Abstract
Understanding the genetic basis of similar phenotypes shared between lineages is a long-lasting research interest. Even though animal evolution offers many examples of parallelism, for many phenotypes little is known about the underlying genes and mutations. We here use a combination of whole-genome sequencing, expression analyses, and comparative genomics to study the parallel genetic origin of ptilopody (Pti) in chicken. Ptilopody (or foot feathering) is a polygenic trait that can be observed in domesticated and wild avian species and is characterized by the partial or complete development of feathers on the ankle and feet. In domesticated birds, ptilopody is easily selected to fixation, though extensive variation in the type and level of feather development is often observed. By means of a genome-wide association analysis, we identified two genomic regions associated with ptilopody. At one of the loci, we identified a 17-kb deletion affecting PITX1 expression, a gene known to encode a transcription regulator of hindlimb identity and development. Similarly to pigeon, at the second loci, we observed ectopic expression of TBX5, a gene involved in forelimb identity and a key determinant of foot feather development. We also observed that the trait evolved only once as foot-feathered birds share the same haplotype upstream TBX5. Our findings indicate that in chicken and pigeon ptilopody is determined by the same set of genes that affect similar molecular pathways. Our study confirms that ptilopody has evolved through parallel evolution in chicken and pigeon.
Keywords: parallel evolution, chicken, foot feathering, PITX1, TBX5, limb development
Introduction
Parallel evolution is the independent development of similar phenotypic traits in separate but related lineages (Davis and Heywood 1963). A defining characteristic of parallel evolution is that the trait is absent in the common ancestor. Animal evolution offers many examples of parallelism. These range from seemingly simple phenotypic changes, such as similar female-limited Batesian mimicry in Papilio butterflies (Iijima et al. 2018), to more complex ones, such as the reduction of the pelvic complex in threespine and ninespine sticklebacks (Shapiro et al. 2006). Even though studies have shown that parallel evolution is not a rare event in animal evolution (Mundy et al. 2004; Shapiro et al. 2006), the genetic bases are still largely unknown for most traits. One hypothesis is that phenotypes that have evolved in parallel and that are extremely similar must have evolved by applying the same genetic mechanism due to developmental constraints. However, at the other extreme, it can be hypothesized that developmental pathways are so complex and can be perturbed in so many ways that essentially an infinite number of combinations of variations could lead to the same outcome.
In vertebrate evolutionary studies, the genomic basis of phenotypic traits that evolved in parallel has not yet been fully understood as appropriate model species are often lacking. Studying the genetic basis of parallelism is further challenged by the fact that the independent evolution of a trait may have occurred many million years ago with very often many genes involved. Domesticated species can provide interesting insights into the genomic architecture of traits that evolved in parallel and the selection that has acted on them (Rubin et al. 2010). Among vertebrates, domestic chicken (Gallus gallus domesticus) is an excellent model, as many of the domesticated phenotypes are common to other domesticated avian species, such as pigeon and duck, and, in some cases, even to more distantly related species, including dog and cattle, presumably due to the desire of humans for particular traits. Examples of similar traits displayed by chicken and other species include the lack of neck feathers (Bartels 2003; Mou et al. 2011), head crest (Wang et al. 2012; Shapiro et al. 2013), feathering rate (Elferink et al. 2008; Derks et al. 2018), and short body stature (Sutter et al. 2007; Boegheim et al. 2017; Wu, Derks, et al. 2018). Many of these phenotypes are determined by a single mutation affecting a single gene that has evolved in parallel in the two species through artificial selection.
In this study, we focus on ptilopody (Pti), a trait observed in domesticated and wild avian species in which the epidermis of the ankle and foot are partially or completely covered with feathers (Boer et al. 2017; Domyan and Shapiro 2017) (fig. 1). The genetic basis of ptilopody has been extensively studied. Previous classical breeding experiments identified a small number of genetic loci (Pti-1, Pti-2, and Pti-3) of large effect in chicken (Somes 1990). However, only recently several candidate genomic regions were identified. Although Dorshorst et al. (2010) identified one quantitative trait locus (QTL) of major effect in Silkie chickens (Dorshorst et al. 2010), Sun et al. (2015) mapped four QTLs, two of which explain more than 20% of the phenotypic variation (Sun et al. 2015). Although recent studies have tried to associate ptilopody to a certain genomic region, it is still unclear which chromosome(s), gene(s), and mutation(s) are directly responsible for the phenotype. A better understanding of the genomic architecture underlying ptilopody comes from a recent study in domestic pigeon, in which two genomic regions containing genes responsible for a partial transformation from hindlimb to forelimb identity were implicated, mainly PITX1 and TBX5 (Domyan et al. 2016). Ptilopody in chicken and pigeon is extremely similar in appearance and this similarity is partly explained by the same genes involved (Domyan et al. 2016). Even though the same genes are involved, the question is whether a similar underlying mutation has enabled the trait to evolve in both lineages. And, even more intriguing, if indeed a similar mutation was to be involved, it becomes relevant to question how the same pathways are altered by the same regulatory mechanisms in both species. This question has never been addressed directly before, neither from a molecular nor an evolutionary perspective.
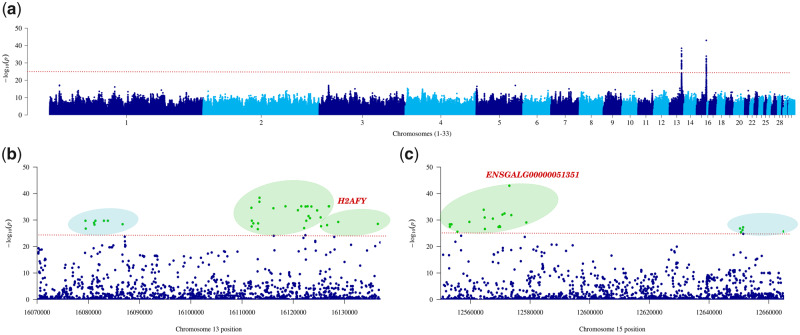
Fig. 2.
Foot feathering is associated with two genomic regions. (a) Genome-wide manhattan plot. The –log10(P) for each variant is shown in the y-axis. Two clear signals can be observed on chromosomes 13 and 15, respectively. (b) Manhattan plot of chromosome 13 (16.0–16.2 Mb). (c) Manhattan plot of chromosome 15 (12.5–12.6 Mb). Significant variants associated with protein-coding genes and lncRNAs are highlighted in green. Intergenic variants are highlighted in light blue. The significant P value threshold (P value <1.0e-25) is identified by the red dotted line.
In this study, we use a combination of whole-genome sequencing (WGS), expression analyses, and comparative genomics to study the genetic basis of foot feathering in chicken. In particular, we identify the underlying causal mutations and affected molecular pathways, while investigating the parallel genetic origin of ptilopody.
