Highlights
-
•
Phenol catabolism among three psychrotolerant yeast strains was examined.
-
•
For this, enzymatic and chromatographic (GC–MS) techniques were applied.
-
•
The impact of cell inoculum preparation on the enzymatic activity was examined.
-
•
All yeast strains utilized phenol via the ortho-cleavage pathway.
-
•
None of the yeast strains utilized phenol via the meta-cleavage pathway.
Keywords: Catechol 1,2- and 2,3-dioxyganeses activity assays; Ortho-cleavage pathway; cis,cis-Muconic acid; Gas chromatography mass spectrometry (GC–MS); Enzymatic assay; Phenol biodegradation by psychrotolerant peatland yeasts
Abstract
In this study, three psychrotolerant phenol-degrading yeast strains Candida subhashii (strain A011), Candida oregonenis (strain B021) and Schizoblastosporion starkeyi-henricii (strain L012) isolated from Rucianka peatland were examined to determine which alternative metabolic pathway for phenol biodegradation is used by these microorganisms. All yeast strains were cultivated in minimal salt medium supplemented with phenol at 500, 750 and 1000 mg l−1 concentration with two ways of conducting phenol biodegradation experiments: with and without the starving step of yeast cells. For studied yeast strains, no catechol 2,3-dioxygenase activities were detected by enzymatic assay and no products of catechol meta-cleavage in yeast cultures supernatants (GC–MS analysis), were detected. The detection of catechol 1,2-dioxygenase activity and the presence of cis,cis-muconic acid in the analyzed samples revealed that all studied psychrotolerant yeast strains were able to metabolize phenol via the ortho-cleavage pathway. Therefore, they may be tested in terms of their use to develop biotechnology for the production of cis,cis-muconic acid, a substrate used in the production of plastics (PET) and other valuable goods.
1. Introduction
Phenol and phenolic compounds are widely used in industry for production of e.g. polymeric resin, oil refining and various cosmetic and medical products [1]. Moreover, these compounds are the main by-products formed during certain industrial processes [2,3], and are detected mostly in plants and industrial effluents, for example from coal conversion – as well as refinery and pharmaceutical industries [4]. Other than many known physical and chemical methods for phenols removal, biological methods play an important role in wastewater and soil treatment. In respect to chemical methods, Advanced Oxidation Processes (AOPs) are widely used for degradation of phenol and its derivatives [[5], [6], [7]]. In comparison with physical and chemical methods, biological methods possess advantageous attributes due to their relatively low processing costs and minimization of secondary pollutant formation [[8], [9], [10]]. Therefore, aerobic biodegradation of phenol has been the subject of numerous investigations [11].
Aerobic biodegradation of phenolic compounds is accomplished via pathways, where catechol or its derivatives play a key role. These intermediates are then further metabolized by intradiol or extradiol dioxygenases [12].
In the case of phenol, the first step of biodegradation involves the transformation of phenol into catechol via phenol hydroxylases, which involves the attachment of a hydroxyl group at the ortho-position of an aromatic ring [13]. Catechol is further degraded via ortho- or meta- cleavage by 1,2-catechol dioxygenases or 2,3-catechol dioxygenases, respectively (Fig. 1 ). These enzymes add two oxygen atoms to the aromatic ring of catechol, followed by a cleavage of chemical bonds and an opening of the aromatic ring [14].
Fig. 1.
Schemes of the ortho- and meta-pathways present in phenol and its derivates-degrading microorganisms; based on Paisio et al. [47].
Catechol 1,2-dioxygenase (C12O) (EC 1.13.11.1) contains Fe3+ as a prosthetic group and belongs to the enzymes that enable cleavage of catechol as intradiol (ortho-cleavage), producing cis,cis-muconic acid [15]. In the case of catechol 2,3-dioxygenase (C23O) (EC 1.13.11.2), it contains Fe2+ as a prosthetic group or the use of other two-valent metal ions to cleave the aromatic ring of catechol as extradiol (meta-cleavage). This leads to the production of 2-hydroxymuconic semialdehyde. During the next steps of the degradation process, these products of ring cleavage are converted to aliphatic compounds which can enter the tricarboxylic acid cycle [15,16].
Over the last decades, catechol 1,2-dioxygenases and catechol 2,3-dioxygenases have been extensively studied due to their key role in the cleavage of catechol’s aromatic ring. Catechol 1,2-dioxygenases play an important role in the degradation pathways of various aromatic compounds and are ubiquitous in microorganisms [17,18]. Furthermore, they are active in some bacterial strains belonging to genus Acinetobacter, Arthrobacter, Pseudomonas, Sphingomonas, Geobacillus and Rhodococcus [[19], [20], [21], [22]]. In the case of yeasts, the activity of these dioxygenases was found, among others, in Candida tropicalis [23,24], Candida albicans [15] and Trichosporon cutaneum [25]. In the light of current research, it is worth noting that Margesin et al. [26,27] isolated and characterized cold-adapted phenol degrading yeasts with C12O enzymatic activity from Alpine glacier cryoconite and petroleum hydrocarbon contained alpine soils in Austria, respectively.
For comparison, catechol 2,3-dioxygenases activity was detected in bacteria from genus Pseudomonas, Sphingomonas, Acinetobacter, Ralstonia, Burkholderia, Rhodococcus and Bacillus [[28], [29], [30], [31], [32], [33], [34], [35]]. However, so far, among yeasts, the activity of catechol 2,3-dioxygenase is very rarely detected. Interestingly, C23Os activity was confirmed in cold-adapted yeast strain belonging to Rhodotorula sp. [26] isolated from Alpine glacier cryoconite.
Summarizing, the literature study indicates that studies on phenol metabolism have been mainly conducted for bacteria. Hence, the information regarding studying enzymatic activities of catechol 1,2- and 2,3-dioxygenases enzymes involved in phenol metabolism and the investigation of their products among yeast strains is rather limited. On the other hand, despite the limited number of studies, we found two different approaches to investigate the activity of these enzymes among yeast strains. The first approach is focused on the examination of enzymatic activities of pure native or recombinant yeast catechol dioxygenases [23,24]. Second approach is focused on determining which of possible catechol dioxygenases activities is present in examined yeast strains. However, in the second approach, the enzymatic activity of specific enzymes or the concentration of the biodegradation products were not examined [26,27].
In this research, we decided to combine these two approaches. Firstly, we determined which of the above mentioned enzymatic activities is present in the tested yeast strains, which allowed us to point out which of the phenol metabolism pathways is used by them. We also decided to determine the presence of products of catechol 1,2- and 2,3-dioxygenases enzymatic activity in yeast cultures, i.e. cis,cis-muconic acid and 2-hydroxymuconic semialdehyde, respectively.
Currently, cis,cis-muconic acid is one of the possible industrial substrates for the production of terephthalic acid - monomer of the high-demand plastic polymer polyethylene terephthalate (PET). Moreover, this compound may be also used for the production of adipic acid, which is a well-known precursor for the production of nylon, lubricants, coatings, plastics, and plasticizers. Nowadays, cis,cis-muconic acid production is based on the two-stage chemical synthesis process which requires a high input of energy and leads to the production of large amounts of greenhouse gas N2O. Compared with the chemical production of cis,cis-muconic acid from catechol, biotechnological processes show the advantages of high productivity and selectivity, with a final yield of 100% in comparison to less than 35% and low purity obtained with the chemical cleavage of catechol [40].
Therefore, recently there is a growing interest in developing microbial technology to synthesize cis,cis-muconic acid as an alternative for currently used technology for its synthesis. For this purpose, the possibility of using genetically modified microorganisms e.g. Pseudomonas putida and Corynebacterium glutamicum utilizing such carbon sources as catechol [41,55] and toluene [42] and Escherichia coli [43] and Saccharomyces cerevisiae utilizing glucose as a carbon source [44,45] has been investigated. More importantly, genetically modified Pseudomonas putida or Corynebacterium glutamicum strains which produce cis,cis-muconic acid from aromatics compounds offer the use of lignin hydrolysates as starting material [41,55]. Lignin (the second most abundant polymer in nature) can be processed into the mixtures of small aromatics compounds, such as e.g. phenol and catechol, through biological and thermochemical depolymerization, respectively [41,55].
