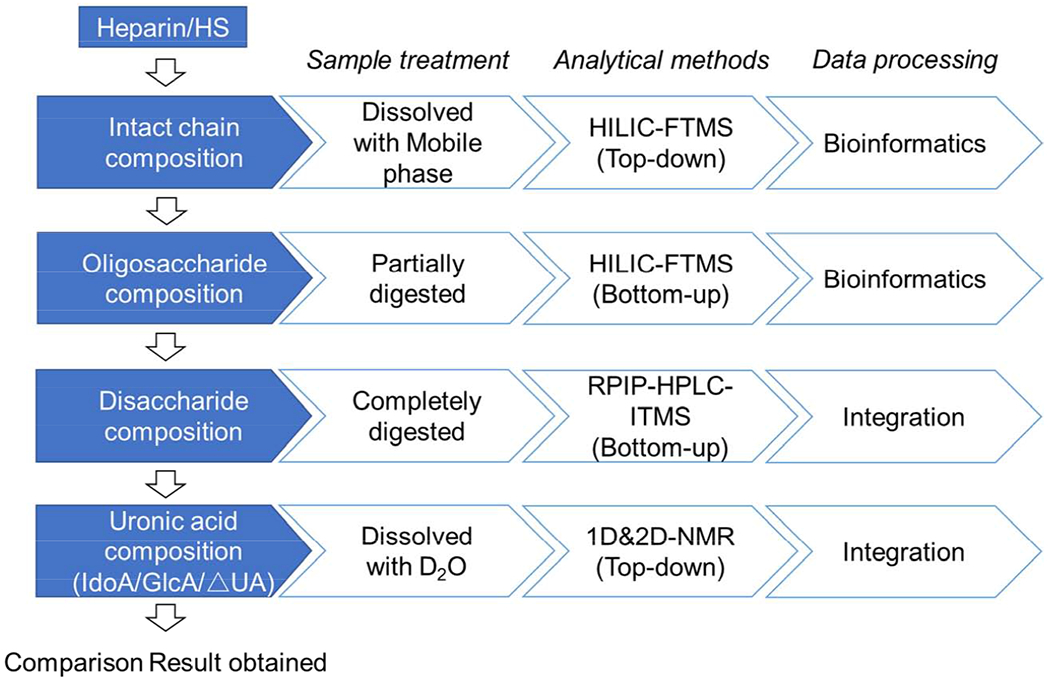
Abstract
Heparan sulfate (HS) is a class of linear, sulfated, anionic polysaccharides, called glycosaminoglycans (GAGs), which present on the mammalian cell surfaces and extracellular matrix. HS GAGs display a wide range of critical biological functions, particularly in cell signaling. HS is composed of repeating units of 1→4 glucosidically linked uronic acid and glucosamine residues. Heparin, a pharmacologically important version of HS, having higher sulfation and a higher content of iduronic acid than HS, is a widely used clinical anticoagulant. However, due to their heterogeneity and complex structure, HS and heparin are very challenging to analyze, limiting biological studies and even resulting in safety concerns in their therapeutic application. Therefore, reliable methods of structural analysis of HS and heparin are critically needed. In addition to the structural analysis of heparin, its concentration in blood needs to be closely monitored to avoid complications such as thrombocytopenia or hemorrhage caused by heparin overdose. This review summarizes the progress in biotechnological approaches in the structural characterization of HS and heparin over the past decade and includes the development of the ultrasensitive approaches for detection and measurement in biological samples.
Keywords: Heparan sulfate, heparin, structure analysis, ultrasensitive analysis
Graphical Abstract
1. Introduction
Heparan sulfate (HS) is a glycosaminoglycan (GAG), a linear, sulfated, polydisperse, structurally complex hetero-co-polysaccharide [1,2]. HS is comprised of a disaccharide repeating unit consisting of β-d-glucɑuronic acid (GlcA) or ɑ-l-iduronic acid (IdoA) and β-d-glucosamine (GlcN) linked by (1→4) glycoside bonds (Figure 1). Modifications of disaccharide units, including the presence of N-acetyl of N-sulfo substituents on the GlcN (GlcNAc or GlcNS), O-sulfo groups at the 6-O-and/or 3-O-positions of GlcN, and 2-O-sulfo substituents on the IdoA positions, making HS diverse structurally (>32 possible units) [3]. HS is widely distributed on animal cell surfaces and in the extracellular matrix (ECM) [2]. There HS binds to many ECM proteins (i.e., growth factors, chemokines, adhesion proteins, etc.,) [4,5], allowing these to act as signaling molecules, playing an important role in cell adhesion, migration, differentiation, signaling and angiogenesis [6,7]. Most importantly, HS has physiological and pharmacological impact in blood coagulation, embryonic development, inflammatory response, and bacteria/viral infections [3,8].
Figure 1.
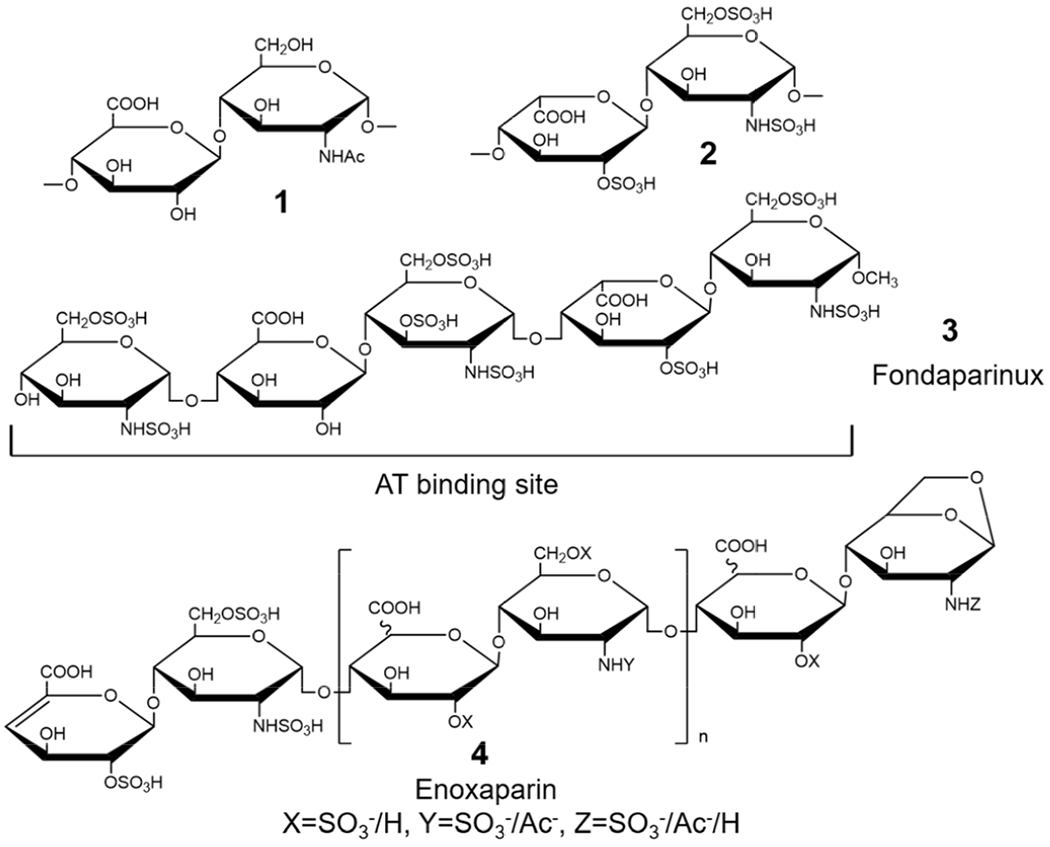
Structures of HS, heparin and heparin derivatives. The major disaccharide repeating units of HS (primarily 1) and heparin (primarily 2) are shown. The structure of an ULMWH, Arixtra® (fondaparinux) 3, containing a pentasaccharide sequence corresponding to an AT-binding site is shown. The LMWH, Lovenox® (enoxaparin) 4 contains a diverse mixture of chain types but only one type of chain present in enoxaparin, containing a 1,6-anhydro group unique to enoxaparin, is shown.
Heparin is a GAG sharing the same biosynthetic pathway as HS but with a higher level of sulfation and prominent repeating trisulfated disaccharides, GlcNS6S(1→4)IdoA2S (Figure 1) [9]. For more than 100 years, heparin has been clinically used as a drug for the treatment of thrombotic disorders [10]. Heparin binds to antithrombin III (AT), an inhibitor of serine proteases, factor Xa and thrombin, resulting a conformational change amplifying the inhibitory activity of AT [3,11]. There are three major forms: the intravenous drug, unfractionated heparin (UFH, MWavg 16,000 Da); several types of subcutaneous low molecular weight heparins (LMWH, MWavg 3500–6000 Da; and subcutaneous ultralow molecular weight heparins (ULMWH, MW< 2000 Da) [3,9]. There is an antidote, cationic peptide drug, protamine, which can reverse the effect of UFH. Life-threatening complications, such as heparin-induced thrombocytopenia (HIT) can occur particularly with UFH [12]. For the past 30 years LMWH such as enoxaparin (Figure 1, 4), generated from UFH by chemical β-elimination, have been widely used because of their longer half-life and subcutaneous bioavailability [13] LMWH binds to AT and acts only on FXa without inhibiting thrombin, facilitating a more subtle regulation of coagulation and an improved therapeutic index [14]. A synthetic ULMWH, fondaparinux, which is the first chemically synthetic antithrombotic pentasaccharide [15], is infrequently used due to its high cost (Figure 1, 3).
Figure 4.
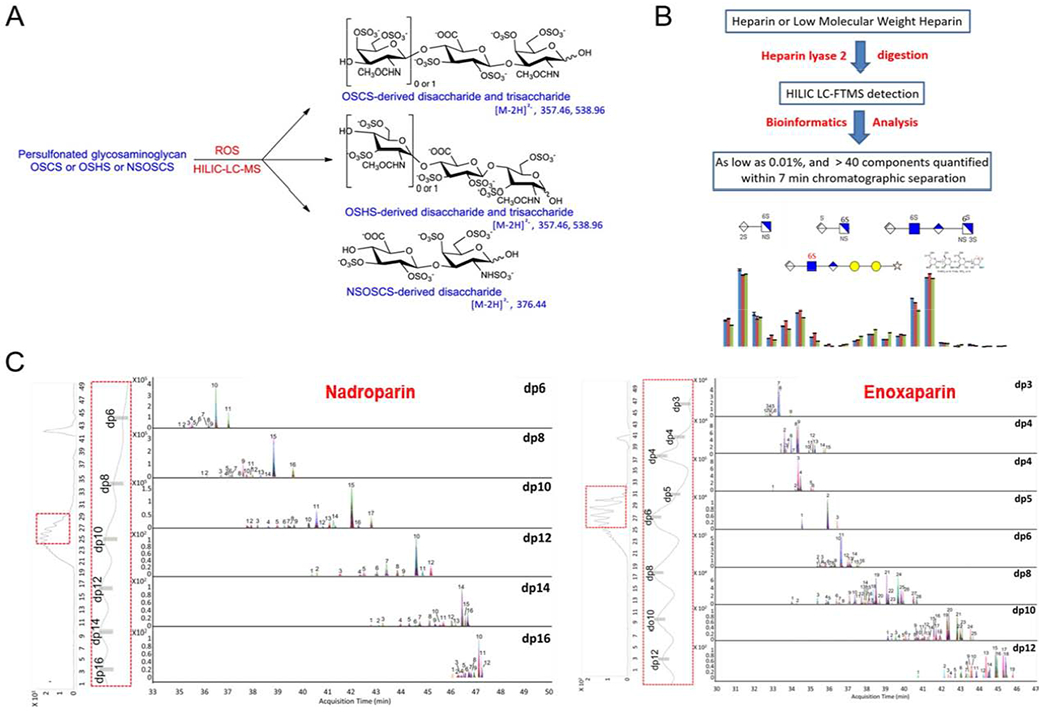
(A) Oxidative depolymerization and HILIC-MS of heparin and oversulfated semi-synthetic GAGs. Copyright 2014 American Chemical Society. (B) Liquid chromatography-Fourier transform mass spectrometry for extensive characterization of low molecular weight heparin. Copyright 2014 American Chemical Society. (C) Profiling analysis of the LMWHs nadroparin and enoxaparin using MHC-2D-LC-Q time-of-flight (TOF)-MS. Copyright 2015 American Chemical Society.
