Significance
Amino acids are involved in metabolism, protein biosynthesis, and signal transduction of humans. Heteromeric amino acid transporters maintain homeostasis of amino acid pools in various tissues and organs representing a unique group among members of the solute carrier superfamily. The neutral and basic amino acid transport complex (b[0,+]AT1-rBAT) is responsible for reabsorption of cystine and dibasic amino acids in kidney and intestine. Misguided trafficking, incomplete maturation, or defective transport activity result in hyperexcretion of cystine, causing cystinuria. While the function of this amino acid transport complex is known, details of its evolution, biogenesis, and mechanism of action have remained elusive. We determined the cryo-EM structure of this protein complex to gain a deeper understanding of its mode of action.
Keywords: human amino acid transporters, SLC7, SLC3, cryo-EM, heteromeric amino acid transporters
Abstract
Heteromeric amino acid transporters (HATs) comprise a group of membrane proteins that belong to the solute carrier (SLC) superfamily. They are formed by two different protein components: a light chain subunit from an SLC7 family member and a heavy chain subunit from the SLC3 family. The light chain constitutes the transport subunit whereas the heavy chain mediates trafficking to the plasma membrane and maturation of the functional complex. Mutation, malfunction, and dysregulation of HATs are associated with a wide range of pathologies or represent the direct cause of inherited and acquired disorders. Here we report the cryogenic electron microscopy structure of the neutral and basic amino acid transport complex (b[0,+]AT1-rBAT) which reveals a heterotetrameric protein assembly composed of two heavy and light chain subunits, respectively. The previously uncharacterized interaction between two HAT units is mediated via dimerization of the heavy chain subunits and does not include participation of the light chain subunits. The b(0,+)AT1 transporter adopts a LeuT fold and is captured in an inward-facing conformation. We identify an amino-acid–binding pocket that is formed by transmembrane helices 1, 6, and 10 and conserved among SLC7 transporters.
Membrane transporters are essential for human physiology with specialized functions in different tissues and organs. More than 60 transporters from 11 human solute carrier (SLC) subfamilies are known to facilitate the transport of different amino acids (1–4). Heterodimeric amino acid transporters (HATs) possess unique structural features among SLCs (5, 6). HATs are composed of two protein components: a light chain subunit from the SLC7 family (SLC7A5-11, -13) and a heavy chain subunit from the SLC3 family (SLC3A1 and -2) (7). The light chain constitutes the transport subunit whereas the heavy chain mediates trafficking to the plasma membrane and maturation of the functional complex (8, 9). The two subunits are linked by a conserved disulfide bond to maintain a distinct heterodimeric stoichiometry at all times (10). Mutation, malfunction, and dysregulation of HATs are associated with a wide range of pathologies or represent the direct cause of inherited and acquired disorders (11–13). The SLC7A9/SLC3A1-encoded neutral and basic amino acid transport complex (b[0,+]AT1-rBAT) is responsible for reabsorption of cystine and dibasic amino acids while exchanging neutral amino acids across the apical membrane of epithelial cells of the renal proximal tubule and small intestine (1, 14–16). Mutations in either or both subunits cause defective transport and thus result in hyperexcretion of cystine in urine and ultimately lead to cystinuria (17–20). Cystinuria is the most common primary inherited amino aciduria among humans with an overall worldwide prevalence of 1:7,000 neonates (7, 21, 22). So far, 122 and 170 clinical mutations in SLC7A9 and SLC3A1 have been identified, respectively (Human Gene Mutation Database) (23). Despite the myriad of clinical data and identified mutations which cause conditions of cystinuria, the molecular details of the b(0,+)AT1-rBAT function and its transport mechanism have remained elusive (24, 25).
Results and Discussion
We stably integrated SLC3A1 and SLC7A9 genes into human embryonic kidney cells (HEK293) and recombinantly produced the b(0,+)AT1-rBAT complex under cell culture conditions. Targeting and insertion into the plasma membrane was confirmed by fluorescence microscopy (SI Appendix, Fig. S1). To assess proper functionality of the membrane-integrated complex, we carried out a radioligand-labeled arginine uptake assay. Our results confirmed an increased uptake of L-arginine in the HEK293 cell line overexpressing simultaneously SLC3A1 and SLC7A9 genes. Cell lines expressing either of the two genes showed no significant difference from wild-type HEK293 cells (SI Appendix, Fig. S1). Digitonin-stabilized complex was purified via affinity chromatography, and its integrity was validated by high-resolution clear native electrophoresis, differential scanning fluorimetry (DSF), and negative-stain electron microscopy (EM) (SI Appendix, Fig. S1). The three-dimensional (3D) structure of the b(0,+)AT1-rBAT complex was determined by electron cryomicroscopy (cryo-EM). A total of 1,780 movies were collected on a Titan Krios G3i microscope using a Gatan K3 camera. Due to the relative flexibility between b(0,+)AT1 and rBAT subunits, several rounds of 3D classification and focused refinement were required to achieve a final reconstruction of rBAT at 2.8 Å and b(0,+)AT1 at 3.4 Å resolutions (SI Appendix, Figs. S2 and S3).
Overall Structure of the b(0,+)AT1-rBAT Complex.
