Abstract
Objectives
Duration of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the upper respiratory tract is extremely variable, but its relation to disease severity is unknown. We investigated this relation in the 530 000 inhabitants of the northeastern Italian province of Udine.
Methods
We analysed real-time RT-PCR tests for SARS-CoV-2 in upper respiratory specimens conducted at the Virology Laboratory of the University Hospital of Udine, Italy (which serves the whole province) from 1 March to 30 April 2020 Specimens were from positive individuals in four groups characterized by different disease severity (critically ill patients admitted to intensive care units, patients admitted to infectious disease units, symptomatic patients visiting the emergency department and not hospitalized, and asymptomatic individuals tested during contact tracing or screening activities). Duration of viral positivity was assessed from the first positive test to the day of the first of two consecutive negative tests. Univariate and multivariate analyses were conducted to investigate differences in the four groups.
Results
From 1 March to 30 April, 39 483 RT-PCR tests for SARS-CoV-2 were conducted on 23 778 individuals, and 974 individuals had a positive test result. Among those with multiple tests (n = 878), mean time to negativity was 23.7 days (standard error 0.3639; median 23, interquartile range 16–30 days). Mean time to negativity was longer in the group admitted to the intensive care unit than in the others, whereas no difference was observed between asymptomatic patients and those with mild disease.
Conclusions
Disease control measures should not be adjusted to account for differences in viral shedding according to symptomatic status.
Keywords: Coronavirus disease 2019, Infectivity, Nasopharyngeal swab, Real-time RT-PCR, Severe acute respiratory syndrome coronavirus 2
Introduction
The coronavirus disease 2019 (COVID-19) outbreak was first identified in Wuhan, the capital of Hubei Province, China, in December 2019 [1] and on 12 March 2020 it was declared a pandemic. Worldwide, identification of symptomatic and asymptomatic carriers of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is possible only by detecting the virus in upper respiratory specimens with real-time RT-PCR tests, as recommended by the WHO [2].
Viral transmission depends mainly on the biological characteristics of pathogens, the potential exposed population and viral shedding. Viral shedding of SARS-CoV-2, as documented in nasopharyngeal secretions of infected patients, is extremely variable, ranging from 7 to 52 days [[3], [4], [5], [6]]. These data are for patients admitted with severe pneumonia, but little is known about the whole clinical spectrum in a general population.
The knowledge of duration of SARS-CoV-2 viral positivity in the whole spectrum of the disease, including pauci-symptomatic and asymptomatic individuals, may have significance in the control and prevention of the disease.
The aim of the research was to study the association between the duration of SARS-CoV-2 in nasopharyngeal swabs and the clinical severity of infection in a general population.
Materials and methods
For this retrospective cohort study, we used the database of the Virology Laboratory of the University Hospital of Udine, Italy, which serves all 530 000 inhabitants of Udine province. The database includes the results of all real-time RT-PCR tests for SARS-CoV-2 on upper respiratory specimens collected by nasopharyngeal swabs since 1 March 2020, the day when the first COVID-19 case was suspected. The database also includes the date of swab collection and the health-care facility where the swab was collected. RT-PCR was conducted as recommended by the WHO for COVID-19 clinical management and outbreak control purposes [2]. In our context, swabs could be collected in hospitalized patients or at the emergency department (ED) for clinical management or by personnel of the prevention department or the hospital management for outbreak control through contact tracing and screening activities, eventually followed by quarantine or isolation.
Individuals with positive test results were identified and the date of the first positive result was recorded for each person as the date of confirmed COVID-19 diagnosis. For those individuals, we linked data from the two hospitals of the province of Udine admitting COVID-19 patients, i.e. the University Hospital of Udine and the Hospital of Palmanova, both including intensive care units (ICU) and infectious diseases units (IDU) for COVID-19 patients.
Individuals with positive RT-PCR tests for SARS-CoV-2 were then divided into four groups, which could be considered as having decreasing disease severity: ICU group, if they had any admission to an ICU from 1 March to 30 April 2020; IDU group, if they were never admitted to ICU but were admitted to an IDU from 1 March to 30 April 2020; ED group, if they were never hospitalized but were visited and had the nasal swab collected at one of the EDs in Udine because of symptoms suggestive of COVID-19; and probably asymptomatic group, if they had nasal swabs collected by the prevention department in the course of outbreak control activities.