Figure 3.
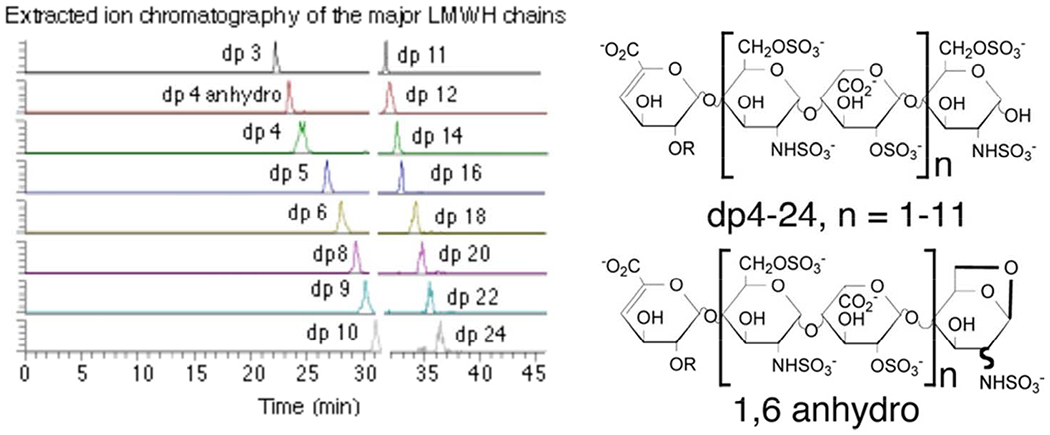
Top-down approach for the direct characterization of low molecular weight heparins using HILIC-LC-FT-MS analysis. Copyright 2012 American Chemical Society.
Recently, biotechnological versions of UFH, LMWH and ULMWH have reached the stage of commercial development [16,17,18]. These products rely on chemoenzymatic synthesis making their preparation more cost competitive than products like fondaparinux, which relies on chemical synthesis. New analytical approaches that are information rich, sensitive, robust and reliable are required in the development of this new generation of biotechnological heparin and HS products. This review focuses on the analytical methods developed over the past decade.
2. Why is analysis of heparin and heparan sulfate important?
The UFH and LMWH currently in clinical use are extracted from animal tissues, such as porcine, bovine intestines and bovine lungs [19]. As a result, these products have purity and homogeneity issues and a variable based on animal species, age, sex, health, animal feed types and other environmental factors [20]. More importantly, there is a risk of viral or prion infection, which poses a serious threat to the safety of these drugs. A global contamination of heparin broke out in 2007, causing over 200 deaths in the US [21]. This crisis resulted from the adulteration of heparin with a toxic and inexpensive oversulfated chondroitin sulfate. Therefore, reliable structural analysis techniques for animal sourced heparin are highly desirable. Biotechnologically prepared heparins and HS offer different but equally challenging analytical objectives. Subtle differences in the structures or process artifacts require new information rich in highly sensitive analytical methods. For instance, the detection of low level process impurities or relatively subtle structural features, such as sugar epimers or sulfo group position or differences in linkage position and anomeric configuration, require more advanced analytical methods. In addition, sensitive, reliable and robust methods to determine heparin levels within biological fluids and tissues are needed to avoid complications such as hemorrhage induced by heparin overdose. Thus, this review will summarize the progress of the structure characterization of heparin/HS in the past decade, and discuss the development of new ultrasensitive approaches to monitor heparin and HS-based therapeutics.
3. Techniques for structure characterization/quantification of heparin/HS
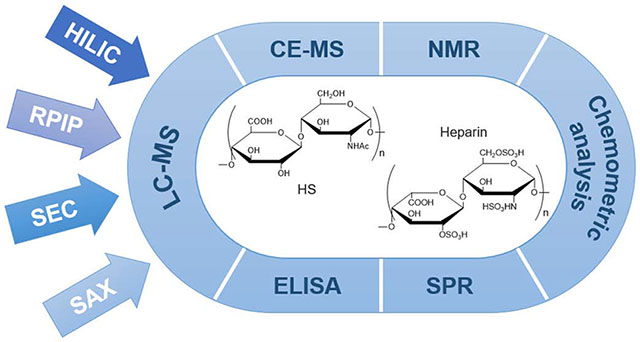
Characterization strategies for heparin/HS, particularly LMWHs, can be generally summarized into two approaches, top-down and bottom-up analysis, and these are based on the Food and Drug Administration (FDA) requirements (Figure 2) [22]. Similar to proteomic strategies, top-down analysis analyzes samples without any chemical or enzymatic pretreatment and can rely on simple methods such as size exclusion chromatography (SEC) to measure the molecular weight, or more complicated methods such as liquid chromatography-mass spectrometry (LC-MS) to obtain fingerprint mapping of intact chains, MS/MS to sequence chains or nuclear magnetic resonance (NMR) to analyze intact chains [23]. Bottom-up strategies rely on the controlled partial depolymerization of polysaccharide chains to afford oligosaccharides for analysis by high-performance liquid chromatography (HPLC), capillary electrophoresis (CE) and LC-MS disaccharide compositional analysis or oligosaccharide sequence analysis [24]. The controlled depolymerization of a heparin/HS can rely on either random or semi-random chemical methods or somewhat more controlled enzymatic methods. Heparinases, either lyases [25] or hydrolases [26] provide a mild method for the controlled enzymatic depolymerization of heparin/HS. The individual methods applied in both top-down and bottom-up will be discussed in detail.
Figure 2.
Representative strategies for heparin/HS structural characterization.
3.1. Liquid chromatography-MS
Liquid chromatography (LC) is a separation and analysis tool relying on a liquid as mobile phase and a stationary solid phase. High-performance liquid chromatography (HPLC) provides faster separation and higher resolution method than LC and has been widely used in the analysis of GAG disaccharides and oligosaccharides [27,28]. Using different mobile and solid phases, based on different separation principles, HPLC can facilitate the separation and analysis of HS/heparin mixtures and provide diverse structural information. HPLC separations of HS/heparin include size exclusion chromatography (SEC) strong anion exchange (SAX)-HPLC, reversed-phase (RP)-HPLC, reversed-phase ion-pairing (RPIP)-HPLC, and hydrophilic interaction chromatography (HILIC) [25,29]. Ultraperformance liquid chromatography (UPLC), microflow, nanoflow, capillary HPLC and UPLC has also been applied [26]. While LC can be used with a variety of detections such as ultraviolet fluorescence and refractive index, it has most recently been used in conjunction with mass spectrometry (MS), providing high-resolution spectra in both top-down and bottom-up analysis affording a stable and fast continuous analysis platform [22,30].
3.1.1. Top-down analysis
Top-down NMR and LC-MS & MS/MS analysis carefully examines the intact HS or heparin chains and can provide excellent results in the analysis of the number and types of sulfo groups, and the types of uronic acid and glucosamine residues present in HS/heparin oligosaccharides and LMWHs [30]. Top-down analysis LC-MS & MS/MS has still not yet been successfully applied to intact chains of UFH because of its relatively high molecular mass (>10,000) and net charge (>−40). There are challenges for using MS analysis with a top-down strategy, in particular, the loss of sulfo groups, and production of multiple adducts. While Fourier transform (FT)-ion cyclotron resonance (ICR) MS offers a high-resolution platform, but the availability of such instrumentation has been problematic. Matrix assisted laser induced ionization (MALDI) MS has limitations in the analysis of highly charged heparin oligosaccharides of large size [31]. The widespread availability of Orbitrap instruments has alleviated this limitation to some extent [32]. RPIP-LC, the first LC-MS method for the analysis of LMWH had problems of instrument contamination with non-volatile ion-pairing reagents, and relatively low resolution and sensitivity [33]. HILIC is a separation methods based on the disparity between the overall polarity of polysaccharides or oligosaccharides, leading to differences in their interaction with the solid phase support [34]. A relatively high-throughput online analyical platform, HILIC LC-FT-MS with Orbitrap detection resulted in relatively high-resolution, excellent separation efficiency, with good reproducibility and stability (Figure 3). The separation relied on a Luna HILIC column, which replaced the standard amine with a cross-linked diol solid-phase support, to improve the resolution of complex LMWH mixtures for MS detection. Bioinformatics software (GlycReSoft) was then used to quickly extract analysis data. This rapid, robust method requires no labeling and can be used to compare different commercial LMWHs, analyzing ~90% of the chains, up to dp18 including minor components [35]. Moreover, small amounts of process byproducts, such as chains with 1,6-anhydro reducing ends could be detected and use to compare generic versions to the innovator drug, Lovenox®. However, HILIC-LC affords lower resolution than RPIP-HPLC, resulting in challenges for this technology to achieve 100% coverage of chains within LMWH. These challenges might one day be overcome to allow the separation of all of the minor components in LMWH through the use of improved HILIC-LC, and higher resolution MS instruments [35].
3.1.2. Bottom-up analysis
A top-down strategy is not currently applicable to the analysis of full chain length HS or UFH polysaccharides. Thus, a bottom-up strategy, with high reproducibility and accuracy, is widely used to infer polysaccharide structure by analyzing disaccharides or oligosaccharides obtained through controlled depolymerization of the intact polysaccharides using enzymatic or chemical methods [30].
3.1.2.1. Hydrophilic interaction chromatography-MS
A HILIC-HPLC-electrospray ionization (ESI)-Fourier transform (FT)-MS platform was established for the sensitive analysis of all chemically sulfonated GAG-based contaminants such as the oversulfated chondroitin sulfate that played a role in the 2007 heparin contamination crisis (Figure 4A) [36]. This semi-synthetic GAG could not be enzymatically depolymerized, so that reactive oxygen species (ROS) were used to prepare oligosaccharides in a highly reproducible and relatively nonselective way, while retaining their primary molecular structure. This method could successfully detect oversulfated chondroitin sulfate and oversulfated heparan sulfate in the presence of heparin. These, and other potential adulterants, such as N-deacetylated/N-sulfonated oversulfated chondroitin sulfate, can be detected even at low levels in heparin active pharmaceutical ingredient (API).
In other studies, HILIC-MS was utilized as a high resolution and fast method to analyze oligosaccharides prepared from depolymerization of heparin using heparin lyase II (Figure 4B). Following heparin lyase II depolymerization of enoxaparin it was possible to quantitatively detect four major 3-O-sulfo group-containing tetrasaccharides (remnants of the AT-binding site), oligosaccharides with 1,6-anhydro-amino residues (process artifacts in enoxaparin preparation) and three linkage region oligosaccharides (where heparin attached to its core protein). This analysis method allows the comparison of the reducing end groups of different commercial LMWHs that cannot easily be achieved using a top-down strategy. This platform has the advantages of fast separation speed (about 7 min), wide dynamic range (>5000) and high sensitivity (detecting 0.01% components), and provides a relatively high throughput and reliable method to control commercial LMWH products [37].
However, HILIC separation still has major limitations in separating isomeric compositions and disaccharides with similar overall polarity having different sulfation patterns. New high-resolution HPLC separation methods compatible with MS detection are critically needed to fully analyze HS/heparin polysaccharides and oligosaccharides [27].
3.1.2.2. RPIP-MS
RPIP involves the interaction between a hydrophobic column (e.g., a C18 column) and analyte ion-paired with lipophilic ion-pairing reagents, such as hexylamine (HXA), tributylamine (TrBA), trimethylamine (TEA), and tetrabutylamine (TBA) [38,39]. A RPIP microflow HPLC-ESI-MS approach has been used to analyze most HS/heparin disaccharides with reliability and high separation efficiency and speed [40]. However, when analyzing biological samples, this method cannot fully resolve the disaccharides because of the impact of impurities such as salts and proteins in the samples. UPLC separation, using the 1.7-μm column under high pressure (up to 108 Pa), retains the principle of HPLC [29] but improves separation speed, resolution, sensitivity and peak capacity (Figure 5A) [38]. This RPIP-UPLC-ESI-MS approach was used with a bottom-up strategy relying on enzymatic depolymerization to analyze small amounts of HS/ heparin in urine, cells and tissues samples and for the structural profiling of the pharmaceutical heparins. This approach allows the qualitative and quantitative detection of eight HS/heparin disaccharides, with short equilibration time and high sensitivity [38]. However, RPIP-HPLC with MS detection still has the disadvantages of complicated preparation, difficult removal of ion-pairing reagent from the mobile phase and low compatibility with on-line MS detection [39,41,42].
