Significance
Plant seedlings depend on efficient development of roots and root hairs for anchorage to the ground and for rapidly reaching nutrients and water for survival and growth. We found that a negative regulator of a small-molecule signaling pathway called “karrikin signaling” plays an important role in regulating root growth of the legume Lotus japonicus. Mutants of this regulator called “SMAX1” have short primary roots and strongly elongated root hairs. This phenotype is caused by enhanced ethylene biosynthesis, which in the wild type is suppressed by SMAX1. Thus, karrikin signaling regulates ethylene biosynthesis to fine-tune root development.
Keywords: Lotus japonicus, karrikin signaling, ethylene, root development, root hairs
Abstract
An evolutionarily ancient plant hormone receptor complex comprising the α/β-fold hydrolase receptor KARRIKIN INSENSITIVE 2 (KAI2) and the F-box protein MORE AXILLARY GROWTH 2 (MAX2) mediates a range of developmental responses to smoke-derived butenolides called karrikins (KARs) and to yet elusive endogenous KAI2 ligands (KLs). Degradation of SUPPRESSOR OF MAX2 1 (SMAX1) after ligand perception is considered to be a key step in KAR/KL signaling. However, molecular events which regulate plant development downstream of SMAX1 removal have not been identified. Here we show that Lotus japonicus SMAX1 is specifically degraded in the presence of KAI2 and MAX2 and plays an important role in regulating root and root hair development. smax1 mutants display very short primary roots and elongated root hairs. Their root transcriptome reveals elevated ethylene responses and expression of ACC Synthase 7 (ACS7), which encodes a rate-limiting enzyme in ethylene biosynthesis. smax1 mutants release increased amounts of ethylene and their root phenotype is rescued by treatment with ethylene biosynthesis and signaling inhibitors. KAR treatment induces ACS7 expression in a KAI2-dependent manner and root developmental responses to KAR treatment depend on ethylene signaling. Furthermore, in Arabidopsis, KAR-induced root hair elongation depends on ACS7. Thus, we reveal a connection between KAR/KL and ethylene signaling in which the KAR/KL signaling module (KAI2–MAX2–SMAX1) regulates the biosynthesis of ethylene to fine-tune root and root hair development, which are important for seedling establishment at the beginning of the plant life cycle.
The most recently discovered putative plant hormone signaling pathway, currently called “karrikin signaling,” has been identified in the context of postfire seed germination (reviewed in ref. 1). Karrikins (KARs) are small butenolide compounds which are found in the smoke of burning vegetation and trigger germination of fire-following plants (2). They also enhance germination of primary dormant Arabidopsis thaliana seeds (3), which enabled the identification of the KAR perception components, the α/β-fold hydrolase receptor KARRIKIN INSENSITIVE 2 (KAI2) and the F-box protein MORE AXILLARY GROWTH 2 (MAX2), by forward and reverse genetics (4, 5). KAI2 is closely related to the strigolactone receptor DWARF14 (D14), which also interacts with MAX2 in strigolactone perception (6). Strigolactones are well-established plant hormones with prominent roles in the suppression of shoot branching and in stimulating the presymbiotic growth of arbuscular mycorrhiza fungi and seed germination of parasitic weeds in the rhizosphere (reviewed in ref. 7). By contrast, the endogenous KAI2 ligand(s) still need(s) to be identified. Nevertheless, the widespread phylogenetic distribution of KAI2 from charophyte algae to all land plants, as well as a number of smoke detection-independent kai2 mutant phenotypes described in Arabidopsis, rice, and petunia, indicates that the original function of KAI2 was to perceive a plant hormone (tentatively called “KAI2 ligand”; KL) while, in fire-following plants, KAI2 was secondarily repurposed for smoke detection (5, 8–13). Duplications of KAI2 occurred during plant diversification, and provided flexibility for additional adaptations. For example, two KAI2 paralogs (KAI2a and KAI2b) of Lotus japonicus and the fire follower Brassica tournefortii diverged in their binding preference to diverse KAR molecules (14, 15), while in parasitic plants and Physcomitrella patens the KAI2 family has expanded to more than 10 members, of which some have retained their ability to recognize KARs whereas others underwent changes in their binding pocket to bind strigolactone or a yet unknown KL-type ligand (16–19).
A key event for triggering plant hormone responses is the inactivation of repressor proteins after hormone perception by ubiquitylation and subsequent proteasomal degradation in most hormone signaling pathways. Forward genetic screens have identified repressors of KAR/KL and strigolactone signaling (20–25). They belong to the same SUPPRESSOR OF MAX2-like (SMXL) family comprising three major clades in seed plants that are in Arabidopsis composed of 1) SMAX1 and SMXL2 for KAR/KL signaling, 2) SMXL6, 7, and 8 for strigolactone signaling, and 3) SMXL3, 4, and 5 acting in a KAR/KL- and strigolactone-independent yet unknown signaling module that controls phloem formation (22–27). The SMXL proteins display similarity to class I Clp ATPases. Arabidopsis SMAX1, SMXL6, 7, and 8, as well as their rice ortholog DWARF53 (D53) were shown to interact with the transcriptional corepressor TOPLESS through ethylene response factor-associated amphiphilic repression (EAR) motifs (20, 21, 23, 25). Furthermore, D53 was described to promote TOPLESS–nucleosome interactions (28). Thus, although the exact biochemical function of the SMXL proteins is yet elusive, they likely act as transcriptional repressors. Ubiquitylation and proteasomal degradation upon ligand perception have been presented for the strigolactone signaling repressors SMXL6, 7, and 8 and D53 in the presence of D14 and MAX2 (20, 21, 23, 25, 29) and very recently also for SMAX1 and SMXL2 in the presence of KAI2 and MAX2 (30, 31).
The role of SMAX1 and SMXL2 in plant development has been genetically addressed in Arabidopsis; smax1 smxl2 double mutants display mild, kai2-opposing phenotypes, such as slightly faster seed germination, shorter hypocotyls, bigger cotyledons, and longer root hairs (12, 24). Molecular events occurring downstream of SMAX1 SMXL2 degradation and regulating these phenotypes are currently unknown. We have previously observed that the legume L. japonicus responds to pharmacological KAR treatment with a reduction in primary root growth (14). Here we analyzed the role of SMAX1 and found that in contrast to Arabidopsis smax1 smxl2 mutant seedlings, L. japonicus smax1 mutant seedlings have a dramatic root phenotype, with 50% shorter primary roots (PRs) than the wild type and a 300% increase in root hair (RH) length. We show that expression of the ethylene biosynthesis gene ACC synthase 7 (ACS7), as well as ethylene release, is increased in smax1 mutants and that the increased ethylene production causes the smax1 root phenotype. Congruously, with degradation of SMAX1 upon KAR/KL perception, KAR treatment enhances ACS7 expression in the wild type. Furthermore, reduced PR growth in response to KAR depends on ethylene signaling. Thus, we reveal a connection between KAR/KL and ethylene signaling, which fine-tunes root development in L. japonicus.
Results
The SMXL Protein Family in L. japonicus.
To identify SMAX1 in L. japonicus, we constructed a phylogenetic tree with SMXL protein sequences of L. japonicus, A. thaliana, Medicago truncatula, Sorghum bicolor, and Oryza sativa. Similar to other non-Brassicaceae dicotyledons, the L. japonicus genome does not contain close homologs to all Arabidopsis SMXL genes, indicative of recent SMXL gene duplications and losses in the dicotyledons (27). In the KAR/KL signaling clade, L. japonicus maintains one SMAX1 homolog (SI Appendix, Fig. S1). There are three SMXLs in the strigolactone signaling clade (SMXL7a, SMXL7b, SMXL8), in which SMXL7a, 7b, and 8 seem to have originated from a common ancestor gene, which has duplicated independently in Arabidopsis and L. japonicus. SMXL5 is not found in the L. japonicus genome, but SMXL3 and 4 as well as an additional SMXL9 are present (27).
