 991–1001 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Hacein-Bey-Abina S. et al., Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest. 118, 3132–3142 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howe S. J. et al., Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J. Clin. Invest. 118, 3143–3150 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.von Levetzow G. et al., Nucleofection, an efficient nonviral method to transfer genes into human hematopoietic stem and progenitor cells. Stem Cells Dev. 15, 278–285 (2006). [DOI] [PubMed] [Google Scholar]
- 13.DiTommaso T. et al., Cell engineering with microfluidic squeezing preserves functionality of primary immune cells in vivo. Proc. Natl. Acad. Sci. U.S.A. 115, E10907–E10914 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie X. et al., Nanostraw-electroporation system for highly efficient intracellular delivery and transfection. ACS Nano 7, 4351–4358 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Cao Y. et al., Nondestructive nanostraw intracellular sampling for longitudinal cell monitoring. Proc. Natl. Acad. Sci. U.S.A. 114, E1866–E1874 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao Y. et al., Universal intracellular biomolecule delivery with precise dosage control. Sci. Adv. 4, eaat8131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu A. M. et al., Quantification of nanowire penetration into living cells. Nat. Commun. 5, 3613 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.VanDersarl J. J., Xu A. M., Melosh N. A., Nanostraws for direct fluidic intracellular access. Nano Lett. 12, 3881–3886 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Nicoletti I., Migliorati G., Pagliacci M. C., Grignani F., Riccardi C., A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J. Immunol. Methods 139, 271–279 (1991). [DOI] [PubMed] [Google Scholar]
- 20.Wiehe J. M. et al., mRNA-mediated gene delivery into human progenitor cells promotes highly efficient protein expression. J. Cell. Mol. Med. 11, 521–530 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diebold S. S., Kaisho T., Hemmi H., Akira S., Reis e Sousa C., Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303, 1529–1531 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Papapetrou E. P., Zoumbos N. C., Athanassiadou A., Genetic modification of hematopoietic stem cells with nonviral systems: Past progress and future prospects. Gene Ther. 12 (suppl. 1), S118–S130 (2005). [DOI] [PubMed] [Google Scholar]
- 23.R Core Team , R: A Language and Environment for Statistical Computing, (R Foundation for Statistical Computing, Vienna, Austria, 2013). [Google Scholar]
- 24.Carvalho B. S., Irizarry R. A., A framework for oligonucleotide microarray preprocessing. Bioinformatics 26, 2363–2367 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huber W. et al., Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 12, 115–121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wickham H., ggplot2: Elegant Graphics for Data Analysis, (Springer-Verlag, New York, 2016). [Google Scholar]
- 27.Ritchie M. E. et al., Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu G., Wang L. G., Han Y., He Q. Y., clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmiderer L., et al. , Gene expression changes in human cord blood-derived hematopoietic stem and progenitor cells upon Centrifugation enhanced Nanostraw Transfection or Conventional Electroporation. Gene Expression Omnibus (GEO) database. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE151027. Deposited 21 May 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray data were deposited in the Gene Expression Omnibus (GEO) database of the National Center for Biotechnology Information (NCBI) under the GEO accession number GSE151027 (29).