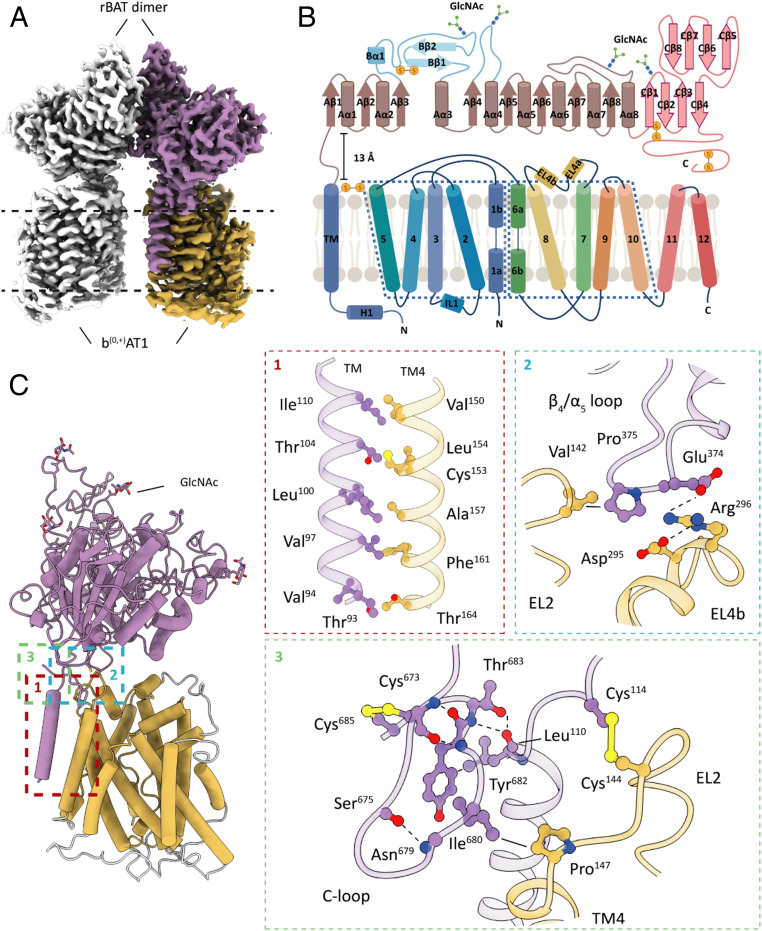
The overall density map reveals a heterotetrameric protein complex composed of two rBAT subunits and two b(0,+)AT1 transporters (Fig. 1A). Our structure shows that rBAT forms a C2 symmetrical homodimer with each protomer attached to a b(0,+)AT1 light-chain subunit. A protein–protein interaction between the membrane-integrated b(0,+)AT1 subunits could not be confirmed. Subunit rBAT is a type II membrane glycoprotein composed of a small N-terminal cytosolic region that is connected to a large extracellular domain (ED) by a single-membrane–spanning helix (transmembrane 1 [TM1]) and a 13-Å-long extracellular linker (26) (Fig. 1B). The overall structure of rBAT shows close resemblance to the cell-surface antigen heavy chain of the LAT1-4F2hc complex (Cα rmsd = 4.5) (27, 28). On the ED surface of each rBAT protomer we were able to identify four N-glycosylation sites including Asn575, which is crucial for maturation and trafficking (29) (Fig. 1B). Despite the fold resemblance, none of the glycosylation sites was conserved between human rBAT and 4F2hc. This difference might indicate alternative trafficking pathways or maturation processes between the two HAT-type transport complexes (6, 26).
Fig. 1.
Cryo-EM structure of the b(0,+)AT1-rBAT transport complex. (A) Cryo-EM density map of the SLC7A9/SLC3A1-encoded neutral and basic amino acid transporter. The b(0,+)AT1-rBAT complex consists of a heterotetrameric assembly of two rBAT and b(0,+)AT1 subunits, respectively. (B) Schematic representation of rBAT and b(0,+)AT1 secondary structure motifs, glycosylation sites, and inter- and intramolecular disulfide bonds. α-Helical and β-sheet elements are shown as cylinders and arrows, respectively. (C) Atomic model of a b(0,+)AT1-rBAT heterodimer and zoom into distinct interaction sites between the two subunits. b(0,+)AT1, yellow; rBAT, purple; hydrogen bonds, dashed lines; van der Waals contacts, straight lines. EL: extracellular loop; TM: transmembrane helix. Flexible protein segments that could not be resolved to side-chain level resolution are indicated in light gray.
Several interaction interfaces were identified between b(0,+)AT1 and rBAT. Within the membrane region, we observed a series of van der Waals contacts formed by nonpolar side chains of b(0,+)AT1 TM4 and the TM of rBAT (Fig. 1C). An analogous contact site is found in the 4F2hc-LAT1 structure (27, 28). On the extracellular surface, rBAT attaches to b(0,+)AT1 via various types of interactions (Fig. 1C). A crucial interface is formed between EL2 of b(0,+)AT1 and the linker region of rBAT where we identify the conserved intermolecular Cys144-Cys114 disulfide bond (Fig. 1 B and C and SI Appendix, Fig. S10A). A second extracellular interaction site is formed by protrusion of the α4/β5 loop of rBAT into the cleft between EL2 and EL4b of b(0,+)AT1. Here, we observed a van der Waals contact between Pro375 of rBAT, and Val142 of b(0,+)AT1, as well as electrostatic interactions mediated by side chains of Glu374 of rBAT, and Arg296 and Asp295 of b(0,+)AT1 (Fig. 1C and SI Appendix, Fig. S10B).
The C-terminal segment of rBAT is a unique structural element known to affect stability and maturation of the b(0,+)AT1-rBAT complex (29). We were able to resolve the entire C terminus (segments 653 to 683) which does not feature previously predicted secondary structure elements. Instead, we identified a loop region (segments 673 to 683) that is stabilized by a critical disulfide bond (Cys673/Cys685) and several electrostatic interactions (Fig. 1 B and C). The C-loop is placed in immediate vicinity to the C-terminal end of the rBAT transmembrane helix and EL2 of b(0,+)AT1. In this local environment, we observed a van der Waals contact between Ile680 of the C-loop and Pro147 of b(0,+)AT1, implying a direct involvement of the C-loop in the heterodimer formation. It is conceivable that C-loop–mediated stabilizing interactions are essential during the maturation process prior to formation of the cysteine-mediated covalent bond between the heavy and light chain subunits. The limited number of interactions on the extracellular surface between rBAT and b(0,+)AT1 translates into a high relative flexibility of these two subunits as observed in our principal component analyses (SI Appendix, Fig. S11 and Movies S1 and S2: multibody analyses).
Structural Insights into rBAT-Mediated Assembly of Tetrameric HAT Complexes.