The L. japonicus SMXL genes differ in their transcript accumulation in the leaf, stem, flower, and root of L. japonicus (SI Appendix, Fig. S2). SMAX1 transcripts accumulate consistently across all tested organs, while SMXL7a, SMXL7b, and SMXL8 transcripts accumulate to higher levels in leaf, stem, and root, and to lower levels in flowers. SMXL3, SMXL4, and SMXL9 transcript levels are generally low.
SMAX1 Is a Proteolytic Target of the KAR Receptor Complex in L. japonicus.
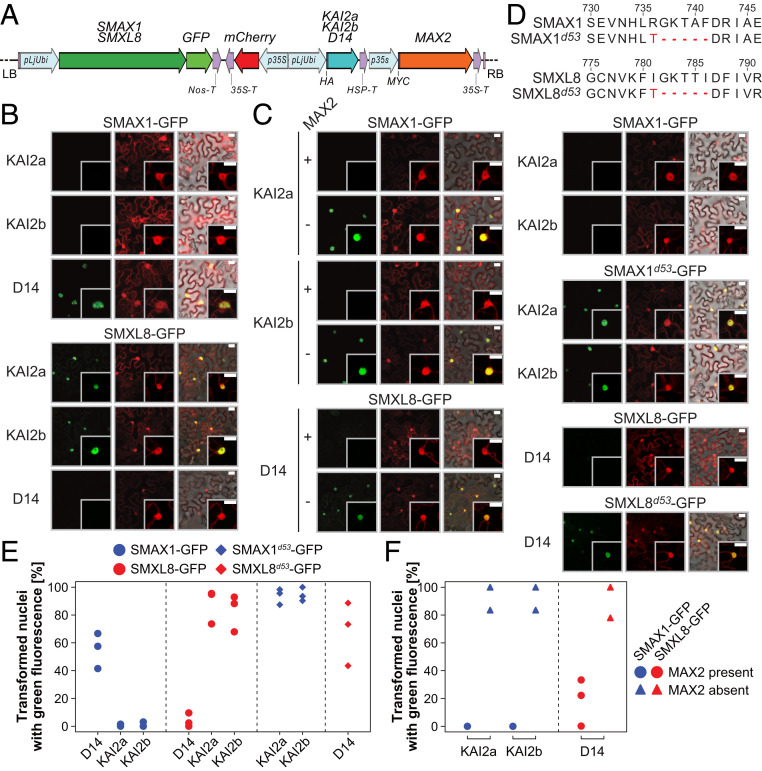
Arabidopsis SMXL6, 7, and 8 and their rice ortholog D53 are degraded after GR24 treatment in a D14- and MAX2-dependent manner (20, 21, 23, 25), and evidence for karrikin-induced and KAI2- and MAX2-dependent degradation of Arabidopsis SMAX1 and SMXL2 has just been published (30, 31). To examine whether L. japonicus SMAX1 is a specific proteolytic target of the KAR/KL receptor complex, we tested its stability in transiently transformed Nicotiana benthamiana leaves in the presence of L. japonicus KAI2 and MAX2. We coexpressed L. japonicus SMAX1-GFP (green fluorescent protein) together with MAX2 and one of the α/β-fold hydrolase receptors KAI2a, KAI2b, or D14 under the control of strong promoters from a single plasmid, which additionally contained an expression cassette for free mCherry as a transformation marker (Fig. 1A). SMAX1-GFP was clearly visible in the nuclei of N. benthamiana leaf epidermal cells coexpressing MAX2 and the strigolactone receptor D14 (Fig. 1 B and E). In contrast, no GFP signal was observed in the presence of MAX2 and the KAR/KL receptors KAI2a or KAI2b, even in the absence of KAR treatment, indicating that SMAX1 is specifically degraded in the presence of the two KAI2 isoforms. We scrutinized the specificity of SMAX1 degradation by testing the stability of other GFP-labeled SMXLs, namely SMXL8 (strigolactone signaling in Arabidopsis) and SMXL3, 4, and 9 (phloem formation in Arabidopsis). As expected for a repressor of strigolactone responses, SMXL8-GFP was not detected in the presence of D14 and MAX2, but it accumulated when D14 was exchanged for KAI2a or KAI2b (Fig. 1 B and E). GFP fusions of SMXL3, 4, and 9 accumulated under all conditions, showing that they are targeted by neither KAI2 nor D14 in the presence of MAX2 (SI Appendix, Fig. S3 A and B). Degradation of both SMAX1 and SMXL8 depends on the presence of L. japonicus MAX2, as both SMXL-GFP fusion proteins accumulated in the nuclei in the absence of L. japonicus MAX2 even in the presence of the cognate α/β-fold hydrolase receptor (Fig. 1 C and F). Thus, N. benthamiana receptor components are insufficient to mediate full removal of L. japonicus SMAX1 and SMXL8. A 5-amino acid deletion in the P loop of the SMXL6,7,8, rice ortholog D53 (GKTGI) renders the resulting d53 mutant resistant to proteasomal degradation (20, 21). At the same location, a similar amino acid motif (GKTAL) has been found by sequence alignment in Arabidopsis SMAX1 (32) and we hypothesized that deletion of this motif may also protect SMAX1 from degradation. Indeed, deletion of the corresponding motifs from L. japonicus SMAX1 (GKTAF) and SMXL8 (GKTTI) allowed SMAX1d53-GFP and SMXL8d53-GFP to accumulate in the nuclei of N. benthamiana leaf epidermal cells despite the presence of MAX2 and the cognate α/β-fold hydrolase receptors (Fig. 1 D and E). The receptor- and P loop-dependent stability patterns of SMAX1, SMXL8, and SMXL3, 4, and 9 were confirmed by Western blot (SI Appendix, Fig. S3C).
Fig. 1.
SMAX1 is specifically degraded in the presence of KAI2 and MAX2. (A) Schematic representation of the expression cassettes in the T-DNA contained in the Golden Gate plasmids used for transient transformation of N. benthamiana leaves for the SMXL degradation assay (LB, left border; RB, right border). All coexpressed proteins were encoded on the same plasmid. (B and C) Confocal microscopy images of N. benthamiana leaves expressing SMAX1 or SMXL8 fused with GFP, a free mCherry transformation marker, and any of the α/β-hydrolase receptors hemagglutinin (HA)-D14, HA-KAI2a, or HA-KAI2b in the presence (B) or presence/absence of MYC-MAX2 (C). For each combination, the green fluorescence of SMXL-GFP fusions (Left), red fluorescence of the mCherry transformation marker (Middle), and an overlay of green and red fluorescence and bright-field images (Right) are shown. (B and C, Insets) A single nucleus at higher magnification. (Scale bars, 25 µm.) (D) Representation of the amino acid deletions which give rise to degradation-resistant SMXL8d53 and SMAX1d53 and the accumulation of SMXL8d53- and SMAX1d53-GFP fusions in nuclei of N. benthamiana leaf epidermal cells in the presence of HA-KAI2a, HA-KAI2b, or HA-D14 and MYC-MAX2. (E and F) Percentage of green fluorescent nuclei (indicating the presence of SMAX1 or SMXL8) per red fluorescent nucleus (indicating successful transformation) in microscopy images of leaf epidermal cells of N. benthamiana in the presence of the HA-tagged α/β-fold hydrolase receptor indicated on the x axis and the presence (E) or presence/absence (F) of MYC-MAX2. In each graph, 23 to 113 (E) or 3 to 9 (F) nuclei were analyzed in n ≥ 2 images per protein combination.
In summary, we show that SMAX1 is specifically degraded in the presence of KAI2a or KAI2b and MAX2, while SMXL8 is specifically degraded in the presence of D14 and MAX2. Thus, SMAX1 is a bona fide target of the KAI2–MAX2 receptor module and the canonical SMAX1–KAI2 and SMXL8–D14 relationships suggested by genetic analysis in Arabidopsis (reviewed in ref. 33) are valid for L. japonicus proteins.
SMAX1 Is Required for Primary Root Elongation and for Restricting Root Hair Growth.
