An updated review of how plant-associated bacterial diazotrophs and fungal endophytes may improve plant N nutrition, with an emphasis on the underlying mechanisms and their exploitation for reducing the use of chemically synthesized N fertilizers.
Keywords: Agroecology, arbuscular mycorrhizal fungi, bacterial diazotrophs, beneficial microbes, nitrogen fertilization, nitrogen use efficiency, symbiosis
Abstract
Nitrogen (N) is an essential element for plant productivity, thus, it is abundantly applied to the soil in the form of organic or chemical fertilizers that have negative impacts on the environment. Exploiting the potential of beneficial microbes and identifying crop genotypes that can capitalize on symbiotic associations may be possible ways to significantly reduce the use of N fertilizers. The best-known example of symbiotic association that can reduce the use of N fertilizers is the N2-fixing rhizobial bacteria and legumes. Bacterial taxa other than rhizobial species can develop associative symbiotic interactions with plants and also fix N. These include bacteria of the genera Azospirillum, Azotobacter, and Bacillus, some of which are commercialized as bio-inoculants. Arbuscular mycorrhizal fungi are other microorganisms that can develop symbiotic associations with most terrestrial plants, favoring access to nutrients in a larger soil volume through their extraradical mycelium. Using combinations of different beneficial microbial species is a promising strategy to boost plant N acquisition and foster a synergistic beneficial effect between symbiotic microorganisms. Complex biological mechanisms including molecular, metabolic, and physiological processes dictate the establishment and efficiency of such multipartite symbiotic associations. In this review, we present an overview of the current knowledge and future prospects regarding plant N nutrition improvement through the use of beneficial bacteria and fungi associated with plants, individually or in combination.
Introduction
After the mid-20th century, world agriculture benefited from unprecedented changes in agronomic practices known as the “Green Revolution”. Consequently, over a 50-year period, there was a 100% increase in the yield of main crops per capita, particularly in some regions of the world such as Asia and South America (Lassaletta et al., 2016; Pretty, 2018). This increase was mainly due to the technological and scientific advances over this period: new crop varieties were bred, inorganic fertilizers and chemically synthesized pesticides and herbicides were developed and extensively used, and their application was greatly facilitated by the modernization of agricultural machinery (Lassaletta et al., 2016; Pretty, 2018). In particular, the use of synthetic inorganic nitrogen (N) fertilizers has increased several fold during the past 50 years (Lassaletta et al., 2016; Pretty, 2018). N fertilizers chemically produced via the industrial Haber–Bosch process for agricultural purposes use 1–2% of the world’s fossil fuel energy output (Chen et al., 2018a). However, because crops do not take up more than 30–50% of the N available in the soil (Wang et al., 2018), the extensive use of N fertilizers has caused major detriments to microbial, animal, and plant biodiversity and to the corresponding non-agricultural ecosystems (Schlesinger, 2009; Withers et al., 2014).
Cereal grains provide 60% of the food necessary to feed the world’s population, either directly as part of the human diet or indirectly as animal feed (Hirel et al., 2007; Lafiandra et al., 2014; Landberg et al., 2019). Cereals such as maize require large inputs of N fertilizers (nitrates in particular) to achieve optimum performance. Achieving optimal economic return in an environmentally friendly manner requires that these crops use N efficiently. N use efficiency (NUE) is most commonly defined as the grain or biomass yield obtained per unit of available N in the soil (Xu et al., 2012; Han et al., 2015; Li et al., 2017; Hawkesford and Griffiths, 2019). Currently, there are three major targets for enhancing NUE in agriculture: (i) plant breeding (Hirel et al., 2007; Amiour et al., 2012; Han et al., 2015; Zhang et al., 2015; Hawkesford and Griffiths, 2019), (ii) employing alternative and improved agronomic practices (Hirel et al., 2011; Chen et al., 2011; Verzeaux et al., 2017a), and (iii) utilizing the beneficial effects of microbes through the study of plant microbiota (Bulgarelli et al., 2013; Reinhold-Hurek et al., 2015; Reis and Teixeira, 2015; Tkacz and Poole, 2015; Mus et al., 2016; Gouda et al., 2018; Rosenblueth et al., 2018; Cordovez et al., 2019; Tao et al., 2019).
It is now possible to conduct research on the nature and composition of plant microbiota with next-generation sequencing (NGS) techniques. NGS has led to the discovery of the huge diversity of microbe communities present inside (endophytic communities) and on the surface of (epiphytic communities) plant organs, as well as in the rhizosphere (Bulgarelli et al., 2013; Reinhold-Hurek et al., 2015; Cordovez et al., 2019). These microbes play several roles during plant development and are precious allies for crop production and adaptation to various biotic and abiotic stresses (Hardoim et al., 2015; Tkacz and Poole, 2015; Liu et al., 2017). In particular, interactions with rhizospheric and endophytic microorganisms can enhance plant mineral nutrition (Jacoby et al., 2017). For instance, phosphate can be solubilized by organic acids or phytases secreted by soil-borne bacteria or fungi, thus favoring its uptake by plants (Sharma et al., 2013; Alori et al., 2017). Siderophores are low-molecular-weight molecules of microbial origin with a high affinity for iron. They are primarily released to solubilize iron for microbial needs, but consequently make this element more accessible to plants (Lemanceau et al., 2009; Ahmed and Holmström, 2014). Regarding N, microbes play major roles in plant mineral nutrition; these roles are diverse, complex and not fully characterized, particularly during the recycling of inorganic N to mineral or gaseous forms (Coskun et al., 2017). Plants can directly take up the different forms of N, such as nitrate (NO3–), ammonium (NH4+), and amino acids, by means of specific transporters (Krapp et al., 2014; Fan et al., 2017; Tegeder and Masclaux-Daubresse, 2018; Wang et al., 2018; Hirel and Krapp, 2020). Indirect uptake of N can also occur following the establishment of beneficial associations with microbes that facilitate plant N acquisition (Santi et al., 2013; Udvardi and Poole, 2013; Courty et al., 2015; Chen et al., 2018b).
In this review, we provide an overview of the major steps involved in N acquisition by plants, with a particular emphasis on the contribution of beneficial microorganisms that extract N from the soil or fix dinitrogen (N2) from the atmosphere before its transfer to the plant. Possible synergistic interactions between these microorganisms is also discussed, as it may offer an avenue for the improvement of plant productivity through the enhancement of N acquisition (Giovannini et al., 2020).
Nitrogen uptake and assimilation in plants
N is present in the soil in the form of NO3–, NH4+, or amino acids, with the availability of the various forms depending upon physical factors such as pH and temperature. Plants such as rice, which are adapted to acidic pH and anaerobic conditions, preferably take up NH4+ (Tabuchi et al., 2007). In contrast, plants that are adapted to more alkaline pH in aerobic soils, as occurs in most arable lands, use mostly NO3– as their N source (Hirel et al., 2007; Masclaux-Daubresse et al., 2010; O’Brien et al., 2016; Xu, 2018). NO3– is taken up by the roots and then transported in the plant by means of low- and high-affinity transporters that are, for the most part, located in the plasma membrane of cells (Krapp et al., 2014; Léran et al., 2014; O’Brien et al., 2016; Kant, 2018; Wang et al., 2018; Zhang et al., 2018). Members of the nitrate transporter 1/peptide transporter family (NRT1/NPF) act as low-affinity NO3– transporters (Fan et al., 2017; Wang et al., 2018). There are two exceptions in the NPF family that have been identified so far: the Arabidopsis NRTNPF6.3/NRT1.1 and the rice NRT1.1B, which display both low and high affinity for NO3– (Ho et al., 2009; Bouguyon et al., 2015). High-affinity NO3– transport systems involve transporters belonging to the NRT2 family, which belongs to the major facilitator superfamily (MFS) (Krapp et al., 2014; Léran et al., 2014; O’Brien et al., 2016; Kant, 2018; Wang et al., 2018; Zhang et al., 2018). Two other families of transporters are involved in intracellular NO3– transport: the chloride channel (CLC) family and the slow-type anion channels (SLAC) (Zifarelli and Pusch, 2010; Krapp et al., 2014; Hedrich and Geiger, 2017; Wang et al., 2018).
