Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is typically very mild and often asymptomatic in children. A complication is the rare multisystem inflammatory syndrome in children (MIS-C) associated with COVID-19, presenting 4–6 weeks after infection as high fever, organ dysfunction, and strongly elevated markers of inflammation. The pathogenesis is unclear but has overlapping features with Kawasaki disease suggestive of vasculitis and a likely autoimmune etiology. We apply systems-level analyses of blood immune cells, cytokines, and autoantibodies in healthy children, children with Kawasaki disease enrolled prior to COVID-19, children infected with SARS-CoV-2, and children presenting with MIS-C. We find that the inflammatory response in MIS-C differs from the cytokine storm of severe acute COVID-19, shares several features with Kawasaki disease, but also differs from this condition with respect to T cell subsets, interleukin (IL)-17A, and biomarkers associated with arterial damage. Finally, autoantibody profiling suggests multiple autoantibodies that could be involved in the pathogenesis of MIS-C.
Keywords: COVID-19, multisystem inflammatory syndrome in children, MIS-C, SARS-CoV-2, Kawasaki disease, hyperinflammation in children, IL-17A, systems immunology, autoantibodies
Graphical Abstract
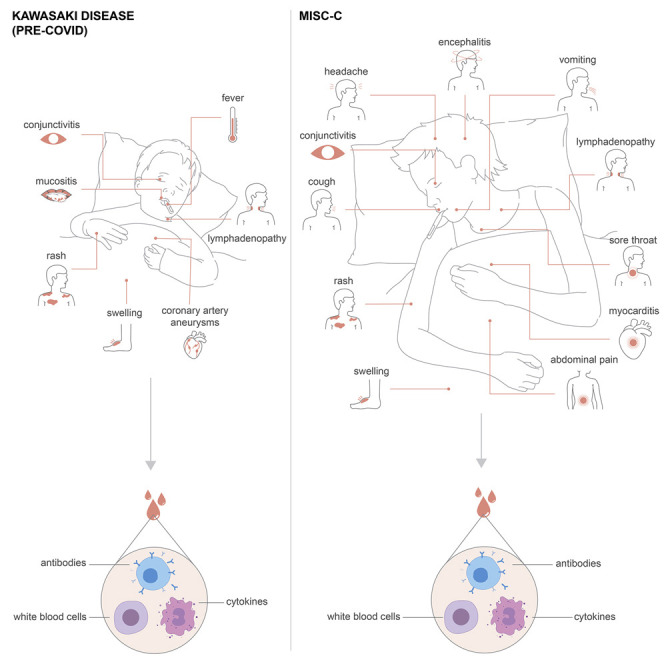
A systems immunology approach describes how multisystem inflammatory syndrome in children (MIS-C) is distinct from Kawasaki disease as well as the cytokine storm associated with severe COVID-19 in terms of its molecular and immune profiles.
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) first emerged in Wuhan in December 2019 (Huang et al., 2020) and then spread rapidly to a number of countries and, in particular, to Europe and the northern regions of Italy in the early weeks of February 2020. The first reports from China showed that children presented with milder symptoms as compared to adults infected by SARS-CoV-2 (Lu et al., 2020). Reasons for this have not been established, but several theories have been discussed, involving immune system differences such as thymic function, cross-reactive immunity to common cold coronaviruses, as well as differences in the expression of the viral entry receptor ACE2, as well as a better overall health status among children as compared to the elderly (Brodin, 2020). The mild COVID-19 in children has been confirmed also in areas with high disease prevalence such as northern Italy (Parri et al., 2020). The view that COVID-19 disease course is always mild in children is now challenged by recent reports of children presenting with a rare, but very severe hyperinflammatory syndrome in the United Kingdom (Riphagen et al., 2020; Whittaker et al., 2020), Italy (Verdoni et al., 2020), Spain (Moraleda et al., 2020), and New York City (Cheung et al., 2020). In these case series, children present with high fevers, and a variable number of symptoms previously associated with Kawasaki disease such as conjunctivitis, lymphadenopathy, mucocutaneous rash, and coronary artery dilation, and in the most severe cases, cardiovascular shock, encephalitis, and multiple organ failure.
Kawasaki disease is a vasculitis affecting medium-sized arteries with highest incidence in children younger than 5 years old, and it is the leading cause of acquired heart disease in developed countries where streptococcal infections are commonly treated and rheumatic fever is rare (Shulman and Rowley, 2015). Kawasaki disease has a particularly high incidence in children with East Asian ancestry and was first described in Japan. The dominating theory for the pathophysiology of Kawasaki disease involves the production of self-reactive antibodies during an acute immune response to a viral infection, probably at mucosal surfaces and focused around IgA-producing plasma cells. Such cells have also been found within the arterial wall in specimens from children with Kawasaki disease (Rowley et al., 2008). Neutrophils infiltrate the arterial wall, and a necrotizing arteritis develops leading to the destruction of the connective tissue and arterial dilation in severe cases (Shulman and Rowley, 2015). Efforts to define the pathogenesis of Kawasaki have also revealed an imbalance of interleukin (IL)-17-producing T cells and regulatory T cells during the acute phase of this disease (Jia et al., 2010).
Given the severity of the multisystem inflammatory syndrome in children (MIS-C) associated with COVID-19 and the uncertain development of the ongoing pandemic, there is an urgent need to understand the pathogenesis of MIS-C, and its similarities and differences with Kawasaki disease, so that optimal treatment strategies can be devised. Here, we have performed a systems-level analysis of immune cells, cytokines, and antibodies in the blood of children presenting with MIS-C as compared to children with mild SARS-CoV-2 infection, children with Kawasaki disease, and healthy children enrolled prior to the COVID-19 pandemic (Figure 1 A). We reveal several details of the hyperinflammatory state in children with MIS-C, contrasting with both the hyperinflammation seen in adults with acute SARS-CoV-2 infection, as well as that of children with Kawasaki disease. We define key cytokine mediators, assess serological responses to SARS-CoV-2 and other viruses, and we identify putative autoantibody targets possibly involved in the pathogenesis of this new disease.
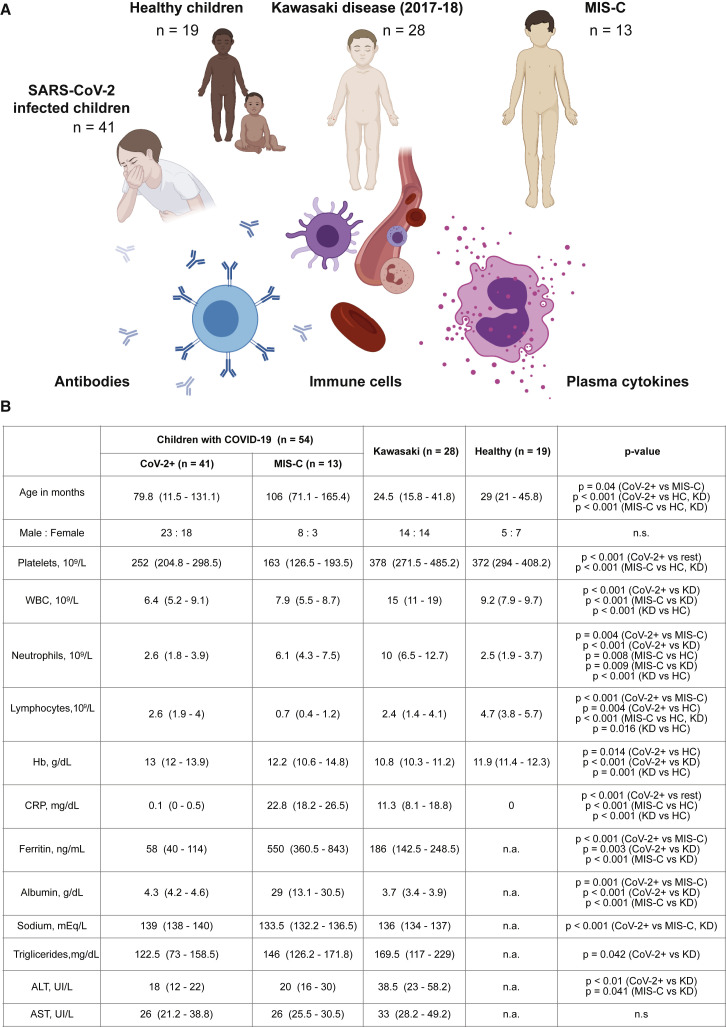
Figure 1.
Systems-Level Analyses in Children with COVID-19, Kawasaki Disease, and MIS-C
(A) Groups of children studied.
(B) Clinical parameters shown as median values (25th to 75th centiles). t test (parametric) and Mann-Whitney test (non-parametric) were used to compare mean values across groups of children.
Results
Children with SARS-CoV-2, MIS-C, and Kawasaki Disease
We enrolled 41 children with acute SARS-CoV-2 infection in Rome, Italy, all with mild disease, and denote these as CoV-2+ children throughout this manuscript (Figure 1B). We also enrolled three children presenting with MIS-C in Rome and 10 children presenting with MIS-C in Stockholm, Sweden. We compare these children with 28 children presenting with Kawasaki disease prior to the COVID-19 pandemic (March 2017 to May 2019). The children with MIS-C were significantly older than children with Kawasaki disease (Figure 1B), in line with previous reports (Whittaker et al., 2020). No significant sex-differences were found among the groups of children. Both MIS-C and CoV-2+ children presented with lower white blood cell counts (WBC) as compared to patients with Kawasaki disease and healthy children (Olin et al., 2018; Figure 1B). Lymphopenia is a hallmark of COVID-19 and was more pronounced in MIS-C than in children with mild SARS-CoV-2 infection or Kawasaki disease (Figure 1B). MIS-C patients also had markedly higher levels of C-reactive protein (CRP) and ferritin and lower platelet counts as compared to both Kawasaki disease patients and CoV-2+ children (Figure 1B). These observations are in line with recent clinical reports on MIS-C (Dufort et al., 2020; Feldstein et al., 2020; Whittaker et al., 2020), suggesting that our cohort is representative of reported patients with MIS-C, Kawasaki disease, and children with mild SARS-CoV-2-infection.
