Abstract
Asian lineage A/H5N1 highly pathogenic avian influenza viruses (HPAIVs) have been responsible for continuous outbreaks in Bangladesh since 2007. Although clades 2.2.2 and 2.3.4.2 HPAIVs have disappeared since poultry vaccination was introduced in 2012, clade 2.3.2.1a viruses have continued to be detected in Bangladesh. In this study, we identified A/H9N2 (n = 15), A/H5N1 (n = 19), and A/H5N1-A/H9N2 (n = 18) mixed viruses from live bird markets, chicken farms, and wild house crows (Corvus splendens) in Bangladesh from 2016 to 2018. We analyzed the genetic sequences of the H5 HPAIVs, to better understand the evolutionary history of clade 2.3.2.1a viruses in Bangladesh. Although seven HA genetic subgroups (B1–B7) and six genotypes (G1, G1.1, G1.2, G2, G2.1, and G2.2) have been identified in Bangladesh, only subgroup B7 and genotypes G2, G2.1, and G2.2 were detected after 2016. The replacement of G1 genotype by G2 in Bangladesh was possibly due to vaccination and viral competition in duck populations. Initially, genetic diversity decreased after introduction of vaccination in 2012, but in 2015, genetic diversity increased and was associated with the emergence of genotype G2. Our phylodynamic analysis suggests that domestic Anseriformes, including ducks and geese, may have played a major role in persistence, spread, evolution, and genotype replacement of clade 2.3.2.1a HPAIVs in Bangladesh. Thus, improvements in biosecurity and monitoring of domestic Anseriformes are needed for more effective control of HPAI in Bangladesh.
Keywords: H5N1, highly pathogenic avian influenza, domestic duck, phylogenetic, genetic evolution
1. Introduction
The A/H5 and A/H7 subtype of low pathogenicity avian influenza viruses (LPAIVs) could evolve into highly pathogenic avian influenza viruses (HPAIVs) by mutations resulting in trypsin-independent replication from insertion of multibasic amino acids at the hemagglutinin (HA) cleavage site (Lee, Criado, and Swayne 2020). While LPAIVs replicate only in the respiratory and digestive tract, HPAIVs replicate systemically damaging vital organs and tissues, which results in high mortality in chickens (Webster et al. 1992). The Goose/Guangdong (Gs/Gd) lineage A/H5 HPAIV that was first detected from geese in Guangdong China in 1996 have caused severe economic loss in the global poultry industry and are a serious threat to public health (Xu et al. 1999; Swayne, Suarez, and Sims 2020). The Gs/Gd lineage A/H5 HPAIVs have evolved into multiple HA clades (0–9) and have been disseminated to various geographic regions (Sonnberg, Webby, and Webster 2013; Lee et al. 2017). At least six countries (Bangladesh, China, Egypt, India, Indonesia, and Vietnam) are endemic for Gs/Gd lineage A/H5 HPAIV in poultry (Sutton 2018).
In Bangladesh, the Gs/Gd lineage A/H5 HPAIV has caused continuous outbreaks since it was first detected in 2007 (Parvin et al. 2018). Clade 2.2.2 virus was introduced into Bangladesh in February 2007, followed by clade 2.3.4.2 and 2.3.2.1a viruses in 2011 (Islam et al. 2012; Marinova-Petkova et al. 2014). These viruses have caused infection in various host species including chickens, ducks, geese, migratory wild waterfowl, quail, pigeons, crows, and humans in Bangladesh (Khan et al. 2014; Chakraborty et al. 2017; Parvin et al. 2018). Since 2012, A/H5 HPAIV vaccination has been implemented for commercial chickens in addition to stamping-out of infected flocks for control of HPAIVs in Bangladesh (Rimi et al. 2019; Hill et al. 2018). In particular, RE-6 inactivated vaccine (Merial, clade 2.3.2.1b) and Potsdam/1986 H5N2 inactivated vaccine (Nobilis®, Intervet, Potsdam/1986) were used in breeders and layers, and rHVT-H5 vectored vaccine (Vectormune®, Ceva, clade 2.2) was used in day-old-chicks with the latter providing protection when administered at hatch or in ovo (OIE 2014). While clade 2.2.2 and 2.3.4.2 viruses disappeared in 2012, clade 2.3.2.1a viruses have continued to circulate and cause outbreaks in Bangladesh despite the use of vaccine (Ansari et al. 2016; Parvin et al. 2018; Barman et al. 2019).
Various subtypes of LPAIV have been identified in poultry in Bangladesh, including H9N2 belonging to G1 lineage which is endemic in chickens (Shanmuganatham et al. 2014; Gerloff et al. 2016; Parvin et al. 2018). Previous phylogenetic studies indicated that domestic ducks in Bangladesh are commonly infected with various LPAIVs that are related to viruses in wild waterfowl populations migrating along the Central Asian flyway (Gerloff et al. 2016; Khan et al. 2018). Particularly, domestic ducks in free-range farms that are in frequent contact with wild waterfowl are vulnerable to introduction of a novel AIV (Barman et al. 2017). Repeatedly novel genotypes of A/H5 HPAIVs have emerged by reassortment with these prevailing LPAIVs in poultry of Bangladesh (Monne et al. 2013; Gerloff et al. 2014; Barman et al. 2017, 2019). In addition, eight human cases for A/H5N1 HPAIV infection have been reported in Bangladesh from 2008 to 2015 (Rimi et al. 2019; WHO 2019). Previous studies indicated that live bird markets (LBM) in Bangladesh have a high incidence of A/H5N1 and A/H9N2 viruses and these LBMs could be a major contact point between humans and infected birds (Ansari et al. 2016; Khan et al. 2018; Kim et al. 2018).
Previous studies indicated that clade 2.3.2.1a HPAIVs are circulating in LBMs and domestic ducks and play a major role in the maintenance and emergence of novel reassortant viruses in Bangladesh (Biswas et al. 2017; Khan et al. 2018; Barman et al. 2019). However, the evolution and transmission dynamics of HPAIV in Bangladesh are poorly understood. In this study, we isolated A/H5 HPAIVs from LBMs, chicken farms, and house crows in Bangladesh during 2016–8 and analyzed the full-genome sequences. The molecular evolution and reassortment patterns of clade 2.3.2.1a A/H5 HPAIVs in Bangladesh were analyzed using comparative phylogenetic analysis. Finally, we characterized the interaction between host species in viral spread and persistence in Bangladesh by incorporating host species into a statistical Bayesian phylogenetic model.
2. Materials and methods
2.1 Detection of AIV
Environmental swab samples from LBMs and oropharyngeal swab samples from poultry on outbreak farms and dead house crows (Corvus splendens) were collected during 2016–8. Environmental swab samples were collected from three areas of each LBM (arrival area, slaughter/processing area, and sales/exposure area) and pooled for the AIV detection test. RNA was extracted using the MagMAX 96 AI/ND Viral RNA isolation kit (Ambion Inc., Austin, TX) with the KingFisher magnetic particle processor (Thermo Scientific, Waltham, MA) and tested for AIV by matrix (M) gene real-time reverse transcription PCR (rRT-PCR) as described previously (Spackman 2014). The AIV positive samples were inoculated into 9- to 11-day-old embryonated chicken eggs and incubated 3–5 days at 37 °C (Spackman and Killian 2020). RNA was extracted from allantoic fluids and was tested for AIV by M gene rRT-PCR. The AIV positive RNAs were used for next generation sequencing (NGS). The subtype of AIV positive samples were determined by H5- and H9-specific rRT-PCR (Spackman and Suarez 2008) and NGS.
2.2 Full-length genome sequencing by NGS
The extracted RNA was amplified by random-priming mediated sequence-independent, single-primer amplification (SISPA) as previously described (Chrzastek et al. 2017). Briefly, first-strand cDNA was synthesized by SuperScript IV Reverse Transcriptase (ThermoFisher scientific) with primer K-8N (GACCATCTAGCGACCTCCACNNNNNNNN) and the first-strand cDNA was converted to double-strand DNA by Klenow polymerase (NEB) and K-8N primer. The dsDNA products were purified using Agencourt AMPure XP beads (Beckman Coulter), and PCR amplification was conducted using primer K (GACCATCTAGCGACCTCCAC) and Phusion DNA polymerase (NEB). PCR products were purified using Agencourt AMPure XP beads (Beckman Coulter), and dsDNA quantification was conducted by Qubit dsDNA HS assay (Invitrogen) according to the manufacturer’s instruction.
The sequencing library was prepared using the Nextera XT DNA Sample Preparation Kit (Illumina, San Diego, CA) and 0.2 ng/μl (1 ng total) of purified PCR product according to the manufacturer’s instruction. We generate dual-indexed libraries by appended unique dual index (i7 and i5) sequences to each sample to allow to be pooled and sequenced together. The libraries were adjusted to 4 nM concentration and equal volumes of 5 μl of each library were pooled. The barcoded multiplexed library sequencing was performed on an Illumina MiSeq platform (Illumina) using 500 cycle MiSeq Reagent Kit v2 (Illumina).
The de novo genome assembly of sequencing reads was performed using MIRA3 algorithm in GALAXY platform (Afgan et al. 2018) and searched the most similar sequence by the BLAST. Using these BLAST results as the reference genome, we repeated genome assemblies by mapping the original NGS reads to the reference genome using Geneious 11 software (https://www.geneious.com/). The consensus sequences of reference mapping results were determined as definitive sequences and used for following genetic analysis. A total of thirty-six HA gene sequences and nineteen full-length genome sequences of A/H5 HPAIVs and sixteen HA gene sequences of A/H9 AIVs were analyzed and deposited in GenBank (accession numbers MN994085–MN994269).
