Abstract
Adenosine triphosphate (ATP) is produced at the early stage of seed germination and provides the energy for metabolism. The source of ATP in seeds may be Perl’s pathway, but this has not yet been confirmed. In this study, using germinating seeds of poplar as the experimental materials, the transcript levels of genes related to Perl’s pathway were determined by real-time PCR. The activities of enzymes in Perl’s pathway were also determined. The results were verified by comparison with RNA-Seq and metabolomics data. The results showed that there were high transcript levels of some genes encoding malate dehydrogenase (MDH), phosphoenolpyruvate carboxykinase (PEPCK), pyruvate decarboxylase (PDC), alcohol dehydrogenase (ADH), and pyruvate kinase (PK) at the early stage of germination (0.75 h). The enzymes MDH, PEPCK, PK, PDC, and ADH showed peaks in activity at around 0.75 h and 6 h during germination. The oxaloacetate concentration was high in poplar seeds at the early stage of germination. This study provides experimental data showing that Perl’s pathway participates in supplying energy during the early stages of poplar seed germination, and lays the foundation for further studies on the complex metabolic processes that function during seed germination.
Electronic supplementary material
The online version of this article (10.1007/s13205-020-02413-z) contains supplementary material, which is available to authorized users.
Keywords: Poplar seed germination, ATP content, qRT-PCR, RNA-Seq, Enzyme activity, Metabolite change
Introduction
Seed germination is a complex process involving a series of changes at morphological, physiological, and molecular levels (Bewley 1997; Finch‐Savage and Leubner‐Metzger 2006; Han and Yang 2015; Holdsworth et al. 2008; Koornneef et al. 2002; Steinbrecher and Leubner-Metzger 2016; Weitbrecht et al. 2011). These changes start at imbibition and continue until the hypocotyl breaks through the seed coat (Nonogaki et al. 2010; Rajjou et al. 2012). According to the change in fresh weight after soaking seeds, seed germination can be divided into three phases: the rapid water absorption period, the slow water absorption period, and the hypocotyl elongation period (Bewley 1997; Koornneef et al. 2002; Weitbrecht et al. 2011). The seed germination stage is the starting point of the life cycle, and it requires energy to meet its metabolic demands (Koornneef et al. 2002; Steinbrecher and Leubner-Metzger 2016).
A few minutes after seed germination begins, there is a sharp increase in oxygen uptake and carbon dioxide release, and the adenosine triphosphate (ATP) content begins to increase rapidly (Benamar et al. 2008; Bewley 1997; Botha et al. 1992; Spoelstra et al. 2002). Analyses of the Arabidopsis transcriptome during seed germination have suggested that glycolysis, fermentation, the tricarboxylic acid (TCA) cycle, and the oxidized pentose phosphate pathway (OPPP) are activated during germination (Weitbrecht et al. 2011). Other studies have revealed that many enzymes involved in major metabolic pathways play important roles in germinating seeds (Fu et al. 2005; Gallardo et al. 2001; Muller et al. 2009; Smiri et al. 2009; Wakao et al. 2008). However, mitochondria in dried seeds require repair and differentiation before they can produce ATP through the oxidative phosphorylation pathway. The process of mitochondrial repair and differentiation requires energy, but ATP is scarce in dry seeds. Therefore, there must be another source of ATP before mitochondria are repaired. Perl (1986) hypothesized that pyruvate kinase produces pyruvate from phosphoenolpyruvate, thereby producing ATP and that oxaloacetate and malate are substrates for phosphoenolpyruvate production. It was proposed that NADH provides the reducing power required for this process and that the NADH levels are balanced by the activities of nicotinamide adenine dinucleotide dehydrogenase (NADH-DH), malate dehydrogenase, and alcohol dehydrogenase (Perl 1986). This hypothesis has been explored in subsequent studies, but there is still little experimental evidence to support it.
Poplar is a model woody plant. Because it is easily propagated asexually, there are relatively few studies on the molecular mechanism of sexual reproduction in this species. However, several recent studies have explored the molecular mechanism of seed germination in poplar. Zhang et al. (2015) conducted a proteomics analysis, which showed that the germination process of poplar seeds is significantly correlated with energy dependence, protein synthesis and degradation, and cell defense- and rescue-related pathways (Zhang et al. 2015). Qu et al. (2019b) monitored changes in gene transcription and metabolism during poplar seed germination and identified some genes that were closely related to changes in primary metabolism through a targeted network correlation analysis (Qu et al. 2019b). Qu et al. (2019e) used a weighted means method to detect changes in primary metabolism and identified which transcription factors were most relevant during parts of the germination period (Qu et al. 2019e). In another study, metabolomic methods revealed the metabolites showing significant changes in abundance at different stages of seed germination (Qu et al. 2019c). Zhang et al. (2019) performed an integrative transcriptome analysis of Populus euphratica and Populus pruinosa at three seed germination phases and identified specifically expressed genes at each phase. In addition, they found that the flavonoid and brassinosteroid pathways were significantly enriched under salinity stress (Zhang et al. 2019). However, the source of energy to fuel these metabolic processes is unknown.
