Abstract
Flesh lignification is a specific chilling response that causes deterioration in the quality of stored red-fleshed loquat fruit (Eribotrya japonica) and is one aspect of wider chilling injury. APETALA2/ETHLENE RESPONSIVE FACTOR (AP2/ERF) transcription factors are important regulators of plant low-temperature responses and lignin biosynthesis. In this study, the expression and action of 27 AP2/ERF genes from the red-fleshed loquat cultivar ‘Luoyangqing’ were investigated in order to identify transcription factors regulating low-temperature-induced lignification. EjERF27, EjERF30, EjERF36, and EjERF39 were significantly induced by storage at 0 °C but inhibited by a low-temperature conditioning treatment (pre-storage at 5 °C for 6 days before storage at 0 °C, which reduces low-temperature-induced lignification), and their transcript levels positively correlated with flesh lignification. A dual-luciferase assay indicated that EjERF39 could transactivate the promoter of the lignin biosynthetic gene Ej4CL1, and an electrophoretic mobility shift assay confirmed that EjERF39 recognizes the DRE element in the promoter region of Ej4CL1. Furthermore, the combination of EjERF39 and the previously characterized EjMYB8 synergistically transactivated the Ej4CL1 promoter, and both transcription factors showed expression patterns correlated with lignification in postharvest treatments and red-fleshed ‘Luoyangqing’ and white-fleshed ‘Ninghaibai’ cultivars with different lignification responses. Bimolecular fluorescence complementation and luciferase complementation imaging assays confirmed direct protein–protein interaction between EjERF39 and EjMYB8. These data indicate that EjERF39 is a novel cold-responsive transcriptional activator of Ej4CL1 that forms a synergistic activator complex with EjMYB8 and contributes to loquat fruit lignification at low temperatures.
Keywords: ERF, ERF–MYB complex, lignification, loquat, low temperature, transcriptional regulation
EjERF39, a novel cold-responsive AP2/ERF transcription factor, is involved in loquat fruit lignin biosynthesis by interacting with EjMYB8 and directly regulating the phenylpropanoid pathway gene Ej4CL1.
Introduction
Low temperature is one of the major abiotic constraints limiting the quality and yield of crops and horticultural products (Liu et al., 1998). The responses of plants to low temperatures involve a range of physiological and biochemical changes, including plasma membrane rigidification, the accumulation of cryoprotective compounds such as soluble sugars, amino acids, and organic acids, and the activation of some branches of the phenylpropanoid pathway (Dixon and Paiva, 1995; Thomashow, 1999; Chinnusamy et al., 2007; Guy et al., 2008), which sometimes leads to lignification. Lignification is a common symptom that occurs in many chilling-sensitive fruits, such as loquat (Cai et al., 2006c), pear (Lu et al., 2015), mangosteen (Dangcham et al., 2008), zucchini (Carvajal et al., 2015), and kiwifruit (Suo et al., 2018) when they are subjected to inappropriate low temperature, and this causes a deterioration of quality that severely limits the storage period. Despite the fact that the understanding of low-temperature-induced lignification could improve fruit quality and prolong postharvest storage time, knowledge of the details of the regulatory mechanism of fruit lignification is very limited.
Lignin is a complex phenylpropanoid-derived polymer (Ralph et al., 2004) that contributes to plant secondary cell wall thickening and enhances adaptation to various abiotic stresses, including low temperature (Moura et al., 2010; Ramakrishna and Ravishankar, 2011; Domon et al., 2013; Shafi et al., 2014; Le et al., 2015; Ployet et al., 2018). The plant lignin biosynthesis pathway involves many enzymatic steps, and the enzymes and corresponding genes have been intensively studied in many species (Boerjan et al., 2003; Bonawitz and Chapple, 2010; Shi et al., 2010; Carocha et al., 2015). The expression of genes encoding phenylalanine ammonia lyase (PAL), 4-coumarate:CoA ligase (4CL), cinnamate 3-hydroxylase, hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase, cinnamoyl-CoA reductase (CCR), and cinnamyl alcohol dehydrogenase (CAD) is dramatically altered by low temperature (El Kayal et al., 2006; Huang et al., 2010; Janska et al., 2011; Ployet et al., 2018).
Loquat (Eriobotrya japonica Lindl.) is a subtropical fruit of high economic value and can be divided into two groups according to the flesh color (Wang et al., 2010). Red-fleshed cultivars are sensitive to chilling and very likely to exhibit chilling-induced lignification if stored at temperatures of 0–4 °C, and the resulting increase in fruit firmness and lignin content occurs mainly during the first 6 days of storage (Cai et al., 2006b, c). White-fleshed cultivars are more tolerant to low temperature, and lignification and fruit firmness are maintained with only limited changes at low temperature (Wang et al., 2010). Loquat flesh lignification, expressed as an increase in fruit firmness, accumulation of lignin content, and reduction of juice yield, also accompanies other symptoms of chilling injury, such as internal browning (Cai et al., 2006c). Due to the significant impact of lignification on fruit quality and marketability, various strategies have been developed to alleviate low-temperature-induced flesh lignification, such as low-temperature conditioning (LTC), heat treatment, and the application of methyl jasmonate and acetylsalicylic acid (Cai et al., 2006a, c; Cao et al., 2008; Zeng et al., 2016). Enzymes within the phenylpropanoid pathway, such as PAL, 4CL, and CAD, show increased activities in response to low temperature, and their transcript levels are correlated with the induction of lignification (Cai et al., 2006c; Shan et al., 2008; Li et al., 2017; Xu et al., 2014). As has been found in model plants, loquat fruit lignification is also regulated by MYB and NAC transcription factors. Three MYB genes, two of the activator type (EjMYB1 and EjMYB8) and one repressor type (EjMYB2), have been shown to have transcriptional effects on the promoters of lignin biosynthesis genes, including EjPAL1, Ej4CL1, and Ej4CL5, and influence low-temperature-induced postharvest flesh lignification (Xu et al., 2014; Wang et al., 2016). EjNAC3, a transcription factor related to the low-temperature response, regulates lignin biosynthesis by directly binding to the promoter of the EjCAD-like gene (Ge et al., 2017).
In addition to these MYB and NAC transcription factors, AP2/ERF members have also been shown to have the potential to regulate genes involved in lignin biosynthesis. For example, overexpression of AtSHN (an ERF member) in rice caused an obvious reduction in total lignin content, particularly the G units of lignin composition (Ambavaram et al., 2011); rice OsERF71, which was involved in drought resistance, controlled lignin biosynthesis by directly binding to the promoter of OsCCR1 (Lee et al., 2016); and Ii049 (a Soloist AP2/ERF member) positively regulated lignin biosynthesis in Isatis indigotica (Ma et al., 2017). In loquat, EjAP2-1, a transcriptional repressor, was shown to interact with EjMYB1/2 and inhibited low-temperature-induced lignification (Zeng et al., 2015). Additionally, transcriptomic analysis of postharvest loquat fruit conducted by Liu et al (2019), involving comparisons between low-temperature storage, LTC, and heat treatment, identified the most differentially expressed genes in the AP2/ERF family, which may potentially play roles in low-temperature-induced lignification of loquat fruit.
In the present study, 27 EjAP2/ERF genes were identified based on RNA-seq data. Their expression in response to low-temperature storage and LTC treatment was analyzed and their potential transcriptional regulatory effects on lignin biosynthesis gene promoters were studied using dual-luciferase and electrophoretic mobility shift assays. The potential interactions between AP2/ERF and transcription factors known from previous work to be involved in low-temperature-induced lignification were also studied.
Materials and methods
Plant materials and treatments
Two loquat fruit cultivars were selected for this study, the red-fleshed cultivar ‘Luoyangqing’ (LYQ) and the white-fleshed cultivar ‘Ninghaibai’ (NHB). LYQ undergoes low-temperature-induced lignification whereas NHB does not.