2.3 Bayesian phylogenetic analysis and effective population size
All available HA gene sequences of A/H5 HPAIVs identified from Bangladesh belonging to clade 2.3.2.1a (n = 270) were downloaded from the Influenza Research Database (IRD) (http://www.fludb.org/) and GISAID Epiflu database (https://www.gisaid.org/) on 1 April 2019. Sequences were aligned with 36A/H5 sequences that isolated and sequenced in this study by MAFFT method (Katoh et al. 2002) and manually trimmed to equal lengths (1,612 bp) using Geneious 11 software (https://www.geneious.com/).
Bayesian phylogenetic tree of 306 HA gene sequences was estimated using BEAST v.1.10.4 (https://beast.community) with an uncorrelated lognormal relaxed molecular clock. The Hasegawa–Kishino–Yano nucleotide substitution model with gamma-distributed rate variation among sites with four rate categories (HKY+G) (Hasegawa, Kishino, and Yano 1985) was used along with a Gaussian Markov random field (GMRF) Bayesian skyride coalescent tree prior (Minin, Bloomquist, and Suchard 2008). The Markov Chain Monte Carlo (MCMC) was run in parallel for three chains, each with 50 million steps and the parameters and trees sampled every 5,000 steps. The resulting log and tree files were combined with LogCombiner v1.10.4 (https://beast.community/logcombiner) after 10 per cent burn-in yielding a total 27,003 parameter states and posterior trees. The parameters were analyzed with TRACER v1.5 (http://tree.bio.ed.ac.uk/software/tracer/) and all parameters had an effective sample size >200. A time-scaled maximum clade credibility (MCC) tree was generated using TreeAnnotator (https://beast.community/treeannotator) in BEAST and visualized using FigTree 1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/). The GMRF Bayesian skyride plot of effective population size over time was reconstructed using TRACER v1.5. To estimate the times for the most common ancestor (tMRCA) of each genotype and substitutional rate, a time-scaled MCC tree for each gene was generated as described above.
2.4 Reassortment analysis
All available complete coding sequences of A/H5 HPAIVs identified from Bangladesh belonging to clade 2.3.2.1a in IRD and GISAID Epiflu databases (n = 171) and A/H5N1 sequences sequenced in this study (n = 19) were used for reassortment analysis. The A/H5N1–A/H9N2 mixed viruses isolated in this study were excluded in this analysis because the genome constellation of mixed viruses was not verifiable. Additionally, all available full-genome sequences of LPAI viruses isolated in Bangladesh after 2011 were downloaded. We excluded the sequences that have a same collection date and identical nucleotide sequences and selected twenty-seven H9N2 viruses and nineteen non-H9N2 LPAIVs. For the NGS gene, we excluded the sequences belonging to B allele. We included the previously reported LPAI viruses that contributed their genes to the genesis of reassortant 2.3.2.1a A/H5N1 viruses in Bangladesh (Monne et al. 2013; Gerloff et al. 2014; Barman et al. 2017, 2019), including forty-six polymerase basic 2 (PB2), polymerase basic 1 (PB1), polymerase acidic (PA), nucleoprotein (NP), and M and forty nonstructural (NS) gene sequences of LPAI viruses isolated in Bangladesh.
Final data sets were aligned by MAFFT method and manually trimmed to equal lengths. We inferred maximum likelihood (ML) phylogenetic trees by the MEGA7 software (https://www.megasoftware.net) using the HKY+G nucleotide substitution model (Hasegawa, Kishino, and Yano 1985). Statistical analysis of the phylogenetic tree was performed by bootstrap analysis carried out on 500 replicates. Backronymed adaptable lightweight tree import code (BALTIC) (https://bedford.io/projects/baltic/) was used for comparison and visualization of ML phylogenetic tree of each gene segment. The phylogenetic position of each strain was traced and colored according to genotypes. Figures were generated by modifying the Jupyter Notebook document used in previous studies (Bell and Bedford 2017; Poen et al. 2019). In Supplementary Fig. S3, the genome constellation of each virus is plotted in the MCC tree of the HA gene using the ggtree R package (Yu et al. 2018).
To identify the evolutionary relationship between genotypes, a median-joining phylogenetic network was constructed for the HA gene using the NETWORK ver. 5.0.1.1 (http://www.fluxus-engineering.com/). In addition, a Bayesian discrete-trait phylodynamic model were adopted on MCC tree of the HA gene to estimate the directional changes of genotypes. We defined genotypes as discrete nominal categories and reconstructed ancestral genotype. MCMC was run in parallel for two chains, each with 50 million steps and the parameters and trees sampled every 5,000 steps. The resulting log and tree files were combined with LogCombiner v1.10.4 (https://beast.community/logcombiner) after 10 per cent burn-in yielding a total of 18,002 parameter states and posterior trees. The parameters were analyzed with TRACER v1.5 (http://tree.bio.ed.ac.uk/software/tracer/) and all parameters had an effective sample size >200. An MCC tree was generated using TreeAnnotator (https://beast.community/treeannotator) in BEAST and visualized using FigTree 1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
2.5 Analysis of selective pressure
The selective pressure was analyzed using the codon-based fixed effects likelihood (FEL) method (Kosakovsky Pond and Frost 2005) on the Datamonkey server (http://www.datamonkey.org/). The FEL method infers nonsynonymous to synonymous substitutions (dN/dS) ratio and P value of selective pressure for each site based on the inference of the ancestral sequence. We compared the selective pressure of each gene segment of genotype G1 and G2 and estimated positively selected amino acid sites showing statistical significance (P value < 0.05).
The nucleotide substitution rates (substitution/site/year) of each gene segment of genotype G1 and G2 were estimated by Bayesian phylogenetic analysis using BEAST v.1.10.4 (https://beast.community) with HKY+G substitution model and uncorrelated lognormal relaxed molecular clock model. To analyze statistical significance in difference of substitution rate (r) between G1 and G2, we estimated the 95 per cent Bayesian credible intervals (BCIs) of mean difference of substitution rate between the groups (i.e., mean of r2 − r1). If the 95 per cent BCI does not contain null hypothesized value zero, it is considered as statistical significance in the difference between genotypes (Hespanhol et al. 2019).
2.6 Host dynamics
Among the HA gene data set, we selected the sequences having information on host species and isolate dates, except environment isolates. The final data set consisted of 249 taxa, which were coded into three host species: wild birds (n = 20), domestic Galliformes (n = 83), and domestic Anseriformes (n = 146).
The Bayesian phylogenetic tree of the HA gene was estimated using BEAST v.1.10.4 (https://beast.community) as above. We reconstructed the ancestral host state and estimated the asymmetric viral exchanges between host species using a nonreversible continuous-time Markov chain model. We applied a Bayesian stochastic search variable selection procedure to support the transition rate between host species statistically and to construct a Bayes factor (BF) test (Lemey et al. 2009). The BF test was conducted for each transition using SPREAD software v1.0.6 (https://beast.community/spread). We identify a transition as significant when posterior probability >0.5 and Bayesian factor >4 (Bahl et al. 2016). We also estimated the rate and number of transitions between hosts (Markov jump) and the time spent in specific hosts (Markov rewards) using stochastic mapping techniques implemented in the BEAST package (Minin and Suchard 2008).
The MCMC was run in parallel for four chains, each with 50 million steps and the parameters and trees sampled every 5,000 steps. The resulting log and tree files were combined with LogCombiner v1.10.4 (https://beast.community/logcombiner) after 10 per cent burn-in yielding a total of 36,004 parameter states and posterior trees. The parameters were analyzed with TRACER v1.5 (http://tree.bio.ed.ac.uk/software/tracer/) and all parameters had an effective sample size >200. An MCC tree was generated using TreeAnnotator (https://beast.community/treeannotator) in BEAST and visualized using FigTree 1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
We analyzed posterior trees using the program PACT (http://www.trevorbedford.com/pact) to compute the number of transition events and the proportion of hosts through time. After resampled the posterior trees every 20,000 states, we broke the posterior trees (n = 9,001) up into multiple temporal sections (0.25 year per section). The host state transition at the tree nodes were counted for each time windows for each posterior tree. The proportion of each host type indicated the proportion of the branches residing in specific host type relative to the total length of the tree over time.
3. Result
3.1 Virus detection and sequence analysis
During AIV surveillance in Bangladesh in 2016–8, a total of 270 samples collected from LBMs (n = 140) and clinical cases from chicken farms (n = 130) were positive for the AIV M gene rRT-PCR, and of these 270 samples, 115 samples (fifty-one from LBM and sixty-four from chicken farm outbreak cases) had virus isolated using embryonating chicken eggs. Fifty-two isolates were randomly selected for subtype analysis and nucleotide sequences for genetic characterization. Of the fifty-two isolates, fifteen were A/H9N2 viruses, nineteen were A/H5N1 viruses, and eighteen were A/H5N1–A/H9N2 mixed viruses and were obtained from environmental samples of LBM (n = 34), chickens farms (n = 12), and house crows (n = 6) (Table 1). The sequence of H5 HA genes of seventeen A/H5N1–A/H9N2 mixed viruses and full genomes of nineteen A/H5N1 viruses were analyzed using an NGS system with Illumina MiSeq (Illumina). All HA genes of A/H5N1 isolates had identical multiple basic amino acids motif at the proteolytic cleavage site (PQRERRRKR/GLF) and shared high nucleotide identities (97.70–100%). Our phylogenetic analysis indicated that all HA genes of A/H5 isolates belong to clade 2.3.2.1a and clustered with the A/H5 viruses identified in Bangladesh during 2015–8 (Supplementary Fig. S1), suggesting that both clade 2.3.2.1a A/H5N1 HPAIV and A/H9N2 LPAIV were cocirculating in poultry in Bangladesh.
Table 1.
