Significance
Arabidopsis ANT controls plant growth and floral organogenesis. We determined the binding motif of maize ANT1 (ZmANT1), showing that ZmANT1 binds promoters of millet SCR1, GNC, and AN3, key regulators of Kranz anatomy, chloroplast development, and plant growth, respectively. We generated mutants of the ANT1 ortholog in the millet Setaria viridis. Two frameshift mutants displayed reduced photosynthesis efficiency and growth rate, smaller leaves, and lower yields than WT plants. Moreover, their leaves sporadically exhibited distorted vein structure. Transcriptomic comparison of WT and mutant leaves identified down-regulated genes enriched in photosynthesis and tetrapyrrole binding in ant1 mutants. Thus, our study identified ANT1 as a master switch functioning upstream of key regulators of signature developmental processes beyond plant growth and floral organogenesis.
Keywords: AINTEGUMENTA, vascular development, Kranz anatomy, chloroplast development, photosynthesis
Abstract
Arabidopsis AINTEGUMENTA (ANT), an AP2 transcription factor, is known to control plant growth and floral organogenesis. In this study, our transcriptome analysis and in situ hybridization assays of maize embryonic leaves suggested that maize ANT1 (ZmANT1) regulates vascular development. To better understand ANT1 functions, we determined the binding motif of ZmANT1 and then showed that ZmANT1 binds the promoters of millet SCR1, GNC, and AN3, which are key regulators of Kranz anatomy, chloroplast development, and plant growth, respectively. We generated a mutant with a single-codon deletion and two frameshift mutants of the ANT1 ortholog in the C4 millet Setaria viridis by the CRISPR/Cas9 technique. The two frameshift mutants displayed reduced photosynthesis efficiency and growth rate, smaller leaves, and lower grain yields than wild-type (WT) plants. Moreover, their leaves sporadically exhibited distorted Kranz anatomy and vein spacing. Conducting transcriptomic analysis of developing leaves in the WT and the three mutants we identified differentially expressed genes (DEGs) in the two frameshift mutant lines and found many down-regulated DEGs enriched in photosynthesis, heme, tetrapyrrole binding, and antioxidant activity. In addition, we predicted many target genes of ZmANT1 and chose 13 of them to confirm binding of ZmANT1 to their promoters. Based on the above observations, we proposed a model for ANT1 regulation of cell proliferation and leaf growth, vascular and vein development, chloroplast development, and photosynthesis through its target genes. Our study revealed biological roles of ANT1 in several developmental processes beyond its known roles in plant growth and floral organogenesis.
Arabidopsis thaliana AINTEGUMENTA gene (AtANT), which belongs to the APETALA 2 (AP2) transcription factor (TF) family, was first discovered for its role in ovule development and floral organ growth (1, 2). Later, AtANT was found to be involved in organ primordium initiation, female gametophyte formation, and organ growth and polarity (1–6). For example, transgenic Arabidopsis plants overexpressing AtANT exhibited large leaves, stems, sepals, and other organs, while its loss-of-function mutants showed a smaller organ size (1–4). It has been suggested that the ANT gene maintains meristematic competence and acts as an organ-size checkpoint (7). Moreover, ANT has been linked to gynoecium development and vascular development in roots in Arabidopsis (8, 9). Recently, Krizek et al. (10) described the roles of ANT in floral growth and patterning in Arabidopsis. According to the Arabidopsis eFP browser (11), the expression of AtANT is predominant in but not restricted to the shoot apex, suggesting that AtANT has other functions in addition to regulating cell proliferation and organogenesis.
In this study, we found from our previous time-course transcriptomes of maize leaf development (12) that ZmANT1 is coexpressed with the SCARECROW1 gene (ZmSCR1, Zm00001d052380), which is a key regulator of Kranz anatomy development in C4 plants (13). This observation suggested that ZmANT1 is a regulator of ZmSCR1. This possibility is worth pursuing because Kranz anatomy, in which the vein is wrapped by one layer of bundle sheath cells and then one layer of mesophyll cells, is the structure that underlies the superior photosynthetic efficiency of C4 plants. We therefore determined the ZmANT1 binding motif and showed that ZmANT1 binds the promoter of ZmSCR1. To further understand the biological function of the ANT1 gene in C4 plants, we pursued a loss-of-function study of ANT1. For this purpose, we used Setaria viridis as the model because it has a shorter generation time and is more amenable to transformation than maize. The SvANT1 gene was then edited via a CRISPR/Cas9 system to generate three homozygous mutant lines for phenotypic, genetic, and molecular analyses to examine the function of ANT1 in S. viridis. These mutant lines displayed a slower growth rate, smaller leaves and spikes, and lower grain yield than the wild type (WT). Moreover, the two mutant lines with a ANT1 frameshift mutation displayed distorted Kranz anatomy and vein spacing in some regions. We used transcriptomic analysis to identify genes that were differentially expressed in these mutant lines compared to WT. We predicted the target genes of SvANT1 and identified those differentially expressed between WT and a mutant line. Of particular interests are SvGNC and SvAN3, which are key regulators of chloroplast development and plant growth, respectively. We further showed that ZmANT1 binds the promoters of SvSCR1, SvGNC, SvAN3, and many other genes involved in leaf vascular development and photosynthesis. Taken together, our data provide evidence that SvANT1 and ZmANT1 play an important role in leaf vascular development, chloroplast development, photosynthesis, plant growth, and grain yield.
Results
ZmANT1 Is a Potential Regulator of ZmSCR1.
While Arabidopsis has only one ANT gene, Zea mays (maize) has four (ZmANT1-4) and S. viridis has three (SvANT1-3). Their phylogenetic relationships are shown in SI Appendix, Fig. S1. Among the four maize ANT genes, only ZmANT1 is coexpressed with ZmSCR1 during the development of maize embryonic leaves (Fig. 1A); the coexpression is also supported by qRT-PCR data (SI Appendix, Fig. S2). We used the DNA affinity purification and sequencing (DAP-seq) technique (14) to determine the binding motif of ZmANT1 (see the position weight matrix [PWM] in Fig. 1B). We mapped this PWM to the promoter of ZmSCR1 and also to the SCR1 orthologs in Brachypodium distachyon, Oryza sativa Japonica, and Sorghum bicolor by FIMO with P value <0.001. We found putative ZmANT1 binding sites in the promoter regions of maize, sorghum, and rice SCR1 genes (Fig. 1C), suggesting good conservation of the putative binding motif in evolution. Electrophoresis mobility shift assay (EMSA) demonstrated that ZmANT1 protein can indeed bind the predicted motif of ZmSCR1 specifically (Fig. 1D).