In this study, we focused on changes in energy production during the early stage of poplar seed germination. The expression patterns of genes related to Perl’s pathway were detected by quantitative real-time PCR (qRT-PCR), and changes in the activities of enzymes and abundance of metabolites in Perl’s pathway were detected. This study provides evidence that Perl’s pathway is the main source of energy at the early stage of poplar seed germination, and provides information about the molecular mechanisms of poplar seed germination.
Materials and methods
Plant materials and treatments
We selected seeds produced in the same year from superior poplar trees (Populus × xiaohei T. S. Hwang et Liang) growing in the greenhouse of Northeast Forestry University (Harbin, Heilongjiang, China). The seeds were placed in a Petri dish with filter paper and kept at a constant temperature of 25 °C in the dark. After removing the surface moisture using absorbent paper, 50 seeds were taken out to measure fresh weight. According to fresh weight and morphological changes after water absorption, The poplar seed germination process was divided into a rapid water absorption stage, a slow water absorption stage, and a hypocotyl elongation stage. In this study, we selected 0.75 h as the time representative of the rapid water absorption stage, 6 h as the time representative of the slow water absorption stage, and 24 h as the time representative of the hypocotyl elongation stage. Seeds at 0 h of germination served as the control. After collection, samples were quickly wrapped in tin foil, frozen in liquid nitrogen, and then stored at − 80 °C until analysis.
RNA isolation and qRT-PCR analysis
Frozen tissue samples (0.1 g) were ground into a fine powder in liquid nitrogen and total RNA was extracted using a total RNA isolation kit (Tiangen, Beijing, China). Extracts were treated with DNase I to remove residual genomic DNA. The cDNAs were synthesized using the PrimeScript™ first-strand cDNA synthesis kit (Takara, Dalian, China) according to the manufacturer’s instructions, with 1 μg total RNA as the template. The qPCR analyses were performed on an ABI7500 RT-PCR system (Applied Biosystems, Carlsbad, CA, USA). The gene-specific primers were designed using Primer Blast online tools based on nucleotide sequences of poplar (taxid:3689) at the NCBI (https://www.ncbi.nlm.nih.gov/). There were three biological replicates for each cDNA. The qRT-PCR reaction conditions were as follows: 10 min activation at 95 °C, followed by 40 cycles of denaturation at 95 °C for 15 s, and then extension at 72 °C for 35 s. To ensure that only single products were generated, a melting curve was acquired for each primer at the end of each run. The relative gene expression levels in different samples were calculated using the comparative 2−ΔΔCT method (Livak and Schmittgen 2001). pnUBQ7 was used as the internal control to compare gene expression among seeds at different developmental stages (Qu et al. 2019d). The relative expression levels of target genes were compared with that at 0 h (control).
Enzyme analyses
We used purchased kits to detect the activities of enzymes (alcohol dehydrogenase, ADH; pyruvate kinase, PK; pyruvate decarboxylase, PDC; phosphoenolpyruvate carboxykinase, PEPCK; and nicotinamide adenine dinucleotide-malate dehydrogenase, NAD-MDH) (Comin, Suzhou, China). The ATP content was detected using an ATP fluorescence kit (Beyotime, Beijing, China). All samples were ground on ice before testing. The assays were performed according to the manufacturer’s instructions. Transpiration rates were measured using a LI-6400 system (LI-COR, Lincoln, NE, USA) according to the manufacturer’s instructions.
RNA-Seq and metabolome data collection and analysis
The RNA-seq and metabolome raw data were collected from the same poplar species, with the same treatments, sampling times, and processing methods as those used in our study, at the State Key Laboratory of Tree Genetics and Breeding (Northeast Forestry University, Harbin, China). These data were generated using the RNA-seq technique on the Illumina HiSeq 2500 platform (Illumina, San Diego, CA, USA) from seeds of poplar at different stages of germination. Each stage had three replicates. All clean reads were mapped to the poplar genome, which was downloaded from the Phytozome website (https://phytozome.jgi.doe.gov/pz/portal.html), and differentially expressed genes (DEGs) were identified with NOISeq with the following thresholds: fold change ≥ 2 and p ≥ 0.8 (Tarazona et al. 2015). Gene expression levels were normalized using the FPKM method, and other details are described in Qu et al. (2019b). Metabolomic analysis was performed using 50 mg poplar seeds for each of the different germination stages. The experimental process was as follows: LC–MS/MS analyses were performed using a UHPLC system (1290, Agilent Technologies, Palo Alto, CA, USA) equipped with a UPLC BEH amide column (1.7 µm 2.1 × 100 mm, Waters, Milford, MA, USA) coupled to a TripleTOF 6600 mass spectrometer (Q-TOF, AB Sciex, Foster City, CA, USA). The mobile phase consisted of 25 mM NH4OAc and 25 mM NH4OH in water (pH = 9.75) (A) and acetonitrile (B). The elution gradient was as follows: 0 min, 85% B; 2 min, 75% B; 9 min, 0% B; 14 min, 0% B; 15 min, 85% B; 20 min, 85% B. The flow rate was 0.3 mL min−1 and the injection volume was 2 µL. The TripleTOF mass spectrometer was used for its ability to acquire MS/MS spectra on an information-dependent basis (IDA) during LC/MS analyses. In this mode, the acquisition software (Analyst TF 1.7, AB Sciex) continuously evaluates the full-scan survey MS data as they are collected, and the acquisition of MS/MS spectra is triggered depending on preselected criteria. In each cycle, six precursor ions with intensity greater than 100 were chosen for fragmentation at collision energy (CE) of 35 V (15 MS/MS events with product ion accumulation time of 50 ms each) (Ivanisevic et al. 2015). Analyses of the metabolomic data revealed 3790 features. After eliminating outliers identified by the interquartile range method, missing values in the metabolic raw data were replaced by numbers corresponding to half of the minimum value. The overall normalization method was used in data analysis (Dunn et al. 2011). Metabolites showing significant differences in abundance among samples were detected using two screening steps: 1. Metabolites with first principal component of variable importance in the projection (VIP) values exceeding 1.0 were selected as those showing changes in abundance; 2. The remaining variables were assessed by Student’s t-test (p > 0.05) (Saccenti et al. 2014). The Mapman tool was used to assign transcript pathway categories and to annotate genes and metabolites (Thimm et al. 2004). All datasets generated or analyzed in this study are available from the corresponding author on reasonable request.