Figure 5.
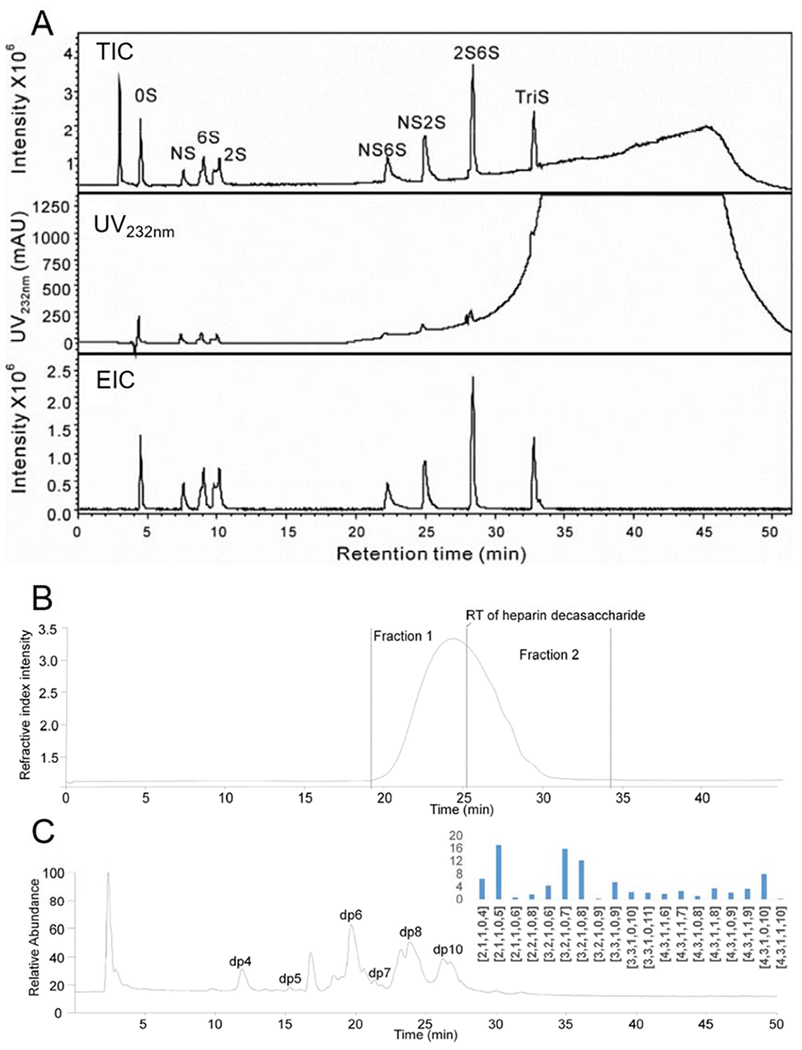
(A) RPIP-UPLC chromatograms of eight heparin/HS-derived disaccharide standards with total ion chromatogram (TIC), UV232 nm, and extracted ion chromatogram (EIC) detection. Copyright 2011 Elsevier. (B) SEC separation of dalteparin; (C) HILIC-ESI-MS of the short dalteparin oligosaccharides collected from SEC and the short oligosaccharide compositions. Copyright 2018 Elsevier.
3.1.2.3. SEC & RPIP-Q/TOF-MS
Different types of HPLC can be used in combination to achieve better separation results[14]. For example, LMWHs contain diverse oligosaccharide chains that cannot be separated using HILIC, IPRP or SAX. By using multiple heart-cutting (MHC) to connect SEC-HPLC and RPIP-HPLC and detecting with quadrupole time-of-flight (Q/TOF) MS it was possible to provide a definitive and systematic analysis of LMWH to obtain detailed structural information (Figure 4C) [43]. The first dimension, SEC, separated the oligosaccharides by molecular size, while the second dimension, RPIP, separated the oligosaccharides on the basis of differences in their charge and polarity. This approach facilitated the analysis of many components having different sizes of similar charge or of the same size but different charge. This relatively high-resolution, stable, easy-to-optimize, high-efficiency, MHC-2D LC-Q/TOF MS approach could detect 84 oligosaccharides in nadroparin and 122 oligosaccharide peaks in enoxaparin. This three-dimensional approach has the potential to control commercial LMWH production and to detect, analyze and sequence large oligosaccharide chains.
3.1.2.4. SEC & SAX-MS/MS
SAX-HPLC is a means of separation relying on the interaction between negatively charged HS/heparin and the positively charged solid state support of an SAX column [44]. This method affords higher resolution than HILIC and can easily separate disaccharides with different sulfate levels, especially gratifyingly in the analysis of disaccharides obtained by the cleavage of heparins and LMWH with lyase [45]. Unfortunately, samples bound to SAX columns typically need to be eluted with MS-unfriendly high-concentration sodium salts. Therefore, when analyzing enoxaparin, this method has often been used in combination with UV detection after separating the disaccharide depolymerization using enzymes. This method is highly accurate and does not require labeling. However, when envisaging the analysis of intact LMWH, this method can only be used with enoxaparin containing double bond as a chromophore, and is not feasible for the LMWH dalteparin sodium due to the lack of chromophore and the higher molecular weight. However, the higher molecular weight and lack of chromophore in dalteparin sodium limits its application. An offline method, which collects, combines, and desalts analytes separated by HPLC allows their analysis by MS, SAX-HPLC-ESI-MS/MS and is useful for sequencing the short-chain oligosaccharides from the LMWH, dalteparin sodium (Figure 5B and 5C) [27]. The short oligosaccharides obtained after preliminary separation by SEC could be further separated by SAX-HPLC using a high resolution ProPac PA1 SAX column. ESI-MS/MS could then provide a sensitive and reliable approach for the direct sequencing of low-purity heparin drug samples. This indispensable and sensitive approach was used for the sequencing of 18 oligosaccharides (from tetrasaccharide to octasaccharide) providing a basis for exploring the precise composition of LMWH.
3.2. CE-MS
CE is an effective technique for separating molecules based on their shape, size, and charge [46,47]. MS can accurately and quickly identify and analyze molecules by measuring the mass-to-charge ratio of ions [48]. CE-MS is becoming one of the most powerful techniques for the separation and identification of HS/heparin oligosaccharide due to a number of advantages including high sensitivity, high resolution, simple operation, high separation efficiency, and a flexible mode of separation [49,50]. However, in the process, an effective interface is essential for combining CE and MS. Insufficient research and development of CE-MS interfaces over the past decade has hindered the application of CE-MS in heparin/HS analysis. A groundbreaking neoteric electrokinetic pump-based CE-MS interface was used to combine a normal polarity CE separation with a positive-ion electrospray ionization MS platform. This interface resulted in great sensitivity, resolution and repeatability allowing the online analysis of heparin disaccharide mixture and LMWH chains (Figure 6A) [51]. This method creatively introduced an uncoated capillary sheath emitter tip as an interface. The negatively charged sugar molecules separated by CE were positively charged with NH4+ in ammonium bicarbonate buffer solution and then sprayed into the mass spectrometer for analysis. Reasonable combination of capillary, emitter, MS, rational constitution and ratio between background electrolyte (BGE) and sheath liquid (SL) reduce the generation of bubbles and ensure stable and steady electrospray at extremely low flow rate (nL/min). This gentle detection environment greatly improves the analysis sensitivity and makes the high-throughput detection of HS oligosaccharides possible. Using CE-MS requires no glycan labeling affording a high signal-to-noise ratio, rapid analysis (< 10 min), on a small amount of sample (picograms) and utilizes little buffer (< 10 microliters).
Figure 6.
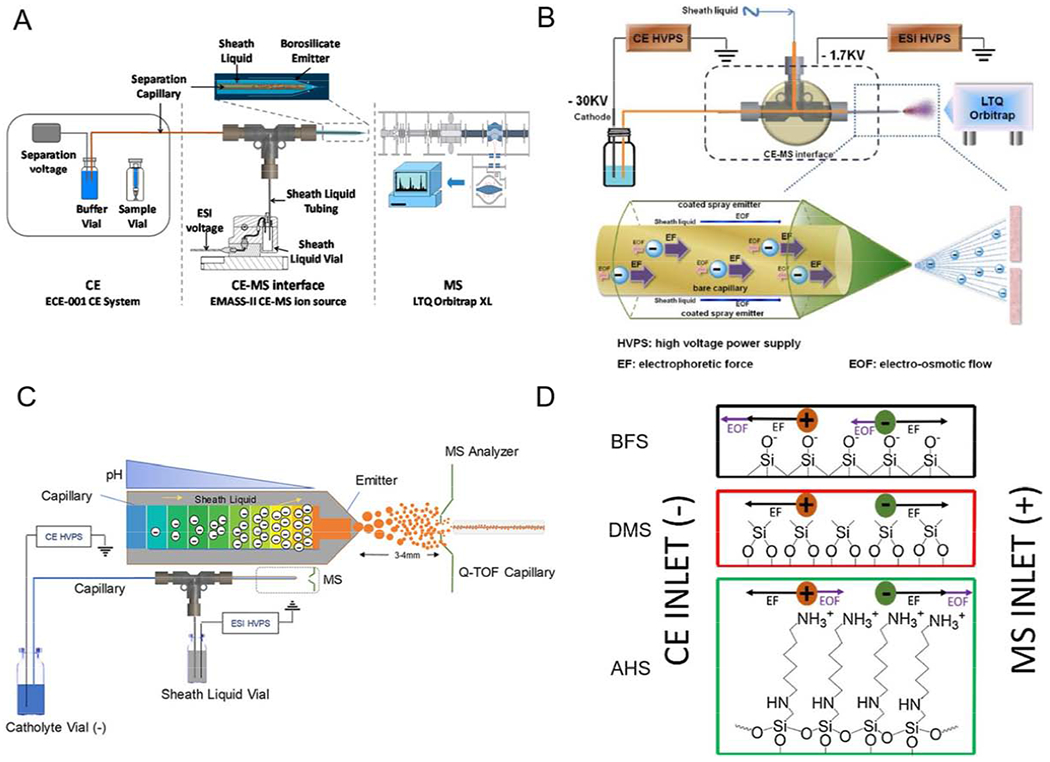
Schematic diagram of CE-MS system under positive-ion mode (A) (Copyright 2016 American Chemical Society.) and negative-ion mode (B) (Copyright 2016 Springer.), respectively. (C) Charge-based separation of acidic oligosaccharides using negative-ion mode capillary isoelectric focusing (cIEF) mass spectrometry. Copyright 2019 American Chemical Society. (D) Diagram depicting the forces of electroosmotic flow and electrophoretic forces that act on analytes during a capillary zone electrophorasis (CZE)-MS experiment with three different inner capillary coatings, BFS (Bare fused silica), DMS (dichlorodimethylsilane) and AHS (N-(6-aminohexyl) aminomethyltriethoxysilane). Copyright 2018 Elsevier.
However, HS is negatively charged and highly sulfated HS chains may undergo desulfation when separated by normal polarity mode CE and analyzing by positive-ion MS. Such decomposition can result reducing analysis accuracy. As an alternative approach, using a protein-coated emitter with positively charged as the interface as well as adjusting the BGE, SL and electrodes allows a reverse polarity CE separation and negative-mode electrospray ionization MS (Figure 6B) [52]. This approach is useful in rapid bottom-up and top-down analysis of LMWH without analyte decomposition. CE-MS has the merits of low false positive results, simple data processing and is capable of detecting and identifying LMWH produced by different manufacturers.