Results
WGS of Scaled and Foot-Feathered Chickens
We unraveled the genetic basis and evolutionary history of ptilopody in chicken by WGS of 169 samples from a variety of domesticated chicken breeds and wild species of Gallus (i.e., G. gallus, G. sonneratii, G. lafayetii, and G. varius). On average, 14.8× coverage was generated for each individual after mapping to the chicken reference genome. Mapping quality was, on average, 33.4 with more than 98% of the reads successfully mapped (supplementary Additional file 1, table S3, Supplementary Material online). Since missing data comprised 12% of the total sites, we imputed missing genotypes and phased haplotypes with high accuracy using 21.0 million single-nucleotide polymorphisms (SNPs) and 1.4 million Insertion/Deletions (InDels) (supplementary Additional file 2, table S2, Supplementary Material online). Variants were also assigned to a range of functional classes, though the vast majority were located in introns (58%) and intergenic regions (30%) similarly to Lawal et al. (2018) (supplementary Additional file 2, table S3, Supplementary Material online). Of the 359,176 protein-coding variants, 217,255 were classified as synonymous, 130,147 as missense, and 11,774 as loss-of-function (supplementary Additional file 2, table S3, Supplementary Material online).
To assess population stratification, we performed a principal component analysis (PCA) and Neighbor-Joining (NJ) analysis on the 19 cases and 150 controls. The PCA did not identify any distinct clustering between the two groups (supplementary Additional file 3, fig. S1, Supplementary Material online) aside from clearly separating all traditional breeds from the individuals of the four wild species of Gallus. However, the NJ tree based on the distance relationship matrix separated the 19 case individuals into three groups, one of the Breda fowl, one of the Dutch booted bantam, and one of the remaining foot-feathered samples (supplementary Additional file 3, fig. S2, Supplementary Material online). Despite that, one sample from the Marans (sample 1283), Sundheimer (sample 1769), and German Faverolles (sample 641) breed did not form any specific cluster.
Two Genomic Regions Control Foot Feathering in Chicken
We conducted a genome-wide association study (GWAS) on the 169 samples using a case/control approach through an adaptive Monte Carlo permutation test with 5,000 replications. The GWAS revealed two significant signals on chromosomes 13 and 15, respectively (fig. 2a). On chromosome 13, we identified 36 significant variants (8 intergenic, 9 upstream gene, and 19 intron variants), which are located between 16.0 and 16.1 Mb (supplementary Additional file 2, table S4, Supplementary Material online). This 57-kb region contains a protein-coding H2A histone family gene, H2AFY, a novel long noncoding RNA, ENSGALG00000048757, and one QTL, QTL127125, which was previously found to be associated with foot feathering (Sun et al. 2015) (fig. 2b). The gene H2AFY is located 145 kb upstream of PITX1, a gene that encodes a homeobox-containing transcription factor that is normally expressed in the vertebrate hindlimb but not the forelimb (Logan et al. 1998; Logan and Tabin 1999; Rodriguez-Esteban et al. 1999; Takeuchi et al. 1999). For this 57-kb region, foot-feathered breeds showed elevated levels of homozygosity relative to scaled birds, a clear signature of positive selection as indicated by the low pooled heterozygosity (ZHp = −3.71 vs. ZHp = 0.80 in control individuals) (supplementary Additional file 3, fig. S4b, Supplementary Material online).
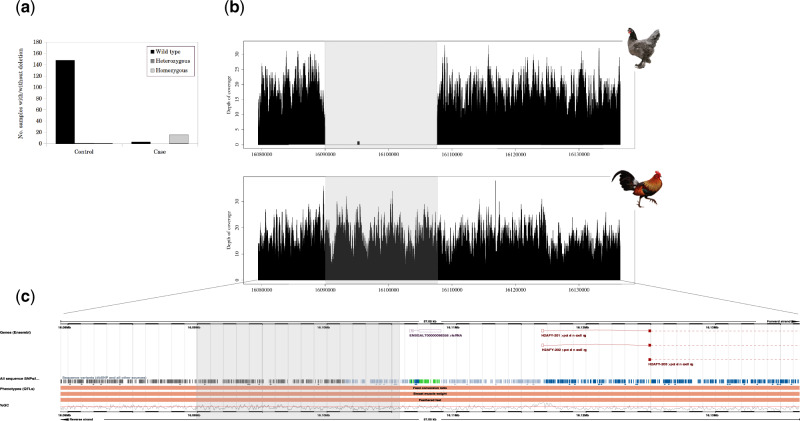
Fig. 3.
Foot-feathered birds share a 17-kb deletion upstream H2AFY. (a) Total number of samples without deletion (wild type), heterozygous or homozygous for the deletion. (b) Genome-wide depth of coverage of chromosome 13 (16.0–16.2 Mb) for a Breda fowl (little feather development) individual and the wild Gallus gallus (scale epidermis). The deletion (13:16,089,992–16,107,660) is visible in the foot-feathered sample by the absence of coverage. (c) Location of deletion (highlighted in gray), lncRNA, H2AFY, and QTL (QTL127125) along the 57-kb significant region on chromosome 13.
On chromosome 15, the 23 significant variants (15 intron noncoding, 3 upstream gene, and 5 intergenic) defined a 112-kb region (12.5–12.6 Mb) (supplementary Additional file 2, table S5, Supplementary Material online), in which we identified a T-box 5 protein-coding gene, TBX5, a novel lncRNA, ENSGALG00000052717, and a previously identified QTL, QTL127126 (Sun et al. 2015) (fig. 2c). Most of the variants were found in the lncRNA. Within this 112-kb region, we also identified one candidate selective sweep (15:12,560,000–12,600,000), which had an average ZHp score of −3.41 (supplementary Additional file 3, fig. S4c, Supplementary Material online). The ZHp score for the same region was above our threshold (ZHp > −3.0) in scaled samples.
Foot-Feathered Birds Share a 17-kb Deletion Upstream H2AFY
We performed a copy-number variation (CNV) analysis to test whether a CNV event is associated with foot feathering. We identified a 17-kb deletion on chromosome 13, 9 kb upstream H2AFY between 16.08 and 16.10 Mb (fig. 3b). The deletion overlapped two 100-kb bins (13:15,964,681–16,217,433) estimated to have an average recombination rate of 4 cM/Mb (supplementary Additional file 3, fig. S7a, Supplementary Material online).
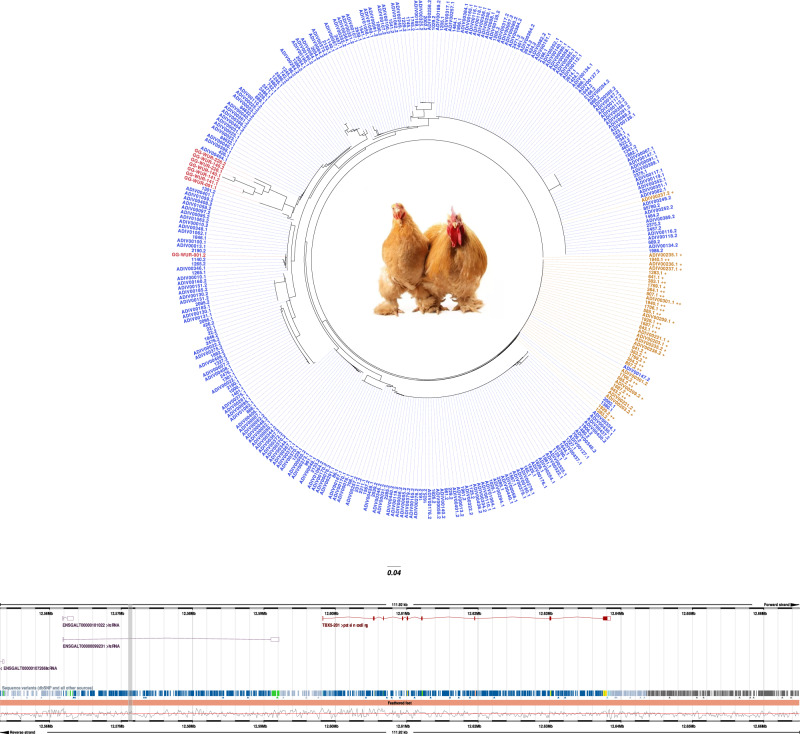
Fig. 4.