Avian influenza viruses isolated in this study, from June 2016 to February 2018.
| Isolates | Collection date | Sample | Location (division) | Subtype | GenBank accession number (HA gene) |
|---|---|---|---|---|---|
| A/environment/Bangladesh/NRL-AI-893/2016 | 6/8/2016 | LBM | Dhaka | H9N2 | MN994085 |
| A/environment/Bangladesh/NRL-AI-894/2016 | 6/9/2016 | LBM | Dhaka | H9N2 | MN994113 |
| A/environment/Bangladesh/NRL-AI-921/2016 | 6/13/2016 | LBM | Dhaka | H9N2 | MN994122 |
| A/environment/Bangladesh/NRL-AI-923/2016 | 6/13/2016 | LBM | Dhaka | H9N2 | MN994088 |
| A/environment/Bangladesh/NRL-AI-933/2016 | 6/14/2016 | LBM | Dhaka | H9N2 | MN994124 |
| A/environment/Bangladesh/NRL-AI-979/2016 | 7/11/2016 | LBM | Dhaka | H9N2 | MN994087 |
| A/environment/Bangladesh/NRL-AI-979-2/2016 | 7/11/2016 | LBM | Dhaka | H9N2 | MN994111 |
| A/environment/Bangladesh/NRL-AI-990/2016 | 7/12/2016 | LBM | Dhaka | H5N1 | MN994265 |
| A/environment/Bangladesh/NRL-AI-1003/2016 | 7/13/2016 | LBM | Dhaka | H5N1 | MN994249 |
| A/environment/Bangladesh/NRL-AI-1005/2016 | 7/13/2016 | LBM | Dhaka | H5N1/H9N2 | MN994110 |
| A/environment/Bangladesh/NRL-AI-1072/2016 | 7/24/2016 | LBM | Dhaka | H9N2 | MN994086 |
| A/environment/Bangladesh/NRL-AI-1085/2016 | 8/17/2016 | LBM | Dhaka | H5N1/H9N2 | MN994116 |
| A/environment/Bangladesh/NRL-AI-1087/2016 | 8/17/2016 | LBM | Dhaka | H9N2 | MN994112 |
| A/environment/Bangladesh/NRL-AI-1088/2016 | 8/17/2016 | LBM | Dhaka | H5N1/H9N2 | MN994089 |
| A/environment/Bangladesh/NRL-AI-1106/2016 | 8/21/2016 | LBM | Dhaka | H9N2 | MN994120 |
| A/environment/Bangladesh/NRL-AI-1117/2016 | 8/22/2016 | LBM | Dhaka | H9N2 | MN994115 |
| A/environment/Bangladesh/NRL-AI-1398/2016 | 11/22/2016 | LBM | Dhaka | H9N2 | MN994109 |
| A/environment/Bangladesh/NRL-AI-1400/2016 | 11/17/2016 | LBM | Dhaka | H5N1/H9N2 | MN994091 |
| A/environment/Bangladesh/NRL-AI-1406/2016 | 11/17/2016 | LBM | Dhaka | H9N2 | MN994105 |
| A/environment/Bangladesh/NRL-AI-1418/2016 | 11/21/2016 | LBM | Dhaka | H9N2 | MN994102 |
| A/environment/Bangladesh/NRL-AI-1423/2016 | 11/21/2016 | LBM | Dhaka | H5N1/H9N2 | MN994106 |
| A/environment/Bangladesh/NRL-AI-1424/2016 | 11/21/2016 | LBM | Dhaka | H5N1/H9N2 | MN994125 |
| A/environment/Bangladesh/NRL-AI-1425/2016 | 11/21/2016 | LBM | Dhaka | H5N1/H9N2 | MN994123 |
| A/environment/Bangladesh/NRL-AI-1447/2016 | 11/23/2016 | LBM | Dhaka | H5N1/H9N2 | MN994104 |
| A/environment/Bangladesh/NRL-AI-1555/2016 | 12/18/2016 | LBM | Dhaka | H5N1/H9N2 | MN994103 |
| A/environment/Bangladesh/NRL-AI-1578/2016 | 12/20/2016 | LBM | Dhaka | H5N1/H9N2 | MN994119 |
| A/environment/Bangladesh/NRL-AI-1579/2016 | 12/21/2016 | LBM | Dhaka | H5N1/H9N2 | MN994101 |
| A/environment/Bangladesh/NRL-AI-1602/2016 | 12/27/2016 | LBM | Dhaka | H5N1/H9N2 | MN994108 |
| A/environment/Bangladesh/NRL-AI-1603/2016 | 12/27/2016 | LBM | Dhaka | H5N1/H9N2 | MN994114 |
| A/environment/Bangladesh/NRL-AI-1607/2016 | 12/28/2016 | LBM | Dhaka | H5N1/H9N2 | MN994090 |
| A/environment/Bangladesh/NRL-AI-1610/2016 | 12/27/2016 | LBM | Dhaka | H5N1 | MN994257 |
| A/environment/Bangladesh/NRL-AI-2553/2017 | 1/15/2017 | LBM | Dhaka | H5N1/H9N2 | MN994117 |
| A/environment/Bangladesh/NRL-AI-2559/2017 | 1/15/2017 | LBM | Dhaka | H5N1/H9N2 | MN994107 |
| A/environment/Bangladesh/NRL-AI-2566/2017 | 1/15/2017 | LBM | Dhaka | H5N1/H9N2 | MN994118 |
| A/crow/Bangladesh/NRL-AI-689/2016 | 2/9/2016 | Crow | Rajshahi | H5N1 | MN994217 |
| A/crow/Bangladesh/NRL-AI-690/2016 | 2/9/2016 | Crow | Rajshahi | H5N1 | MN994225 |
| A/crow/Bangladesh/NRL-AI-1615/2017 | 1/17/2017 | Crow | Rajshahi | H5N1 | MN994201 |
| A/crow/Bangladesh/NRL-AI-1616/2017 | 1/17/2017 | Crow | Rajshahi | H5N1 | MN994209 |
| A/crow/Bangladesh/NRL-AI-8471/2017 | 12/31/2017 | Crow | Dhaka | H5N1 | MN994233 |
| A/crow/Bangladesh/NRL-AI-8473/2017 | 12/31/2017 | Crow | Dhaka | H5N1 | MN994241 |
| A/chicken/Bangladesh/NRL-AI-1613/2017 | 1/17/2017 | Chicken farm | Dhaka | H5N1 | MN994129 |
| A/chicken/Bangladesh/NRL-AI-1614/2017 | 1/17/2017 | Chicken farm | Dhaka | H5N1 | MN994137 |
| A/chicken/Bangladesh/NRL-AI-8251/2017 | 1/30/2017 | Chicken farm | Rajshahi | H5N1 | MN994169 |
| A/Chicken/Bangladesh/NRL-AI-2109/2017 | 4/3/2017 | Chicken farm | Dhaka | H5N1 | MN994095 |
| A/chicken/Bangladesh/NRL-AI-3237/2017 | 4/23/2017 | Chicken farm | Dhaka | H5N1 | MN994145 |
| A/chicken/Bangladesh/NRL-AI-3238/2017 | 4/23/2017 | Chicken farm | Dhaka | H9N2 | MN994100 |
| A/chicken/Bangladesh/NRL-AI-8051/2017 | 8/22/2017 | Chicken farm | Dhaka | H5N1 | MN994153 |
| A/chicken/Bangladesh/NRL-AI-8052/2017 | 8/22/2017 | Chicken farm | Dhaka | H5N1 | MN994161 |
| A/chicken/Bangladesh/NRL-AI-8323/2017 | 12/8/2017 | Chicken farm | Dhaka | H5N1 | MN994177 |
| A/chicken/Bangladesh/NRL-AI-8468/2017 | 12/31/2017 | Chicken farm | Mymensingh | H5N1 | MN994185 |
| A/chicken/Bangladesh/NRL-AI-8469/2017 | 12/31/2017 | Chicken farm | Dhaka | H5N1 | MN994193 |
| A/chicken/Bangladesh/NRL-AI-8682/2018 | 2/18/2018 | Chicken farm | Dhaka | H5N1/H9N2 | MN994121 |
In addition, we isolated A/H5N1 viruses from dead house crows found near waste deposit areas that contained poultry internal organs discarded from LBM. The ingestion of infected poultry internal organs was suspected as a source for house crow infection.
3.2 Bayesian phylogenetic analysis of HA gene
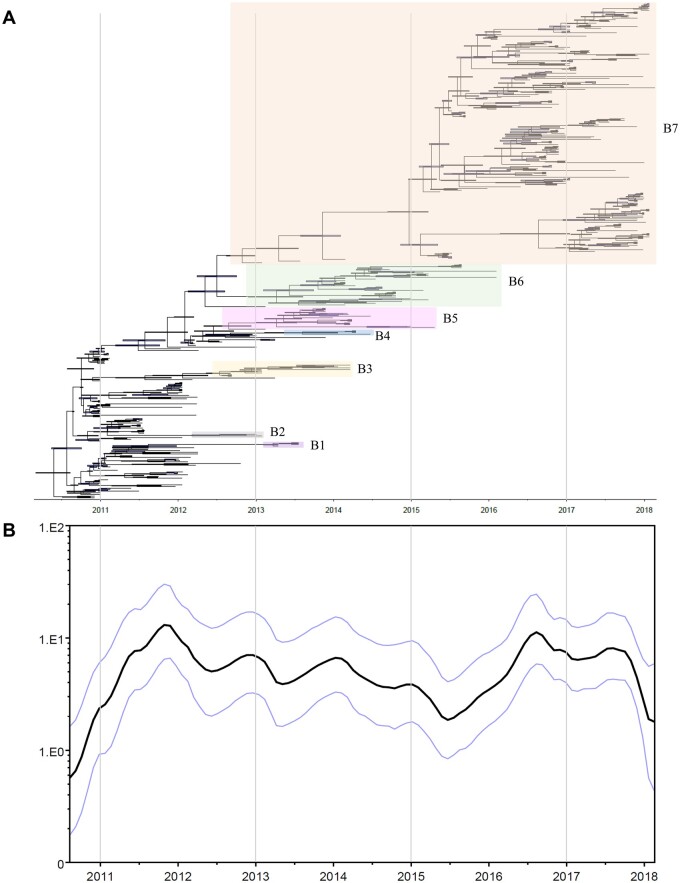
Phylogenetic tree of HA genes showed that A/H5 clade 2.3.2.1a viruses in Bangladesh evolved into at least seven distinct genetic subgroups (B1–B7) in the latter half of 2012, which was supported by high posterior probability in Bayesian phylogenetic tree (>99%) (Fig. 1A and Supplementary Fig. S1). Most of the subgroups (B1–B6) have not been detected since March 2016, but the B7 subgroup was continuously identified in poultry of Bangladesh since March 2016. All A/H5N1 viruses isolated in this study belonged to subgroup B7.