The extracellular domains of rBAT and 4F2hc share a common architecture of a (β/α)8 TIM-barrel fold (subdomain A) followed by eight antiparallel β-sheets (subdomain C) (30) (Figs. 1B and 2A). The cystinuria-causing mutations Met467Thr/Lys demonstrate the critical role of atomic interactions between these subdomains (31) (SI Appendix, Fig. S9B). In a functional wild-type complex, the side chain of Met467 participates in a network of van der Waals contacts along the interface of subdomains A and C. Mutations to polar or charged residues most likely disrupt this hydrophobic environment and thus result in intracellular trafficking defects (32).
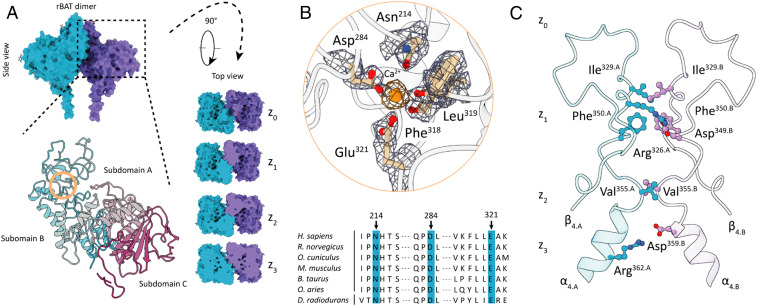
Fig. 2.
Extracellular domain organization of rBAT. (A) Surface representation of the rBAT dimer in side and top view orientations. The top view panel indicates dimerization interfaces along a z axis that is perpendicular to the membrane plane. A close-up of the ED as a ribbon model indicates the existence of three subdomains: subdomain A is depicted in gray, subdomain B in cyan, and domain C in magenta. The orange circle indicates the area of the identified cation-binding site. (B) A close-up of the Ca2+-binding site showing coordinating side-chain and backbone atoms of subdomain B. Map densities of amino acid side chains are shown as blue volumes. The Ca2+ density is illustrated as orange volume. A sequence alignment of mammalian rBAT homologs and the bacterial trehalose synthase demonstrates the close relationship between rBAT and the GH13 family. Blue boxes indicate conserved residues participating in Ca2+ binding and coordination. (C) Atomic interactions of C2 symmetry-related β4/α4 extensions of subdomain B mediate the dimer formation between the two rBAT protomers along the z axis. Symmetry-related electrostatic interactions are shown only once for clarity. Capital letters A and B indicate the individual rBAT protomers.
The rBAT ectodomain possesses an additional structural motif (subdomain B) that is formed by extension of loops connecting β3/α3 (segment 212 to 290) and β4/α4 (segment 314 to 356) of the TIM barrel. A distinguishing feature of rBAT is the existence of a cation-binding site that is formed within subdomain B (Fig. 2A). Our structure shows a coordination pocket formed by side chains of Glu321, Asp284, Asn214 (originally postulated as N-glycosylation site), and the two backbone carbonyl groups of Leu319 and Phe318 (Fig. 2B). In the center of this pocket we observed a prominent nonproteinogenous density indicating the presence of a cation. Analyses of related domain folds and ion-binding sites revealed that members of the glycoside hydrolase family (GH13) share an even closer fold homology to rBAT than human 4F2hc (SI Appendix, Figs. S7 and S12). Structure-based alignments showed that the bacterial trehalose synthase from Deinococcus radiodurans (Cα rmsd = 2.4) contains a calcium-binding site within a region that is homologous to rBAT subdomain B. Coordinating side-chain residues and positions of backbone carbonyls of the ion-binding sites are fully conserved between these two structures and thus indicate that the observed density in subdomain B of our structure represents a Ca2+ ion (Fig. 2B and SI Appendix, Fig. S8). It is conceivable that the calcium-mediated interaction between β3/α3 and β4/α4 is a stabilizing property of subdomain B (33). The close structural relationship between rBAT and members of the glycoside hydrolase family suggests that rBAT might have evolved from a bacterial homolog of this family but lost its enzymatic activity in favor of gaining a trafficking function in eukaryotic cells (34, 35) (SI Appendix, Fig. S12). Structural differences between rBAT and 4F2hc, most prominently the loss of subdomain B, support the hypothesis that 4F2hc has been experiencing an accelerated evolution and has thus gained multiple physiological roles in humans (36). While it is generally accepted that the function of rBAT is limited to HAT biogenesis and trafficking, 4F2hc is described as acting as a trafficking chaperone of multiple membrane transporters, as well as an interaction partner of integrins and basigin for cell signaling, immune response, and metabolic regulation (37–39). The importance of the structural integrity of subdomain B is highlighted by the highly frequent cystinuria causative mutation Thr216Met (40, 41). As this mutation site is in immediate adjacency to Asn214, it might have an influence on the Ca2+-binding site, presumably destabilizing the interaction network due to displacement of the protein backbone segment or introducing local flexibility of the surrounding environment due to the exchange of the originally polar threonine residue to a nonpolar methionine group (SI Appendix, Fig. S9C).
The dimerization between the rBAT protomers is mediated by an interaction network of symmetry-related side chains of the β4/α4 extension within subdomain B (Fig. 2A). A z axis cross-section shows that three levels (z1 to z3) of interaction interfaces form the dimerization site. The z1 level is characterized by van der Waals contacts (Phe350.A-Phe350.B, Ile329.A-Ile329.B) and charge interactions (Arg326.A/B-Asp349.B/A), while z2 solely shows van der Waals (Val355.A-Val355.B) and z3 only charged interactions (Arg362.A/B-Asp359.A/B) (Fig. 2C). The existence of this dimerization motif is unique for rBAT-associated HATs and constitutes a structural property that separates human HATs into dimeric and tetrameric subgroups. Based on previously reported data and the insights gained from the 3D structure, we conclude that the dimerization of rBAT might represent a checkpoint event in the maturation process rather than being indispensable for transport activity of b(0,+)AT1 (42).
Structure of b(0,+)AT1.