Having unequivocally identified SMAX1 in L. japonicus through phylogenetic analysis and degradation assays, we searched for smax1 retrotransposon (LORE1) insertion mutants (34) to investigate the role of SMAX1 in L. japonicus development. We found three mutants (smax1-1, smax1-2, smax1-3) with insertions in the first exon of the gene (SI Appendix, Fig. S4A). Because segregating seeds carrying the smax1-1 insertion had a strong germination defect, we focused on smax1-2 and smax1-3. As previously shown in Arabidopsis smax1 mutants (22), transcripts of the KAR/KL marker gene DLK2 (5, 14) accumulate at high levels in the roots of both allelic mutants, confirming that SMAX1 is nonfunctional (see Fig. 3B).
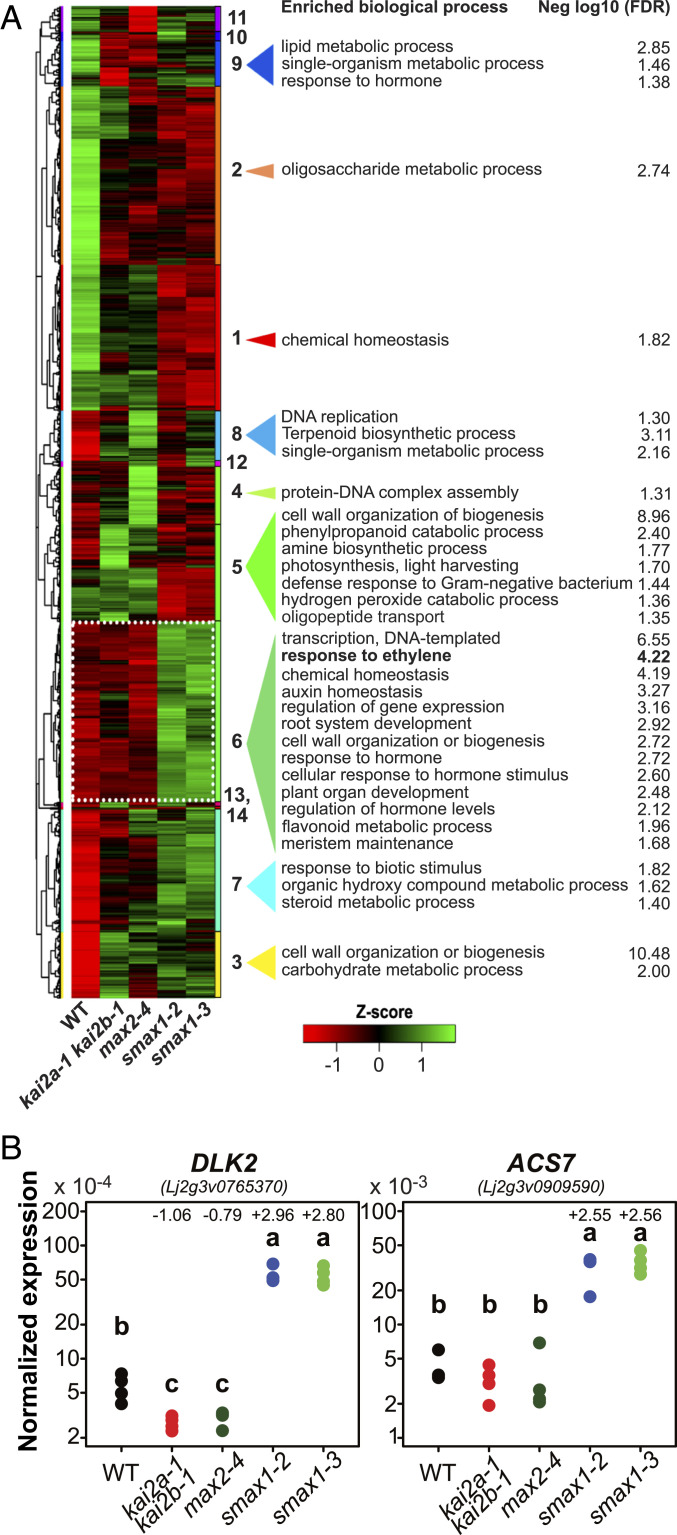
Fig. 3.
Differential gene expression in L. japonicus KAR/KL signaling mutants. (A) Hierarchical clustering of 3,748 unique DEGs (comparing each mutant vs. WT). Z scores were obtained by scaling the r–log-transformed count data. Red represents lower and green indicates higher transcript accumulation. The clusters are indicated by a dendrogram (Left) and illustrated by different colors and a number (Right). Gene ontology enrichment analysis was performed using AgriGO for each cluster separately. Negative log10(FDR) represents the statistical significance (FDR, false discovery rate) of the enrichment, with higher values representing stronger enrichment (cutoff, −log10FDR ≥ 1.3). (B) Transcript accumulation of D14like2 (DLK2) and ACS7 genes in roots of the indicated genotypes as determined by qRT-PCR. Expression values were normalized to the expression of ubiquitin. Letters indicate statistical differences between genotypes (ANOVA, post hoc Tukey test (n = 3 or 4). Numbers above the data points indicate, if significant, the log2 fold change for the mutant vs. wild type comparison as determined by RNA-seq analysis.
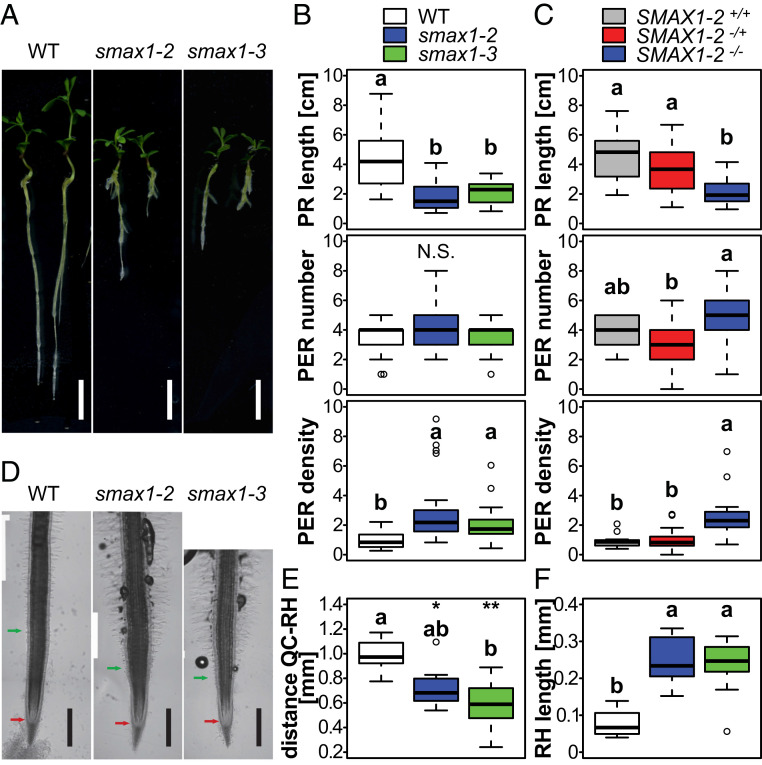
Interestingly, both allelic smax1 mutants display a strong seedling root phenotype (Fig. 2). The PR length is strongly reduced, whereas the number of postembryonic roots (PERs), including lateral and adventitious roots (14), is similar to the wild type, resulting in an increased PER density (Fig. 1 A and B). Association of this phenotype with the homozygous LORE1 insertion in SMAX1 was confirmed by cosegregation analysis with a population segregating for the smax1-2 insertion (binomial linear regression for PR length, P = 0.001144 and for PER density, P = 6.59 × 10−5; Fig. 1C). From a population of 72 individuals, 13 seedlings were homozygous wild type for the SMAX1-2 locus, 44 were heterozygous, and 15 were homozygous mutant, respecting a Mendelian segregation (χ2 = 3.67, P = 0.16).
Fig. 2.