In summary, the presence of C12Os and C23Os activities in cold-adapted yeast strains [26,27] and the biotechnological potential of microorganisms metabolizing catechol via ortho-cleavage pathway for their future developing in the plastic industry, encouraged us to conduct the presented study on cold-adapted yeasts strains isolated from Rucianka peatland. These yeast strains were characterized as microorganisms which enable the effective utilization of phenol [36] and catechol [46] as a sole source of carbon. Therefore, the main purpose of this study was the identification of catechol dioxygenases activities (C12Os or C23Os activity) and the products of the enzymatic reaction of catechol ring cleavage (Fig. 1) catalyzed by them (cis,cis-muconic acid and 2-hydroxymuconic semialdehyde, respectively) and hence, which metabolic pathway (ortho-cleavage or meta-cleavage pathway) is involved in phenol biodegradation by Candida subhashii strain A011, Candida oregonensis strain B021 and Schizoblastosporion starkeyi-henricii strain L012.
Moreover, according to the literature, during the biodegradation experiments examined microorganisms are preincubated in basal medium without any carbon source (starving step) and then used as an inoculum of a culture medium with phenol as the sole source of carbon [28,37]. However, in analogous studies, the starving step is not involved as a part of phenol biodegradation experiments [38,39]. Therefore, this study also includes a comparison of the different preparation types of yeast cells inoculum and evaluation of their possible impact on catechol dioxygenases activities in the examined yeast strains.
2. Materials and methods
2.1. Microorganisms
The phenol degrading psychrotolerant yeast strains, including Candida subhashii (strain A011), Candida oregonenis (strain B021) and Schizoblastosporion starkeyi-henricii (strain L012), were examined during this study and isolated from water and soil samples collected from Rucianka peatland. All strains were characterized in our previous publication, see ref. [36]. A011 and L012 strains were able to degrade phenol as a sole source of carbon at 500, 750 and 1000 mg l−1 in culture medium. B021 strain was able to degrade phenol also at 500 and 750 mg l−1 but not at 1000 mg l−1.
2.2. Culture growth conditions
2.2.1. Culture growth of yeast strains without the starving step
In this type of cultivation, the first step of an experiment involved the preparation of inoculum for each analyzed yeast strains. This was achieved by the inoculation of 3 mL of YPD (Yeast extract Peptone Dextrose) medium with the single colony of the examined yeast strain. In this study, YPD contained (per liter): glucose 20 g, yeast extract 10 g and casein peptone 20 g, and was also supplemented with 1 mL (per liter) of each antibiotic: chloramphenicol (stock solution: 34 mg ml−1) and ampicillin (stock solution: 100 mg ml−1). The addition of antibiotics was aimed at inhibiting bacterial growth caused by an accidental contamination of YPD medium.
Then, test tubes were incubated at 18 °C on the rotary shaker at 170 rpm for 1 day. After this period, 80 μL of each yeast culture was transferred to 20 mL of sterile YPD medium in 200 mL Erlenmeyer flasks. The flasks were incubated at 18 °C on the rotary shaker at 170 rpm for 3 days. Next, these cultivated yeast cells were used as an inoculum. In all experiments 5% of subculture was inoculated into sterile 80 mL of mineral salt medium (MSM) in 500 mL Erlenmeyer flasks, supplemented with phenol (Sigma Aldrich, USA) at a concentration of 500 mg l−1, 750 mg l−1, and 1000 mg l−1, respectively. Mineral salt medium contained (per liter); 0.4 g of (NH4)2SO4, 0.4 g of K2HPO4, 0.2 g of KH2PO4, 0.1 g of NaCl, 0.1 g of MgSO4, 0.01 g of MnSO4·H2O, 0.01 g of Fe2(SO4)3·nH2O, (all purchased from POCH S.A., Poland) 0.01 g of Na2MoO4·2H2O (Todini Europe Sp. z o.o., Kraków, Poland), and 0.25 g of casein peptone (BTL Sp. z o.o., Łódź, Poland), supplemented with 1 mL (per liter) of each antibiotics: chloramphenicol (stock solution: 34 mg ml−1) and ampicillin (stock solution: 100 mg ml−1). The pH of the medium was adjusted to 6.0.
All yeast strains were cultivated until yeast cultures obtained the stationary phase of growth.
2.2.2. Culture growth of yeast strains with starving step
In this type of cultivation, an inoculum of each yeast strain was prepared by cultivation of microorganisms in Yeast Nitrogen base without an amino acid medium (Sigma Aldrich, USA) which was not supplemented with any source of carbon. YNB w/a contained (per liter): 6.7 g of YNB w/a base, supplemented with 1 mL (per liter) of each antibiotic: chloramphenicol (stock solution: 34 mg ml−1) and ampicillin (stock solution: 100 mg ml−1).
For the inoculum preparation, 4 mL of YNB w/a medium was inoculated with a single colony of each analyzed yeast strain. Then, test tubes with all examined yeast strains were incubated at 18 °C on a rotary shaker at 170 rpm for 4 days. Next, the cultivated yeast cells were used as an inoculum. In all the experiments 5% of subculture was inoculated into sterile 80 mL of mineral salt medium in 500 mL Erlenmeyer flasks, supplemented with phenol at a concentration of 500, 750, and 1000 mg l−1, respectively.
All yeast strains were cultivated until yeast cultures obtained the stationary phase of growth.
To avoid repeating the “initial phenol concentration in culture medium” term, abbreviations ipc 500, ipc 750 and ipc 1000 (for 500, 750 and 1000 mg l−1 initial phenol concentration, respectively) were employed throughout the manuscript.
2.3. Preliminary screening of catechol dioxygenases activities in phenol degrading yeast cultures
To determine the presence of catechol 1,2- or 2,3-dioxygenases activities, respectively, the analyzed yeast cultures were tested using 96-well plates (qualitative assay). Catechol 1,2-dioxygenase activity was tested according to Neidle and Ornston [48] and Birger et al. [49]. The first step of examination involved the preparation of a solution containing 0.004% phenol red (Sigma Aldrich, USA), 1 mM EDTA (POCH S.A., Poland) and 10 mM catechol (Sigma Aldrich, USA) (pH = 7.5, adjusted with ammonium hydroxide). To 150 μl of this solution, 50 μl of the liquid yeast culture of each phenol-degrading yeasts (OD600 was adjusted to 1) was added. The color of the analyzed samples changed from red to yellow/orange occurred within 10 min when placed in the dark at room temperature, which was due to the presence of catechol 1,2-dioxygenase.
Catechol 2,3-dioxygenase activity was determined as described by Morgan et al. [50] and Birger et al. [49]. 150 μL of solution containing 90 mM catechol in 50 mM tris-acetate buffer (pH 7.5) was added to 50 μl of the liquid culture of each yeast strains (OD600 was adjusted to 1). The formation of a green-brownish color occurred within 2 h in the analyzed samples when placed in the dark at room temperature, which was associated with catechol 2,3-dioxygenase activity. Reagents for tris-acetate preparation were purchased from POCH S.A., Poland.
2.4. Preparation of crude enzyme extracts
Crude enzyme extracts were obtained by yeast cells disruption with glass beads and then removal of cell debris. Cells of each yeast strain were cultivated in the conditions presented above, respectively, until the yeast strains obtained the stationary phase of cultures growth. Then, the yeast cultures were harvested and centrifuged at 10,000 rpm for 10 min. Subsequently, the cell pellets were suspended in sterile deionized water and centrifuged at conditions presented above. The “cells washing” step was repeated twice. Then, the cell pellets were suspended in 50 mM Tris−HCl (pH 8.0) buffer and disrupted with glass beads with 425−600 μm diameter (Sigma Aldrich, USA). Disruption was performed ten times: 30 s incubation on ice and 30 s of vortexing of each analyzed sample. Next, the cell debris was removed by centrifugation at 10,000 rpm for 10 min at 4 °C. The cleared crude enzyme extracts were used for the enzymatic assays, total protein concentration assay and GC–MS analysis. Reagents for Tris−HCl preparation were purchased from POCH S.A., Poland.
2.5. Enzymatic assays
Enzyme activities for catechol 1,2-dioxygenases (C12Os) and catechol 2,3-dioxygenases (C23Os) were determined for both, yeast cultures and crude enzyme extracts, respectively.
C12Os and C23Os activities were determined according to the methods described in ref. [27,51,52] with some modifications. 100 μL of crude enzyme extracts or yeast cultures were mixed with 750 μL of 50 mM Tris−HCl (pH 8.0) and 50 μL of 4 mM catechol. The activity of C12Os was determined by increasing the absorbance values at 260 nm using a spectrophotometer due to the cleavage of catechol into cis,cis-muconic acid. In the case of C23Os, the activity was determined by increasing the absorbance values at 375 nm due to the conversion of catechol into 2-hydroxymuconic semialdehyde (2-HMS). The formation of cis,cis-muconic acid or 2-HMS was monitored for 5 min. Control reactions without yeast cultures or crude enzyme extracts, respectively, were performed for each assay.
For crude enzyme extracts, total protein concentration was determined using the Bradford assay [53].