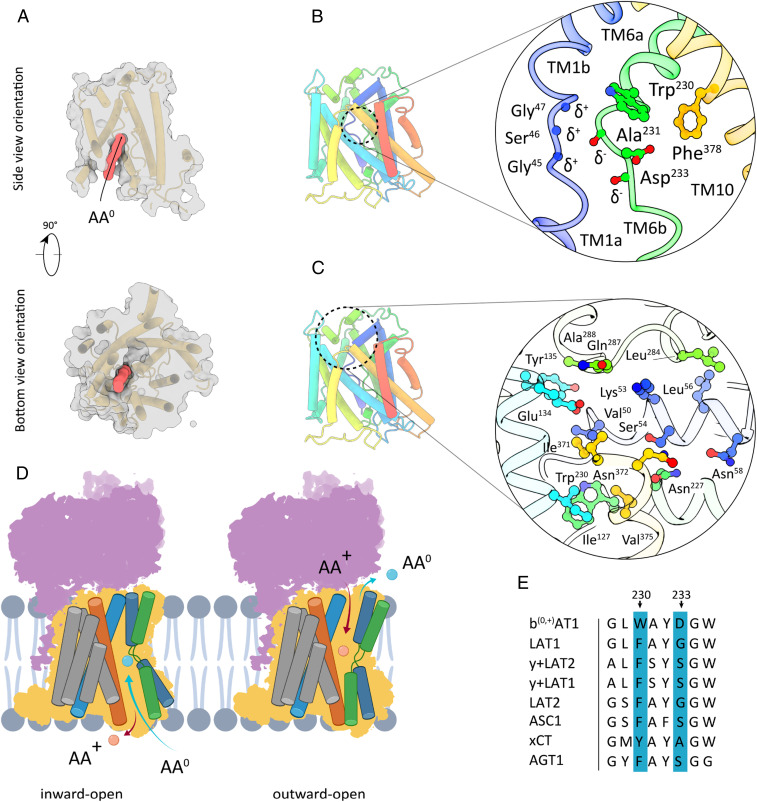
The membrane-integrated b(0,+)AT1 subunit is a solute carrier which belongs to the Amino acid, Polyamine, and Organocation (APC) superfamily (9). Its 12 transmembrane segments feature a canonical LeuT fold of 10 core TMs with a 5+5 inverted repeat topology forming a cylindrical barrel, and two additional TMs (11, 12) are located peripherally (43). TMs 1 and 6 are divided into two discontinuous helices, disrupted within the membrane by short Gly-containing regions (Fig. 1B). These helix break motifs are conserved in most APC transporters and known to act as a toggle switch playing a key role in orchestrating the conformational changes required for the alternating access mechanism (44, 45). In our structure, b(0,+)AT1 adopts an inward open conformation as judged by the general TM topology and the presence of a 26-Å-long solvent-accessible cavity exposed to the cytoplasm (Fig. 3A). We identified a putative AA0-binding pocket at the apex of the cytoplasmic channel where TMs 1, 6, and 10 are positioned very closely to each other. At this site, Trp230, Asp233, Phe378, and the conserved GSG motif of TM1 form a local environment that could accommodate a neutral amino acid group (Fig. 3B and SI Appendix, Fig. S6). Structures of cognate SLC7 homologs GkApcT, AdiC, and LAT1 with amino acids or competitive inhibitors bound in analogous pockets suggest that in b(0,+)AT1 the backbone amide groups of Gly45, Ser46, and Gly47 most likely participate in hydrogen bond interactions with the backbone carboxyl groups of neutral amino acid substrates (27, 44, 46) (SI Appendix, Fig. S4). In a reciprocal manner, the carbonyl oxygen atoms of Ala231 and Asp233 might form hydrogen bonds with the amino groups of the substrate molecules. While coordination of substrate backbones appears to be conserved among SLC7 members, specificities for distinct amino acid classes are most likely modulated by the chemical nature and steric properties of the amino acid side chains constituting the binding pocket (Fig. 3E).
Fig. 3.
Substrate-binding site and extracellular barrier of b(0,+)AT1. (A) Surface representation of b(0,+)AT1 in side and top view orientations. The red volume indicates a substrate-accessible channel connecting the cytoplasm and the AA0-binding pocket. (B) Tube model depiction of the inward-open b(0,+)AT1 conformation and close-up view of the putative AA0-binding pocket formed by TMs 1 (blue), 6 (green), and 10 (yellow). (C) General location and close-up view of the extracellular barrier formed by TMs 1b (blue), 3 (turquoise), and EL4 (green). (D) Schematic presentation of transmembrane segments involved in cytoplasmic and extracellular barrier formation during a AA+/AA0 transport cycle. (E) Sequence comparison of residues involved in formation of substrate-binding sites between selected members of HAT-associated human SLC7 transporters. AA+: cationic amino acid. AA0: neutral amino acid.
The location of Trp230 corresponds to that of Phe252 in human LAT1, which plays a crucial role in substrate binding by participating in van der Waals contacts and forming an occlusion gate to the extracellular surface (27, 28). Based on sequence alignments, we find that, of eight HAT-associated light chain subunits of the human SLC7 family, only SLC7A9 possesses a Trp residue at this site (Fig. 3 B and E). Impaired transport activity of b(0,+)AT1 in cystinuria patients with a Trp230Arg mutation emphasizes the importance of this residue (47) (SI Appendix, Fig. S9D). Whether the role of Trp230 is to form a tight gate to constrain diffusion of small nonpolar amino acids from the cytoplasm to the extracellular space or to provide an additional electrostatic interaction site for substrate coordination remains a subject for future research.
The interrupted helix segment of TM6 further contains Asp233, which orients its propionate group toward the AA0-binding site. Sequence analyses indicate that the presence of the functionally relevant Asp233 is unique for SLC7A9 and absent in other human SLC7 members (Fig. 3 B and E). Within the binding pocket, the side chain carboxylate (pKa 6.5) of this residue could participate in a weak CH-O hydrogen bond with side-chain atoms of nonpolar substrate amino acids (48). Such an interaction would ensure a rapid release of the neutral amino acid upon conformational transition to the outward open state. As part of the extracellularly accessible AA+ pocket, Asp233 could be involved in binding and coordination of L-Arg and thus play a dual role in the transport cycle.