Fig. 1.
Testing the binding of maize ANT1 the SCR1 promoter. (A) Expression profiles of maize ANT and SCR1 genes under natural light/dark cycles and Pearson correlation coefficients (PCCs) between the expression levels of maize SCR1 and ANT genes in the time-series transcriptomes of maize embryonic leaves (12). The time course was from T00 (dry seed) to hour 72 (T72) postimbibition. (B) The transcription factor binding site (TFBS) of maize ANT1 (Zm00001d052380) determined by DAP-seq. The TFBS is represented by a PWM. (C) Alignment of a conserved cis motif within the promoter region of ZmSCR1, SbSCR1, OsSCR1, and BdSCR1 to the ZmANT1 TFBS (consensus sequence) (bold and colored). Abbreviations: Zm, Zea mays; Sb, Sorghum bicolor; Os, Oryza sativa; and Bd, Brachypodium distachyon. (D) Gel shift showing binding of maize ANT1 to the motif sequence (hot probe) in the ZmSCR1 promoter region. Lane 1 contained no protein. Lane 2 contained the ANT1 protein and the hot probe. Lane 3 contained the protein and the cold probe. Lane 4 contained the protein and a mutant probe. The same amount of ANT1 protein was used in lanes 1 through 4. Red and black arrowheads indicate TF-probe complexes and free probe, respectively.
ANT1 Is Expressed in Vasculatures of Developing Leaf Primordia.
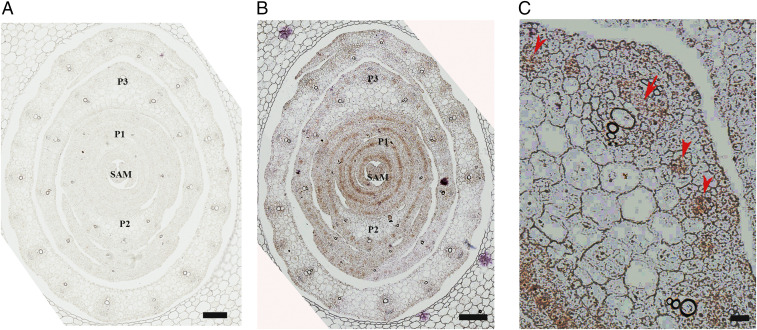
Tissue- or cell-type expression patterns of ZmANT1 may help to infer its biological functions. We therefore conducted an in situ hybridization study of ZmANT1 by selecting a 3′-UTR region unique to ZmANT1 to synthesize a probe. We found that ZmANT1 transcripts accumulate in vasculatures of developing leaf primordia (Fig. 2), similar to the expression of AtANT in cells in an internal zone extending over the length of a cotyledon at the torpedo stage (1). Transverse sections through the shoot apex at the level of the leaf founder cells revealed a punctuated accumulation of ZmANT1 transcripts that preceded the differentiation of vascular bundles in developing leaves (Fig. 2B). Accumulation of ZmANT1 transcripts was initially localized as a single spot within each developing vascular bundle of young leaf primordia and then occurred in vascular tissues of secondary veins in P3 leaf primordia (Fig. 2C). This pattern of ZmANT1 transcript accumulation is consistent with the view that ZmANT1 plays a general role in organ primordium initiation and organ growth during shoot development (4).
Fig. 2.
In situ hybridization assays of ZmANT1 transcript accumulation in the maize seedling shoot. (A and B) Transverse sections of wild-type seedling apices. The sections reveal a punctuated, interspersed accumulation pattern of ZmANT1 transcripts (purple) over the vasculature and in the shoot apical meristem (SAM), and primordia 1 (P1), P2, and P3 primordia prior to the development of vasculature. Gradients of ZmANT1 expression are also noted between vascular bundles in the leaf (A) sense and (B) antisense. (C) Accumulation of ZmANT1 transcript occurs in vascular tissues of veins in P3 leaf primordia (arrows) in the undifferentiated vascular bundle of the fourth leaf from the SAM. P1, P2, P3, and P4 denote leaf primordia. (Scale bar in A and B, 100 µm; in C, 20 µm.)
CRISPR/Cas9-Mediated Targeted Mutagenesis of SvANT1 in S. viridis.
To further assess the function of ANT1, we generated ANT1 gene mutants in S. viridis, a close relative of maize, using a CRISPR/Cas9 gene editing protocol. Our mutagenesis targeted the first exon of SvANT1, which contains restriction sites that can be used for screening mutations using a PCR-based restriction enzyme digestion assay (Fig. 3A). We obtained three S. viridis mutant lines. Among them, Svant1-1 carries an in-frame 3-bp deletion, whereas Svant1-2 and Svant1-3 are both frameshift mutants, carrying 2-bp and 1-bp deletions, respectively (Fig. 3B). The deduced amino acid (aa) sequences of Svant1-2 and Svant1-3 mutants contain no AP2 domain, so they are unlikely to have retained the ANT1 function (Fig. 3C).
Fig. 3.
CRISPR/Cas9 mutagenesis of the SvANT1 gene. (A) The 20-bp target sequence of sgRNA is indicated by a yellow box, including a NGG protospacer adjacent motif (PAM) sequence. (B) Sequences of the first SvANT1 exon in the WT and three transgenic lines. The yellow dashes and letters indicate the identified mutations. The sgRNA targeting sequence and PAM are indicated by red boxes. The sizes of deletions are shown at the Right of each mutated sequence: Δ1, Δ2, and Δ3 denote the deletions of 1, 2, and 3 nt, respectively. The numbers on Top of the sequence are the nucleotide numbers. (C) Schematic presentation of the deduced SvANT amino acid sequences of the WT and three mutants. The orange rectangles indicate AP2 domains. SvANT1-1 mRNA encodes an ANT1 protein with a single amino acid deletion because of an in-frame 3-bp deletion. Svant1-2 and Svant1-3 each may generate a protein without any AP2 domain because of the frameshift mutations.
Using the web tool CRISPOR (crispor.tefor.net) (15), we predicted nine putative off targets of the sgRNA used (SI Appendix, Table S1). Six of the putative off-target sites were located in coding regions, containing three or four mismatches within the sgRNA1-PAM sequence. We sequenced the region of each predicted off-target site in the wild-type and mutant lines, but did not find any sequence changes (SI Appendix, Fig. S3).
Svant Mutations Affect Plant Growth, Development, and Yield.
Svant1-2 and -3 plants showed significantly shorter plant height (from soil surface to the flag leaf) and reduced leaf length compared to WT (SI Appendix, Fig. S4 A–C), possibly due to the predicted truncations of the SvANT1 protein caused by the frameshift mutations (Fig. 3C). In contrast, Svant1-1 has phenotypes resembling those of WT, implying the 1-aa deletion in Svant1-1 protein does not seriously compromise its biological functions.