Hyperinflammation during MIS-C Differs from that of Severe, Acute COVID-19
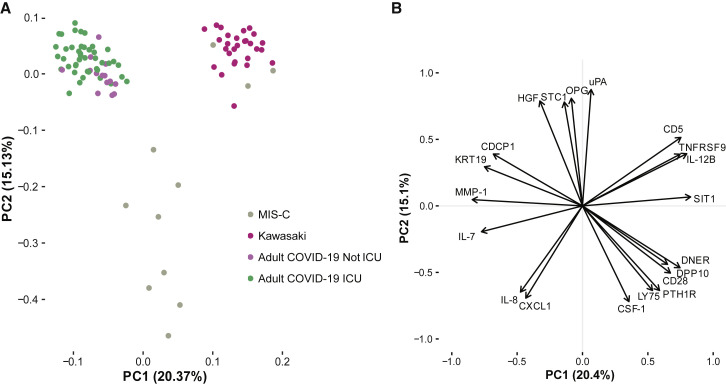
In adults and the elderly, the main cause of severe COVID-19 disease and death is uncontrolled immune activation, hyperinflammation, and immunopathology (Vardhana and Wolchok, 2020). This life-threatening response requires urgent management, and the immunological aspects of this process are under intense investigation (Kuri-Cervantes et al., 2020; Mathew et al., 2020). A number of clinical trials are ongoing to test immunomodulatory strategies that can calm the cytokine storm in severe COVID-19. Because MIS-C is also a hyperinflammatory condition associated with COVID-19, albeit with a delayed presentation, we wondered whether the hyperinflammatory response in MIS-C is similar to that seen in adults with severe COVID-19 disease. We measured 180 plasma proteins involved in immune response and inflammation in serum samples from children with mild MIS-C and Kawasaki disease and compared these to the cytokine profiles in adults with severe acute COVID-19 and hyperinflammation (Rodriguez et al., 2020). After filtering out proteins with >30% measurements below the threshold of detection, we performed principal component analysis (PCA) including 120 unique proteins (Figure 2 A; Table S1). Adults with acute COVID-19, both patients in the intensive care unit (ICU) and floor (non-ICU), but all severely ill, had very different cytokine profiles from those seen in children with either MIS-C or Kawasaki disease (Figure 2A). The main contributing features explaining this difference included IL-8 (Figure 2B), a chemokine recently shown to be associated with lymphopenia in severe COVID-19 cases (Zhang et al., 2020). IL-7 was also higher in acute COVID-19 hyperinflammation than in MIS-C and Kawasaki disease, IL-7 is a cytokine involved in T cell maintenance and associated with lymphocyte counts (Figure 2B). The MIS-C and Kawasaki hyperinflammatory states partially overlapped and both differed from adults with acute COVID-19 hyperinflammation (Figure 2A).
Figure 2.
MIS-C Hyperinflammation Differs from Severe Acute COVID-19 Hyperinflammation
(A) Principal components 1 and 2 show variation in cytokine profiles among adult COVID-19 patients with severe disease treated in intensive care units (ICU) or not, and children with MIS-C or Kawasaki disease. n = 97 samples included, and 112 unique proteins included in the analysis.
(B) Top 20 proteins mostly contributing to the PCs 1–2.
See also Table S1.
Differences in T Cell Subsets in MIS-C and Kawasaki Disease
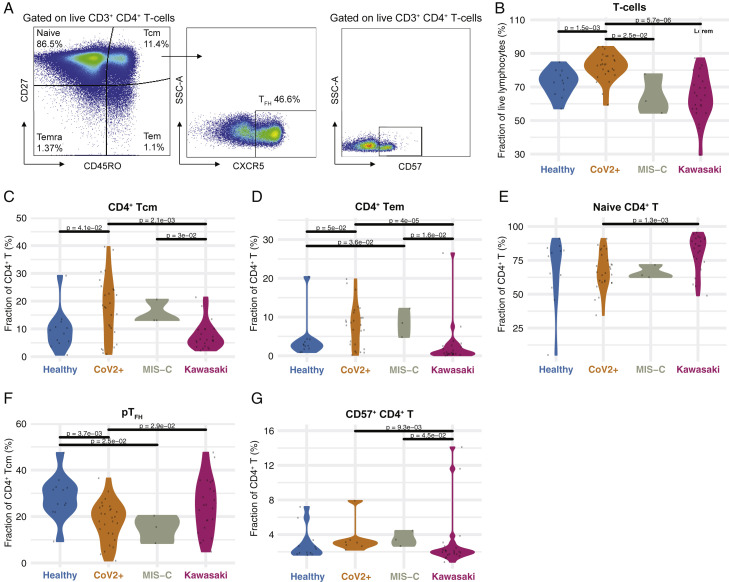
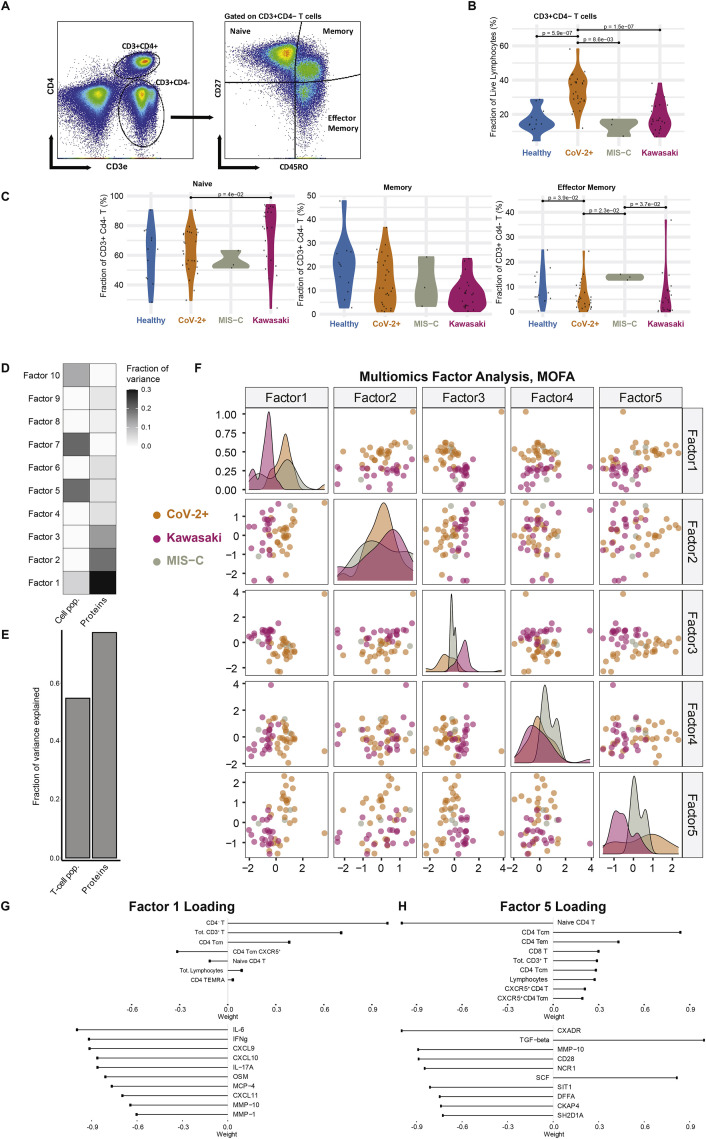
To better understand the hyperinflammation in MIS-C and Kawasaki disease, we assessed peripheral blood mononuclear cell (PBMC) phenotypes by flow cytometry. Samples were collected at hospital admission, prior to treatment, and reflect cellular states during the hyperinflammatory immune response. We found differences in the distributions of subpopulations of CD4+ T cells as defined by the expression of CD45RO and CD27, and the frequency of T-follicular helper cells (TFH) expressing the chemokine receptor CXCR5 (Figure 3 A). Total T cell frequencies were lower in both types of hyperinflammatory patients, MIS-C, and Kawasaki disease as compared to healthy children (Figure 3B). Within the CD4+ T cell compartment, MIS-C patients and children with mild SARS-CoV-2+ infection had similar subset distributions, indicating that differences seen in relation to healthy children could be related to the SARS-CoV-2 infection itself (Figures 3C–3E). Both groups of patients that experienced SARS-CoV-2 had higher abundances of central memory (CM) and effector memory (EM) CD4+ T cells, but fewer naive CD4+ T cells as compared to Kawasaki disease patients (Figures 3C–3E). Follicular helper T cells, important players in germinal center reactions and supporters of B cell responses, were reduced in SARS-CoV-2 experienced children, both with and without MIS-C, but not in Kawasaki disease patients (Figure 3F). CD57 marks terminally differentiated effector CD4+ T cells and these have been shown to be reduced in adult patients with severe acute COVID-19 and acute respiratory distress syndrome (Anft et al., 2020). In children with mild COVID-19 and children with MIS-C, we find higher levels of these terminally differentiated cells when comparing to Kawasaki disease patients and healthy children (Figure 3G). We find that CD4− T cells (mostly CD8+ T cells) were significantly lower in MIS-C children as compared to children with mild SARS-CoV-2 infection (Figures S1 A–S1D). These results further emphasize that the hyperinflammation seen in acute, severe COVID-19 in adults differs from that seen in MIS-C and also indicates some specific differences between immune cell responses in MIS-C and patients with Kawasaki disease occurring prior to the COVID-19 pandemic.
Figure 3.
CD4+ T Cell Subsets in COVID-19 and MIS-C
(A) Gating schema to identify CD4+ T cell subsets from peripheral blood mononuclear cells.
(B) CD4+ T cells as a fraction of lymphocytes. Black lines indicate statistical tests and p values across indicated populations.
(C–G) Fraction of (C) CD4+ Tcm, (D) CD4+ Tem, (E) naive CD4+, (F) pTFH, and (G) CD57+ CD4+ T cell subsets (%) in the indicated patient group. Black lines indicate statistical tests and p values across indicated populations.
See also Figure S1.
Figure S1.
Multiomics Immune System Differences among Disease Groups, Related to Figure 3
(A) Gating of CD4- T cell subsets. (B) Frequency of CD4- T cells across disease groups. (C) Naive and memory phenotype CD4- T cells. (D) Fraction of variance explained by 10 latent factors, (E) Fraction of variance explained by data type, (F) Samples distributed across the top five latent factors. (G) Top contributing features for factor 1 and (H) factor 5 respectively.