Foot feathering has a single haplotype origin. Each individual is identified by two haplotypes, one labeled after the name of the individual with the suffix “.1” and the second with the suffix “.2.” Individuals from the control group (i.e., scaled epidermis) are colored in blue, whereas samples from the case group (i.e., feathered feet) are shown in orange. The individual from the Gallus gallus, G. sonneratii, G. lafayetii, and G. varius is labeled in red. Samples with little feather development are identified by the “+” symbol following the haplotypes name, otherwise by a “++” symbol if heavily feathered. Haplotypes were defined by taking 2 kb upstream and 2 kb downstream the most significant variant (15:12,571,054). The genomic location of the reconstructed haplotype with respect to the lncRNA and TBX5 protein-coding gene is highlighted in gray.
The breakpoints of the deletion are both within the significant peak identified by the GWAS (fig. 3c). In addition to that, the deletion fully overlaps the 44-kb deletion reported in pigeon on scaffold 79 (supplementary Additional file 3, fig. S5, Supplementary Material online). Of the 19 foot-feathered chickens, 16 were homozygous for the deletion, whereas no deletion was observed for the Sundheimer, Marans, and German Faverolles breed (fig. 3a and supplementary Additional file 1, table S4, Supplementary Material online). Of the 150 controls, 148 lacked the deletion, whereas the Phoenix and Toutenkou breed were homozygous and heterozygous, respectively (fig. 3a and supplementary Additional file 1, table S4, Supplementary Material online). Despite that, all four wild species of Gallus did not have the deletion, therefore exhibiting a normal coverage distribution (fig. 3b and supplementary Additional file 1, table S4, Supplementary Material online). The polymerase chain reaction (PCR) validated our CNV analysis, confirming the presence/absence of a deletion in our samples (supplementary Additional file 4, Supplementary Material online). We used the UCSC RepeatMasker track to check for presence of repetitive and transposable elements at the deletion breakpoints in the chicken reference genome and identified four long interspersed nuclear elements (supplementary Additional file 2, table S6, Supplementary Material online). Even though we observed two long interspersed nuclear elements very close to each other in a genome that is not particularly enriched for them, their location (1 kb upstream and 4 kb downstream the deletion breakpoint) suggests that is unlikely that the deletion is caused by transposable elements.
We further identified a 7-bp microhomology at the deletion breakpoint junction (supplementary Additional file 3, fig. S6, Supplementary Material online). The nucleotide sequence of the microhomology flanking the first deletion breakpoint (13:16,089,992) is conserved in many other bird species, including duck, pigeon, collared flycatcher, white-throated sparrow, and medium ground finch (supplementary Additional file 1, table S5, Supplementary Material online). On the contrary, the nucleotide sequence of the microhomology adjacent to the second breakpoint (13:16,107,660) is conserved only between chicken and the two other species belonging to the same Galloanserae subgroup, being turkey and duck (supplementary Additional file 1, table S5, Supplementary Material online).
Foot Feathering Has a Single Haplotype Origin
We also performed a CNV analysis on chromosome 15 but did not observe any CNV potentially associated with the phenotype. We therefore decided to reconstruct haplotypes for each individual by taking 2 kb upstream and 2 kb downstream the intron noncoding variant with the lowest P value (15:12,573,054) (supplementary Additional file 1, table S6, Supplementary Material online). The total 4-kb region (15:12,571,054–12,575,054) included 44 biallelic variants, 3 of which were significantly associated with foot feathering. All 44 biallelic variants are located in the lncRNA ENSGALG00000052717. The 4-kb haplotype also overlapped two 100-kb bins (15:12,388,192–12,633,066) estimated to have an average recombination rate of 7 cM/Mb (supplementary Additional file 3, fig. S7b, Supplementary Material online).
The phylogenetic analysis of the haplotypes clearly separated scaled from feathered samples, indicating an identical origin of haplotypes for the foot-feathered samples (fig. 4). The clear separation between scaled and foot-feathered samples was further confirmed by the fixation index (Fst) analysis performed on the 44 biallelic variants found in the same 4-kb region. We reported the highest Fst value for the noncoding variant used to reconstruct the haplotypes (Fst = 0.97) (supplementary Additional file 2, table S7, Supplementary Material online).
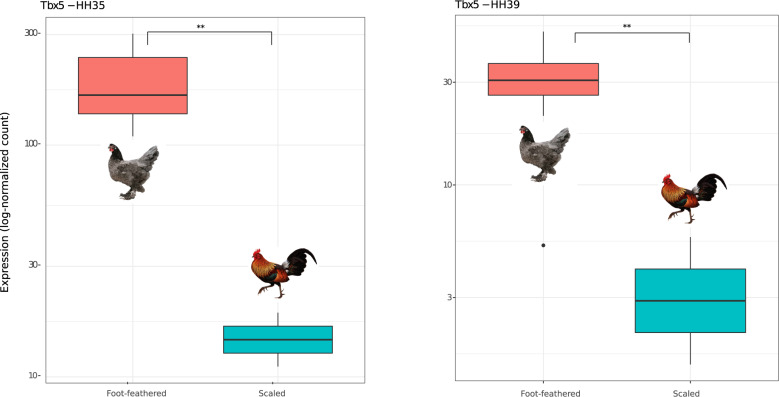
Fig. 5.
TBX5 is upregulated in the hindlimb of foot-feathered embryos. Expression values of TBX5 are shown in the y-axis as log-normalized counts at HH35 (left) and HH39 (right). Differences in expression between foot-feathered and scaled birds were significant based on the adjusted P value at stage HH35 (q-value: 2.49e-14) and HH39 (q-value: 6.87e-03). **q-value <0.05.
Variants Associated with Foot Feathering Are in Highly Conserved Regions
To understand the evolution of foot feathering, we looked for presence of conserved elements (CEs) in the 23 sauropsids multiple sequence alignment, which includes 15 birds, 3 crocodilians, 4 turtles, and anole lizard (supplementary Additional file 3, fig. S8, Supplementary Material online). On chromosome 13, 115 CEs were found within the GWAS peak. Of the 36 significant variants, two (one upstream gene and one intron variant) overlapped a CE (table 1) and both were associated with the H2AFY gene. In the peak region of chromosome 15, we identified 275 CEs, although only the lncRNA intronic variant with the lowest P value was found in a CE of considerable size (table 1).