Following uptake, NO3– is reduced to nitrite (NO2–) by the enzyme nitrate reductase (NR) located in the cytosol. Then, NO2– is further reduced to NH4+ by a plastidic nitrite reductase. The resulting NH4+ is used to synthesize glutamine using glutamate as a substrate (Masclaux-Daubresse et al., 2010; Wang et al., 2018). NH4+ can also be taken up by plants directly from the soil. However, its availability mainly depends on the soil characteristics (Nieder et al., 2011). Ammonium can be present in its neutral form (ammonia; NH3) or its cationic form (NH4+) depending on the pH of the soil or growth medium; the latter is predominant under most environmental or growth conditions (Ludewig et al., 2007). When used as the sole N source, NH4+ generally induces physiological and growth perturbations in most plant species (Marino and Moran, 2019). Nevertheless, NH4+ can be directly taken up by means of high-affinity NH4+ transporters (AMT) and non-saturable low-affinity transporters (aquaporin or cation channels) (Tegeder and Masclaux-Daubresse, 2018). NH4+ transport is mediated by proteins belonging to the AMMONIUM TRANSPORTER/METHYLAMMONIUM PERMEASE RHESUS (AMT/MEP/Rh) family, located in the root cell plasma membrane. These proteins allow the transport of NH4+ to the different parts of the plant (Ludewig et al., 2007; Yuan et al., 2007; Tegeder and Masclaux-Daubresse, 2018). NH4+ derived from NO3– reduction or from direct uptake is finally incorporated into organic molecules via the combined activity of two enzymes, glutamine synthase (GS) and glutamate synthase (GOGAT) (Masclaux-Daubresse et al., 2010; Krapp et al., 2014; Wang et al., 2018; Hirel and Krapp, 2020).
The processes associated with the metabolization and recycling of N are referred to as assimilation and remobilization, respectively. N transport, assimilation, and remobilization contribute to plant NUE (Hirel et al., 2007; Lea and Miflin, 2011), of which the two main components are N uptake efficiency (NupE) and N utilization efficiency (NutE) (Han et al., 2015; Li et al., 2017). This implies that both NupE and NutE need to be considered individually or together to improve NUE, which largely depends on the species, the genotype, and the environmental conditions. Intensive research has thus been conducted over the past three decades to find ways to improve NUE, mostly using transgenic plants or mutants altered for the expression of genes involved in N transport and assimilation (Han et al., 2015; Li et al., 2017; Wang et al., 2018). However, these approaches generally have not led to marked and reproducible positive effects on plant growth and development. Moreover, the positive effects that have been achieved have rarely been confirmed, particularly when plants are grown under agronomic conditions (Plett et al., 2017, Hirel and Krapp, 2020; Cañas et al., 2020).
Due to the limited success of such approaches in terms of agronomic applications, as well as the restrictions regarding the use of genetically modified plants in many countries and the growing importance of sustainable agriculture practices, research has been reoriented to evaluate the impact of beneficial microbes on plant N acquisition (Han et al., 2015; Tao et al., 2019). In line with these changes in research strategies, reports suggest that plant NUE can be a key determinant favoring the establishment of beneficial microbes in the rhizosphere of rice (Zhang et al., 2019). Using an elegant genetic and metagenomics approach, Zhang et al. (2019) showed that the rice NRT1.1b nitrate transceptor is associated with the recruitment of bacterial taxa that display enzymatic functions with high N-cycling potential. Thus, the use of beneficial microbes appears to be an attractive alternative to genetic engineering for the implementation of new breeding strategies, although they require several years of development before they can be validated and marketed.
Plant nitrogen nutrition mediated by beneficial microbes
Microbes play essential roles in the conversion of different forms of N. These different forms of N can be used by plants following chemical reactions carried out by living organisms, and contribute to the N cycle that occurs between the soil and the atmosphere. These reactions involve bacteria, archaea, and fungi. First, atmospheric N2 enters the N cycle following its reduction to NH4+, a reaction catalyzed by the enzyme nitrogenase (Nase). Nase is present in N2-fixing symbiotic and associative symbiotic microorganisms called diazotrophs, which are mostly represented by bacteria and archaea (Coskun et al., 2017; Lehnert et al., 2018a). An estimated 50% of land N originates from biological N2 fixation (agricultural and natural) that occurs mostly in the Rhizobium–legume symbiosis and generates inorganic N mainly in the form of NH4+ (Fowler et al., 2013; Lehnert et al., 2018a). Then, NH4+ is incorporated into carbon-containing molecules for the synthesis of amino acids or oxidized by microorganisms such as nitrifying bacteria that produce nitric oxides and nitrates (Coskun et al., 2017; Lehnert et al., 2018a). These oxidized forms of N can be reduced by denitrifying bacteria, archaea, or fungi that convert NO3– to NO2–, then to nitric oxide (NO), nitrous oxide (N2O), and finally back to N2 (Fowler et al., 2013; Coskun et al., 2017). Although most microbes involved in the N cycle do not directly interact with plants, part of the microbial community present in the soil is associated with plant roots. These microbes can have several beneficial effects, in particular on plant growth and for the acquisition of nutrients such as N, a process that is tightly linked to their biological characteristics and their ability to fix N2.
Improved plant nitrogen nutrition with diazotrophs
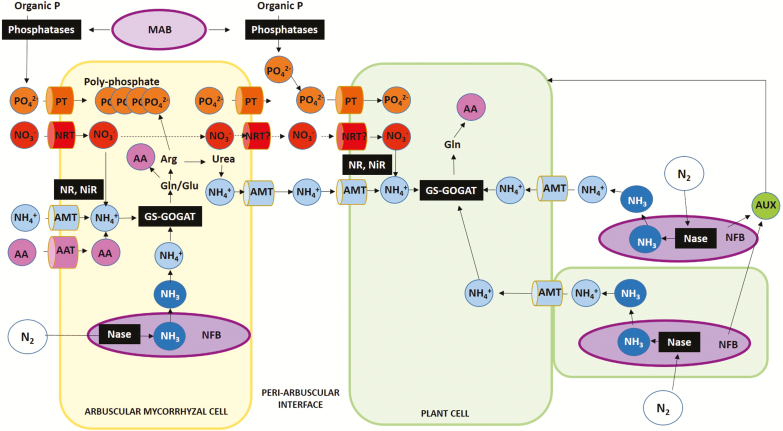
Biological N2 fixation occurs only in prokaryotes. Converting N2 to NH4+ is an energy-consuming process catalyzed by the multimeric enzyme Nase, which is known to be inhibited by oxygen (Fig. 1) (Hoffman et al., 2014). There are two main types of symbiotic associations with N2-fixing bacteria (Table 1). The first involves a symbiotic association between plants and diazotrophic bacteria, leading to highly efficient atmospheric N2 fixation and the transfer of the fixed N to the host plant. The second type leads to much less specific and less efficient endophytic associative symbiotic interactions.
Fig. 1.
Nutritional exchanges between plant, arbuscular mycorrhizal fungi, and bacteria that help improve plant nutrition, including nitrogen (N) acquisition. Plant N acquisition can be improved in the presence of N2-fixing symbiotic and associative symbiotic bacteria and arbuscular mycorrhizal fungi (AMF). Minerals are acquired from the soil by the fungal extraradical mycelium and transported to the arbuscular cells. Minerals are then excreted by the fungal cells at the peri-arbuscular interface and taken up by plant cells. This transport involves specific transporters, which have not yet been fully characterized in either plants or fungi. Following solubilization by phosphatases secreted by mycorrhizae-associated bacteria (MAB), inorganic phosphate is acquired by fungal and plant cells via phosphate transporters (PT). Polyphosphate can be stored in the AMF intra- and extraradical mycelium. Polyphosphate is hydrolyzed and then inorganic phosphate is transported to the peri-arbuscular interface. Fungal cells can also take up N, ammonium (NH4+), amino acids, and small peptides. In the fungal cells, nitrate (NO3–) is reduced to NH4+ by two successive reactions catalyzed by the enzymes nitrate reductase (NR) and nitrite reductase (NiR). NH4+ is then incorporated into organic molecules via the combined action of the enzymes glutamine synthase (GS) and glutamate synthase (GOGAT), leading to the production of glutamine (Gln) and glutamate (Glu), two amino acids that foster fungal cell growth. N is finally stored in the form of arginine (Arg), which is bound to polyphosphate in the vacuolar fungal cells. Arginine is broken down into urea and then into NH4+, which is finally transferred to the peri-arbuscular interface. NH4+ is considered to be the major form of N transferred from the fungus to the plant, although it is likely that NO3– can also be transported. Symbiotic diazotrophs such as rhizobia or Frankia sp. can establish highly efficient N2-fixing symbiosis via nitrogenase (Nase) activity and transfer N to the plant, particularly legumes, in specialized organs called nodules. A number of associative symbiotic N2-fixing bacteria (NFB) interact with root cells or colonize plant roots and shoots. Some of these bacteria can also secrete auxins (AUX) that promote plant growth. Diazotrophic bacteria associated with a symbiotic fungus can also contribute AMF and thus plant N acquisition. AAT, Amino acid transporter; AMT, ammonium transporter; NRT, nitrate transporter.