Unraveling the Cytokine Storm in MIS-C and Kawasaki Disease
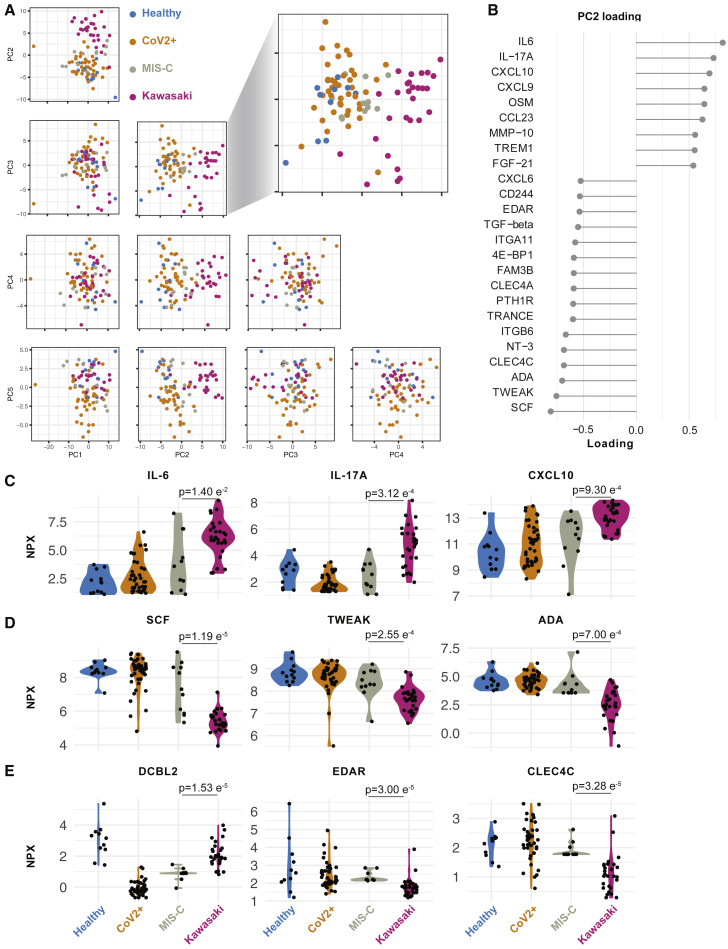
To further investigate the hyperinflammatory immune states in MIS-C and Kawasaki disease patients, we performed Olink assays on plasma samples from 11 children with MIS-C and 28 children with Kawasaki disease. We measured 180 unique proteins using two Olink panels (Key Resources Table) and filtered out proteins with >30% measurements below the threshold of detection, resulting in 125 plasma proteins used for PCA (Figure 4 A; Table S2). We find that healthy children and children infected with SARS-CoV-2, without MIS-C, mostly overlapped, illustrating the mild infection and low-grade inflammatory response in these CoV-2+ children (Figure 4A). As shown by the blown up PC2 versus 3 plot, MIS-C hyperinflammatory states differ from that of Kawasaki disease (Figure 4A). PC2 best discriminated these hyperinflammatory states in MIS-C and Kawasaki disease, and we focused on the contributing features (loadings) of PC2 (Figure 4B). We find that elevated IL-6, IL-17A, CXCL10 contributed the most to the cytokine storm (Figure 4C). IL-17A is important in Kawasaki disease (Jia et al., 2010), but was significantly lower in MIS-C patients, indicating a difference in the underlying immunopathology (Figure 4C). In contrast, the top negative contributors were adenosine deaminase (ADA), stem cell factor (SCF), and TWEAK, a negative regulator of interferon (IFN)γ and Th1-type immune response (Maecker et al., 2005). As a regulator of angiogenesis, TWEAK is also an interesting finding given the vasculitis component of Kawasaki disease and probably also MIS-C (Figure 4D).
Figure 4.
Cytokine Profiles in MIS-C and Kawasaki Disease
(A) Principal component analysis of top 5 components explains 58.3% of the variance in concentrations of 133 plasma proteins. PC2 versus 3 separate children with Kawasaki (n = 28) from MIS-C (n = 11), healthy children (n = 12), and SARS-CoV-2+ children without hyperinflammation (n = 41).
(B) Proteins explaining (highest loading for) PC2.
(C and D) Raw (NPX) values for top (C) positive and (D) negative proteins (loading) in PC2. p values for mean comparisons between MIS-C and Kawasaki disease children.
(E) Raw (NPX) values of three proteins significantly different between Kawasaki and MIS-C. p values for mean comparisons between MIS-C and Kawasaki disease children.
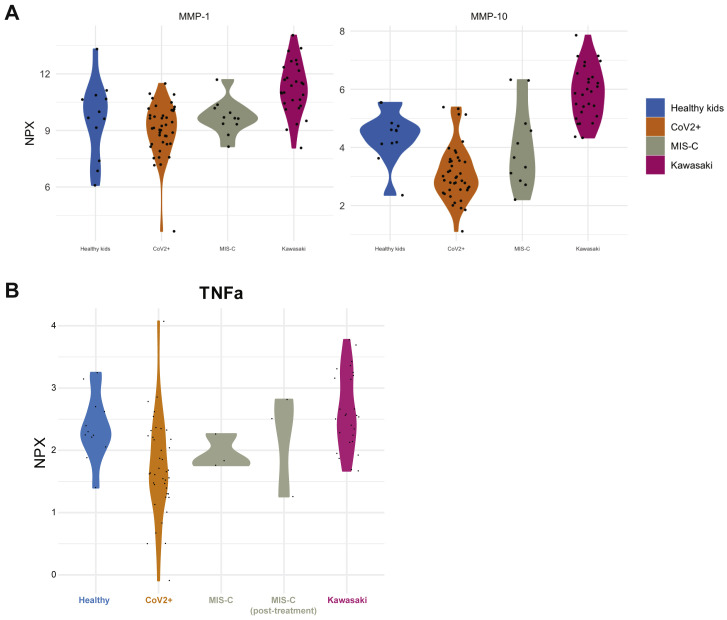
We also identified additional plasma proteins distinguishing MIS-C from Kawasaki. Apart from IL-17A, DCBLD2, also called ESDN, was more elevated in Kawasaki disease than MIS-C. This protein has been shown to be secreted by endothelial cells attacked by allogeneic immune cells after heart transplantation (Sadeghi et al., 2007) and in balloon-dilated carotid arteries (Kobuke et al., 2001) suggestive of more pronounced arterial damage in Kawasaki than in MIS-C. Collectively, these findings indicate more arterial involvement in Kawasaki than MIS-C and also an IL-17A-driven cytokine storm in Kawasaki disease but not in MIS-C. IL-17-blocking agents are used in psoriasis and other diseases and could also be of interest for future clinical trials in these patients. Moreover, other key proteins differing among these groups are MMP-1 and MMP-10 (Figure S2 A), proteins involved in arterial disease (Martinez-Aguilar et al., 2015), which is intriguing given the likely arterial inflammation component of both MIS-C and Kawasaki disease.
Figure S2.
Related to Figure 4
(A) Plasma MMP-1 and MMP-10 across disease groups. (B) Plasma TNFα levels in patient groups. NPX is Normalized expression level.
MIS-C Immune Profiles Intersect Mild COVID-19 and Kawasaki Disease and Change upon Immunomodulation
To understand the interaction among different immune system components and their coregulation, we applied multiomics factor analysis (MOFA) (Argelaguet et al., 2019) to integrate T cell subset frequencies and plasma protein concentrations from MIS-C patients, Cov-2+ children, and children with Kawasaki disease. We identified 10 latent factors that explain the combined variance across cell and protein datasets (Figures S1D and S1E). The first factor separated Kawasaki disease samples from both groups of SARS-CoV-2 infected children (MIS-C and CoV-2+) (Figure S1F). Conversely, factor 5 discriminated the extremes of Kawasaki versus mild SARS-CoV-2+ infections and placed MIS-C samples in between these polar opposites (Figures S1G and S1H), indicating that the immunopathology in MIS-C shares features with both acute SARS-CoV-2 infection and the post-infectious response associated with Kawasaki disease.
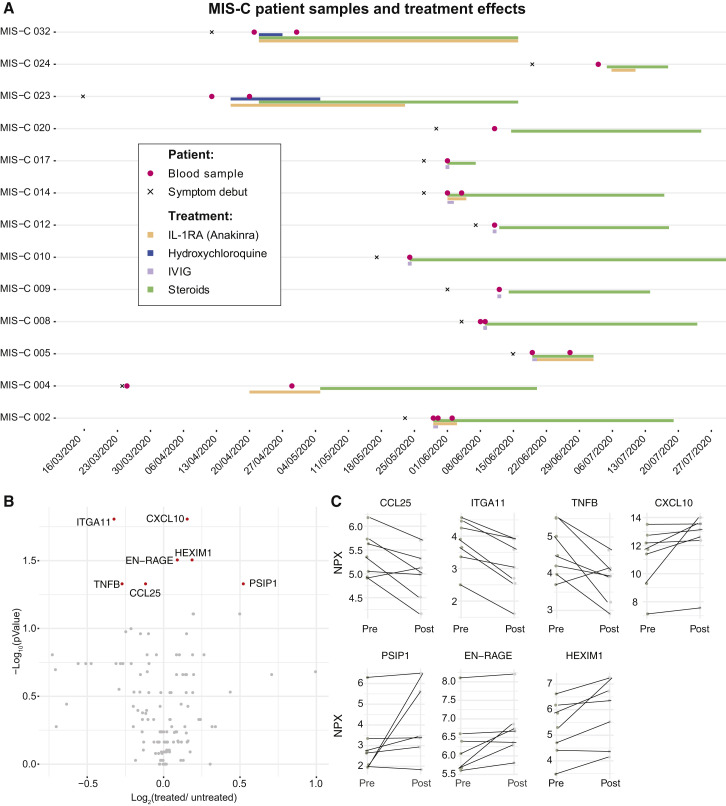
MIS-C patients are treated with strong immunomodulatory agents. The treatment regimens for the 13 MIS-C patients in our cohort are shown in Figure 5 A. Most patients were treated by high dose steroids in combinations with intravenous immunoglobulins (IVIG) to override the effect of autoantibodies and stimulate inhibitory Fc-receptors (Figure 5A). A number of children were also given recombinant IL1RA (anakinra) to block IL-1a/b-mediated inflammation (Figure 5A). We assessed the cytokine changes upon treatment and found that tumor necrosis factor beta (TNF-β), ITGA11, and CCL25 levels decreased, whereas HEXIM1, PSP1, and CXCL10 levels increased in response to treatment in the seven MIS-C patients with available pre/post samples (Figures 5B and 5C). These results indicate that there is a particular cytokine profile associated with MIS-C, which differs from Kawasaki disease hyperinflammation and that changes in response to immunomodulatory treatment.
Figure 5.
Immunomodulatory Treatment in Patients with MIS-C
(A) Timelines for individual MIS-C patients (n = 13) indicating date of symptom debut, immunomodulatory treatment regimen, and date of blood sampling. Two patients lacking clinical treatment data were blood sampled prior to treatment.
(B) Volcano plot showing fold-change (post-/pre-treatment) versus p value for 7 patients with multiple samples.
(C) Top markers of treatment response in MIS-C patients. Fold-change pre/post treatment (p < 0.05, false discovery rate [FDR], 1%).
See also Figure S2.