Table 1.
Variants Associated with Foot Feathering Are Highly Conserved.
| Chr | CE Start | CE End | Size | Strand | Variant | Maj/Min Allele | P Value |
|---|---|---|---|---|---|---|---|
| 13 | 16,112,206 | 16,112,215 | 9 | + | 16,112,207 | C/T | 1.822e-29 |
| 13 | 16,128,762 | 16,128,774 | 12 | + | 16,128,768 | G/A | 5.395e-30 |
| 15 | 12,572,741 | 12,573,218 | 477 | + | 12,573,054 | C/T | 1.141e-43 |
Note.—The allele associated with the phenotype is underlined. Chr, chromosome; CE, conserved element; Maj, major allele; Min, minor allele. The P value is that of the genome-wide association analysis.
TBX5 Is Upregulated in the Hindlimb of Foot-Feathered Birds
We generated high quality RNA-seq data from 21 chicken embryos sacrificed at Hamburger–Hamilton (HH) stages 35 and 39 to test whether our candidate genes and lncRNA are significantly differentially expressed in foot-feathered samples (supplementary Additional file 2, table S8, Supplementary Material online). Overall, more than 90% of the reads were uniquely mapped with an average size of ∼300 bp (supplementary Additional file 2, table S9, Supplementary Material online). As expected, clustering of samples based on read counts followed the embryonic HH stage (supplementary Additional file 3, fig. S10, Supplementary Material online).
PITX1 was significantly downregulated in the hindlimb of foot-feathered birds at HH35 (q-value: 1.79e-03), but not at HH39 (q-value: 0.38) (supplementary Additional file 3, fig. S11, Supplementary Material online). We observed a similar pattern in expression for H2AFY at HH35 (q-value: 0.016) compared with HH39 (q-value: 0.88) (supplementary Additional file 3, fig. S12, Supplementary Material online). On the contrary, TBX5 was always significantly upregulated in foot-feathered birds at both embryonic stages (HH35 q-value: 2.49e-14; HH39 q-value: 6.87e-03) (fig. 5). At HH35, among the first top ten most significant differentially expressed genes, we also identified ZIC1 (q-value: 2.42e-21), a transcription factor acting as scale-feather converter (Wu, Yan, et al. 2018). The FEELnc program classified our candidate lncRNA ENSGALG00000052717 as belonging to the set of mRNAs, since its coding potential was above the cutoff estimated by a 10-fold cross validation procedure that maximizes both sensitivity and specificity (Wucher et al. 2017). We think that the low lncRNA read counts observed in our samples may have affected the FEELnc classification.
Fig. 1.
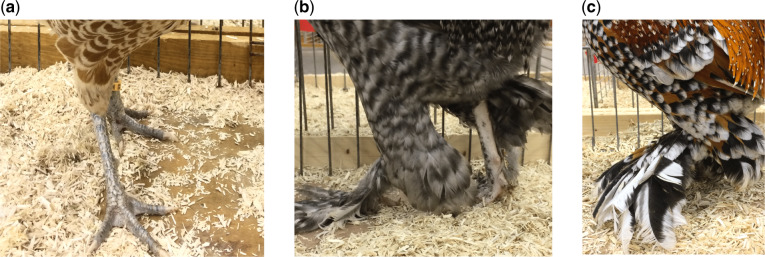
WGS of scaled and foot-feathered chickens. The scale epidermis is the common phenotype in wild and most chickens (a). However, feet of some birds display short and tight feathers on the metatarsus and digits (b), which in some cases appear like long flight-like feathers (c).
Discussion
Foot Feathering Has a Parallel Genetic Origin
Researchers have repeatedly questioned the genetic basis of parallel evolution. Despite this long-lasting interest, for many traits that evolved in parallel little is known whether these are mirrored in underlying genes or mutations. Foot feathering is an interesting case since, although it is a very recognizable trait that can be very easily selected to fixation in breeds, it is in fact not a monogenic trait.
The molecular basis of foot feathering has so far only been studied in detail in domestic pigeon (Domyan et al. 2016; Boer et al. 2019). Chicken and pigeon diverged more than 89 Ma (Jarvis et al. 2014) and are currently classified as belonging to two separate subgroups within the Neognathae clade, the Galloanserae and Neoaves, respectively (Jarvis et al. 2014; Brusatte et al. 2015). Since domestication, both species have experienced selection for a variety of traits that are remarkably different or absent in the wild ancestor (Tixier-Boichard et al. 2011; Domyan and Shapiro 2017). Foot feathering has been under artificial selection since ancient times and nowadays extensive variation can be observed among breeds (i.e., groused feet, slippered legs, muffed legs, and vulture hocks). As for many other traits, ptilopody has been artificially selected to fixation and has become a breed characteristic in both species (Bartels 2003). For parallel evolution to occur, loci associated with similar regulatory pathways and likely to generate variation should be targeted by selection. Our findings and those of Domyan et al. (2016) corroborate this hypothesis, as the independent evolution of foot feathering in chicken and pigeon involves a similar genetic basis and set of genes that not only generate outstanding variation, but this variation is also targeted for recurring artificial selection. In this study, we showed that artificial selection has left clear signatures of positive selection in the genome of foot-feathered birds, as indicated by the negative ZHp scores. These results also illustrate how combining genome-wide association studies and signature of selection analyses forward our understanding of the genomic basis of traits, providing support for the role of, in this case, chromosomes 13 and 15 in foot feather development. Similar signatures of selection were also identified in pigeon by Domyan et al. (2016). However, compared with chicken, pigeons homozygous for the deletion on scaffold 79 showed elevated levels of haplotype homozygosity relative to scaled birds, whereas positive selection was only observed among heavily feathered birds (i.e., muff phenotype) on scaffold 70 (Domyan et al. 2016).
Interestingly, foot feathering is also observed in avian wild species, including snowy owl, golden eagle, and rock ptarmigan (Bartels 2003; Boer et al. 2017). Even though in raptor and boreal species ptilopody has entirely evolved by natural selection, the occurrence of the phenotype suggests that the same underlying genes and mutations can evolve in different species under different types of selection and selection pressure. However, studies on both wild and domesticated avian species are required to further validate this hypothesis.
Foot Feathering Is Associated with a Single, Identical Haplotype in Chicken
As we showed, foot feathering has evolved independently in chicken and pigeon as a result of human-driven selection and this selection pressure has resulted in similar causal mutations. In chicken, foot-feathered birds were also found to share an identical 4-kb haplotype on chromosome 15 independently on whether the individual has the 17-kb deletion on chromosome 13. Sharing of an identical haplotype was also reported in pigeon (i.e., scaffold 70), though a clear clustering was only observed among heavily feathered birds (Domyan et al. 2016). The presence of a single, identical underlying haplotype suggests that foot feathering is caused by mutations that occurred only once in the domestication history of chicken. These causal mutations have then been selected in multiple breeds, in many cases by deliberately crossing foot-feathered birds with scaled birds of a different breed. Because of repeated crossing, the causal mutations underlying ptilopody have been recycled many times since domestication and because the genetic basis is strikingly the same among breeds, the underlying genes can easily be detected by an across-breed GWAS. The mutations found in the 4-kb haplotype, which are clearly related to domesticated populations, are likely to have first appeared in Asia and have later been introgressed into Europe through human migration.