Statistical analyses
All qRT-PCR and enzyme activity data were subjected to one-way analysis of variance. Significant differences among samples were detected by Duncan’s test using SPSS v. 20.0 (SPSS Inc., Chicago, IL, USA). The level of significance was p < 0.05.
Results
Physiological changes during seed germination
The changes in fresh weight during different stages of seed germination are shown in Fig. 1. From the beginning of imbibition, the fresh weight of the seeds began to increase. The increase in fresh weight continued until about 1 h after the start of imbibition, and then the seed weight entered a relatively stable phase. These results show that poplar seeds absorb water rapidly up to 1 h of germination, and then absorb water more slowly after 1 h of germination. Referring to a previous study (Qu et al. 2019b), we selected seeds at 0.75 h after the start of imbibition as being representative of the rapid water absorption period (early germination period) and those at 6 h after the start of imbibition as being representative of the slow water absorption period (germination period).
Fig. 1.
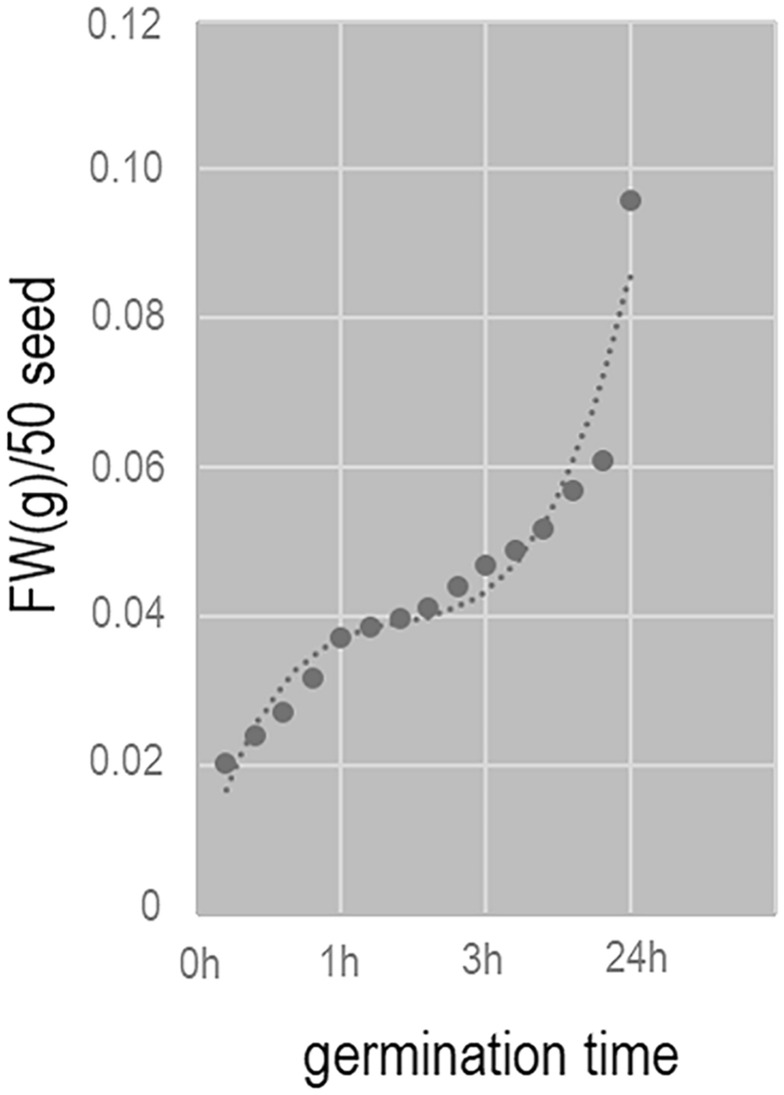
Changes in fresh weight of poplar seeds during germination. FW: Fresh weight
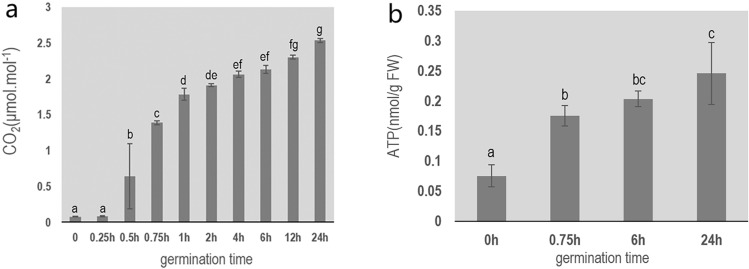
The changes in respiration rate during poplar seed germination are shown in Fig. 2a. The respiration rate began to increase at 0.5 h, and reached 1.387 µmol mol−1 at 0.75 h, 2.131 µmol mol−1 at 6 h, and 2.536 µmol mol−1 at 24 h of germination (around the time of hypocotyl extension, indicative of the end of seed germination). These results show that ATP synthesis began to recover at 0.75 h of seed germination.