L. japonicus smax1 mutants have a reduced primary root and increased root hair length. (A) Representative images of wild type, smax1-2, and smax1-3 grown on Petri dishes at 10 d postgermination. (Scale bars, 1 cm.) (B and C) Primary root length, postembryonic root number, and PER density of wild type, smax1-2, and smax1-3 (n ≥ 23) (B) and a population segregating for the smax1-2 LORE1 insertion (C) (n ≥ 13). (D) Representative images of the root tip. Red arrows indicate the position of the quiescent center (QC); green arrows indicate the nearest root hair. (Scale bars, 500 µm.) (E and F) Distance of the first RH from the QC (E) and RH length at 1.5 to 2 mm from the apex (F) in the wild type, smax1-2, and smax1-3 (n ≥ 6). (E) Asterisks indicate significant differences compared with the wild type (ANOVA, post hoc Dunnett test; N.S., not significant, P > 0.05; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001). (B, C, E, and F) Letters indicate significant differences (ANOVA, post hoc Tukey test).
In addition to a reduced PR length, the smax1 mutants display elongated root hairs, which are on average three times longer than those of the wild type (Fig. 2 D and F). Furthermore, the first root hairs emerge 400 µm closer to the root tip quiescent center (Fig. 2E). We examined whether the shorter PR length and distance between quiescent center and first root hairs of smax1 mutants are caused by a defect in root cell elongation. Longitudinal sections at the root tip region revealed that the transition zone above the root tip of smax1 mutants is swollen with compact-looking cells (SI Appendix, Fig. S5A). We determined the cumulative length for the 25 first observable cortical cells situated directly below the epidermis starting from the meristematic zone, because these are most continuously observable in the longitudinal sections (SI Appendix, Fig. S5B). In the wild type, the cells above cell 6 or 7 start to elongate whereas, in the two smax1 mutants, cell elongation is delayed, and starts only at cell numbers 14 and 15. In addition, smax1 mutants display enhanced cortical cell and root widths as compared with the wild type (SI Appendix, Fig. S5 C–E), suggesting that the cells may be defective in anisotropic growth.
To examine whether smax1 is epistatic over kai2a and kai2b for root phenotypes, we compared smax1 with the kai2a kai2b smax1 triple mutant. The triple mutant recapitulated the PR and root hair phenotypes of smax1, showing that smax1 is fully epistatic over kai2a kai2b (SI Appendix, Fig. S6). Interestingly, and in contrast to previously reported Arabidopsis kai2 mutant phenotypes (12), L. japonicus kai2a kai2b mutant roots resembled the wild type and did not display longer PRs and had only marginally shorter root hairs.
The smax1 Root Phenotype Is Not Caused by Low Sugar or Phosphate Supply.
Seed nutrient reserves are essential for early seedling development and growth (35). We investigated whether the smaller root system of smax1 mutants may be caused by reduced seed reserves, and weighed the seeds from homozygous and heterozygous smax1 parents and measured their two-dimensional area, which is a good proxy for L. japonicus seed weight (SI Appendix, Fig. S7 A–C). Seeds from homozygous smax1 mutants are around 25% lighter and smaller than wild-type seeds, while segregating seeds from heterozygous parents have on average an intermediate weight, size, and size distribution (SI Appendix, Fig. S7). This raises the possibility that growth of smax1 mutant seedlings suffers from a reduced nutrient (for example, carbon) supply from their seed.
However, addition of 1% sugar to the medium did not rescue root growth of smax1, although it strongly improved primary root growth of the wild type (SI Appendix, Fig. S8A). Furthermore, it positively affected PER number, such that PER density remained higher in the smax1 mutants as compared with the wild type, together making it unlikely that reduced sugar supply is causative of the smax1 root phenotype.
Decreased PR length can also be caused by phosphate starvation (36) and, since the smax1 root phenotype was observed on a medium with a low concentration of phosphate (2.5 µM), the smax1 root phenotype may be a symptom of hypersensitivity to phosphate starvation. However, smax1 mutant root growth cannot be restored on medium containing a high phosphate concentration (2.5 mM). In this condition, L. japonicus wild-type plants responded with reduced PR length whereas PR length of smax1 remained unchanged (SI Appendix, Fig. S8B). The phosphate conditions did not affect the PER number and PER density of both genotypes. Together, this indicates that the smax1 root architecture phenotype is not caused by low phosphate availability. Thus, the root phenotype does not seem to be determined by seed reserves, but the small seed size may be a result of the smaller size of the mother plant.
Transcriptome Patterns of KAR/KL Signaling Mutants.
To identify pathways which may be deregulated in smax1 mutants and responsible for their primary root and root hair phenotypes, we performed Illumina RNA sequencing (RNA-seq) from roots of the two allelic smax1 mutants and the KAR/KL perception mutants kai2a kai2b and max2. After read mapping to the L. japonicus MG20 messenger RNA reference (version 3.0), exploratory analysis for assessing genotype-specific transcriptome variation, and differential expression analysis comparing each mutant with the wild-type transcriptome (adjusted P value ≤ 0.01 and log2FC ≥ |0.48|; FC, fold change), we found a total of 7,759 unique differentially expressed genes (DEGs) (SI Appendix, Figs. S9–S11 and Datasets S1, S2, and S3). Since the current version of the L. japonicus reference genome contains a considerable number of false positive gene annotations or duplicate annotations, a LegumeMine search was performed for each mutant DEG list to filter for bona fide genes conserved among legumes. This reduced the number of original DEGs to a total of 5,340 unique DEGs (SI Appendix, Fig. S9 and Dataset S2). Most DEGs (4,420 DEGs) were found in the smax1 mutants and 57% (2,511) of them overlapped between the two smax1 mutant alleles (SI Appendix, Fig. S12A and Tables S8 and S9). Since these 2,511 “smax1 DEGs” were robustly confirmed in two independent allelic mutants, we used them for further analyses. In kai2a kai2b and max2 roots, we found a total of 2,431 DEGs (SI Appendix, Fig. S12B and Tables S8 and S9). Between these mutants, 765 (31.5%) DEGs overlapped and are therefore robust candidates for targets of KAR/KL signaling. A total of 506 DEGs overlapped for all mutants (SI Appendix, Fig. S12C). Unexpectedly, only 4 DEGs were significantly regulated in opposite directions in kai2a kai2b and max2 versus the smax1 mutants, although SMAX1 is a proteolytic target of the KAR receptor complex (SI Appendix, Fig. S12E). The well-known KAR signaling marker gene DLK2 (Lj2g3v0765370; Fig. 3B), Lj3g3v0139639 (unknown), and Lj4g3v2400850 (coatomer subunit beta′-2-like isoform X1) were up-regulated in the smax1 mutants and down in kai2a kai2b and max2, whereas Lj2g3v1155500 (peptide transporter 5) showed the opposite expression pattern. Low fold-change value in the comparison of the mutant versus the wild type may prevent many genes from passing the significance threshold for a fraction of mutants. Alternatively, yet unknown feedback mechanisms may cause a range of genes (275 up, 226 down; SI Appendix, Fig. S12D) to be regulated in all mutants in the same direction. A number of genes were dysregulated in the smax1 mutants but in the kai2a kai2b and max2 mutants their transcript accumulation did not differ from the wild type. These may not be regulated in the wild type under our growth conditions or they could be candidates for secondary response genes to the removal of SMAX1. All transcript accumulation patterns were validated by qRT-PCR on a subset of 18 genes (Fig. 3B and SI Appendix, Fig. S13).
Increased Transcript Accumulation of the Ethylene Biosynthesis Gene ACS7 in smax1 Mutants.
Hierarchical clustering on the 5,340 DEGs (using the r–log-transformed read count dataset) followed by gene ontology (GO) enrichment of each cluster (Fig. 3A, SI Appendix, Fig. S14, and Dataset S4) as well as GO enrichment analysis for each mutant versus the wild type (SI Appendix, Fig. S15 and Dataset S5) revealed enrichment of the term “cell wall metabolic process” for all mutants. smax1 mutant transcriptomes specifically displayed a deregulation of secondary metabolite processing, such as flavonoid, steroid, and phenylpropanoid metabolic processes, whereas kai2a kai2b and max2 transcriptomes displayed an enrichment of isoprenoid and terpenoid biosynthetic processes. Furthermore, transcripts overaccumulating in smax1 mutants were enriched for GO terms related to transcriptional regulation, response to biotic stimulus, meristem maintenance, and responses to hormone stimuli, especially to auxin and ethylene.