Serologic Responses and Pre-existing Humoral Immunity in MIS-C
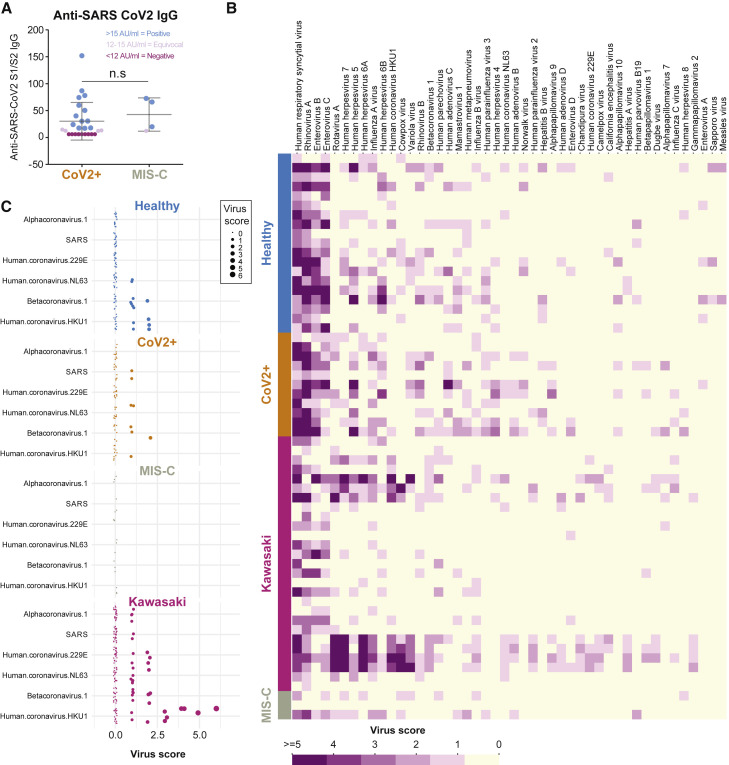
Given that Kawasaki disease and possibly also MIS-C display autoantibody-mediated immunopathology, we decided to test immunoglobulin G (IgG) responses to the SARS-CoV-2 virus in these patients. We measured IgG binding of the receptor-binding domain (RBD) of the spike protein from SARS-CoV-2. We found that the majority of children in our cohort testing positive for infection by PCR also seroconverted (Figure 6 A). We also found that 3 out of 4 tested MIS-C patients seroconverted and had comparable levels of SARS-CoV-2 IgG antibodies to the spike protein as the children with mild SARS-CoV-2 infection and no MIS-C (Figure 6A). This result is in line with data from other groups studying antibody responses in MIS-C patients (Gruber et al., 2020; Rostad et al., 2020). A possible hypothesis for why some children develop MIS-C is that prior immunity to other viruses could modulate their responses to SARS-CoV-2 infection and give rise to hyperinflammation either by antibody-mediated enhancement or other mechanisms (Tetro, 2020). To assess this, we applied VirScan, a phage display method for testing IgG binding of 93,904 epitopes from 206 different viruses able to infect human cells (Xu et al., 2015). We found that the children in our cohort had IgG antibodies to common viruses such as respiratory syncytial virus (RSV) and rhinovirus as well as viruses of the herpesvirus family (Figure 6B). This is in line with our previous results (Pou et al., 2019). The common cold coronaviruses are particularly interesting as possible modulators of immune responses to SARS-CoV-2 given their similarities. We focused on IgG antibodies to these viruses in MIS-C, CoV-2+, healthy, and Kawasaki-disease children. We found that IgG antibodies to human coronavirus HKU1 was commonly observed, as were antibodies to betacoronavirus 1, but the MIS-C patients were the only ones lacking antibodies to either of these common coronaviruses (Figure 6C). The relevance of this difference remains to be determined; this could either reflect the fact that the MIS-C patients are older than the other groups of children analyzed, but it is also possible that the lack of IgG antibodies to common coronaviruses modulates the immune response to SARS-CoV-2 infection and plays a role in the pathogenesis of MIS-C.
Figure 6.
Serological Responses and Prior Immunity in Kawasaki and MIS-C
(A) Anti-SARS-CoV-2 S1/S2 IgG antibodies in patients with mild infection (n = 26) and MIS-C (n = 4).
(B) VirScan analysis of IgG responses to 93,904 epitopes of 206 viruses in healthy (n = 19), CoV-2 infected (n = 11), Kawasaki disease (n = 27), and children with MIS-C (n = 3). Virus scores is a function of the number of targeted epitopes per virus, and the heatmap shows the top 46 viruses with top average virus scores.
(C) The virus scores of coronaviruses available in the VirScan library, shown across individual children divided by disease groups.
Proteome Array Profiling Reveals Potentially Pathogenic Autoantibodies in MIS-C
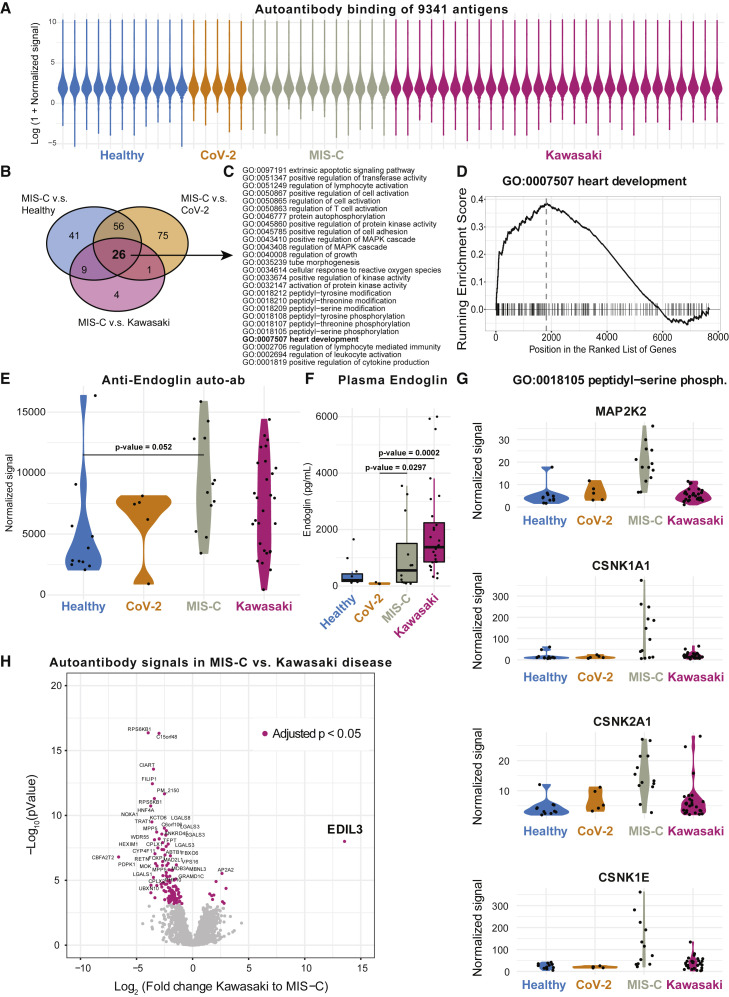
Several studies have proposed autoantibodies as an element of the immunopathology of Kawasaki disease (Sakurai, 2019). An external trigger, likely in the form of a virus, is indicated by epidemics of Kawasaki disease, and direct evidence of antiviral antibody responses, particularly targeting viral particles in inclusion bodies in the respiratory epithelium, have been reported (Rowley et al., 2008). Autoantibodies targeting endothelial cells can activate IL-6 production by such cells in vitro (Grunebaum et al., 2002), but a possible role of autoantibodies in MIS-C is unknown. To search for such autoantibodies in MIS-C, we screened serum samples from children with MIS-C (n = 12), Kawasaki disease (n = 28), SARS-CoV-2+ infection (n = 5), as well as healthy control children (n = 11). We probed serum samples against 9,341 human antigens from 7,669 unique human proteins using protein arrays (Zhu et al., 2001) (ProtoArray v5.1, PAH05251020, ThermoFisher, Waltham, MA). As expected, the autoantibody signals were low for the vast majority of antigenic targets (Landegren et al., 2016; Figure 7 A). All autoantibody binding intensities across all samples tested are provided in Table S3. We ranked autoantibody targets using fold-change calculations between the MIS-C group and each of the other groups of samples and looked for enriched Gene Ontology (GO)-terms among the targets using gene set enrichment analysis (GSEA) (Subramanian et al., 2005). There were 26 GO-terms that were enriched in MIS-C samples when compared to all other groups (Figure 7B), and these involved lymphocyte activation processes, phosphorylation signaling pathways, and heart development (Figure 7C). The latter was interesting given that myocarditis and impaired cardiac function are hallmarks of MIS-C clinical presentation. We studied individual autoantibody targets within this GO-term (Figure 7D) and identified endoglin, a glycoprotein expressed by endothelial cells and necessary for structural integrity of arteries to be differentially regulated among groups of samples (Figure 7E). Loss of endoglin leads to the disease, hereditary hemorrhagic telangiectasia, a disease characterized by multisystemic vascular dysplasia (McAllister et al., 1994). Several, but not all, of the MIS-C patients had elevated levels of autoantibodies targeting endoglin above the average levels seen in healthy controls with a couple of exceptions (Figure 7E). A subset of Kawasaki disease patients also had elevated levels of autoantibodies to endoglin (Figure 7E). Endoglin protein expression is seen predominantly in the vascular endothelium, with the heart muscle having the highest mRNA expression of all tissues (Uhlén et al., 2015). We also measured plasma levels of endoglin, but these levels were elevated in Kawasaki and MIS-C patients as compared to healthy children, possibly indicating that autoantibodies to endoglin are not the cause of tissue damage, but rather a consequence thereof (Figure 7F). It is interesting to investigate a possible role for this protein and antibodies targeting it, in the context of MIS-C and Kawasaki disease pathogenesis or as a possible biomarker of endothelial damage that could be useful to monitor in these patients.
Figure 7.
Autoantibodies in MIS-C, Kawasaki, and Healthy Children
(A) Overall antibody binding intensities against 9,341 antigens from 7,669 human proteins in healthy children (n = 11), CoV-2+ (n = 5), MIS-C (n = 12), and Kawasaki (n = 28) violin plots colored by sample group.
(B) Venn diagram showing 26 enriched GO-terms across MIS-C versus healthy, Cov-2+, and Kawasaki disease children.
(C) The 26 GO terms enriched in MIS-C versus all other groups listed.
(D) GSEA plot for GO:0007507 heart development.
(E) Autoantibodies targeting the glycoprotein endoglin (CD105). p value comparing means in healthy children and MIS-C (FDR, 1%).
(F) Plasma endoglin levels measured by ELISA in plasma samples.
(G) Four candidate antigens bound by autoantibodies across the four patient groups but at highest levels in MIS-C children.
(H) Volcano plot showing fold-change differences in autoantibody signals between Kawasaki disease (n = 28) and MIS-C (n = 13). Purple and annotated target antigens have p < 0.05 (FDR, 1%). EDIL3 is the single most overrepresented protein in Kawasaki disease.