Svant1-2 and -3 plants had significantly shorter panicles than those of WT and Svant1-1 plants (Fig. 4 A and C), but similar grain sizes (Fig. 4B). Smaller tillers and total seed weight per plant compared to WT were observed for both Svant1-2 and Svant1-3 mutants (Fig. 4 D and F), although Svant1-2 plants have more tillers (Fig. 4E). Thus, Svant1-2 and especially Svant1-3 plants tend to produce a lower grain yield than WT plants.
Fig. 4.
Effects of SvANT1 mutations on teller number, panicle number, and grain yield. (A and B) Representative panicles and seeds of 8-wk-old WT and mutant plants (Scale bar in A, 1 cm; in B, 1 mm.) (C) Panicle lengths. (D) The number of tillers per plant at week 7. (E) The number of panicles per plant at week 7. (F) The total seed weight per plant. ***P value ≤0.001, *P value ≤0.05 (Student’s t test). The error bars represent SDs obtained from 10 replicates.
As ZmANT1 appears to regulate ZmSCR1, a key regulator of vascular development, we compared the vascular patterns in SvANT1 mutant and WT leaves by Lugol staining (Fig. 5 A–C). Cross-section histological analyses revealed sporadically two minor veins with merged bundle sheath (BS) cells, smaller Kranz anatomy, or only one mesophyll (M) cell between two veins (Fig. 5 D–F). The distorted vascular patterning and reduced veins appeared significantly more frequently in Svant1-2 and -3 plants than in WT and Svant1-1 plants (Fig. 5G). Taken together, the above observations suggest that SvANT1 functions in plant growth, leaf development, vein patterning, and grain yield.
Fig. 5.
Leaf vascular phenotypes of 5-wk-old WT and SvANT1 mutant plants. (A–C) Light micrographs of Lugol’s staining leaf tissue [I2KI (iodine-potassium iodide)-stained to reveal starch] of WT, Svant1-2, and Svant1-3 plants. Small arrowheads indicate merged veins. (D–F) Transverse sections of WT, Svant1-2, and Svant1-3 leaves. Svant1-2 showed an example of two minor veins with merged bundle sheath cells. Svant1-3 also showed two minor veins with merged bundle sheath cells. (Scale bar, 20 µm.) Small arrowheads indicate merged veins. Large arrowheads indicate minor veins with merged BS cells, smaller Kranz anatomy, or only one M cell between two veins. (G) Percentage of distorted veins in a middle cross-section of the first leaf. **P < 0.01,***P < 0.001. Error bar represents SD (n = 9 leaves).
Transcriptome Analysis of Svant Mutant Lines.
We compared the leaf transcriptional profiles of WT and the three SvANT mutant lines (Svant1-1, -2, and -3). Total RNA samples were extracted from 10-d-old seedling leaves and sequenced to identify differentially expressed genes (DEGs) (Dataset S1). We found 520, 700, and 784 up-regulated and 289, 636, and 418 down-regulated DEGs in Svant1-1, -2, and -3, respectively, compared to WT (Fig. 6 A and C). To identify the biological processes in which DEGs are involved, we performed a singular enrichment analysis (SEA) using agriGO (16) (Dataset S2). Svant1-1 did not show enriched gene ontology (GO) terms in up-regulated DEGs, and showed significant enrichments only in “biosynthetic process” and “transcription factor activity” among down-regulated DEGs. In contrast, in Svant1-2 and -3 the GO terms of “carboxylic acid biosynthetic process,” “single-organism cellular process,” “coenzyme metabolic process,” “chromatin,” and “intracellular non-membrane-bounded organelle” were enriched among the up-regulated DEGs (Fig. 6B). Moreover, down-regulated DEGs were enriched in “photosynthesis,” “response to stress,” “response to stimulus,” “iron-sulfur cluster binding,” “metal cluster binding,” “thylakoid part,” “thylakoid,” “photosynthetic membrane,” and “photosystem” (Fig. 6D).
Fig. 6.
DEGs, functional enrichment analysis, and reduced photosynthesis rates in mutants. (A) Venn diagrams of up-regulated DEGs. The numbers of up-regulated DEGs were 520 in Svant1-1, 700 in Svant1-2, and 784 in Svant1-3. (B) GO enrichment analysis of up-regulated DEGs. (C) Venn diagrams of down-regulated DEGs. The numbers of down-regulated DEGs were 289 in Svant1-1, 636 in Svant1-2, and 418 in Svant1-3. (D) GO enrichment analysis of down-regulated DEGs. Fisher's exact test was applied to identify the significant GO categories and false discovery rate (FDR) was used to correct the P value. The P values are presented in a heat map. Red and yellow colors represent significant GO enrichment, while gray color represents nonsignificant enrichment. (E and F) Heat maps of DEGs in WT and mutant lines. (E) Genes related to heme and tetrapyrrole binding, and oxidoreductase activity. (F) Genes related to photosynthesis and light-harvesting complex, and TF genes. The bar represents the scale of the expression levels for each gene in WT and mutant lines as indicated by red and blue rectangles. Red indicates up-regulation (1.0) and blue indicates down-regulation (−1.0) as compared with the wild type. (G) Photosynthetic rate (Pn). Data comparison between transgenic and wild-type plants was performed using R (Student’s t test, P = 0.05); values are expressed as means (n > 5 plant for each test); error bars indicate SD. ***P < 0.001 for t test.
Photosynthesis and Chloroplast-Related Genes.
The DEGs identified above included many genes involved in heme and tetrapyrrole binding, or antioxidant activity. Tetrapyrroles provide critical functions to important processes, such as light signaling, detoxification of reactive oxygen species, assimilation of nitrate and sulfate, respiration, programmed cell death, and light-harvesting reactions of photosynthesis (17). Twenty-seven DEGs that participate in heme binding, tetrapyrrole binding, and/or oxidoreductase activity were down-regulated in both Svant1-2 and -3 mutants, including Hema1 (Sevir.9G250700), which is involved in the early steps of chlorophyll biosynthesis, and in plastid-to-nucleus retrograde signal transduction, respectively (Fig. 6E). Moreover, the DEGs identified eight genes involved in photosynthesis that were down-regulated in both Svant1-2 and -3 lines (Upper part of Fig. 6F). These included three light harvesting genes: LHCA2 (Sevir.2G377600), LHB1B2-1 (Sevir.3G146000), and LHB1B2-2 (Sevir.5G222600) and five photosystem genes: PSAG (Sevir.2G259000), PSI-H (Sevir.3G151700), PSBW1 (Sevir.3G189600), PSBW2 (Sevir.5G340900), and PSAF (Sevir.9G068500) (Upper part of Fig. 6F).