Table 1.
Categories of interactions with microbes that can improve plant N acquisition and associated biological processes
| Phylum | Family | N-associated biological process | Specificity | Efficiency of plant N nutrition improvement | Intracellular versus Extracellular | Specific cellular structure | Bacterial taxa | Plant taxa |
|---|---|---|---|---|---|---|---|---|
| Bacteria | Rhizobia | N fixation | High | High | Intracellular | Nodule | Rhizobium (alpha proteobacteria) Gram-negative | Fabaceae |
| Rhizobia | N fixation | High | High | Intracellular | Nodule | Rhizobium (alpha proteobacteria) Gram-negative | Parsaponia sp. | |
| Frankia | N fixation | High | High | Intracellular | Nodule | Frankia sp. | Actinorhizal plants (8 taxa) | |
| Cyanobacteria | N fixation | Wide range | High | Intracellular/ extracellular | Heterocyst | Nostoc sp. | Marine, aquatic and terrestrial plants (bryophytes, pteridophytes, gymnosperms, and angiosperms) | |
| Other diazotrophs | N fixation | Wide range | Low/high | Intracellular/ extracellular | no | Azospirillum sp., Herbaspirillum sp., Paenibacillus sp., etc. | Wide range including angiosperms and crops | |
| PGPR (including some diazotrophs) | Growth stimulation | Wide range | Low | Extracellular | no | Wide range including angiosperms and crops | ||
| Fungi | AMF | Growth stimulation/N uptake stimulation | Wide range | Low/high | Intracellular/ extracellular | Arbuscules | Glomeromycota | Wide range including angiosperms and crops |
AMF, Arbuscular mycorrhizal fungi; PGPR, plant-growth-promoting rhizobacteria.
The highly efficient N2-fixing symbiotic associations involve bacteria belonging to the family Rhizobiaceae and Actinorhizae such as Frankia sp. Rhizobia are alpha-proteobacteria that establish an N2-fixing symbiosis mostly with legumes (Fabaceae). This is the best characterized process of endosymbiosis in plants involving N2-fixing bacteria. Upon molecular recognition of bacterial nodulation factors, root epidermal cells begin to differentiate into an organ called a nodule that hosts modified bacterial cells called bacteroids. Bacteroids perform N2 fixation in the root nodules, allowing NH4+ to be transferred to the plant via its vascular system. Biological N2 fixation occurring in the Rhizobium–legume symbiosis has been extensively reviewed (Oldroyd et al., 2011; Udvardi and Poole, 2013) and will not be covered here. Some rhizobial species are able to induce the formation of N2-fixing root nodules in the non-legume plant Parasponia sp. (Behm et al., 2014; van Velzen et al., 2018). In this type of symbiotic association, the bacteroids are not fully differentiated as they are in legumes. In turn, both Nase activity and the number of bacterial cells per nodule are much lower compared with that in legumes (Behm et al., 2014; van Velzen et al., 2018). Filamentous actinobacteria of the genus Frankia establish symbioses with species from eight plant families belonging to the orders Fagales, Rosales, and Cucurbitales. Compared with rhizobia, the specificity of Frankia to their host plant is less strict (Froussart et al., 2016). Interestingly, Frankia cells are able to fix N2 regardless of the symbiotic or non-symbiotic conditions, whereas rhizobia fix N2 only under symbiotic conditions (Santi et al., 2013). Frankia host species are shrubs and trees growing under a large variety of environmental and climatic conditions.
Some N2-fixing cyanobacteria can associate with plants and provide NH4+ to their hosts without forming specialized organs such as nodules. Generally, these symbiotic N2-fixing cyanobacteria belong to the genus Nostoc. They have the capacity to differentiate specialized cells called heterocysts that carry out N2 fixation. These cyanobacteria can associate with a wide range of bryophytes (liverworts and hornworts), pteridophytes (Azolla sp.) gymnosperms (Cycadaceae), and angiosperms (Gunneraceae) (Santi et al., 2013; Mus et al., 2016). Cyanobacteria colonize specific structures in their hosts, which can be glands located on the dorsal lobe of leaves (Azolla) or cavities in stems (Gunnera) (Santi et al., 2013). Most interactions between plants and cyanobacteria are facultative, except for Azolla sp., which has been used as an N biofertilizer for thousands of years in rice paddy fields (Kollah et al., 2016). Many plant species can associate with associative symbiotic free-living N2-fixing bacteria (AS-NFB). AS-NFB are present in a number of taxa, including alpha-proteobacteria, beta-proteobacteria, gamma-proteobacteria, and firmicutes. They include Azoarcus, Azospirillum, Herbaspirillum, Azotobacter, Burkholderia, Klebsiella, Gluconobacter, Acetobacter, and Pseudomonas spp. (Santi et al., 2013; Reinhold-Hurek et al., 2015; Mus et al., 2016). A number of these microorganisms are present in the rhizosphere and remain at the root surface, whereas others can colonize the plant endosphere; they are classified as plant-interacting or endophytic bacteria, respectively. Diazotrophic bacteria are present in the rhizosphere of many wild and cultivated plant species, such as cereals. Due to their ability to reduce N2 to NH4+, a form of N readily utilizable by the host plant, there is growing interest in determining the physiological and molecular mechanisms underlying this plant–bacterial association (Rosenblueth et al., 2018). Therefore, several studies have been undertaken to isolate and characterize these diazotrophic bacteria, particularly when the associated or host plants are grown under low N fertilization conditions. The characterization of these bacteria is primarily based on the detection of nif gene sequences encoding Nase.
Studies have demonstrated that N2 fixation occurs in bacteria harboring nif genes and that there is a subsequent transfer of NH4+ to the plant. Nase activity can be measured using the acetylene reduction assay (ARA) and by means of 15N dilution experiments. The ARA is based on the capacity of Nase to reduce acetylene to ethylene, a molecule that is easily quantified by gas chromatography (Kifle and Laing, 2015; Brusamarello-Santos et al., 2017). 15N dilution experiments can estimate the contribution of diazotrophic 14N2 fixation to total plant N acquisition following 15N labelling (Chalk, 2016). A few studies have shown that AS-NFB can fix N2 whether they are associated with or colonize the plant, and that the resulting inorganic N is transferred to the plant (Sevilla et al., 2001; Hurek et al., 2002; Iniguez et al., 2004; Pankievicz et al., 2015). To increase the efficiency of AS-NFB in terms of their ability to transfer N to plants, genetic manipulations (mutations or introduction of constitutive promoters) can be conducted to deregulate NH4+ production and excretion for direct incorporation by shoots and roots (Ambrosio et al., 2017; Santos et al., 2017). Wheat inoculation with a spontaneous mutant excreting large amounts of NH4+ resulted in an increase in plant yield (Santos et al., 2017).
In addition to providing N to the plant, several AS-NFB have the ability to promote root and shoot growth of wheat, maize, rice, and sorghum through the production of hormones, such as auxins, known to promote organ growth (Dobbelaere et al., 1999; Santi et al., 2013; Vacheron et al., 2013). These bacteria are referred to as plant-growth-promoting rhizobacteria (PGPR) (Cassán and Diaz-Zorita, 2016; Kudoyarova et al., 2019). Because the effect of PGPR can be multifactorial, it has been described by the “Multiple Mechanisms Theory” (Bashan and de-Bashan, 2010).