Ethylene is known to inhibit primary root cell elongation and to promote root hair elongation in Arabidopsis (reviewed in ref. 37), thus causing similar phenotypes as observed in the L. japonicus smax1 mutants. Several AP2 transcription factor genes annotated as ethylene response factors (ERFs) are present in cluster 6 (Fig. 3A), which contains genes that are up-regulated in smax1 mutants, as well as an ACC-SYNTHASE gene (Lj2g3v0909590), which is homologous to Arabidopsis ACS7 (Fig. 3B and SI Appendix, Fig. S16). ACSs are rate-limiting enzymes in ethylene biosynthesis and it has been demonstrated that transcriptional regulation of ACS genes is an important factor in regulating ethylene biosynthesis (reviewed in ref. 38). We identified all ACS genes (and corresponding protein sequences) in the L. japonicus genome and assessed their transcript read count in the RNA-seq dataset. Only ACS7 transcripts and those of the closely related but weakly expressed duplicated gene ACS7-like overaccumulated in smax1 mutants (SI Appendix, Fig. S16 A–C). In Arabidopsis, overexpression of ACS7 causes short primary roots and elongated root hairs, resembling the L. japonicus smax1 root phenotype (39). Furthermore, 14 N-terminal amino acid residues, which make AtACS7 susceptible to proteasomal degradation (39, 40), are not conserved in LjACS7 (SI Appendix, Fig. S16D). Together, this bolsters the hypothesis that the increase in ACS7 transcripts may explain the smax1 root phenotypes.
Root Growth Inhibition in smax1 Is Caused by Increased Ethylene Biosynthesis.
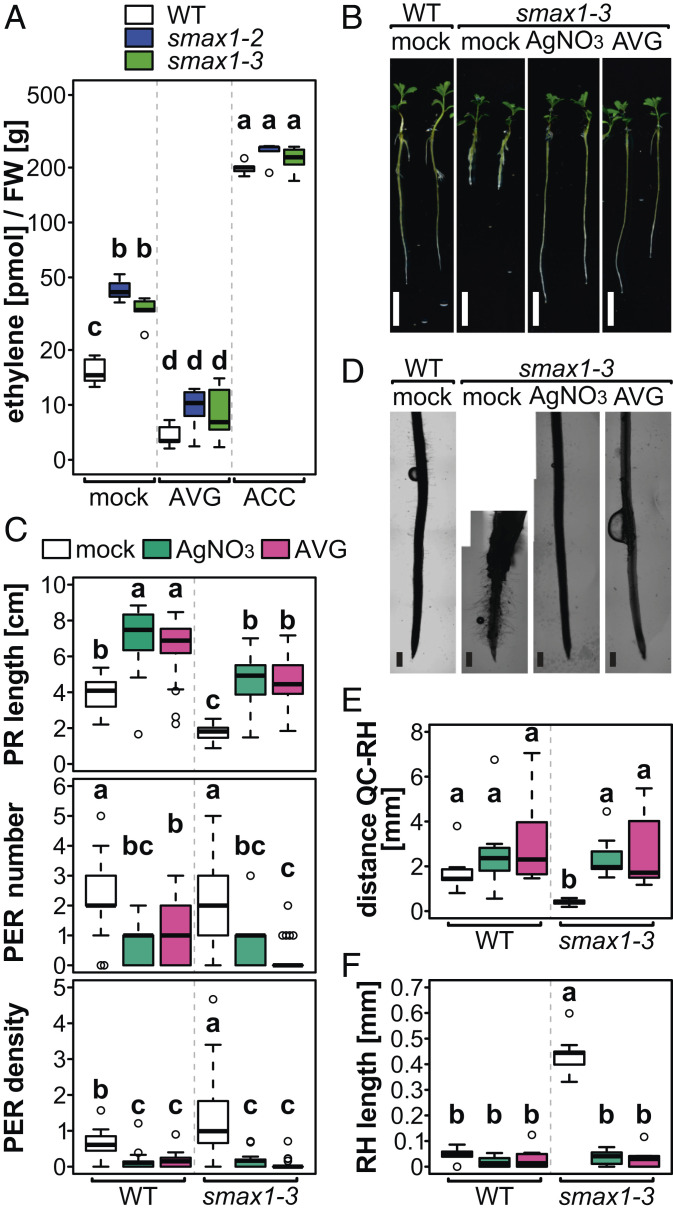
We measured by gas chromatography whether increased ACS7 expression results in elevated ethylene biosynthesis in the smax1 mutants. Both smax1 mutants released at least 2.5 times more ethylene than the wild type (Fig. 4A). Nevertheless, the smax1 mutants were responsive to pharmacological perturbation of ethylene biosynthesis through the ethylene precursor ACC (1-aminocyclopropane-1-carboxylic acid) and the ACC biosynthesis inhibitor AVG (aminoethoxyvinylglycine) in a similar manner as the wild type (Fig. 4A).
Fig. 4.
Primary root and root hair phenotypes of smax1 mutants are caused by increased ethylene production. (A) Ethylene released by L. japonicus wild type, smax1-2, and smax1-3 seedlings and in response to treatment with 0.1 µM AVG or 1 µM ACC as determined by gas chromatography (n = 5). (B and C) Representative images (B) and quantification (C) of PR length, PER number, and PER density of 10-d-old wild-type and smax1-3 seedlings grown in the presence of 50 µM silver nitrate (AgNO3) or 0.1 µM AVG (n ≥ 24). (Scale bars, 1 cm.) (D–F) Representative images of the root tip (D) and quantification of the distance between the first RH and the QC (E) and of the RH length (F) of wild-type and smax1-3 seedlings in the presence of 50 µM silver nitrate or 0.1 µM AVG (n ≥ 7). (Scale bars, 500 µm.) (A, C, E, and F) Letters indicate significant differences (ANOVA, post hoc Tukey test, P ≤ 0.001).
To understand whether increased ethylene levels can cause short primary roots and elongated root hairs in L. japonicus, we treated wild-type seedlings with the ethylene precursor ACC and the ethylene-releasing chemical ethephon. With this treatment, the wild type recapitulated smax1 root and root hair phenotypes (SI Appendix, Fig. S17). Importantly, treatment with AVG, which specifically blocks the synthesis of the ethylene precursor ACC through inhibition of ACS (41), or with silver nitrate, which blocks multiple ethylene receptors such as ETHYLENE RECEPTOR 1 (ETR1) and thereby ethylene perception (42), restores wild type-like primary root and root hair development in both allelic smax1 mutants (Fig. 4 B–F and SI Appendix, Fig. S18). Thus, the overproduction of ethylene in the smax1 mutants causes the defects in primary root and root hair development.
Increased Ethylene Biosynthesis Does Not Affect Hypocotyl Development in smax1 Mutants.
To understand whether increased ethylene biosynthesis in smax1 mutants also affects the seedling shoot, we performed hypocotyl triple-response assays [for reduced hypocotyl growth, increased hypocotyl thickening, and exaggerated apical hook curvatures (43)] with etiolated seedlings (SI Appendix, Fig. S19 A–C). smax1 mutants do not show a constitutive triple response, but they respond to increasing concentrations of added ACC in a dose-dependent manner, similar to the wild type. However, the dark-grown (SI Appendix, Fig. S19 A–C) as well as light-grown (SI Appendix, Fig. S19 D and E) smax1 hypocotyls are shorter than those of the wild type, and this is not rescued by AVG or silver nitrate (SI Appendix, Fig. S19 D and E). Nevertheless, ACS7 transcript accumulation was increased in smax1 mutants (SI Appendix, Fig. S19F). This suggests that the increased ethylene biosynthesis in L. japonicus smax1 mutants specifically affects the root, possibly because hypocotyls are less sensitive to ethylene than roots. Furthermore, mechanisms other than ethylene signaling seem to influence hypocotyl elongation in smax1 mutants.