The underlying alternating access mechanism of APC family transporters is predominantly based on the formation of intracellular and extracellular barriers to restrict access of substrate molecules to one respective surrounding at a time (28, 45). In the inward-facing conformation of b(0,+)AT1, the extracellular barrier is formed by side-chain residues of TMs 1b, 3, and 10 located above the substrate-binding pocket close to the membrane surface (Fig. 3C). In addition, EL4 plugs the extracellular surface of this closed gateway by forming interactions between the loop residues Ala288, Gln287, and Leu284 and the TM residues Tyr135, Glu134, Lys53, and Leu56 (Fig. 3C). Conversely, the cytoplasmic gate remains closed in the inward-facing state (SI Appendix, Fig. S5). In our structure, the accessibility of the AA0-binding pocket from the cytoplasm is most likely achieved by a swing-open motion of TM1a, TM6b, and TM8 in between which a solvent-exposed cavity is formed. Assuming that b(0,+)AT1 will adopt a LeuT-type outward facing conformation, it is to be expected that IL1 will plug the intracellular barrier in a similar manner as EL4 does in the current structure (49, 50). A clear functional implication of IL1 is highlighted by defective transport of b(0,+)AT1 in patients with prevalent Gly105Arg/Glu mutations (19, 41, 47) (SI Appendix, Fig. S9D). Hence, we conclude that the gate-sealing loops are critical for stabilization of membrane barriers and regulation of transport activity.
Materials and Methods
Generation of Inducible HEK293 Stable Cell Lines.
The complementary DNAs of full-length b(0,+)AT1 (human SLC7A9, NIH Mammalian Gene Collection (MGC) Project BC017962) and rBAT (human SLC3A1, NIH MGC Project BC093624) were cloned into pcDNA5/FRT/TO (Invitrogen) and pACMV-TetO vectors, respectively. The gene for b(0,+)AT1 was modified by a C-terminal His10 or eGFP-His10 fusion tag. The gene for rBAT was modified by a C-terminal Strep-II tag.
The recombinant Flp-In T-REx293-b(0,+)AT1-rBAT cell line was generated by using a tetracycline-inducible and commercially available Flp-In T-REx host-cell line system from Invitrogen. Flp-In T-REx293 cells were cultured in high-glucose Dulbecco’s Modified Eagle Medium (DMEM) (Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS) (Gibco), 1× Pen/Strep (Gibco), 1 µg/mL Zeocin (Invitrogen), and 10 µg/mL blasticidin S hydrochloride (Invitrogen) at 37 °C in an atmosphere of 5% CO2. Cells were not tested for mycoplasma contamination. For stable integration, the pcDNA-5/FRT-SLC7A9-(eGFP-)His10 vector was first cotransfected with the Flp recombinase encoding expression vector pOG44 (Invitrogen) in a 1:13 ratio. Stable clones were selected by cultivation in growth medium containing 100 µg/mL hygromycine B. Subsequently, the pACMV-TetO-SLC3A1-Strep-II vector was transfected into the stable clones of the Flp-In REx293-b(0,+)AT1 cell line. All transfection procedures were performed with Lipofectamine 2000 reagent according to the manufacturer’s instructions (Thermo Fisher). To select for a stable Flp-In T-REx293-b(0,+)AT1-rBAT cell line, transfected cells were cultivated in the presence of G418 (Geneticin) (1,000 µg/mL).
Production and Purification of the b(0,+)AT1-rBAT Complex.
For heterologous protein production, the Flp-In T-REx293-b(0,+)AT1-rBAT cell line was cultured in roller bottles (Greiner Bio-One) in growth media containing 100 µg/mL hygromycine B and 500 µg/mL G418 for 7 d under the above-mentioned conditions. Gene expression was induced at 100% confluence by adding a final concentration of 2 µg/mL doxycycline hydrochloride. Production was carried out for 72 h. Cells were harvested with Accutase solution (Sigma-Aldrich) and stored at −80 °C until further use.
Harvested cells were suspended in cold lysis buffer containing 25 mM Tris (pH 8.0), 150 mM NaCl, and 0.1 g/mL SigmaFast ethylenediaminetetraacetic acid (EDTA) free Protease inhibitor (Sigma-Aldrich) and disrupted by stirring under high-pressure nitrogen atmosphere (750 MPa) for 45 min at 4 °C in a cell-disruption vessel (Parr Instrument). The cell lysate was centrifuged at 8,000 × g at 4 °C for 15 min. Subsequently, the low-velocity supernatant was centrifuged at 220,000 × g at 4 °C for 60 min. Pelleted membranes were resuspended and stored in a storage buffer containing 25 mM Tris (pH 8.0), 150 mM NaCl, 10% glycerol (vol/vol), and 0.1 g/mL SigmaFast EDTA free Protease inhibitor (Sigma-Aldrich).
All purification steps of the b(0,+)AT1-rBAT complex were performed at 4 °C. Isolated membranes were solubilized with 1% (wt/vol) 4-trans(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside (t-PCCαM) (GLYCON Biochemicals) with gentle stirring for 1 h. The insoluble membrane fraction was removed via ultracentrifugation at 220,000 × g for 1 h. A final concentration of 30 mM imidazole was added to the supernatant. Subsequently, the mixture was incubated with Ni-NTA Agarose resin (Qiagen) for 1 h. The resin was pre-equilibrated with buffer containing 25 mM Tris (pH 8.0), 150 mM NaCl, 0.5% t-PCCαM (wt/vol), and 30 mM imidazole. Detergent exchange was performed by washing the resin with 20 column volumes (CVs) of a digitonin-containing buffer (25 mM Tris [pH 8.0], 150 mM NaCl, and 0.1% digitonin [Serva]). The protein was eluted from the Ni-NTA resin with 10 CVs of the same buffer supplemented with 300 mM imidazole. The eluate was diluted 1:1 with digitonin exchange buffer and applied to a hand-packed Strep-TactinXT Superflow high-capacity column. After washing the column with 5 CVs of the digitonin buffer, the protein was eluted with 5 CVs of a biotin-containing buffer (25 mM Tris [pH 8.0], 150 mM NaCl, 0.1% digitonin [Serva], and 50 mM biotin). The eluted sample was subjected to a Superose 6 Increase 10/300 column (GE Healthcare) equilibrated with size exclusion chromatography (SEC) buffer (25 mM Tris [pH 8.0], 150 mM NaCl, and 0.1% digitonin [Serva]). Peak fractions were pooled, concentrated to 2 mg/mL, and used for further analysis.
