Abstract
With a population of around 4 000 individuals, the Kalash people have been living in the Hindu-Kush mountain valleys of present-day northern Pakistan for centuries. Due to their mysterious origin and fairer European complexion, the genetic history of this ethnic group has been investigated previously using different markers. To date, however, the maternal genetic architecture has not been systematically dissected based on high-resolution complete mitochondrial genomes (mitogenomes), making their maternal genetic history, especially their genetic connection with Europeans from a matrilineal perspective, unclear. To unravel this issue, we analyzed mitogenome data of 34 Kalash samples together with 6 075 individuals from across Eurasia. Our results indicated exclusive western Eurasian origin of the Kalash people, represented by eight haplogroups. Among these haplogroups, J2b1a7a and R0a5a (accounting for ~50% of the Kalash gene pool) displayed in situ differentiations in the Kalash and could be traced to the Mediterranean region. Age estimations suggested these haplogroups arose in the Kalash population ~2.26 and 3.01 thousand years ago (kya), a time frame consistent with the invasion of Alexander III of Macedon to the region. One possible explanation for the maternal genetic contribution from Europeans to the Kalash people would be the involvement of women in foreign campaigns of ancient Greek warfare, followed by a founder effect. Our study thus sheds important light on the genetic origin of the Kalash community of Pakistan.
Keywords: Kalash, Mediterranean, Mitochondrial genome, Founder effect
DEAR EDITOR,
With a population of around 4 000 individuals, the Kalash people have been living in the Hindu-Kush mountain valleys of present-day northern Pakistan for centuries. Due to their mysterious origin and fairer European complexion, the genetic history of this ethnic group has been investigated previously using different markers. To date, however, the maternal genetic architecture has not been systematically dissected based on high-resolution complete mitochondrial genomes (mitogenomes), making their maternal genetic history, especially their genetic connection with Europeans from a matrilineal perspective, unclear. To unravel this issue, we analyzed mitogenome data of 34 Kalash samples together with 6 075 individuals from across Eurasia. Our results indicated exclusive western Eurasian origin of the Kalash people, represented by eight haplogroups. Among these haplogroups, J2b1a7a and R0a5a (accounting for ~50% of the Kalash gene pool) displayed in situ differentiations in the Kalash and could be traced to the Mediterranean region. Age estimations suggested these haplogroups arose in the Kalash population ~2.26 and 3.01 thousand years ago (kya), a time frame consistent with the invasion of Alexander III of Macedon to the region. One possible explanation for the maternal genetic contribution from Europeans to the Kalash people would be the involvement of women in foreign campaigns of ancient Greek warfare, followed by a founder effect. Our study thus sheds important light on the genetic origin of the Kalash community of Pakistan.
The Kalash or Kalasha people are an ancient Indo-European speaking indigenous group with unique culture and traditions, living restrictively in the Hindu-Kush mountain range of present-day northern Pakistan. The enigmatic origin of the Kalash and interestingly their distinct European complexion, e.g., lighter skin tone and blue eyes, in addition to certain customs and beliefs have so far reinforced their claim to be Greek descents following the invasion of Alexander III of Macedon to the region (Cacopardo, 2011). In the past several decades, various genetic studies have been carried out to investigate the genetic structure and history of the Kalash people, in particular their genetic connection with western Eurasians. For example, several studies have indicated that this ethnic group originated from either the Middle East or Europe, followed by a population bottleneck (Qamar et al., 2002; Rosenberg et al., 2002). It is also widely concerned whether the Kalash were genetically isolated for more than 10 kya (Ayub et al., 2015) or received genetic admixture from western Eurasia during 990 and 210 BCE (Hellenthal et al., 2014). Moreover, the possible genetic connection between Greeks and the Kalash remains controversial (Cacopardo, 2011; Firasat et al., 2007; Mansoor et al., 2004; Qamar et al., 2002).
Many previous genetic studies have been based on nuclear genome or Y chromosome data, while the maternal genetic structure of the Kalash had only been dissected based on mitochondrial DNA (mtDNA) restricted fragment length polymorphism (RFLP) and control region variations (Quintana-Murci et al., 2004), thus greatly limiting our understanding of the maternal genetic landscape of this ethnic group. Therefore, whether there is a substantial maternal genetic contribution from Europeans to the Kalash, and when this genetic contact was established, remain unclear.