One unit (U) of C12Os or C23Os activity was defined as the amount of enzyme that generates 1 μmol of specific product, respectively, per min under the assay conditions. Specific activities were expressed as units each enzymatic activity (U) per mg of total cell proteins.
2.6. Detection of phenol degradation intermediates by GC–MS technique
2.6.1. Extraction and derivatization of phenol biodegradation intermediates
Derivatization and detection of phenol degradation intermediates were performed for crude cell extracts and supernatants of yeast cultures. Crude cell extracts after derivatization were labeled as “derivatized cell extracts”.
After 24, 48 and 96 h of yeast cultures growth without the starving step and after 24, 48, 96 and 168 h of cultures growth with this step, all yeast cultures were centrifuged at 12,000 rpm for 10 min, and supernatants collected. In the case of using crude cell extracts, after the cultures were centrifuged, cell pellets were suspended in 10 mL of MSM medium and disrupted with glass beads. The same procedure as described in section 2.4. was used for this purpose.
Next, 10 mL of each supernatant or crude cell extract was acidified to pH 2–3 with 6 M HCl and extracted with 10 mL ethyl acetate (POCH S.A., Poland). Next, each of the collected ethyl acetate phases were dried over anhydrous Na2SO4 and concentrated using a rotary evaporator to a volume of 1 mL. Derivatization was performed at 60 °C for 30 min using 0.1 mL N,O-bis(trimethylsilyl) acetamide [54]. 25 μL of 4-chlorophenol (Sigma Aldrich, Poland) internal standard solution was added to each sample before derivatization (IS, final concentration: 0.006 mg/mg H2O).
2.6.2. Detection of phenol degradation intermediates by gas chromatography - GC–MS analysis
To confirm the presence of phenol biodegradation intermediates and measure their concentration, derivatized cell extracts or derivatized supernatants of each yeast culture were analyzed by GC–MS technique. 1 mL of each sample after derivatization was placed in a 2 mL glass vial (SHIM-POL A. M. Borzymowski, Poland). Samples were analyzed using a QP2010 GC–MS SE gas chromatograph-mass spectrometer (Shimadzu, Japan) equipped with a combi-PAL AOC 5000 autosampler (Shimadzu, Japan) and DB-624 ms capillary column (60 m x0.25 mm x1.40 μm) (Restek, USA).
Chromatographic conditions:
Temperature program: 50 °C (5 min) – ramped at 5 °C/min to 250 °C (5 min); injection port temperature 300 °C; purge off time 2 min (splitless mode); 1 μL of the extract was injected into the GC system; detector temperature 280 °C; ion source temperature (EI, 70 eV) 220 °C; GC–MS transfer line temperature 280 °C; the carrier gas was hydrogen (1 mL/min). The mass spectrometer was operated in single ion monitoring mode (SIM). Quantitative analysis: internal standard method.
Phenol, catechol and cis,cis-muconic acid in the derivatized samples obtained from yeast culture supernatants and cell extracts were identified by GC–MS technique on the basis of retention time values and characteristic ratio of fragmentation ions (mass to charge ratio values, m/z) determined for each compound during the method development stage using analytical standards. The concept of additional identification in SIM mode based on calculated ratio of multiple m/z value specific to each analyte was described in our previous paper [3]. The GC–MS spectrum of main derivatized products of phenol degradation has been presented on Fig. 2 .
Fig. 2.
GC–MS spectra (Electron impact ionization) of derivatized analytes – a) phenol TMS derivative; b) catechol TMS derivative; c) cis,cis-muconic acid TMS derivative.
3. Result and discussion
3.1. Preliminary screening of catechol dioxygenases activities in cultures of phenol degrading yeast strains
C12Os activity was detected in cultures of all analyzed yeast strains, independently of the initial phenol concentration in the culture medium and the presence or absence of starving step during phenol biodegradation experiments. A yellow or orange color appeared in all wells (96-well plates), which contained the samples of phenol-degrading yeast cultures mixed with the reaction solution. Hence, all analyzed yeast strains were probably able to utilize phenol via ortho-cleavage pathway.
However, C23Os activity was not detected in the cultures of any examined yeast strains, under the same experimental conditions as C12Os, as shown above. The examined yeast strains displayed no change in the reaction mixtures color in the wells of 96-well plates during the preliminary screening. Furthermore, these yeast strains were probably not capable of utilizing phenol via meta-cleavage pathway.
3.2. Enzymatic assays
In general, for yeast cultures, independently of initial phenol concentration in medium or the presence or absence of starving step during phenol biodegradation experiments, no activity of catechol 2,3-dioxygenases was detected (data not shown). In the case of the second examined enzyme, all analyzed yeast cultures exhibited low catechol 1,2-dioxygenase activities (Table 1 and Table 3 ). However, according to Tsai et al. [15], enzymes involved in phenol degradation should be not secret to the culture medium but remain inside of the yeast cells. Hence, the same assays for both enzymes were conducted for crude enzyme extracts of analyzed yeast strains (Table 2 and Table 4 ). The comparison of the total 1,2-dioxygenase activity value for each strain of yeast (Table 1 with Table 2 and Table 3 with Table 4, respectively) confirmed the conclusions presented by Tsai et al. [15].
Table 1.
Total enzymatic C12O activity [U] of yeast strain cultures where inoculum was prepared without the starving step.
|
Yeast strain (initial phenol concentration [mg l−1]) |
Total enzymatic C12O activity [U] |
|---|---|
| A011 (500) | 0.007 ± 0.0003 |
| A011 (750) | 0.014 ± 0.0007 |
| A011 (1000) | 0.007 ± 0.0004 |
| B021 (500) | 0.005 ± 0.0003 |
| B021 (750) | 0.001 ± 0.0001 |
| L012 (500) | 0.006 ± 0.0003 |
| L012 (750) | 0.001 ± 0.0001 |
| L012 (1000) | 0.0001 ± 0.000 |
Table 3.
Total enzymatic C12O activity [U] of yeast strains cultures where inoculum was prepared with the starving step.
|
Yeast strain (initial phenol concentration [mg l−1]) |
Total enzymatic C12O activity [U] |
|---|---|
| A011 (500) | 0.001 ± 0.0001 |
| A011 (750) | 0.010 ± 0.0005 |
| A011 (1000) | 0.008 ± 0.0004 |
| B021 (500) | 0.002 ± 0.0001 |
| B021 (750) | 0.003 ± 0.0002 |
| L012 (500) | 0.002 ± 0.0001 |
| L012 (750) | 0.004 ± 0.0002 |
| L012 (1000) | 0.051 ± 0.0025 |
Table 2.
Total [U] and specific C12O activity [U mg −1] of crude enzymatic extracts obtained from yeast cultures where inoculum was prepared without the starving step.
|
Yeast strain (initial phenol concentration [mg l−1]) |
Total enzymatic C12O activity [U] |
Specific enzymatic C12O activity [U mg−1] |
|---|---|---|
| A011 (500) | 0.071 ± 0.0036 | 0.084 ± 0.0042 |
| A011 (750) | 0.16 ± 0.0081 | 0.14 ± 0.0070 |
| A011 (1000) | 0.19 ± 0.0092 | 0.25 ± 0.012 |
| B021 (500) | 0.16 ± 0.0081 | 0.41 ± 0.021 |
| B021 (750) | 0.13 ± 0.0067 | 2.17 ± 0.11 |
| L012 (500) | 0.023 ± 0.0011 | 0.044 ± 0.0022 |
| L012 (750) | 0.13 ± 0.0067 | 0.16 ± 0.0082 |
| L012 (1000) | 0.0051 ± 0.00062 | 0.058 ± 0.0029 |
Table 4.