Ethylene-Dependent and -Independent Transcriptional Regulation in smax1 Mutants.
We examined whether transcript regulation in smax1 roots is a secondary response to ethylene by testing the expression of eight of the qRT-PCR–confirmed DEGs (SI Appendix, Fig. S13) after treatment with AVG and silver nitrate (SI Appendix, Fig. S20). The elevated transcript accumulation of three genes in the smax1 mutants, annotated as Germin-like (Lj3g3v2601420), IAMT1-like (Lj2g3v3222870), and Auxin-Induced-5NG4-like (Lj6g3v2244450), was suppressed by both AVG and silver nitrate treatment, demonstrating that their increased expression is ethylene-dependent. Interestingly, an Expansin gene (Lj0g3v0287409) is repressed only upon AVG but not silver nitrate treatment, suggesting that this gene is likely not regulated in response to ethylene but possibly to ACC perception (44, 45). In contrast, transcripts of DLK2 as well as of a gene of unknown function (Lj0g3v0127589), a gene annotated as serotonin receptor-like (Lj4g3v0496580), and an AP2 transcription factor gene annotated as ERF (Lj2g3v1068730) overaccumulated in smax1 mutants independent of ethylene inhibitor treatment. We called the ERF gene ERIK (for ERF INDUCED BY KARRIKIN; see below).
To test if the ethylene signaling-independent smax1 DEGs are early targets of KAR/KL signaling, we analyzed their expression after 2- and 6-h treatments by KAR1 in roots. KAR1 caused a significant induction of DLK2, ERIK, and the serotonin receptor-like gene in a KAI2a KAI2b- and MAX2-dependent manner (SI Appendix, Fig. S21). As expected, the ethylene-dependent Germin-like gene, included as a control, was not induced by KAR1 treatment. Thus, we confirm that DLK2 is an early KAR response gene and, in addition, we present two KAR/KL marker genes in L. japonicus, the ethylene-independent ERF gene ERIK (Lj2g3v1068730) and the serotonin receptor-like gene (Lj4g3v0496580).
Ethylene Signaling Is Required for the Effect of KAR1 on Root Development.
If increased ACS7 expression is a direct effect of SMAX1 removal, then expression of this gene should also be induced by treatment of wild-type roots with KAR. Indeed, KAR1 induced a small increase of ACS7 transcript accumulation after 2 h and a significant increase after 6 h of treatment in a KAR/KL receptor-dependent manner (SI Appendix, Fig. S22 A and B). However, we were unable to detect increased ethylene release in response to KAR treatment of wild-type roots (SI Appendix, Fig. S22C), possibly because ethylene release may be triggered in the wild type only in a small number of cells, thus remaining under the detection limit. We have previously observed that treatment of wild-type roots by KAR1 leads to a mild inhibition of PR length and a resulting increase in PER density (14). We examined whether this effect is dependent on ethylene signaling by cotreating wild-type plants with silver nitrate and with KAR1. KAR1 treatment alone led to inhibition of PR growth and an increase of PER density but this was not observed when plants were cotreated with silver nitrate (SI Appendix, Fig. S21D). Furthermore, an ein2a ein2b ethylene perception mutant (46) did not respond to KAR1 treatment with reduced PR growth (SI Appendix, Fig. S22E), corroborating that ethylene signaling is required for the response. Together, these data indicate that increased ethylene production is a bona fide downstream response to KAR perception and demonstrate that ethylene signaling is required for the effect of KAR1 on L. japonicus PR growth.
A. thaliana ACS7 Is Required for KAR-Induced Root Hair Growth.
Ectopic expression of ACS7 causes increased root hair growth in Arabidopsis (39). Furthermore, we previously reported that Arabidopsis smax1 smxl2 double mutants display an increased root hair length and that KAR induces root hair elongation in the wild type (12). smax1 smxl2 double mutants also had slightly shorter PRs than the wild type, but the phenotype was very mild as compared with L. japonicus (SI Appendix, Fig. S23 A and B). Nevertheless, we examined whether the negative impact of SMAX1 SMXL2 on ACS7 transcript accumulation is conserved in Arabidopsis roots. Along with DLK2, ACS7 transcripts accumulated to higher levels in smax1 smxl2 mutants as compared with the wild type, although this was only significant at the 90% level (SI Appendix, Fig. S23C). However, Arabidopsis acs7-1 as well as the ein2-1 mutants failed to respond with increased root hair elongation to KAR treatment (SI Appendix, Fig. S23D). Thus, we demonstrate that ACS7-mediated ethylene production as well as ethylene perception are required for root hair elongation downstream of KAR/KL signaling in A. thaliana, and that the KAR/KL ethylene signaling module is conserved between L. japonicus and A. thaliana, despite some observed differences in phenotypic output.
Discussion
A hallmark of most plant hormone signaling pathways is the proteasomal degradation of negative regulators after hormone perception to release downstream responses from repression. The mechanistic functions and regulatory roles of SMAX1, the negative regulator of KAR/KL signaling (22), are largely elusive. Here we demonstrate that L. japonicus smax1 mutant roots overaccumulate ACS7 transcripts, as well as ethylene, causing typical ethylene-related phenotypes such as inhibition of primary root (cell) elongation and increased root hair length (47–49), and these phenotypes are rescued by treatment with inhibitors of ethylene biosynthesis and perception. We confirm that transcriptional activation of ACS7 is a downstream response of KAR/KL signaling because the gene is induced by KAR1 in a KAI2a KAI2b- and MAX2-dependent manner. Therefore, we propose that in the L. japonicus wild type, the KAI2–MAX2–SMAX1 module regulates primary root and root hair development by controlling the homeostasis of ACS7 gene expression and of ethylene biosynthesis (Fig. 5). In nature, this signaling module thus likely plays an important role in supporting the establishment of small seedlings, which quickly need to reach and maintain access to water. This is also the case in Arabidopsis, in which a short-root hair phenotype of max2 mutants, which we recently showed to be a consequence of disruption of KAR/KL signaling (12), was rescued by ACC treatment (50). Here we demonstrate that KAR-induced root hair elongation in Arabidopsis depends on ACS7 and does not seem to rely on additional ACS isoforms. Given the evidence presented here, this sole dependence on ACS7 (and possibly ACS7-like) is likely conserved in L. japonicus. Ethylene signaling downstream of KAR signaling may play a role also in seed germination since an increase in ACS activity and ethylene release from germinating Brassica oleracea seeds upon exogenous KAR1 treatment was recently reported (51).
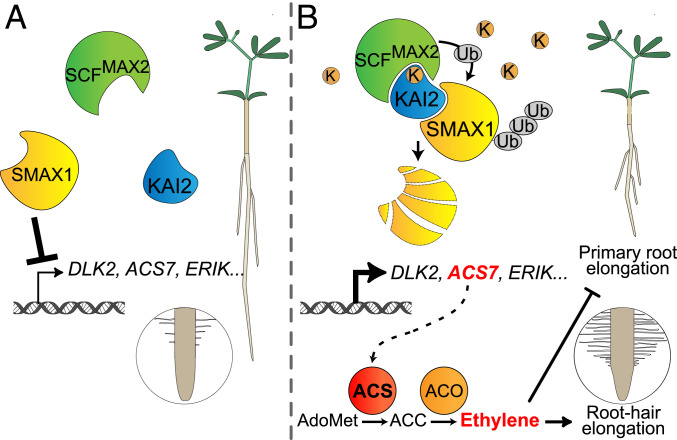
Fig. 5.