The difference in age among groups was assessed using analysis of variance after checking the normality of its distribution with a Kruskal–Wallis test. The difference in sex distribution among groups was assessed with a χ2 test. To assess whether the time from the first positive test to return to negativity (i.e. date of the first of two consecutive negative tests) differed depending on the severity of the SARS-CoV-2 infection, a survival analysis was conducted. Each participant was followed from the date of the first positive RT-PCR test result to the day of the second consecutive negative test, up to 30 April 2020. Individuals who did not return to negativity by 30 April 30 were censored either at the date of the most recent positive test or at the date of death (if any). Individuals with no further records after the first positive test were excluded from further analyses.
In each group, time of return to negativity was described through the mean and standard error (SE) and the quartiles. Kaplan–Meier curves were used to describe time to negativity in each group. Log-rank test and Wilcoxon's test were used to assess whether the groups differed significantly. Values of p < 0.05 were considered statistically significant. Finally, Cox regression was used to assess whether time to return to negativity was affected by the severity of infection, adjusting for age and sex of the patient. The hazard ratio and 95% CI were calculated. All analyses were conducted using SAS v9.4 (SAS Institute Inc., Cary, NC, USA).
Ethics statement
This study was approved by the Ethics Committee of the Friuli Venezia Giulia Region (CEUR-2020-Os-088; 24 April 2020).
Results
From 1 March to 30 April, the Virology Laboratory of the University Hospital of Udine processed 39 483 RT-PCR tests for SARS-CoV-2 from 23 778 persons. In all, 974 individuals had a positive RT-PCR test result. Of them, 36 (3.7%) were admitted to ICU, 204 (20.9%) were hospitalized in IDU, 50 (5.1%) were not hospitalized but were visited at the ED, and 684 (70.2%) were neither hospitalized nor attended the ED. Thirty-five individuals died (nine from the ICU group and 26 from the IDU group), but two of them had two consecutive negative tests before their deaths. In all, 878 individuals had at least one further event after the first positive test and were included in further analyses: 34 (3.9%) in the ICU group, 202 (23.0%) in the IDU group, 44 (5.0%) in the ED group and 598 (68.1%) in the asymptomatic group.
The demographic characteristics of all individuals included in the analysis and the range of dates of the first positive tests are shown in Table 1 . Individuals admitted to hospital, and in particular those admitted to the ICU, were older than the others and they were more often male. Although in all groups there were patients who had their first positive tests in April, it should be noted that 75% of all individuals had their first positive test from 1 March to 30 March, and 95% had the first positive test before 10 April. Overall, 809 individuals had returned a negative test by 30 April 2020.
Table 1.
Demographic characteristics of 878 COVID-19 confirmed cases at the University Hospital of Udine
| Group |
||||
|---|---|---|---|---|
| Asymptomatic | ED | IDU | ICU | |
| Total (n) | 598 | 44 | 202 | 34 |
| Female n (%)a | 364 (60.9%) | 23 (52.3%) | 86 (42.6%) | 9 (26.5%) |
| Age (years), mean ± SDb | 53.8 ± 22.0 | 51.3 ± 18.9 | 66.3 ± 16.7 | 64.6 ± 10.7 |
| Date of first positive result (min; max) | 4 March; 25 April | 6 March; 16 April | 3 March; 27 April | 8 March; 3 April |
Abbreviations: COVID-19, coronavirus disease 2019; ED, emergency department; ICU, intensive care unit; IDU, infectious diseases unit.
a χ2 test, p < 0.0001.
b Analysis of variance test, p < 0.0001.