Fruit of uniform size with no visible signs of wounding or disease were selected for postharvest treatments. Commercially mature LYQ loquat fruit were harvested from an orchard at Luqiao (Zhejiang, China) in 2013 and 2015. In 2013, LYQ loquat fruit were stored at two different temperatures (approximately 450 fruit in each batch), one at room temperature (stored at 20 °C for 6 days) and the other at low temperature (0 °C for 6 days). Fruit were sampled at 0, 1, 2, 4, and 6 days of room-temperature and low-temperature storage. For the low-temperature storage experiment conducted in 2015, the fruit were divided into two batches of approximately 450 fruit. The first batch of fruit was kept at 5 °C for 6 days and then transferred to 0 °C (LTC); the second batch was immediately stored at 0 °C for 6 days and used as a control. In the 2015 experiment, fruit were sampled at 0, 0.5, 1, 2, and 6 days of postharvest storage. The white-fleshed NHB fruit were obtained in 2012 from Zhenhai (Zhejiang, China) and stored at 0 °C for 6 days as a postharvest treatment, and fruit were sampled at 0, 1, 2, and 6 d.
All treatments described above were performed with three biological replicates. Fruit flesh (three replicates, four fruit per replicate) without skins and stones were sliced and collected at each sampling point and stored at –80 °C for further use.
Fruit firmness
Fruit firmness was measured using the TA-XT plus Texture Analyzer (Stable Micro Systems, UK) with a 5 mm diameter probe. After removing a small piece of peel, the fruit flesh was penetrated at a rate of 1 mm s–1 to a depth of 4 mm. Firmness measurements of each fruit were taken at two positions 90° apart at the fruit equator and the results were averaged (Wang et al. 2010). Fruit firmness was expressed as Newtons (N) and measured from 10 individual fruit replicates at each sampling point.
Gene isolation and sequence analysis
Sequenced fragments of the unigenes annotated as AP2/ERF transcription factors were obtained from the postharvest LYQ loquat fruit RNA-seq database (Liu et al., 2019). Full-length sequences of each gene were isolated using a SMART RACE cDNA amplification Kit (Clontech, CA, USA). The RACE primers are described in Supplementary Tables S1 and S2 at JXB online. The sequences of full-length AP2/ERF genes were amplified and confirmed with primers listed in Supplementary Table S3 and assembled with the previously reported EjAP2/ERF gene data (Zeng et al., 2016) to avoid the inclusion of redundant sequences. Non-redundant loquat and Arabidopsis AP2/ERF sequences were aligned using the neighbor-joining method in ClustalX (v. 1.8.1), and a phylogenetic tree was constructed with Figtree (v.1.3.1).
RNA extraction and real-time PCR analysis
Total RNA was extracted from frozen loquat fruit flesh samples using the cetyltrimethylammonium bromide method described by Shan et al. (2008). Total RNA was treated with a TURBO DNA-free kit (Ambion) to remove genomic DNA, and 1 µg DNA-free RNA was used to synthesize first-strand cDNA with the iScriptTM cDNA Synthesis Kit (Bio-Rad).
The gene-specific oligonucleotide primer pairs for real-time PCR were designed with Primer3 (http://primer3.ut.ee/); primers are described in Supplementary Table S4. The reaction mixture (20 µl total volume) for real-time PCR consisted of 10 µl SYBR PCR supermix (Bio-Rad), 6 µl diethylpyrocarbonate-treated H2O, 2 µl diluted cDNA template, and 1 µl of each primer (10 µM). Real-time PCR was performed with a Bio-Rad CFX96 instrument (Bio-Rad) using the following PCR procedure: a pre-denaturation step of 95 °C for 30 s, followed by 95 °C for 10 s and 60 °C for 10 s for 45 cycles, completed with a melting curve analysis.
Dual-luciferase assay
To investigate the transactivation activity of EjAP2/ERFs on promoters of loquat lignin biosynthesis genes, dual-luciferase assays were performed according to the protocol described by Yin et al. (2010). Full-length EjAP2/ERF genes were amplified with primers listed in Supplementary Table S5 and integrated into the pGreenII 0029 62-SK vector. The constructs of fruit lignification-related EjMYBs and promoters of lignin biosynthesis genes (EjPAL1, Ej4CL1/5, EjCAD-like, EjCAD3, and Arabidopsis lignin biosynthetic pathway genes) used for the dual-luciferase assay were originally prepared by Xu et al. (2014) and Wang et al. (2016).
Agrobacterium tumefaciens strain GV3101 transformed with SK- and LUC- constructs were collected and resuspended in infiltration buffer (10 mM MES, 10 mM MgCl2, and 0.2 mM acetosyringone) and adjusted to a standard concentration (OD600=0.75). The dual-luciferase assays were performed in Nicotiana benthamiana leaves with transcription factors and promoters combined in a ratio of 10:1 (v:v). Three days after infiltration, discs from the infiltrated tobacco leaves were collected and the ratio of enzyme activities of firefly luciferase (LUC) and renilla luciferase (REN) was measured. The result obtained with empty vector SK mixed with the promoters was set to a value of 1, as a calibrator. To assess the synergistic effect of two transcription factors on the target promoter, the two transcription factors and promoters were combined in a ratio of 5:5:1 (v:v:v). The effect of each transcription factor with empty vector SK was also tested with the target promoter as a control. Each analysis was carried out with at least three replicates.
Purification of EjERF39 and electrophoretic mobility shift assay
The full-length cDNA sequence of EjERF39 without stop codon was inserted into the pGEX-4t-1 (GE) vector and introduced into Escherichia coli strain BL21 (Novagen). Expression of the recombinant pGEX-EjERF39 protein in BL21 was fully induced by the addition of IPTG at a final concentration of 1 mM at 16 °C. The target protein was purified following the instructions of the GST-tag Protein Purification Kit (Beyotime), and checked and visualized using SDS-PAGE with Coomassie blue staining. The 40 bp Ej4CL1 promoter probes containing either the wild-type C-repeat/dehydration-responsive element (DRE) (GCCGAC) or a mutated DRE (AAAAAA) were synthesized and 3′ end labeled with biotin (GeneBio), and cold competitor probes were generated without biotinylation. Details of the electrophoretic mobility shift assay (EMSA) experiments were described by Ge et al. (2017).
Subcellular localization of transcription factors
Agrobacterium containing 35S-EjERF39-GFP and 35S-EjMYB8-GFP were resuspended in medium and transiently expressed in leaves of 4-week-old RFP-H2B transgenic N. benthamiana plants (Huang et al., 2013). The green fluorescent protein (GFP) and red fluorescent protein (RFP) fusion proteins were examined and imaged 30 h after infiltration using a Zeiss LSM710NLO confocal laser scanning microscope. GFP and RFP fluorescence was detected at laser wavelengths of 488 nm and 543 nm, respectively.
Bimolecular fluorescence complementation
Bimolecular fluorescence complementation (BiFC) assays were performed to analyze the protein–protein interaction between EjERF39 and EjMYB8 using the protocols described by Li et al. (2017) with some modifications. Full-length EjERF39 and EjMYB8 sequences, without termination codons, were constructed in vectors containing C-terminal or N-terminal fragments of yellow fluorescent protein (YFP), respectively, and transiently expressed by Agrobacterium-mediated infiltration into leaves of 5-week-old N. benthamiana plants. Primers used are listed in Supplementary Table S6. The YFP fluorescence of tobacco leaves was imaged 2 days after infiltration using a Zeiss LSM710NLO confocal laser scanning microscope.
Firefly luciferase complementation imaging
A luciferase complementation imaging (LCI) assay with pCAMBIA-nLUC and pCAMBIA-cLUC vectors was used to validate the protein–protein interaction between EjERF39 and EjMYB8 (Li et al., 2017). The LUC enzyme was divided into the N-terminal (nLUC) and C-terminal (cLUC) portions. EjMYB8 was fused with nLUC in the pCAMBIA-nLUC vector and EjERF39 was fused with cLUC in the pCAMBIA-cLUC vector. Primer pairs used for vector construction are shown in Supplementary Table S7.
Agrobacterium strains harboring either nLUC, cLUC, nLUC-EjMYB8, or cLUC-EjERF39 were resuspended in infiltration buffer at a final concentration of OD600=0.75. Equal volumes of each strain were mixed and incubated for 3 h at room temperature before infiltration. Luciferase activity was observed 2 days after infiltration with luciferin (0.2 mM) sprayed on to the infiltrated position of the leaves and kept in the dark for 30 min. The experiment was imaged with a NightSHADE LB 985 imaging system (Berthold) and repeated with three to five independent biological replicates.