See also Table S3.
Among the other enriched GO-terms, the majority involved signal transduction and, in particular, signals propagated by phosphorylation of tyrosine and serine residues. GO: 0018100105 is one example with clear overrepresentation among MIS-C autoantibody profiles as compared to children with mild SARS-CoV-2 infection. We found several autoantibodies in this category that were exclusively seen in MIS-C-patient samples and none of the other groups. Antibodies to MAP2K2, and antibodies to three members of the Casein kinase family (CSNK1A1, CSNK2A1, CSNK1E1) are notable examples (Figure 7G). The latter groups of autoantibody targets are interesting in the context of SARS-CoV-2 infection, because a recently published analysis of global phosphorylation patterns in SARS-CoV-2 infected cells reported strong upregulation of casein-kinase 2 activity, and potent antiviral activity of a drug, silmitasertib, inhibiting this kinase (Bouhaddou et al., 2020). This finding, in combination with our results here, suggests that the casein-kinase 2 pathway is important in viral replication, and possibly, autoantibodies to intermediates within these related pathways arise during acute SARS-CoV-2 infection and have play a role in MIS-C development thereafter in a subset of previously infected children. These possible autoantibody targets are also broadly expressed across tissues, which is in line with the diffuse, multi-organ involvement in patients with MIS-C.
Next, we decided to contrast autoantibody profiles in MIS-C and Kawasaki disease patients given that there are similarities in the presentation of these conditions, but there are also clear differences. Kawasaki disease involves inflammation of medium-sized arteries such as the coronary arteries, whereas MIS-C patients seem to have more diffuse presentation involving intestine, myocardium, and brain. In our cohort, we found that Kawasaki disease patients have elevated plasma biomarkers of arterial damage as compared to MIS-C patients. We compared the autoantibodies profiles to identify the most different autoantibody targets between Kawasaki and MIS-C patients (Figure 7H). Although there were a number of differentially regulated targets, autoantibodies to EDIL3 were most clearly overexpressed in Kawasaki disease patients as compared to MIS-C (Figure 7H). The gene encoding this protein has previously been associated with Kawasaki disease (Jiang et al., 2017), and the target protein is a structural glycoprotein in arterial vessel walls that is regulated in response to vascular injury. One function of EDIL3 is to inhibit inflammatory cell recruitment and extravasation across the endothelium, and when EDIL3 is disrupted in mice, there is increased tissue infiltration of neutrophils and elevated inflammatory responses (Choi et al., 2008). Whether such autoantibodies to EDIL3 are pathogenic in Kawasaki disease remains to be determined.
Collectively, our findings show that MIS-C is a hyperinflammatory disease with a qualitatively different inflammatory response as compared to what is seen in acute SARS-CoV-2 infection and is also different from the hyperinflammation in children with Kawasaki disease. T cell differences can be seen, but are uncertain, given differences in age between MIS-C and Kawasaki disease patients. Serological responses to SARS-CoV-2 are seemingly normal in MIS-C patients, but pre-existing immunity to related coronaviruses is lower. A number of candidate autoantibodies are identified for future studies of MIS-C pathogenesis.
Discussion
The SARS-CoV-2 virus infects children at a similar rate as adults (Jones et al., 2020), but fortuitously, the disease COVID-19 is very mild in the overwhelming majority of infected children (Brodin, 2020). Despite the mild COVID-19 in children, recent reports of severe hyperinflammatory disorders developing 1–2 months after the acute infection with SARS-CoV-2 are cause for much concern. This MIS-C associated with COVID-19, also called pediatric multisystem inflammatory syndrome, temporally associated with COVID-19 (PIMS-TS), has now been reported in hundreds of children worldwide (Cheung et al., 2020; Moraleda et al., 2020; Riphagen et al., 2020; Verdoni et al., 2020; Whittaker et al., 2020). The pathogenesis is under intense investigation but so far remains elusive. One study has found evidence of microangiopathy in MIS-C (Diorio et al., 2020), and a recent autopsy study of a child who died from MIS-C revealed abundant viral particles across a number of tissues, possibly suggestive of a “second-hit” virus-mediated pathology (Dolhnikoff et al., 2020). At the same time, children with MIS-C typically respond well to aggressive immunomodulatory and anti-inflammatory therapies, which is unlikely to be effective if pathology was mediated by the virus rather than the immune system.
Here, we have combined several high-dimensional analysis methods to uncover multiple aspects of the hyperinflammatory response in children with MIS-C. We find similarities with the inflammatory response seen in children with Kawasaki disease, but also important differences, such as the IL-17A-mediated hyperinflammation in Kawasaki disease, but not MIS-C. In addition, differences in T cell subsets and cytokine mediators place MIS-C at the intersection of Kawasaki disease and acute SARS-CoV-2 infection immune states in children as well as the hyperinflammation seen in adults with severe COVID-19. We also find higher levels of biomarkers associated with arteritis and coronary artery disease in Kawasaki disease than in MIS-C, suggesting a more diffuse endothelial involvement and immunopathology in MIS-C than in Kawasaki disease. Finally, we perform global autoantibody screening and find binding of autoantibodies to proteins involved particularly in immune cell signaling, and structural proteins in heart and blood vessels. These data suggest possible targets of autoimmune attack. The hypothesis that autoantibodies contribute to the pathology in MIS-C is supported by the efficacy of intravenous immunoglobulin in MIS-C, a common approach to activate inhibitory Fc-receptors and prevent membrane-attack complexes by complement factors and thereby mitigating autoantibody-mediated pathology (Kazatchkine et al., 2001).
Our results showing lower naive CD4+ T cell, TFH, and increases in central and effector memory subpopulations in MIS-C as compared to Kawasaki disease patients could be partially explained by differences in patient age, because immune cell proportions change with age, even in healthy children (Olin et al., 2018; Schatorjé et al., 2012). We did, however, see that these cell population differences remained even when comparing against additional older healthy controls, comparable in age to the MIS-C patients. Several studies have indicated defective T cell responses as a key element of severe COVID-19 disease in adult patients (Vardhana and Wolchok, 2020). We find expanded CD57+ CD4+ T cells often representing immunosenescent and dysregulated T cells in both MIS-C and SARS-CoV-2+ children with mild infections, suggesting that this might be a consequence of the SARS-CoV-2 infection. Additional studies in mild, and especially asymptomatic, COVID-19 cases will be required to investigate this further.
Treatments of MIS-C have mostly followed protocols used in atypical Kawasaki disease given the overlap in presentation between these groups of patients. Our data present a more complex picture with both shared features and stark differences that should influence treatment strategies for these conditions. Kawasaki disease, but not MIS-C, is associated with robust IL-17A-mediated inflammation, and IL-17A blocking agents, such as secukinumab (Hueber et al., 2010), could be considered in severe Kawasaki disease patients in future trials but perhaps less relevant to MIS-C. Children with MIS-C in this study were treated with a combination of intravenous immunoglobulins (IVIGs), corticosteroids, and recombinant IL-1-receptor antagonist, IL1RA (anakinra). The profiles of seven cytokines indicated treatment responses and can be useful to monitor in MIS-C patients undergoing treatment (Figures 5B and 5C). Autoantibodies represent a possible pathogenic mechanism in MIS-C and Kawasaki disease. IVIGs can neutralize some of the immunopathological effects of autoantibodies, whereas IL-1RA neutralizes strong IL-1 response elicited by endothelial cells that are damaged by autoantibodies and complement. Corticosteroids provide more general immunosuppression but the mechanism of action in MIS-C for these treatments remains to be determined. Other treatment strategies reported by other groups include TNF-α blockade (infliximab) (Whittaker et al., 2020), but when comparing plasma TNF-α levels in MIS-C patients to adults with acute COVID-19, TNF-α levels are much lower in MIS-C (Figure 2), and comparable to healthy children, we find no elevated levels (Figure S2B), suggesting that TNF-α-blockade might not mitigate the hyperinflammation in MIS-C. IL-6 blockade has been used in acute COVID-19 and in MIS-C, but in our MIS-C patients, plasma levels of IL-6 were highly variable and often normal (Figure 4C).
The autoantibody profiling presented here revealed a number of possible autoantibody candidates. It is worth noting that the result is not as clear, because the pathogenic autoantibodies seen in well-established autoimmune disorders, such as autoimmune thyroiditis or Addison’s disease, or in patients with loss of tolerance due to AIRE deficiency (autoimmune polyendocrine syndrome type-1 [APS1]) (Landegren et al., 2016). The results here are more diffuse, with many antigens targeted and only a few shared across MIS-C patients as compared to other patient groups. One caveat of these types of arrays is the incorrect structure of some target antigens, and another caveat is the inability of the secondary antibody to detect pathogenic autoantibodies of different classes than IgG, such as IgA. Given that the initial immune response is likely elicited in the respiratory or intestinal mucosa, IgA antibodies are of interest, and IgA antibodies have been directly implicated in Kawasaki disease pathogenesis (Shulman and Rowley, 2015). Despite these caveats, there are a number of possible autoantibodies with pathogenic potential detected in the MIS-C patient cohort herein. Overall, the data presented here suggest novel directions for future work toward more mechanistic understanding of the immunopathology in MIS-C, its underlying immune perturbation, and development of better immunomodulatory therapies for mitigating the hyperinflammatory disease and long-term tissue damage in such rare children severely affected by COVID-19.