Many TF genes regulating chloroplast development were down-regulated in the Svant1-2 and -3 lines (Lower part of Fig. 6F), including two master regulators of chloroplast development GLK1 (Sevir.4G123200) and GNC (Sevir.4G246900) (18) and also regulators of chlorophyll biosynthesis by light, such as GBF1 (Sevir.4G207900) and GBF3 (Sevir.3G143000) (19). Another down-regulated TF gene is the RNA POLYMERASE SIGMA SUBUNIT 1 (Sevir.6G047300) (Lower part of Fig. 6F), which is the most abundant sigma factor that binds to the promoters of photosystem I and II genes and may be a key factor that regulates the expression of the above genes (20).
The enrichment of DEGs in chloroplast function and photosynthesis prompted us to examine the photosynthetic rate of mutant and WT plants. The net photosynthetic rate (Pn) in mature leaves were significantly reduced in the Svant transgenic plants (Fig. 6G). Taken together, these and the above results indicate that SvANT is involved in the regulation of chloroplast development and photosynthesis and that mutations in SvANT can reduce photosynthesis efficiency and grain yield.
Predicting and Validating SvANT1 Target Genes.
Using the S. viridis genome, the DAP-seq data and the binding motif (PWM) of SvANT1, which was predicted to be the same as that of ZmANT1 (Fig. 1C), we predicted potential target genes of SvANT1, i.e., genes that possess SvANT1 binding motif in their promoters (Materials and Methods and Dataset S1). Those potential target genes with “differential expression” between wild-type and mutants were considered candidate differentially expressed target genes (DETGs). A set of 247 “putative DETGs” of SvANT1 was identified by adding the evolutionary conservation test of Yu et al. (21) (Dataset S1). From these putative DETGs, we identified enriched GO terms mainly in “photosynthesis,” “thylakoid part,” “thylakoid,” “photosynthetic membrane,” and “photosystem” (Dataset S2). We are particularly interested in the down-regulated DETGs because many of them are photosynthesis and photosystem genes that have not been reported to be regulated by ANT1. Among the 247 putative DETGs, 26 Arabidopsis homologs showed aberrant phenotypes when defective (22). In addition, by comparing against a list of down-regulated DETGs, we found six DETGs that are involved in vascular development or photosynthesis: SvHB7 (Sevir.7G194700) and SvVIN2 (Sevir.3G333700), which are involved in vascular development (23, 24); SvBBX22 (Sevir.5G141601), which is involved in photomorphogenesis (25); and SvHEMA1 (Sevir.9G250700), SvSIG1 (Sevir.6G047300), and SvRBCL (Sevir.9G044690), which are involved in the early steps of chlorophyll biosynthesis and photosynthesis (26–29). SI Appendix, Table S2 lists the above 32 putative target genes.
EMSA was used to show that ZmANT1 can physically bind to the promoters of 13 candidate target genes, which are classified into four groups in terms of function (SI Appendix, Table S3 and Fig. S5). Group I. Vascular development: 1) SvSCR1, which is a key Kranz anatomy regulator (13); 2) SvHB7, which is homologous to Arabidopsis HB7 (AtHB7), which is involved in vascular development (23, 30); and 3) SvVIN2, which is a NAC domain transcription factor that negatively regulates xylem vessel formation in Arabidopsis (24). Group II. Plant growth: 1) SvAN3, which is a transcriptional coactivator involved in cell proliferation during leaf and flower development; its loss-of-function mutations exhibit narrow, pointed leaves and narrow floral organs (31, 32). Group III. Chloroplast development: 1) SvGNC, which regulates the development of the chloroplast from the proplastid and controls chloroplast growth and division (33, 34); 2) SvHEMA1, which is involved in the early steps of chlorophyll biosynthesis (28); and 3) SvSIG1, which is the most abundant sigma factor that accumulates during leaf development and binds to promoters of photosystem I and II genes (29). Group IV. Photosynthesis: 1) SvABCI8, which encodes an iron-stimulated ATPase and is involved in Fe-S cluster assembly (35); 2) SvPSBW1, which encodes the photosystem II reaction center subunit W; in Arabidopsis, loss of PSBW destabilizes the supramolecular organization of PSII (36); 3) SvPSBW2; 4) SvPSAG, which encodes subunit G of photosystem I, an 11-kDa membrane protein that plays an important role in electron transport between plastocyanin and PSI and is involved in the stability of the PSI complex (37); 5) SvPSI-H, which is an intrinsic membrane protein that is a subunit of photosystem I; PSI-H is required for stable accumulation of PSI and efficient electron transfer in the complex (38); and 6) SvCHL, which is involved in the protection of thylakoidal membrane lipids against reactive oxygen species, especially singlet oxygen, produced upon excess light (39). CHL (chloroplastic lipocalin) is required for sustained photoprotective energy dissipation or NPQ (qH) to occur (39). As the binding motif of SvANT1 is predicted to be the same as that of ZmANT1, the S. viridis orthologs of these 13 genes are expected to also be the target genes of SvANT1.
These 13 putative target genes are further supported by their down-regulation in Svant1-2 and -3 developing leaves compared to WT, except that SvAN3 was not significantly down-regulated in Svant1-2 (SI Appendix, Fig. S6A). Conducting a qRT-PCR analysis, we found that the expression level of SvAN3 was significantly lower in Svant1-2 and Svant1-3 than WT (SI Appendix, Fig. S6B). Previously, AtAN3 has been identified as a potential target gene of AtANT (10).
Discussion
Nole-Wilson and Krizek (40) used the systematic evolution of ligands by exponential enrichment (SELEX) technique and Krizek et al. (10) used chromatin immunoprecipitation sequencing (ChIP-seq) to determine the binding motif (PWM) of AtANT (Arabidopsis ANT), and in this study we used DAP-seq to determine the PWM of ZmANT1 (SI Appendix, Fig. S7A). We found substantial differences between the two PWMs. The Pearson correlation coefficients (PCCs) are only 0.54 to 0.64 between our PWM and those of the other two studies (SI Appendix, Fig. S7B). ANT is an AP2 TF that contains two DNA binding domains and its binding motif can be divided into the left and right motif cores (SI Appendix, Fig. S7A). If we restrict the comparison to the left and right core motifs, then the PCCs become 0.73 between our PWM and those of Nole-Wilson and Krizek (40) and meme-3 of Krizek et al. (10). The PCC between our left motif core and meme-1 is 92%, but meme-1 does not possess a right motif core (SI Appendix, Fig. S7A). The low correlations between our motif of ZmANT1 and that of AtANT and the differences in the tissues used between the two studies (developing leaves vs. developing flowers) may explain why there were only 23 overlaps between our millet ANT1 and Krizek et al.’s Arabidopsis ANT candidate target genes (Dataset S1).