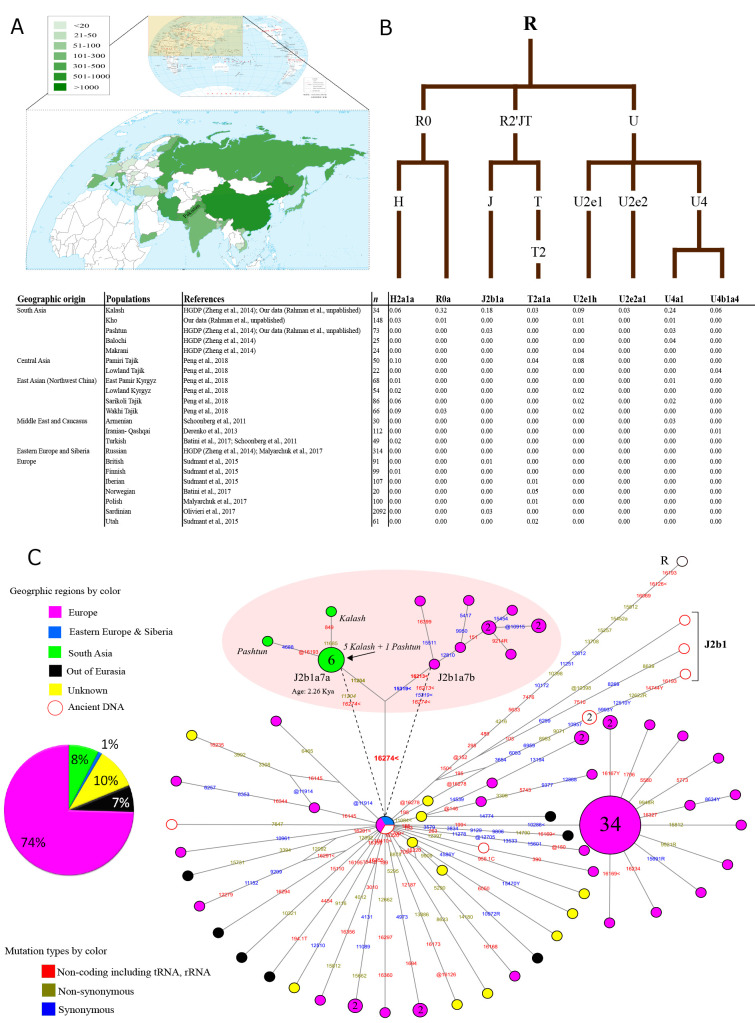
To provide more insight into the genetic history of the Kalash from a matrilineal perspective, we collected and analyzed available complete mitochondrial genome (mitogenome) data of 34 Kalash individuals (25 from the CEPH Human Genome Diversity Project (HGDP) panel (Cann et al., 2002) and nine from this work), as well as 6 075 individuals sampled from Europe and Asia (Figure 1A; Supplementary Table S1). As showed in our results, a total of eight mtDNA haplogroups were identified in the Kalash, including R0a, U4a1, J2b1a, U2e1h, H2a1a, U4b1a4, T2a1a, and U2e2a1, all of which exclusively arise from the Eurasian macro haplogroup R, an observation in agreement with previous study (Quintana-Murci et al., 2004). Comparison of the maternal composition between Kalash and other Eurasian populations (Supplementary Table S1) showed that most of the identified haplogroups in the Kalash were substantially shared with neighboring Dardic group (Kho), as well as being ubiquitous in other western Eurasians (Figure 1B), indicating a western origination of this ethnic group. This is consistent with previous studies that were based on both uniparental markers and whole-genome data (Hellenthal et al., 2014; Qamar et al., 2002; Quintana-Murci et al., 2004). Phylogeographic analysis based on all available complete mitogenomes retrieved from the online platform MitoTool (http://mitotool.kiz.ac.cn/) (Fan & Yao, 2011) as well as from published literature further suggested that most haplogroups identified in Kalash, like R0a, U2e1h, U4a1, H2a1a, T2a1a, and U2e2a1, had sub-branches (e.g., R0a5a, U2e1h1, U4a1f, H2a1a3, etc.) distributed restrictively in northern Pakistan and shared by the Kalash and other Indo-European-speaking populations in the area (Supplementary Figure S1; Supplementary Table S2). Interestingly, the Kalash individuals distributed sporadically in the terminal positions of the sub-branches, strongly suggesting traces of recent gene flow from other groups into the Kalash (Supplementary Figure S1). Moreover, these haplogroups also showed prevalence in the Mediterranean region (e.g., U2e2a1, J2b1a1, and R0a) or in Eurasian Steppe (e.g., H2a1a, T2a1a, U2e1h, U4a1, and U4b1a4), thus possibly reached the Hindu-Kush region in different periods and further introgressed into the Kalash by recent gene flow.