Haplotype length can provide important information on the age of the haplotype. This means that longer haplotypes are of more recent origin (“younger”), as recombination events did not break them down into smaller tracks over time. A negative correlation is, therefore, expected between haplotype length and recombination. The relative small size of the haplotype reported on chromosome 15 supports our conclusions on the single, and likely old, occurrence of the mutations underlying ptilopody on chromosome 15. In fact, it is likely that repeated crossing has not only allowed the spreading of the causal mutation but also contributed to breaking down the original haplotype at each generation, which, by means of artificial selection, is now fixed in all foot-feathered birds considered in this study. The intact haplotype length is also explained by the local recombination rate reported in bins of 100 kb on chromosome 15, if we consider that recombination rate in microchromosomes (50–100 kb/cM) is nearly three times higher than that of macrochromosomes (∼300 kb/cM) (Megens et al. 2009). The limited number of SNPs found in the 4-kb haplotype makes, however, the estimation of the substitution model and mutation rate required to infer the haplotype age a challenging task. A possible solution would be to analyze the same candidate region in ancient samples to better estimate the age of the haplotype.
The long-noncoding variant (15:12,573,054) used for the haplotype analysis is found upstream TBX5, a gene encoding a key transcriptional regulator of forelimb identity and development (Logan et al. 1998; Logan and Tabin 1999; Rodriguez-Esteban et al. 1999; Takeuchi et al. 1999). Interestingly, the same mutation was associated with foot feathering in a parallel study in chicken by Li et al. (2020). In chicken embryos, TBX5 is normally expressed in the forelimb, but its misexpression in the hindlimb at early embryonic stages can induce a partial wing-like transformation, including the formation and development of feathers on the feet (Takeuchi et al. 1999). Similarly to pigeon, the absence of fixed nonsynonymous coding mutations in TBX5 confirms the role of expression changes in the determination of feathered versus scaled feet in chicken as well. We could observe misexpression in the hindlimb of feathered embryos at both HH35 and HH39, meaning that misexpression starts at a very early embryonic stage (in the study of Domyan, similar expression changes were observed at HH25) and is maintained almost up to the end of the embryonic development. Among our most significantly upregulated genes in the hindlimb at HH35, we also found the novel scale-feather converter ZIC1, a transcription factor whose overexpression in feather forming regions (i.e., wings and tail feathers) is sufficient to initiate the invagination step required to form the follicle, but not to form mature follicles (Wu, Yan, et al. 2018).
The 17-kb Deletion on Chromosome 13 Likely Acts as Qualitative Molecular Driver
Even though ectopic expression of TBX5 is associated with foot feathering in chicken, extensive variation in feather type and distribution is often observed. In our study, the 17-kb deletion on chromosome 13 was homozygous in 16 birds, whereas absent in three birds (supplementary Additional file 1, table S4, Supplementary Material online). The absence of the deletion suggests that this locus may affect the qualitative variation in epidermal appendages. This means that the deletion is important for an individual to display variation in the type and extent of feathers, such as enlarged feathers on the feet or wing-like feathers on the feet and toes, but is not essential to determine the localized development of feathers on the feet, which seems the function of TBX5. Therefore, the qualitative role of the 17-kb deletion reasonably explains the discrepancies observed in the Marans, Sundheimer, and German Faverolles breed.
Strikingly, in pigeon a similar deletion, 44 kb in size, in the exact same region as the 17-kb deletion in chicken, is present (Domyan et al. 2016). The 7-bp microhomology we identified at the deletion breakpoints in chicken indicates that this structural variant (SV) has emerged in both species multiple times independently. However, contrary to chicken, based on their QTL and WGS analyses Domyan et al. (2016) concluded that the deletion is sufficient for the development of small feathers (grouse phenotype), whereas the development of large feathers (muff phenotype) is mostly driven by the TBX5 locus.
In chicken, the 17-kb deletion is ∼9 kb upstream H2AFY and ∼200 kb upstream PITX1, a gene encoding a key transcriptional regulators of hindlimb identity and development. PITX1 is normally expressed in the hindlimb, but not in the forelimb, as an abnormal expression in the forelimb blocks feather development (Logan and Tabin 1999). Interestingly, in pigeon the peak on scaffold 79 is also ∼200 kb upstream PITX1. Compared with H2AFY, which was downregulated at HH35 and upregulated at HH39, PITX1 was always downregulated. The key role of PITX1 in limb-type morphology determination has been demonstrated across multiple species (Logan and Tabin 1999; Takeuchi et al. 1999; DeLaurier et al. 2006; Ouimette et al. 2010; Kragesteen et al. 2018) and in all species investigated the relationship between PITX1 and H2AFY is maintained. In pigeon, the 44-kb deletion spans an element orthologous to a known human limb enhancer, hs1473, which shows a strong limb-specific activity (Spielmann et al. 2012; Domyan et al. 2016). As limb patterning and morphogenesis are regulated by highly conserved networks (Boer et al. 2019), it is reasonable to assume that, as in pigeon, also in chicken the deletion encompasses this enhancer, causing loss of PITX1 expression, thus resulting in a partial leg-to-wing homeotic transformation. However, further molecular analysis, including ChiP-seq data, is required to formally confirm this conclusion.
Conclusions
Foot feathering is an interesting example of a polygenic trait that has evolved by parallel evolution as its parallel evolution is mirrored in almost every detail at the molecular and, most likely, developmental level. In this study, we showed that, although chicken and pigeon diverged more than 89 Ma, in both avian species the exact same number of loci containing the exact same set of genes are involved. This similarity is even more striking as a similar deletion at one of the loci has the same outcome in regulating gene expression.
Even though genetic variants arose independently millions of years after the species divergence, it is remarkable to see that not only are the exact same genes involved, but they are affected in very similar ways, despite the many ways in which a similar phenotype conceivably could have arisen. Therefore, even under different types of selection and selection pressure, the same genes and causal mutations underlying major phenotypic changes can evolve in different lineages. Our findings provide support for the hypothesis that only a limited number of evolutionary trajectories at the molecular level are open to generate a specific outcome if developmental pathways are sufficiently constrained.
Materials and Methods
Blood Collection and Animal Experiments
Collections of blood samples was done in accordance with the German Animal Protection Law and was approved by the Committee of Animal Welfare at the Institute of Farm Animal Genetics (Friedrich-Loeffler-Institut) and the Lower Saxony State Office for Consumer Protection and Food Safety (No. 33.9-42502-05-10A064). Sample collection and data recording were also conducted strictly according to the Dutch law on animal protection and welfare (Gezondheids- en welzijnswet voor dieren).