We have shown that ZmANT1 binds the promoter of ZmSCR1 (Fig. 1), which is a key regulator of Kranz anatomy development. However, using FIMO to scan the promoter sequence of the Arabidopsis SCR gene (AtSCR), we cannot find a binding site of AtANT on the promoter with P value <10−3. If we relax the P value threshold to 1.2 × 10−3, we can find a potential binding site on the promoter of AtSCR. However, this site is not present in the orthologous SCR genes in its related species, including Arabidopsis lyrata, Brassica oleracea, and Brassica rapa. Thus, it is unlikely that AtANT can bind the promoter of AtSCR. Therefore, it is likely that the binding of ZmANT1 to the promoter of ZmSCR1 is an evolved function.
Previous studies focused on the roles of Arabidopsis ANT in floral organogenesis and plant growth (1–6, 10), while in this study we studied the roles of maize and millet ANT1 not only in plant growth but also in vascular development, chloroplast development, and photosynthesis. From our study, we propose four regulatory pathways of SvANT1 in Fig. 7. These pathways and their relevance to observations in this study are discussed below.
Fig. 7.
A model for ANT1 regulation of vascular and Kranz anatomy development, leaf growth, chloroplast development, and photosynthesis in developing leaves. Pathway 1: SvANT1 regulates vascular and Kranz anatomy development through its regulation of SvSCR1, SvVIN2, and SvHB7. Pathway 2: SvANT1 regulates SvAN3 and thus modulates cell proliferation and leaf growth. Moreover, SvANT1 also regulates chloroplast development because it regulates SvCRF2 and SvGRF5; SvGRF5 in turn regulates SvGLK1. Pathway 3: SvANT1 regulates chloroplast development through its regulation of SvGNC, SvHEMA1, and SvSIG1. Pathway 4: SvANT1 regulates photosynthesis through its regulation of SvABCI8, SvCHL, PSI genes (SvPSAH2 and SvPSAG), and PSII genes (SvPSBW1 and SvPSAW2).
ANT1 Regulates Kranz Anatomy and Vascular Development.
Our observation that ZmANT1 is a regulator of ZmSCR1 may explain why loss-of-function mutations of SvANT1 caused defects in Kranz anatomy (merged BS cells, smaller Kranz anatomy, and only one M cell between two veins). The observed defects in Svant1-2 and -3 plants were mild and occurred only sporadically likely because there are two other ANT genes in S. viridis and there might be other regulators yet to be identified. We found that SvANT1 also likely regulates SvHB7 and SvVIN2 (SI Appendix, Figs. S5 and S6), which are involved in vascular development (23, 24). We propose that, through SvHB7, SvVIN2, and SvSCR1, SvANT1 regulates vascular and Kranz anatomy development (Fig. 7). The expression of ANT1 in leaf primordia (Fig. 2) suggests that its role in vascular development is likely at an early stage of leaf development.
SvANT1 Modulates Plant Growth.
It is known that Arabidopsis AtANT regulates AtAN3, which is a transcriptional activator of the GIF family and is involved in leaf growth (10, 41). Loss-of-function mutants of AtAN3 exhibited smaller and narrower leaves due to reduced cell numbers (31, 32), while ectopic overexpression of AtAN3 resulted in enlarged leaf size (32, 42). In this study, SvAN3 was down-regulated in Svant1-2 and -3 and our EMSA assay indicated that ZmANT1 binds the promoter of SvAN3 (SI Appendix, Figs. S5 and S6). Thus, SvANT1 likely directly regulates SvAN3, and through SvAN3 it regulates plant growth (the second pathway in Fig. 7). This may explain why Svant1-2 and -3 mutants had shorter leaves and reduced stature (SI Appendix, Fig. S4). There are other potential growth-related target genes of SvANT1 (SI Appendix, Table S2).
SvANT1 Regulates Chloroplast Development.
Our study suggests that SvANT1 regulates chloroplast development via activating genes known to regulate chloroplast development. GATA transcription factor GNC is known to regulate chloroplast development (43, 44). Our target gene predictions and EMSA validation indicated that GNC is an ANT1 target gene. Thus, a possible pathway for regulating chloroplast development is that ANT1 directly regulates GNC (the third pathway in Fig. 7) and triggers the transition of proplastid to chloroplast growth and division (33, 34, 45).
In addition, we showed that ANT1 regulates AN3 in S. viridis. In Arabidopsis, the transcription factor AtGRF5 is a direct target of AtAN3 (46). AtGLK1 was shown to be up-regulated by AtGRF5 overexpression during early leaf development (47). Therefore, a second possible route for SvANT1 to regulate chloroplast development is that it indirectly regulates SvGLK1 (Sevir.4G123200) (Fig. 7). In addition, it has been shown that AtCRF2, an AP2 transcription factor, is a direct target of AtAN3 and it regulates chloroplast development (47). Thus, a third possible route for SvANT1 to modulate chloroplast development is through SvAN3 and SvCRF2 (Sevir.5G014500) (Fig. 7).
Taken together, our findings imply that in maize and millet, ANT1 directly or indirectly regulates three master regulators (GNC, GLK1, and CRF2) of chloroplast development (Fig. 7).
SvANT1 Modulates Photosynthesis.
Six of the 13 direct target genes of SvANT1 are photosynthesis genes, including SvABCI8 and SvCHL, and photosystem I and II genes, including SvPSBW1, SvPSBW2, SvPSI-H, and SvPSAG. These 6 genes were all down-regulated in Svant1-2 and Svant1-3 mutants (SI Appendix, Fig. S6). Therefore, SvANT1 likely modulates photosynthesis by activating the expression of these genes (the fourth pathway in Fig. 7). This may explain the reduced expression of 1) genes involved in the early steps of chlorophyll biosynthesis and in plastid-to-nucleus signal transduction such as SvHema1, and SvSIG1 in Svant1-2 and -3 mutants (Fig. 6 E and F); 2) three light harvesting genes: SvLHCA2, SvLHB1B2-1, and SvLHB1B2-2 and five photosystem genes: SvPSAG, SvPSI-H, SvPSBW1, SvPSBW2, and SvPSAF (Fig. 6F); and 3) regulators of chlorophyll biosynthesis by light, such as GBF1 and GBF3 (Fig. 6F). It may also explain why Svant1-2 and -3 mutant plants showed reduced photosynthesis efficiency (Fig. 6G). Some other potential target genes of SvANT1 are related to photosynthesis (SI Appendix, Table S2).