Total [U] and specific C12O activity [U mg −1] of crude enzymatic extracts obtained from yeast cultures where inoculum was prepared with the starving step.
|
Yeast strain (initial phenol concentration [mg l−1]) |
Total enzymatic C12O activity [U] |
Specific enzymatic C12O activity [U mg−1] |
|---|---|---|
| A011 (500) | 0.071 ± 0.0035 | 0.17 ± 0.0086 |
| A011 (750) | 0.0041 ± 0.00022 | 0.0064 ± 0.00032 |
| A011 (1000) | 0.27 ± 0.00092 | 0.57 ± 0.029 |
| B021 (500) | 0.0034 ± 0.00014 | 0.0047 ± 0.00023 |
| B021 (750) | 0.016 ± 0.00083 | 0.023 ± 0.0012 |
| L012 (500) | 0.0093 ± 0.00052 | 0.36 ± 0.018 |
| L012 (750) | 0.35 ± 0.0052 | 0.47 ± 0.023 |
| L012 (1000) | 0.0032 ± 0.00014 | 0.012 ± 0.00060 |
In the case of crude enzyme extracts obtained after experiments without the starving step, higher specific activities of catechol 1,2-dioxygenases were observed for two yeast strains as well as increased phenol concentration (Table 2). This was observed for strain Candida subhashii A011 and Candida oregonensis B021 in which the highest activities were for 1000 mg l−1 and 750 mg l−1 initial phenol concentration and reached almost 0.25 U mg−1 and 2.17 U mg−1, respectively. In the case of strain Schizoblastosporion starkeyi-henricii L012, the highest specific enzyme activity (0.16 U mg−1) was observed for ipc 750, however, the abject ipc 500 and ipc 1000 obtained enzymatic activities were comparable but much lower than for ipc 750 (Table 2). Among all examined yeast strains and different initial phenol concentration in the culture medium, the highest C12O enzyme activity was detected for strain B021 and phenol at 750 mg l−1 concentration – 2.17 U mg−1.
For experiments including the starving step, no clear correlation between increasing initial phenol concentrations in the culture medium and higher specific enzymatic activities of C12Os was determined (Table 4). All yeast strains performed at the highest enzymatic activity for specific concentration – A011 for ipc 1000, both B021 and L012 for ipc 750.
Unfortunately, at this point it was impossible to unambiguous determine the exact way that yeast cultures cultivation promoted higher enzymatic activity or to point out the one common pattern according which yeasts’ enzymatic activity changed during the cultivation with different conditions and initial phenol concentration. Other conclusions determined for the analysis were: i) in the case of inoculums prepared without the starving step, almost always, other than one yeast strains – L012, increasing initial phenol concentration was associated with increasing specific activity values of C12Os; ii) for strain A011 in the case of ipc 500 and 1000, specific C12O activities of crude enzyme extracts obtained for this strain cultivated with the starving step were at least twice as high than those cultivated without this step; iii) the highest activity among all variants of yeast cultivation were observed for strain B021 cultivated without the starving step and ipc 750; iv) for strain B021, catechol 1,2-dioxygenese activity was almost 100 times higher for its cultivation without the starving step.
As it was mentioned in the Introduction section, because of the limited number of studies dedicated for characterization of yeast catechol dioxygenases [15,[23], [24], [25], [26], [27]] the comparison of our results with previously published research results offers a limited possibility for a discussion. Nevertheless, we decided to show the similarity between results for specific enzymatic activities of yeast catechol 1,2-dioxygeneses obtained during our study and with analogous results obtained by Tsai et al. [15] for yeast Candida albicans strain TL3. The compared values of specific C12O enzymatic activities, for example for strains A011 (ipc 750), B021 (ipc 500) presented in Table 2 and for A011 (ipc 500) and L012 (ipc 500) in Table 4, had the same order of magnitude (10−1) as the analogous values of yeast specific C12O enzymatic activities from C. albicans strain TL3 revealed by Tsai for initial phenol concentration range from 5 mM to 22 mM. Moreover, we may conclude that the order of magnitude 10-2 was observed for enzymatic activity of catechol 1,2-dioxygenase for C. albicans TL3 for initial phenol concentration 24 mM and e.g. strain A011 (ipc 500) and L012 (ipc 1000), presented in Table 2. One fact which is worthy of consideration is that in case of experiments conducted by Tsai et al., the highest enzymatic activity was equal to 0.734 U mg−1 (ipc 10 mM), whereas in our study the highest enzymatic activities obtained for the cultures conducted without the starving step were: 2.17 U mg−1 (ipc 750) for strain B021, 0.25 U mg−1 (ipc 1000) for strain A011 and 0.16 U mg−1 (ipc 750) for strain L012 (Table 2). In case of the cultures conducted with the starving step, the highest obtained enzymatic activities were: 0.57 U mg−1 (ipc 1000) for strain A011, 0.023 U mg −1 (ipc 750) for strain B021 and 0.47 U mg −1 (ipc 750) for strain L012 (Table 4).
3.3. Detection of phenol degradation intermediates by GC–MS
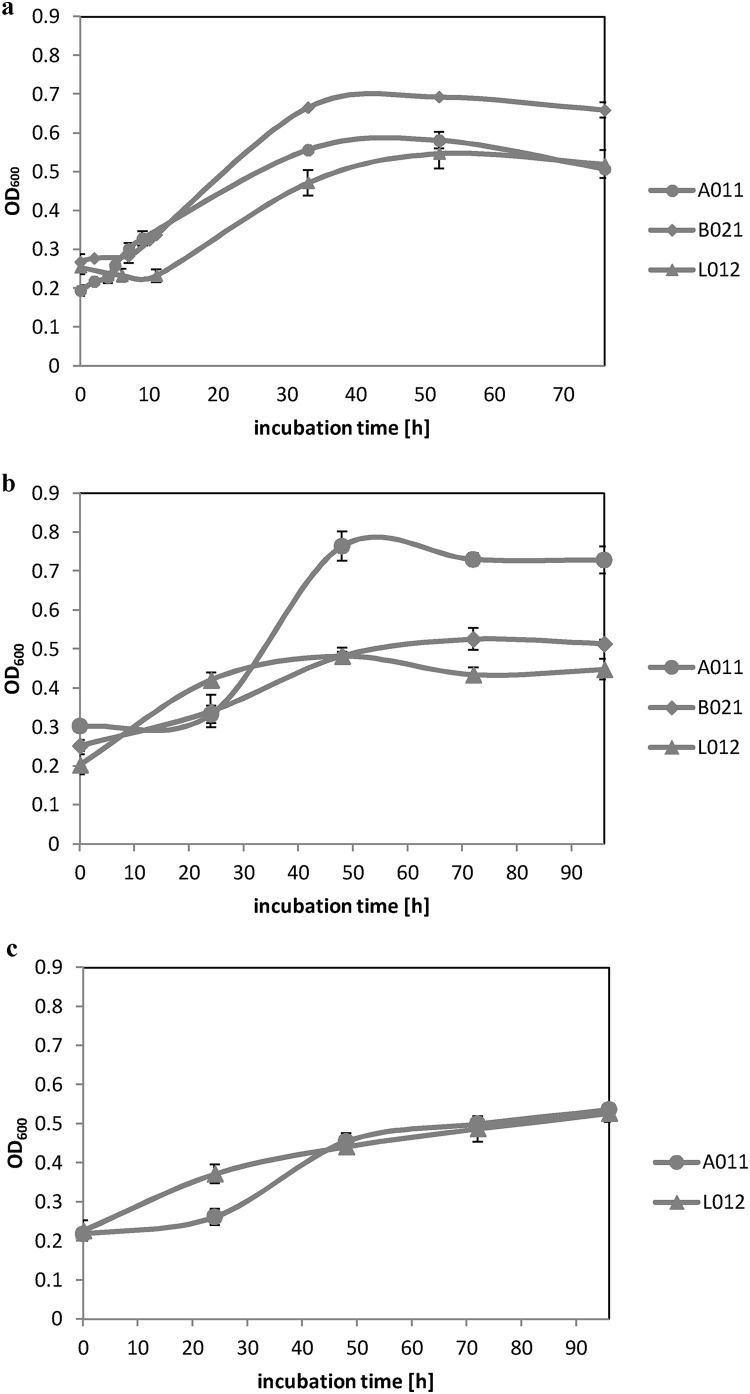
The presence of selected possible intermediates, which may be produced during the catabolism of phenol in yeast cells was analyzed by GC–MS [54]. For this purpose, the presence of catechol, cis,cis-muconic acid and 2-hydroxymouconic semialdehyde was examined in samples taken from yeast cultures without the starving step after 24, 48 and 96 h of cultures growth, as well as those taken from yeast cultures with the starving step after 24, 48, 96 and 168 h of cultures growth, respectively. These times of sampling correspond to the growth rate of yeast strains in the experiments without the starving step (Fig. 3 a, 3b and 3c) and in the experiments which included the starving step (Fig. 4 a, 4b and 4c), respectively. The detection of phenol degradation intermediates was performed for both derivatized yeast cultures supernatants and cell extracts. For derivatized cell extracts, no characteristic products for ortho- or meta-cleavage were observed (data not shown).
Fig. 3.
a Growth curves for examined yeast strains in MSM medium without the starving step, supplemented with phenol at 500 mg l−1. b Growth curves for examined yeast strains in MSM medium without the starving step, supplemented with phenol at 750 mg l−1. c Growth curves for examined yeast strains in MSM medium without the starving step, supplemented with phenol at 1000 mg l−1.