In the study period, 1611 swabs were conducted on the 598 persons of the asymptomatic group, 124 on the 44 patients of the ED group, 563 on the 202 patients of the IDU group, and 145 on the 34 patients of the ICU group. Time interval between consecutive swabs decreased from the asymptomatic group (mean 8.7 ± 6.6 days, median 7 days) to the ED group (mean 8.1 ± 6.2 days, median 7 days) to the IDU group (mean 6.9 ± 4.9 days, median 7 days) to the ICU group (mean 6.0 ± 4.6 days, median 6 days; Kruskal–Wallis test p < 0.0001).
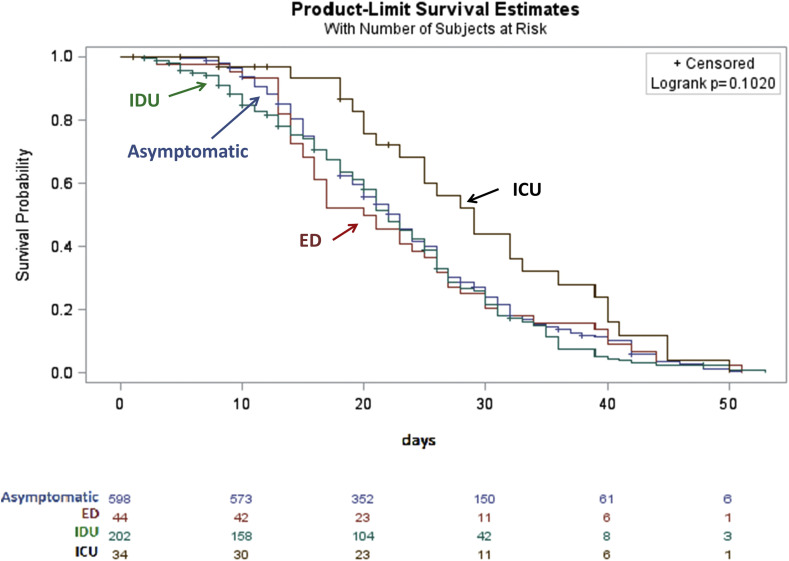
Overall, mean time to negativity was 23.7 days (SE 0.3639), median 23 days, interquartile range 16–30 days. Table 2 shows the distributions of time to negativity in the four groups. Mean, median and maximum values were similar in the asymptomatic, ED and IDU groups, but they were higher in the ICU group. Fig. 1 shows Kaplan–Meier curves for time to negativity in the four groups. The curves were not significantly different according to the log-rank test (p 0.1020), but they were significantly different according to Wilcoxon's test, which is more sensitive to earlier time points (p 0.0242).
Table 2.
Proportion of individuals returning to negativity at different times and time to negativity (from first positive RT-PCR test result to the first of two consecutive negative results) among 878 confirmed COVID-19 cases at the University Hospital of Udine
| n | Censored | % reaching negativity |
Mean time to negativity | SE | 25th centile | Median | 75th centile | Min. | Max. | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| at 10 days | at 20 days | at 30 days | ||||||||||
| Asymptomatic | 598 | 26 | 6.4 | 44.3 | 76.1 | 23.8 | 0.4273 | 16 | 23 | 30 | 4 | 51 |
| ED | 44 | 0 | 6.8 | 50.0 | 79.5 | 22.8 | 1.6655 | 14 | 20.5 | 29 | 3 | 51 |
| IDU | 202 | 35 | 15.1 | 41.8 | 78.3 | 22.6 | 0.7982 | 15 | 22 | 30 | 2 | 53 |
| ICU | 34 | 8 | 3.1 | 24.3 | 55.9 | 29.6 | 2.0058 | 21 | 29 | 39 | 8 | 50 |
Abbreviations: COVID-19, coronavirus disease 2019; ED, emergency department; ICU, intensive care unit; IDU, infectious diseases unit.
Log-rank test p 0.1020; Wilcoxon test p 0.0242.
Fig. 1.
Kaplan–Meier curves for time to negativity among 878 individuals with confirmed COVID-19 at the University Hospital of Udine.
The results of Cox regression are illustrated in Table 3 . After adjusting for sex and age, at any time-point, individuals in the ICU group were less likely to return to negativity than the other individuals. Female sex and increasing age were associated with a reduced likelihood of returning to negativity.
Table 3.