Limitations of Study
One caveat of this study was the limited sample size for children with MIS-C (n = 13), and analyses with children with SARS-CoV-2 infection (n = 5) in Figure 7. We also had a limited amount of paired pre-/post-treatment samples from children with MIS-C, which limited our power to understand MIS-C recovery post-treatment. Future work will need to focus on understanding the pathogenesis of MIS-C and Kawasaki disease, respectively, and a more definitive understanding of the relevant autoantigens involved. We have not been able to investigate a possible “second hit” of viral replication in MIS-C or possible viral reservoirs driving hyperinflammation. This will require more work in the future.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Flow cytometry | ||
| Marker/Fluorophore | Clone/Vendor | Cat# |
| CD3/PE-CF594 | UCHT1/BD | Cat# 562280, RRID:AB_11153674 |
| CD4/BV510 | SK3/BD | Cat# 562970, RRID:AB_2744424 |
| CD45RO/PE-Cy5 | UCHL1/Biolegend | Cat# 304208, RRID:AB_314424 |
| CD57/APC | NK-1/BD | Cat# 560845, RRID:AB_10563760 |
| CD127/PE-Cy7 | EbioRDR5/eBioscience | Cat# 25-1278-42, RRID:AB_1659672 |
| CD185 (CXCR5)/BV605 | RF8B2/BD | Cat# 740379, RRID:AB_2740110 |
| CD25/PE | M-A251/BD | Cat# 555432, RRID:AB_395826 |
| CD279 (PD1)/BV711 | EH12.2H7/Biolegend | Cat# 329927, RRID:AB_11218612 |
| CD27/V450 | M-T271/BD | Cat# 561408, RRID:AB_10682577 |
| LIVE/DEAD Fixable Near-IR Dead Cell Stain Kit | Invitrogen | L34976 |
| Protein array-screening | ||
| ProtoArray v5.1 | ThermoFisher | PAH05251020 |
| Alexa Fluor 647 goat anti-human IgG antibody | ThermoFisher | Cat# A21445, RRID:AB_2535862 |
| Dylight 550 goat anti-GST | Cayman Chemicals | DY550011-13-001 |
| Blocking buffer | Life Technologies | PA055 |
| Biological Samples | ||
| PBMCs and plasma from healthy controls, patients with Kawasaki disease, mild SARS-CoV-2 infection, and MIS-C | N/A | N/A |
| Critical Commercial Assays | ||
| Immune Response panel (article # 95320, panel versions v.3202 and v.3203) | Olink AB (Uppsala, Sweden) | N/A |
| Inflammation panel (article #95302, panel version v.3022) | Olink AB (Uppsala, Sweden) | N/A |
| Endoglin (CD105) Human ELISA Kit | ThermoFisher | EHENG |
| Software and Algorithms | ||
| R (version 3.6.2) | https://cran.r-project.org/ | N/A |
| MOFA2 R package (version 1.0) | https://github.com/bioFAM/MOFA2 | N/A |
| OlinkAnalyze R package | https://github.com/Olink-Proteomics/OlinkRPackage | N/A |
| limma R package | https://bioconductor.org/packages/release/bioc/html/limma.html | Ritchie et al., 2015 |
| DESeq2 R package | https://github.com/mikelove/DESeq2 | Love et al., 2014 |
| clusterProfiler R package | https://github.com/YuLab-SMU/clusterProfiler | Yu et al., 2012 |
| Custom analysis scripts | https://github.com/Brodinlab/MIS-C_manuscript | N/A |
| Additional Supplementary Items and Datasets | https://doi.org/10.17632/ds6g796xyg.1 | N/A |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Petter Brodin (petter.brodin@ki.se).
Materials Availability
No new reagents were developed during the study. Patient samples can be made available within the limits of the approved Ethical permits. Such requests should be directed to the lead contact author.
Data and Code Availability
Original data have been deposited to Mendeley Data: http://dx.doi.org/10.17632/ds6g796xyg.1. Scripts to reproduce the analyses presented in each figure of the paper are available at https://github.com/Brodinlab/MIS-C_manuscript.
Experimental Model and Subject Details
Study Participants and Sample Collection
44 SARS-CoV-2 infected children were enrolled within the CACTUS study at Bambino Gesù children hospital between March 17th and May 15th 2020. Diagnostic tests for SARS-CoV-2 infection are reported below. Three of the 44 Italian children found to be SARS-CoV-2+ by PCR herein were classified as having MIS-C. In addition, we enrolled 10 children at the Karolinska University Hospital fulfilling the same MIS-C WHO criteria (https://www.who.int/news-room/commentaries/detail/multisystem-inflammatory-syndrome-in-children-and-adolescents-with-covid-19). A total of 41 children with CoV2+ (23 males, 18 female) and 13 children with MIS-C (8 males, 3 female) were analyzed in this study. In summary this states that children and adolescents between 0 and 19 years old, with fever > 3 days, and displaying two of the following signs: i) rash or bilateral non purulent conjunctivitis or mucocutaneous inflammation signs, ii) hypotension or shock, iii) Features of myocardial dysfunction, pericarditis, valvulitis, or coronary abnormalities (including ECHO findings or elevated Troponin/NT-proBNP), iv) evidence of coagulopathy (by PT, PTT, elevated d-Dimers), v) acute gastrointestinal problems (diarrhea, vomiting, or abdominal pain); AND elevated markers of inflammation such as ESR, C-reactive protein, or procalcitonin; AND no other obvious microbial cause of inflammation, including bacterial sepsis, staphylococcal or streptococcal shock syndromes; AND evidence of SARS-CoV-2 infection (RT-PCR or serology positive), or likely exposure to COVID-19 patient. Twenty-eight children diagnosed with Kawasaki disease (14 male, 14 female) were enrolled during acute phase at the Academic Department of Pediatrics, Division of Immune and Infectious Diseases at Bambino Gesù Children’s Hospital (OPBG) between September 2017 and June 2019, well before the COVID-19 pandemic hit Europe. KD diagnosis was made according to the 2017 criteria of the American Heart association, including both the complete and incomplete types. Fever onset was considered as the first day of the acute KD phase. Nineteen healthy controls (HC) were included in the study. Demographic and clinical data of all the patients are summarized in Figure 1B. Swedish patients are included in accordance with the Ethical permit (EPM ID: 2020-01911). Italian children are included in accordance with a study protocol approved by the OPBG Ethics Committee. Informed consent was obtained from parents or guardians of all patients both in Sweden and Italy. APS-1 patient samples were included as positive controls in autoantibody analyses and their sampling and analyses were covered by Ethical permit (EPM ID: 2016/2553-31/2).
Method Details
Confirmation of SARS-CoV-2 Infection
All patients enrolled in the CACTUS study with nasopharyngeal swab tested positive for SARS-CoV-2 nucleic acid using reverse-transcriptase qualitative PCR assay were considered confirmed cases of SARS-CoV-2 infection. In addition, all serum samples of COVID-19 patients in Italy were investigated for presence of SARS-CoV-2 antibodies. IgG antibodies were quantitatively tested by LIAISON® SARS-CoV-2 S1/S2 IgG test (Diasorin).
Serum Protein Profiling
Serum proteins were analyzed using a multiplex technology based upon proximity-extension assays (Lundberg et al., 2011). We measured 180 unique proteins using the Olink panels of Immune Response and Inflammation (Key Resources Table). Briefly, each kit consisted of a microtiter plate for measuring 92 protein biomarkers in all 88 samples and each well contained 96 pairs of DNA-labeled antibody probes. To minimize inter- and intra-run variation, the data were normalized using both an internal control (extension control) and an inter-plate control, and then transformed using a pre-determined correction factor. The pre-processed data were provided in the arbitrary unit Normalized Protein Expression (NPX) on a log2 scale and where a high NPX represents high protein concentration.
VirScan
We investigated and compared antiviral antibody repertoires (IgG) of MIS-C, CoV-2+, Kawasaki and healthy children with VirScan, a viral epitope scanning method that relies on bacteriophage display and immunoprecipitation and we used the method as previously described (Pou et al., 2019; Xu et al., 2015). Briefly, VirScan uses a library of bacteriophages presenting 56-amino-acid-long linear peptides that overlap by 28 amino acids to collectively encompass the entire genomes of 1,276 viral strains from 206 viral species known to infect human cells. After virus inactivation and normalization to total IgG concentration, plasma samples were incubated with the phage library to form IgG-phage immunocomplexes, later captured by magnetic beads. Collected bacteriophages were lysed and sequenced to identify the IgG-targeted epitopes. VirScore is the output given by VirScan and corresponds to the number of peptide hits that do not share epitopes, and it is incremented by 1 when a hit is enriched in the ‘output’ compared with the ‘input’. Background noise is commonly observed in bacteriophage display assays. In order to minimize false-positive hits, we removed hits occurring as a consequence of unspecific binding to beads only (blank reactions without antibodies). Moreover, we also removed cross-reactive antibodies by ignoring hits that share a subsequence of at least seven amino acids with any other enriched hit found in the same sample.
Autoantibody Profiling
We performed autoantibody profiling on samples from healthy children (n = 11), children with CoV-2+ (n = 5), children with MIS-C (n = 12) and children with Kawasaki (n = 28) in two batches. Serum autoantibody reactivity was studied using full-length human protein arrays (ProtoArray v5.1, PAH05251020, ThermoFisher) (Zhu et al., 2001). Protein arrays were probed with serum at a dilution of 1:2000, and otherwise followed the protocol provided by the manufacturer for immune response biomarker profiling. Protein arrays were first incubated with blocking buffer (PA055, Life Technologies) for 1 hour, followed by 90 min incubation with serum at 1:2000 dilution, and 90 min incubation with detection antibodies: Alexa Fluor 647 goat anti-human IgG antibody (A21445, ThermoFisher) at 1:2000 dilution and Dylight 550 goat anti-GST (#DY550011-13-001, Cayman Chemicals) at 1:10,000 dilution. The LuxScan HT24 (BioCapital) microarray scanner was used.
ELISA
Serum endoglin levels (CD105) were determined using manufacturer’s instruction (Key Resources Table). Briefly, samples were heat inactivated (56°C for 30 min) and ∼50mL was used for ELISA-based quantification of Endoglin levels. Samples with out-of-range values were diluted and measured again.
Flow Cytometry
Blood samples from KD and SARS-CoV-2+ patients were collected at the time of diagnosis and with regards to KD, always before Intravenous immunoglobulin administration. After ficoll, PBMCs and plasma samples were stored in liquid nitrogen or at −20°C, respectively, in Nunc Cryotubes (Merk KGaA, Darmstadt, Germany). Flow cytometry was performed for 30 patients, and isolated PBMCs were stained with LIVE/DEAD Fixable Near-IR Dead Cell Stain Kit (for 633 or 635 nm excitation, ThermoFisher, Waltham, Massachusetts, US) for 15 minutes at 4°C. Then, the cells were washed in wash buffer (phosphate-buffer saline with 1% bovine serum albumin) and stained for 30 minutes at 4°C with anti-hCD3 PE-CF594, anti-hCD4 BV510, anti-hCD25 PE, anti-hCD45RO-PerCP-Cy 5.5, anti-hCD27 V450, anti-hCD57 APC, anti-hCD185(CXCR5) BV605 (all from BD Biosciences, Milan, Italy), anti-hCD127 PE-Cy7, anti-hPD1 BV711 (all from Biolegend, San Diego, CA). Data acquired by CytoFLEX cytometer (Beckman Coulter, Milan, Italy) were analyzed by FlowJo software v.10 (Treestar Software, Ashland, Oregon, USA).