Although a CE-MS interface offers a rapid analytical method and has been commercialized, the sensitivity and reproducibility of this method needs further improvement. Online reverse-polarity negative-ion mode capillary isoelectric focusing (cIEF)-MS based on an electrokinetically pumped sheath liquid nanospray CE-MS for the separation and detection of HS disaccharides (Figure 6C) [53]. Using a high voltage a pH gradient with low pH at the anode and high pH at the cathode is formed within the capillary. Therefore, an analyte with a low isoelectric point (pI) will flow first out of the anode side of the capillary and spray into the mass spectrometer for detection. This method enables separation of positional isomers of disaccharides with the same number of O-sulfo groups and also discriminates between N-sulfo groups and O-sulfo groups. Unfortunately, this method has some limitations in separating GAG analytes with very similar pI values.
By combing reverse-polarity capillary zone electrophoresis with negative-ion mode mass spectrometry (CZE-MS) an effective and sensitive tool has been developed for the analysis of GAGs mixtures (Figure 6D) [49]. Using a cation coated capillary, structurally similar sulfated GAG oligosaccharides and complex mixtures of the same were successfully separated and analyzed with CZE-MS in a fast and reproducible manner. The analysis method was applied to a mixture of heparin/HS oligosaccharides, varying chain length from trisaccharide to dodecasaccharides resulting in more than 80 molecular compositions being identified by accurate mass measurement.
Since the diameter capillary is very small it is prone to blockage and the capillary length has not been optimized or standardized so that the reproducibility of CE-MS analysis still has certain limitations. In addition, as with most liquid phase analyses, standards are required [49].
3.3. Nuclear magnetic resonance
Nuclear magnetic resonance (NMR) spectroscopy is based on the principle that an NMR signal changes base on the position of the detected nucleus within a molecule. NMR widely used in analyzing the molecular structure of HS/heparin (including conformation) and detecting sample content and purity [54,55,56]. Heparin contaminants and impurities are difficult to detect as they differ only in sulfo group position, degree of sulfation, and sugar epimers. Although a database of pure HS/heparin oligosaccharides is currently lacking, NMR is considered an advantageous means of determining the fine structure of heparin. A library consisting of 66 types of HS/heparin oligosaccharides of different sizes, sulfation levels and positions were by a biotechnological chemoenzymatic synthesis (Figure 7A–7C) [57]. The library contained diverse structures, including rare GlcNS3S6S, Glc2S and IdoA residues, which are found in low abundance in HS/heparin natural products but are important for a variety of biological activities. In the NMR analysis of this oligosaccharide library prepared using biotechnology, the unique anomeric protons showed broad peaks in the 1H NMR spectra and low signal intensity in 2D the spectra. The chemical shifts anomeric protons and carbons of the various uronic acid and glucosamine residues could be assembled in a table representing a valuable database. Signal strength and chemical shifts of residues such as, IdoA2S, could be significantly changed by adding ethylenediaminetetraacetic acid (EDTA) to remove trace amounts of divalent cations greatly improving the accuracy of NMR detection of HS/heparin oligosaccharides. This systematic NMR study paves the way for the characterization of more complicated HS/heparin polysaccharides (UFH) helping to safeguard the quality of UFH and ensuring the similarity of bioengineered heparin products with the current used animal-derived heparins.
Figure 7.
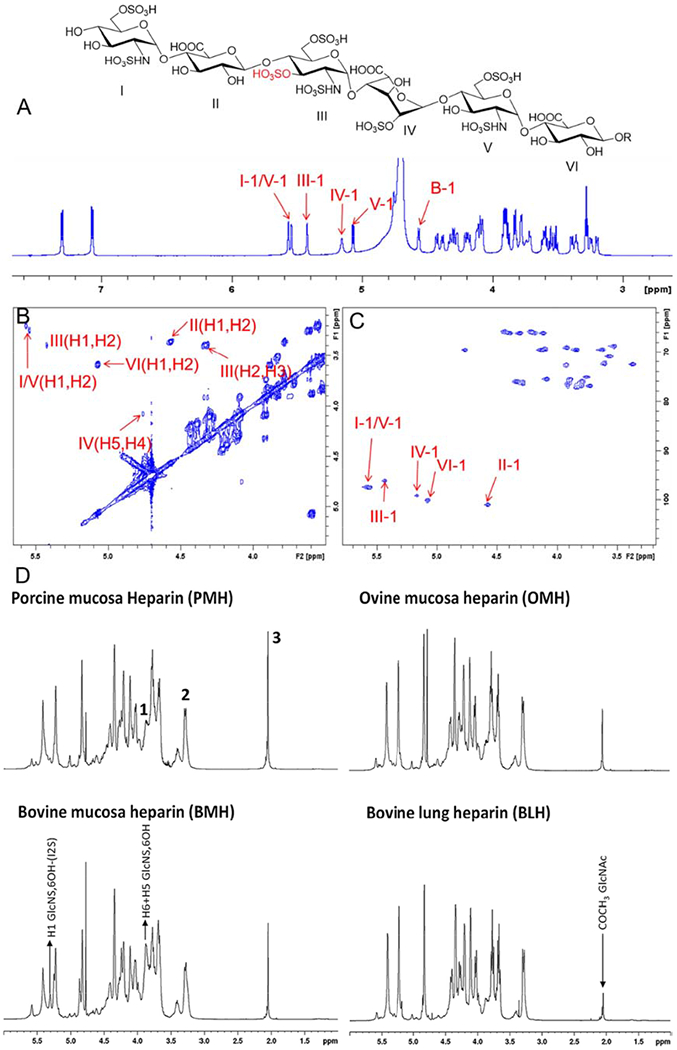
(A-C) Example of full NMR characterization of a heparin hexasaccharide prepared through chemoenzymatic synthesis. Copyright 2017 The Royal Society of Chemistry. (D) Heparin samples extracted from different species/organ. Signals 1, 2, and 3, corresponding to H5 and H6 of 6-O-desulfated glucosamine, H2 of N-sulfated glucosamine, and acetyl group of N-acetylated glucosamine, respectively, were used as diagnostic peaks for the determination of their origin. Copyright 2019 Frontiers.
In recent years, 2D NMR has shown unique advantages in the analysis of disaccharides and the quantitative detection of heparin and LMWH [58,59,60], and is capable of the detection of contaminants, such as oversulfated chondroitin sulfate (OSCS) in heparin [61,62], as well as the analysis of danaparoid sodium salt, an anticoagulant drug containing HS, DS and CS [63]. Furthermore, 2D NMR can be combined with SAX-HPLC to expand the range of detectable structural features, and become a powerful tool for distinguishing sources, monitoring manufacture and ensuring safety and quality of commercial prepared heparin [45]. In response to the call of FDA, which suggesting introducing bovine heparins to address the shortage of heparin drugs, a combination of 1D and 2D-HSQC NMR methods have been used to detect the origin or blending of heparins from different animals and tissues (Figure 7D) [64]. In addition to testing the purity and safety of HS/heparin oligosaccharide products, NMR can also be used to control the synthesis of heparin in standard and biotechnological production processes. 1H NMR was used to quantify heparosan, a precursor of bioengineered heparin, in a fermentation supernatant during its production from E. coli K5 and to optimize fermentation parameters and calculate the yield [65]. However the sensitivity of NMR analysis was insufficiently high to directly monitor heparosan, thus, there are still some limitations for the detection of GAGs.
3.4. Chemometric analysis
There are inevitably limitations when using a single analytical method such as NMR, HPLC-MS to analyze HS/heparin. Multiple assays can be used with chemometric analysis to build a more complete analytical method for HS/heparin analysis. 1H NMR analysis of intact heparin was used to determine structural differences among heparin products, and combined HPLC-MS analysis on disaccharides treated with heparin lyases to analyze disaccharide concentrations and bottom-up HPLC-MS analysis heparin lyase 2- resistant tetrasaccharides to determine differences in the anticoagulant activity on 30 heparin samples from different sources(Figure 8) [66]. Comprehensive analysis non-intersecting clusters obtained by principal component analysis (PCA) and the K-means, obtained from the visual data mining software, established a detailed database for meticulous analysis of heparin structure. This database was useful in analyzing the biological source of heparin and could also detect mixtures of these heparins prohibited by US FDA regulations. It is worth noting that the NMR spectrum, which provides an array of data of great significance as a reference, is particularly important in PCA and has important applications in many aspects, such as the identification of animal and tissue source, the purity and safety of biologically sourced heparins [67,68], the structural characterization of heparin derivatives [69], the identification of contaminants [70], the analysis of anticoagulant activity [71], and the detection of commercial heparin sources [72].
Figure 8.
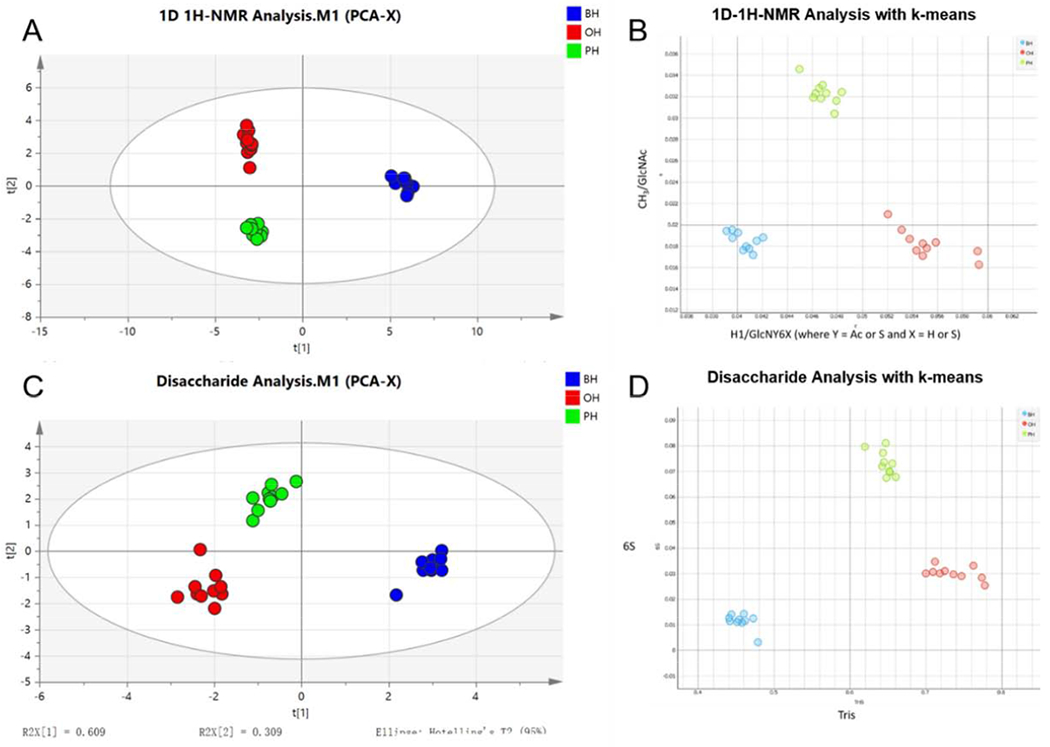
Statistical analysis of bovine, ovine, and porcine UFHs based on 1H NMR information (A,B) and LC-MS disaccharide analysis (C,D). PCA score plot of two first principle components and K-means clustering analysis of the NMR data and disaccharide data are presented. Copyright 2019 Elsevier.