Results
Isolation of loquat AP2/ERF genes and phylogenetic analysis
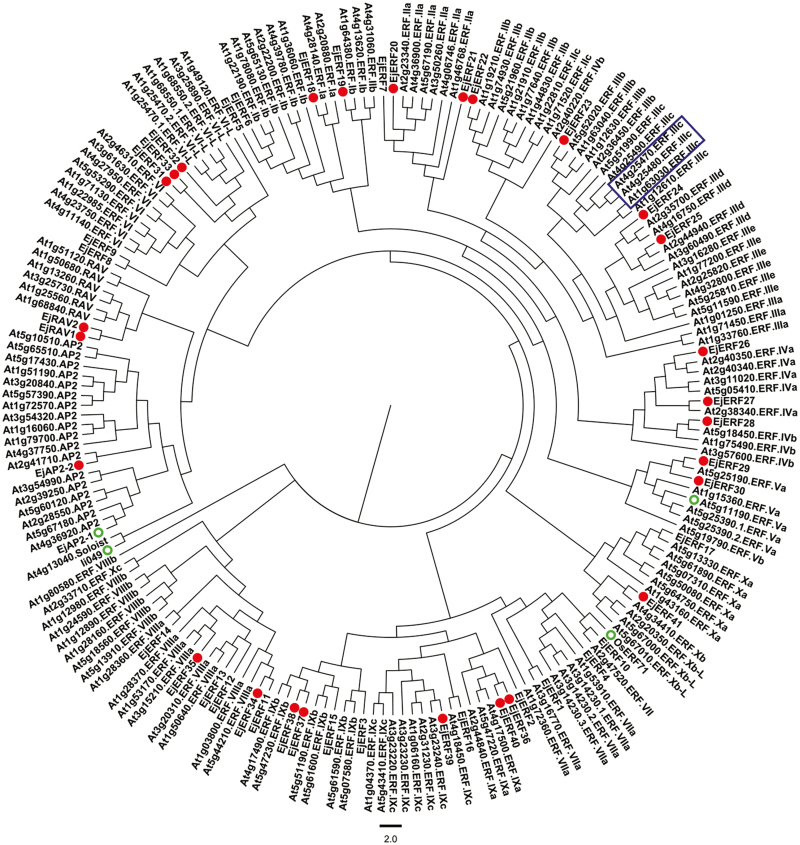
Twenty-seven EjAP2/ERF genes, assigned to either the ERF family (EjERF18-EjERF41, MH753387-410), AP2 family (EjAP2-2, MH753413), or RAV family (EjRAV1 and EjRAV2, MH753411-2), were isolated based on the loquat RNA-seq database (Liu et al., 2019). Phylogenetic analysis indicated that 24 EjERF genes were distributed into nine subfamilies, including subfamilies I, II, III, IV, V, VIII, IX, X and VI-L (Fig. 1), according to Nakano et al. (2006).
Fig. 1.
Phylogenetic analysis of EjAP2/ERF genes. The deduced amino acid sequences of Arabidopsis AtAP2/ERF genes were obtained from NCBI and TAIR. The newly identified loquat EjAP2/ERF genes are marked with solid circles and lignin-related AP2/ERF genes are marked with open circles. Arabidopsis DREB1/CBF members are enclosed with a box.
Relationship between EjAP2/ERF expression and low-temperature-induced lignification
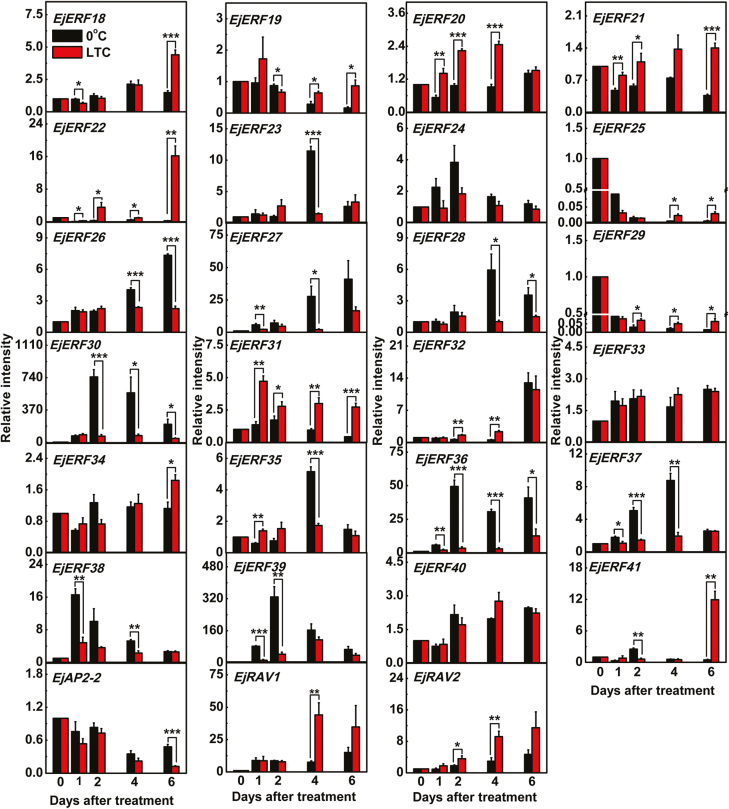
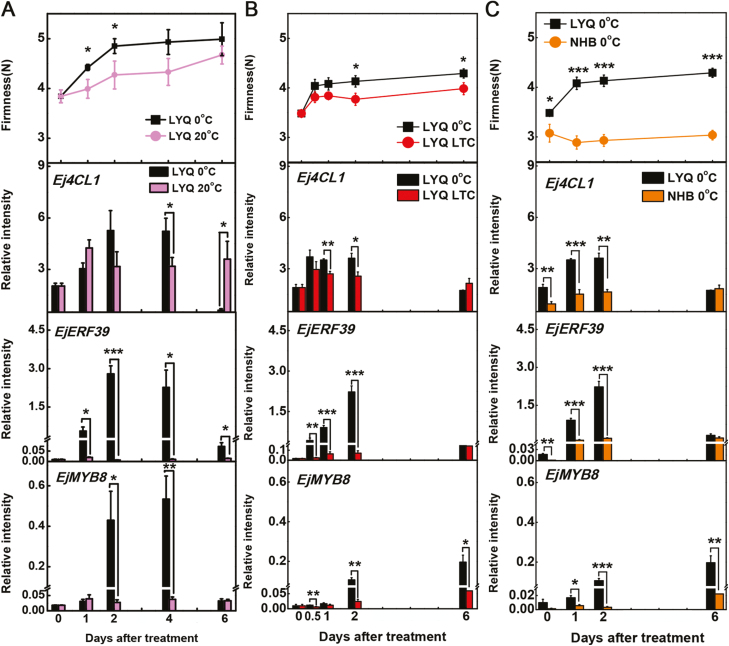
The relationship between the 27 newly isolated EjAP2/ERF genes and fruit lignification was evaluated in LYQ loquat fruit using immediate low-temperature storage (0 °C) and LTC treatment. Fruit firmness and lignin content rapidly increased during 0 °C storage, and prior LTC treatment effectively alleviated this process (Xu et al., 2014; Zeng et al., 2015). Transcript analysis indicated that the 27 EjAP2/ERF genes responded differentially to 0 °C storage and LTC treatment. EjERF27, EjERF30, EjERF36, and EjERF39 were most responsive to 0 °C. The transcript abundance of EjERF27 increased during the whole storage period and reached a maximum abundance of 41-fold relative to day 0 by day 6, while EjERF30, EjERF36, and EjERF39 showed greater abundance at day 2 compared with day 0 (748-, 50- and 329-fold increase, respectively) (Fig. 2). The enrichment of transcripts of the same four EjERF genes was positively correlated with flesh lignification at 0 °C, and their transcript levels were inhibited in fruit that had previously received LTC treatment (Fig. 2). This reduction in transcript levels by LTC treatment suggested that these transcription factors could play a role in low-temperature-induced flesh lignification, since this is also reduced by LTC.
Fig. 2.
Expression levels of EjAP2/ERF genes in response to 0 °C and LTC treatments in LYQ loquat fruit. mRNA levels are presented as a ratio relative to the harvest time point (0 days), the value of which was set as 1. Error bars indicate the SE from three replicates. Statistical analyses were performed using Student’s t-test: *P<0.05, **P<0.01, ***P<0.001. The materials were collected and treatments conducted with three biological replicates as described by Zeng et al. (2015).