Plasma Membrane Localization of b(0,+)AT1-rBAT.
Flp-In T-REx293-b(0,+)AT1-eGFP-His10-rBAT cells were cultured in high-glucose DMEM containing 10% (vol/vol) FBS, 1× penicillin/streptomycin (Pen/Strep), 100 µg/mL hygromycine B, 10 µg/mL blasticidin S hydrochloride, and 1,000 µg/mL G418 in an atmosphere of 5% CO2 at 37 °C for 48 h. Heterologous protein production was induced using a final concentration of 2 µg/mL doxycycline and carried out for 72 h. For cell imaging, the medium was gently removed, and the cells were rinsed and covered in 5 mL prewarmed Live Cell Imaging Solution (LCIS) from Thermo Fisher Scientific (140 mM NaCl, 2.5 mM KCl, 1.8 mM CaCl2, 1.0 mM MgCl2 buffered with 20 mM Hepes, pH 7.4, 300 mOsm). Cells were examined using an EVOS FL cell imaging system with a light cube for green fluorescent protein (Ex(nm) 470/22, Em(nm) 510/42) from Thermo Fisher Scientific at a magnification of 40×.
Differential Scanning Fluorimetry.
Purified protein samples were diluted with SEC buffer (see above) to 1 mg/mL and used for DSF measurements. A volume of 10 µL protein solution was used per Prometheus NT.48 Series nanoDSF Grade Standard capillary (NanoTemper Technologies). A temperature gradient of 1 °C min−1 from 20 to 95 °C was applied while the intrinsic protein fluorescence at 330 and 350 nm was recorded. Five technical replicates were recorded for Tm calculation.
Transport Measurements Using Stable Cell Lines.
For transport measurements, the cell lines Flp-In T-REx293, Flp-In T-REx293-b(0,+)AT1, Flp-In T-REx293-rBAT, and Flp-In T-REx293-b(0,+)AT1-rBAT (5 × 104 cells/well) were seeded into Nunclon Delta 12-well plates (NUNC) and cultured in high-glucose DMEM containing 10% (vol/vol) FBS, and 1× Pen/Strep in an atmosphere of 5% CO2 at 37 °C for 48 h. Subsequently, protein production was induced with a final concentration of 2 µg/mL doxycycline. After 48 h of protein production, the medium was removed and the cells were equilibrated for 5 min in prewarmed LCIS. Uptake measurements were initiated by incubating the cells in 0.5 mL LCIS supplemented with 50 µM L-arginine (18.35 nM L-[3H]-arginine, 54.5 Ci/mmol, 1 mCi/mL; arginine monohydrochloride L-[2,3,4-3H] [NET1123250UC], PerkinElmer) for different time periods (1, 3, 5, 10, and 30 min). For cystine competition assays, cells were incubated for 30 min in 0.5 mL LCIS supplemented with 50 µM L-arginine (18.35 nM L-[3H]-arginine, 54.5 Ci/mmol, 1 mCi/mL; arginine monohydrochloride L-[2,3,4-3H] [NET1123250UC], PerkinElmer) as well as the indicated L-cystine concentrations (Sigma-Aldrich). After incubation, the experiment was terminated by removing the uptake solution on ice and gently washing the cells twice with 0.5 mL cold LCIS. The cells were subsequently detached using 0.5 mL Accutase solution (Sigma-Aldrich) and transferred into pony vials (PerkinElmer) containing 0.5 mL 2% sodium dodecyl sulfate in LCIS for cell lysis (5 min at room temperature). After addition of 4 mL Rotiszint Eco Plus scintillation liquid (Carl Roth), radioactivity levels were analyzed via liquid scintillation (TriCarb1500 Liquid Scintillation Analyzer, Packard). All experiments were performed in triplicate.
The intrinsic [3H]-arginine uptake has not been subtracted from the data in this work. Our data are presented as mean ± SE values.
Single-Particle Cryo-EM Sample Vitrification and Data Acquisition.
Sample vitrification.
The purified protein sample was incubated with 300 µM cystine for 1 h before cryo-EM specimen preparation. C-flat R1.2/1.3 copper grids (400 mesh) were glow-discharged three times with a PELCO easiGlow device at 15 mA for 45 s. The purified b(0,+)AT1-rBAT complex was vitrified at 10 °C, 100% humidity, and a blot force of −1 using a Vitrobot IV device (Thermo Fischer). Blotting was carried out for 9 s before plunge-freezing in liquid ethane.
Image recording.
A total of 1780 movies were recorded in Energy-Filtered Transmission Electron Microscopy mode using a Titan Krios G3i microscope operated at 300 kV (Thermo Fisher). Data were acquired automatically using EPU software (Thermo Fisher) in electron-counting mode with a K3 direct electron detector in combination with a postcolumn energy-imaging filter (Gatan) at a nominal magnification of 105,000×, corresponding to a calibrated pixel size of 0.837 Å. Dose-fractionated movies were recorded for 2 s at an electron flux of 15 e− × pixel−1 × s−1 corresponding to a total dose of ∼40 e−/Å2. A defocus range of −1.1 to −2.2 µm was applied over the entire dataset.
Data Processing.
MotionCor2 (51) was used to correct for beam-induced motion and to generate dose-weighted images. Gctf (52) was used to determine the contrast transfer function (CTF) parameters and perform correction steps. Images with estimated poor resolution (>4 Å) were removed at this step. Particles were initially picked by using the Laplacian-of-Gaussian picking function of RELION-3.1 (53) to generate a trained reference for further picking with Topaz (54). A total of 223,706 particles were picked and used for all further processing steps. Initial model building, 3D classification, CTF refinement, Bayesian polishing, multibody 3D refinement, and final map reconstructions were performed with RELION-3.1 (53). A detailed overview of our processing workflow is given in SI Appendix, Fig. S2.