Schematic model of the ethylene-mediated influence of KAR/KL signaling on root and root hair development. (A) When KAR/KL levels are low, SMAX1 represses the transcription of KAR/KL response genes such as DLK2, ERIK, and ACS7 through an unknown mechanism. Primary root growth is not repressed, and root hairs develop normally. (B) At high KAR/KL levels, most KAI2 proteins bind KAR/KL, likely undergo a conformational change, and interact with MAX2 and SMAX1, thereby triggering ubiquitylation and degradation of SMAX1 and releasing transcriptional repression of KAR/KL response genes. Among them, the ACC synthase gene ACS7 catalyzes the biosynthesis of ethylene, causing repression of primary root elongation and promotion of root hair growth.
Interestingly, untreated L. japonicus kai2a kai2b double mutants do not display obvious root phenotypes, although by extrapolation from the smax1 root phenotype, increased primary root growth and shorter root hairs may be expected. Nevertheless, L. japonicus seedlings respond to KAR1 treatment with a suppression of primary root elongation in an ethylene signaling-dependent manner (14) (this study). Perhaps the biosynthesis of endogenous KL or yet unknown properties of the KL receptor complex change dynamically in response to environmental conditions, and the condition, which would reveal differences between the wild type and kai2a kai2b, is not met in our experimental system.
In Arabidopsis, under standard growth conditions, kai2 and smax1 smxl2 root and root hair phenotypes manifest in an opposite fashion: kai2 mutants display clear phenotypic differences from the wild type, with longer primary roots and shorter root hairs, while smxl2 and smax1 smxl2 double mutants show only a small difference in primary root growth (this study) and also a slighter increase in root hair length as compared with L. japonicus (12). It is possible that Arabidopsis and L. japonicus diverge in their ethylene sensitivity, amount of ethylene produced, molecular wiring of ethylene signaling, or other physiological optima. Any of these may cause differences in phenotypic consequences of mutations in KAR/KL signaling genes. In this context, it is worth highlighting that both species also respond differently to mutation of the ethylene signaling gene EIN2 with respect to lateral root formation. Arabidopsis ein2 mutant seedlings develop increased numbers of lateral roots (52), while L. japonicus ein2a ein2b double-mutant seedlings rarely develop any postembryonic roots (SI Appendix, Fig. S20D). Our study emphasizes the need to study and understand signaling modules across diverse plant species to 1) grasp the full phenotypic space controlled by these modules and 2) allow agricultural application of molecular knowledge to a diversity of crops.
We demonstrate that L. japonicus SMAX1 and SMXL8 are degraded in N. benthamiana leaves in the presence of MAX2 and their cognate α/β-fold hydrolase receptors KAI2a/KAI2b or D14, respectively. Surprisingly, the presence/absence of nuclear SMAX1- and SMXL8-GFP was observed without adding artificial ligands such as KAR or the synthetic strigolactone analog rac-GR24. This contrasts with other recently published studies performed with Arabidopsis SMAX1 and SMXL2, in which degradation was only observed in the presence of ligands (30, 31). However, the experimental setting in these studies differed from ours, as they expressed SMAX1 and SMXL2 in the homologous Arabidopsis background as well as in Arabidopsis protoplasts [SMXL2 (31)] or N. benthamiana leaves without coexpressing Arabidopsis KAI2 and MAX2 [SMAX1 (30)]. Possibly, in our study, the L. japonicus α/β-fold hydrolase receptors respond to a ligand present in N. benthamiana leaves. Alternatively, crowding of highly expressed proteins in the nuclei allows protein complex formation and SMAX1/SMXL8 degradation in the absence of a ligand. Whatever the case, the result of the assay suggests specific interaction surfaces between the repressors and their cognate receptors, since SMAX1- and SMXL8-GFP disappeared only in the presence of KAI2a/b and D14, respectively, while SMXL3, 4, and 9 remained stable in the presence of all receptors. The specificity of the assay is further supported by the accumulation of SMAX1- and SMXL8-GFP in the absence of L. japonicus MAX2 or, when the P-loop motifs GKTAF (SMAX1) or GKTTI (SMXL8) are deleted, like in rice d53 (SI Appendix, Fig. S3 B and C) (20, 21, 29).
We generated genome-wide transcriptional profiles of KAR/KL signaling mutant roots, which will be a useful resource to mine for pathways and functions downstream of KAR/KL signaling. Compared with other hormone signaling pathways such as auxin or jasmonate, the amplitude of transcriptional responses to treatment with strigolactone or KAR has been reported to be low (3, 14, 53, 54). This is also reflected in transcriptional differences between L. japonicus KAR/KL signaling mutants and the wild type (Dataset S3) and may be due to transcriptional changes in only a limited number of cells. Nevertheless, in addition to revealing the link between KAR/KL and ethylene biosynthesis (ACS7 expression), we identified additional early KAR response genes (ERIK and a Serotonin Receptor-like gene) in L. japonicus (Fig. 4) complementing the well-established marker gene DLK2 (5, 14). Like DLK2, they are characterized by a higher expression in smax1 mutants as well as a KAR/KL receptor-dependent induction by a 2-h KAR treatment. It will be interesting to determine whether the AP2 transcription factor ERIK regulates secondary target genes of KAR/KL signaling and whether it regulates any important KAR/KL-related phenotype(s).
To understand the KAR/KL signaling pathway and the regulation of its downstream targets, it is critical to identify the transcription factor(s), which is directly inhibited by SMAX1 and which regulates primary KAR/KL response genes. In rice, one target of D53 in strigolactone signaling, the SQUAMOSA PROMOTER BINDING PROTEIN-LIKE family transcription factor IDEAL PLANT ARCHITECTURE 1 (IPA1), has been identified. D53 interacts with IPA1 and represses its transcriptional activity (55). However, one early strigolactone response gene encoding CYTOKININ OXIDASE/DEHYDROGENASE 9 in rice is induced by rac-GR24 in an IPA1-independent manner. This indicates that D53 likely targets more than one transcription factor (56). Given the diversity of phenotypes regulated by KAR/KL signaling (7), it is possible that also SMAX1 inhibits more than one target protein.
Supplementary Material
Acknowledgments
We are grateful to Marcos Castellanos Uribe for making a personal effort to rapidly send us A. thaliana acs7-1 mutant seeds from the Nottingham Arabidopsis Stock Centre during the coronavirus disease 2019 lockdown. We thank Christine Wurmser at NGS@TUM for high-quality Illumina sequencing, Brigitte Poppenberger for A. thaliana ein2-1 seeds, Dugald Reid and Jens Stougaard for L. japonicus ein2a ein2b double-mutant seeds, and the LotusBase team for providing L. japonicus LORE1 insertion lines. The study was supported by the Emmy Noether Program of the Deutsche Forschungsgemeinschaft (Grant GU1423/1-1 to C.G.) and DAAD (German Academic Exchange Service) Doctoral Student Fellowship 57381412 (to K.V.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2006111117/-/DCSupplemental.
Data Availability.
The RNA-seq data reported in this manuscript have been deposited in the National Center for Biotechnology Information database, https://dataview.ncbi.nlm.nih.gov/object/PRJNA591291?reviewer=vhn4av2bafiiutcnb53pd72ego (BioProject no. PRJNA591291). All study data are included in the article and SI Appendix.