Figure 1.
Bayesian phylogenetic tree and changes of effective population size of clade 2.3.2.1a H5 HPAIVs in Bangladesh. (A) Bayesian phylogenetic MCC tree of HA gene of clade 2.3.2.1a H5N1 HPAIVs in Bangladesh. Coding region of HA was used for reconstruction of phylogenetic tree. Horizontal bars on the nodes indicate the 95 per cent BCIs of divergence time estimates. (B) GMRF Bayesian skyride plots indicating effective population size (relative genetic diversity) over time.
The analysis of the virus population dynamic revealed a gradual decrease in genetic diversity with fluctuations through time from the beginning of 2012 to the middle of 2015, followed by a sudden increase from late 2015 to early 2016 (Fig. 1B). The evolutionary rate estimated for the HA gene of clade 2.3.2.1a A/H5N1 viruses identified during 2010–8 was 5.517 × 10−3 substitution/site/year (95% BCI: 4.781 × 10−3–6.247 × 10−3).
3.3 Emergence of novel reassortant genotypes
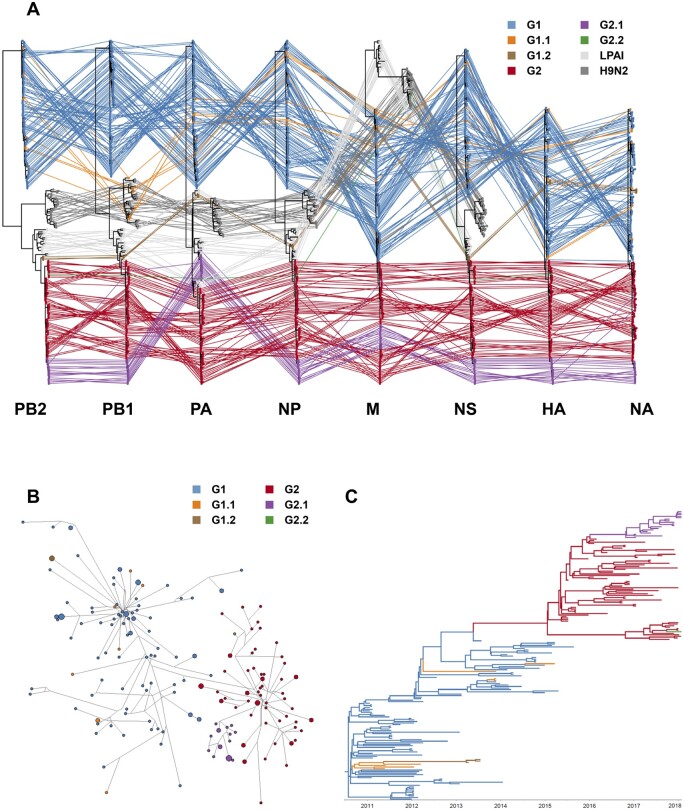
ML phylogenetic trees and time-scaled MCC trees for eight genes of clade 2.3.2.1a Bangladesh A/H5 viruses were constructed and a total of six genotypes (G1, G1.1, G1.2, G2, G2.1, and G2.2) were detected (Figs 2A, 3 and Supplementary Figs S2–S4). The Genotype G1.1 was detected during 2011–5, which had PB1 derived from H9N2 LPAIV circulating in Bangladesh. Only three strains isolated in 2013 (A/duck/Bangladesh/14VIR1121_16/2013, A/duck/Bangladesh/14VIR1121_17/2013, and A/duck/Bangladesh/14VIR1121_18/2013) belong to Genotype G1.2, which had PB2, PB1, PA, NP, and NS genes derived from LPAIVs of domestic ducks in Bangladesh. From 2015, the Genotype G2, which had PB2, PB1, PA, NP, and NS genes of other LPAIVs of domestic ducks, were detected. All genes of genotype G2.1 had identical genome constellation with genotype G2 except the PA gene that clustered with other domestic duck LPAIVs. An A/H5N2 isolate (A/chicken/Bangladesh/34722/2018), which has the PB2, PB1, HA, NP, and NS genes derived from G2 and the PA gene from G2.1 and neuraminidase (NA) and M gene of Bangladesh H9N2 LPAIV, was classified as the G2.2.
Figure 2.
Reassortemnt of clade 2.3.2.1a H5 HPAIVs in Bangladesh. (A) Phylogenetic incongruence analysis. Maximum likelihood phylogenetic trees of eight gene segments compared and the gene segments from equivalent strains were connected across the trees. Tips and connecting lines are colored according to genotypes. (B) Median-joining phylogenetic network for HA gene of clade 2.3.2.1a H5N1 HPAIVs in Bangladesh. Each unique sequence is represented by a circle sized relative to its frequency in the data set. Branch length is proportional to the number of mutations. Isolates are colored according to genotypes. (C) Bayesian phylogenetic MCC tree for HA gene of clade 2.3.2.1a H5N1 HPAIVs in Bangladesh. Branches are colored according to genotypes.
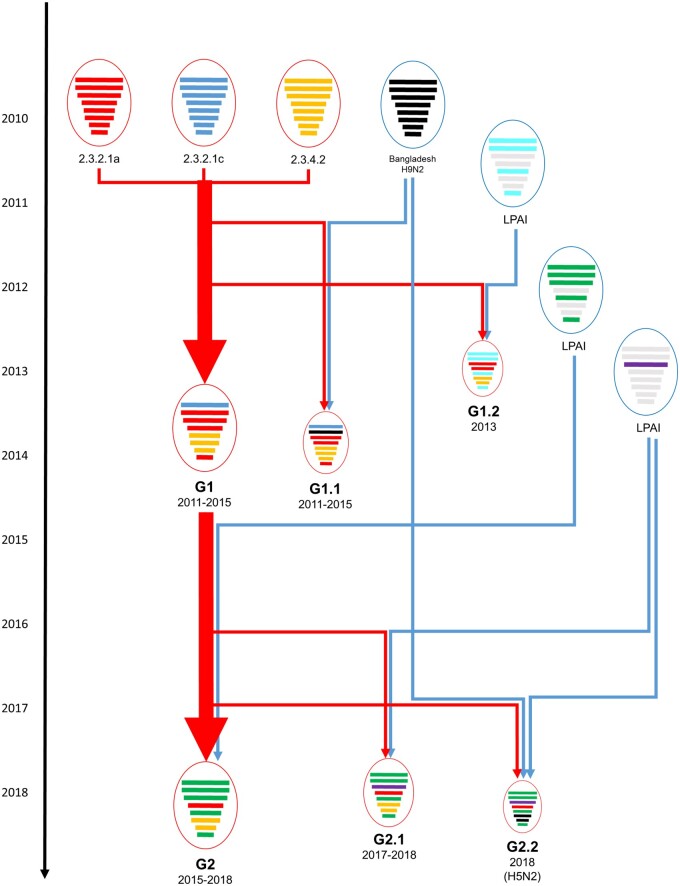
Figure 3.
Schematic diagram for probable genesis of genotypes of clade 2.3.2.1a H5 HPAIV identified in Bangladesh. Eight gene segments in each of the schematic virus particles are arranged from top to bottom to represent the PB2, PB1, PA, HA, NP, NA, M, and NS genes. The circle and arrow of HPAI viruses are colored in red, and LPAI viruses are colored in blue. For each gene segment, different phylogenetic groupings are in different colors.
To identify the evolutionary relationship between genotypes, we constructed the median-joining phylogenetic network (Fig. 2B). In addition, to identify the ancestral genotype for each genotype, the ancestral genotype was reconstructed in MCC trees for the HA gene (Fig. 2C and Supplementary Fig. S4). These phylogenetic analyses indicated that G1.1, G1.2, and G2 genotypes emerged from G1, and G2.1 and G2.2 genotypes emerged from G2 by reassortment with local LPAIVs. All genes of G1.1 except PB1 did not cluster together in phylogenetic analysis, indicating that multiple independent reassortments repeatedly occurred between the genotype G1 A/H5N1 viruses and PB1 gene of H9N2 viruses (Fig. 2 and Supplementary Figs S3 and S4). In our analysis, at least four independent reassortment events were detected during 2011–5 (Fig. 2 and Supplementary Figs S3 and S4). Although the PA genes of G2.1 and G2.2 genotypes clustered together in phylogenetic analysis, the HA gene of G2.2 was phylogenetically distinct from G2.1 in MCC tree and network analysis indicating that G2.2 genotype emerged through independent reassortment between G2 and LPAIVs of domestic ducks. The time-scaled phylogenetic analysis indicated that the dominant genotype was replaced from G1 to G2 after 2015 and HA genes of genotype G2, G2.1, and G2.2 belong to HA subgroup B7 (Fig. 2C and Supplementary Fig. S3).
To identify the time of reassortment, the tMRCA of each gene segment was inferred (Table 2 and Supplementary Fig. S4). G1 genotype emerged between September 2009 and November 2010 and G1.2 emerged between May 2012 and March 2013. The time for emergence of new dominant genotype G2 was between May 2013 and March 2015 and the time for further reassortment for G2.1 genotype was between October 2015 and November 2016. The genotype G1.1 and G2.2 were excluded from tMRCA analysis because multiple independent reassortments were detected for genotype G1.1, and only one full-genome sequence was available for genotype G2.2.