Fig. 2.
Changes in ATP content and carbon dioxide release rate during the early stage of poplar seed germination. a Carbon dioxide release rate during poplar seed germination. b Changes in ATP content during poplar seed germination
Some ATP was detected at the early stage of poplar seed germination, but the ATP content increased significantly at 0.75 h and continued to increase after 6 h (Fig. 2b), consistent with the changes in the respiration rate.
qRT-PCR analyses
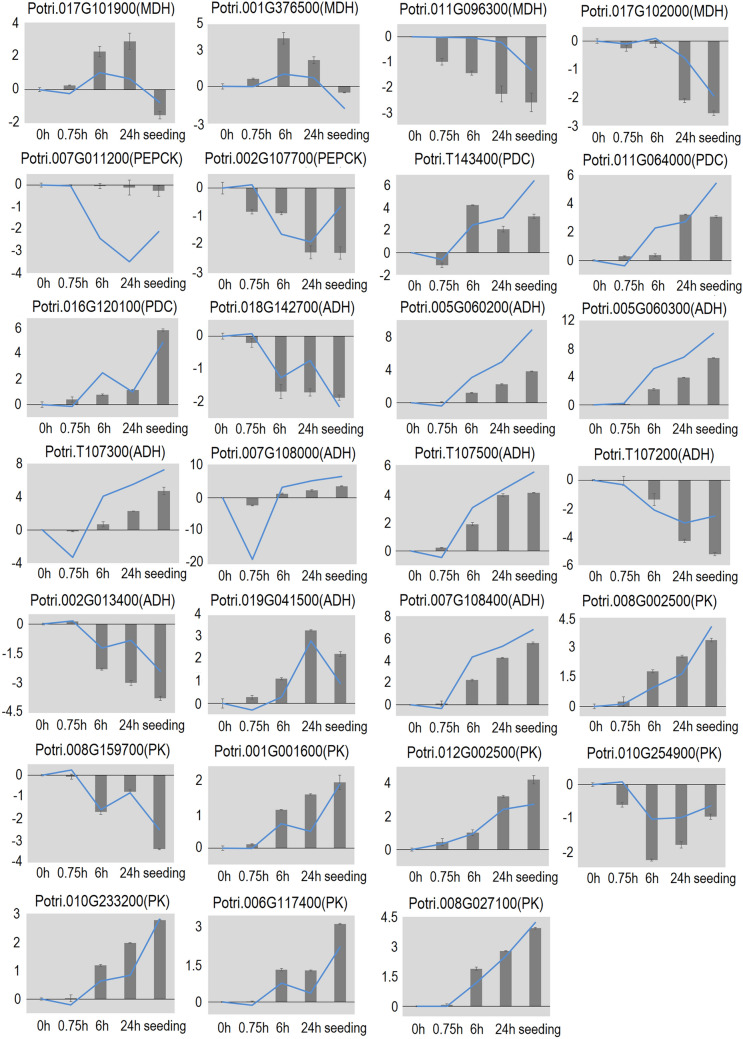
According to annotations assigned using Mapman tools, we identified four genes encoding MDH, two genes encoding PEPCK, three genes encoding PDC, ten genes encoding ADH, and eight genes encoding PK. The results of the qRT-PCR analyses indicated that the transcript levels of potri.017G101900 and potri.001G376500 (encoding MDH) were relatively low between 0 and 0.75 h, and reached higher levels at around 6 h. The other two genes encoding MDH, potri.011G096300 and potri.017G102000, showed relatively stable transcript levels at the early stage of seed germination (0.75 h), and lower transcript levels after 24 h of germination.
The two PEPCK genes showed similar trends in expression. Their transcript levels were higher between 0 and 0.75 h and decreased to relatively low levels by 6 h and 24 h of germination. The three genes encoding PDC showed relatively stable expression at 0–0.75 h and increased transcript levels at 6 h of germination. Among the 10 genes encoding ADH, potri.018G142700, potri.T107200, and potri.002G013400 showed higher transcript levels at 0.75 h, while potri.005G060200, potri.005G060300, and potri.T107300 showed higher transcript levels at 6 h and 24 h of germination. Among the eight genes encoding PK, potri.010G254900 and potri.008G159700 had higher transcript levels before 0.75 h, while the other six genes showed significant increases in transcript levels at 6 h of germination. These results suggest that different members of gene families in Perl’s pathway are regulated differently and have different functions.