Fig. 4.
a Growth curves for examined yeast strains in MSM medium with the starving step, supplemented with phenol at 500 mg l−1. b Growth curves for examined yeast strains in MSM medium with the starving step, supplemented with phenol at 750 mg l−1. c Growth curves for examined yeast strains in MSM medium with the starving step, supplemented with phenol at 1000 mg l−1.
However, for samples of derivatized yeast cultures supernatants, independently of presence or absence of the starving step during the growth of yeast cultures, GC–MS analysis revealed the presence of cis,cis-muconic acid (Fig. 5 b and 6b). Cis,cis-muconic acid is an intermediate characteristic of phenol catabolism via ortho-cleavage pathway [14] (Fig. 1). In contrast, the examined samples did not show the presence of 2-hydroxymuconic semialdehyde, the product of meta-cleavage pathway (data not show). Therefore, this may support the enzymatic assay results, which displayed the lack of catechol 2,3-dioxygenase activity in all analyzed samples.
Fig. 5.
a Catechol detected in examined yeast strains cultures - experiments conducted without the starving step (initial phenol concentration [mg l−1]). bCis,cis-muconic acid detected in examined yeast strains cultures - experiments conducted without the starving step (initial phenol concentration [mg l−1]).
Moreover, for all studied yeast strains, independently of initial phenol concentration in the culture medium and the presence or absence of the starving step during the growth of yeast cultures, production of catechol was observed (Fig. 5a and 6a). These results were consistent with published reports [[13], [14], [15], [16]], which indicated that under aerobic conditions, phenol degradation was initiated by hydroxylation of the aromatic ring and catechol formation. The formation of catechol is tantamount with the presence of phenol hydroxylase during phenol degradation in cells of the studied yeast strains belonging to Candida spp. and Trichosporum spp. [13].
According to our previous report [36], all examined yeast strains in which inoculums were prepared without the starving step, were able to degrade phenol at 500 mg l−1 below detectable level after 24 h of cultivation. For all yeast strains, the highest concentrations of produced catechol were detected after 24 h of cultivation were 0.369 mg l−1, 0.553 mg l−1 and 0.007 mg l−1 for strains A011, B021 and L012, respectively (Fig. 5a). Furthermore, the highest cis,cis-muconic acid concentrations were detected after the first day of cultivation for strains A011 and L012. Hence, at initial phenol concentration equal to 500 mg l−1, these yeast strains, were able to simultaneously produce both compounds. In comparison, strain B021 produced cis,cis-muconic acid only after 96 h of cultivation. This suggests that catechol was formed at earlier stages of phenol biodegradation (Fig. 5b). The result is consistent with those under aerobic conditions, in which the first step in the metabolic pathway of phenol is its conversion to catechol [13,16].
In the case of phenol at 750 mg l−1 initial concentration in culture medium, selected yeast strains were able to utilize phenol after 24 h of cultivation for A011 and 48 h for B021 and L012 [36]. In terms of catechol production, A011 produced very low concentrations (0.002 mg l−1) after the first day of cultivation, whereas B021 and L012 generated 0.441 mg l−1 and 0.519 mg l−1 catechol concentrations, respectively. After 48 h, besides strain L012, catechol was detected at low levels (Fig. 5a).
In the case of cis,cis-muconic acid, it was detected for yeast strains A011 and L012 after the first and second day of cultivation, but decreased and increased after prolonged cultivation time, respectively. For yeast strain B021, cis,cis-muconic acid was detected after 48 h and 96 h of cultivation and its concentration was almost the same, respectively.
For 1000 mg l−1 phenol initial concentration in culture medium, phenol degradation was observed after 48 h for A011 and L012, however, strain B021 did not exhibit any phenol degradation at such high concentrations [36]. Production of catechol was detected after 24 and 48 h, for strain A011 and after 48 and 96 h of cultivation for strain L012. However, catechol concentration values in culture of strain L012 were at low levels. Production of cis,cis-muconic acid was detected only for strain A011 after 48 h and 96 h of cultivation and increased over prolonged cultivation time (Fig. 5a and 5b).
As was mentioned above, in the case of experiments that included yeast cultures with the starving step, independently from the initial phenol concentration, production of catechol and cis,cis-muconic acid were also detected for all analyzed yeast strains (Fig. 6 a and 6b). However, when we compared, the data presented on Fig. 5a and Fig. 6a, and also Fig. 5b and Fig. 6b, respectively, it seems that the change in inoculum preparation method had noticeable impact on the both compounds production be examined yeast strains.
Fig. 6.
a Catechol detected in examined yeast strains cultures - experiments conducted with the starving step (initial phenol concentration [mg l−1]). bCis,cis-muconic acid detected in examined yeast strains cultures – experiments conducted with the starving step (initial phenol concentration [mg l−1]).
All studied yeast strains were able to utilize phenol at 500 mg l−1 concentration after 48 h of cultivation. Catechol was detected after 24 h of cultivation for strains A011 and L012 and after 48 h for strain B021. Cis,cis-muconic acid was detected after 168 h of cultivation for strain A011 and B021 and after 48 and 168 h for L012 (Fig. 6a and 6b). Hence, catechol was produced at the earlier stages of phenol biodegradation and there was no simultaneous production of catechol and cis,cis-muconic acid.
In the case of phenol at 750 mg l−1 initial concentration of growth medium, total phenol degradation was observed after 48 h of cultivation for strain L012 and after 72 h for A011 and B021. The highest catechol values were detected after 48 h for A011 and B021, whereas for L012 was after 24 h of cultivation (Fig. 6a).
Moreover, cis,cis-muconic acid was detected after 168, 48 h and 168 h of cultivation for strains A011 and L012, respectively. However, cis,cis-muconic acid formation was not detected for B021 even after 168 h of cultivation (Fig. 6b), which may indicate that rapid transformation of phenol occurs.
In the case of phenol at 1000 mg l−1 initial concentration in culture medium, degradation was only observed for strains A011 and L012 after 168 h of cultivation. For both strains, production of cis,cis-muconic acid was observed after 48 h of cultivation, whereas catechol was detected after 24 and 48 h of cultivation, for A011 and after 24, 48 and 96 h for L012 (Fig. 6a and 6b).
To sum up, the production of catechol was observed for all tested yeast strains when their inoculums were prepared with or without starving step, independently of initial phenol concentration and changed during the their cultures incubation time. In the most of examined strains and different initial phenol concentrations, catechol was produced at the earlier stages of degradation and its production was no simultaneous with the production of cis,cis-muconic acid. However, during the culture of the studied yeast strains with the starving step, measured catechol concentrations after 24 h of cultivation for A011 and B021 were lower than those without the starving step, but it was not observed for strain L012. In this case, the starving step seemed to induce the production of a remarkable higher level of catechol after 24 h of this yeast strain cultivation. Moreover, in the case of the experiments included the starving step, for all yeast strains, no production of catechol after the first and fourth day of cultivation was observed.
In general, the comparison of the concentration of cis,cis-muconic acid in all analyzed samples revealed that the tested strains may be able to more efficient production this compound (at higher concentrations) where they are cultivated with the use of inoculums prepared without the starving step (Fig. 5b). For experiments without the starving step, among tested yeast strains, the highest value of produced cis,cis-muconic acid level was observed for strain B021 (ipc 750) and was almost 10 mg l−1 (Fig. 5b). Also, for this culture condition and for this yeast strain and the initial phenol concentration, we measured the highest specific activity of catechol 1,2-dioxygenase (Table 2, 2.17 U mg-1), enzyme responsible for production of cis,cis-muconic acid via mechanism of ortho-cleavage ring of catechol. However, for experiments with the starving step, among tested yeast strains, the highest level of produced cis,cis-muconic acid were detected for strain L012 (ipc 500) and was almost 1.7 mg l-1.