Figure 1. Sample locations, distribution of haplogroups identified in Kalash people, and phylogeographic structure of haplogroup J2b1a.
A: Geographic locations of populations with complete mitogenome sequences in Pakistan and surrounding countries are shown in the inset. Number of mitogenomes available from each region is proportional to color intensity in respective regions defined in figure legends (comprising 6 075 complete mitogenome sequences; see Supplementary Table S1). B: Schematic tree showing eight west Eurasian haplogroups identified in Kalash and their frequency in other local and west Eurasian populations. C: Phylogeographic reconstruction using median-joining network for haplogroup J2b1a from complete mitogenomes (comprising 106 complete mitogenome sequences belonging to haplogroup J2b1a and five ancient mitogenomes belonging to J2b1; see Supplementary Table S3). Each circle represents one individual sample, unless represented by a number in the circle. Dotted line shows case in which J2b1a7a and J2b1a7b emanate from root of J2b1a independently, with position 16274 (in italics) serving as a parallel mutation on both branches. Mutated positions are shown on branches with different colors for each type of mutation, as seen in legend. Specific clade shared between Sardinians and Kalash is enclosed in red circle; Kalash and Pashtun samples are shown in italics on different branches of node. Geographic affiliations of samples are shown in different colors, as defined in legends. Red circles represent ancient mitogenomes included in network construction. R or Y indicate heteroplasmic states. @ represents reverse mutation, < represents parallel mutation on branches.
Different from the above lineages in which the Kalash samples distributed sporadically in different branches, haplogroup J2b1a had a sub-branch (defined by a non-synonymous transition at position 11204 and tentatively named as J2b1a7a) occupied by six Kalash and two Pashtun individuals, a neighboring group previously shown to have had a limited European connection based on Y chromosome study (Firasat et al., 2007). Further phylogeographic analysis showed that the root types of J2b1a7a were predominantly found in Kalash, whereas a Pashtun individual positioned in one terminal branch, indicating an in-situ differentiation of this lineage in the region and further spread into the Pashtuns. Importantly, J2b1a7a shared substitution 16274 with its sister haplogroup (defined by substitutions 15319 and 16213 and tentatively named as J2b1a7b) from Europe (nine Sardinians) (Figure 1C; Supplementary Table S3), indicating a close genetic connection between the Kalash and Europeans. Together with the relatively high proportion of J2b1a7a in the Kalash samples (17.6%), this haplogroup sheds important light on the European ancestry of this ethnic group.
Moreover, considering that the shared position 16274 between the Kalash and Sardinians is hypervariable, it is also probable that the two lineages J2b1a7a and J2b1a7b were derived from the root of J2b1a independently, with 16274 serving as a parallel mutation on both branches. We therefore turned our attention to the ancestral node, J2b1a. Coincidently, the majority (74%) of J2b1a samples, as well as its ancient root type J2b1, were found in Europe, especially in Sardinia (Figure 1C; Supplementary Table S3). This evidence therefore implies an origination of J2b1a in Europe (probably around the Mediterranean region), in agreement with previous study (Pala et al., 2012). Additional support comes from the observation of haplogroup J2b1a in bones of ancient Europeans (Figure 1C, Supplementary Table S3). Further age estimations using mitogenome rate (Soares et al., 2009) revealed that the major haplogroup J2b1a can be traced back to 10.59±1.28 kya, a timeframe within the Neolithization and Bronze Age processes in the Mediterranean region (Marcus et al., 2020), with the Kalash branch (J2b1a7a) 2.26±1.44 kya reflecting a recent split from its European counterpart, followed by independent differentiation in the Hindu-Kush region. Similarly, haplogroup R0a5a, with root types found around the Mediterranean region and a coalescent age of ~3.01±1.5 kya in the Kalash, would also have been introduced into the Kalash gene pool during these recent times. Taken together, about ~50% of the Kalash maternal genetic components were derived from haplogroups J2b1a7a and R0a5a, thus documenting recent genetic introgression (likely from the ancestors of modern Sardinians) to the Kalash, around the time when migration to Sardinia was active from the northern and eastern Mediterranean regions (starting ~1 000 BCE) (Fernandes et al., 2020).