3.5. SPR techniques
Surface plasmon resonance (SPR), analyzes the interaction between putative ligands and receptor molecules (i.e., enzymes, peptides, antibodies, and DNA) immobilized on a thin metal layer (i.e., silver, gold, or aluminum films) by detecting refractive index changes of the medium near the nanometal film based on optical biosensor technology. SPR is a label-free, high sensitivity, high repeatability, real time detection method[73,74]. With over three decades of use, SPR is considered to be one of the most efficient techniques for identifying affinity, specificity, and kinetic parameters between macromolecules (i.e., protein-polysaccharide, receptor-drug, enzyme-substrate, etc.) [75]. SPR can be used to detect the interaction of viruses with GAGs to understand viral pathogenesis and develop treatments. SPR technology for the study of Zika virus (ZIKV), recognized in 2016 by the World Health Organization (WHO) to cause severe congenital defects, was used to explain the role of GAGs in placenta and brain host cells for ZIKV entry (Figure 9A) [76]. ZIKV envelope protein (ZIKV E) has a strong affinity for long-chain heparin oligosaccharides (octasaccharide to octadecasaccharides) and heparins with higher sulfate levels. Human placenta chondroitin sulfate (CS) also binds more tightly to ZIKV E than does porcine brain HS, suggesting that placenta CS may be the most likely receptor mediating ZIKV invasion through the placenta. The porcine brain HS was enriched in trisulfated disaccharide presumably resulting in tighter binding ability ZIKV E. The exploration of the interaction of HS and CS with ZIKV E, using SPR, not only lays a foundation for further understanding ZIKV pathogenesis, but also provides a new approach studying the mechanism of virual infection. SPR is also useful for assessing the anticoagulant activity of heparin. Competitive SPR to test the ability of soluable heparin to compete with AT preventing its binding to heparin immobilized on a chip (Figure 9B–9E) [77]. This competitive binding method correlates well with standard bio assay method, and it reduces the required steps, shortens analysis time, and reduces assay costs while maintaining repeatability. However, the current instrument costs for performing SPR are extremely high limiting this method in points-of-care (PoC) assay until miniaturized and inexpensive SPR sensors become available [78].
Figure 9.
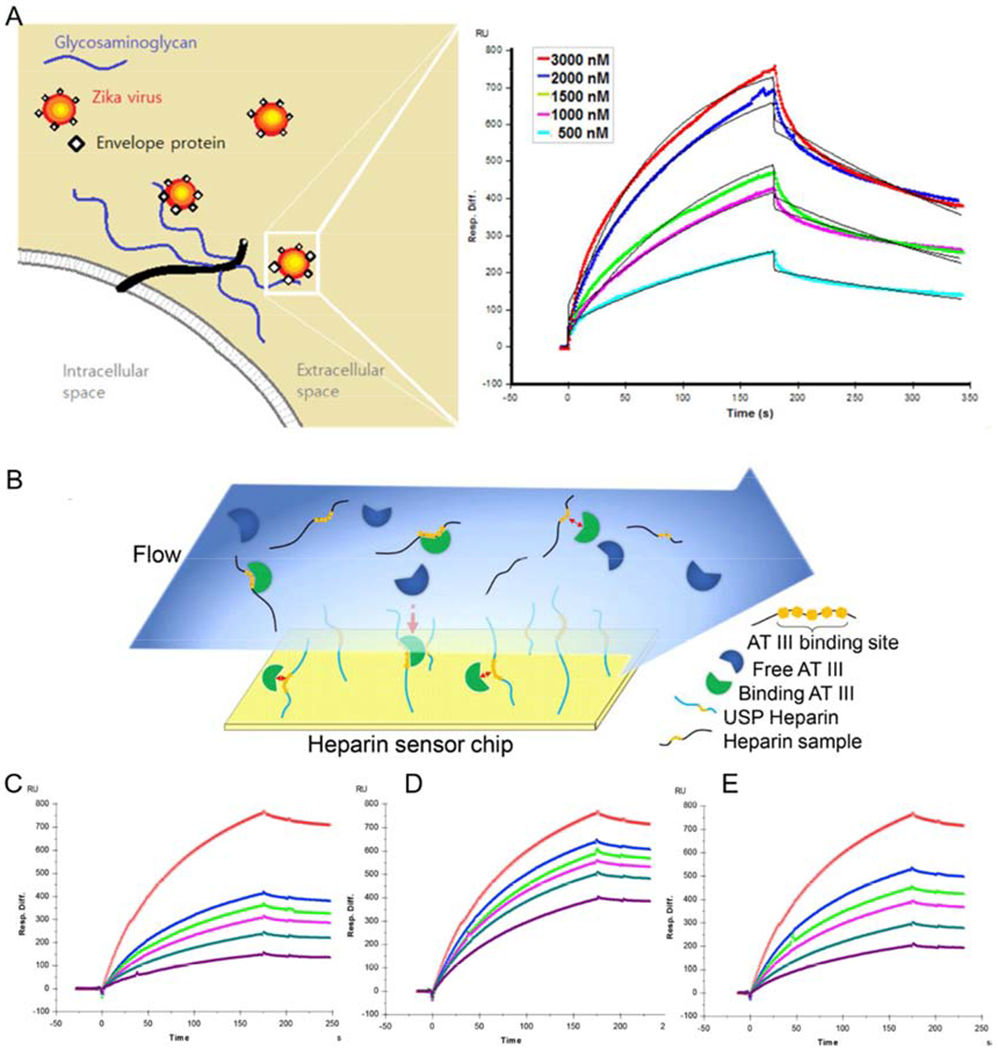
(A) Sensorgrams of interactions between ZIKV E protein and UFH. Copyright 2017 American Chemical Society. (B) Diagram of SPR solution competition experiment for anticoagulant activity measurement of heparin samples; (C) to (E): SPR sensorgrams of AT binding to heparin surface competing with different heparin samples, where (C): USP UFH standard; (D): LMWH (E): bovine lung derived UFH. Copyright 2017 Elsevier.
3.6. Enzyme-linked immunosorbent assay
The enzyme-linked immunosorbent assay (ELISA) is a highly sensitive bioassay method based on antigen-antibody binding that can be used in both qualitative and quantitative determinations[79,80]. ELISA with enzyme signal amplification has been used as a high-throughput platform for the detection of 3-OST sulfation of a bioengineered heparin precursor (a non-anticoagulant heparin missing its 3-O-sulfo group made in a biotechnological chemoenzymatic process (Figure 10A) [81]. Biotin-labeled UFH is immobilized on the inner surface of a 96-well plate coated with streptavidin. After the addition of AT and bioengineered heparin precursor for incubation and AT antibody was added, a secondary antibody horseradish peroxidase (HRP)-conjugated IgG, and 3,3’,5,5’-tetramethylbenzidine (TMB) substrate were then added for enzymatic signal amplification. The anticoagulant activity of the bioengineered heparin could then be determined by UV measurement to assess the progress of 3-OST catalyzed reaction. After improving detection performances such as shortening time and reducing costs by continuously optimizing the main factors affecting signal response. This sensitive and accurate detection method provides a new approach for ensuring the consistent biotechnology process for the industrial production of bioengineered heparin.
Figure 10.
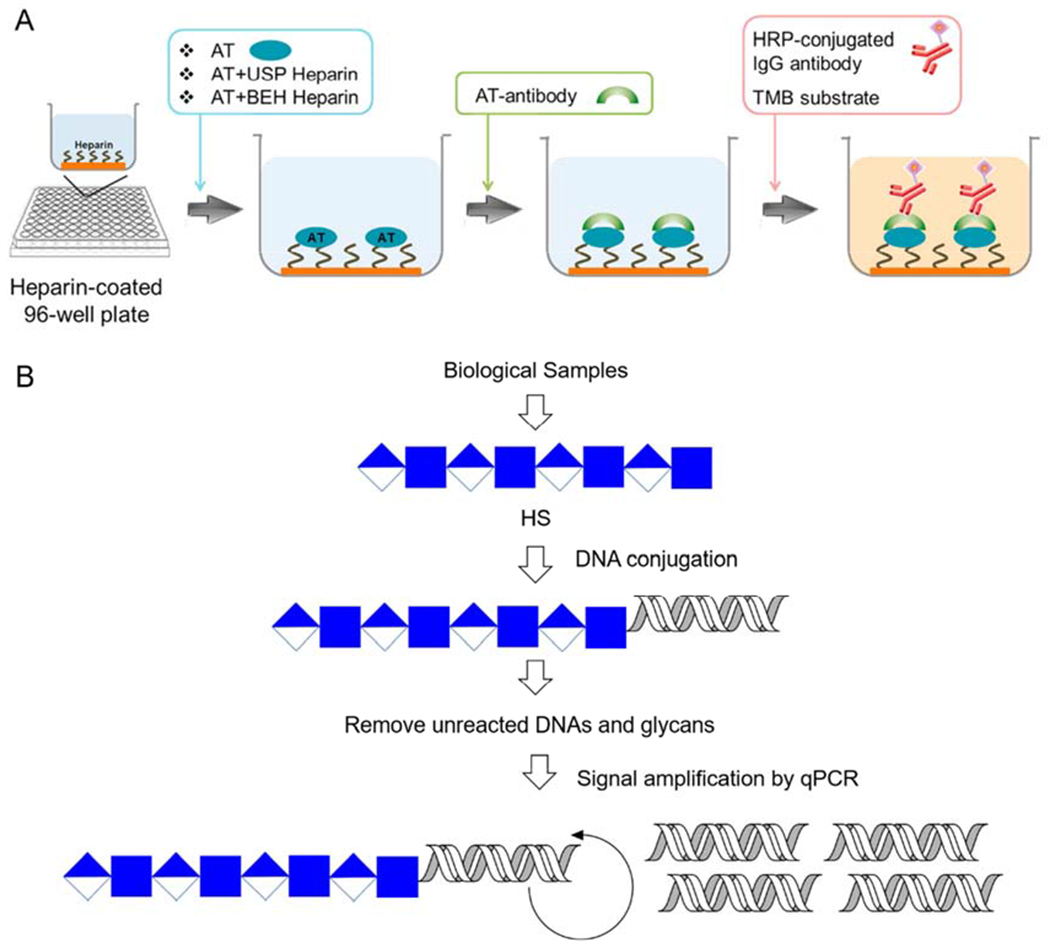
(A) Schematic representation of high-throughput method for in-process monitoring 3-OST sulfation in bioengineered heparin synthesis. Copyright 2019 Elsevier. (B) Schematic representation of glyco-qPCR. Target carbohydrates (GAGs) from biological samples can be conjugated with different DNA “bar code”, followed by removing unreacted DNA and detecting the corresponding GAG-DNA conjugates with amplified signals (qPCR).
3.7. Ultrasensitive analysis of heparin and heparan sulfate
During clinical procedures such as cardiovascular surgery and kidney dialysis, heparin levels need to be closely monitored to avoid complications such as hemorrhage induced by heparin overdose and HIT. The therapeutic dosing level of heparin is 2-8 U mL−1 (17-67 mM) during cardiovascular surgery and 0.2-1.2 U mL−1 (1.7-10 mM) in postoperative and long-term care [82]. Thus, simple, real-time continuous measurements of heparin levels in serum during cardiovascular surgery and the postoperative therapy period, is desirable. There is also a need for detection methods to monitor heparin in infusion solutions to avoid human dosing errors, particularly in pediatric patients. Traditional clinical methods for heparin detection rely on bioassays such as activated clotting time (ACT) or activated partial thromboplastin time (aPPT) [83]. These methods are often not unreliable and inaccurate are subject to interference and are difficult to perform in clinical settings. Thus, developing new methods for heparin detection, especially the ultrasensitive determination is desirable, not only facilitate clinic studies, but also improve our understanding of the biological roles of heparin involved in the critical biological processes that it regulates. However, ultrasensitive analysis of heparin is very challenging due to the low concentrations present in complex biological fluids (i.e., plasma or serum) as well as the chemical complexity of glycan chains present in UFH and LMWH drugs. Heparin has no natural chromophores or fluorophores, and because of its highly negative charge often shows poor ionization efficiency in MS analysis. Heparins cannot be detected by amplification methods such as those used to detect nucleic acids, and in contrast to proteins, there are few available carbohydrate-specific antibodies.
Polymerase chain reaction (PCR) is a technique capable of amplifying DNA fragments exponentially for ultrasensitive detection [84,85]. HS/heparin detection cannot be achieved signal amplification used to detect DNA. A biotin-labeled oligosaccharide conjugated to DNA “bar code” constructed and glyco-qPCR was explored as an ultrasensitive online detection platform giving higher sensitivity than MS and fluorescence detection (Figure 10B) [86]. Biotinylated CS disaccharide (as a model system) obtained by reductive amination with a DNA “bar code”, and unreacted DNA completely removed. The CS-DNA conjugate was then used as a template for Qpcr. The results showed that the platform could detect CS oligosaccharides at concentrations < 1 zmol. However, this platform can only analyze to detect the total content of a particular GAG. If complex detection, such as composition, is required, glycol-qPCR requires a micro-separation step. When coupled with CE, glyco-qPCR could detect as few as 500 molecules of CS present in Chinese hamster ovary (CHO) cells. These results suggest the possibility of ultrasensitive detection and the single-cell analysis of GAGs. This approach can also be used for the sensitive analysis of GAG-protein interactions providing a reliable basis for exploring the mechanism of GAGs actions and in drug development. Although these experiments only applied glyco-qPCR to the CS GAG it should be possible to apply this ultrasensitive method to the detection of HS/heparin.