From the biotechnological point of view, the presence of catechol 1,2-dioxygenase activity and the presence of cis,cis-muconic acid in cultures of examined yeast strains provide a starting point for research focused on obtaining a new cis,cis-muconic acid producer through metabolic engineering of yeast. To date, to the best of our knowledge, the studies regarding microbial production of cis,cis-muconic acid employ two main strategies of the “construction” of cis,cis-muconic acid producers: i) genetic modification of microorganisms which do not possess the natural ability to the production of cis,cis-muconic acid e.g. yeast Saccharomyces cerevisiae [44,45] and E. coli [43], ii) metabolic engineering bacterial strains naturally capable of utilizing phenol, catechol and other small aromatic compounds [41,42]. The second of the presented strategies led to obtaining recombinant strains of microorganisms capable of very efficient production of cis,cis-muconic acid. Becker et al. [41] reported that the metabolically engineered strain Corynebacterium glutamicum MA-2 was able to produce from catechol around 85 g l−1 of cis,cis-muconic acid after the 60 h of fed-batch cultivation and from small aromatics obtained after softwood lignin hydrolysis around 1.8 g l−1 of this acid. Kohlstedt et al. [55] also reported the biotechnological production of cis,cis-muconic acid from catechol using a genetically engineered Pseudomonas putida strain MA-6 which was able to produce from catechol around 64 g l−1 of cis,cis-muconic acid within 55 h of cultivation. In the presented studies, the high rate of the production of cis,cis-muconic acid seemed to be connected with: i) deletion/inactivation of genes encoding an enzyme of muconate cycloisomerase enables the production of MA via the catechol branch of the β-ketoadipate pathway from small aromatics and ii) overexpression of gene encoding catechol 1,2-dioxygenase in bacterial cells. Hence, the presented metabolic engineering strategies of bacterial strains naturally utilizing phenol, catechol or other small aromatics compounds seem to be also an attractive starting point for the developing of analogous strategies of metabolic engineering for one of our psychrotolerant peatland yeast strains utilizing phenol and catechol, selected for further study.
4. Conclusions
Although Margesin et al. [26] has identified the cold-adapted yeast strain belonging to Rhodotorula sp. isolated from Alpine glacier cryoconite which utilizes phenol via the meta-cleavage pathway, in this study, all tested cold-adapted yeast strains promoted phenol degradation via the ortho-cleavage pathway. This conclusion is based both on the results of the relevant enzyme tests for detection of C12O or C23O enzymatic activities (also preliminary tests), as well as on the results of the GC–MS tests. In this study, GC–MS analysis was performed to examine possible products of phenol metabolism obtained during yeast cultivations under different conditions. This approach was applied to the detection of products produced by ortho-cleavage or meta-cleavage, which led to the formation and detection of cis,cis-muconic acid or 2-hydroxymuconic semialdehyde, respectively. To the best of our knowledge, this is the first report of identification of ortho-cleavage pathway in cold-adapted yeast strains isolated from peatland. Previously, the identification of ortho-cleavage pathway was reported as the metabolic pathway of phenol utilization among alpine cold-adapted phenol-degrading yeasts isolated from Alpine glacier cryoconite and petroleum hydrocarbon contained alpine soils in Austria, respectively [26,37]. On the other hand, by analogy, this is the first report of the identification of phenol utilizing yeasts via the ortho-cleavage pathway belonging to the species: Candida subhashii (strain A011), Candida oregonenis (strain B021) and Schizoblastosporion starkeyi-henricii (strain L012). Interestingly, on the base of the results of our study and the literature study, it appears that, so far, the ortho-cleavage pathway seems to be the predominant metabolic pathway of phenol-degrading yeast [15,[23], [24], [25], [26],37].
It is worth noting that in all analyzed samples, regardless of whether yeast cultures were prepared with or without the starving step, only 1,2-dioxygenase enzyme activity was observed. On the other hand, all analyzed yeast strains reach higher OD values in a shorter time of cultivation (biomass growth is more effective) in cultures prepared without starving step (Fig. 3–Fig. 4).
However, summing up the discussion of results presented in Section 3.2, we may also conclude that the individual character of examined yeast strain and also the conditions of conducting experiments (e.g. experiment conducted with or without the starving step), may have a noticeable impact on the results of C12O enzymatic activity assay. The inoculum preparation method, in the cases of two out of three tested yeast strains, seemed to influence on the obtained results of measuring the specific enzymatic activity of their C12O enzymes. For inoculum preparation method without starving step, the measured specific enzymatic C12O activity values for strain B021 are noticeable higher, and for strain L012 (ipc 500 and ipc 750) are lower than those measured for both yeast strains when their inoculums were prepared with the starving step.
Summarizing the results of the GC–MS analyzes, catechol and cis,cis-muconic acid production were observed for all studied yeast strains, independently of initial phenol concentration in culture medium and changed during the cultivation time. The majority of examined yeast strains and different initial concentration of phenol in the culture medium, catechol was detected at the earlier stages of degradation, whereas cis,cis-muconic acid was produced at later stages. For analyzed yeast strains and different initial phenol concentration in culture medium (Fig. 5a and 6a), during the cultivation time, catechol concentration levels decreased, with exception A011 (ipc 1000) (inoculum prepared without the starving step, Fig. 5a) and B021 (ipc 500 and ipc 750) (inoculum prepared with the starving step, Fig. 6a), respectively. Previously publicized reports have shown that ortho-cleavage is induced by cis,cis-muconate acid, the product of the catechol ring fission [56,57]. In case of cultivations where inoculum was prepared without the starving step, increasing the initial phenol concentration caused an increase in cis,cis-muconic acid concentrations (except strain L012 at ipc 1000). Additionally, increasing cis,cis-muconic acid concentration was correlated with increasing specific activity of the obtained crude enzyme extracts (Table 2) within each specific strain. However, this correlation is not highly observed in the case of the experiments which included the starving step. It may be caused by the fact that during culture growth of analyzed yeast strains cis,cis-muconic acid was transformed to muconolactone, which was the next metabolite of the ortho-cleavage pathway and was not assayed during our study. Considering all factors, this suggests that the study of biodegradation pathways of phenol should focus not only on enzymatic assays or chromatographic techniques but combine both techniques. This point of view may be useful for a better understanding of the mechanism of biodegradation and for an unequivocal identification of biodegradation phenol pathways by microorganisms.
Because cis,cis-muconic acid is produced by microorganisms in only one of the known main metabolic pathways which they use to utilize such environmental aromatic pollutants like e.g. catechol, phenol, toluene and benzene as carbon sources [46] the research carried out in this work allowed us to answer an important question: “If the psychrotolerant phenol-degrading yeast strains isolated by us from the peatland may be interesting in terms of their further research on the development of a biotechnological method of obtaining cis,cis-muconic acid?” In this research, we found that all examined yeast strains utilize phenol via the ortho-cleavage pathway, where cis,cis-muconic acid is one of the intermediates. The highest value of cis,cis-muconic acid concentration in all analyzed yeast cultures was detected for Candida oregonensis strain B021 (initial phenol concentration 750 mg l−1, inoculum preparation without the starving step) and was almost 10 mg l−1. Hence, in our opinion, this yeast strain seems to be the most promising for achieving a metabolically engineered Candida oregonensis strain capable of efficient production of cis,cis-muconic acid by biotechnological biotransformation of phenol, catechol and other small aromatic compounds resulting from lignin hydrolysis.
Author agreement statement
We the undersigned declare that this manuscript is original, has not been published before and is not currently being considered for publication elsewhere.
We confirm that the manuscript has been read and approved by all named authors and that there are no other persons who satisfied the criteria for authorship but are not listed. We further confirm that the order of authors listed in the manuscript has been approved by all of us.
We understand that the Corresponding Author is the sole contact for the Editorial process. He/she is responsible for communicating with the other authors about progress, submissions of revisions and final approval of proofs.
Due to the pandemic of COVID-19 there is no possibility to obtain each author’s hand signature. Therefore, we are submitting email addresses. In case of a need, each author may confirm this statement via email.
Funding
This research did not receive any specific grants from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT authorship contribution statement
Natalia Filipowicz: Conceptualization, Methodology, Investigation, Writing - original draft, Writing - review & editing. Malwina Momotko: Methodology, Investigation, Writing - original draft. Grzegorz Boczkaj: Methodology, Resources, Writing - review & editing. Hubert Cieśliński: Resources, Writing - review & editing.
Declaration of Competing Interest
None.