Interestingly, this genetic connection echoes well with the close genetic affinity found between Sardinians and Kalash from studies based on eye-color informative single nucleotide polymorphisms (SNPs) (Walsh et al., 2011), thus probably underlying the similarities in physical features, e.g., lighter complexion of Kalash and Europeans. Moreover, given that the age of J2b1a7a fell within the Macedonian advancement towards northern Pakistan (327 BCE) (Olivieri et al., 2019), and the existence of J2b1c, J2b1a1, and J2b1a3 (sister and sub-type lineages of J2b1a) in ancient and modern Greeks (Lazaridis et al., 2017; Pala et al., 2012), including evidence of eastern Mediterranean immigrants in South Asia (Harney et al., 2019), it is also probable that this genetic connection was mediated by the Greeks. In fact, according to historical records, limited females participated in foreign campaigns of ancient Greek warfare (Loman, 2004), making it likely that the females also took part in this occupation, thus contributing to the Kalash gene pool. This scenario is further supported by evidence of human mobility towards mainland Greece and islands like Sardinia, especially from the Mediterranean, via both sea and land routes during the Mesolithic and even more recent times (Demand, 2012; Fernandes et al., 2020; Marcus et al., 2020). However, the absence of J2b1a in other regions that had been occupied by Alexander’s ancient empire (especially Greece), as well as its prevalence in Sardinian and Kalash people, should not be ignored. One probable explanation would be limited female migration along with Alexander’s siege into other regions, or genetic dilution by later demographic events. Additionally, genetic isolation, followed by bottlenecks in both Sardinians (Di Gaetano et al., 2014) and Kalash (Ayub et al., 2015), further played likely roles in the increase of this lineage in these two regions. Moreover, the limited number of reported mitogenome sequences available from Greece so far could also result in this observation. More studies will be carried out to explain whether this maternal genetic connection between the Kalash and Sardinians was mediated by Greek expansion.
In summary, our analysis observed a genetic ancestry from Europe (probably around the Mediterranean) within the Kalash people from about 3.01±1.5 and 2.26±1.4 kya. This recent genetic contribution from Europe, as revealed in this study, accounts for a significant proportion (~50%) of the Kalash, thus playing an important role in the formation of the maternal gene pool of this ethnic group. Thus, our study sheds important light on the genetic history of the Kalash people of northern Pakistan.
DATA AVAILABILITY
The mitogenome sequences of nine Kalash individuals were obtained from our unpublished dataset (GenBank accession Nos. MN595835-MN595843). Additionally, the 11 mitogenome sequences from northern Pakistan were retrieved from our unpublished work (GenBank accession Nos. MN595685, MN595706, MN595718, MN595749, MN595751, MN595765, MN595769, MN595807, MN595818, MN595820, and MN595890).
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
Q.P.K., Y.C.L., and Z.U.R. designed the research; Z.U.R. collected samples; Z.U.R. and J.Y.T. collected and analyzed the data; Z.U.R., Y.C.L., and Q.P.K. wrote the paper. All authors read and approved the final version of the manuscript.
Funding Statement
This work was supported by the Strategic Priority Research Program (XDA20040102), Second Tibetan Plateau Scientific Expedition and Research (STEP) (2019QZKK0607), National Natural Science Foundation of China (31620103907), Chinese Academy of Sciences (QYZDB-SSW-SMC020), and Yunnan Applied Basic Research Project (2017FB044)
Contributor Information
Yu-Chun Li, Email: liyuchun@mail.kiz.ac.cn.
Qing-Peng Kong, Email: kongqp@mail.kiz.ac.cn.
References
- 1.Ayub Q, Mezzavilla M, Pagani L, Haber M, Mohyuddin A, Khaliq S, et al The Kalash genetic isolate: ancient divergence, drift, and selection. The American Journal of Human Genetics. 2015;96(5):775–783. doi: 10.1016/j.ajhg.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cacopardo AS Are the Kalasha really of Greek origin? The Legend of Alexander the Great and the Pre-Islamic World of the Hindu Kush. Acta Orientalia. 2011;72:47–92. [Google Scholar]
- 3.Cann HM, de Toma C, Cazes L, Legrand MF, Morel V, Piouffre L, et al A human genome diversity cell line panel. Science. 2002;296(5566):261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- 4.Demand NH. 2012. The Mediterranean Context of Early Greek History. New York: John Wiley & Sons.