Transactivation of promoters of loquat lignin biosynthesis genes by EjAP2/ERFs
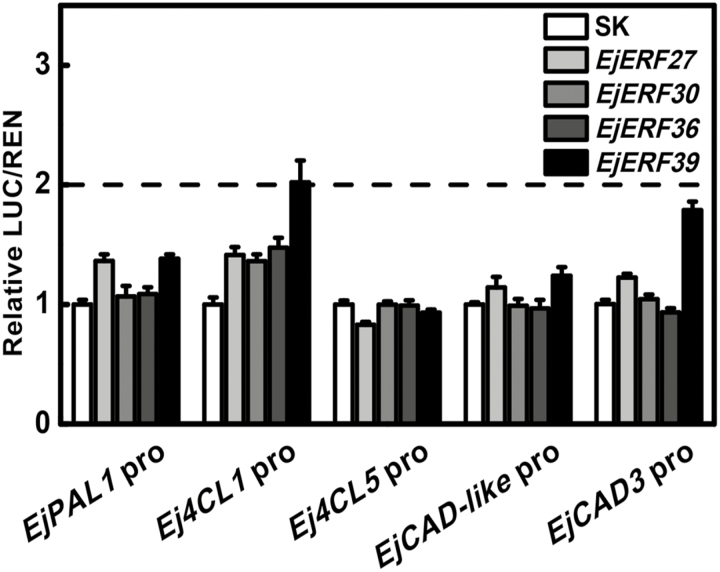
In order to investigate the possible regulatory role of EjERF27, EjERF30, EjERF36, and EjERF39 in controlling loquat lignin biosynthesis genes, their activities on the promoters of key genes previously implicated in the control of low-temperature-induced lignification, EjPAL1, Ej4CL1, Ej4CL5, EjCAD-like, and EjCAD3 (Xu et al., 2014; Wang et al., 2016; Ge et al., 2017), were tested. Dual-luciferase assays indicated that EjERF39 could transactivate the Ej4CL1 promoter, with 2.02-fold enhancement, but had limited or no effect on the other promoters (less than 2-fold increase) (Fig. 3). Despite the correlation between EjERF27, EjERF30, and EjERF36 expression and loquat flesh lignification (Fig. 2), these transcription factors had very limited effects on the promoters of any of the loquat lignin biosynthesis genes tested (Fig. 3).
Fig. 3.
Regulatory effects of EjAP2/ERF genes on promoters of loquat lignin biosynthesis genes in loquat identified by a dual-luciferase assay. The LUC/REN ratio of the empty vector (SK) plus promoter was used as a calibrator (value set to 1). Error bars indicate the SE from three replicates.
The regulatory effect of EjERF39 was also tested with promoters of genes involved in the Arabidopsis phenylpropanoid pathway. As shown in Supplementary Fig. S1, EjERF39 selectively transactivated the promoters of AtPAL3 and AtCAD5 over 2-fold.
Interaction between EjERF39 and the Ej4CL1 promoter
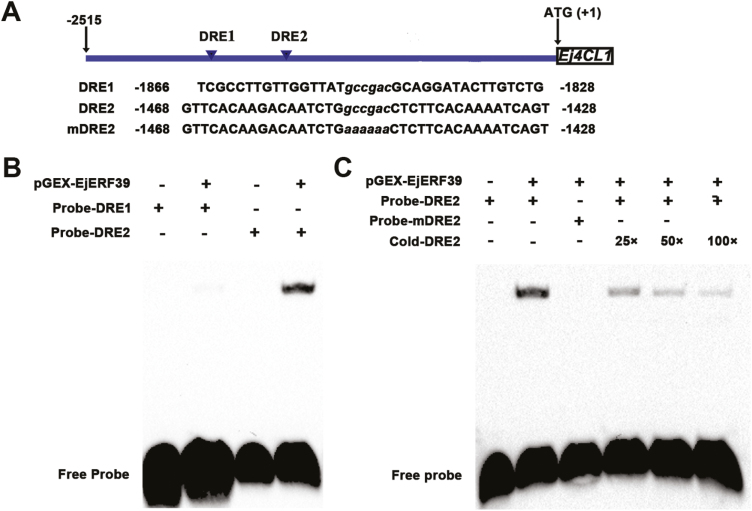
It is well established that AP2/ERF transcription factors preferentially bind to the DRE element of their target gene promoters in response to abiotic stresses such as low temperature (Stockinger et al., 1997). Sequence analysis identified two DRE (GCCGAC) elements in the promoter region of the Ej4CL1 gene (Fig. 4A), suggesting that it might be the direct target of EjERF39, and EMSA was conducted to validate the interaction of EjERF39 with the Ej4CL1 promoter. DNA fragments containing the DRE elements (DRE1, DRE2) and a relevant base mutant (mDRE2) were used as probes (Fig. 4A). The results showed that recombinant pGEX-EjERF39 fusion protein could bind one of the DRE elements used as probes (DRE2) but not the other (DRE1), which suggested that EjERF39 physically binds to the promoter of Ej4CL1 (DRE2 fragment) (Fig. 4B). The specificity of the interaction between EjERF39 and DRE2 but not the DRE1 fragment in the Ej4CL1 promoter was confirmed by using the mutant probe and cold competitors (Fig. 4C).
Fig. 4.
Electrophoretic mobility shift assay (EMSA) of EjERF39 binding to the DRE element in the promoter of Ej4CL1 in vitro. (A) DRE elements in the Ej4CL1 promoter and oligonucleotides used for the EMSA, with the core sequences and base mutant indicated with lower-case letters. (B, C) Biotin-labeled DNA probe from the wild-type promoter or mutant probe was incubated with pGEX-EjERF39 protein. - and + represent the absence or presence of the indicated constructs.
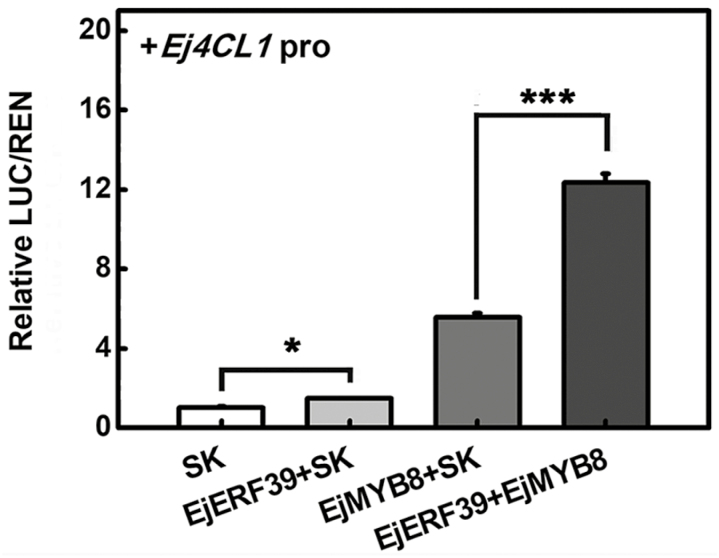
Synergistic effects of EjERF39 and EjMYB8 on Ej4CL1
The results of gene expression and the dual-luciferase assay indicated that EjERF39 could transactivate the promoter of Ej4CL1 and was a candidate for participation in the cold-induced lignification of loquat fruit. Our previous studies indicated that some other transcription factors were also involved in loquat fruit lignification via direct interaction with the Ej4CL1 promoter (Xu et al., 2014; Wang et al., 2016). Accordingly, the effects of EjERF39 and these previously characterized transcription factors were investigated in combination, using the dual-luciferase assay. EjERF39 and EjMYB8 together triggered a substantially higher induction of the Ej4CL1 promoter, with a LUC/REN ratio of 12.31, compared with transfection of EjERF39 (1.47-fold activation) or EjMYB8 (5.55-fold activation) with the empty vector (Fig. 5). The combination of EjERF39 with each of the other transcription factors showed no significant additive effects (Supplementary Fig. S2).
Fig. 5.
Synergistic transactivation effect of EjERF39 and EjMYB8 on the Ej4CL1 promoter. The LUC/REN ratio of the empty vector (SK) plus promoter was used as a calibrator (value set to 1). Statistical analyses were performed using Student’s t-test: *P<0.05, ***P<0.001.