Table 2.
tMRCA for each gene segment of the four genotypes of clade 2.3.2.1a H5N1 viruses identified in Bangladesh.
| Gene | tMRCA (95% BCI) |
|||
|---|---|---|---|---|
| G1 | G1.2 | G2 | G2.1 | |
| PB2 | May 10 (Jan 10–Sep 10) | Feb 13 (Dec 12–Mar 13) | Jul 14 (Jan 14–Jan 15) | Jul 16 (Mar 16–Nov 16) |
| PB1 | Apr 10 (Nov 09–Sep 10) | Sep 12 (May 12–Jan 13) | Jul 14 (Jan 14–Dec 14) | Jul 16 (Mar 16–Oct 16) |
| PA | Jun 10 (Mar 10–Sep 10) | Jan 13 (Oct 12–Mar 13) | Sep 14 (May 14–Jan 15) | Apr 16 (Oct 15–Oct 16) |
| HA | Jun 10 (Mar 10–Sep 10) | Feb 13 (Jan 13–Mar 13) | Dec 14 (Sep 14–Mar 15) | Sep 16 (Jul 16–Nov 16) |
| NP | Jun 10 (Feb 10–Sep 10) | Jan 13 (Oct 12–Mar 13) | Aug 14 (Feb 14–Jan 15) | Jul 16 (Feb 16–Oct 16) |
| NA | Sep 10 (Jul 10–Nov 10) | Jan 13 (Dec 12–Mar 13) | Nov 14 (Jun 14–Mar 15) | Jun 16 (Mar 16–Oct 16) |
| M | Mar 10 (Sep 09–Sep 10) | Jan 13 (Aug 12–Mar 13) | Jul 14 (Dec 13–Jan 15) | Aug 16 (Mar 16–Nov 16) |
| NS | Mar 10 (Oct 09–Aug 10) | Feb 13 (Dec 12–Mar 13) | Jan 14 (May 13–Sep 14) | Jun 16 (Feb 16–Oct 16) |
3.4 Comparing evolutionary rates and selective pressures between genotypes
We compared evolutionary rate and selective pressure between genotype G1 and G2, including related genotype such as G1.1, G1.2, G2.1, and G2.2, to determine if this genotype replacement in Bangladesh related to positive selection by vaccination (Table 3). All genes of clade 2.3.2.1a HPAIVs are under purifying selection (dN/dS ratio < 1), indicating that the nonsynonymous mutations affecting their function have been removed by purifying selection. Although there was no significant difference in dN/dS ratio between genotypes, amino acid residues under positive selection were more frequently identified in genotype G1 than G2. Two amino acid residues in HA gene (position 123 and 189 in H5 numbering), two amino acid residues in NA gene (position 320 and 362), and one amino acid residue in PA gene (position 618) were under positive selection in genotype G1, but only one amino acid residue in PA gene (position 538) was under positive selection in genotype G2 (Table 3), respectively. In selective pressure analysis of all HA gene, two amino acid residue, 123 and 136, were identified to be under positive selection and these sites were located in the H5 antigenic site A (Yang et al. 2016). Notably, the RE-6 vaccine strain had different amino acid (P123 and S136) compared with recent Bangladesh viruses at these positions. While G1 genotype viruses had P (7/105, 6.7%) or S (95/105, 90.5%) at position 123 and P (95/105, 90.5%) or S (10/105, 9.5%) at position 136, most of genotype G2 viruses had S123 (84/85, 98.8%) and P136 (83/85, 97.6%). The rHVT-H5 vaccine (A/Swan/Hungary/4999/2006) and Potsdam/1986 H5N2 inactivated vaccine had S123 and P136. In addition, the nucleotide substitution rate of HA of genotype G1 viruses (6.09 × 10−3, 95% BCI: 4.98 × 10−3–7.21 × 10−3) was significantly higher than that of genotype G2 viruses (4.22 × 10−3, 95% BCI: 3.22 × 10−3–5.28 × 10−3). However, there was no significant difference in the nucleotide substitution rate of other genes between genotype G1 and G2. These results indicated that genotype G1 viruses were under stronger positive selection than recent genotype G2 viruses, and RE-6 vaccine may have contributed to the positive selection of clade 2.3.2.1a A/H5N1 viruses in Bangladesh.
Table 3.
Nucleotide substitution rate and selection pressures in the clade 2.3.2.1a H5N1 viruses in Bangladesh.
| Gene | Genotype | Substitution rate (10−3 subs/site/year) |
Mean dN/dS ratio | Positively selected site (P < 0.05) | |
|---|---|---|---|---|---|
| Mean | 95% BCI | ||||
| HA | All | 5.18 | 4.44–5.90 | 0.139 | 123, 136b |
| G1, G1.1, G1.2a | 6.09 | 4.98–7.21 | 0.149 | 123, 189 | |
| G2, G2.1, G2.2 | 4.22 | 3.22–5.28 | 0.130 | NDc | |
| NA | All | 6.41 | 5.47–7.44 | 0.246 | 320, 362 |
| G1, G1.1, G1.2 | 7.36 | 6.21–8.54 | 0.251 | 320, 362 | |
| G2, G2.1 | 6.12 | 4.24–8.33 | 0.239 | ND | |
| PB2 | G1, G1.1 | 5.34 | 4.40–6.24 | 0.113 | ND |
| G2, G2.1, G2.2 | 4.83 | 3.62–6.12 | 0.123 | ND | |
| PB1 | G1 | 4.44 | 3.70–5.22 | 0.089 | ND |
| G2, G2.1, G2.2 | 4.41 | 3.36–5.54 | 0.124 | ND | |
| PA | G1, G1.1 | 5.12 | 4.36–6.03 | 0.164 | 618 |
| G2 | 4.46 | 3.38–5.68 | 0.172 | 538 | |
| G2.1, G2.2 | 7.53 | 2.37–14.30 | 0.175 | ND | |
| NP | G1, G1.1 | 4.96 | 3.97–5.93 | 0.047 | ND |
| G2, G2.1, G2.2 | 4.29 | 3.22–5.58 | 0.058 | ND | |
| M | All | 5.27 | 3.74–6.97 | M1 = 0.049, M2 = 0.553 | ND |
| G1, G1.1, G1.2 | 5.16 | 3.10–7.44 | M1 = 0.041, M2 = 0.955 | ND | |
| G2, G2.2 | 5.66 | 3.14–8.57 | M1 = 0.035, M2 = 0.478 | ND | |
| NS | G1, G1.1 | 5.72 | 4.45–7.06 | NS1 = 0.424, NEP = 0.230 | ND |
| G2, G2.1, G2.2 | 4.7 | 3.36–6.22 | NS1 = 9.464; NEP= 0.254 | ND | |
The nucleotide substitution rate of HA gene of genotype G1 and related viruses is significantly higher than that of genotype G2 and related viruses.
H5 numbering used for HA protein.
ND, not detected.
3.5 Host dynamics
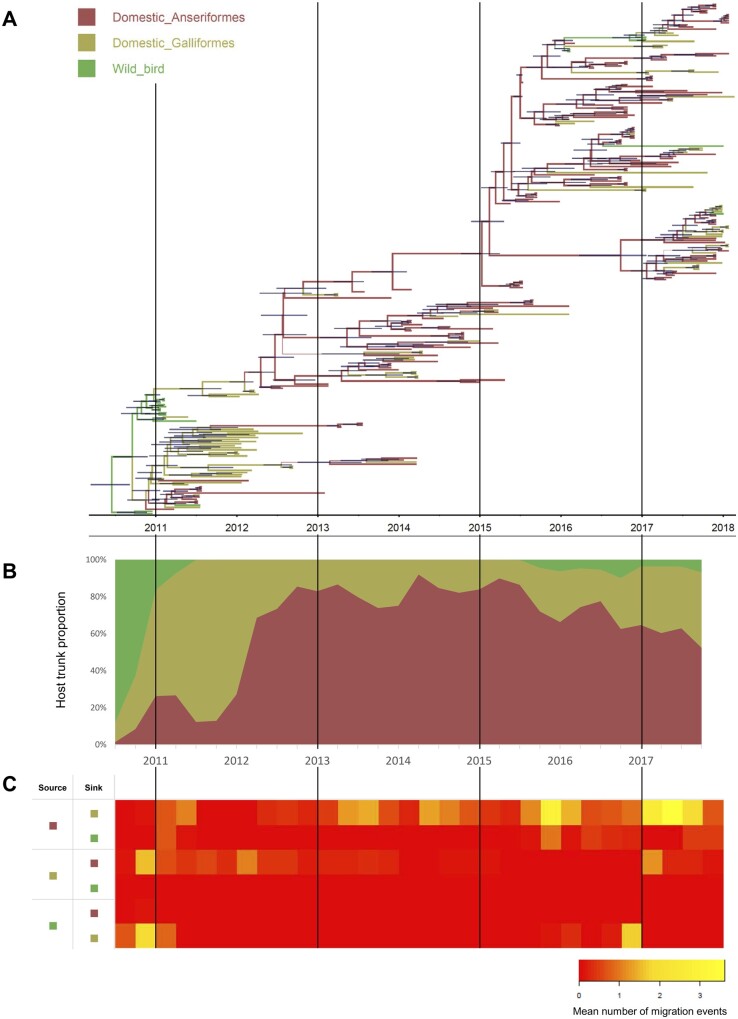
To evaluate the contribution of host species on viral transmission and circulation, the number of host transition (Markov jump) were estimated by incorporating host species in Bayesian phylogenetic analysis and ancestral host reconstruction (Fig. 4A, Supplementary Fig. S5, and Table 4). The frequent bidirectional host transition between domestic Anseriformes and domestic Galliformes was detected with high BF (>100) and posterior probability (1.00). The number of host transition from domestic Anseriformes to domestic Galliformes (27.81) was higher than from domestic Galliformes to domestic Anseriformes (10.18). The transitions from domestic Anseriformes to wild birds and from wild birds to domestic Galliformes were also well supported with high BF and posterior probability, but the number of transitions was relatively low (4.23 and 6.12).