References
- 1.Yao J., Waters M. T., Perception of karrikins by plants: A continuing enigma. J. Exp. Bot. 71, 1774–1781 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flematti G. R., Ghisalberti E. L., Dixon K. W., Trengove R. D., A compound from smoke that promotes seed germination. Science 305, 977 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Nelson D. C. et al., Karrikins enhance light responses during germination and seedling development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 107, 7095–7100 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson D. C. et al., F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 108, 8897–8902 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waters M. T. et al., Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139, 1285–1295 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Hamiaux C. et al., DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol. 22, 2032–2036 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Waters M. T., Gutjahr C., Bennett T., Nelson D. C., Strigolactone signaling and evolution. Annu. Rev. Plant Biol. 68, 291–322 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Delaux P. M. et al., Origin of strigolactones in the green lineage. New Phytol. 195, 857–871 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Gutjahr C. et al., Rice perception of symbiotic arbuscular mycorrhizal fungi requires the karrikin receptor complex. Science 350, 1521–1524 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Conn C. E., Nelson D. C., Evidence that KARRIKIN-INSENSITIVE2 (KAI2) receptors may perceive an unknown signal that is not karrikin or strigolactone. Front. Plant Sci. 6, 1219 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Y. K., Flematti G. R., Smith S. M., Waters M. T., Reporter gene-facilitated detection of compounds in Arabidopsis leaf extracts that activate the karrikin signaling pathway. Front. Plant Sci. 7, 1799 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Villaécija-Aguilar J. A. et al., SMAX1/SMXL2 regulate root and root hair development downstream of KAI2-mediated signalling in Arabidopsis. PLoS Genet. 15, e1008327 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu G. et al., Strigolactones play an important role in shaping exodermal morphology via a KAI2-dependent pathway. iScience 17, 144–154 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carbonnel S., et al. , Duplicated KAI2 receptors with divergent ligand-binding specificities control distinct developmental traits in Lotus japonicus. bioRxiv:10.1101/754937 (3 September 2019). [Google Scholar]
- 15.Sun Y. K. et al., Divergent receptor proteins confer responses to different karrikins in two ephemeral weeds. Nat. Commun. 11, 1264 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conn C. E. et al., PLANT EVOLUTION. Convergent evolution of strigolactone perception enabled host detection in parasitic plants. Science 349, 540–543 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Toh S. et al., Structure-function analysis identifies highly sensitive strigolactone receptors in Striga. Science 350, 203–207 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Tsuchiya Y. et al., PARASITIC PLANTS. Probing strigolactone receptors in Striga hermonthica with fluorescence. Science 349, 864–868 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Burger M. et al., Structural basis of karrikin and non-natural strigolactone perception in Physcomitrella patens. Cell Rep. 26, 855–865.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang L. et al., DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504, 401–405 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou F. et al., D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504, 406–410 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stanga J. P., Smith S. M., Briggs W. R., Nelson D. C., SUPPRESSOR OF MORE AXILLARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis. Plant Physiol. 163, 318–330 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soundappan I. et al., SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell 27, 3143–3159 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanga J. P., Morffy N., Nelson D. C., Functional redundancy in the control of seedling growth by the karrikin signaling pathway. Planta 243, 1397–1406 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L. et al., Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53-like SMXL repressor proteins for ubiquitination and degradation. Plant Cell 27, 3128–3142 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wallner E. S. et al., Strigolactone- and karrikin-independent SMXL proteins are central regulators of phloem formation. Curr. Biol. 27, 1241–1247 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker C. H., Siu-Ting K., Taylor A., O’Connell M. J., Bennett T., Strigolactone synthesis is ancestral in land plants, but canonical strigolactone signalling is a flowering plant innovation. BMC Biol. 17, 70 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma H. et al., A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex. Sci. Adv. 3, e1601217 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang Y., Ward S., Li P., Bennett T., Leyser O., SMAX1-LIKE7 signals from the nucleus to regulate shoot development in Arabidopsis via partially EAR motif-independent mechanisms. Plant Cell 28, 1581–1601 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khosla A., et al. , Structure-function analysis of SMAX1 reveals domains that mediate its karrikin-induced proteolysis and interaction with the receptor KAI2. Plant Cell 32, 2639–2659 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L. et al., Strigolactone and karrikin signaling pathways elicit ubiquitination and proteolysis of SMXL2 to regulate hypocotyl elongation in Arabidopsis thaliana. Plant Cell 32, 2251–2270 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wallner E. S., López-Salmerón V., Greb T., Strigolactone versus gibberellin signaling: Reemerging concepts? Planta 243, 1339–1350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Machin D. C., Hamon-Josse M., Bennett T., Fellowship of the rings: A saga of strigolactones and other small signals. New Phytol. 225, 621–636 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Małolepszy A. et al., The LORE1 insertion mutant resource. Plant J. 88, 306–317 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Kennedy P. G., Hausmann N. J., Wenk E. H., Dawson T. E., The importance of seed reserves for seedling performance: An integrated approach using morphological, physiological, and stable isotope techniques. Oecologia 141, 547–554 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Müller J. et al., Iron-dependent callose deposition adjusts root meristem maintenance to phosphate availability. Dev. Cell 33, 216–230 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Vandenbussche F., Van Der Straeten D., “The role of ethylene in plant growth and development” in The Plant Hormone Ethylene, McManus M. T., Ed. (Wiley-Blackwell, 2012), Vol. vol. 44, pp. 219–241. [Google Scholar]
- 38.Chae H. S., Kieber J. J., Eto Brute? Role of ACS turnover in regulating ethylene biosynthesis. Trends Plant Sci. 10, 291–296 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Sun G. et al., N-terminus-mediated degradation of ACS7 is negatively regulated by senescence signaling to allow optimal ethylene production during leaf development in Arabidopsis. Front. Plant Sci. 8, 2066 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiong L., Xiao D., Xu X., Guo Z., Wang N. N., The non-catalytic N-terminal domain of ACS7 is involved in the post-translational regulation of this gene in Arabidopsis. J. Exp. Bot. 65, 4397–4408 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Yu Y. B., Yang S. F., Auxin-induced ethylene production and its inhibition by aminoethoxyvinylglycine and colbalt ion. Plant Physiol. 64, 1074–1077 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McDaniel B. K., Binder B. M., Ethylene receptor 1 (ETR1) is sufficient and has the predominant role in mediating inhibition of ethylene responses by silver in Arabidopsis thaliana. J. Biol. Chem. 287, 26094–26103 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guzmán P., Ecker J. R., Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2, 513–523 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoon G. M., Kieber J. J., 1-Aminocyclopropane-1-carboxylic acid as a signalling molecule in plants. AoB Plants 5, plt017 (2013). [Google Scholar]
- 45.Van de Poel B., Van Der Straeten D., 1-Aminocyclopropane-1-carboxylic acid (ACC) in plants: More than just the precursor of ethylene!. Front. Plant Sci. 5, 640 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reid D. et al., Dynamics of ethylene production in response to compatible Nod factor. Plant Physiol. 176, 1764–1772 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Růzicka K. et al., Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19, 2197–2212 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feng Y. et al., Ethylene promotes root hair growth through coordinated EIN3/EIL1 and RHD6/RSL1 activity in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 114, 13834–13839 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harkey A. F. et al., Identification of transcriptional and receptor networks that control root responses to ethylene. Plant Physiol. 176, 2095–2118 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kapulnik Y. et al., Strigolactones interact with ethylene and auxin in regulating root-hair elongation in Arabidopsis. J. Exp. Bot. 62, 2915–2924 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Sami A., Riaz M. W., Zhou X., Zhu Z., Zhou K., Alleviating dormancy in Brassica oleracea seeds using NO and KAR1 with ethylene biosynthetic pathway, ROS and antioxidant enzymes modifications. BMC Plant Biol. 19, 577 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Negi S., Ivanchenko M. G., Muday G. K., Ethylene regulates lateral root formation and auxin transport in Arabidopsis thaliana. Plant J. 55, 175–187 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mashiguchi K. et al., Feedback-regulation of strigolactone biosynthetic genes and strigolactone-regulated genes in Arabidopsis. Biosci. Biotechnol. Biochem. 73, 2460–2465 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Mayzlish-Gati E. et al., Strigolactones are positive regulators of light-harvesting genes in tomato. J. Exp. Bot. 61, 3129–3136 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song X. et al., IPA1 functions as a downstream transcription factor repressed by D53 in strigolactone signaling in rice. Cell Res. 27, 1128–1141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Duan J. et al., Strigolactone promotes cytokinin degradation through transcriptional activation of CYTOKININ OXIDASE/DEHYDROGENASE 9 in rice. Proc. Natl. Acad. Sci. U.S.A. 116, 14319–14324 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data reported in this manuscript have been deposited in the National Center for Biotechnology Information database, https://dataview.ncbi.nlm.nih.gov/object/PRJNA591291?reviewer=vhn4av2bafiiutcnb53pd72ego (BioProject no. PRJNA591291). All study data are included in the article and SI Appendix.