The expression of Ej4CL1, EjERF39, and EjMYB8 in response to low temperature was measured in different cultivars (Fig. 6). As shown in Fig.6A, Ej4CL1, EjERF39, and EjMYB8 were all cold responsive, and fruit exposed to low temperature underwent a corresponding change in firmness. Consistent with the previous results, the expression of EjERF39 and EjMYB8 was substantially enhanced by storage at 0 °C and reduced by prior LTC treatment, and the accumulation of Ej4CL1 transcripts was highly correlated with fruit firmness (Fig. 6B).
Fig. 6.
Relationship between firmness (flesh lignification) and the accumulation of Ej4CL1, EjERF39, and EjMYB8 transcripts during postharvest treatments of LYQ fruit (which shows low-temperature-induced lignification) (A, B) and in LYQ and the cultivar NHB (which does not show lignification in response to low temperature) (C). Error bars indicate the SE from three replicates. Statistical analyses were performed using Student’s t-test: *P<0.05, **P<0.01, ***P<0.001.
The positive correlations between Ej4CL1, EjERF39, and EjMYB8 expression and firmness was also examined in fruit of the red-fleshed cultivar LYQ (which is sensitive to low-temperature-induced lignification) and the white-fleshed cultivar NHB (which does not show low-temperature-induced lignification). The firmness of NHB fruit remained unchanged from 3.07 N at day 0 to 3.04 N at day 6 during storage at 0 °C, and there was no obvious enrichment of Ej4CL1, EjERF39, and EjMYB8 transcripts in NHB fruit (Fig.6C). The coordinated expression of Ej4CL1, EjERF39, and EjMYB8 and the increase in firmness of LYQ fruit undergoing cold-induced lignification, and the absence of any increase in firmness in the low-temperature lignification-free NHB cultivar, suggested that these genes could play vital roles in the low-temperature-induced lignification of red-fleshed loquat fruit.
Protein–protein interaction between EjERF39 and EjMYB8
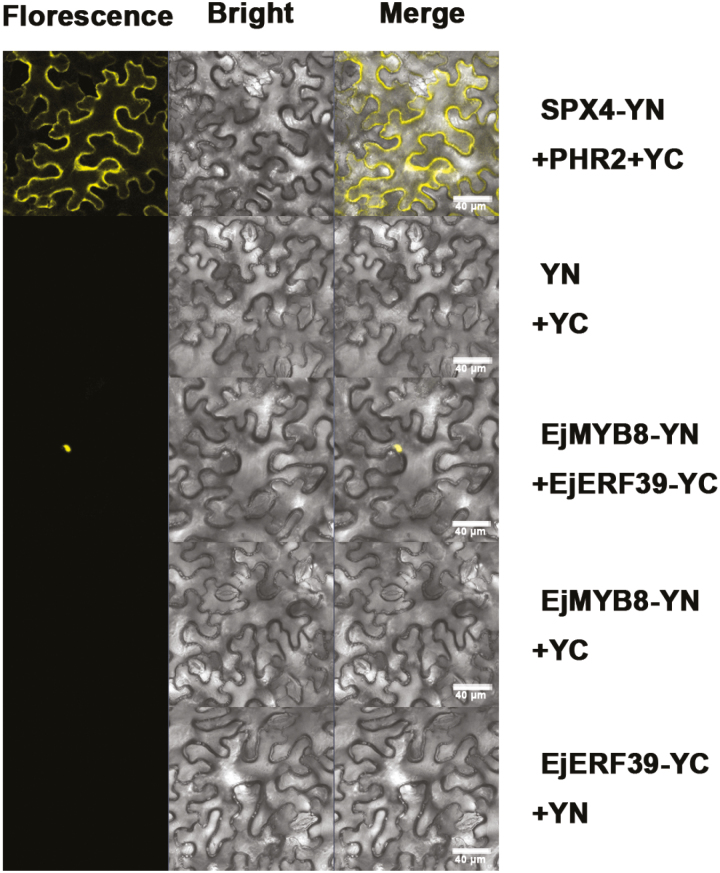
Subcellular localization of GFP-EjERF39 showed a strong fluorescence signal in the nucleus, while GFP-EjMYB8 may locate to both the nucleus and the cytoplasm or near the plasma membrane (Supplementary Fig. S3). The protein–protein interaction between EjERF39 and EjMYB8 was studied using BiFC. No YFP signal was observed in tobacco leaves expressing the combination of a single construct and the corresponding empty vector or bidirectional empty vectors (negative controls, Fig. 7). Tobacco leaves co-transformed with PHR2-YC and SPX4-YN showed an obvious YFP fluorescent signal in the cytoplasm (positive controls, Fig. 7; Lv et al., 2014). EjERF39-YC co-transformed with EjMYB8-YN also showed a YFP fluorescent signal in the nucleus; these results indicated that EjERF39 could interact in the nucleus with EjMYB8 (Fig. 7).
Fig. 7.
Protein–protein interaction between EjERF39 and EjMYB8 demonstrated using bimolecular fluorescence complementation in tobacco leaves. EjERF39 and EjMYB8 proteins were fused to the C- and N-termini of YFP (YC and YN), respectively. PHR2-YC and SPX4-YN were used as positive controls; YC and YN were used as negative controls. Bars=40 μm.
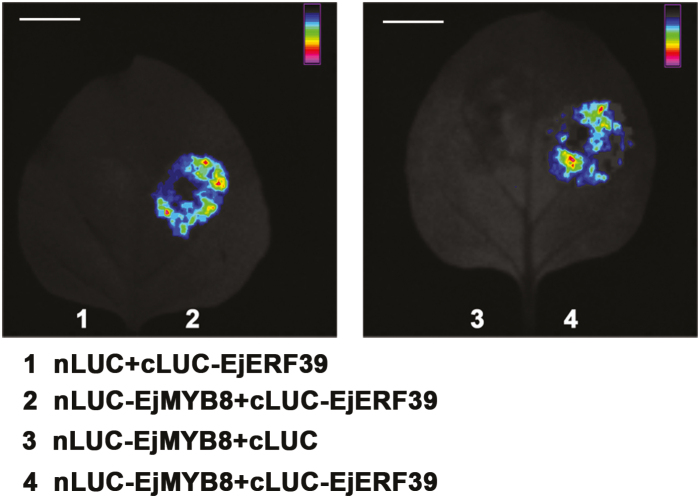
In order to verify the results obtained by BiFC, the EjERF39 and EjMYB8 interaction was analyzed by LCI. Co-infiltration of nLUC-EjERF39 and cLUC-EjMYB8 in tobacco leaves led to a strong luminescence signal of LUC (Fig. 8), whereas no obvious LUC activity was detected in negative controls (the combinations of nLUC + cLUC-EjMYB8 or nLUC-EjERF39 + cLUC) (Fig. 8).
Fig. 8.
Protein–protein interaction between EjERF39 and EjMYB8 demonstrated using luciferase complementation imaging in tobacco leaves. Bars=5 cm.
Discussion
Lignification has enabled the long-term adaptation of vascular plants to the terrestrial environment (Campbell and Sederoff, 1996; Bonawitz and Chapple, 2010; Moura et al., 2010). Apart from vegetative tissues, lignin accumulation also occurs in fruit flesh and is widely related to stress conditions such as wounding (Kamdee et al., 2014) and low temperature (Dangcham et al., 2008; Xu et al., 2014), and significantly reduces fruit quality and marketability. Loquat is an ideal fruit for exploring the mechanism of flesh lignification, given the existence of two different types that exhibit distinct low-temperature responses and texture characteristics. Red-fleshed loquat undergoes chilling-induced lignification, while white-fleshed loquat does not (Wang et al., 2010). LTC and heat treatment have been reported to be effective strategies to alleviate low-temperature-induced lignification of red-fleshed loquat (Cai et al., 2006c; Zeng et al., 2016). Investigation of the molecular basis for loquat flesh lignification has identified several lignin biosynthesis structural genes (EjPAL1, Ej4CL1/5, EjCAD-like, and EjCAD3) and transcription factors (EjMYB1/2/8, EjAP2-1, and EjNAC3) involved in low-temperature-induced lignification (Xu et al., 2014; Wang et al., 2016; Zeng et al., 2016; Li et al., 2017). However, compared with model plants, the regulatory mechanism of stress-induced flesh lignification remains unclear.