Figure 4.
Phylogenetic estimation of viral transitions between host species. (A) Bayesian phylogenetic MCC tree of HA gene of clade 2.3.2.1a H5N1 HPAIVs in Bangladesh. Branches are colored according to host type and thickness of branches indicate posterior probabilities of the ancestral host type. Horizontal bars on the nodes indicate the 95 per cent BCIs of divergence time estimates. (B) The proportion of the trunk belonging to each host type over time. (C) Heat maps showing the number of transition events between each host type.
Table 4.
The transition rate, number of Markov jump, and statistical support value between domestic Anseriformes, domestic Galliformes, and wild bird populations in Bangladesh during the year 2010–8.
| Transition from | Transition to | Mean actual transition ratea (95% BCI) | Mean number of Markov jump (95% BCI) | BF | Posterior probability |
|---|---|---|---|---|---|
| Domestic Anseriformes | Domestic Galliformesb | 1.85 (0.28–3.79) | 27.81 (23, 32) | >100 | 1.00 |
| Wild birdsb | 0.33 (0.02–0.8) | 4.23 (3, 6) | >100 | 1.00 | |
| Domestic Galliformes | Domestic Anseriformesb | 1.06 (0.13–2.28) | 10.18 (4, 15) | >100 | 1.00 |
| Wild birds | 0.03 (0–0.19) | 0.09 (0, 1) | 0.24 | 0.16 | |
| Wild birds | Domestic Anseriformes | 0.09 (0–0.53) | 0.25 (0, 2) | 0.45 | 0.27 |
| Domestic Galliformesb | 1.03 (0.12–2.29) | 6.12 (4, 8) | >100 | 1.00 |
Actual transition rates were calculated as rate × indicator.
Well-supported viral transitions, a posterior probability >0.5 and a BF >3.
The ancestral host species of Bangladesh clade 2.3.2.1a viruses were estimated as wild birds with high posterior probability (99.63%), indicating this virus was introduced into Bangladesh by wild birds (Fig. 4A). The host transition from wild birds to domestic Galliformes was detected in late 2010, and the transition from domestic Galliformes to domestic Anseriformes was detected during late 2010 to early 2012 (Fig. 4A and C). After 2012, the host trunk proportion of domestic Anseriformes was increased and domestic Anseriformes became the dominant species infected by HPAIV in Bangladesh (Fig. 4B). The high number of host transitions from domestic Anseriformes to domestic Galliformes was detected from 2013 to 2017 (Fig. 4A, C). The contribution of the house crow to viral transmission was detected in the latter half of 2015 in this study (Fig. 4A, C). However, considering these house crows were found near LBM waste deposit areas, undetected viruses in LBM were suspected as a source of these viral transmissions rather than house crows. These results suggested that while clade 2.3.2.1a viruses were introduced into domestic Galliformes in Bangladesh by wild birds during late 2010 and early 2012, domestic Anseriformes played a major role in long-term circulation and dissemination of viruses in Bangladesh after 2013.
4. Discussion
Clade 2.3.2.1a HPAIVs have caused continuous outbreaks in Bangladesh and evolved into multiple subgroups and genotypes since 2011, despite the use of vaccine. Although seven HA gene subgroups and six genotypes were identified in this study, six subgroups (B1–B6) and three genotypes (G1, G1.1, and G1.2) have disappeared since 2016. Novel reassortant genotype G2 became the dominant genotype since 2015, and two novel genotypes, G2.1 and G2.2, emerged by additional reassortment of genotype G2 with local LPAIVs. The decreasing genetic diversity corresponded to the use of vaccination since 2012, while the sudden increase after the middle of 2015 indicated rapid spread and diversification of viruses despite the use of vaccine.
Our data indicate the genotype G1 was under stronger positive selective pressure than the new genotype (G2), possibly due to the vaccination. Previous studies demonstrated that vaccination in poultry could accelerate genetic evolution and nucleotide substitution rate of the HA gene (Cattoli et al. 2011; Wang et al. 2012). The general nucleotide substitution rate of HA gene of Bangladesh clade 2.3.2.1a HPAIVs (5.517 × 10−3 substitution/site/year) was faster than those of viruses of nonvaccinated poultry populations (China 1996–4: 3.37 × 10−3, Thailand 2004–8: 2.69 × 10−3, Turkey 2005–8: 4.04 × 10−3, and Nigeria 2006–8: 5.20 × 10−3) and was similar or slower than those of viruses of vaccinated poultry populations (China 2005–0: 7.28 × 10−3, Indonesia 2003–9: 6.13 × 10−3, and Egypt 2006–0: 5.36 × 10−3) (Cattoli et al. 2011; Wang et al. 2012). However, the nucleotide substitution rate of the HA gene of recent viruses was significantly lower than previous viruses. In addition, genetic diversity has increased again following the emergence and expansion of novel G2 genotype in 2015 in Bangladesh. These results indicated that vaccination in commercial chickens in Bangladesh has accelerated the evolution of clade 2.3.2.1a viruses during early stages of outbreak in Bangladesh, but recent viruses are rapidly spreading in Bangladesh with, respectively, low positive selection pressure despite the use of vaccine. Thus, the antigenicity of the vaccine against recent viruses needs to be reevaluated.
Because vaccine-induced evolution has typically impacted mutation of external proteins of influenza virus, such as HA and NA protein (Wang et al. 2012; Youk et al. 2019), other factors may also contribute to the genotype replacement in Bangladesh. The viral competition in duck populations is also suspected as another factor for genotype replacement. During early stage of the outbreak, 2010–2, chickens were a major source of HPAIV, but domestic ducks became the dominant species for HPAIV infection in Bangladesh after 2012. This host change and cocirculating of LPAIVs could have induced replacement of the dominant genotype. For example, previous studies indicated that A/H5N1 HPAIV was replaced by A/H5N6 HPAIV in ducks, and these A/H5N6 viruses showed higher replicability and transmissibility in ducks than previous A/H5N1 viruses in China (Bi et al. 2016; Sun et al. 2016; Kwon et al. 2019). The viruses that have high replicability and transmissibility in ducks may be selected in duck populations, and this selection may have contributed to genotype replacement. However, these biological characteristics of Bangladesh viruses in duck species have not been fully evaluated.
In our phylodynamic analysis, domestic Anseriformes, especially domestic duck, became the dominant host species for clade 2.3.2.1a from 2012 and transmitted the virus to domestic Galliformes during 2013–7. In Bangladesh, the A/H5 vaccination was focused on the commercial layer and breeder chickens, and domestic ducks were not a major target for vaccination and biosecurity for HPAIV control (Rimi et al. 2019). Although HPAIV induce 100 per cent mortality in unvaccinated chickens, the mortality has varied and usually has been low in duck species (Pantin-Jackwood et al. 2017; Kwon et al. 2019). Bangladesh clade 2.3.2.1a viruses also caused low mortality (around 10%) in infected duck farms during the early stage of the outbreak (Nooruzzaman et al. 2019a). Further, nonclinically infected domestic ducks were suspected as a reservoir for clade 2.3.2.1a viruses in Bangladesh. In addition, prevailing LPAIVs in domestic ducks provided internal genes for novel reassortant of HPAIVs. Previous studies indicated that domestic ducks in Bangladesh had various LPAIVs originated from wild waterfowl migrating along the Central Asian flyway and these viruses phylogenetically were related to the recent new genotypes of HPAIVs (Barman et al. 2017, 2019; Hassan et al. 2017; Nooruzzaman et al. 2019b). These results indicated that domestic ducks played a key role in the maintenance and emergence of new genotypes of clade 2.3.2.1a A/H5N1 viruses in Bangladesh.
Since 2002, H5N1 viruses have been isolated from dead birds of several wild terrestrial species, including magpie, tree sparrow, pigeon, and crow. In Bangladesh, the detection of A/H5N1 HPAIV in a crow found dead was initially reported in 2011 (Khan et al. 2014). The dead crow was found within three kilometers of an LBM, which sold chickens, ducks, geese, and pigeons (Khan et al. 2014). In this study, A/H5N1 infected house crows were also found near LBM waste deposit areas. Based on these results, we concluded that the viruses circulating in LBM may have been the source of these two crow outbreaks, highlighting crows as a possible important host in HPAIV ecology in Bangladesh.
Although the method used in this study was less susceptible to sampling bias, biased collection and sequencing could potentially have affected the results (Bahl et al. 2016; Kwon et al. 2019). The A/H5 HPAI vaccination has been focused on chickens in Bangladesh and vaccinated chickens could be infected with HPAIV without detectable clinical signs but shed low titers of viruses (Swayne 2006; Rauw et al. 2012; Swayne et al. 2014; Zeng et al.2016). Therefore, HPAIV infection in vaccinated chicken could be less frequently detected than infection in ducks. In addition, sampling biases could exist over both space and time. Despite our efforts to minimize the effect of sampling biases, we cannot exclude the potential role of unsampled or less sampled populations in the spread and maintenance of HPAIV. However, the emergence of new G2 genotype by reassortment with LPAIVs in ducks and the decrease in genetic diversity after vaccination of chickens support the of our model that the domestic duck played an important role in the maintenance and spread of HPAIV after vaccination in Bangladesh. In addition, previous studies indicated a high geographical correlation between HPAI outbreaks and domestic duck density in South Asia, including Bangladesh (Gilbert et al. 2010; Hill et al. 2015). Continuous and regular surveillance and field epidemiological investigation will be needed for a better understanding of HPAIV ecology in Bangladesh especially the undefined amplification role and maintenance of AIVs from unvaccinated village poultry that move in and out of LBM, interfacing with Anseriformes and vaccinated commercial chickens.