- 5.Di Gaetano C, Fiorito G, Ortu MF, Rosa F, Guarrera S, Pardini B, et al Sardinians genetic background explained by runs of Homozygosity and genomic regions under positive selection. PLoS One. 2014;9(3):e91237. doi: 10.1371/journal.pone.0091237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan L, Yao YG MitoTool: a web server for the analysis and retrieval of human mitochondrial DNA sequence variations. Mitochondrion. 2011;11(2):351–356. doi: 10.1016/j.mito.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 7.Fernandes DM, Mittnik A, Olalde I, Lazaridis I, Cheronet O, Rohland N, et al The spread of steppe and Iranian-related ancestry in the islands of the western Mediterranean. Nature Ecology & Evolution. 2020;4(3):334–345. doi: 10.1038/s41559-020-1102-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Firasat S, Khaliq S, Mohyuddin A, Papaioannou M, Tyler-Smith C, Underhill PA, et al Y-chromosomal evidence for a limited Greek contribution to the Pathan population of Pakistan. European Journal of Human Genetics. 2007;15(1):121–126. doi: 10.1038/sj.ejhg.5201726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harney É, Nayak A, Patterson N, Joglekar P, Mushrif-Tripathy V, Mallick S, et al Ancient DNA from the skeletons of Roopkund Lake reveals Mediterranean migrants in India. Nature Communication. 2019;10(1):3670. doi: 10.1038/s41467-019-11357-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hellenthal G, Busby GBJ, Band G, Wilson JF, Capelli C, Falush D, et al A genetic atlas of human admixture history. Science. 2014;343(6172):747–751. doi: 10.1126/science.1243518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lazaridis I, Mittnik A, Patterson N, Mallick S, Rohland N, Pfrengle S, et al Genetic origins of the Minoans and Mycenaeans. Nature. 2017;548(7666):214–218. doi: 10.1038/nature23310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loman P No woman no war: women’s participation in ancient Greek warfare. Greece & Rome. 2004;51(1):34–54. [Google Scholar]
- 13.Mansoor A, Mazhar K, Khaliq S, Hameed A, Rehman S, Siddiqi S, et al Investigation of the Greek ancestry of populations from northern Pakistan. Human Genetics. 2004;114(5):484–490. doi: 10.1007/s00439-004-1094-x. [DOI] [PubMed] [Google Scholar]
- 14.Marcus JH, Posth C, Ringbauer H, Lai L, Skeates R, Sidore C, et al Genetic history from the Middle Neolithic to present on the Mediterranean island of Sardinia. Nature Communications. 2020;11(1):939. doi: 10.1038/s41467-020-14523-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olivieri LM, Marzaioli F, Passariello I, Iori E, Micheli R, Terrasi F, et al A new revised chronology and cultural sequence of the Swat valley, Khyber Pakhtunkhwa (Pakistan) in the light of current excavations at Barikot (Bir-kot-ghwandai) Nuclear Instruments and Methods in Physics Research Section B: Beam Interactions with Materials and Atoms. 2019;456:148–156. doi: 10.1016/j.nimb.2019.05.065. [DOI] [Google Scholar]
- 16.Pala M, Olivieri A, Achilli A, Accetturo M, Metspalu E, Reidla M, et al Mitochondrial DNA signals of late glacial recolonization of Europe from near eastern refugia. The American Journal of Human Genetics. 2012;90(5):915–924. doi: 10.1016/j.ajhg.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qamar R, Ayub Q, Mohyuddin A, Helgason A, Mazhar K, Mansoor A, et al Y-chromosomal DNA variation in Pakistan. The American Journal of Human Genetics. 2002;70(5):1107–1124. doi: 10.1086/339929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quintana-Murci L, Chaix R, Wells RS, Behar DM, Sayar H, Scozzari R, et al Where west meets east: the complex mtDNA landscape of the southwest and Central Asian corridor. The American Journal of Human Genetics. 2004;74(5):827–845. doi: 10.1086/383236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, et al Genetic structure of human populations. Science. 2002;298(5602):2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 20.Soares P, Ermini L, Thomson N, Mormina M, Rito T, Röhl A, et al Correcting for purifying selection: an improved human mitochondrial molecular clock. The American Journal of Human Genetics. 2009;84(6):740–759. doi: 10.1016/j.ajhg.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walsh S, Liu F, Ballantyne KN, van Oven M, Lao O, Kayser M IrisPlex: a sensitive DNA tool for accurate prediction of blue and brown eye colour in the absence of ancestry information. Forensic Science International: Genetics. 2011;5(3):170–180. doi: 10.1016/j.fsigen.2010.02.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.
Data Availability Statement
The mitogenome sequences of nine Kalash individuals were obtained from our unpublished dataset (GenBank accession Nos. MN595835-MN595843). Additionally, the 11 mitogenome sequences from northern Pakistan were retrieved from our unpublished work (GenBank accession Nos. MN595685, MN595706, MN595718, MN595749, MN595751, MN595765, MN595769, MN595807, MN595818, MN595820, and MN595890).