This method is not yet sufficiently mature to apply to the analysis of more complex oligosacchene and polysaccharide analytes. The automation and integration of glyco-qPCR for the ultrasensitive analysis of heparin/HS oligosaccharides and polysaccharides should one day be possible. One major challenge is the complete removal of the unreacted DNA as residual DNA can adversely impact on the accuracy and sensitivity of the analysis.
Fluorescence resonance energy transfer (FRET), a technique that utilizes a phenomenon of fluorescence energy transfer that occurs at a suitable distance between the donor and acceptor fluorophores [87,88,89,90], has been applied to the ultrasensitive analysis of HS/heparin [91,92]. An approach for the detection of a chondroitin sulfate disaccharide as a model GAG oligosaccharide was developed based on FRET using a CdSe-ZnS core-shell nanocrystal quantum dot (QD) donor and a Cy5 acceptor [93]. This method allows the ultrasensitive detection of GAGs disaccharides with a sensitivity more than 1,000 times higher than CE-laser-induced fluorescence (LIF) (Figure 11A–11C). In addition, there are currently studies using fluorescent probes to detect heparin in plasma. Warttinger et al. using Heparin Red, a commercial fluorescent probe that can oligomerize polyanionic polysaccharides to quench fluorescence, can result in the quantitative detection of non-anticoagulant heparin in human plasma, such as N-desulfated-N-acetylated heparin and LMWH derivative, tafoxiparin (Figure 11D) [94].
Figure 11.
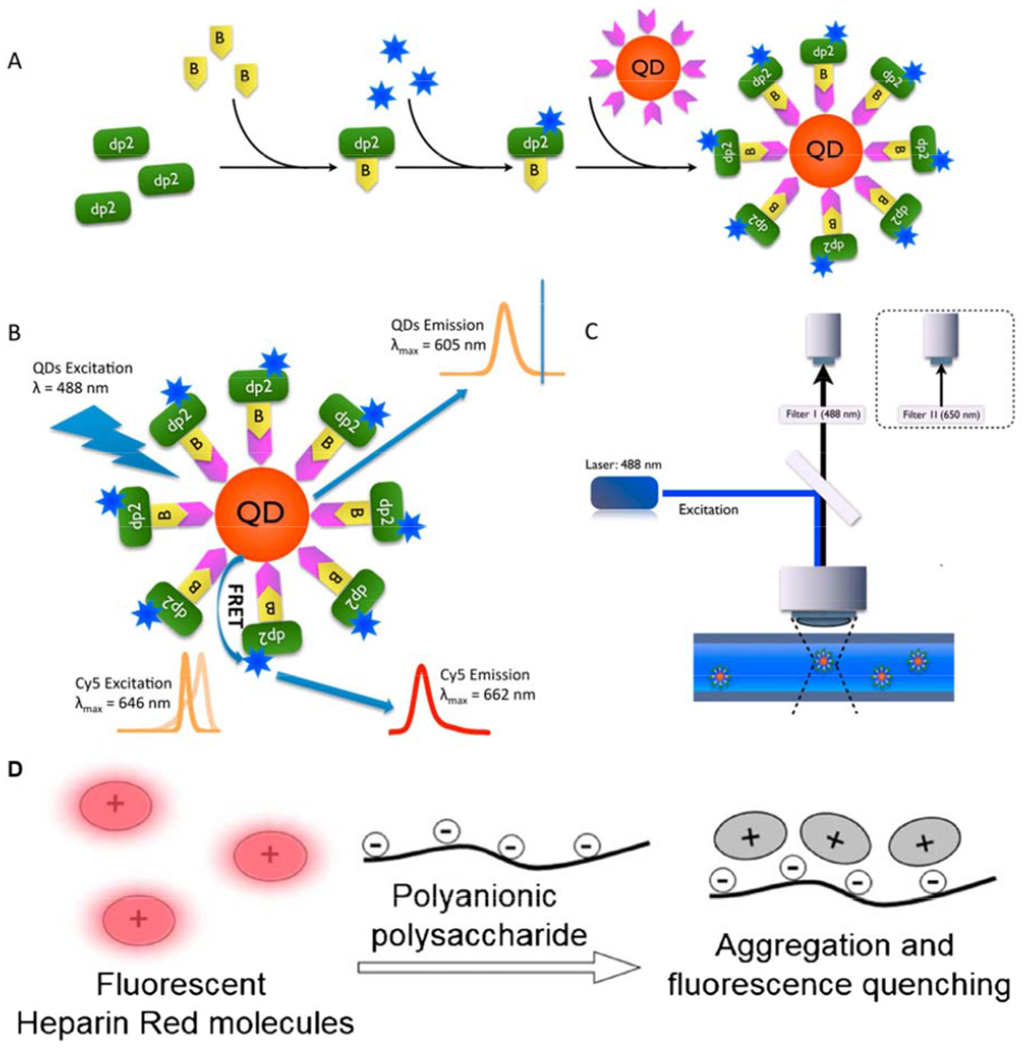
Scheme of the FRET system construction in this study. (A) Conjugation scheme of the FRET system. (B) The FRET donor, QD, is excited with a laser at 488 nm; because Cy5 dye, located on the same disaccharide, is close, FRET occurs and Cy5 is excited by emission from the QD, and the emission of Cy5 is then detected. (C) CE-LIF instrumental setup for FRET detection. A 488 nm (filter I, donor-acceptor channel) and 650 nm (filter II, acceptor channel) band-pass filter was used for FRET detection. Copyright 2013 American Chemical Society. (D) Schematic representation of fluorescence quenching of the molecular probe Heparin Red in the presence of polyanionic polysaccharides. Copyright 2016 Springer.
4. Conclusions and perspectives
HS and heparins are important GAGs that are responsible for a range of physiological and pharmacological functions. However, due to the high heterogeneity and complex structures, their analysis is very challenging, limiting their biotechnological synthesis, biological studies, and preventing the adequate addressing of safety concerns. After the heparin contamination crisis of 2007, the US FDA has promoted the diversification of heparin sources such as the reintroduction of bovine heparin or the introduction of bioengineered heparin into the US market. These urgings have stimulated the study of heparin analysis resulting in the steady improvement in this research field. MS is still the most widely used analytical tool particularly when coupled to separation methods, providing high sensitivity, high resolution, and high specificity. In particular, the development of MS and MS/MS in recent years has provided more comprehensive information on the fine structures and domain structures of HS/heparin and has greatly improved detection sensitivity. In addition, combined with stable isotope labeling, LC-MS/MS represents one of the most promising methods to investigate the roles of heparin/HS in metabolic pathways to distinguish exogenously administered stable isotope labeled heparin/HS from their endogeneously biosynthesized counterparts [95]. CE-MS has recently gained popularity with the introduction of new CE-MS interfaces due to its short separation time, high efficiency, and the small amounts of sample required. NMR is able to analysis disaccharide, characterize heparin and LMWH and distinguish contaminants present in UFH or LMWH drugs. Despite its relatively low sensitivity, NMR affords high resolution capable of assessing glycosidic bonds, uronic acid epimers and sulfo group substitution. Chemometric analysis can be used to combine a variety of analytical methods and apply statistics and machine learning to provide more comprehensive structural information. SPR uniquely and rapidly analyzes HS/heparin interactions with proteins in real time. The various analytical techniques introduced in this review can be applied to specific analytical problems or coupled to one another to facilitate the heparin/HS analysis. Reliable ultrasensitive analysis techniques for heparin are also needed clinically to avoid complications such as hemorrhage and HIT.
In the future, the analysis of HS/heparin will continue to develop resulting in even higher sensitivity, higher resolution, shorter analysis times, lower cost, more user-friendliness, higher throughput and improved automation. The development of analytical technology should promote a better understanding of the functions, pharmacodynamic characteristics, and pharmacokinetics of HS/heparin, the analysis of commercial drugs, and the characterization of biotechnological products such as bioengineered heparins and chemoenzymatically synthesized HS/heparin oligosaccharides.
Table 1.
Structures of heparin/HS disaccharide standards are shown where ΔUA corresponds to deoxy-α-l-threo-hex-4-enopyranosyluronic acid.
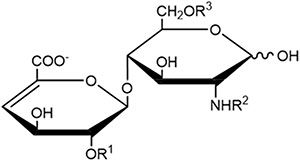 |
|||||
|---|---|---|---|---|---|
| HS Disaccharides | Structure | R1 | R2 | R3 | Molecular Weight |
| 0S | ΔUA(1,4)GlcNAc | H | Ac | H | 379.1115 |
| 2S | ΔUA2S(1,4)GlcNAc | SO3− | Ac | H | 459.0683 |
| 6S | ΔUA(1,4)GlcNAc6S | H | Ac | SO3− | 459.0683 |
| NS | ΔUA(1,4)GlcNS | H | SO3− | H | 417.0577 |
| 2S6S | ΔUA2S(1,4)GlcNAc6S | SO3− | Ac | SO3− | 539.0251 |
| NS2S | ΔUA2S(1,4)GlcNS | SO3− | SO3− | H | 497.0145 |
| NS6S | ΔUA(1,4)GlcNS6S | H | SO3− | SO3− | 497.0145 |
| TriS | ΔUA2S(1,4)GlcNS6S | SO3− | SO3− | SO3− | 576.9713 |
Highlights.