Consistent with the results of this study, the reassortment events of clade 2.3.2.1a A/H5N1 HPAIVs and LPAIVs were reported in previous studies (Monne et al. 2013; Gerloff et al. 2014; Barman et al. 2017, 2019; Nooruzzaman et al. 2019b). However, the phylogenetic relationship between genotypes, the timing of emergence of new genotypes, and the number of reassortment events were not fully determined in previous studies. Although a previous study indicated that new dominant genotype (G2 in this study) was derived from 2013 reassortants (G1.2 in this study) (Nooruzzaman et al. 2019b), our Bayesian phylogenetic analysis showed that G2 genotype emerged by separate reassortment during September–December 2014. In addition, the results of this study demonstrated the replacement of dominant genotype in Bangladesh and the evolutionary relationship between genotypes.
A previous study indicated that HPAIV could replicate and cause clinical illness in vaccinated chicken farms in Bangladesh, indicating current vaccines did not confer complete protection in chickens against clade 2.3.2.1a A/H5N1 HPAIVs in Bangladesh (Ansari et al. 2016; Rimi et al. 2019). In 2017, novel clade 2.3.4.4 A/H5N6 HPAIVs showing different antigenicity were detected in wild waterfowl in Bangladesh (Yang et al. 2019). To minimize the economic loss by future HPAI outbreaks, biosecurity needs to be improved and current vaccine strategy updated, including assessment of licensed vaccine efficacy against recent strains. Our data suggest that domestic Anseriformes, including ducks and goose, most likely played a key role in maintenance, spread, and reassortment of clade 2.3.2.1a HPAIVs in Bangladesh. To prevent and control HPAIV in Bangladesh, the control strategies should be improved and include a new focused on domestic Anseriformes.
Supplementary Material
Acknowledgements
We gratefully acknowledge the authors, originating and submitting laboratories of the sequences from GISAID’s EpiFlu (https://www.gisaid.org/) and IRD (http://www.fludb.org/) database. Data are available in Supplementary Material. The sequence acknowledgment tables for laboratory contributions are shown in Supplementary Table S1. The authors thank the USAID Emerging Pandemic Threats program and the Emergency Centre for Transboundary Animal Diseases (ECTAD) Food and Agricultural Organization of the United Nations (FAO) for financial and technical support to the National Reference Laboratory for Avian Influenza in avian influenza surveillance of poultry. We thank the Institute of Epidemiology, Disease Control and Research (IEDCR) and the EcoHealth Alliance for their support in the crow outbreak investigation. The crow mortality events investigation was supported by USAID’s Emerging Pandemic Threats PREDICT program (Cooperative Agreement No.AID-OAA-A-14-00102) through EcoHealth Alliance. The Department of Livestock Service is thanked for providing clinical samples.
Funding
This research was supported by United States Department of Agriculture, Agricultural Research Service (ARS) project (No. 6040-32000-066-00D) and the United States Department of Agriculture, Animal and Plant Health Inspection Service and Agricultural Research Service Interagency Agreement (No. 60-6040-6-005).
Conflict of interest: None declared.
Contributor Information
Jung-Hoon Kwon, U.S. National Poultry Research Center, Agricultural Research Service, U.S. Department of Agriculture, 934 College Station Road, Athens, GA 30605, USA; College of Veterinary Medicine, Kyungpook National University, 80 Daehakro, Bukgu, Daegu 41566, Republic of Korea.
Dong-Hun Lee, Department of Pathobiology and Veterinary Science, University of Connecticut, 61 N. Eagleville Road, Storrs, CT 06269, USA.
Miria Ferreira Criado, U.S. National Poultry Research Center, Agricultural Research Service, U.S. Department of Agriculture, 934 College Station Road, Athens, GA 30605, USA.
Lindsay Killmaster, U.S. National Poultry Research Center, Agricultural Research Service, U.S. Department of Agriculture, 934 College Station Road, Athens, GA 30605, USA.
Md Zulfekar Ali, Animal Health Research Division, National Reference Laboratory for Avian Influenza, Bangladesh Livestock Research Institute, Savar, Dhaka 1341, Bangladesh.
Mohammad Giasuddin, Animal Health Research Division, National Reference Laboratory for Avian Influenza, Bangladesh Livestock Research Institute, Savar, Dhaka 1341, Bangladesh.
Mohammed A Samad, Animal Health Research Division, National Reference Laboratory for Avian Influenza, Bangladesh Livestock Research Institute, Savar, Dhaka 1341, Bangladesh.
Md. Rezaul Karim, Animal Health Research Division, National Reference Laboratory for Avian Influenza, Bangladesh Livestock Research Institute, Savar, Dhaka 1341, Bangladesh.
Mahmudul Hasan, Animal Health Research Division, National Reference Laboratory for Avian Influenza, Bangladesh Livestock Research Institute, Savar, Dhaka 1341, Bangladesh.
Eric Brum, Emergency Centre for Transboundary Animal Diseases, Food and Agriculture Organization of the United Nations (FAO), Dhaka, Bangladesh.
Tanzinah Nasrin, Emergency Centre for Transboundary Animal Diseases, Food and Agriculture Organization of the United Nations (FAO), Dhaka, Bangladesh.
David E Swayne, U.S. National Poultry Research Center, Agricultural Research Service, U.S. Department of Agriculture, 934 College Station Road, Athens, GA 30605, USA.
References
- Afgan E. et al. (2018) ‘The Galaxy Platform for Accessible, Reproducible and Collaborative Biomedical Analyses: 2018 Update’, Nucleic Acids Research, 46: W537–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari W. K. et al. (2016) ‘Surveillance, Epidemiological, and Virological Detection of Highly Pathogenic H5N1 Avian Influenza Viruses in Duck and Poultry from Bangladesh’, Veterinary Microbiology, 193: 49–59. [DOI] [PubMed] [Google Scholar]
- Bahl J. et al. (2016) ‘Ecosystem Interactions Underlie the Spread of Avian Influenza a Viruses with Pandemic Potential’, PLoS Pathogens, 12: e1005620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barman S. et al. (2017) ‘Role of Domestic Ducks in the Emergence of a New Genotype of Highly Pathogenic H5N1 Avian Influenza a Viruses in Bangladesh’, Emerging Microbes & Infections, 6: e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barman S. et al. (2019) ‘Continuing Evolution of Highly Pathogenic H5N1 Viruses in Bangladeshi Live Poultry Markets’, Emerging Microbes & Infections, 8: 650–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S. M., Bedford T. (2017) ‘Modern-Day SIV Viral Diversity Generated by Extensive Recombination and Cross-Species Transmission’, PLoS Pathogens, 13: e1006466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi Y. et al. (2016) ‘Genesis, Evolution and Prevalence of H5N6 Avian Influenza Viruses in China’, Cell Host & Microbe, 20: 810–21. [DOI] [PubMed] [Google Scholar]
- Biswas P. K. et al. (2017) ‘Biosecurity and Circulation of Influenza A (H5N1) Virus in Live-Bird Markets in Bangladesh, 2012’, Transboundary and Emerging Diseases, 64: 883–91. [DOI] [PubMed] [Google Scholar]
- Cattoli G. et al. (2011) ‘Evidence for Differing Evolutionary Dynamics of a/H5N1 Viruses among Countries Applying or Not Applying Avian Influenza Vaccination in Poultry’, Vaccine, 29: 9368–75. [DOI] [PubMed] [Google Scholar]
- Chakraborty A. et al. (2017) ‘Mild Respiratory Illness among Young Children Caused by Highly Pathogenic Avian Influenza A (H5N1) Virus Infection in Dhaka, Bangladesh, 2011’, The Journal of Infectious Diseases, 216: S520–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrzastek K. et al. (2017) ‘Use of Sequence-Independent, Single-Primer-Amplification (SISPA) for Rapid Detection, Identification, and Characterization of Avian RNA Viruses’, Virology, 509: 159–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerloff N. A. et al. (2014) ‘Multiple Reassortment Events among Highly Pathogenic Avian Influenza A(H5N1) Viruses Detected in Bangladesh’, Virology, 450–1: 297–307. [DOI] [PubMed] [Google Scholar]
- Gerloff N. A. et al. (2016) ‘Genetically Diverse Low Pathogenicity Avian Influenza A Virus Subtypes Co-circulate among Poultry in Bangladesh’, PLoS One, 11: e0152131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M. et al. (2010) ‘Flying over an Infected Landscape: Distribution of Highly Pathogenic Avian Influenza H5N1 Risk in South Asia and Satellite Tracking of Wild Waterfowl’, EcoHealth, 7: 448–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M., Kishino H., Yano T. (1985) ‘Dating of the Human-Ape Splitting by a Molecular Clock of Mitochondrial DNA’, Journal of Molecular Evolution, 22: 160–74. [DOI] [PubMed] [Google Scholar]
- Hassan M. M. et al. (2017) ‘Are Poultry or Wild Birds the Main Reservoirs for Avian Influenza in Bangladesh?’ EcoHealth, 14: 490–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hespanhol L. et al. (2019) ‘Understanding and Interpreting Confidence and Credible Intervals around Effect Estimates’, Brazilian Journal of Physical Therapy, 23: 290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill E. M. et al. (2018) ‘The Impact of Surveillance and Control on Highly Pathogenic Avian Influenza Outbreaks in Poultry in Dhaka Division, Bangladesh’, PLoS Computational Biology, 14: e1006439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill S. C. et al. (2015) ‘Wild Waterfowl Migration and Domestic Duck Density Shape the Epidemiology of Highly Pathogenic H5N8 Influenza in the Republic Of Korea’, Infection, Genetics and Evolution, 34: 267–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M. R. et al. (2012) ‘New Introduction of Clade 2.3.2.1 Avian Influenza Virus (H5N1) into Bangladesh’, Transboundary and Emerging Diseases, 59: 460–3. [DOI] [PubMed] [Google Scholar]