In a symbiotic Rhizobium–legume association, the plant benefits from the increased availability of reduced N2, while the bacteria utilize carbohydrates provided by the host plant (Udvardi and Poole, 2013; Hoffman et al., 2014). When there is an interaction between a plant and diazotrophic bacteria, the vicinity of the roots is enriched with carbohydrates present in the root exudates. It is thus likely that these carbohydrates are used to provide the energy required for N2 fixation. In line with this hypothesis, reports indicate that, in a tropical maize genotype, aerial roots that produce a mucilage constitute a chemoattractive niche for diazotrophic bacteria (Van Deynze et al., 2018). This mucilage provides substantial amounts of carbohydrates to the bacteria, leading to a remarkably active N2 fixation process that is greatly favored in this sugar-enriched environment (Knee et al., 2001; Van Deynze et al., 2018). Future agronomic developments utilizing the ability of plant genotypes to release carbohydrates in root exudates not only to attract AS-NFB but also to boost beneficial bacterial colonization and N2 fixation thus requires further research (Coskun et al., 2017; Jacoby et al., 2017).
Improved plant nitrogen nutrition in plant–fungus associations
Plant N acquisition can also be enhanced when plants develop symbiotic mycorrhizal associations with fungi of the order Glomeromyta by means of mycelium that colonizes root cells (endomycorrhiza) or is attached extracellularly to them (ectomycorrhiza). This type of symbiotic association makes it possible to capture nutrients in the surrounding rhizosphere. In particular, the fungi provide minerals to the host plant and improve plant tolerance to biotic and abiotic stresses (Wang et al., 2017; Choi et al., 2018; Ferlian et al., 2018). Arbuscular mycorrhizal fungi (AMF) are the most widespread symbiotic association: they colonize 80–90% of terrestrial plants (Gianinazzi et al., 2010). Following spore germination, AMF colonize intra- and inter-cellular spaces and then develop highly branched structures called arbuscules in the cortical cells of the host plant. These structures are surrounded by a peri-arbuscular interface where nutrient exchanges with the host plant cells take place. The arbuscules are connected to an intraradical mycelium that is present inside the root tissues. The intraradical mycelium is also connected to an extraradical mycelium that spreads in the soil, allowing the uptake of minerals such as phosphate and N (Fig. 1) (Chen et al., 2018b; Choi et al., 2018; Ferrol et al., 2019).
Following colonization by AMF, plant transcripts of genes coding for NO3– and NH4+ transporters are up-regulated (Hildebrandt et al., 2002; Courty et al., 2015; Garcia et al., 2016; Koegel et al., 2017). Similarly, fungal NO3– and NH4+ transporters are up-regulated during the plant colonization process (Courty et al., 2015; Koegel et al., 2017). So far, most of the studies aiming to understand the mechanisms involved in plant N nutrition in the presence of AMF have been based on gene transcriptional activation (Guether et al., 2009; Pérez-Tienda et al., 2014; Garcia et al., 2016; Chen et al., 2018b). Interestingly, specific up-regulation of some transporters has been observed in roots colonized by AMF, indicating that under these conditions the transport mechanism is modified compared with that found in non-colonized plants (Garcia et al., 2016). For instance, the AMT3.1 plant NH4+ transporter transcript is specifically up-regulated in cereals such as sorghum, maize, and rice colonized by AMF (Koegel et al., 2017). The same study proposed that AMT3.1 is the major driver of N and phosphorus (P) transfer to the plant colonized by AMF (Koegel et al., 2017). Thus, the whole N uptake machinery available to the plant in the absence of mycorrhizae is not entirely recruited during these interactions.
Part of the N acquired by the fungal cells is used for their own metabolism. Storage of N as arginine (Arg) is indicated by the large amounts of Arg found in the extraradical mycelium and by the up-regulation of the genes and enzymes involved in the biosynthesis of Arg in the extraradical mycelium and its breakdown into urea in the intraradical mycelium (Cruz et al., 2007; Chen et al., 2018b). Arg is strongly bound to polyphosphate in vacuolar fungal cells, which may reflect a connection between P and N metabolisms (Fig. 1) (Woods and Ferré, 2005; Kikuchi et al., 2014). NH4+ is considered to be the major form of N transfer from the fungus to the plant, although there may be NO3– transfer as well because NRT genes from both the plant and the fungus have been found to be up-regulated (for review, see Courty et al., 2015; Garcia et al., 2016; Chen et al., 2018b). Carbohydrates and long-chain fatty acids are excreted by plant cells in the peri-arbuscular interface and then acquired by fungal cells via specific transporters (MacLean et al., 2017, Choi et al., 2018). Further investigations are now needed to determine whether improved plant N acquisition is due to the transfer of N from the AMF or to an increase in N uptake triggered by the fungus, considering at the same time that the fungus and the plant can compete for limited resources (Makarov, 2019). Developing such studies may be a means to further assess whether AMF symbiosis has a direct or an indirect impact on plant N nutrition. In addition, it is necessary to determine whether there is competition for N acquisition between the plant and the fungus, which might imply that, under agronomic conditions, N fertilization needs to be optimized for the plant to benefit from the symbiotic association.
Improvement in plant nitrogen nutrition by combining bacterial and mycorrhizal colonization
A promising strategy to enhance plant nutrition and thus plant performance is through the development of tripartite associations with bacteria and mycorrhizal fungi (Wu et al., 2005, Artursson et al., 2006; Bonfante and Anca, 2009, Giovannini et al., 2020). AMF are in some cases associated with other microbes in the rhizosphere. Such associations can be either beneficial or detrimental to the AMF (Jansa et al., 2013). Although these tripartite associations are not very well characterized yet, they seem to strongly depend on nutritional exchanges between AMF and bacteria. These exchanges involve the production of exudates by the fungi that attract the bacteria, and the ability of the bacteria to facilitate the access to nutrients of the fungi (Kaiser et al., 2015; Zhang et al., 2016). For instance, bacteria can more easily solubilize phosphate than fungi, thereby improving both fungal and plant phosphate nutrition (Zhang et al., 2016). Interestingly, bacteria of the genus Paenibacillus isolated from the roots of Sorghum bicolor are able to stimulate mycorrhizal colonization of the plant by Glomus mosseae (Budi et al., 1999). Moreover, Paenibacillus sp. has been detected inside cells of the ectomycorrhizal fungus Laccaria bicolor (Bertaux et al., 2003). Paenibacillus validus is a species that can promote the growth in vitro and the formation of spores in Rhizophagus irregularis (Hildebrandt et al., 2006). This stimulating effect on spore production may be due to the presence of sugars secreted by the bacterium that enhance fungal growth until spores form. Thus, enhanced fungal growth may be beneficial to the development of a mycorrhizal association with a plant (Hildebrandt et al., 2006). In addition, a number of Paenibacillus species are potential N2 fixers that can solubilize phosphate and iron and secrete phytohormones (Grady et al., 2016). Several plant-associated fungi are colonized by endosymbiotic diazotrophs that can provide N to the fungi (Minerdi et al., 2001; Sharma et al., 2008, Torres-Cortés et al., 2015; Paul et al., 2020).
These various tripartite associations can favor the establishment of more efficient fungal and N2-fixing symbioses, potentially leading to better acquisition of N by the plant (Paul et al., 2020). Thus, further investigations to identify bacteria favoring the establishment of a symbiotic association between plants and AMF are worth conducting to determine whether this tripartite association can boost plant N acquisition, particularly under reduced fertilization conditions (Giovannini et al., 2020). Associating plants with more complex microbial consortia is another strategy that can boost plant performance, because AMF inoculation in combination with a microbial consortium isolated from non-fertilized soils increase N uptake in Brachypodium dystachion (Hestrin et al., 2019).