The FPKM values provide an estimate of transcript abundance. The FPKM values were highly consistent with the qRT-PCR results, thus confirming the credibility of the experimental data (Fig. 3).
Fig. 3.
qRT-PCR analyses of transcript levels of 27 genes related to Perl’s pathway during seed germination. Columns represent relative gene expression levels. Data are mean ± SD (n = 3). Broken lines represent RNA-Seq results
Changes in enzyme activity
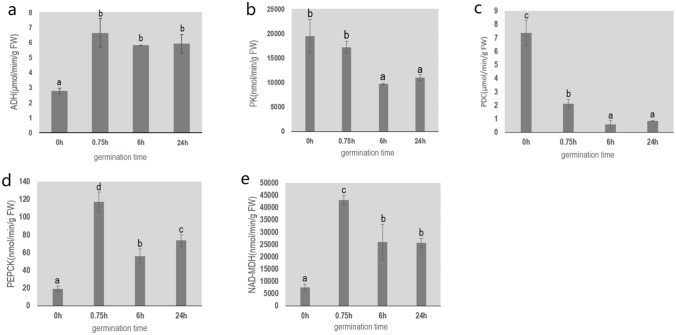
The changes in activities of enzymes in Perl’s pathway during germination are shown in Fig. 4. The activity of MDH at 0.75 h was 43,081 nmol min−1 g−1, approximately double that at 6 h. The activity of PEPCK was also higher at 0.75 h (117 nmol min−1 g−1) than at 6 h (56 nmol min−1 g−1). The activity of PK at 0–0.75 h (approx. 17,000–19,000 nmol min−1 g−1) was higher than that at 6 h (about 10,000 nmol min−1 g−1). Similar to the trend in PK activity, PDC activity was also higher at the early stage of seed germination but showed a significant decrease at 0.75 h to about 2.12 µmol min−1 g−1, and even lower activity between 6 and 24 h of germination. At the dry seed stage, ADH activity was about 2.77 µmol min−1 g−1 (if enzymes are active in dry seeds), and then it increased and remained relatively stable at about 6 µmol min−1 g−1 from 0.75 h to 24 h of germination. In general, these enzymes were active at about 0.75 h of seed germination, and some enzymes had higher activity during this period than at the later stage of seed germination.
Fig. 4.
Changes in activities of enzymes related to Perl’s pathway. a–e ADH (a), PK (b), PDC (c) PEPCK (d), NAD-MDH (e). In each plot, different lowercase letters indicate significant differences
Changes in abundance of metabolites during germination
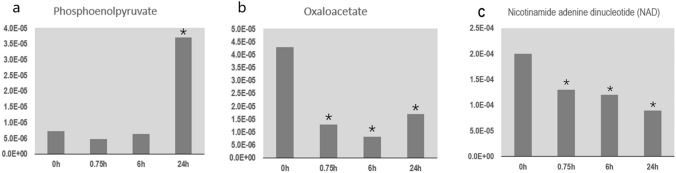
We detected metabolites related to Perl’s pathway at different stages of seed germination (Fig. 5). The phosphoenolpyruvate concentration decreased slightly at 0.75 h, but this decrease was not significant, and then it increased significantly after 24 h. The oxaloacetate concentration was high in dry seeds but decreased significantly by 0.75 h, reached its lowest level around 6 h of germination, and then started to increase. The NAD concentration was high in dry seeds and significantly decreased by 0.75 h of germination.
Fig. 5.
Changes in contents of metabolites related to Perl’s pathway during poplar seed germination: Phosphoenolpyruvate (a), Oxaloacetate (b), NAD (c). Columns represent relative abundance of metabolites. In each plot, asterisks indicate a significant difference
Discussion
In this study, we analyzed poplar seeds at different stages of germination to determine which pathway(s) provided energy for metabolism. The starting point for these analyses was the pathway proposed by Perl (1986). The sources of energy to fuel early seed germination were explored by qRT-PCR and enzyme activity analyses. The results were validated by comparison with transcriptome and metabolome data. We found that some members of Perl’s pathway-related gene families had higher relative expression levels at the early stage of germination and that the enzymes in this pathway were either more active in dry seeds or showed high activity at the early stage of germination. Oxaloacetate may be the main substrate for this pathway. The results of this study lay a theoretical foundation for exploring the energy sources for poplar seed germination.