In conclusion, our study not only has confirmed ANT1’s roles in plant growth by showing that SvANT1 regulates SvAN3, but also has revealed its roles in vascular development, chloroplast development, and photosynthesis. Thus, ANT1 is a key gene in plant development and growth. Further study of its paralogs in maize and millet may reveal even more functions.
Materials and Methods
Plant Materials and Growth Conditions.
Seeds of S. viridis accessions A10.1 and Svant mutant lines were germinated in pots containing soil and Metro mix 360 potting mix. Plants were grown under a 16-h light/8-h dark photoperiod, 450 μmol m−2 s−1 light intensity, 25 ± 2 °C, and 50 to 60% relative humidity to maturity. Agronomic traits shown in SI Appendix, Fig. S4 were examined. At least 10 plants were scored for each genotype in each experiment.
Sequence Analysis.
All sequence analyses were performed using MEGA 7 (48). Amino acid sequence alignment was done using Muscle (49). The phylogenetic tree was constructed using full-length A. thaliana, rice, Setaria italica, S. viridis, and maize ANT protein sequences obtained from Phytozome v12.1 (https://phytozome.jgi.doe.gov/). The evolutionary history was inferred using the neighbor-joining method (MEGA 7). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to each branch. The evolutionary distances were computed using the Poisson correction method and are in the units of number of amino acid substitutions per site. All positions containing alignment gaps and missing data were eliminated.
Construction of the CRISPR/Cas9 Expression Vector and sgRNA Design.
To construct a plasmid vector carrying both sgRNA and Cas9 cassettes, assembled and fused with ZmUbi1 (Z. mays ubiquitin 1) promoter. Sequence information for S. viridis were downloaded from the Phytozome (https://phytozome.jgi.doe.gov/). sgRNAs were designed using the web tool CRISPOR (crispor.tefor.net/) (15), which displayed all optional sgRNA sequences (20 bp) immediately followed by 5′‐NGG (PAM, protospacer adjacent motif) in the forward or reverse strand (SI Appendix, Table S5). Sequence corresponding to sgRNA was integrated upstream of the sgRNA scaffolds in the pBUN411 vector (50). The vector also harbored a customized sgRNA driven by the rice U3 promoter and a bar gene driven by a CaMV 35S promoter as a screening marker.
Transformation of S. viridis and Screening for Mutations.
Agrobacterium tumefaciens strain AGL1 carrying CRISPR/Cas9 expression vectors was used to transform S. viridis (accession A10.1) by following the procedures described by Brutnell et al. (51). Briefly, S. viridis seeds with seed coats mechanically removed were sterilized before plating on callus induction medium (CIM). After 4 wk in the dark at 24 °C, any seedling structures or gelatinous calli were removed and the remaining calli were transferred to fresh CIM for 2 wk. After dividing calli and incubating them on fresh CIM for 1 wk, these calli were ready for transformation as described below.
AGL1 harboring the construct of interest was grown in Luria-Bertani (LB) medium with 50 µg l−1 kanamycin and 25 µg l−1 rifampicin with shaking at 28 °C to OD600 = 0.5, spun down, and resuspended in CIM without Gelzan and hormones. Acetosyringone (200 µM) and synperonic (0.01% [wt/vol]) were added to the Agrobacterium culture before incubating with the calli in the medium for 5 min at room temperature. The calli were blotted dry on sterile filter paper, and transferred to selective CIM and incubated at 22 °C for 3 d in the dark. The calli were then transferred to selective CIM (CIM containing 3 mg l−1 glufosinate, 150 mg l−1 timentin) and incubated in the dark at 24 °C for 16 d. Calli were then transferred to selective plant regeneration medium (PRM) and maintained at 24 °C under a 16-h light/8-h dark photoperiod with a light intensity of 60 µmol m−2 s−1. Developing shoots were transferred to selective rooting medium (RM). Shoots that survived and developed roots were genotyped using primers against the bar gene by PCR, and positive transformants were transplanted to soil (SI Appendix, Table S5).
Genotyping was conducted by genomic PCR with the DNA isolated from each individual T0 plant and with the specific primers listed in SI Appendix, Table S5. The PCR products were digested with the BslI and resolved on 1.5% agarose gel. The nature of edited SvANT1 in each T0 line was confirmed by sequencing. T0 plants with desirable mutations were propagated and screened for homozygous lines.
RNA-Seq Library Construction, Sequencing, and Analysis.
Ten-day-old WT and mutant seedlings were snap-frozen in liquid nitrogen and ground into fine powder using 3-mm tungsten-carbide beads in a Tissue Lyser-II (Qiagen). Total RNA was extracted using the Direct-Zol RNA Purification Kit (Zymo Research) with on-column DNase I digest following the manufacturer’s protocols. The RNA samples were quantified using a Qubit RNA HS Assay Kit (Thermo Fisher Scientific) and assessed for purity and quality using NanoDrop (Thermo Fisher Scientific) and BioAnalyzer (Agilent). RNA-seq libraries were constructed using the TruSeq Stranded mRNA Library Prep Kit (Illumina) following the manufacture’s protocol. RNA-seq reads were generated by paired-end 2 × 151 bp sequencing on the Illumina HiSEq. 2500 platform at the High Throughput Genomics Core in Academia Sinica, Taiwan.
The read processing procedure was the same as in Liu et al. (12). The processed reads were mapped to the S. viridis A10.1 reference genome (https://phytozome.jgi.doe.gov/; v1.1) using TopHat (52) (version 2.0.10) and its embedded aligner Bowtie2 (53) (version 2.1.0). The expression level (reads per kilobase of transcript per million mapped reads [RPKM]) of each gene was estimated using Cufflinks (54) (v2.1.1). To compare the RPKMs of the selected genes across time points in a set of transcriptomes, we applied the upper quartile normalization procedure (55). Gene-level expression values were represented by RPKMs and a consensus RPKM was determined for each gene based on its representation across two biological replicates. Differential expression was determined using NOI-sEq (56, 57). with q ≧ 0.8. Gene ontology enrichment analysis was performed using the agriGO v2.0 (16).
Measurement of Photosynthetic Rates.
Leaf photosynthetic rate was measured with a LI-6400 XT portable photosynthesis system (LI-COR) under 1,500 µmol m−2 s−1 photosynthetic active radiation (PAR) by using an LED red-blue light source. Net photosynthetic rate Pn (mol m−2 s−1) of leaves were determined between 9:00 and 11:00 from fully expanded first blades.
EMSA Assays.