References
- 1.Michalowicz J., Duda W. Phenols - sources and toxicity. Pol. J. Environ. Stud. 2007;16:347–362. [Google Scholar]
- 2.Boczkaj G., Przyjazny A., Kamiński M. New procedures for control of industrial effluents treatment processes. Ind. Eng. Chem. Res. 2014;53:1503–1514. doi: 10.1021/ie402126d. [DOI] [Google Scholar]
- 3.Boczkaj G., Makoś P., Przyjazny A. Application of dispersive liquid-liquid microextraction and gas chromatography with mass spectrometry for the determination of oxygenated volatile organic compounds in effluents from the production of petroleum bitumen. J. Sep. Sci. 2016;39:2604–2615. doi: 10.1002/jssc.201501355. [DOI] [PubMed] [Google Scholar]
- 4.Boczkaj G., Fernandes A., Makoś P. Study of different advanced oxidation processes for wastewater treatment from petroleum bitumen production at basic pH. Ind. Eng. Chem. Res. 2017;56:8806–8814. doi: 10.1021/acs.iecr.7b01507. [DOI] [Google Scholar]
- 5.Boczkaj G., Fernandes A. Wastewater treatment by means of advanced oxidation processes at basic pH conditions: a review. Chem. Eng. J. 2017;320:608–633. doi: 10.1016/j.cej.2017.03.084. [DOI] [Google Scholar]
- 6.Fernandes A., Makoś P., Boczkaj G. Treatment of bitumen post oxidative effluents by sulfate radicals based advanced oxidation processes (S-AOPs) under alkaline pH conditions. J. Clean. Prod. 2018;195:374–384. doi: 10.1016/j.jclepro.2018.05.207. [DOI] [Google Scholar]
- 7.Gągol M., Przyjazny A., Boczkaj G. Wastewater treatment by means of advanced oxidation processes based on cavitation – a review. Chem. Eng. J. 2018;338:599–627. doi: 10.1016/j.cej.2018.01.049. [DOI] [Google Scholar]
- 8.Aksu Z., Akpinar D., Kabasakal E., Köse B. Simultaneous biosorption of phenol and nickel(II) from binary mixtures onto dried aerobic activated sludge. Process Biochem. 1999;35:301–308. doi: 10.1016/S0032-9592(99)00072-2. [DOI] [Google Scholar]
- 9.Yuan R., Jiang M., Gao S., Wang Z., Wang H., Boczkaj G., Liu Z., Ma J., Li Z. 3D mesoporous A-Co(OH)2 nanosheets electrodeposited on nickel foam: a new generation of macroscopic cobalt-based hybrid for peroxymonosulfate activation. Chem. Eng. J. 2020;380:1–11. doi: 10.1016/j.cej.2019.122447. [DOI] [Google Scholar]
- 10.Wang C.C., Lee C.M., Lu C.J., Chuang M.S., Huang C.Z. Biodegradation of 2,4,6-trichlorophenol in the presence of primary substrate by immobilized pure culture bacteria. Chemosphere. 2000;41:1873–1879. doi: 10.1016/s0045-6535(00)00090-4. http://www.ncbi.nlm.nih.gov/pubmed/11061309 (accessed December 20, 2016) [DOI] [PubMed] [Google Scholar]
- 11.Al-Khalid T., El-Naas M.H. Aerobic biodegradation of phenols: a comprehensive review. Crit. Rev. Environ. Sci. Technol. 2012;42:1631–1690. doi: 10.1080/10643389.2011.569872. [DOI] [Google Scholar]
- 12.Caposio P., Pessione E., Giuffrida G., Conti A., Landolfo S., Giunta C., Gribaudo G. Cloning and characterization of two catechol 1,2-dioxygenase genes from Acinetobacter radioresistens S13. Res. Microbiol. 2002;153:69–74. doi: 10.1016/S0923-2508(01)01290-6. [DOI] [PubMed] [Google Scholar]
- 13.Krastanov A., Alexieva Z., Yemendzhiev H. Microbial degradation of phenol and phenolic derivatives. Eng. Life Sci. 2013;13:76–87. doi: 10.1002/elsc.201100227. [DOI] [Google Scholar]
- 14.Whiteley C.G., Lee D.-J. Enzyme technology and biological remediation. Enzyme Microb. Technol. 2006;38:291–316. doi: 10.1016/J.ENZMICTEC.2005.10.010. [DOI] [Google Scholar]
- 15.Tsai S.C., Tsai L.D., Li Y.K. An isolated Candida albicans TL3 capable of degrading phenol at large concentration. Biosci. Biotechnol. Biochem. 2005;69:2358–2367. doi: 10.1271/bbb.69.2358. [DOI] [PubMed] [Google Scholar]
- 16.Zeyaullah M., Ahmad R., Naseem A., Islam B., Hasan H.M.I., Abdelkafe A.S., Benkhayal F.A., Rizvi M.A., Ali A. Catechol biodegradation by Pseudomonas strain : a critical analysis. Int. J. Chem. Sci. 2009;7:2211–2221. http://www.tsijournals.com/articles/catechol-biodegradation-by-pseudomonas-strain-a-critical-analysis.pdf (accessed January 5, 2018) [Google Scholar]
- 17.Broderick J.B., O’Halloran T.V. Overproduction, purification, and characterization of chlorocatechol dioxygenase, a non-heme iron dioxygenase with broad substrate tolerance. Biochemistry. 1991;30:7349–7358. doi: 10.1021/bi00243a040. http://www.ncbi.nlm.nih.gov/pubmed/1649626 (accessed August 20, 2018) [DOI] [PubMed] [Google Scholar]
- 18.Latus M., Seitz H., Eberspacher J., Lingens F. Purification and characterization of hydroxyquinol 1,2-Dioxygenase from Azotobacter sp. Strain GP1. Appl. Environ. Microbiol. 1995;61:2453–2460. doi: 10.1128/aem.61.7.2453-2460.1995. http://www.ncbi.nlm.nih.gov/pubmed/16535063 (accessed August 20, 2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barbe V., Vallenet D., Fonknechten N., Kreimeyer A., Oztas S., Labarre L., Cruveiller S., Robert C., Duprat S., Wincker P., Ornston L.N., Weissenbach J., Marlière P., Cohen G.N., Médigue C. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 2004;32:5766–5779. doi: 10.1093/nar/gkh910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strachan P.D., Freer A.A., Fewson C.A. Purification and characterization of catechol 1,2-dioxygenase from Rhodococcus rhodochrous NCIMB 13259 and cloning and sequencing of its catA gene. Biochem. J. 1998;333:741–747. doi: 10.1042/bj3330741. http://www.ncbi.nlm.nih.gov/pubmed/9677336 (accessed August 20, 2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saxena P., Thakur I.S. 2009. Purification and Characterization of Catechol 1, 2-dioxygenase of Pseudomonas fluorescens for Degradation of 4-chlorobenzoic Acid.https://www.semanticscholar.org/paper/Purification-and-characterization-of-catechol-1-%2C-Saxena-Thakur/83ef735b3256ada26df9ccf5e0afb36dda1e13f8 (accessed August 20, 2018) [Google Scholar]
- 22.Giedraityte G., Kalėdienė L., Catechol 1,2-dioxygenase from α-naphthol degrading thermophilic Geobacillus sp. strain: purification and properties. Open Life Sci. 2009;4 doi: 10.2478/s11535-008-0049-y. [DOI] [Google Scholar]
- 23.Long Y., Yang S., Xie Z., Cheng L. Cloning, expression, and characterization of catechol 1,2-dioxygenase from a phenol-degrading Candida tropicalis JH8 strain. Prep. Biochem. Biotechnol. 2016;46:673–678. doi: 10.1080/10826068.2015.1135449. [DOI] [PubMed] [Google Scholar]
- 24.Vilímková L., Páca J., Kremláčková V., Páca J., Stiborová M. Isolation of cytoplasmic NADPH-dependent phenol hydroxylase and catechol-1,2-dioxygenase from Candida tropicalis yeast. Interdiscip. Toxicol. 2008;1:225–230. doi: 10.2478/v10102-010-0046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aleksieva Z., Ivanova D., Godjevargova T., Atanasov B. Degradation of some phenol derivatives by Trichosporon cutaneum R57. Process Biochem. 2002;37:1215–1219. doi: 10.1016/S0032-9592(01)00336-3. [DOI] [Google Scholar]
- 26.Margesin R., Gander S., Zacke G., Gounot A.M., Schinner F. Hydrocarbon degradation and enzyme activities of cold-adapted bacteria and yeasts. Extremophiles. 2003;7:451–458. doi: 10.1007/s00792-003-0347-2. [DOI] [PubMed] [Google Scholar]
- 27.Margesin R., Fonteyne P.-A., Redl B. Low-temperature biodegradation of high amounts of phenol by Rhodococcus spp. And basidiomycetous yeasts. Res. Microbiol. 2005;156:68–75. doi: 10.1016/j.resmic.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 28.Fialová A., Boschke E., Bley T. Rapid monitoring of the biodegradation of phenol-like compounds by the yeast Candida maltosa using BOD measurements. Int. Biodeterior. Biodegradation. 2004;54:69–76. doi: 10.1016/j.ibiod.2004.02.004. [DOI] [Google Scholar]