Association between EjAP2/ERF expression and low-temperature-induced lignification of loquat fruit
EjAP2-1 was previously reported as a negative regulator of low-temperature-induced lignification in loquat (Zeng et al., 2015). However, the possible roles of other AP2/ERF genes in positively regulating lignification induced by low temperature have not been examined. Accordingly, in this study the relationship between the expression of members of the AP2/ERF family and low-temperature-induced lignification in loquat fruit was analyzed, and four transcription factors (EjERF27, EjERF30, EjERF36, and EjERF39) were identified whose expression was positively correlated with lignification induced by low temperature (Fig. 2). LTC, similar to cold acclimation, is an effective method for prolonging storage time and alleviating symptoms of chilling injury for diverse chilling-sensitive fruits, such as grapefruit (Chaudhary et al., 2014), mango (Zhang et al., 2017), peach (Wang et al., 2017), pomegranate (Kashash et al., 2016), and zucchini (Carvajal et al., 2018). LTC can suppress subsequent lignin accumulation during cold storage and reduce the accompanying internal browning of loquat fruit that occurs at low temperature (Cai et al., 2006c; Tucker et al., 2017). The reduction in expression of EjERF27, EjERF30, EjERF36, and EjERF39 in response to LTC pretreatment was also correlated with a reduction in firmness (Fig. 2), thus these EjERFs may contribute to the lignification process.
The AP2/ERF superfamily plays a pivotal role in various biotic and abiotic stresses, including pathogen infection, wounding, salt, drought, hypoxia, and temperature stress, and responses to several stress-related hormones, such as ethylene, jasmonic acid, and abscisic acid (Mizoi et al., 2012; Licausi et al., 2013). For example, ERF-IX subgroup members have been extensively characterized in plant pathogen responses (Lorenzo et al., 2002; Moffat et al., 2012), and ERF-VII group genes are involved in the hypoxia and submergence response (Licausi et al., 2010). At low temperatures, members of the DREB1/CBF subfamily (subfamily III), AtDREB1B/CBF1, AtDREB1C/CBF2, and AtDREB1A/CBF3, directly bind to DRE elements in the promoter region of cold-responsive related (COR) genes and modulate their expression to enhance chilling/freezing tolerance (Stockinger et al., 1997; Gilmour et al., 1998; Liu et al., 1998; Nakashima et al., 2009). These DREBs are major regulators of the cold stress response, and their homologs have been identified in numerous plant species, such as rice (Dubouzet et al., 2003), wheat (Vagujfalvi et al., 2003), maize (Qin et al., 2004), and tomato (Zhang et al., 2004). Based on phylogenetic analysis, EjERF27 is a member of subfamily IV, EjERF30 is a member of subfamily V, and EjERF36 and EjERF39 are members of subfamily IX, and thus all belong to clades other than the classical cold-responsive DREB1/CBF members (Fig. 1). This finding indicates that EjERF27, EjERF30, EjERF36, and EjERF39 may be new members of a larger group of cold-responsive genes and candidates for involvement in low-temperature-induced lignification.
EjERF39 is a direct activator of lignin biosynthesis via Ej4CL1 modulation
Ej4CL1 is an important target of loquat flesh lignification, with its expression being positively correlated with fruit firmness and flesh lignin accumulation (Fig. 6; Li et al., 2017). Transient overexpression of Ej4CL1 in tobacco leaves significantly induced lignin content (Li et al., 2017). Ej4CL1 is a direct target of several low-temperature-lignification related transcription factors (EjMYB1/2/8 and EjAP2-1; Xu et al., 2014; Zeng et al., 2015; Wang et al., 2016). Using the dual-luciferase system, we found that EjERF39 was capable of transactivating the promoter of Ej4CL1 (with the response being significantly above 2-fold) (Fig. 3), suggesting its potential role in the lignin biosynthesis pathway. The roles of EjERF27, EjERF30, and EjERF36, were not investigated further. The relationship between EjERF39 and Arabidopsis lignin biosynthetic structural genes was further tested and the results indicated that EjERF39 transactivated the promoters of AtPAL3 and AtCAD6 in the Arabidopsis lignin biosynthetic pathway (Supplementary Fig. S1). EMSA indicated that EjERF39 could directly bind to one of the DRE elements of the Ej4CL1 promoter (Fig. 4), which suggested that EjERF39 is a direct activator of the in vivo expression of Ej4CL1, resulting in fruit lignification.
The observation that EjERF39 preferentially bound only one DRE element rather than both of them is consistent with the behavior of the Arabidopsis homolog AtERF1 (AT3G23240). AtERF1 is involved in salt, drought, and heat stress as well as pathogen resistance (Moffat et al., 2012; Cheng et al., 2013). Under different stress conditions, AtERF1 activates stress-response genes by targeting specific cis-elements (GCC boxes during biotic stress and DRE elements during abiotic stress), and AtERF1 specifically binds to only one of the DRE elements in the promoters of abiotic stress-related genes (e.g. RD20, RD29B, COR47, HSP101) (Cheng et al., 2013). AtERF1 and its homologs belonging to the ERF-IX subgroup have been reported to be involved in regulating several aspects of plant secondary metabolism, such as the biosynthesis of monoterpenoid indole alkaloids (CrORAC2 and CrORAC3 in Catharanthus roseus; Menke et al., 1999; van der Fits et al., 2000, 2001), nicotine alkaloids (seven clustered ERF-IX members are located at the NIC-2 locus in tobacco; Shoji et al., 2010; De Boer et al., 2011), and artemisinin (AaERF1 and AaERF2 in Artemisia annua; Yu et al., 2012). Lignin is also an important secondary metabolite, but the previously reported lignin-related AP2/ERF genes, such as AtSHN (a member of ERF subfamily V; Ambavaram et al., 2011), EjAP2-1 (an AP2 member; Zeng et al., 2015), OsERF71 (a member of ERF subfamily VII; Lee et al., 2016) and Ii049 (a Soloist member; Ma et al., 2017), belong to different clades from EjERF39. All of these data indicate that EjERF39 is a novel regulator of lignification.
EjERF39 complexes with EjMYB8 and synergistically activates the Ej4CL1 promoter
Individual transcription factors can be effective regulators of lignin biosynthesis but some also operate by forming complexes with other proteins. For instance, AtMYB75 interacts with AtKNAT7 (a member of the KNOX family) and inhibits lignin synthesis in Arabidopsis stems (Bhargava et al., 2013), and complexes between maize MYB11 and ZML2 (a member of the TIFY family) are involved in wound-induced lignification (Velez-Bermudez et al., 2015). In loquat, EjAP2-1 regulates lignin biosynthesis by interacting with MYB (EjMYB1/2; Zeng et al., 2015). However, no such interactions between two transcriptional activators have been investigated in relation to low-temperature-induced lignification.