- Katoh K. et al. (2002) ‘MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform’, Nucleic Acids Research, 30: 3059–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. U. et al. (2014) ‘Investigating a Crow Die-Off in January-February 2011 During the Introduction of a New Clade of Highly Pathogenic Avian Influenza Virus H5N1 into Bangladesh’, Archives of Virology, 159: 509–18. [DOI] [PubMed] [Google Scholar]
- Khan S. U. et al. (2018) ‘Avian Influenza Surveillance in Domestic Waterfowl and Environment of Live Bird Markets in Bangladesh, 2007-2012’, Scientific Reports, 8: 9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. et al. (2018) ‘Prevalence of Avian Influenza A(H5) and A(H9) Viruses in Live Bird Markets’, Emerging Infectious Diseases, 24: 2309–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond S. L., Frost S. D. (2005) ‘Not So Different after All: A Comparison of Methods for Detecting Amino Acid Sites under Selection’, Molecular Biology and Evolution, 22: 1208–22. [DOI] [PubMed] [Google Scholar]
- Kwon J. H. et al. (2020) ‘Domestic Ducks Play a Major Role in the Maintenance and Spread of H5N8 Highly Pathogenic Avian Influenza Viruses in South Korea’, Transboundary and Emerging Diseases, 67: 844–51. [DOI] [PubMed] [Google Scholar]
- Kwon J. H. et al. (2019) ‘Different Pathogenicity of Two Strains of Clade 2.3.4.4c H5N6 Highly Pathogenic Avian Influenza Viruses Bearing Different PA and NS Gene in Domestic Ducks’, Virology, 530: 11–8. [DOI] [PubMed] [Google Scholar]
- Lee D. H. et al. (2017) ‘Evolution, Global Spread, and Pathogenicity of Highly Pathogenic Avian Influenza H5Nx Clade 2.3.4.4’, Journal of Veterinary Science, 18: 269–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. H., Criado M. F., Swayne D. E. (2020) ‘Pathobiological Origins and Evolutionary History of Highly Pathogenic Avian Influenza Viruses’, Cold Spring Harbor Perspectives in Medicine. doi: 10.1101/cshperspect.a038679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P. et al. (2009) ‘Bayesian Phylogeography Finds Its Roots’, PLoS Computational Biology, 5: e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinova-Petkova A. et al. (2014) ‘Multiple Introductions of Highly Pathogenic Avian Influenza H5N1 Viruses into Bangladesh’, Emerging Microbes & Infections, 3: e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minin V. N., Bloomquist E. W., Suchard M. A. (2008) ‘Smooth Skyride through a Rough Skyline: Bayesian Coalescent-Based Inference of Population Dynamics’, Molecular Biology and Evolution, 25: 1459–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minin V. N., Suchard M. A. (2008) ‘Fast, Accurate and Simulation-Free Stochastic Mapping’, Philosophical Transactions of the Royal Society B: Biological Sciences, 363: 3985–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monne I. et al. (2013) ‘Reassortant Avian Influenza A(H5N1) Viruses with H9N2-PB1 Gene in Poultry’, Emerging Infectious Diseases, 19: 1630–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nooruzzaman M. et al. (2019. a) ‘Pathology of Clade 2.3.2.1 Avian Influenza Virus (H5N1) Infection in Quails and Ducks in Bangladesh’, Avian Pathology, 48: 73–9. [DOI] [PubMed] [Google Scholar]
- Nooruzzaman M. et al. (2019. b) ‘A New Reassortant Clade 2.3.2.1a H5N1 Highly Pathogenic Avian Influenza Virus Causing Recent Outbreaks in Ducks, Geese, Chickens and Turkeys in Bangladesh’, Transboundary and Emerging Diseases, 66: 2120–33. [DOI] [PubMed] [Google Scholar]
- OIE (World Organisation for Animal Health) (2014) Regional Workshop on Enhancing Influenza A Viruses National Surveillance Systems. Paris: OIE <https://rr-asia.oie.int/uploads/tx_oiefiles/Workshop_Summary_Influenza.pdf> accessed 1 Feb 2020.
- Pantin-Jackwood M. J. et al. (2017) ‘Infectivity, Transmission and Pathogenicity of H5 Highly Pathogenic Avian Influenza Clade 2.3.4.4 (H5N8 and H5N2) United States Index Viruses in Pekin Ducks and Chinese Geese’, Veterinary Research, 48: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvin R. et al. (2018) ‘Review Analysis and Impact of Co-circulating H5N1 and H9N2 Avian Influenza Viruses in Bangladesh’, Epidemiology and Infection, 146: 1259–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poen M. J. et al. (2019) ‘Co-circulation of Genetically Distinct Highly Pathogenic Avian Influenza a Clade 2.3.4.4 (H5N6) Viruses in Wild Waterfowl and Poultry in Europe and East Asia, 2017’, Virus Evolution, 5: vez004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauw F. et al. (2012) ‘Efficacy of rHVT-AI Vector Vaccine in Broilers with Passive Immunity against Challenge with Two Antigenically Divergent Egyptian Clade 2.2.1 HPAI H5N1 Strains’, Avian Diseases, 56: 913–22. [DOI] [PubMed] [Google Scholar]
- Rimi N. A. et al. (2019) ‘A Decade of Avian Influenza in Bangladesh: Where Are We Now?’ Tropical Medicine and Infectious Disease, 4: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmuganatham K. et al. (2014) ‘Genesis of Avian Influenza H9N2 in Bangladesh’, Emerging Microbes & Infections, 3: e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnberg S., Webby R. J., Webster R. G. (2013) ‘Natural History of Highly Pathogenic Avian Influenza H5N1’, Virus Research, 178: 63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spackman E. (2014) ‘Avian Influenza Virus Detection and Quantitation by Real-Time RT-PCR’, Methods in Molecular Biology (Clifton, N.J.), 1161: 105–18. [DOI] [PubMed] [Google Scholar]
- Spackman E., Killian M. L. (2020) ‘Avian Influenza Virus Isolation, Propagation, and Titration in Embryonated Chicken Eggs’, Methods in Molecular Biology (Clifton, N.J.), 2123: 149–64. [DOI] [PubMed] [Google Scholar]
- Spackman E., Suarez D. L. (2008) ‘Detection and Identification of the H5 Hemagglutinin Subtype by Real-Time RT-PCR’, Methods in Molecular Biology (Clifton, N.J.), 436: 27–33. [DOI] [PubMed] [Google Scholar]
- Sun H. et al. (2016) ‘Characterization of Clade 2.3.4.4 Highly Pathogenic H5 Avian Influenza Viruses in Ducks and Chickens’, Veterinary Microbiology, 182: 116–22. [DOI] [PubMed] [Google Scholar]
- Sutton T. C. (2018) ‘The Pandemic Threat of Emerging H5 and H7 Avian Influenza Viruses’, Viruses, 10: 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swayne D. E. (2006) ‘Principles for Vaccine Protection in Chickens and Domestic Waterfowl against Avian Influenza: Emphasis on Asian H5N1 High Pathogenicity Avian Influenza’, Annals of the New York Academy of Sciences, 1081: 174–81. [DOI] [PubMed] [Google Scholar]
- Swayne D. E., Suarez D. L., Sims L. (2020) ‘Influenza’, in Swayne D. E., Boulianne M., Logue C., McDougald L. D., Nair V., Suarez D. L. (eds.) Diseases of Poultry, pp. 210–256. Ames: Wiley. [Google Scholar]
- Swayne D. E., Spackman E., Pantin-Jackwood M. (2014) ‘Success Factors for Avian Influenza Vaccine Use in Poultry and Potential Impact at the Wild Bird-Agricultural Interface’, EcoHealth, 11: 94–108. [DOI] [PubMed] [Google Scholar]
- Wang Z. et al. (2012) ‘Increased Substitution Rate in H5N1 Avian Influenza Viruses During Mass Vaccination of Poultry’, Chinese Science Bulletin, 57: 2419–24. [Google Scholar]
- Webster R. G. et al. (1992) ‘Evolution and Ecology of Influenza a Viruses’, Microbiological Reviews, 56: 152–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO). (2019) Cumulative Number of Confirmed Human Cases for Avian Influenza A(H5N1) Reported to WHO, 2003–2019. Geneva: WHO <https://www.who.int/influenza/human_animal_interface/2019_06_24_tableH5N1.pdf? ua=1> accessed 1 Feb 2020.
- Xu X. et al. (1999) ‘Genetic Characterization of the Pathogenic Influenza a/Goose/Guangdong/1/96 (H5N1) Virus: Similarity of Its Hemagglutinin Gene to Those of H5N1 Viruses from the 1997 Outbreaks in Hong Kong’, Virology, 261: 15–9. [DOI] [PubMed] [Google Scholar]
- Yang G. et al. (2019) ‘Detection of Highly Pathogenic Avian Influenza A(H5N6) Viruses in Waterfowl in Bangladesh’, Virology, 534: 36–44. [DOI] [PubMed] [Google Scholar]
- Yang H. et al. (2016) ‘Molecular Characterizations of Surface Proteins Hemagglutinin and Neuraminidase from Recent H5Nx Avian Influenza Viruses’, Journal of Virology, 90: 5770–84. doi: 10.1128/JVI.00180-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youk S. et al. (2019) ‘Rapid Evolution of Mexican H7N3 Highly Pathogenic Avian Influenza Viruses in Poultry’, PLoS One, 14: e0222457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G. et al. (2018) ‘Two Methods for Mapping and Visualizing Associated Data on Phylogeny Using Ggtree’, Molecular Biology and Evolution, 35: 3041–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X. et al. (2016) ‘Protective Efficacy of the Inactivated H5N1 Influenza Vaccine Re-6 against Different Clades of H5N1 Viruses Isolated in China and the Democratic People’s Republic of Korea’, Avian Disease, 60: 238–40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.