Our results show that gene transcriptional patterns varied among different members of Perl’s pathway-related gene families. This result indicates that different members of the same family might be differently regulated and have different functions. For example, among the genes encoding MDH, potri.011G096300 and potri.017G102000 showed higher transcript levels at the early stage of seed germination, but potri.017G101900 and potri.001G376500 showed higher transcript levels after 6 h. Among the genes encoding PK, potri.010G254900 and potri.008G159700 had higher transcript levels at 0–0.75 h of seed germination, while the other members of this family showed gradual increases in transcription during germination. The same phenomenon was observed among genes encoding ADH. Gallardo (2001) analyzed the transcriptome of dry seeds and found that genes encoding PDC2 and ADH had the highest transcript levels of all the genes in the fermentation pathway. They found that after the start of seed germination, the transcript levels of genes encoding PDC2 and ADH decreased rapidly but the ADH protein levels remained unchanged. The activity of ADH reflects the sum of activities of stored and/or newly synthesized ADH accumulated during seed maturation (Fu et al. 2005; Gallardo et al. 2001; Rajjou et al. 2006). The results of those studies suggest that there are complex patterns of activation for genes in particular pathways and that different members of gene families cooperate to maintain a constant level of total protein. Given the different expression patterns of gene family members detected in our study, we believe that such complex regulatory processes also affect members of gene families in Perl’s pathway.
Our enzyme activity analyses indicated that some of the enzymes associated with Perl’s pathway, such as PK and PDC, may be active in dry seeds. Enzymes may be present in an inactive form in dry seeds with a low moisture content, and then regain activity upon seed imbibition. In our study, the enzymes ADH, PEPCK, and MDH showed significantly increased activity by 0.75 h of germination. This observation indicates that these enzymes may play important roles during the early stage of poplar seed germination.
The Perl’s pathway starts with the dehydrogenation of malate, and a reaction catalyzed by PEPCK and PK to form ATP. In these reactions, NADH provides the reducing power (Fig. 6). As shown in Fig. 5, the oxaloacetate content was higher in dry seeds and dropped significantly by 0.75 h of germination, while the phosphoenolpyruvate content also decreased slightly but not significantly. We hypothesize that oxaloacetate may serve as the substrate for the production of phosphoenolpyruvate at 0.75 h of germination. Although the phosphoenolpyruvate content fluctuated from 0 to 6 h of germination, the changes were not significant. These fluctuations may be related to the role of phosphoenolpyruvate as an intermediate in Perl’s pathway, and the significant decrease in NAD content may be related to its mobilization. Previous studies have shown that the transcription of genes encoding glycolytic and tricarboxylic acid cycle-related enzymes begins to recover by 6 h of germination (Qu et al. 2019e). We propose that the Perl’s pathway may provide energy until 6 h of germination, before activation of glycolysis and the TCA cycle.
Fig. 6.
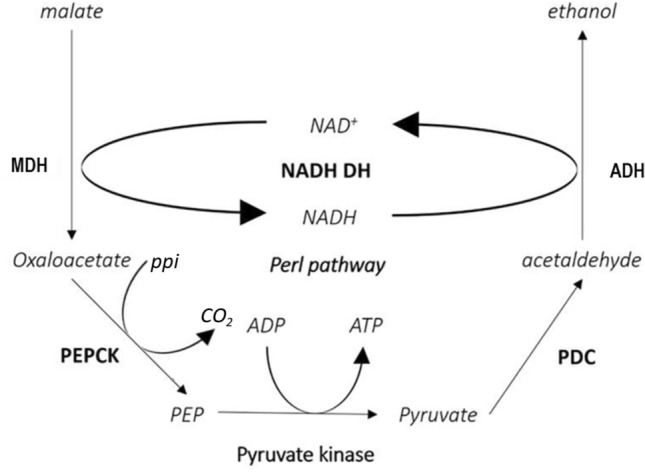
Schematic representation of Perl’s pathway. MDH Malate dehydrogenase, PEPCK Phosphoenolpyruvate carboxylase, PEP Phosphoenolpyruvate, PDC Pyruvate decarboxylase, ADH Alcohol dehydrogenase, NADH-DH Nicotinamide adenine dinucleotide dehydrogenase
It has been proposed that Perl’s pathway provides energy for seed germination. Our results show that the activities of some enzymes in this pathway increased within 1 h of seed imbibition, and then decreased at the later stage of germination. These results indicate that Perl’s pathway only plays an important role during the early stage of seed germination. Consistent with this idea, most of the enzymes in Perl’s pathway showed higher activities at the early stage of seed germination than at the later stage. Because this pathway only plays an important role in seed germination for a short period of time, and because of technical limitations, few studies have focused on Perl’s pathway and its contribution to seed germination. With the recent development of various -omics technologies, more studies have tried to unravel the molecular mechanism of seed germination. Some recent studies have provided information about the mechanism of Perl’s pathway and its function from different perspectives. For example, Liu (2018) found that wheat AspAT contributes to the formation of oxaloacetate, the substrate for Perl’s pathway. Zhang (2018) found that the small RNA miR5141 targets a gene encoding ATP synthase, which is involved in ATP synthesis via Perl’s pathway, during the early stage of seed germination in Paeonia ostii. In future studies, we intend to conduct further research on poplar seeds to screen and study the signaling pathways that regulate Perl’s pathway.
Conclusions
The starting point for this study was the pathway hypothesized by Perl (1986). To test whether this pathway is the main source of energy during the early stage of seed germination, we analyzed germinating poplar seeds. We used qRT-PCR to quantify the transcript levels of genes in Perl’s pathway, detected the activity levels of various enzymes, and verified our results by comparison with RNA-Seq and metabolomics data. Our findings indicate that Perl’s pathway is the main source of energy at the early stage of poplar seed germination and that oxaloacetate and other substances play a role in the supply of raw materials. This study provides new information about the metabolic processes that function during the early stage of poplar seed germination.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author contributions
CQ and GL conceived and designed this study. CQ, SZ and ZX performed the experiments. HZ, JC and ZZ performed the data collection and statistical analysis. CQ and GL wrote the manuscript. YC and ZX contributed suggestions. All authors gave the final approval of the paper.