Concluding remarks
To fully exploit the beneficial effect of soil microbiota to improve plant N acquisition, more research is required to study and thus optimize the associations between plants, bacteria, and fungi in order to use them for the implementation of more sustainable agricultural practices (Hirel et al., 2011; Verzeaux et al., 2017a). First, it will be necessary to perform large-scale surveys and experiments to isolate bacterial and fungal species or strains associated with crop species or crop varieties and determine in which area in the world improved plant performance is observed, not only under laboratory conditions but also in the field (Cassán and Diaz-Zorita, 2016). This will allow the identification of microorganisms (or combinations of microorganisms) and crop genotypes (races collected around the world, lines, hybrids, etc.) that develop efficient interactions, particularly when N fertilization is lowered or limiting.
Furthermore, there clearly is genetic diversity among crop varieties for beneficial plant–microorganism associations, which suggests that, both among different species and within the same species, there are favorable alleles involved in the control of such associations (Sawers et al., 2018; Valente et al., 2020, Chandra et al., 2020). More ambitious studies at the genome level could be undertaken to identify which genes or loci are involved in the genetic control of plant performance in response to inoculation with N2-fixing bacteria and AMF (Lehnert et al., 2018b; Vidotti et al., 2019).
Another important issue will be to assess the persistence and competitiveness of beneficial microbes when there is a positive interaction with plants, particularly when microbial inoculants are used to improve plant performance (Romano et al., 2020). In parallel, fundamental research needs to be conducted to characterize the interactions regarding N acquisition between plants and beneficial microorganisms at the physiological and molecular levels. This type of study can help identify markers that can further help breeders to screen or develop new crop varieties for their responsiveness to microorganisms, either already present in the rhizosphere (van der Heijden et al., 2008) or provided as bio-inoculants. The agronomic practices adapted to or developed for these new varieties must be “microbe friendly” so that they favor and do not reduce soil-borne beneficial microbes (Fig. 2) (Hirel et al., 2011; Verzeaux et al., 2017a). Because fungicides are detrimental to soil microbial populations, the reduction in pesticide use can promote the benefits of soil microbiota ecosystemic services (Jacobsen and Hjelmsø, 2014). No-till farming, which is based on sowing seeds without disturbing the soil, is being adopted by growers because it allows the conservation of mycorrhizal hyphal soil networks (Verzeaux et al., 2017a, b). The use of cover crops, grown between the main crop, mown, and then left in place before sowing the next crop, helps conserve soil organic matter, favoring soil microbe proliferation (Verzeaux et al., 2016). A wise combination of all these strategies will help reduce the use of N fertilizers (Verzeaux et al., 2017a).
Fig. 2.
Strategies to optimize interactions with beneficial microbes to improve plant nitrogen (N) acquisition. Various strategies can be combined to reduce the use of N fertilizers and improve crop N acquisition. They include (i) capitalizing on the beneficial impact of microbes either individually or when they are present in tripartite associations; (ii) utilizing plant genetic deversity to select the most efficient interactions in terms of the acquisition of nutrients, N in particular; and (iii) developing agronomic practices such as no-till and those based on the use of cover crops that favor plant–microbe interactions (Hirel et al., 2011).
Acknowledgements
This research was supported by a grant from the French National Research Agency (no. ANR-16-CE04-0007) and a grant from the Institut Carnot Plant2 Pro, Micro-Organismes Maïs Azote.
References
- Ahmed E, Holmström SJ. 2014. Siderophores in environmental research: roles and applications. Microbial Biotechnology 7, 196–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alori ET, Glick BR, Babalola OO. 2017. Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Frontiers in Microbiology 8, 971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosio R, Ortiz-Marquez JCF, Curatti L. 2017. Metabolic engineering of a diazotrophic bacterium improves ammonium release and biofertilization of plants and microalgae. Metabolic Engineering 40, 59–68. [DOI] [PubMed] [Google Scholar]
- Amiour N, Imbaud S, Clément G, et al. 2012. The use of metabolomics integrated with transcriptomic and proteomic studies for identifying key steps involved in the control of nitrogen metabolism in crops such as maize. Journal of Experimental Botany 63, 5017–5033. [DOI] [PubMed] [Google Scholar]
- Artursson V, Finlay RD, Jansson JK. 2006. Interactions between arbuscular mycorrhizal fungi and bacteria and their potential for stimulating plant growth. Environmental Microbiology 8, 1–10. [DOI] [PubMed] [Google Scholar]
- Bashan Y, de-Bashan LE. 2010. How the plant growth-promoting bacterium Azospirillum promotes plant growth—a critical assessment. In: Sparks DL, ed. Advances in agronomy, vol 108 San Diego: Academic Press, 177–136. [Google Scholar]
- Behm JE, Geurts R, Kiers ET. 2014. Parasponia: a novel system for studying mutualism stability. Trends in Plant Science 19, 757–763. [DOI] [PubMed] [Google Scholar]
- Bertaux J, Schmid M, Prevost-Boure NC, Churin JL, Hartmann A, Garbaye J, Frey-Klett P. 2003. In situ identification of intracellular bacteria related to Paenibacillus spp. in the mycelium of the ectomycorrhizal fungus Laccaria bicolor S238N. Applied and Environmental Microbiology 69, 4243–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfante P, Anca I-A. 2009. Plants, mycorrhizal fungi, and bacteria: a network of interactions. Annual Review of Microbiology 63, 363–383. [DOI] [PubMed] [Google Scholar]
- Bouguyon E, Brun F, Meynard D, et al. 2015. Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nature Plants 1, 15015. [DOI] [PubMed] [Google Scholar]
- Brusamarello-Santos LC, Gilard F, Brulé L, Quilleré I, Gourion B, Ratet P, Maltempi de Souza E, Lea PJ, Hirel B. 2017. Metabolic profiling of two maize (Zea mays L.) inbred lines inoculated with the nitrogen fixing plant-interacting bacteria Herbaspirillum seropedicae and Azospirillum brasilense. PLOS ONE 12, e0174576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budi SW, van Tuinen D, Martinotti G, Gianinazzi S. 1999. Isolation from the Sorghum bicolor mycorrhizosphere of a bacterium compatible with arbuscular mycorrhiza development and antagonistic towards soilborne fungal pathogens. Applied and Environmental Microbiology 65, 5148–5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgarelli D, Schlaeppi K, Spaepen S, van Themaat EVL, Schulze-Lefert P. 2013. Structure and functions of the bacterial microbiota of plants. Annual Review of Plant Biology 64, 807–838. [DOI] [PubMed] [Google Scholar]
- Cañas RA. Yesbergenova-Cuny Z. Belanger L. Rouster J, Brulé L. Gilard F. Quilleré I. Sallaud C. Hirel B. 2020. NADH-GOGAT overexpression does not improve maize (Zea mays L.) performance even when pyramiding with NAD-IDH, GDH and GS. Plants 9,130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassán F, Diaz-Zorita M. 2016. Azospirillum sp. in current agriculture: from the laboratory to the field. Soil Biology and Biochemistry 103, 117–130. [Google Scholar]
- Chalk P. 2016. The strategic role of 15N in quantifying the contribution of endophytic N2 fixation to the N nutrition of non-legumes. Symbiosis 69, 63. [Google Scholar]