Heparan sulfates are structurally complex bioactive polysaccharides
The analysis of complex heterogeneous polysaccharides is difficult
Mass spectral analysis is a detailed and sensitive method for analysis
Hyphenated techniques and chemometric analyses are playing a greater role
Ultrasensitive methods are required for analyzing biological samples
Acknowledgments
Disclosure statement
The authors report no declarations of interest. The authors were supported in part by grants from Open Project Program of the Key Laboratory of Tropical Marine Bio-resources and Ecology, Chinese Academy of Sciences (LMB20201006 to XZ), Natural Science Research Project of Jiangsu Higher Education Institutions (19KJB150012 to LL) and the National Institutes of Health (CA231074 to RJL).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Zhang X, Lin L, Huang H, Linhardt RJ, Chemoenzymatic synthesis of glycosaminoglycans, Acc. Chem. Res 53 (2020) 335–346. [DOI] [PubMed] [Google Scholar]
- [2].Li JP, Kusche-Gullberg M, Heparan sulfate: Biosynthesis, structure, and function, in: Jeon KW(Eds.), International review of cell and molecular biology, Elsevier Academic Press Inc, San Diego, 2016, pp. 215–273. [DOI] [PubMed] [Google Scholar]
- [3].Fu L, Suflita M, Linhardt RJ, Bioengineered heparins and heparan sulfates, Adv. Drug Deliver. Rev 97 (2016) 237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Capila I, Linhardt RJ, Heparin - protein interactions, Angew. Chem. Int. Ed 41 (2002) 391–412. [DOI] [PubMed] [Google Scholar]
- [5].Xu D, Esko JD, Demystifying heparan sulfate-protein interactions, in: Kornberg RD(Eds.), Annu. Rev. Biochem, Annual Reviews, Palo Alto, 2014, pp. 129–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].van Wijk XMR, van Kuppevelt TH, Heparan sulfate in angiogenesis: A target for therapy, Angiogenesis. 17 (2014) 443–462. [DOI] [PubMed] [Google Scholar]
- [7].Zhang X, Linhardt RJ, Chemoenzymatic synthesis of low-molecular-weight heparin and heparan sulfate, in: Tan Z, Wang LX(Eds.), Chemical biology of glycoproteins, The Royal Society of Chemistry Cambridge, 2017, pp. 233–252. [Google Scholar]
- [8].Kamhi E, Joo EJ, Dordick JS, Linhardt RJ, Glycosaminoglycans in infectious disease, Biol. Rev 88 (2013) 928–943. [DOI] [PubMed] [Google Scholar]
- [9].Liu J, Linhardt RJ, Chemoenzymatic synthesis of heparan sulfate and heparin, Nat. Prod. Rep 31 (2014) 1676–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hemker HC, A century of heparin: Past, present and future, J. Thromb. Haemost 14 (2016) 2329–2338. [DOI] [PubMed] [Google Scholar]
- [11].Spadarella G, Di Minno A, Donati MB, Mormile M, Ventre I, Di Minno G, From unfractionated heparin to pentasaccharide: Paradigm of rigorous science growing in the understanding of the in vivo thrombin generation, Blood Rev. 39 (2020) 14. [DOI] [PubMed] [Google Scholar]
- [12].Lee GM, Arepally GM, Heparin-induced thrombocytopenia, Hematol.-AM. Soc. Hemat (2013) 668–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Babin JL, Traylor KL, Witt DM, Laboratory monitoring of low-molecular-weight heparin and fondaparinux, Semin. Thromb. Hemost 43 (2017) 261–269. [DOI] [PubMed] [Google Scholar]
- [14].Mulloy B, Hogwood J, Gray E, Lever R, Page CP, Pharmacology of heparin and related drugs, Pharmacol. Rev 68 (2016) 76–141. [DOI] [PubMed] [Google Scholar]
- [15].Petitou M, van Boeckel CAA, A synthetic antithrombin iii binding pentasaccharide is now a drug! What comes next?, Angew. Chem.-Int. Edit 43 (2004) 3118–3133. [DOI] [PubMed] [Google Scholar]
- [16].Cress BF, Bhaskar U, Vaidyanathan D, Williams A, Cai C, Liu XY, Fu L, M-Chari V, Zhang FM, Mousa SA, Dordick JS, Koffas MAG, Linhardt RJ, Heavy heparin: A stable isotope-enriched, chemoenzymatically-synthesized, poly-component drug, Angew. Chem. Int. Ed 58 (2019) 5962–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Xu YM, Chandarajoti K, Zhang X, V Pagadala, Dou WF, Hoppensteadt DM, Sparkenbaugh EM, Cooley B, Daily S, Key NS, Severynse-Stevens D, Fareed J, Linhardt RJ, Pawlinski R, Liu J, Synthetic oligosaccharides can replace animal-sourced low-molecular weight heparins, Sci. Transl. Med 9 (2017) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Xu YM, Masuko S, Takieddin M, Xu HM, Liu RP, Jing J, S.A. Mousa, Linhardt RJ, Liu J, Chemoenzymatic synthesis of homogeneous ultralow molecular weight heparins, Science. 334 (2011) 498–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Keire D, Manufacturing heparin with equivalent chemical composition from different animal sources, Thromb. Haemostasis 119 (2019) 688–688. [DOI] [PubMed] [Google Scholar]
- [20].Yu YL, Chen Y, Mikael P, Zhang FM, Stalcup AM, German R, Gould F, Ohlemacher J, Zhang H, Linhardt RJ, Surprising absence of heparin in the intestinal mucosa of baby pigs, Glycobiology. 27 (2017) 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Szajek AY, Chess E, Johansen K, Gratzl G, Gray E, Keire D, Linhardt RJ, Liu J, Morris T, Mulloy B, Nasr M, Shriver Z, Torralba P, Viskov C, Williams R, Woodcock J, Workman W, Al-Hakim A, The us regulatory and pharmacopeia response to the global heparin contamination crisis, Nat. Biotechnol 34 (2016) 625–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Liu XY, St Ange K, Lin L, Zhang FM, Chi LL, Linhardt RJ, Top-down and bottom-up analysis of commercial enoxaparins, J. Chromatogr. A 1480 (2017) 32–40. [DOI] [PubMed] [Google Scholar]
- [23].Beni S, Limtiaco JF, Larive CK, Analysis and characterization of heparin impurities, Anal. Bioanal. Chem 399 (2011) 527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Volpi N, Maccari F, Suwan J, Linhardt RJ, Electrophoresis for the analysis of heparin purity and quality, Electrophoresis. 33 (2012) 1531–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Tripathi CKM, Banga J, Mishra V, Microbial heparin/heparan sulphate lyases: Potential and applications, Appl. Microbiol. Biot 94 (2012) 307–321. [DOI] [PubMed] [Google Scholar]
- [26].Yates EA, Rudd TR, Recent innovations in the structural analysis of heparin, Int. J. Cardiol 212 (2016) S5–S9. [DOI] [PubMed] [Google Scholar]
- [27].Wang ZJ, Zhang TJ, Xie SS, Liu XY, Li HM, Linhardt RJ, Chi LL, Sequencing the oligosaccharide pool in the low molecular weight heparin dalteparin with offline hplc and esi-ms/ms, Carbohydr. Polym 183 (2018) 81–90. [DOI] [PubMed] [Google Scholar]
- [28].Zhang FM, Yang B, Ly M, Solakyildirim K, Xiao ZP, Wang ZY, Beaudet JM, Torelli AY, Dordick JS, Linhardt RJ, Structural characterization of heparins from different commercial sources, Anal. Bioanal. Chem 401 (2011) 2793–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Yang B, Solakyildirim K, Chang YQ, Linhardt RJ, Hyphenated techniques for the analysis of heparin and heparan sulfate, Anal. Bioanal. Chem 399 (2011) 541–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Sun XJ, Sheng AR, Liu XY, Shi F, Jin L, Xie SS, Zhang FM, Linhardt RJ, Chi LL, Comprehensive identification and quantitation of basic building blocks for low-molecular weight heparin, Anal. Chem 88 (2016) 7738–7744. [DOI] [PubMed] [Google Scholar]
- [31].Laremore TN, Murugesan S, Park TJ, Y Avci F, V Zagorevski D, Linhardt RJ, Matrix-assisted laser desorption/ionization mass spectrometric analysis of uncomplexed highly sulfated oligosaccharides using ionic liquid matrices, Anal. Chem 78 (2006) 1774–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Eliuk S, Makarov A, Evolution of orbitrap mass spectrometry instrumentation, in: Cooks RG, Pemberton JE(Eds.), Annual review of analytical chemistry, Annual Reviews, Palo Alto, 2015, pp. 61–80. [DOI] [PubMed] [Google Scholar]
- [33].Thanawiroon C, Rice KG, Toida T, Linhardt RJ, Liquid chromatography/mass spectrometry sequencing approach for highly sulfated heparin-derived oligosaccharides, J. Biol. Chem 279 (2004) 2608–2615. [DOI] [PubMed] [Google Scholar]
- [34].Hitchcock AM, Yates KE, Costello CE, Zaia J, Comparative glycomics of connective tissue glycosaminoglycans, Proteomics. 8 (2008) 1384–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Li L, Zhang F, Zaia J, Linhardt RJ, Top-down approach for the direct characterization of low molecular weight heparins using lc-ft-ms, Anal. Chem 84 (2012) 8822–8829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Y. Li G, Cai C, Li LY, Fu L, Chang YQ, Zhang FM, Toida T, Xue CH, Linhardt RJ, Method to detect contaminants in heparin using radical depolymerization and liquid chromatography-mass spectrometry, Anal. Chem 86 (2014) 326–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Y Li G, Steppich J, Wang ZY, Sun Y, Xue CH, Linhardt RJ, Li LY, Bottom-up low molecular weight heparin analysis using liquid chromatography-fourier transform mass spectrometry for extensive characterization, Anal. Chem 86 (2014) 6626–6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Yang B, Weyers A, Y. Baik J, Sterner E, Sharfstein S, Mousa SA, Zhang FM, Dordick JS, Linhardt RJ, Ultra-performance ion-pairing liquid chromatography with on-line electrospray ion trap mass spectrometry for heparin disaccharide analysis, Anal. Biochem 415 (2011) 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Doneanu CE, Chen WB, Gebler JC, Analysis of oligosaccharides derived from heparin by ion-pair reversed-phase chromatography/mass spectrometry, Anal. Chem 81 (2009) 3485–3499. [DOI] [PubMed] [Google Scholar]
- [40].Zhang ZQ, Xie J, Liu HY, Liu J, Linhardt RJ, Quantification of heparan sulfate disaccharides using ion-pairing reversed-phase microflow high-performance liquid chromatography with electrospray ionization trap mass spectrometry, Anal. Chem 81 (2009) 4349–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Campo GM, Campo S, Ferlazzo AM, Vinci R, Calatroni A, Improved high-performance liquid chromatographic method to estimate aminosugars and its application to glycosaminoglycan determination in plasma and serum, J. Chromatogr. B: Analyt. Technol. Biomed. Life Sci 765 (2001) 151–160. [DOI] [PubMed] [Google Scholar]
- [42].Thanawiroon C, Rice KG, Toida T, Linhardt RJ, Liquid chromatography/mass spectrometry sequencing approach for highly sulfated heparin-derived oligosaccharides, J. Biol. Chem 279 (2004) 2608–2615. [DOI] [PubMed] [Google Scholar]
- [43].Ouyang YL, Zeng YY, Rong YX, Song Y, Shi L, Chen B, Yang XL, Xu NY, Linhardt RJ, Zhang ZQ, Profiling analysis of low molecular weight heparins by multiple heart-cutting two dimensional chromatography with quadruple time-of-flight mass spectrometry, Anal. Chem 87 (2015) 8957–8963. [DOI] [PubMed] [Google Scholar]
- [44].Yi L, Zhang QH, Meng Y, Hao J, Xie BY, Gan H, Li DX, Dong K, Zhang ZQ, Qualitative and quantitative analysis of 2, 5-anhydro-d-mannitol in low molecular weight heparins with high performance anion exchange chromatography hyphenated quadrupole time of flight mass spectrometry, J. Chromatogr. A 1569 (2018) 160–167. [DOI] [PubMed] [Google Scholar]
- [45].Spelta F, Liverani L, Peluso A, Marinozzi M, Ursa E, Guerrini M, Naggi A, Sax-hplc and hsqc nmr spectroscopy: Orthogonal methods for characterizing heparin batches composition, Front. Med 6 (2019) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Stickney M, Sanderson P, Leach FE, Zhang F, Linhardt RJ, Amster IJ, Online capillary zone electrophoresis negative electron transfer dissociation tandem mass spectrometry of glycosaminoglycan mixtures, Int. J. Mass Spectrom 445 (2019) 116209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ucakturk E, Cai C, Li L, Li G, Zhang F, Linhardt RJ, Capillary electrophoresis for total glycosaminoglycan analysis, Anal. Bioanal. Chem 406 (2014) 4617–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Solakyildirim K, Recent advances in glycosaminoglycan analysis by various mass spectrometry techniques, Anal. Bioanal. Chem 411 (2019) 3731–3741. [DOI] [PubMed] [Google Scholar]