Funding
This research was supported by Science Fund Project of Heilongjiang Province of China (LH2020C043), Heilongjiang Touyan Innovation Team Program (Tree Genetics and Breeding Innovation Team), the Fundamental Research Funds for the Central Universities (2572019CT02) and the 111 project (B16010). The funding was funded by Innovative Research Group Project of the National Natural Science Foundation of China (CN) (C2018009), the National Natural Science Foundation of China (31600534).
Data availability
The data and materials relevant to this study may be obtained from the corresponding author on reasonable request.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
Not applicable.
Informed consent
Written informed consent for publication was obtained from all participants.
Contributor Information
Chunpu Qu, Email: qcp_0451@163.com.
Shuang Zhang, Email: 15046637546@163.com.
Hancheng Zhao, Email: 243258848@qq.com.
Jinyuan Chen, Email: 15546522295@163.com.
Zhuang Zuo, Email: 504627511@qq.com.
Xue Sun, Email: sx329330132@163.com.
Yuxiang Cheng, Email: chengyuxiang@nefu.edu.cn.
Zhiru Xu, Email: xuzhiru2003@126.com.
Guanjun Liu, Email: liuguanjun2013@nefu.edu.cn.
References
- Benamar A, Rolletschek H, Borisjuk L, Avelange-Macherel MH, Curien G, Mostefai HA, Andriantsitohaina R, Macherel D. Nitrite-nitric oxide control of mitochondrial respiration at the frontier of anoxia. Biochem Biophys Acta. 2008;1777:1268–1275. doi: 10.1016/j.bbabio.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Bewley JD. Seed germination and dormancy. Plant Cell. 1997;9:1055–1066. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botha F, Potgieter G, Botha A-M. Respiratory metabolism and gene expression during seed germination. Plant Growth Regul. 1992;11:211–224. doi: 10.1007/BF00024560. [DOI] [Google Scholar]
- Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-McIntyre S, Anderson N, Brown M, Knowles JD, Halsall A, Haselden JN, Nicholls AW, Wilson ID, Kell DB, Goodacre R, C HSMH Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc. 2011;6:1060–1083. doi: 10.1038/nprot.2011.335. [DOI] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G. Seed dormancy and the control of germination. New Phytol. 2006;171:501–523. doi: 10.1111/j.1469-8137.2006.01787.x. [DOI] [PubMed] [Google Scholar]
- Fu Q, Wang BC, Jin X, Li HB, Han P, Wei KH, Zhang XM, Zhu YX. Proteomic analysis and extensive protein identification from dry, germinating Arabidopsis seeds and young seedlings. J Biochem Mol Biol. 2005;38:650–660. doi: 10.5483/bmbrep.2005.38.6.650. [DOI] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SP, Puype M, Demol H, Vandekerckhove J, Job D. Proteomic analysis of arabidopsis seed germination and priming. Plant Physiol. 2001;126:835–848. doi: 10.1104/pp.126.2.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C, Yang P. Studies on the molecular mechanisms of seed germination. Proteomics. 2015;15:1671–1679. doi: 10.1002/pmic.201400375. [DOI] [PubMed] [Google Scholar]
- Holdsworth MJ, Finch-Savage WE, Grappin P, Job D. Post-genomics dissection of seed dormancy and germination. Trends Plant Sci. 2008;13:7–13. doi: 10.1016/j.tplants.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Ivanisevic J, Elias D, Deguchi H, Averell PM, Kurczy M, Johnson CH, Tautenhahn R, Zhu ZJ, Watrous J, Jain M, Griffin J, Patti GJ, Siuzdak G. Arteriovenous blood metabolomics: a readout of intra-tissue metabostasis. Sci Rep. 2015 doi: 10.1038/srep12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Bentsink L, Hilhorst H. Seed dormancy and germination. Curr Opin Plant Biol. 2002;5:33–36. doi: 10.1016/j.cub.2017.05.050. [DOI] [PubMed] [Google Scholar]
- Liu Y, Han C, Deng X, Liu D, Liu N, Yan Y. Integrated physiology and proteome analysis of embryo and endosperm highlights complex metabolic networks involved in seed germination in wheat (Triticum aestivum L.) J Plant Physiol. 2008;229:63–76. doi: 10.1016/j.jplph.2018.06.011. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Muller K, Carstens AC, Linkies A, Torres MA, Leubner-Metzger G. The NADPH-oxidase AtrbohB plays a role in Arabidopsis seed after-ripening. New Phytol. 2009;184:885–897. doi: 10.1111/j.1469-8137.2009.03005.x. [DOI] [PubMed] [Google Scholar]
- Nonogaki H, Bassel GW, Bewley JD. Germination-still a mystery. Plant Sci. 2010;179:574–581. doi: 10.1016/j.plantsci.2010.02.010. [DOI] [Google Scholar]