- Chandra AK, Kumar A, Bharati A et al. 2020. Microbial-assisted and genomic-assisted breeding: a two way approach for the improvement of nutritional quality traits in agricultural crops. Biotech 10, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Crooks RM, Seefeldt LC, et al. 2018a. Beyond fossil fuel–driven nitrogen transformations. Science 360, eaar6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X-P, Cui Z-L, Vitousek PM, et al. 2011. Integrated soil–crop system management for food security. Proceedings of the National Academy of Sciences, USA 108, 6399–6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Gu M, Wang S, Chen J, Xu G. 2018. Transport properties and regulatory roles of nitrogen in arbuscular mycorrhizal symbiosis. Seminars in Cell & Developmental Biology 74, 80–88. [DOI] [PubMed] [Google Scholar]
- Choi J, Summers W, Paszkowski U. 2018. Mechanisms underlying establishment of arbuscular mycorrhizal symbioses. Annual Review of Phytopathology 56, 135–160. [DOI] [PubMed] [Google Scholar]
- Cordovez V, Dini-Andreote F, Carrión VJ, Raaijmakers JM. 2019. Ecology and evolution of plant microbiomes. Annual Review of Microbiology. 20,11911–11929. [DOI] [PubMed] [Google Scholar]
- Coskun D, Britto DT, Shi W, Kronzucker HJ. 2017. How plant root exudates shape the nitrogen cycle. Trends in Plant Science 22, 661–673. [DOI] [PubMed] [Google Scholar]
- Courty PE, Smith P, Koegel S, Redecker D, Wipf D. 2015. Inorganic nitrogen uptake and transport in beneficial plant root-microbe interactions. Critical Reviews in Plant Sciences 34, 4–16. [Google Scholar]
- Cruz C, Egsgaard H, Trujillo C, Ambus P, Requena N, Martins-Loução MA, Jakobsen I. 2007. Enzymatic evidence for the key role of arginine in nitrogen translocation by arbuscular mycorrhizal fungi. Plant Physiology 144, 782–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deynze A, Zamora P, Delaux PM, et al. 2018. Nitrogen fixation in a landrace of maize is supported by a mucilage-associated diazotrophic microbiota. PLOS Biology 16, e2006352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere S, Croonenborghs A, Thys A, Vande Broek A, Vanderleyden J. 1999. Phytostimulatory effect of Azospirillum brasilense wild type and mutant strains altered in IAA production on wheat. Plant and Soil 212, 153–162. [Google Scholar]
- Fan X, Naz M, Fan X, Xuan W, Miller AJ, Xu G. 2017. Plant nitrate transporters: from gene function to application. Journal of Experimental Botany 68, 2463–2475. [DOI] [PubMed] [Google Scholar]
- Ferlian O, Biere A, Bonfante P, et al. 2018. Growing research networks on Mycorrhizae for mutual benefits. Trends in Plant Science 23, 975–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrol N, Azcón-Aguilar C, Pérez-Tienda J. 2019. Review: Arbuscular mycorrhizas as key players in sustainable plant phosphorus acquisition: an overview on the mechanisms involved. Plant Science 280, 441–447. [DOI] [PubMed] [Google Scholar]
- Fowler D, Coyle M, Skiba U, et al. 2013. The global nitrogen cycle in the twenty-first century. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 368, 20130164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froussart E, Bonneau J, Franche C, Bogusz D. 2016. Recent advances in actinorhizal symbiosis signaling. Plant Molecular Biology 90, 613–622. [DOI] [PubMed] [Google Scholar]
- Garcia K, Doidy J, Zimmermann SD, Wipf D, Courty PE. 2016. Take a trip through the plant and fungal transportome of mycorrhiza. Trends in Plant Science 21, 937–950. [DOI] [PubMed] [Google Scholar]
- Gianinazzi S, Gollotte A, Binet MN, van Tuinen D, Redecker D, Wipf D. 2010. Agroecology: the key role of arbuscular mycorrhizas in ecosystem services. Mycorrhiza 20, 519–530. [DOI] [PubMed] [Google Scholar]
- Giovannini L, Palla M, Agnolucci M, Avio L, Sbrana C, Turrini A, Giovanetti M. 2020. Arbuscular mycorrhizal fungi and associated microbiota as plant biostimulants: research strategies for the selection of the best performing inocula; Agronomy 10, 106. [Google Scholar]
- Gouda S, Kerry RG, Das G, Paramithiotis S, Shin HS, Patra JK. 2018. Revitalization of plant growth promoting rhizobacteria for sustainable development in agriculture. Microbiological Research 206, 131–140. [DOI] [PubMed] [Google Scholar]
- Grady EN, MacDonald J, Liu L, Richman A, Yuan ZC. 2016. Current knowledge and perspectives of Paenibacillus: a review. Microbial cell Factories 15, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guether M, Balestrini R, Hannah M, He J, Udvardi MK, Bonfante P. 2009. Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus. New Phytologist 182, 200–212. [DOI] [PubMed] [Google Scholar]
- Han M, Okamoto M, Beatty PH, Rothstein SJ, Good AG. 2015. The genetics of nitrogen use efficiency in crop plants. Annual Review of Genetics 49, 269–289. [DOI] [PubMed] [Google Scholar]
- Hardoim PR, van Overbeek LS, Berg G, Pirttilä AM, Compant S, Campisano A, Döring M, Sessitsch A. 2015. The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiology and Molecular Biology Reviews 79, 293–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkesford MJ, Griffiths S. 2019. Exploiting genetic variation in nitrogen use efficiency for cereal crop improvement. Current Opinion in Plant Biology 49, 35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrich R, Geiger D. 2017. Biology of SLAC1-type anion channels – from nutrient uptake to stomatal closure. New Phytologist 216, 46–61. [DOI] [PubMed] [Google Scholar]
- Hestrin R, Hammer EC, Mueller CW, Lehmann J. 2019. Synergies between mycorrhizal fungi and soil microbial communities increase plant nitrogen acquisition. Communications Biology 2, 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt U, Ouziad F, Marner FJ, Bothe H. 2006. The bacterium Paenibacillus validus stimulates growth of the arbuscular mycorrhizal fungus Glomus intraradices up to the formation of fertile spores. FEMS Microbiology Letters 254, 258–267. [DOI] [PubMed] [Google Scholar]
- Hildebrandt U, Schmelzer E, Bothe H. 2002. Expression of nitrate transporter genes in tomato colonized by an arbuscular mycorrhizal fungus. Physiologia Plantarum 115, 125–136. [DOI] [PubMed] [Google Scholar]
- Hirel B, Krapp A. 2020. Nitrogen utilization in plants I. Biological and agronomic importance. In: Encyclopedia of biological chemistry. 3rd edition Elsevier, doi: 10.1016/B978-0-12-809633-8.21265-X. [Google Scholar]
- Hirel B, Le Gouis J, Ney B, Gallais A. 2007. The challenge of improving nitrogen use efficiency in crop plants: towards a more central role for genetic variability and quantitative genetics within integrated approaches. Journal of Experimental Botany 58, 2369–2387. [DOI] [PubMed] [Google Scholar]
- Hirel B, Tétu T, Lea PJ, Dubois F. 2011. Improving nitrogen use efficiency in crops for sustainable agriculture. Sustainability 3,1452–1485. [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF. 2009. CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194. [DOI] [PubMed] [Google Scholar]
- Hoffman BM, Lukoyanov D, Yang ZY, Dean DR, Seefeldt LC. 2014. Mechanism of nitrogen fixation by nitrogenase: the next stage. Chemical Reviews 114, 4041–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurek T, Handley LL, Reinhold-Hurek B, Piché Y. 2002. Azoarcus grass endophytes contribute fixed nitrogen to the plant in an unculturable state. Molecular Plant-Microbe Interactions 15, 233–242. [DOI] [PubMed] [Google Scholar]
- Iniguez AL, Dong Y, Triplett EW. 2004. Nitrogen fixation in wheat provided by Klebsiella pneumoniae 342. Molecular Plant-Microbe Interactions 17, 1078–1085. [DOI] [PubMed] [Google Scholar]
- Jacobsen CS, Hjelmsø MH. 2014. Agricultural soils, pesticides and microbial diversity. Current Opinion in Biotechnology 27, 15–20. [DOI] [PubMed] [Google Scholar]