Samples and Phenotype
DNA of 169 samples from 87 traditional chicken breeds and one individual from each of the four living wild species of Gallus (i.e., G. gallus, G. sonneratii, G. lafayetii, and G. varius) was used for WGS on an Illumina HiSeq 3000 (supplementary Additional file 1, table S1, Supplementary Material online). WGS data of the 97 birds sampled in the Netherlands were previously deposited in the European Nucleotide Archive under accession number PRJEB34245 (Bortoluzzi et al. 2020). WGS data of the remaining 68 birds sampled in Germany have been deposited in ENA under accession number PRJEB36674. Detailed information on the sequenced reads can be found in supplementary Additional file 1, table S2, Supplementary Material online. Information on foot feathering was collected during sampling and confirmed by the breeding associations. The phenotype was observed in 11 breeds, for a total of 19 samples. Of these, nine showed short and tight feathers on the metatarsus and digits, whereas ten samples had extensive feather development, as well as long flight-like feathers on the posterior toes (fig. 1; supplementary Additional file 2, table S1, Supplementary Material online). The remaining 150 samples, which have scaled epidermis, were used as control.
Whole-Genome Sequencing
Library preparation and sequencing were performed at the Institut national de la recherche agronomique, France, following their established protocols. Reads were mapped with the Burrows–Wheeler alignment (BWA-MEM) algorithm v0.7.17 (Li and Durbin 2009) to the chicken GRCg6a reference genome (GenBank accession: GCA_000002315.5) with default settings. Duplicate reads were removed with the markdup option in Sambamba v0.6.3 (Tarasov et al. 2015). Sites with mapping quality <30 and base quality <20 were discarded from further analyses (supplementary text, Supplementary Material online).
Phasing, Imputation, and Annotation
Genotypes were imputed and phased into haplotypes with Beagle v4.0 (Browning and Browning 2013) by considering in each of the ten independent cycles 20,000 markers in each sliding window, allowing 1,000 markers to overlap between sliding windows. Imputation accuracy was estimated by masking 10% of the known sites (supplementary text, Supplementary Material online). Imputed variants were annotated to the Ensembl’s G. gallus annotation database using the Ensembl Variant Effect Predictor (release 95) tool (McLaren et al. 2016) (supplementary text, Supplementary Material online).
Population Stratification
Genetic distances were analyzed with a PCA and a NJ tree, both based on a subset of phased variants filtered for a minor allele frequency <0.05. Filtering and PCA were performed in PLINK v1.9 (Purcell et al. 2007). The phylogenetic tree was generated in PHYLIP v3.696 (Felsenstein 2005) from the distance relationship matrix estimated in PLINK.
Association Study and Annotation of Genes
We performed a standard case/control association analysis using the Fisher’s exact test to generate uncorrected and corrected P values, subsequently applying an adaptive Monte Carlo permutation test with 5,000 replications. Variants with a P value <1.0e-25 were considered to be significantly associated with the phenotype. Manhattan plots were generated using the qqman library in R v3.2.0 (R Core Team 2013; Turner 2014). Genes in genomic regions showing significant association with the phenotype were identified using the Ensembl Genes 95 Database in BioMart (Kinsella et al. 2011). Chicken QTLs were downloaded from the Animal QTL Database (Hu et al. 2013). Genomic coordinates were converted to the GRCg6a assembly in LiftOver (Rhead et al. 2009).
Signatures of Selection
Screening for signatures of selection was performed on the control (n = 150) and case (n = 19) group, separately, and only on the autosomes that showed a significant association with the phenotype. For each pool and identified SNP, we determined the number of reads corresponding to the most (nMAJ) and least abundant allele (nMIN). The pooled heterozygosity (Hp) was calculated in sliding 40-kb windows following Rubin et al. (2010):
To resemble a normal distribution, Hp values were normalized into ZHp scores as . Windows with at least 300 SNPs and a ZHp ≤ −3 were retained, as windows below this threshold represent the extreme lower end of the distribution (supplementary Additional file 3, fig. S3, Supplementary Material online).
Recombination Rate
We used the linkage map of Elferink et al. (2010) to estimate the recombination rate, expressed as the genetic length in centimorgans (cM) divided by the physical genomic distance in mega base pairs. Recombination rate was calculated in bins of ∼100 kb after converting the genomic positions of all SNPs to the GRCg6a genome assembly.
Structural Variants
SVs calling and genotyping were performed using Smoove (https://github.com/brentp/smoove, last accessed November 11, 2019). Smoove makes use of various existing tools to call SVs and improves specificity by removing noise from spurious alignment signals. First, discordantly mapped and split reads were extracted from the alignment by Samblaster (Faust and Hall 2014). Next, Lumpy software (Layer et al. 2014) was used to call SVs, and genotyping was performed by SVtyper (Chiang et al. 2015). To further filter SV calls, Mosdepth (Pedersen and Quinlan 2018) was used to discard reads from regions where the sequence depth of split or discordant reads was >1,000 to remove regions that contribute to spurious calls. Duphold (Pedersen and Quinlan 2019) was subsequently used to annotate depth changes within and on the breakpoints of SVs.
PCR-Based Screens for Genomic Rearrangement
A set of four PCR primers was designed to amplify two bands around the deletion breakpoints and two bands over the deletion. More information on the primers and PCR protocol are reported in supplementary Additional file 4, Supplementary Material online. Gel image with the presence (control, scale-footed samples) or absence (case, feather-footed samples) of PCR product is reported in supplementary Additional file 4, Supplementary Material online.
Phylogenetic Analysis of Haplotypes
For each sample, we extracted and considered the two alternative haplotypes as separate haplotypes, so that haplotypes belonging to the same individual did not necessarily cluster together. No missing alleles were present in the phased haplotypes since missing sites were imputed with Beagle. Haplotypes were reconstructed considering only biallelic sites. We then constructed a NJ tree based on the distance matrix estimated in PLINK from all haplotypes.
DNA Sequence Conservation
CEs were predicted from the 23 sauropsids multiple whole-genome alignment generated by Green et al. (2014) (supplementary text, Supplementary Material online). CEs were predicted using PhastCons (Siepel et al. 2005) from a neutral evolutionary model estimated from 114,709 four-fold degenerate (4D) sites in Phylofit (Siepel and Haussler 2004; Green et al. 2014). After filtering for assembly gaps, a total of 1.14 million CEs covering 73 Mb of the chicken genome were retained (supplementary text, Supplementary Material online).
Tissue Collection, RNA Isolation, and RNA Sequencing
Forelimb and hindlimb buds were harvested from 21 chicken embryos sacrificed at HH stage 35 (n = 11) (Hamburger and Hamilton 1951) and HH39 (n = 10) (supplementary Additional file 3, fig. S9, Supplementary Material online). Detailed information on the breeds and samples can be found in supplementary Additional file 2, table S8, Supplementary Material online. Sequencing was performed at BGI, China, following the manufacturer’s protocol.
RNA-seq Analysis
Clean reads were mapped to the chicken GRCg6a reference genome using STAR v2.4.0 (Dobin et al. 2013) with the chicken reference genome and its annotation file as guide, both downloaded from Ensembl (release 95). Quality of mapped RNA-seq data was assessed using Deeptools v3.3.1 (Ramírez et al. 2014). RSEM was used to quantify expression of RNA transcripts and genes (Li and Dewey 2011), whereas StringTie v2.0.3 (Pertea et al. 2015) was used for gene modeling using the Ensembl gene annotation file as reference. Transcripts of all samples were afterward combined using the merge option in StringTie and used as input file in the FlExible Extraction of Long noncoding RNA (FEELnc) program (Wucher et al. 2017) to predict and annotate lncRNAs (SI Text).