- 29.Jiang Y., Yang X., Liu B., Zhao H., Cheng Q., Cai B. Catechol 2.,3-Dioxygenase from Pseudomonas sp. Strain ND6: gene sequence and enzyme characterization. Biosci. Biotechnol. Biochem. 2004;68:1798–1800. doi: 10.1271/bbb.68.1798. [DOI] [PubMed] [Google Scholar]
- 30.Kasuga I., Nakajima F., Furumai H. Diversity of catechol 2,3-dioxygenase genes of bacteria responding to dissolved organic matter derived from different sources in a eutrophic lake. FEMS Microbiol. Ecol. 2007;61:449–458. doi: 10.1111/j.1574-6941.2007.00347.x. [DOI] [PubMed] [Google Scholar]
- 31.Li W., Shi J., Wang X., Han Y., Tong W., Ma L., Liu B., Cai B. Complete nucleotide sequence and organization of the naphthalene catabolic plasmid pND6-1 from Pseudomonas sp. Strain ND6. Gene. 2004;336:231–240. doi: 10.1016/j.gene.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 32.Müller C., Petruschka L., Cuypers H., Burchhardt G., Herrmann H. Carbon catabolite repression of phenol degradation in Pseudomonas putida is mediated by the inhibition of the activator protein PhlR. J. Bacteriol. 1996;178:2030–2036. doi: 10.1128/jb.178.7.2030-2036.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sei K., Asano K., Tateishi N., Mori K., Ike M., Fujita M. Design of PCR primers and gene probes for the general detection of bacterial populations capable of degrading aromatic compounds via catechol cleavage pathways. J. Biosci. Bioeng. 1999;88:542–550. doi: 10.1016/s1389-1723(00)87673-2. [DOI] [PubMed] [Google Scholar]
- 34.Viggiani A., Siani L., Notomista E., Birolo L., Pucci P., Di Donato A. The role of the conserved residues His-246, His-199, and Tyr-255 in the catalysis of catechol 2,3-dioxygenase from Pseudomonas stutzeri OX1. J. Biol. Chem. 2004;279:48630–48639. doi: 10.1074/jbc.M406243200. [DOI] [PubMed] [Google Scholar]
- 35.Wei J., Zhou Y., Xu T., Lu B. Rational design of Catechol-2, 3-dioxygenase for improving the enzyme characteristics. Appl. Biochem. Biotechnol. 2010;162:116–126. doi: 10.1007/s12010-009-8720-y. [DOI] [PubMed] [Google Scholar]
- 36.Filipowicz N., Momotko M., Boczkaj G., Pawlikowski T., Wanarska M., Cieśliński H. Isolation and characterization of phenol-degrading psychrotolerant yeasts. Water Air Soil Pollut. 2017;228:210. doi: 10.1007/s11270-017-3391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Margesin R., Bergauer P., Gander S. Degradation of phenol and toxicity of phenolic compounds: a comparison of cold-tolerant Arthrobacter sp. And mesophilic Pseudomonas putida. Extremophiles. 2004;8:201–207. doi: 10.1007/s00792-004-0378-3. [DOI] [PubMed] [Google Scholar]
- 38.Jiang Y., Ren N., Cai X., Wu D., Qiao L., Lin S. Biodegradation of phenol and 4-Chlorophenol by the mutant strain CTM 2. Chinese J. Chem. Eng. 2008;16:796–800. doi: 10.1016/S1004-9541(08)60158-5. [DOI] [Google Scholar]
- 39.Yan J., Jianping W., Hongmei L., Suliang Y., Zongding H. The biodegradation of phenol at high initial concentration by the yeast Candida tropicalis. Biochem. Eng. J. 2005;24:243–247. doi: 10.1016/j.bej.2005.02.016. [DOI] [Google Scholar]
- 40.Khalil I., Quintens G., Junkers T., Dusselier M. Muconic acid isomers as platform chemicals and monomers in the biobased economy. Green Chem. 2020;22:1517–1541. doi: 10.1039/C9GC04161C. [DOI] [Google Scholar]
- 41.Becker J., Kuhl M., Kohlstedt M., Starck S., Wittmann C. Metabolic engineering of Corynebacterium glutamicum for the production of cis,cis-muconic acid from lignin. Microb. Cell Fact. 2018;17 doi: 10.1186/s12934-018-0963-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chua J.W., Hsieh J.H. Oxidative bioconversion of toluene to 1,3-butadiene-1,4-dicarboxylic acid (cis,cis-muconic acid) World J. Microbiol. Biotechnol. 1990;6:127–143. doi: 10.1007/BF01200932. [DOI] [PubMed] [Google Scholar]
- 43.J.W. Frost, A. Miermont, D. Schweitzer, V. Bui, US Patent. (2013) US8426639B2.
- 44.Pyne M.E., Narcross L., Melgar M., Kevvai K., Mookerjee S., Leite G.B., Martin V.J.J. An engineered Aro1 protein degradation approach for increased cis,cis-Muconic acid biosynthesis in Saccharomyces cerevisiae. App. Environ. Microbiol. 2018;84 doi: 10.1128/AEM.01095-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang G., Øzmerih S., Guerreiro R., Meireles A.C., Carolas A., Milne N., Jensen M.K., Ferreira B.S., Borodina I. Improvement of cis,cis-Muconic acid production in Saccharomyces cerevisiae through biosensor-aided genome engineering. ACS Synth. Biol. 2020;9:634–646. doi: 10.1021/acssynbio.9b00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Filipowicz N., Cieśliński H. A rapid and simple method for screening microorganisms with a potential for catechol biodegradation. Int. J. Environ. Res. 2020;14:87–92. doi: 10.1007/s41742-019-00239-z. [DOI] [Google Scholar]
- 47.Paisio C.E., Oller A.L.W., Ibanez S.G., Talano M.A., Gonzalez P.S., Medina M.I., Agostini E. In: Bioremediation as a useful biotechnological strategy for the treatment of phenolics: advances, challenges and future prospects. Velazgues-Fernandes J.B., Muniz-Hernandes S., editors. Nova Science Publishers; New York: 2014. pp. 57–79. [Google Scholar]
- 48.Neidle E.L., Ornston L.N. Cloning and expression of Acinetobacter calcoaceticus catechol 1,2-dioxygenase structural gene catA in Escherichia coli. J. Bacteriol. 1986;168:815–820. doi: 10.1128/jb.168.2.815-820.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Birger A., Krauss G., Kiesel B., Dermietzel J., Gläßer W. Abbaupotential für aliphatische und aromatische kohlenwasserstoffe in bakteriellen und heterotrophen communities differenter grundwasser-biozönosen. In: Kreysa G., Wiesner J., editors. Möglichkeiten Und Grenzen Der Reinigung Kontaminierter Gewässer, Dechema, Frankfurt. 1997. pp. 571–581. [Google Scholar]
- 50.Morgan J.A., Winstanley C., Pickup R.W., Jones J.G., Saunders J.R. Direct phenotypic and genotypic detection of a recombinant pseudomonad population released into lake water. Appl. Environ. Microbiol. 1989;55:2537–2544. doi: 10.1128/aem.55.10.2537-2544.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakazawa T., Nakazawa A. Pyrocatechase (Pseudomonas) Methods Enzymol. 1970;17:518–522. doi: 10.1016/0076-6879(71)17234-5. [DOI] [Google Scholar]
- 52.Nozaki M. Metapyrocatechase (Pseudomonas) Methods Enzymol. 1970;17:522–525. doi: 10.1016/0076-6879(71)17235-7. [DOI] [Google Scholar]
- 53.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. http://www.ncbi.nlm.nih.gov/pubmed/942051 (accessed August 20, 2018) [DOI] [PubMed] [Google Scholar]
- 54.Nie H., Nie M., Yang Y., Zhao J., Zhang X., Guo Y., Wan Y., Zi J. Characterization of phenol metabolization by P. stutzeri N2. Polycycl. Aromat. 2016;36:587–600. doi: 10.1080/10406638.2015.1033434. [DOI] [Google Scholar]
- 55.Kohlstedt M., Starck S., Barton N., Stolzenberger J., Selzer M., Mehlmann K., Schneider R., Pleissner D., Rinkel J., Dickschat J.S., Venus J., van Duuren J.B.J.H., Wittmann C. From lignin to nylon: cascaded chemical and biochemical conversion using metabolically engineered Pseudomonas putida. Metab. Eng. 2018;47(2018):279–293. doi: 10.1016/j.ymben.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 56.Cao B., Geng A., Loh K.-C. Induction of ortho- and meta-cleavage pathways in Pseudomonas in biodegradation of high benzoate concentration: MS identification of catabolic enzymes. Appl. Microbiol. Biotechnol. 2008;81:99–107. doi: 10.1007/s00253-008-1728-3. [DOI] [PubMed] [Google Scholar]
- 57.Song J., Sung J., Kim Y.M., Zylstra G.J., Kim E. Roles of the meta- and ortho-cleavage pathways for the efficient utilization of aromatic hydrocarbons by Sphingomonas yanoikuyae B1. J. Microbiol. 2000;38:245–249. [Google Scholar]