The protocol is as described in Yu et al. (21) Ten nanograms of biotin-labeled promoter probes (SI Appendix, Table S4) were incubated with ∼50 ng of GST or recombinant TF protein expressed in and purified from Escherichia coli Rosetta (DE3) for 20 min at 22 °C. Competition experiments were performed with 100 ng of unlabeled or mutated promoter probes (Fig. 1 and SI Appendix, Fig. S6). The EMSA mixture was separated by a 4% polyacrylamide native gel and transferred to a Hybond N+ membrane (GE) by semidry transfer cell (Bio-Rad). The biotin-labeled probe and the TF-probe complexes were detected by streptavidin-horseradish peroxidase (HRP) conjugates (Life Technologies) with substrates from ECL Plus (GE). The chemiluminescent signals were visualized by the BioSpectrum imaging system (UVP).
DAP-Seq Workflow and Inference of TF Binding Motif.
Our protocol is as described in O’Malley et al. (14) and Bartlett et al. (58) with minor modifications. Genomic DNA was extracted from 4-d-old millet (S. italica) leaves, fragmented, and ligated with the truncated adaptor from NEBNext Ultra II DNA Library Prep Kit for Illumina (NEB). For protein expression, the TF coding sequence was cloned into the pIX in vitro expression vector with N-terminal HALO-Tag (from Ecker Laboratory) using LR Clonase (Thermo Fisher Scientific) and expressed using the TNT T7 Coupled Wheat Germ Extract System (Promega). The GST sequence was cloned into the pIX and used as an experimental protein control. HALO-tagged TF and GST proteins were immobilized on Magne HaloTag beads (Promega), washed, and incubated with 350 ng of the adapter-ligated gDNA library. After bead washing, the bead-bound gDNA fragments were eluted and amplified with the NEB indexing primers. Sequencing was performed on Illumina HiSEq. 2500 with paired-end 2 × 101 bp at the High Throughput Genomics Core, Academia Sinica, Taiwan.
To infer the binding motif of a TF from DAP-seq data, the paired-end (PE) reads were preprocessed to remove adapters, and low-quality bases were trimmed off using Trimmomatic (v0.39) (59) with options: ILLUMINACLIP:TruSeq3-PE.fa:2:40:12:8:true LEADING:10 SLIDINGWINDOW:4:15 MINLEN:50. The trimmed reads were aligned using Bowtie2 (v2) (53) to the S. viridis genome (v2.1) in which the repetitive sequences were masked. Those reads aligned to multiple loci and those duplicate reads aligned to the same locus were filtered out using SAMtools (60). The enriched binding events were identified by comparing reads coverage between GST-ZmANT1 treatment and GST-only control using MACS2 (v2.1.2) (61) with q-value <0.05. The top 500 peaks (summits.bed by MACS2) of binding regions based on the MACS2 q-values were selected to extract sequences from two 100-bp flanking regions of the peaks, 200 bp in total. The top five motifs (PWMs) were then identified using MEME-ChIP (v5.0.5) (62). The PWM having the largest number of occurrences (at least 100 sites) among the top 500 peak sequences was selected as the PWM of the TF under study.
TF Target Gene Prediction.
We used the PWM of a TF to find its potential target genes by mapping the PWM to the promoter sequences of all millet genes using FIMO (63) with P value <0.005. The promoter sequence of a gene was defined as the region from −1,000 bp to +200 bp relative to the transcription start site (TSS) of the gene. We then added the genes with promoters covered by DAP-seq peaks (as described above), even though they did not pass the search of PWM sequence. From this set of genes, we selected those that showed differential gene expression to form a set of candidate DETGs. Then, following the method of Yu et al. (64), we required that a PWM sequence be found in at least two of the orthologous promoters in the following four species: O. sativa Japonica, B. distachyon, S. bicolor, and Z. mays. These genes were considered the putative DETGs (Dataset S1).
Supplementary Material
Acknowledgments
This study was supported by Academia Sinica (AS-TP-106-L14, AS-TP-109-L10, AS-KPQ-109-ITAR-TD05). We thank Dr. Beth Krizek for suggestions. S.-H.S. was partially supported by NSF Grants IOS-1546617 and DEB-1655386 and Department of Energy Great Lakes Bioenergy Research Center (BER DE-SC0018409). The millet genome sequence data were produced by the Department of Energy Joint Genome Institute.
Footnotes
The authors declare no competing interest.
Data deposition: The transcriptome data and DAP-seq data have been deposited to the National Center for Biotechnology Information Sequence Read Archive (NCBI SRA) with accession number PRJNA633079.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2012245117/-/DCSupplemental.
Data Availability.
The transcriptome data and DAP-seq data have been deposited to the NCBI SRA with accession numbers PRJNA633079.