Cox regression for time to negativity among 878 confirmed COVID-19 cases at the University Hospital of Udine
| HR | 95% CI | P value | |
|---|---|---|---|
| Female (vs male) | 0.803 | 0.695–0.927 | 0.0027 |
| Age (per increasing year) | 0.990 | 0.987–0.994 | <0.0001 |
| ED group (vs asymptomatic) | 0.922 | 0.677–1.256 | 0.6061 |
| IDU (vs asymptomatic) | 1.102 | 0.922–1.316 | 0.2850 |
| ICU group (vs asymptomatic) | 0.603 | 0.404–0.899 | 0.0131 |
Abbreviations: COVID-19, coronavirus disease 2019; ED, emergency department; HR, hazard ratio; ICU, intensive care unit; IDU, infectious diseases unit.
Discussion
The main finding of our study is the long duration of SARS-CoV-2 positivity in a population including patients ranging from asymptomatic to acute respiratory distress syndrome requiring ICU care. For the whole cohort, the median duration was 23 days (with a range from 2 to 53 days).
Another important finding is the lack of a significant difference in duration of nasopharyngeal swab test results among asymptomatic patients and symptomatic patients not requiring ICU care. So, apart from critically ill patients, virus clearance from the upper respiratory tract is not influenced by the degree of inflammatory/immune response, and from another perspective, milder disease seems not to be related to a shorter persistence of virus in the upper respiratory tract. On the other hand, individuals with very serious disease had longer viral shedding (approximately 1 week longer) and, at any time point, they were 10%–60% less likely than asymptomatic person to return to negativity.
Contradictory results on virus shedding duration are reported, but most research findings regard hospitalized patients, and data on viral shedding in asymptomatic individuals are still missing. Initial studies reported a median duration of viral shedding in nasopharyngeal swab of 12 days and 15 patients (83%) had viral shedding from the nasopharynx detected for 7 days or longer [3]. Subsequent studies showed durations of viral shedding ranging between 8 and 37 days. The median duration of viral shedding was 20 days in survivors but continued until death in fatal cases [4].
In COVID-19, data on the association between viral shedding and clinical severity (for example, inflammatory and immune responses) are lacking. Cereda et al. [7], analysing the first 5830 laboratory-confirmed cases in Italy, did not observe significantly different viral loads in nasal swabs between symptomatic and asymptomatic persons. In Middle East respiratory syndrome coronavirus infection, viral load and shedding duration were associated with clinical severity [8,9], with persistence of viral RNA in respiratory secretions beyond 21 days from symptom onset only in severely ill patients.
Recently, Lin et al. [6] found, in a cohort of 137 hospitalized patients, a median duration of nasopharyngeal swab positivity of 12 days (range 4–45 days). In this study, a significantly shorter SARS-CoV-2 viral positivity duration was associated with clinical and laboratory characteristics of less severity: age <47 years (mean 12.7 days versus 16.6 days), not severe disease (13.9 days versus 18.4 days), higher lymphocyte count (13.1 days versus 16.8 days), higher eosinophil count (12.3 days versus 16.3 days), and higher CD8+ T-cell count (11.8 days versus 16.8 days), and lower levels of interleukin-6 (11.9 days versus 16.2 days). However, the cohort only included selected patients not representing the whole span of clinical severity. Our finding that older individuals were less likely to return to negativity is consistent with other studies [6,10]. On the other hand, in contrast to other studies, which found no difference in viral shedding between sexes [10], we observed prolonged time to negativity among women compared with men. The slower clearance among women apparently conflicts with their reduced susceptibility and disease severity reported in observational studies [11]. Our result might indicate that women are infectious for longer times than men, but this needs to be confirmed as it has important public health implications.
A merit of our study was that it includes all consecutive SARS-Cov-2-positive individuals from ‘case 0’ up to 27 April 2020 in a province of 530 000 people served by a large University Hospital, including asymptomatic and critically ill individuals.
In addition, a standardized method of nasopharyngeal swab collection was adopted. Bronchoalveolar lavage or gastric tubage were never used to assess the non-infectiveness of the individual during the healing phase.