- [49].Sanderson P, Stickney M, Leach FE, Xia QW, Yu YL, Zhang FM, Linhardt R, Amster IJ, Heparin/heparan sulfate analysis by covalently modified reverse polarity capillary zone electrophoresis-mass spectrometry, J. Chromatogr. A 1545 (2018) 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Yang B, Solakyildirim K, Chang Y, Linhardt RJ, Hyphenated techniques for the analysis of heparin and heparan sulfate, Anal. Bioanal. Chem 399 (2011) 541–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Sun X, Lin L, Liu X, Zhang F, Chi L, Xia Q, Linhardt RJ, Capillary electrophoresis-mass spectrometry for the analysis of heparin oligosaccharides and low molecular weight heparin, Anal. Chem 88 (2016) 1937–1943. [DOI] [PubMed] [Google Scholar]
- [52].Lin L, Liu X, Zhang F, Chi L, Amster IJ, Leach FE, Xia Q, Linhardt RJ, Analysis of heparin oligosaccharides by capillary electrophoresis-negative-ion electrospray ionization mass spectrometry, Anal. Bioanal. Chem 409 (2016) 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Ouyang YL, Han XR, Xia QW, Chen JL, Velagapudi S, Xia K, Zhang ZQ, Linhardt RJ, Negative-ion mode capillary isoelectric focusing mass spectrometry for charge-based separation of acidic oligosaccharides, Anal. Chem 91 (2019) 846–853. [DOI] [PubMed] [Google Scholar]
- [54].Hricovini M, Guerrini M, Bisio A, Torri G, Petitou M, Casu B, Conformation of heparin pentasaccharide bound to antithrombin iii, Biochem. J 359 (2001) 265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Guerrini M, Naggi A, Guglieri S, Santarsiero R, Torri G, Complex glycosaminoglycans: Profiling substitution patterns by two-dimensional nuclear magnetic resonance spectroscopy, Anal. Biochem 337 (2005) 35–47. [DOI] [PubMed] [Google Scholar]
- [56].Mauri L, Boccardi G, Torri G, Karfunkle M, Macchi E, Muzi L, Keire D, Guerrini M, Qualification of hsqc methods for quantitative composition of heparin and low molecular weight heparins, J. Pharmaceut. Biomed 136 (2017) 92–105. [DOI] [PubMed] [Google Scholar]
- [57].Zhang X, Pagadala V, Jester HM, Lim AM, Pham TQ, Goulas AMP, Liu J, Linhardt RJ, Chemoenzymatic synthesis of heparan sulfate and heparin oligosaccharides and nmr analysis: Paving the way to a diverse library for glycobiologists, Chem. Sci 8 (2017) 7932–7940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Mauri L, Boccardi G, Torri G, Karfunkle M, Macchi E, Muzi L, Keire D, Guerrini M, Qualification of HSQC methods for quantitative composition of heparin and low molecular weight heparins, J. Pharm. Biomed. Anal 136 (2017) 92–105. [DOI] [PubMed] [Google Scholar]
- [59].Langeslay DJ, Beecher CN, Naggi A, Guerrini M, Torri G, Larive CK, Characterizing the microstructure of heparin and heparan sulfate using N-sulfoglucosamine H-1 and H-5 NMR chemical shift analysis, Anal. Chem 85 (2013) 1247–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Guerrini M, Rudd TR, Mauri L, Macchi E, Fareed J, Yates EA, Naggi A, Torri G, Differentiation of generic enoxaparins marketed in the united states by employing nmr and multivariate analysis, Anal. Chem 87 (2015) 8275–8283. [DOI] [PubMed] [Google Scholar]
- [61].Guerrini M, Zhang ZQ, Shriver Z, Naggi A, Masuko S, Langer R, Casu B, Linhardt RJ, Torri G, Sasisekharan R, Orthogonal analytical approaches to detect potential contaminants in heparin, Proc. Natl. Acad. Sci. U. S. A 106 (2009) 16956–16961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Sassaki GL, Riter DS, Santana AP, Guerrini M, Lima MA, Cosentino C, Souza LM, Cipriani TR, Rudd TR, Nader HB, Yates EA, Gorin PAJ, Torri G, Iacomini M, A robust method to quantify low molecular weight contaminants in heparin: Detection of tris(2-n-butoxyethyl) phosphate, Analyst. 136 (2011) 2330–2338. [DOI] [PubMed] [Google Scholar]
- [63].Gardini C, Urso E, Guerrini M, van Herpen R, de Wit P, Naggi A, Characterization of danaparoid complex extractive drug by an orthogonal analytical approach, Molecules. 22 (2017) 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Mauri L, Marinozzi M, Phatak N, Karfunkle M, St Ange K, Guerrini M, Keire DA, Linhardt RJ, 1D and 2D-HSQC NMR: Two methods to distinguish and characterize heparin from different animal and tissue sources, Front. Med.-Lausanne 6 (2019) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Wang Z, Zhang Z, McCallum SA, Linhardt RJ, Nuclear magnetic resonance quantification for monitoring heparosan k5 capsular polysaccharide production, Anal. Biochem 398 (2010) 275–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Ouyang YL, Han XR, Yu YL, Chen JL, Fu L, Zhang FM, Linhardt RJ, Fareed J, Hoppensteadt D, Jeske W, Kouta A, Zhang ZQ, Xia K, Chemometric analysis of porcine, bovine and ovine heparins, J. Pharmaceut. Biomed 164 (2019) 345–352. [DOI] [PubMed] [Google Scholar]
- [67].Lima MA, Rudd TR, de Farias EHC, Ebner LF, Gesteira TF, de Souza LM, Mendes A, Cordula CR, Martins JRM, Hoppensteadt D, Fareed J, Sassaki GL, Yates EA, Tersariol ILS, Nader HB, A new approach for heparin standardization: Combination of scanning UV spectroscopy, nuclear magnetic resonance and principal component analysis, PLoS One 6 (2011) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Mauri L, Marinozzi M, Mazzini G, Kolinski RE, Karfunkle M, Keire DA, Guerrini M, Combining nmr spectroscopy and chemometrics to monitor structural features of crude heparin, Molecules. 22 (2017) 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Rudd TR, Skidmore MA, Guimond SE, Cosentino C, Torri G, Fernig DG, Lauder RM, Guerrini M, Yates EA, Glycosaminoglycan origin and structure revealed by multivariate analysis of NMR and CD spectra, Glycobiology. 19 (2009) 52–67. [DOI] [PubMed] [Google Scholar]
- [70].Rudd TR, Gaudesi D, Skidmore MA, Ferro M, Guerrini M, Mulloy B, Torri G, Yates EA, Construction and use of a library of bona fide heparins employing H-1 NMR and multivariate analysis, Analyst. 136 (2011) 1380–1389. [DOI] [PubMed] [Google Scholar]
- [71].Monakhova YB, Fareed J, Yao YM, Diehl BWK, Anticoagulant activity of porcine heparin: Structural-property relationship and semi-quantitative estimation by nuclear magnetic resonance (nmr) spectrometry, J. Pharm. Biomed. Anal 174 (2019) 639–643. [DOI] [PubMed] [Google Scholar]
- [72].Monakhova YB, Fareed J, Yao YM, B.W.K. Diehl, Improving reliability of chemometric models for authentication of species origin of heparin by switching from 1D to 2D nmr experiments, J. Pharm. Biomed. Anal 153 (2018) 168–174. [DOI] [PubMed] [Google Scholar]
- [73].Zhang F, Lee KB, Linhardt RJ, Spr biosensor probing the interactions between timp-3 and heparin/gags, Biosensors. 5 (2015) 500–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Zhang F, Beaudet JM, Luedeke DM, Walker RG, Thompson TB, Linhardt RJ, Analysis of the interaction between heparin and follistatin and heparin and follistatin-ligand complexes using surface plasmon resonance, Biochemistry. 51 (2012) 6797–6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Nguyen HH, Park J, Kang S, Kim M, Surface plasmon resonance: A versatile technique for biosensor applications, Sensors (Basel). 15 (2015) 10481–10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Y Kim S, Zhao J, Liu X, Fraser K, Lin L, Zhang X, Zhang F, Dordick JS, Linhardt RJ, Interaction of zika virus envelope protein with glycosaminoglycans, Biochemistry. 56 (2017) 1151–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Zhao J, Liu X, Malhotra A, Li Q, Zhang F, Linhardt RJ, Novel method for measurement of heparin anticoagulant activity using spr, Anal. Biochem 526 (2017) 39–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Prabowo BA, Purwidyantri A, Liu KC, Surface plasmon resonance optical sensor: A review on light source technology, Biosensors. 8 (2018) 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Gan SD, Patel KR, Enzyme immunoassay and enzyme-linked immunosorbent assay, J. Invest. Dermatol 133 (2013) e12. [DOI] [PubMed] [Google Scholar]
- [80].Dobrovolskaia E, Gam A, Slater JE, Competition enzyme-linked immunosorbant assay (elisa) can be a sensitive method for the specific detection of small quantities of allergen in a complex mixture, Clin. Exp. Allergy 36 (2006) 525–530. [DOI] [PubMed] [Google Scholar]
- [81].Lin L, Yu Y, Zhang F, Zhang X, Linhardt RJ, High-throughput method for in process monitoring of 3-o-sulfotransferase catalyzed sulfonation in bioengineered heparin synthesis, Anal. Biochem 586 (2019) 113419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Y. Zhan R, Fang Z, Liu B, Naked-eye detection and quantification of heparin in serum with a cationic polythiophene, Anal. Chem 82 (2010) 1326–1333. [DOI] [PubMed] [Google Scholar]
- [83].Murray DJ, Brosnahan WJ, Pennell B, Kapalanski D, Weiler JM, Olson J, Heparin detection by the activated coagulation time: A comparison of the sensitivity of coagulation tests and heparin assays, J. Cardiothor. Vasc. An 11 (1997) 24–28. [DOI] [PubMed] [Google Scholar]
- [84].Pecorini S, Camurri G, Torrini L, Ferraresi R, Highly sensitive real-time pcr method to identify species origin in heparinoids, Anal. Bioanal. Chem 412 (2020) 289–298. [DOI] [PubMed] [Google Scholar]
- [85].Auguste C, Dereux S, Martinez C, Anger P, New developments in quantitative polymerase chain reaction applied to control the quality of heparins, Anal. Bioanal. Chem 399 (2011) 747–755. [DOI] [PubMed] [Google Scholar]
- [86].Kwon SJ, Lee KB, Solakyildirim K, Masuko S, Ly M, Zhang F, Li L, Dordick JS, Linhardt RJ, Signal amplification by glyco-qpcr for ultrasensitive detection of carbohydrates: Applications in glycobiology, Angew. Chem. Int. Ed 51 (2012) 11800–11804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Dai JN, Ma CG, Zhang P, Fu YQ, Shen BX, Recent progress in the development of fluorescent probes for detection of biothiols, Dyes Pigment. 177 (2020) 13. [Google Scholar]
- [88].Shen BX, Qian Y, Red emission cysteine probe with high selectivity based on fluorescent protein chromophores and turn-on fluorescence in cell cultures, Dyes Pigment. 166 (2019) 350–356. [Google Scholar]
- [89].Shen BX, Wang LF, Zhi X, Qian Y, Construction of a red emission bodipy-based probe for tracing lysosomal viscosity changes in culture cells, Sens. Actuator B-Chem 304 (2020) 9. [Google Scholar]
- [90].Shen BX, Zhu WY, Zhi X, Qian Y A lysosome targeting probe based on fluorescent protein chromophore for selectively detecting GSH and Cys in living cells, Talanta 208 (2020) 7. [DOI] [PubMed] [Google Scholar]
- [91].Long Q, Zhao JN, Yin BD, Li HT, Zhang YY, Yao SZ, A novel label-free upconversion fluorescence resonance energy transfer-nanosensor for ultrasensitive detection of protamine and heparin, Anal. Biochem 477 (2015) 28–34. [DOI] [PubMed] [Google Scholar]
- [92].Xu JJ, Takai A, Takeuchi M, Red-green-blue trichromophoric nanoparticles with dual fluorescence resonance energy transfer: Highly sensitive fluorogenic response toward polyanions, Chem.-Eur. J 22 (2016) 13014–13018. [DOI] [PubMed] [Google Scholar]
- [93].Chang YQ, Cai C, Li LY, Miao JJ, Ucakturk E, Li GY, Ly M, Linhardt RJ, Ultrasensitive detection and quantification of acidic disaccharides using capillary electrophoresis and quantum dot-based fluorescence resonance energy transfer, Anal. Biochem 85 (2013) 9356–9362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Warttinger U, Giese C, Harenberg J, Holmer E, Kramer R, A fluorescent probe assay (heparin red) for direct detection of heparins in human plasma, Anal. Bioanal Chem 408 (2016) 8241–8251. [DOI] [PubMed] [Google Scholar]
- [95].Zhang X, Han XR, Xia K, Xu YM, Yang YM, Oshima K, Haeger SM, Perez MJ, McMurtry SA, Hippensteel JA, Ford JA, Herson PS, Liu J, Schmidt EP, Linhardt RJ, Circulating heparin oligosaccharides rapidly target the hippocampus in sepsis, potentially impacting cognitive functions, Proc. Natl. Acad. Sci. USA 116 (2019) 9208–9213. [DOI] [PMC free article] [PubMed] [Google Scholar]