We used DESeq2 (Love et al. 2014) to test whether the genes/lncRNAs identified by the genome-wide association analysis were differentially expressed in foot-feathered birds (case) compared with scaled birds (control). The differential expression analysis was performed for each embryonic stage and for the forelimb (F) and hindlimb (H), separately, considering only genes with at least 20 reads. Protein-coding genes and lncRNAs were considered to be significantly differentially expressed only if their adjusted P value was <0.05 (Benjamini–Hochberg adjustment).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We would like to sincerely thank all hobby breeders and breeders associations for their availability and collaboration during sampling. A special thanks to the breeders and people of the Nederlands Pluimveemuseum for their help, support, and enthusiasm during the sampling of eggs that were used for the RNA-seq analyses. We would also like to sincerely thank Zhou Wu for her help in the lab and for providing us the pictures of the chicken embryos used in this study. And last, but not least, we would also like to thank the two anonymous reviewers for their comments and suggestions, which have considerably improved our manuscript. The research leading to some of these results has been conducted as part of the IMAGE project, which received funding from the European Union’s Horizon 2020 Research and Innovation Programme under the Grant Agreement No. 677353.
Author Contributions
C.B., M.B., and H.-J.M. conceived the study. C.B. performed the analysis. M.F.L.D. performed the structural variant analysis. B.D., K.L., and S.W. performed the PCR. S.W. performed the blood collection of the German samples. C.B., B.D., K.L., and R.P.M.A.C. performed the sampling of eggs used for the extraction of limb buds. M.B., H.-J.M., M.A.M.G., and R.P.M.A.C. supervised the project. C.B., M.B., and H.-J.M. wrote the manuscript. All coauthors read and contributed to the manuscript. All coauthors agreed on the final manuscript.
Whole-genome sequencing data of the 97 birds sampled in the Netherlands were previously deposited at the European Nucleotide Archive under accession number PRJEB34245 (Bortoluzzi et al. 2020). Whole-genome sequencing data of the 68 birds sampled in Germany and RNA-seq data of the 21 chicken embryos have been deposited under accession number PRJEB36674.
References
- Bartels T. 2003. Variations in the morphology, distribution, and arrangement of feathers in domesticated birds. J Exp Zool. 298B(1):91–108. [DOI] [PubMed] [Google Scholar]
- Boegheim IJM, Leegwater PAJ, van Lith HA, Back W.. 2017. Current insights into the molecular genetic basis of dwarfism in livestock. Vet J. 224:64–75. [DOI] [PubMed] [Google Scholar]
- Boer EF, Van Hollebeke HF, Park S, Infante CR, Menke DB, Shapiro MD.. 2019. Pigeon foot feathering reveals conserved limb identity networks. Dev Biol. 454(2):128–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer EF, Van Hollebeke HF, Shapiro MD.. 2017. Genomic determinants of epidermal appendage patterning and structure in domestic birds. Dev Biol. 429(2):409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortoluzzi C, Bosse M, Derks MFL, Crooijmans R, Groenen MAM, Megens HJ.. 2020. The type of bottleneck matters: insights into the deleterious variation landscape of small managed populations. Evol Appl. 13(2):330–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR.. 2013. Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics 194(2):459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusatte SL, O’Connor JK, Jarvis ED.. 2015. The origin and diversification of birds. Curr Biol. 25(19):R888–R898. [DOI] [PubMed] [Google Scholar]
- Chiang C, Layer RM, Faust GG, Lindberg MR, Rose DB, Garrison EP, Marth GT, Quinlan AR, Hall IM.. 2015. SpeedSeq: ultra-fast personal genome analysis and interpretation. Nat Methods. 12(10):966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis PH, Heywood VH.. 1963. Principles of angiosperm taxonomy. Edinburgh/London: Oliver & Boyd. [Google Scholar]
- DeLaurier A, Schweitzer R, Logan M.. 2006. Pitx1 determines the morphology of muscle, tendon, and bones of the hindlimb. Dev Biol. 299(1):22–34. [DOI] [PubMed] [Google Scholar]
- Derks MFL, Herrero-Medrano JM, Crooijmans R, Vereijken A, Long JA, Megens HJ, Groenen M.. 2018. Early and late feathering in Turkey and chicken: same gene but different mutations. Genet Sel Evol. 50(1):3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domyan ET, Kronenberg Z, Infante CR, Vickrey AI, Stringham SA, Bruders R, Guernsey MW, Park S, Payne J, Beckstead RB, et al. 2016. Molecular shifts in limb identity underlie development of feathered feet in two domestic avian species. Elife 5:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domyan ET, Shapiro MD.. 2017. Pigeonetics takes flight: evolution, development, and genetics of intraspecific variation. Dev Biol. 427(2):241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorshorst B, Okimoto R, Ashwell C.. 2010. Genomic regions associated with dermal hyperpigmentation, polydactyly and other morphological traits in the silkie chicken. J Hered. 101(3):339–350. [DOI] [PubMed] [Google Scholar]
- Elferink MG, Vallée AAA, Jungerius AP, Crooijmans R, Groenen M.. 2008. Partial duplication of the PRLR and SPEF2 genes at the late feathering locus in chicken. BMC Genomics. 9(1):391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elferink MG, van As P, Veenendaal T, Crooijmans R, Groenen M.. 2010. Regional differences in recombination hotspots between two chicken populations. BMC Genet. 11(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust GG, Hall IM.. 2014. SAMBLASTER: fasta duplicate marking and structural variant read extraction. Bioinformatics 30(17):2503–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 2005. PHYLIP (Phylogeny Inference Package) version 3.6.