- Perl M. ATP synthesis and utilization in the early stage of seed germination in relation to seed dormancy and quality. Physiol Plant. 1986;66:177–182. doi: 10.1111/j.1399-3054.1986.tb01253.x. [DOI] [Google Scholar]
- Qu C, Zhao H, Chen J, Zuo Z, Sun X, Huang J, Yang C, Zhang X, Zhang P, Quan X, Xu Z, Liu G. The transcriptional events and their relationship to physiological changes during poplar seed germination and post-germination. BMC Genomics. 2019;20:801. doi: 10.1186/s12864-019-6180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu C, Zuo Z, Cao L, Huang J, Sun X, Zhang P, Yang C, Li L, Xu Z, Liu G. Comprehensive dissection of transcript and metabolite shifts during seed germination and post-germination stages in poplar. BMC Plant Biol. 2019;19:279. doi: 10.1186/s12870-019-1862-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu CP, Chen JY, Cao LN, Teng XJ, Li JB, Yang CJ, Zhang XL, Zhang YH, Liu GJ, Xu ZR. Non-targeted metabolomics reveals patterns of metabolic changes during poplar seed germination. Forests. 2019 doi: 10.3390/F10080659. [DOI] [Google Scholar]
- Qu CP, Hao BQ, Xu XY, Wang YC, Yang CJ, Xu ZR, Liu GJ. Functional research on three presumed asparagine synthetase family members in poplar. Genes-Basel. 2019 doi: 10.3390/Genes10050326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu CP, Zhang YQ, Chen JY, Zhang S, Yu JJ, Yang CJ, Zhang XL, Xu ZR, Liu GJ. A weighted mean value analysis to identify biological pathway activity changes during poplar seed germination. Forests. 2019 doi: 10.3390/F10080664. [DOI] [Google Scholar]
- Rajjou L, Belghazi M, Huguet R, Robin C, Moreau A, Job C, Job D. Proteomic investigation of the effect of salicylic acid on Arabidopsis seed germination and establishment of early defense mechanisms. Plant Physiol. 2006;141:910–923. doi: 10.1104/pp.106.082057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou L, Duval M, Gallardo K, Catusse J, Bally J, Job C, Job D. Seed germination and vigor. Annu Rev Plant Biol. 2012;63:507–533. doi: 10.1146/annurev-arplant-042811-105550. [DOI] [PubMed] [Google Scholar]
- Saccenti E, Hoefsloot HCJ, Smilde AK, Westerhuis JA, Hendriks MMWB. Reflections on univariate and multivariate analysis of metabolomics data. Metabolomics. 2014;10:361–374. doi: 10.1007/s11306-013-0598-6. [DOI] [Google Scholar]
- Smiri M, Chaoui A, El Ferjani E. Respiratory metabolism in the embryonic axis of germinating pea seed exposed to cadmium. J Plant Physiol. 2009;166:259–269. doi: 10.1016/j.jplph.2008.05.006. [DOI] [PubMed] [Google Scholar]
- Spoelstra P, Joosen RV, Van der Plas LH, Hilhorst HW. The distribution of ATP within tomato (Lycopersicon esculentum Mill.) embryos correlates with germination whereas total ATP concentration does not. Seed Sci Res. 2002;12:231–238. doi: 10.1079/SSR2002114. [DOI] [Google Scholar]
- Steinbrecher T, Leubner-Metzger G. The biomechanics of seed germination. J Exp Bot. 2016;68:erw428. doi: 10.1093/jxb/erw428. [DOI] [PubMed] [Google Scholar]
- Tarazona S, Furio-Tari P, Turra D, Pietro AD, Nueda MJ, Ferrer A, Conesa A. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res. 2015;43:e140. doi: 10.1093/nar/gkv711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. Mapman: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- Wakao S, Andre C, Benning C. Functional analyses of cytosolic glucose-6-phosphate dehydrogenases and their contribution to seed oil accumulation in Arabidopsis. Plant Physiol. 2008;146:277–288. doi: 10.1104/pp.107.108423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitbrecht K, Muller K, Leubner-Metzger G. First off the mark: early seed germination. J Exp Bot. 2011;62:3289–3309. doi: 10.1093/jxb/err030. [DOI] [PubMed] [Google Scholar]
- Zhang H, Wang WQ, Liu SJ, Moller IM, Song SQ. Proteome analysis of poplar seed vigor. PLoS ONE. 2015;10:e0132509. doi: 10.1371/journal.pone.0132509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang Y, Gao X, Liu C, Gai S. Identification and characterization of microRNAs in tree peony during chilling induced dormancy release by high-throughput sequencing. Sci Rep. 2018;8:4537. doi: 10.1038/s41598-018-22415-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Luo W, Li Y, Zhang X, Bai X, Niu Z, Zhang X, Li Z, Wan D. Transcriptomic Analysis of Seed Germination Under Salt Stress in Two Desert Sister Species (Populus euphratica and P pruinosa) Front Genet. 2019;10:231. doi: 10.3389/fgene.2019.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and materials relevant to this study may be obtained from the corresponding author on reasonable request.