- Jacoby R, Peukert M, Succurro A, Koprivova A, Kopriva S. 2017. The role of soil microorganisms in plant mineral nutrition—current knowledge and future directions. Frontiers in Plant Science 8, 1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansa J, Bukovská P, Gryndler M. 2013. Mycorrhizal hyphae as ecological niche for highly specialized hypersymbionts – or just soil free-riders? Frontiers in Plant Science 4, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C, Kilburn MR, Clode PL, Fuchslueger L, Koranda M, Cliff JB, Solaiman ZM, Murphy DV. 2015. Exploring the transfer of recent plant photosynthates to soil microbes: mycorrhizal pathway vs direct root exudation. New Phytologist 205, 1537–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S. 2018. Understanding nitrate uptake, signaling and remobilisation for improving plant nitrogen use efficiency. Seminars in Cell & Developmental Biology 74, 89–96. [DOI] [PubMed] [Google Scholar]
- Kifle MH, Laing MD. 2015. Isolation and screening of bacteria for their diazotrophic potential and their influence on growth promotion of maize seedlings in greenhouses. Frontiers in Plant Science 6, 1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y, Hijikata N, Yokoyama K, Ohtomo R, Handa Y, Kawaguchi M, Saito K, Ezawa T. 2014. Polyphosphate accumulation is driven by transcriptome alterations that lead to near-synchronous and near-equivalent uptake of inorganic cations in an arbuscular mycorrhizal fungus. New Phytologist 204, 638–649. [DOI] [PubMed] [Google Scholar]
- Knee EM, Gong FC, Gao M, Teplitski M, Jones AR, Foxworthy A, Mort AJ, Bauer WD. 2001. Root mucilage from pea and its utilization by rhizosphere bacteria as a sole carbon source. Molecular Plant-Microbe Interactions 14, 775–784. [DOI] [PubMed] [Google Scholar]
- Koegel S, Mieulet D, Baday S, et al. 2017. Phylogenetic, structural, and functional characterization of AMT3;1, an ammonium transporter induced by mycorrhization among model grasses. Mycorrhiza 27, 695–708. [DOI] [PubMed] [Google Scholar]
- Kollah B, Patra AK, Mohanty SR. 2016. Aquatic microphylla Azolla: a perspective paradigm for sustainable agriculture, environment and global climate change. Environmental Science and Pollution Research International 23, 4358–4369. [DOI] [PubMed] [Google Scholar]
- Krapp A, David LC, Chardin C, Girin T, Marmagne A, Leprince AS, Chaillou S, Ferrario-Méry S, Meyer C, Daniel-Vedele F. 2014. Nitrate transport and signalling in Arabidopsis. Journal of Experimental Botany 65, 789–798. [DOI] [PubMed] [Google Scholar]
- Kudoyarova G, Arkhipova T, Korshunova T, Bakaeva M, Loginov O, Dodd IC. 2019. Phytohormone mediation of interactions between plants and non-symbiotic growth promoting bacteria under edaphic stresses. Frontiers in Plant Science 10, 1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafiandra D, Riccardi G, Shewry PR. 2014. Improving cereal grain carbohydrates for diet and health. Journal of Cereal Science 59, 312–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landberg R, Hanhineva K, Tuohy K, Garcia-Aloy M, Biskup I, Llorach R, Yin X, Brennan L, Kolehmainen M. 2019. Biomarkers of cereal food intake. Genes & Nutrition 14, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassaletta L, Billen G, Garnier J, Bouwman L, Velazquez E, Mueller ND, Gerber JS. 2016. Nitrogen use in the global food system: past trends and future trajectories of agronomic performance, pollution, trade, and dietary demand. Environmental Research Letters. 11, 095007. [Google Scholar]
- Lea PJ, Miflin BJ. 2011. Nitrogen assimilation and its relevance to crop improvement. In: Foyer CH, Zhang H,ed. Nitrogen metabolism in plants in the post‐genomic era. Annual plant reviews, vol 42 Oxford: Wiley-Blackwell, 1–40. [Google Scholar]
- Lehnert N, Dong HT, Harland JB, Hunt AP, White CJ. 2018a. Reversing nitrogen fixation. Nature Reviews Chemistry 2, 278–289. [Google Scholar]
- Lehnert H, Serfling A, Friedt W, Ordon F. 2018. Genome-wide association studies reveal genomic regions associated with the response of wheat (Triticum aestivum L.) to mycorrhizae under drought stress conditions. Frontiers in Plant Science 9, 1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemanceau P, Expert D, Gaymard F, Bakker PAHM, Briat J-F. 2009. Role of iron in plant–microbe interactions. In: Van Loon LC, ed. Advances in botanical research, vol 51 San Diego: Academic Press, 491–549. [Google Scholar]
- Léran S, Varala K, Boyer JC, et al. 2014. A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends in Plant Science 19, 5–9. [DOI] [PubMed] [Google Scholar]
- Li H, Hu B, Chu C. 2017. Nitrogen use efficiency in crops: lessons from Arabidopsis and rice. Journal of Experimental Botany 68, 2477–2488. [DOI] [PubMed] [Google Scholar]
- Liu H, Carvalhais LC, Crawford M, Singh E, Dennis PG, Pieterse CMJ, Schenk PM. 2017. Inner plant values: diversity, colonization and benefits from endophytic bacteria. Frontiers in Microbiology 8, 2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludewig U, Neuhäuser B, Dynowski M. 2007. Molecular mechanisms of ammonium transport and accumulation in plants. FEBS Letters 581, 2301–2308. [DOI] [PubMed] [Google Scholar]
- MacLean AM, Bravo A, Harrison MJ. 2017. Plant signaling and metabolic pathways enabling arbuscular mycorrhizal symbiosis. The Plant Cell 29, 2319–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov MI. 2019. The role of mycorrhiza in transformation of nitrogen compounds in soil and nitrogen nutrition of plants: a review. Eurasian Soil Science 52, 193–205. [Google Scholar]
- Marino D, Moran JF. 2019. Can ammonium stress be positive for plant performance? Frontiers in Plant Science 10, 1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A. 2010. Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Annals of Botany 105, 1141–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minerdi D, Fani R, Gallo R, Boarino A, Bonfante P. 2001. Nitrogen fixation genes in an endosymbiotic Burkholderia strain. Applied and Environmental Microbiology 67, 725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mus F, Crook MB, Garcia K, et al. 2016. Symbiotic nitrogen fixation and the challenges to its extension to nonlegumes. Applied and Environmental Microbiology 82, 3698–3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieder R, Benbi DK, Scherer HW. 2011. Fixation and defixation of ammonium in soils: a review. Biology and Fertility of Soils 47, 1–14. [Google Scholar]
- O’Brien JA, Vega A, Bouguyon E, Krouk G, Gojon A, Coruzzi G, Gutiérrez RA. 2016. Nitrate transport, sensing, and responses in plants. Molecular Plant 9, 837–856. [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Murray JD, Poole PS, Downie JA. 2011. The rules of engagement in the legume-rhizobial symbiosis. Annual Review of Genetics 45, 119–144. [DOI] [PubMed] [Google Scholar]
- Pankievicz VC, do Amaral FP, Santos KF, et al. 2015. Robust biological nitrogen fixation in a model grass–bacterial association. The Plant Journal 81, 907–919. [DOI] [PubMed] [Google Scholar]
- Paul K, Saha C, Nag M, et al. 2020. A tripartite interaction among the basidiomycete Rhodotorula mucilaginosa, N2-fixing endobacteria, and rice improves plant nitrogen nutrition. The Plant Cell 32, 486–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Tienda J, Corrêa A, Azcón-Aguilar C, Ferrol N. 2014. Transcriptional regulation of host NH4+ transporters and GS/GOGAT pathway in arbuscular mycorrhizal rice roots. Plant Physiology and Biochemistry 75, 1–8. [DOI] [PubMed] [Google Scholar]
- Plett D, Garnett T, Okamoto M. 2017. Molecular genetics to discover and improve nitrogen use efficiency in crop plants. In: Hossain MA, Kamiya T, Burritt DJ, Tran LP, Fujiwara T, eds. Plant macronutrient use efficiency. London: Academic Press, 93–122. [Google Scholar]
- Pretty J. 2018. Intensification for redesigned and sustainable agricultural systems. Science 362, 6417. [DOI] [PubMed] [Google Scholar]
- Reinhold-Hurek B, Bünger W, Burbano CS, Sabale M, Hurek T. 2015. Roots shaping their microbiome: global hotspots for microbial activity. Annual Review of Phytopathology 53, 403–424. [DOI] [PubMed] [Google Scholar]