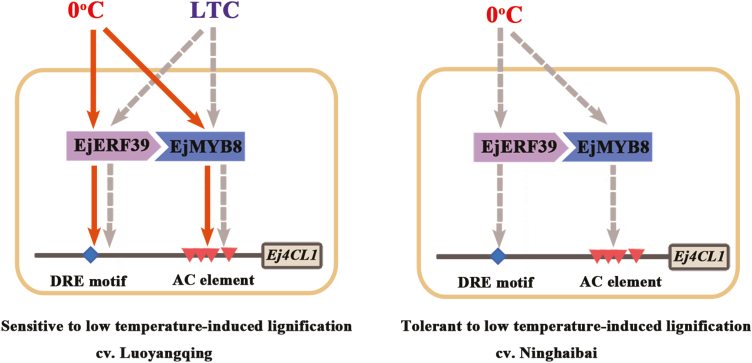
Here, the analysis of synergistic effects in promoter activation and protein–protein interaction studies (Figs 5–8) indicate that EjERF39 is not only a direct regulator of loquat fruit lignification but also interacts with another direct activator, EjMYB8. EjERF39 was coordinately expressed with EjMYB8 in response to low temperature, postharvest LTC treatment, and also in different cultivars with distinct texture characteristics (Fig. 6), and the combination of EjERF39 and EjMYB8 greatly enhanced the activation of the Ej4CL1 promoter (12-fold; Fig. 5). The synergistic effect of EjERF39 and EjMYB8 may be due not only to the fact that they have different cis-element targets, with EjERF39 binding to a DRE motif and EjMYB8 binding to the AC element (Fig. 4; Wang et al., 2016) but also to protein–protein interaction between EjERF39 and EjMYB8 (Figs 7 and 8). A proposed model incorporating these findings is shown in Fig. 9. In low-temperature-sensitive LYQ loquat, low temperature (0 °C) up-regulates the expression of EjERF39 and EjMYB8, which are involved in low-temperature-induced lignification by directly activating the promoter of lignification-related Ej4CL1 and also form protein–protein complexes that lead to flesh lignification and quality deterioration (Fig. 9). LTC treatment blocks the up-regulation of EjERF39 and EjMYB8 expression and suppresses low-temperature-induced lignin accumulation (Fig. 9). In the low-temperature-tolerant NHB loquat, expression of EjERF39 and EjMYB8 remains low and low-temperature-induced lignification is virtually abolished (Fig. 9). The mechanism underlying the differences in low-temperature response and lignin accumulation between the red-fleshed and white-fleshed cultivars requires further investigation. AP2/ERF–MYB complexes have also been reported to regulate other aspects of fruit quality, including color [PyERF3 and PyMYB114 (Yao et al., 2017), and Pp4ERF24, Pp12ERF96, and PpMYB114 (Ni et al., 2019) in anthocyanin biosynthesis] and volatile compound production (FaERF#9 and FaMYB98 in furaneol synthesis; Zhang et al., 2018). Our results provide a new example of the role of AP2/ERF–MYB complexes in plants and expand our understanding of their roles in plant biology.
Fig. 9.
A proposed regulatory model of EjERF39 and EjMYB8 in low-temperature- induced lignification of loquat fruit. Low-temperature (0 °C) storage activates the expression of EjERF39 (a direct activator of lignin biosynthesis via the DRE motif) and EjMYB8 (a previously reported lignin-related activator; Wang et al. 2016). EjERF39 and EjMYB8, which are involved in the regulation of lignification, can also form protein–protein complexes. Solid arrows indicate significant activations; dashed arrows indicate the absence of activations.
In conclusion, the present study has identified 4 AP2/ERF transcription factors that are correlated with low-temperature-induced loquat flesh lignification. EjERF39 belongs to a different subgroup from the previously reported lignin-related AP2/ERF transcription factors, and acts as a direct activator of the Ej4CL1 promoter. Moreover, EjERF39 also forms a protein–protein complex with EjMYB8 and enhances the activation of the Ej4CL1 promoter.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Primer sequences for 3′RACE.
Table S2. Primer sequences for 5′RACE.
Table S3. Primer sequences for full-length amplification.
Table S4. Primer sequences for real-time PCR analysis.
Table S5. Primer sequences for dual-luciferase assays.
Table S6. Primer sequences for BiFC.
Table S7. Primer sequences for LCI.
Fig. S1. Regulatory effects of EjERF39 on promoters of Arabidopsis lignin biosynthesis genes using dual-luciferase assay.
Fig. S2. Synergistic transactivation effect of EjERF39 and EjMYB1/2 on the Ej4CL1 promoter.
Fig. S3. Subcellular localization of EjMYB8 and EjERF39.
Acknowledgements
This research was supported by the National Natural Science Foundation of China (31630067, 31722042), the National Key Research and Development Program (2016YFD0400102), the Natural Science Foundation of Zhejiang Province, China (LR16C150001), and the 111 Project (B17039).
Glossary
Abbreviations
- AP2/ERF
APETALA2/ETHLENE RESPONSIVE FACTOR
- BiFC
bimolecular fluorescence complementation
- CAD
cinnamyl alcohol dehydrogenase
- CCR
cinnamoyl-CoA reductase
- DRE
C-repeat/dehydration-responsive element
- EMSA
electrophoretic mobility shift assay
- LCI
luciferase complementation imaging
- LTC
low-temperature conditioning
- PAL
phenylalanine ammonia lyase
- 4CL
4-coumarate:CoA ligase.
References
- Ambavaram MM, Krishnan A, Trijatmiko KR, Pereira A. 2011. Coordinated activation of cellulose and repression of lignin biosynthesis pathways in rice. Plant Physiology 155, 916–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava A, Ahad A, Wang S, Mansfield SD, Haughn GW, Douglas CJ, Ellis BE. 2013. The interacting MYB75 and KNAT7 transcription factors modulate secondary cell wall deposition both in stems and seed coat in Arabidopsis. Planta 237, 1199–1211. [DOI] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M. 2003. Lignin biosynthesis. Annual Review of Plant Biology 54, 519–546. [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Chapple C. 2010. The genetics of lignin biosynthesis: connecting genotype to phenotype. Annual Review of Genetics 44, 337–363. [DOI] [PubMed] [Google Scholar]
- Cai C, Li X, Chen KS. 2006a. Acetylsalicylic acid alleviates chilling injury of postharvest loquat (Eriobotrya japonica Lindl.) fruit. European Food Research and Technology 223, 533–539. [Google Scholar]
- Cai C, Xu CJ, Li X, Ferguson I, Chen KS. 2006b. Accumulation of lignin in relation to change in activities of lignification enzymes in loquat fruit flesh after harvest. Postharvest Biology and Technology 40, 163–169. [Google Scholar]
- Cai C, Xu CJ, Shan LL, Li X, Zhou CH, Zhang WS, Ferguson I, Chen KS. 2006c. Low temperature conditioning reduces postharvest chilling injury in loquat fruit. Postharvest Biology and Technology 41, 252–259. [Google Scholar]
- Campbell MM, Sederoff RR. 1996. Variation in lignin content and composition (mechanisms of control and implications for the genetic improvement of plants). Plant Physiology 110, 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao SF, Zheng YH, Wang KT, Rui HJ, Tang SS. 2008. Effect of methyl jasmonate on cell wall modification of loquat fruit in relation to chilling injury after harvest. Food Chemistry 118, 641–647. [Google Scholar]
- Carocha V, Soler M, Hefer C, Cassan-Wang H, Fevereiro P, Myburg AA, Paiva JA, Grima-Pettenati J. 2015. Genome-wide analysis of the lignin toolbox of Eucalyptus grandis. New Phytologist 206, 1297–1313. [DOI] [PubMed] [Google Scholar]
- Carvajal F, Palma F, Jamilena M, Garrido D. 2015. Cell wall metabolism and chilling injury during postharvest cold storage in zucchini fruit. Postharvest Biology and Technology 108, 68–77. [Google Scholar]
- Carvajal F, Rosales R, Palma F, Manzano S, Cañizares J, Jamilena M, Garrido D. 2018. Transcriptomic changes in Cucurbita pepo fruit after cold storage: differential response between two cultivars contrasting in chilling sensitivity. BMC Genomics 19, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary PR, Jayaprakasha GK, Porat R, Patil BS. 2014. Low temperature conditioning reduces chilling injury while maintaining quality and certain bioactive compounds of ‘Star Ruby’ grapefruit. Food Chemistry 153, 243–249. [DOI] [PubMed] [Google Scholar]
- Cheng MC, Liao PM, Kuo WW, Lin TP. 2013. The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiology 162, 1566–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu J, Zhu JK. 2007. Cold stress regulation of gene expression in plants. Trends in Plant Science 12, 444–451. [DOI] [PubMed] [Google Scholar]
- Dangcham S, Bowen J, Ferguson IB, Ketsa S. 2008. Effect of temperature and low oxygen on pericarp hardening of mangosteen fruit stored at low temperature. Postharvest Biology and Technology 50, 37–44. [Google Scholar]
- De Boer K, Tilleman S, Pauwels L, Vanden Bossche R, De Sutter V, Vanderhaeghen R, Hilson P, Hamill JD, Goossens A. 2011. APETALA2/ETHYLENE RESPONSE FACTOR and basic helix-loop-helix tobacco transcription factors cooperatively mediate jasmonate-elicited nicotine biosynthesis. The Plant Journal 66, 1053–1065. [DOI] [PubMed] [Google Scholar]