Quantification and Statistical Analysis
Serum Protein Analyses
Plasma protein analyses in Figure 2 were performed using samples from children with MIS-C (n = 13), children with Kawasaki (n = 28), adult CoV-2+ non-ICU (n = 7 subjects, 14 time points total), and adult CoV-2+ ICU (n = 10 subjects, 43 time points total). In order to compare Adult CoV-2+ cases with Kawasaki and MIS-C children, bridge normalization was used due to available bridge samples between assays regarding children. Bridge normalization was done with the function olink_normalization() from OlinkAnalyze R package. A 30% LOD cutoff was used to filter out proteins for all Olink plates. Then both plates were filtered for matching proteins across. The normalized combined plate with children cases as well as the plate containing the Adult CoV-2+ cases were scaled within each set to unit variance (z-score) before merging. For the PCA and PC contribution plots, both were done using the library factoextra and only the top 20 contributions were displayed in the contribution plot. Plasma protein analyses in Figure 4 were performed using samples from healthy children (n = 12), children with SARS-CoV-2 without hyperinflammation (n = 41), children with MIS-C (n = 21, pre-treatment n = 11, post-treatment n = 7), and children with Kawasaki (n = 28). The four proteins (IL10, IL6, CCL11, IL5) that overlapped between Olink Immune Response and Inflammation panels were averaged. Proteins with > 30% measurements below the threshold of detection were filtered out, resulting in 120 and 133 plasma proteins used for PCA in Figures 2 and 4 respectively. Because Olink data from Figure 4 was run in two batches, bridge normalization was applied using function olink_normalization from R package OlinkAnalyze, followed by batch correction using the function removeBatchEffect from R package limma. The analyzed Olink parameters for Figures 2 and 4, and their mean NPX and standard deviations are provided (Tables S1 and S2). For analyses in Figure 5, seven paired pre/post treatment MIS-C samples were compared.
Virscan Data Analysis
For each sample, sequencing reads were first mapped to the original library sequences with Bowtie, and the number of reads of each peptide in the original library were counted with SAMtools. A zero-inflated generalized Poisson distribution was applied to fit the frequency of peptides, and was calculated for each peptide as the probability of enrichment. Peptides with larger than 2.3 in both technical replicates were considered to be significantly enriched. Among the significantly enriched peptides, those appear in at least 3 of the beads samples were removed as nonspecific bindings. We also filtered out the peptides that were enriched in only one sample. The remaining enriched peptides were used to calculate the virus scores. To remove the hits caused by cross-reactive antibodies, we first sorted the virus by their total number of enriched peptides in descending order. For each virus in this order, we iterated through all the enriched peptides and removed those that shared a sequence of more than 7 amino acids with any previously observed peptides in any virus of the same sample. The remaining enriched peptides were considered specific and their number is the virus score for the virus. Afterward, we filtered out the outlier viruses with Virus Scores > 1 in only sample and negative Virus Scores in all others. We also filtered out viruses with Virus Scores < 2 in all samples.
Autoantibody Analyses
All analyses of autoantibody reactivities were performed using background-subtracted mean signal values of protein duplicates of the human IgG channel. Samples were quantile-normalized, and technical duplicates were averaged. For comparisons of autoantibody target reactivity between MIS-C and other groups (healthy control, CoV-2+ and Kawasaki), differential expression was run using deseq function of the R package DESeq2. Ranked lists were obtained for each comparison to MIS-C (contrasts) and used for GSEA. GSEA was run on the ranked list of unique targets using the gseGO() function of the clusterProfiler R package.
Flow Cytometry Data Analysis
Statistical comparisons between two groups on flow cytometric frequencies were performed with t test if both distributions were approximately normal or, conversely, with the Wilcoxon nonparametric test. In both cases, the normality of the distributions was tested with the D’Agostino-Pearson test. P values less than 0.05 were considered to be statistically significant. MOFA was performed with the MOFA+ package (Argelaguet et al., 2019) (version 1.0) in the R statistical environment. Data analysis was performed using R (version 3.6.2).
Consortia
The members of the CACTUS Consortium at the Children’s Hospital “Bambino Gesù” are Stefania Bernardi, MD, Emma Concetta Manno, MD, Paola Zangari, MD, Lorenza Romani, MD, Carlo Concato, Bsc, Paola Pansa, MD, Sara Chiurchiu, MD, Andrea Finocchi, MD, PhD, Caterina Cancrini, MD, PhD, Laura Lancella, MD, Laura Cursi, MD, Maia De Luca, MD, Renato Cutrera, MD, Libera Sessa, PhD, Elena Morrocchi, PhD, Lorenza Putignani, PhD, Francesca Calò Carducci, MD, Maria A. De Ioris, MD, Patrizia D’Argenio, MD, Marta Ciofi degli Atti, MD, and Carmen D’Amore, MD.
Acknowledgments
We would like to acknowledge all patients and guardians who decided to participate to the study. We thank all the CACTUS study nurses team of the COVID-19 Center of Bambino Gesù Children’s Hospital and Jennifer Faudella, Sonya Martin, and Giulia Neccia for their precious administrative assistance. We thank SciLifeLab Plasma Profiling Facility for generating Olink data and the SciLifeLab autoimmunity profiling facility for instrument support. This work was supported by a grant from Knut and Alice Wallenberg Foundation (KAW) to SciLifeLab as well as donations from Bure Equity AB and Jonas and Christina af Jochnick Foundation to Karolinska Institutet and P.B. We also acknowledge funding provided by SciLifeLab/KAW national COVID-19 research program project grant. The authors are also grateful for support from Children’s Hospital Bambino Gesù, 5 X mille 2019, ricerca corrente 2020 to N.C. and ricerca corrente 2019 to P.P.
Author Contributions
P.B., P.P., and N.C. conceived the study. C.R.C. analyzed the data and generated most figures. F.S. and N.L. performed autoantibody analyzes with support from D.E. L.R. analyzed adult COVID-19 data. C.P. performed VirScan experiments and Z.T. processed this data. T.C. and Y.B. supported P.B. in MIS-C patient enrollment in Stockholm. J.W. and T.L. provided experimental support. G.R.P. analyzed FACS data. D.A., S.Z., A.R., V.S., A.M., A.C., and A.V. enrolled, sampled, and performed experiments on children enrolled in Rome with support from the entire CACTUS consortium. P.B., C.R.C., P.P., and N.C. wrote the paper with input from all co-authors.
Declaration of Interests
P.B. and T.L. are founders and shareholders of Cytodelics AB (Stockholm, Sweden). P.B. is an advisor Scailyte AG (Switzerland).
Published: September 6, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.09.016.
Supplemental Information
Median NPX value (Standard Deviation).
Median NPX value (Standard Deviation).
References
- Anft M., Paniskaki K., Blazquez-Navarro A., Doevelaar A.A.N., Seibert F., Hoelzer B., Skrzypczyk S., Kohut E., Kurek J., Zapka J., et al. COVID-19 progression is potentially driven by T cell immunopathogenesis. medRxiv. 2020 doi: 10.1101/2020.04.28.20083089. [DOI] [Google Scholar]
- Argelaguet R., Arnol D., Bredikhin D., Deloro Y., Velten B., Marioni J.C., Stegle O. MOFA+: a probabilistic framework for comprehensive integration of structured single-cell data. bioRxiv. 2019 doi: 10.1101/837104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhaddou M., Memon D., Meyer B., White K.M., Rezelj V.V., Marrero M.C., Polacco B.J., Melnyk J.E., Ulferts S., Kaake R.M., et al. The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell. 2020;182:685–712. doi: 10.1016/j.cell.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin P. Why is COVID-19 so mild in children? Acta Paediatr. 2020;109:1082–1083. doi: 10.1111/apa.15271. [DOI] [PubMed] [Google Scholar]
- Cheung E.W., Zachariah P., Gorelik M., Boneparth A., Kernie S.G., Orange J.S., Milner J.D. Multisystem Inflammatory Syndrome Related to COVID-19 in Previously Healthy Children and Adolescents in New York City. JAMA. 2020;324:294–296. doi: 10.1001/jama.2020.10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E.Y., Chavakis E., Czabanka M.A., Langer H.F., Fraemohs L., Economopoulou M., Kundu R.K., Orlandi A., Zheng Y.Y., Prieto D.A., et al. Del-1, an endogenous leukocyte-endothelial adhesion inhibitor, limits inflammatory cell recruitment. Science. 2008;322:1101–1104. doi: 10.1126/science.1165218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diorio C., Henrickson S.E., Vella L.A., McNerney K.O., Chase J.M., Burudpakdee C., Lee J.H., Jasen C., Balamuth F., Barrett D.M., et al. Multisystem inflammatory syndrome in children and COVID-19 are distinct presentations of SARS-CoV-2. J. Clin. Invest. 2020 doi: 10.1172/JCI140970. Published online July 30, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolhnikoff M., Ferranti J.F., Monteiro R.A. de A., Duarte-Neto A.N., Gomes-Gouvêa M.S., Degaspare N.V., Delgado A.F., Fiorita C.M., Leal G.N., Rodrigues R.M., et al. SARS-CoV-2 in cardiac tissue of a child with COVID-19-related multisystem inflammatory syndrome. Lancet Child Adolesc. Health. 2020 doi: 10.1016/S2352-4642(20)30257-1. Published online August 20, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufort E.M., Koumans E.H., Chow E.J., Rosenthal E.M., Muse A., Rowlands J., Barranco M.A., Maxted A.M., Rosenberg E.S., Easton D., et al. New York State and Centers for Disease Control and Prevention Multisystem Inflammatory Syndrome in Children Investigation Team Multisystem Inflammatory Syndrome in Children in New York State. N. Engl. J. Med. 2020;383:347–358. doi: 10.1056/NEJMoa2021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldstein L.R., Rose E.B., Horwitz S.M., Collins J.P., Newhams M.M., Son M.B.F., Newburger J.W., Kleinman L.C., Heidemann S.M., Martin A.A., et al. Overcoming COVID-19 Investigators and the CDC COVID-19 Response Team Multisystem Inflammatory Syndrome in U.S. Children and Adolescents. N. Engl. J. Med. 2020;383:334–346. doi: 10.1056/NEJMoa2021680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber C., Patel R., Trachman R., Lepow L., Amanat F., Krammer F., Wilson K.M., Onel K., Geanon D., Tuballes K., et al. Mapping Systemic Inflammation and Antibody Responses in Multisystem Inflammatory Syndrome in Children (MIS-C) medRxiv. 2020 doi: 10.1101/2020.07.04.20142752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunebaum E., Blank M., Cohen S., Afek A., Kopolovic J., Meroni P.L., Youinou P., Shoenfeld Y. The role of anti-endothelial cell antibodies in Kawasaki disease - in vitro and in vivo studies. Clin. Exp. Immunol. 2002;130:233–240. doi: 10.1046/j.1365-2249.2002.02000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueber W., Patel D.D., Dryja T., Wright A.M., Koroleva I., Bruin G., Antoni C., Draelos Z., Gold M.H., Group P.S., et al. Effects of AIN457, a Fully Human Antibody to Interleukin-17A, on Psoriasis, Rheumatoid Arthritis, and Uveitis. Sci. Transl. Med. 2010;2:52ra72. doi: 10.1126/scitranslmed.3001107. [DOI] [PubMed] [Google Scholar]