Model Building and Map Validation.
All model-building steps were performed using COOT (version 0.8) (55). The model of rBAT was de novo built into the EM density map. For building the b(0,+)AT1 model, we used the Protein Data Bank (PDB) submission of LAT1 (PDB 6JMQ) as a template structure. After backbone fitting and placement of side chains in the respective map densities, we performed real-space refinement using Phenix (version 1.14) (56). Refinement results were manually inspected and corrected if needed. The finalized models were validated by the MolProbity online server (57). A summary of the model parameters and the corresponding cryo-EM map statistics is found in SI Appendix, Table S1. The finalized models are visualized using Chimerax (58) and Chimera (59). To avoid overinterpretation of data, we did not build segments without distinct side-chain densities in our submission models. For completeness, the visualized models include polyalanine backbone traces of these regions. Tunnels and interior cavities were mapped with MOLE 2.5 (60).
Map-to-model and cross-validation of cryo-EM half-maps was performed as previously described (61, 62). In brief, the refined final model was converted to a density map with Chimera, and both unfiltered half-maps were summed. The resulting maps were used to calculate FSCmap-to-model curves by Phenix (version 1.14) (56). FSC0.5 was used as the cutoff to define resolution. All plots were generated with Graphpad Prism 8.0.
Supplementary Material
Acknowledgments
We thank Sofia Vater for technical assistance; Susann Kaltwasser and Mark Linder for technical support in electron microscopy; Yongchan Lee for valuable discussions and suggestions for data processing; and Hao Xie for discussions of experiments.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008111117/-/DCSupplemental.
Data Availability.
The cryo-EM maps presented in this work have been deposited in the Electron Microscopy Data Bank (EMDB) and can be found under accession numbers EMD-10933, EMD-10936, and EMD-10940. Model files of human b(0,+)AT1 and rBAT were deposited under the PDB under accession numbers 6YUP, 6YUZ, and 6YV1. (b(0,+)AT1-rBAT heterotetramer, rBAT homodimer, and b(0,+)AT1 respectively). Electron micrograph data have been deposited in the EMDB under accession number EMD-10933. All other study data are included in the article and supporting information.
References
- 1.Kandasamy P., Gyimesi G., Kanai Y., Hediger M. A., Amino acid transporters revisited: New views in health and disease. Trends Biochem. Sci. 43, 752–789 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Bhutia Y. D., Ganapathy V., Glutamine transporters in mammalian cells and their functions in physiology and cancer. Biochim. Biophys. Acta 1863, 2531–2539 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin L., Yee S. W., Kim R. B., Giacomini K. M., SLC transporters as therapeutic targets: Emerging opportunities. Nat. Rev. Drug Discov. 14, 543–560 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colas C., Ung P. M.-U., Schlessinger A., SLC transporters: Structure, function, and drug discovery. MedChemComm 7, 1069–1081 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mastroberardino L. et al., Amino-acid transport by heterodimers of 4F2hc/CD98 and members of a permease family. Nature 395, 288–291 (1998). [DOI] [PubMed] [Google Scholar]
- 6.Verrey F. et al., CATs and HATs: The SLC7 family of amino acid transporters. Pflugers Arch. 447, 532–542 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Fotiadis D., Kanai Y., Palacín M., The SLC3 and SLC7 families of amino acid transporters. Mol. Aspects Med. 34, 139–158 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Wagner C. A., Lang F., Bröer S., Function and structure of heterodimeric amino acid transporters. Am. J. Physiol. Cell Physiol. 281, C1077–C1093 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Palacín M., Errasti-Murugarren E., Rosell A., Heteromeric amino acid transporters. In search of the molecular bases of transport cycle mechanisms. Biochem. Soc. Trans. 44, 745–752 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Bröer S., Wagner C. A., Structure-function relationships of heterodimeric amino acid transporters. Cell Biochem. Biophys. 36, 155–168 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Bröer S., Palacín M., The role of amino acid transporters in inherited and acquired diseases. Biochem. J. 436, 193–211 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Chillarón J., Roca R., Valencia A., Zorzano A., Palacín M., Heteromeric amino acid transporters: Biochemistry, genetics, and physiology. Am. J. Physiol. Renal Physiol. 281, F995–F1018 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Palacín M., Bertran J., Zorzano A., Heteromeric amino acid transporters explain inherited aminoacidurias. Curr. Opin. Nephrol. Hypertens. 9, 547–553 (2000). [DOI] [PubMed] [Google Scholar]
- 14.Mizoguchi K. et al., Human cystinuria-related transporter: Localization and functional characterization. Kidney Int. 59, 1821–1833 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Verrey F. et al., Novel renal amino acid transporters. Annu. Rev. Physiol. 67, 557–572 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Kudo Y., Boyd C. A., Human placental amino acid transporter genes: Expression and function. Reproduction 124, 593–600 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Calonge M. J. et al., Cystinuria caused by mutations in rBAT, a gene involved in the transport of cystine. Nat. Genet. 6, 420–425 (1994). [DOI] [PubMed] [Google Scholar]
- 18.Brauers E., Schmidt C., Zerres K., Eggermann T., Functional characterization of SLC7A9 polymorphisms assumed to influence the cystinuria phenotype. Clin. Nephrol. 65, 262–266 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Feliubadaló L. et al.; International Cystinuria Consortium , Non-type I cystinuria caused by mutations in SLC7A9, encoding a subunit (bo,+AT) of rBAT. Nat. Genet. 23, 52–57 (1999). [DOI] [PubMed] [Google Scholar]