References
- 1.Elliott R. C. et al., AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8, 155–168 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klucher K. M., Chow H., Reiser L., Fischer R. L., The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8, 137–153 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krizek B. A., Ectopic expression of AINTEGUMENTA in Arabidopsis plants results in increased growth of floral organs. Dev. Genet. 25, 224–236 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Mizukami Y., Fischer R. L., Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. U.S.A. 97, 942–947 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krizek B., AINTEGUMENTA and AINTEGUMENTA-LIKE6 act redundantly to regulate Arabidopsis floral growth and patterning. Plant Physiol. 150, 1916–1929 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamaguchi N. et al., A molecular framework for auxin-mediated initiation of flower primordia. Dev. Cell 24, 271–282 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Mizukami Y., A matter of size: Developmental control of organ size in plants. Curr. Opin. Plant Biol. 4, 533–539 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Nole-Wilson S., Azhakanandam S., Franks R. G., Polar auxin transport together with aintegumenta and revoluta coordinate early Arabidopsis gynoecium development. Dev. Biol. 346, 181–195 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Randall R. S. et al., AINTEGUMENTA and the D-type cyclin CYCD3;1 regulate root secondary growth and respond to cytokinins. Biol. Open 4, 1229–1236 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krizek B. A., Blakley I. C., Ho Y. Y., Freese N., Loraine A. E., The Arabidopsis transcription factor AINTEGUMENTA orchestrates patterning genes and auxin signaling in the establishment of floral growth and form. Plant J. 103, 752–768 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winter D. et al., An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2, e718 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu W. Y. et al., Anatomical and transcriptional dynamics of maize embryonic leaves during seed germination. Proc. Natl. Acad. Sci. U.S.A. 110, 3979–3984 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slewinski T. L., Anderson A. A., Zhang C., Turgeon R., Scarecrow plays a role in establishing Kranz anatomy in maize leaves. Plant Cell Physiol. 53, 2030–2037 (2012). [DOI] [PubMed] [Google Scholar]
- 14.O’Malley R. C. et al., Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165, 1280–1292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haeussler M. et al., Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian T. et al., agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45, W122–W129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larkin R. M., Tetrapyrrole signaling in plants. Front. Plant Sci. 7, 1586 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zubo Y. O. et al., Coordination of chloroplast development through the action of the GNC and GLK transcription factor families. Plant Physiol. 178, 130–147 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi K., Obayashi T., Masuda T., Role of the G-box element in regulation of chlorophyll biosynthesis in Arabidopsis roots. Plant Signal. Behav. 7, 922–926 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macadlo L. A., Ibrahim I. M., Puthiyaveetil S., Sigma factor 1 in chloroplast gene transcription and photosynthetic light acclimation. J. Exp. Bot. 71, 1029–1038 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu C. P. et al., Transcriptome dynamics of developing maize leaves and genomewide prediction of cis elements and their cognate transcription factors. Proc. Natl. Acad. Sci. U.S.A. 112, E2477–E2486 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lloyd J., Meinke D., A comprehensive dataset of genes with a loss-of-function mutant phenotype in Arabidopsis. Plant Physiol. 158, 1115–1129 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cabello J. V., Chan R. L., Arabidopsis and sunflower plants with increased xylem area show enhanced seed yield. Plant J. 99, 717–732 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Yamaguchi M. et al., VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 22, 1249–1263 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gangappa S. N. et al., The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell 25, 1243–1257 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masuda T., Fujita Y., Regulation and evolution of chlorophyll metabolism. Photochem. Photobiol. Sci. 7, 1131–1149 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Shimizu M. et al., Sigma factor phosphorylation in the photosynthetic control of photosystem stoichiometry. Proc. Natl. Acad. Sci. U.S.A. 107, 10760–10764 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmied J., Hedtke B., Grimm B., Overexpression of HEMA1 encoding glutamyl-tRNA reductase. J. Plant Physiol. 168, 1372–1379 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Hanaoka M., Kato M., Anma M., Tanaka K., SIG1, a sigma factor for the chloroplast RNA polymerase, differently associates with multiple DNA regions in the chloroplast chromosomes in vivo. Int. J. Mol. Sci. 13, 12182–12194 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ré D. A., Capella M., Bonaventure G., Chan R. L., Arabidopsis AtHB7 and AtHB12 evolved divergently to fine tune processes associated with growth and responses to water stress. BMC Plant Biol. 14, 150 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim J. H., Kende H., A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 13374–13379 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horiguchi G., Kim G. T., Tsukaya H., The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 43, 68–78 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Bi Y. M. et al., Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J. 44, 680–692 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Mara C. D., Irish V. F., Two GATA transcription factors are downstream effectors of floral homeotic gene action in Arabidopsis. Plant Physiol. 147, 707–718 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu X. M., Adams S., Chua N. H., Møller S. G., AtNAP1 represents an atypical SufB protein in Arabidopsis plastids. J. Biol. Chem. 280, 6648–6654 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.García-Cerdán J. G. et al., The PsbW protein stabilizes the supramolecular organization of photosystem II in higher plants. Plant J. 65, 368–381 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Varotto C. et al., Single and double knockouts of the genes for photosystem I subunits G, K, and H of Arabidopsis. Effects on photosystem I composition, photosynthetic electron flow, and state transitions. Plant Physiol. 129, 616–624 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naver H., Haldrup A., Scheller H. V., Cosuppression of photosystem I subunit PSI-H in Arabidopsis thaliana. Efficient electron transfer and stability of photosystem I is dependent upon the PSI-H subunit. J. Biol. Chem. 274, 10784–10789 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Malnoë A. et al., The plastid lipocalin LCNP is required for sustained photoprotective energy dissipation in Arabidopsis. Plant Cell 30, 196–208 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nole-Wilson S., Krizek B. A., DNA binding properties of the Arabidopsis floral development protein AINTEGUMENTA. Nucleic Acids Res. 28, 4076–4082 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krizek B. A. et al., RNA-seq links the transcription factors AINTEGUMENTA and AINTEGUMENTA-LIKE6 to cell wall remodeling and plant defense pathways. Plant Physiol. 171, 2069–2084 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee B. H. et al., The Arabidopsis GRF-INTERACTING FACTOR gene family performs an overlapping function in determining organ size as well as multiple developmental properties. Plant Physiol. 151, 655–668 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hudson D. et al., GNC and CGA1 modulate chlorophyll biosynthesis and glutamate synthase (GLU1/Fd-GOGAT) expression in Arabidopsis. PLoS One 6, e26765 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiang Y. H. et al., Functional characterization of the GATA transcription factors GNC and CGA1 reveals their key role in chloroplast development, growth, and division in Arabidopsis. Plant Physiol. 160, 332–348 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bastakis E., Hedtke B., Klermund C., Grimm B., Schwechheimer C., LLM-domain B-GATA transcription factors play multifaceted roles in controlling greening in Arabidopsis. Plant Cell 30, 582–599 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vercruyssen L. et al., ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development. Plant Cell 26, 210–229 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vercruyssen L. et al., GROWTH REGULATING FACTOR5 stimulates Arabidopsis chloroplast division, photosynthesis, and leaf longevity. Plant Physiol. 167, 817–832 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar S., Stecher G., Tamura K., MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xing H. L. et al., A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14, 327 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brutnell T. P. et al., Setaria viridis: A model for C4 photosynthesis. Plant Cell 22, 2537–2544 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trapnell C., Pachter L., Salzberg S. L., TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trapnell C. et al., Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bullard J. H., Purdom E., Hansen K. D., Dudoit S., Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics 11, 94 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tarazona S., García-Alcalde F., Dopazo J., Ferrer A., Conesa A., Differential expression in RNA-seq: A matter of depth. Genome Res. 21, 2213–2223 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tarazona S. et al., Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res. 43, e140 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bartlett A. et al., Mapping genome-wide transcription-factor binding sites using DAP-seq. Nat. Protoc. 12, 1659–1672 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li H. et al.; 1000 Genome Project Data Processing Subgroup , The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y. et al., Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Machanick P., Bailey T. L., MEME-ChIP: Motif analysis of large DNA datasets. Bioinformatics 27, 1696–1697 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grant C. E., Bailey T. L., Noble W. S., FIMO: Scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu C. P., Lin J. J., Li W. H., Positional distribution of transcription factor binding sites in Arabidopsis thaliana. Sci. Rep. 6, 25164 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The transcriptome data and DAP-seq data have been deposited to the NCBI SRA with accession numbers PRJNA633079.