- Jia S., Li C., Wang G., Yang J., Zu Y. The T helper type 17/regulatory T cell imbalance in patients with acute Kawasaki disease. Clin. Exp. Immunol. 2010;162:131–137. doi: 10.1111/j.1365-2249.2010.04236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Cai Y., Li Z., Huang L., Chen J., Tian L., Wu Z., Li X., Chen Z., Chen C., Yang Z. Screening of differentially expressed genes associated with Kawasaki disease by microarray analysis. Exp. Ther. Med. 2017;14:3159–3164. doi: 10.3892/etm.2017.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.C., Mühlemann B., Veith T., Biele G., Zuchowski M., Hoffmann J., Stein A., Edelmann A., Corman V.M., Drosten C. An analysis of SARS-CoV-2 viral load by patient age. medRxiv. 2020 doi: 10.1101/2020.06.08.20125484. [DOI] [Google Scholar]
- Kazatchkine M.D., Kaveri S.V., Kazatchkine M.D., Kaveri S.V. Immunomodulation of autoimmune and inflammatory diseases with intravenous immune globulin. N. Engl. J. Med. 2001;345:747–755. doi: 10.1056/NEJMra993360. [DOI] [PubMed] [Google Scholar]
- Kobuke K., Furukawa Y., Sugai M., Tanigaki K., Ohashi N., Matsumori A., Sasayama S., Honjo T., Tashiro K. ESDN, a novel neuropilin-like membrane protein cloned from vascular cells with the longest secretory signal sequence among eukaryotes, is up-regulated after vascular injury. J. Biol. Chem. 2001;276:34105–34114. doi: 10.1074/jbc.M105293200. [DOI] [PubMed] [Google Scholar]
- Kuri-Cervantes L., Pampena M.B., Meng W., Rosenfeld A.M., Ittner C.A.G., Weisman A.R., Agyekum R., Mathew D., Baxter A.E., Vella L., et al. Immunologic perturbations in severe COVID-19/SARS-CoV-2 infection. bioRxiv. 2020 doi: 10.1101/2020.05.18.101717. [DOI] [Google Scholar]
- Landegren N., Sharon D., Freyhult E., Hallgren Å., Eriksson D., Edqvist P.-H., Bensing S., Wahlberg J., Nelson L.M., Gustafsson J., et al. Proteome-wide survey of the autoimmune target repertoire in autoimmune polyendocrine syndrome type 1. Sci. Rep. 2016;6:20104. doi: 10.1038/srep20104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Zhang L., Du H., Zhang J., Li Y.Y., Qu J., Zhang W., Wang Y., Bao S., Li Y., et al. Chinese Pediatric Novel Coronavirus Study Team SARS-CoV-2 Infection in Children. N. Engl. J. Med. 2020;382:1663–1665. doi: 10.1056/NEJMc2005073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg M., Eriksson A., Tran B., Assarsson E., Fredriksson S. Homogeneous antibody-based proximity extension assays provide sensitive and specific detection of low-abundant proteins in human blood. Nucleic Acids Res. 2011;39:e102. doi: 10.1093/nar/gkr424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maecker H., Varfolomeev E., Kischkel F., Lawrence D., LeBlanc H., Lee W., Hurst S., Danilenko D., Li J., Filvaroff E., et al. TWEAK attenuates the transition from innate to adaptive immunity. Cell. 2005;123:931–944. doi: 10.1016/j.cell.2005.09.022. [DOI] [PubMed] [Google Scholar]
- Martinez-Aguilar E., Gomez-Rodriguez V., Orbe J., Rodriguez J.A., Fernández-Alonso L., Roncal C., Páramo J.A. Matrix metalloproteinase 10 is associated with disease severity and mortality in patients with peripheral arterial disease. J. Vasc. Surg. 2015;61:428–435. doi: 10.1016/j.jvs.2014.09.002. [DOI] [PubMed] [Google Scholar]
- Mathew D., Giles J.R., Baxter A.E., Greenplate A.R., Wu J.E., Alanio C., Oldridge D.A., Kuri-Cervantes L., Pampena M.B., D’Andrea K., et al. Deep immune profiling of COVID-19 patients reveals patient heterogeneity and distinct immunotypes with implications for therapeutic interventions. bioRxiv. 2020 doi: 10.1101/2020.05.20.106401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister K.A., Grogg K.M., Johnson D.W., Gallione C.J., Baldwin M.A., Jackson C.E., Helmbold E.A., Markel D.S., McKinnon W.C., Murrell J., et al. Endoglin, a TGF-β binding protein of endothelial cells, is the gene for hereditary haemorrhagic telangiectasia type 1. Nat. Genet. 1994;8:345–351. doi: 10.1038/ng1294-345. [DOI] [PubMed] [Google Scholar]
- Moraleda C., Serna-Pascual M., Soriano-Arandes A., Simó S., Epalza C., Santos M., Grasa C., Rodríguez M., Soto B., Gallego N., et al. Multi-Inflammatory Syndrome in Children related to SARS-CoV-2 in Spain. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1042. Published online July 25, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olin A., Henckel E., Chen Y., Lakshmikanth T., Pou C., Mikes J., Gustafsson A., Bernhardsson A.K., Zhang C., Bohlin K., Brodin P. Stereotypic Immune System Development in Newborn Children. Cell. 2018;174:1277–1292. doi: 10.1016/j.cell.2018.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parri N., Lenge M., Buonsenso D., Coronavirus Infection in Pediatric Emergency Departments (CONFIDENCE) Research Group Children with Covid-19 in Pediatric Emergency Departments in Italy. N. Engl. J. Med. 2020;383:187–190. doi: 10.1056/NEJMc2007617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pou C., Nkulikiyimfura D., Henckel E., Olin A., Lakshmikanth T., Mikes J., Wang J., Chen Y., Bernhardsson A.K., Gustafsson A., et al. The repertoire of maternal anti-viral antibodies in human newborns. Nat. Med. 2019;25:591–596. doi: 10.1038/s41591-019-0392-8. [DOI] [PubMed] [Google Scholar]
- Riphagen S., Gomez X., Gonzalez-Martinez C., Wilkinson N., Theocharis P. Hyperinflammatory shock in children during COVID-19 pandemic. Lancet. 2020;395:1607–1608. doi: 10.1016/S0140-6736(20)31094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez L., Pekkarinen P.T., Lakshmikanth T., Tan Z., Consiglio C.R., Pou C., Chen Y., Mugabo C.H., Nguyen N.A., Nowlan K., et al. Systems-level immunomonitoring from acute to recovery phase of severe COVID-19. Cell Rep. Med. 2020;1:100078. doi: 10.1016/j.xcrm.2020.100078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostad C.A., Chahroudi A., Mantus G., Lapp S.A., Teherani M., Macoy L., Rostad B.S., Milla S.S., Tarquinio K.M., Basu R.K., et al. Serology in Children with Multisystem Inflammatory Syndrome (MIS-C) associated with COVID-19. medRxiv. 2020 doi: 10.1101/2020.07.10.20150755. [DOI] [PubMed] [Google Scholar]
- Rowley A.H., Baker S.C., Orenstein J.M., Shulman S.T. Searching for the cause of Kawasaki disease--cytoplasmic inclusion bodies provide new insight. Nat. Rev. Microbiol. 2008;6:394–401. doi: 10.1038/nrmicro1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghi M.M., Esmailzadeh L., Zhang J., Guo X., Asadi A., Krassilnikova S., Fassaei H.R., Luo G., Al-Lamki R.S.M., Takahashi T., et al. ESDN is a marker of vascular remodeling and regulator of cell proliferation in graft arteriosclerosis. Am. J. Transplant. 2007;7:2098–2105. doi: 10.1111/j.1600-6143.2007.01919.x. [DOI] [PubMed] [Google Scholar]
- Sakurai Y. Autoimmune Aspects of Kawasaki Disease. J. Investig. Allergol. Clin. Immunol. 2019;29:251–261. doi: 10.18176/jiaci.0300. [DOI] [PubMed] [Google Scholar]
- Schatorjé E.J.H., Gemen E.F.A., Driessen G.J.A., Leuvenink J., van Hout R.W.N.M., de Vries E. Paediatric reference values for the peripheral T cell compartment. Scand. J. Immunol. 2012;75:436–444. doi: 10.1111/j.1365-3083.2012.02671.x. [DOI] [PubMed] [Google Scholar]
- Shulman S.T., Rowley A.H. Kawasaki disease: insights into pathogenesis and approaches to treatment. Nat. Rev. Rheumatol. 2015;11:475–482. doi: 10.1038/nrrheum.2015.54. [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetro J.A. Is COVID-19 receiving ADE from other coronaviruses? Microbes Infect. 2020;22:72–73. doi: 10.1016/j.micinf.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Vardhana S.A., Wolchok J.D. The many faces of the anti-COVID immune response. J. Exp. Med. 2020;217:e20200678. doi: 10.1084/jem.20200678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdoni L., Mazza A., Gervasoni A., Martelli L., Ruggeri M., Ciuffreda M., Bonanomi E., D’Antiga L. An outbreak of severe Kawasaki-like disease at the Italian epicentre of the SARS-CoV-2 epidemic: an observational cohort study. Lancet. 2020;395:1771–1778. doi: 10.1016/S0140-6736(20)31103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker E., Bamford A., Kenny J., Kaforou M., Jones C.E., Shah P., Ramnarayan P., Fraisse A., Miller O., Davies P., et al. PIMS-TS Study Group and EUCLIDS and PERFORM Consortia Clinical Characteristics of 58 Children With a Pediatric Inflammatory Multisystem Syndrome Temporally Associated With SARS-CoV-2. JAMA. 2020;324:259–269. doi: 10.1001/jama.2020.10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G.J., Kula T., Xu Q., Li M.Z., Vernon S.D., Ndung’u T., Ruxrungtham K., Sanchez J., Brander C., Chung R.T., et al. Viral immunology. Comprehensive serological profiling of human populations using a synthetic human virome. Science. 2015;348:aaa0698. doi: 10.1126/science.aaa0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Tan Y., Ling Y., Lu G., Liu F., Yi Z., Jia X., Wu M., Shi B., Xu S., et al. Viral and host factors related to the clinical outcome of COVID-19. Nature. 2020;583:437–440. doi: 10.1038/s41586-020-2355-0. [DOI] [PubMed] [Google Scholar]
- Zhu H., Bilgin M., Bangham R., Hall D., Casamayor A., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T., et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Median NPX value (Standard Deviation).
Median NPX value (Standard Deviation).
Data Availability Statement
Original data have been deposited to Mendeley Data: http://dx.doi.org/10.17632/ds6g796xyg.1. Scripts to reproduce the analyses presented in each figure of the paper are available at https://github.com/Brodinlab/MIS-C_manuscript.