- Reis VM, Teixeira KR. 2015. Nitrogen fixing bacteria in the family Acetobacteraceae and their role in agriculture. Journal of Basic Microbiology 55, 931–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano I, Ventorino V, Pepe O. 2020. Effectiveness of plant beneficial microbes: overview of the methodological approaches for the assessment of root colonization and persistence. Frontiers in Plant Science 11, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblueth M, Ormeño-Orrillo E, López-López A, Rogel MA, Reyes-Hernández BJ, Martínez-Romero JC, Reddy PM, Martínez-Romero E. 2018. Nitrogen fixation in cereals. Frontiers in Microbiology 9, 1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi C, Bogusz D, Franche C. 2013. Biological nitrogen fixation in non-legume plants. Annals of Botany 111, 743–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos KFDN, Moure VR, Hauer V, Santos ARS, Donatti L, Galvão CW, Pedrosa FO, Souza EM, Wassem R, Steffens MBR. 2017. Wheat colonization by an Azospirillum brasilense ammonium-excreting strain reveals upregulation of nitrogenase and superior plant growth promotion. Plant and Soil 415, 245–255. [Google Scholar]
- Sawers RJH, Ramírez-Flores MR, Olalde-Portugal V, Paszkowski U. 2018. The impact of domestication and crop improvement on arbuscular mycorrhizal symbiosis in cereals: insights from genetics and genomics. New Phytologist 220, 1135–1140. [DOI] [PubMed] [Google Scholar]
- Schlesinger WH. 2009. On the fate of anthropogenic nitrogen. Proceedings of the National Academy of Sciences, USA 106, 203–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevilla M, Burris RH, Gunapala N, Kennedy C. 2001. Comparison of benefit to sugarcane plant growth and 15N2 incorporation following inoculation of sterile plants with Acetobacter diazotrophicus wild-type and nif¯ mutant strains. Molecular Plant-Microbe Interactions 14, 358–366. [DOI] [PubMed] [Google Scholar]
- Sharma SB, Sayyed RZ, Trivedi MH, Gobi TA. 2013. Phosphate solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. SpringerPlus 2, 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M, Schmid M, Rothballer M, Hause G, Zuccaro A, Imani J, Kämpfer P, Domann E, Schäfer P, Hartmann A, Kogel KH. 2008Detection and identification of bacteria intimately associated with fungi of the order Sebacinales. Cellular Microbiology 10, 2235–2246. [DOI] [PubMed] [Google Scholar]
- Tabuchi M, Abiko T, Yamaya T. 2007. Assimilation of ammonium ions and reutilization of nitrogen in rice (Oryza sativa L.). Journal of Experimental Botany 58, 2319–2327. [DOI] [PubMed] [Google Scholar]
- Tao K, Kelly S, Radutoiu S. 2019. Microbial associations enabling nitrogen acquisition in plants. Current Opinion in Microbiology 49, 83–89. [DOI] [PubMed] [Google Scholar]
- Tegeder M, Masclaux-Daubresse C. 2018. Source and sink mechanisms of nitrogen transport and use. New Phytologist 217, 35–53. [DOI] [PubMed] [Google Scholar]
- Tkacz A, Poole P. 2015. Role of root microbiota in plant productivity. Journal of Experimental Botany 66, 2167–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Cortés G, Ghignone S, Bonfante P, Schüßler A. 2015. Mosaic genome of endobacteria in arbuscular mycorrhizal fungi: transkingdom gene transfer in an ancient mycoplasma-fungus association. Proceedings of the National Academy of Sciences, USA 112, 7785–7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi M, Poole PS. 2013. Transport and metabolism in legume-rhizobia symbioses. Annual Review of Plant Biology 64, 781–805. [DOI] [PubMed] [Google Scholar]
- Vacheron J, Desbrosses G, Bouffaud ML, Touraine B, Moënne-Loccoz Y, Muller D, Legendre L, Wisniewski-Dyé F, Prigent-Combaret C. 2013. Plant growth-promoting rhizobacteria and root system functioning. Frontiers in Plant Science 4, 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente J, Gerin F, Le Gouis J, Moënne-Loccoz Y, Prigent-Combaret C. 2020. Ancient wheat varieties have a higher ability to interact with plant growth-promoting rhizobacteria. Plant, Cell & Environment 43, 246–260. [DOI] [PubMed] [Google Scholar]
- van der Heijden MG, Bardgett RD, van Straalen NM. 2008. The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecology Letters 11, 296–310. [DOI] [PubMed] [Google Scholar]
- van Velzen R, Holmer R, Bu F, et al. 2018. Comparative genomics of the nonlegume Parasponia reveals insights into evolution of nitrogen-fixing rhizobium symbioses. Proceedings of the National Academy of Sciences, USA 115, E4700–E4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzeaux J. Alahmad A. Habbib H, et al. 2016. Cover crops prevent from deleterious effect of nitrogen fertilization on bacterial diversity by maintaining carbon concentration in plowed soil. Geoderma 281, 49–57. [Google Scholar]
- Verzeaux J, Hirel B, Dubois F, Lea PJ, Tétu T. 2017a. Agricultural practices to improve nitrogen use efficiency through the use of arbuscular mycorrhizae: basic and agronomic aspects. Plant Science 264, 48–56. [DOI] [PubMed] [Google Scholar]
- Verzeaux J., Nivelle E., Roger R., Hirel B., Dubois F., Tetu T. 2017bSpore density of arbuscular mycorrhizal fungi is fostered by 6 years of no-till and is correlated with environmental parameters in a silty loam soil. Agronomy 7, 38. [Google Scholar]
- Vidotti MS, Matias FI, Alves FC, Pérez-Rodríguez P, Beltran GA, Burgueño J, Crossa J, Fritsche-Neto R. 2019. Maize responsiveness to Azospirillum brasilense: insights into genetic control, heterosis and genomic prediction. PLOS ONE 14, e0217571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YY, Cheng YH, Chen KE, Tsay YF. 2018. Nitrate transport, signaling, and use efficiency. Annual Review of Plant Biology 69, 85–122. [DOI] [PubMed] [Google Scholar]
- Wang W, Shi J, Xie Q, Jiang Y, Yu N, Wang E. 2017. Nutrient exchange and regulation in arbuscular mycorrhizal symbiosis. Molecular Plant 10, 1147–1158. [DOI] [PubMed] [Google Scholar]
- Withers PJA, Neal C, Jarvie HP, Doody DG. 2014. Agriculture and eutrophication: where do we go from here? Sustainability 6, 5853–5875. [Google Scholar]
- Woods AS, Ferré S. 2005. Amazing stability of the arginine–phosphate electrostatic interaction. Journal of Proteome Research 4, 1397–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SC, Cao ZH, Li ZG, Cheung KC, Wong MH. 2005. Effects of biofertilizer contianing N-fixer, P and K solubilizers and AM fungi on maize growth: a greenhouse trial. Geoderma 125, 155–156. [Google Scholar]
- Xu G. 2018. Sensing and transport of nutrients in plants. Seminars in Cell & Developmental Biology 74, 78–79. [DOI] [PubMed] [Google Scholar]
- Xu G, Fan X, Miller AJ. 2012. Plant nitrogen assimilation and use efficiency. Annual Review of Plant Biology 63, 153–182. [DOI] [PubMed] [Google Scholar]
- Yuan L, Loqué D, Kojima S, Rauch S, Ishiyama K, Inoue E, Takahashi H, von Wirén N. 2007. The organization of high-affinity ammonium uptake in Arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1-type transporters. The Plant Cell 19, 2636–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Davidson EA, Mauzerall DL, Searchinger TD, Dumas P, Shen Y. 2015. Managing nitrogen for sustainable development. Nature 528, 51–59. [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu YX, Zhang N, et al. 2019. NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nature Biotechnology 37, 676–684. [DOI] [PubMed] [Google Scholar]
- Zhang G-B, Meng S, Gong J-M. 2018. The expected and unexpected roles of nitrate transporters in plant abiotic stress resistance and their regulation. International Journal of Molecular Sciences 19, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Xu M, Liu Y, Zhang F, Hodge A, Feng G. 2016. Carbon and phosphorus exchange may enable cooperation between an arbuscular mycorrhizal fungus and a phosphate-solubilizing bacterium. New Phytologist 210, 1022–1032. [DOI] [PubMed] [Google Scholar]
- Zifarelli G, Pusch M. 2010. CLC transport proteins in plants. FEBS Letters 584, 2122–2127. [DOI] [PubMed] [Google Scholar]