- 20.Fernández E. et al., rBAT-b(0,+)AT heterodimer is the main apical reabsorption system for cystine in the kidney. Am. J. Physiol. Renal Physiol. 283, F540–F548 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Eggermann T., Venghaus A., Zerres K., Cystinuria: An inborn cause of urolithiasis. Orphanet J. Rare Dis. 7, 19 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chillarón J. et al., Pathophysiology and treatment of cystinuria. Nat. Rev. Nephrol. 6, 424–434 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Stenson P. D. et al., Human gene mutation database (HGMD): 2003 update. Hum. Mutat. 21, 577–581 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Sahota A., Tischfield J. A., Goldfarb D. S., Ward M. D., Hu L., Cystinuria: Genetic aspects, mouse models, and a new approach to therapy. Urolithiasis 47, 57–66 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martell H. J. et al., Associating mutations causing cystinuria with disease severity with the aim of providing precision medicine. BMC Genomics 18 (Suppl. 5), 550 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palacín M., Kanai Y., The ancillary proteins of HATs: SLC3 family of amino acid transporters. Pflugers Arch. 447, 490–494 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Yan R., Zhao X., Lei J., Zhou Q., Structure of the human LAT1-4F2hc heteromeric amino acid transporter complex. Nature 568, 127–130 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Lee Y. et al., Cryo-EM structure of the human L-type amino acid transporter 1 in complex with glycoprotein CD98hc. Nat. Struct. Mol. Biol. 26, 510–517 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Rius M., Sala L., Chillarón J., The role of N-glycans and the C-terminal loop of the subunit rBAT in the biogenesis of the cystinuria-associated transporter. Biochem. J. 473, 233–244 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Janecek S., Svensson B., Henrissat B., Domain evolution in the α-amylase family. J. Mol. Evol. 45, 322–331 (1997). [DOI] [PubMed] [Google Scholar]
- 31.Dello Strologo L. et al., Comparison between SLC3A1 and SLC7A9 cystinuria patients and carriers: A need for a new classification. J. Am. Soc. Nephrol. 13, 2547–2553 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Chillarón J. et al., An intracellular trafficking defect in type I cystinuria rBAT mutants M467T and M467K. J. Biol. Chem. 272, 9543–9549 (1997). [DOI] [PubMed] [Google Scholar]
- 33.Kobayashi M. et al., Calcium ion-dependent increase in thermostability of dextran glucosidase from Streptococcus mutans. Biosci. Biotechnol. Biochem. 75, 1557–1563 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Gabriško M., Janeček S., Looking for the ancestry of the heavy-chain subunits of heteromeric amino acid transporters rBAT and 4F2hc within the GH13 α-amylase family. FEBS J. 276, 7265–7278 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Wang Y. L. et al., Structures of trehalose synthase from Deinococcus radiodurans reveal that a closed conformation is involved in catalysis of the intramolecular isomerization. Acta Crystallogr. D Biol. Crystallogr. 70, 3144–3154 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janeček Š., Gabriško M., Remarkable evolutionary relatedness among the enzymes and proteins from the α-amylase family. Cell. Mol. Life Sci. 73, 2707–2725 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu D., Hemler M. E., Metabolic activation-related CD147-CD98 complex. Mol. Cell. Proteomics 4, 1061–1071 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cantor J. M., Ginsberg M. H., CD98 at the crossroads of adaptive immunity and cancer. J. Cell Sci. 125, 1373–1382 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feral C. C. et al., CD98hc (SLC3A2) mediates integrin signaling. Proc. Natl. Acad. Sci. U.S.A. 102, 355–360 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lahme S., Bichler K.-H., Eggermann T., Lang F., Genomic and functional investigations of mutations of the SLC3A1 gene in cystinuria. Urol. Int. 69, 207–211 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Koulivand L. et al., Mutation analysis of SLC3A1 and SLC7A9 genes in patients with cystinuria. Urolithiasis 43, 447–453 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Fernández E. et al., The structural and functional units of heteromeric amino acid transporters. The heavy subunit rBAT dictates oligomerization of the heteromeric amino acid transporters. J. Biol. Chem. 281, 26552–26561 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Zhou Z. et al., LeuT-desipramine structure reveals how antidepressants block neurotransmitter reuptake. Science 317, 1390–1393 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jungnickel K. E. J., Parker J. L., Newstead S., Structural basis for amino acid transport by the CAT family of SLC7 transporters. Nat. Commun. 9, 550 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krishnamurthy H., Piscitelli C. L., Gouaux E., Unlocking the molecular secrets of sodium-coupled transporters. Nature 459, 347–355 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao X. et al., Mechanism of substrate recognition and transport by an amino acid antiporter. Nature 463, 828–832 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Font M. A. et al.; International Cystinuria Consortium , Functional analysis of mutations in SLC7A9, and genotype-phenotype correlation in non-Type I cystinuria. Hum. Mol. Genet. 10, 305–316 (2001). [DOI] [PubMed] [Google Scholar]
- 48.Gu Y., Kar T., Scheiner S., Fundamental properties of the CH···O interaction: Is it a true hydrogen bond? J. Am. Chem. Soc. 121, 9411–9422 (1999). [Google Scholar]
- 49.Yamashita A., Singh S. K., Kawate T., Jin Y., Gouaux E., Crystal structure of a bacterial homologue of Na+/Cl-dependent neurotransmitter transporters. Nature 437, 215–223 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Weyand S. et al., Structure and molecular mechanism of a nucleobase-cation-symport-1 family transporter. Science 322, 709–713 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng S. Q. et al., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zivanov J. et al., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bepler T. et al., Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 16, 1153–1160 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adams P. D. et al., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen V. B. et al., MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goddard T. D. et al., UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pettersen E. F. et al., UCSF Chimera: A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Pravda L. et al., MOLEonline: A web-based tool for analyzing channels, tunnels and pores (2018 update). Nucleic Acids Res. 46, W368–W373 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Amunts A. et al., Structure of the yeast mitochondrial large ribosomal subunit. Science 343, 1485–1489 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brown A. et al., Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D Biol. Crystallogr. 71, 136–153 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM maps presented in this work have been deposited in the Electron Microscopy Data Bank (EMDB) and can be found under accession numbers EMD-10933, EMD-10936, and EMD-10940. Model files of human b(0,+)AT1 and rBAT were deposited under the PDB under accession numbers 6YUP, 6YUZ, and 6YV1. (b(0,+)AT1-rBAT heterotetramer, rBAT homodimer, and b(0,+)AT1 respectively). Electron micrograph data have been deposited in the EMDB under accession number EMD-10933. All other study data are included in the article and supporting information.