- Green RE, Braun EL, Armstrong J, Earl D, Nguyen N, Hickey G, Vandewege MW, St John JA, Capella-Gutiérrez S, Castoe TA, et al. 2014. Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science 346(6215):1254449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL.. 1951. A series of normal stages in the development of the chick embryo. J Morphol. 88(1):49–92. [PubMed] [Google Scholar]
- Hu ZL, Park CA, Wu XL, Reecy JM.. 2013. Animal QTLdb: an improved database tool for livestock animal QTL/association data dissemination in the post-genome era. Nucleic Acids Res. 41:871–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima T, Rei K, Shinya K, Chung-Ping L, Teiji S, Takehiko I, Haruhiko F.. 2018. Parallel evolution of Batesian mimicry supergene in two Papilio butterflies. Sci Adv. 4(4):eaao5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED, Mirarab S, Aberer AJ, Li B, Houde P, Li C, Ho SYW, Faircloth BC, Nabholz B, Howard JT, et al. 2014. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346(6215):1320–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsella RJ, Kähäri A, Haider S, Zamora J, Proctor G, Spudich G, Almeida-King J, Staines D, Derwent P, Kerhornou A, et al. 2011. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database 2011(0):bar030–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragesteen BK, Spielmann M, Paliou C, Heinrich V, Schöpflin R, Esposito A, Annunziatella C, Bianco S, Chiariello AM, Jerković I, et al. 2018. Dynamic 3D chromatin architecture contributes to enhancer specificity and limb morphogenesis. Nat Genet. 50(10):1463–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawal RA, Al-Atiyat RM, Aljumaah RS, Silva P, Mwacharo JM, Hanotte O. 2018. Whole-genome resequencing of red junglefowl and indigenous village chicken reveal new insights on the genome dynamics of the species. Front Genet. 9:264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer RM, Chiang C, Quinlan AR, Hall IM.. 2014. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 15(6):R84–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN.. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12(1):323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lee M, Davis BW, Lamichhaney S, Dorshorst BJ, Siegel PB, Andersson L. Forthcoming 2020. Mutations upstream of the TBX5 and PITX1 transcription factor genes are associated with feathered legs in the domestic chicken. Mol Biol Evol. doi:10.1093/molbev/msaa093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan M, Simon HG, Tabin C.. 1998. Differential regulation of T-box and homeobox transcription factors suggests roles in controlling chick limb-type identity. Development 125(15):2825–2835. [DOI] [PubMed] [Google Scholar]
- Logan M, Tabin CJ.. 1999. Role of Pitx1 upstream of Tbx4 in specification of hindlimb identity. Science 283(5408):1736–1739. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15(12):1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, Flicek P, Cunningham F.. 2016. The Ensembl Variant Effect Predictor. Genome Biol. 17(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megens H-J, Crooijmans RP, Bastiaansen JW, Kerstens HH, Coster A, Jalving R, Vereijken A, Silva P, Muir WM, Cheng HH, et al. 2009. Comparison of linkage disequilibrium and haplotype diversity on macro- and microchromosomes in chicken. BMC Genet. 10(1):86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou C, Pitel F, Gourichon D, Vignoles F, Tzika A, Tato P, Yu L, Burt DW, Bed’hom B, Tixier-Boichard M, et al. 2011. Cryptic patterning of avian skin confers a developmental facility for loss of neck feathering. PLoS Biol. 9(3):e1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy NI, Nichola SB, Tom H, Kim S, Kirstin J, Nicola JN.. 2004. Conserved genetic basis of a quantitative plumage trait involved in mate choice. Science 303(5665):1870–1873. [DOI] [PubMed] [Google Scholar]
- Ouimette J-F, Jolin ML, L’honoré A, Gifuni A, Drouin J.. 2010. Divergent transcriptional activities determine limb identity. Nat Commun. 1(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen BS, Quinlan AR.. 2018. Mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics 34(5):867–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen BS, Quinlan AR.. 2019. Duphold: scalable, depth-based annotation and curation of high-confidence structural variant calls. Gigascience 8(4):1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL.. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 33(3):290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2013. R: A language and environment for statistical computing.
- Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T.. 2014. DeepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42:187–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. 2009. The UCSC genome browser database: update. Nucleic Acids Res. 38(2010):613–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Esteban C, Tsukui T, Yonei S, Magallon J, Tamura K, Belmonte JCI.. 1999. The T-box genes Tbx4 and Tbx5 regulate limb outgrowth and identity. Nature 398(6730):814–818. [DOI] [PubMed] [Google Scholar]
- Rubin C-J, Zody MC, Eriksson J, Meadows JRS, Sherwood E, Webster MT, Jiang L, Ingman M, Sharpe T, Ka S, et al. 2010. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature 464(7288):587–591. [DOI] [PubMed] [Google Scholar]
- Shapiro MD, Bell MA, Kingsley DM.. 2006. Parallel genetic origins of pelvic reduction in vertebrates. Proc Natl Acad Sci U S A. 103(37):13753–13758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro MD, Kronenberg Z, Li C, Domyan ET, Pan H, Campbell M, Tan H, Huff CD, Hu H, Vickrey AI, et al. 2013. Genomic diversity and evolution of the head crest in the rock pigeon. Science 339(6123):1063–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LDW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15(8):1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Haussler D.. 2004. Phylogenetic estimation of context-dependent substitution rates by maximum likelihood. Mol Biol Evol. 21(3):468–488. [DOI] [PubMed] [Google Scholar]
- Somes RG., Jr. 1990. Mutations and major variants of plumage and skin in chickens In: Crawford RD, editor. Poultry breeding and genetics. Amsterdam: Elsevier; p. 169–208. [Google Scholar]
- Spielmann M, Brancati F, Krawitz PM, Robinson PN, Ibrahim DM, Franke M, Hecht J, Lohan S, Dathe K, Nardone AM, et al. 2012. Homeotic arm-to-leg transformation associated with genomic rearrangements at the PITX1 locus. Am J Hum Genet. 91(4):629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Liu R, Zhao G, Zheng M, Sun Y, Yu X, Li P, Wen J.. 2015. Genome-wide linkage analysis identifies loci for physical appearance traits in chickens. G3 (Bethesda) 5:2037–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter NB, Bustamante CD, Chase K, Gray MM, Zhao K, Zhu L, Padhukasahasram B, Karlins E, Davis S, Jones PG, et al. 2007. A single IGF1 allele is a major determinant of small size in dogs. Science 316(5821):112–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi JK, Koshiba-Takeuchi K, Matsumoto K, Vogel-Höpker A, Naitoh-Matsuo M, Ogura K, Takahashi N, Yasuda K, Ogura T.. 1999. Tbx5 and Tbx4 genes determine the wing/leg identity of limb buds. Nature 398(6730):810–814. [DOI] [PubMed] [Google Scholar]
- Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P.. 2015. Sambamba: fast processing of NGS alignment formats. Bioinformatics 31(12):2032–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tixier-Boichard M, Bed’Hom B, Rognon X.. 2011. Chicken domestication: from archeology to genomics. C R Biol. 334(3):197–204. [DOI] [PubMed] [Google Scholar]
- Turner SD. 2014. qqman: an R package for visualizing GWAS results using QQ and Manhattan plots. Biorxiv. 005165. [Google Scholar]
- Wang Y, Gao Y, Imsland F, Gu X, Feng C, Liu R, Song C, Tixier-Boichard M, Gourichon D, Li Q, et al. 2012. The crest phenotype in chicken is associated with ectopic expression of hoxc8 in cranial skin. PLoS One 7(4):e34012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Yan J, Lai YC, Ng CS, Li A, Jiang X, Elsey RM, Widelitz R, Bajpai R, Li WH, et al. 2018. Multiple regulatory modules are required for scale-to-feather conversion. Mol Biol Evol. 35(2):417–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Derks MFL, Dibbits B, Megens HJ, Groenen MAM, Crooijmans R.. 2018. A novel loss-of-function variant in transmembrane protein 263 (TMEM263) of autosomal dwarfism in chicken. Front Genet. 9:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wucher V, Legeai F, Hédan B, Rizk G, Lagoutte L, Leeb T, Jagannathan V, Cadieu E, David A, Lohi H, et al. 2017. FEELnc: a tool for long non-coding RNA annotation and its application to the dog transcriptome. Nucleic Acids Res. 45:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.