This research has a number of limitations. Our source of data did not include information on medications used by patients, so we cannot say whether any therapeutic agent might have influenced viral shedding. At the time the study was conducted, however, no pharmacological treatment had proven effective on viral clearance, so our results should not be distorted by treatments received by patients. In addition, therapies administered to all critical patients were almost identical, with no inclusion in randomized trials. As more treatment options become available, it will be important to assess their effect on viral shedding.
In addition, true time to negativity might be shorter than measured in this study, as it was affected by the interval between consecutive swabs. However, although the difference among groups was statistically significant, median intervals were similar across groups. Slightly longer intervals were observed among outpatients, probably because they were not usually tested during weekends. This bias should be more evident among asymptomatic individuals as they had, on average, longer intervals between swabs, making even more implausible the hypothesis that asymptomatic individuals have longer duration of positivity.
Another limit of the study is that viral cultures were not used to assess whether the virus was viable in the different patient categories, Finally, because of the very heavy workload of the laboratory personnel employed in SARS-CoV-2 testing, information on starting viral loads, inversely related to cycle threshold (Ct) values of the RT-PCR assay [5], has not been made available yet for research purposes.
Despite these limits, our study indicates that critically ill patients have longer viral shedding than those with milder or asymptomatic infections. In contrast, in individuals not requiring ICU care, our results do not support that disease control measures should be adjusted to account for differences in viral shedding according to symptomatic status.
Transparency declaration
The authors declare that they have no conflicts of interest.
Funding : None.
Author contributions
FV designed the study, analysed and interpreted the data and wrote the manuscript; ADC conceived the study, interpreted the results and wrote the manuscript.
Acknowledgements
None.
Editor: A. Kalil
References
- 1.World Health Organization Coronavirus disease 2019 (COVID-19) situation report: 28. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200217-sitrep-28-covid-19.pdf Published February 17, 2020. Available from:
- 2.World Health Organization Laboratory testing for coronavirus disease (COVID-19) in suspected human cases. https://www.who.int/publications-detail/laboratory-testing-for-2019-novel-coronavirus-in-suspected-human-cases-20200117 Available from:
- 3.Young B.E., Ong S.W.X., Kalimuddin S., Low J.G., Tan S.Y., Loh J. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA. 2020 doi: 10.1001/jama.2020.3204. Mar 3. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin A., He Z.B., Zhang S.B., Zhang J.G., Zhang X., Yan W.H. Early risk factors for the duration of SARS-CoV-2 viral positivity in COVID-19 patients. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa490. Apr 27. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cereda D., Tirani M., Rovida F., Demicheli V., Ajelli M., Poletti P. The early phase of the COVID-19 outbreak in Lombardy, Italy. https://arxiv.org/ftp/arxiv/papers/2003/2003.09320.pdf Available from: [DOI] [PMC free article] [PubMed]
- 8.Corman V.M., Albarrak A.M., Omrani A.S., Albarrak M.M., Farah M.E., Almasri M. Viral shedding and antibody response in 37 patients with Middle East respiratory syndrome coronavirus infection. Clin Infect Dis. 2016;62:477–483. doi: 10.1093/cid/civ951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oh M.D., Park W.B., Choe P.G., Choi S.J., Kim J.I., Chae J. Viral load kinetics of MERS coronavirus infection. N Engl J Med. 2016;375:1303–1305. doi: 10.1056/NEJMc1511695. [DOI] [PubMed] [Google Scholar]
- 10.Du X., Yu X., Li Q., Li X., Qin T., Luo Q. Duration for carrying SARS-CoV-2 in COVID-19 patients. J Infect. 2020;81:e78–e79. doi: 10.1016/j.jinf.2020.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ueyama H., Kuno T., Takagi H., Krishnamoorthy P., Vengrenyuk Y., Sharma S.K. Gender difference is associated with severity of coronavirus disease 2019 infection: an insight from a meta-analysis. Crit Care Explor. 2020;2 doi: 10.1097/CCE.0000000000000148. [DOI] [PMC free article] [PubMed] [Google Scholar]