- Dixon RA, Paiva NL. 1995. Stress-induced phenylpropanoid metabolism. The Plant Cell 7, 1085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domon JM, Baldwin L, Acket S, et al. . 2013. Cell wall compositional modifications of Miscanthus ecotypes in response to cold acclimation. Phytochemistry 85, 51–61. [DOI] [PubMed] [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. 2003. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. The Plant Journal 33, 751–763. [DOI] [PubMed] [Google Scholar]
- El Kayal W, Keller G, Debayles C, Kumar R, Weier D, Teulieres C, Marque C. 2006. Regulation of tocopherol biosynthesis through transcriptional control of tocopherol cyclase during cold hardening in Eucalyptus gunnii. Physiologia Plantarum 126, 212–223. [Google Scholar]
- Ge H, Zhang J, Zhang YJ, Li X, Yin XR, Grierson D, Chen KS. 2017. EjNAC3 transcriptionally regulates chilling-induced lignification of loquat fruit via physical interaction with an atypical CAD-like gene. Journal of Experimental Botany 68, 5129–5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF. 1998. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. The Plant Journal 16, 433–442. [DOI] [PubMed] [Google Scholar]
- Guy C, Kaplan F, Kopka J, Selbig J, Hincha DK. 2008. Metabolomics of temperature stress. Physiologia Plantarum 132, 220–235. [DOI] [PubMed] [Google Scholar]
- Huang C, Hu G, Li F, Li Y, Wu J, Zhou X. 2013. NbPHAN, a MYB transcriptional factor, regulates leaf development and affects drought tolerance in Nicotiana benthamiana. Physiologia Plantarum 149, 297–309. [DOI] [PubMed] [Google Scholar]
- Huang J, Gu M, Lai Z, Fan B, Shi K, Zhou YH, Yu JQ, Chen Z. 2010. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiology 153, 1526–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janská A, Aprile A, Zámečník J, Cattivelli L, Ovesná J. 2011. Transcriptional responses of winter barley to cold indicate nucleosome remodelling as a specific feature of crown tissues. Functional & Integrative Genomics 11, 307–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamdee C, Imsabai W, Kirk R, Allan AC, Ferguson IB, Ketsa S. 2014. Regulation of lignin biosynthesis in fruit pericarp hardening of mangosteen (Garcinia mangostana L.) after impact. Postharvest Biology and Technology 97, 68–76. [Google Scholar]
- Kashash Y, Mayuoni-Kirshenbaum L, Goldenberg L, Choi HJ, Porat R. 2016. Effects of harvest date and low-temperature conditioning on chilling tolerance of ‘Wonderful’ pomegranate fruit. Scientia Horticulturae 209, 286–292. [Google Scholar]
- Le MQ, Pagter M, Hincha DK. 2015. Global changes in gene expression, assayed by microarray hybridization and quantitative RT-PCR, during acclimation of three Arabidopsis thaliana accessions to sub-zero temperatures after cold acclimation. Plant Molecular Biology 87, 1–15. [DOI] [PubMed] [Google Scholar]
- Lee DK, Jung H, Jang G, Jeong JS, Kim YS, Ha SH, Do Choi Y, Kim JK. 2016. Overexpression of the OsERF71 transcription factor alters rice root structure and drought resistance. Plant Physiology 172, 575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SJ, Yin XR, Wang WL, Liu XF, Zhang B, Chen KS. 2017. Citrus CitNAC62 cooperates with CitWRKY1 to participate in citric acid degradation via up-regulation of CitAco3. Journal of Experimental Botany 68, 3419–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Xu Y, Zhang L, Ji Y, Tan D, Yuan H, Wang A. 2017. The jasmonate-activated transcription factor MdMYC2 regulates ETHYLENE RESPONSE FACTOR and ethylene biosynthetic genes to promote ethylene biosynthesis during apple fruit ripening. The Plant Cell 29, 1316–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zang C, Ge H, Zhang J, Grierson D, Yin XR, Chen KS. 2017. Involvement of PAL, C4H, and 4CL in chilling injury-induced flesh lignification of loquat fruit. Hortscience 52, 127–131. [Google Scholar]
- Licausi F, Ohme-Takagi M, Perata P. 2013. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytologist 199, 639–649. [DOI] [PubMed] [Google Scholar]
- Licausi F, van Dongen JT, Giuntoli B, Novi G, Santaniello A, Geigenberger P, Perata P. 2010. HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. The Plant Journal 62, 302–315. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. 1998. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. The Plant Cell 10, 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Zhang J, Jiao C, Yin X, Fei Z, Wu Q, Chen K. 2019. Transcriptome analysis provides insights into the regulation of metabolic processes during postharvest cold storage of loquat (Eriobotrya japonica) fruit. Horticulture Research 6, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sánchez-Serrano JJ, Solano R. 2002. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. The Plant Cell 15, 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G, Li Z, Zhang X, Wang R, Yang S. 2015. Expression analysis of lignin associated genes in hard end pear (Pyrus pyrifolia Whangkeumbae) and its response to calcium chloride treatment conditions. Journal of Plant Growth Regulation 34, 251–262. [Google Scholar]
- Lv Q, Zhong Y, Wang Y, et al. . 2014. SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. The Plant Cell 26, 1586–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma R, Xiao Y, Lv Z, et al. . 2017. AP2/ERF transcription factor, Ii049, positively regulates lignan biosynthesis in Isatis indigotica through activating salicylic acid signaling and lignan/lignin pathway genes. Frontiers in Plant Science 8, 1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke FL, Champion A, Kijne JW, Memelink J. 1999. A novel jasmonate- and elicitor-responsive element in the periwinkle secondary metabolite biosynthetic gene Str interacts with a jasmonate- and elicitor-inducible AP2-domain transcription factor, ORCA2. The EMBO Journal 18, 4455–4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. 2012. AP2/ERF family transcription factors in plant abiotic stress responses. Biochimica et Biophysica Acta 1819, 86–96. [DOI] [PubMed] [Google Scholar]
- Moffat CS, Ingle RA, Wathugala DL, Saunders NJ, Knight H, Knight MR. 2012. ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against Botrytis cinerea in Arabidopsis. PLoS One 7, e35995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura JC, Bonine CA, de Oliveira Fernandes Viana J, Dornelas MC, Mazzafera P. 2010. Abiotic and biotic stresses and changes in the lignin content and composition in plants. Journal of Integrative Plant Biology 52, 360–376. [DOI] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. 2006. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiology 140, 411–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Ito Y, Yamaguchi-Shinozaki K. 2009. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiology 149, 88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Bai S, Zhao Y, Qian M, Tao R, Yin L, Gao L, Teng Y. 2019. Ethylene response factors Pp4ERF24 and Pp12ERF96 regulate blue light-induced anthocyanin biosynthesis in ‘Red Zaosu’ pear fruits by interacting with MYB114. Plant Molecular Biology 99, 67–78. [DOI] [PubMed] [Google Scholar]
- Ployet R, Soler M, Carocha V, et al. . 2018. Long cold exposure induces transcriptional and biochemical remodelling of xylem secondary cell wall in Eucalyptus. Tree Physiology 38, 409–422. [DOI] [PubMed] [Google Scholar]
- Qin F, Sakuma Y, Li J, Liu Q, Li YQ, Shinozaki K, Yamaguchi-Shinozaki K. 2004. Cloning and functional analysis of a novel DREB1/CBF transcription factor involved in cold-responsive gene expression in Zea mays L. Plant & Cell Physiology 45, 1042–1052. [DOI] [PubMed] [Google Scholar]
- Ralph J, Lundquist K, Brunow G, et al. . 2004. Lignins: natural polymers from oxidative coupling of 4-hydroxyphenylpropanoids. Phytochemistry Reviews 3, 29–60. [Google Scholar]
- Ramakrishna A, Ravishankar GA. 2011. Influence of abiotic stress signals on secondary metabolites in plants. Plant Signaling & Behavior 6, 1720–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafi A, Dogra V, Gill T, Ahuja PS, Sreenivasulu Y. 2014. Simultaneous over-expression of PaSOD and RaAPX in transgenic Arabidopsis thaliana confers cold stress tolerance through increase in vascular lignifications. PLoS One 9, e110302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan LL, Li X, Wang P, Cai C, Zhang B, Sun CD, Zhang WS, Xu CJ, Ferguson I, Chen KS. 2008. Characterization of cDNAs associated with lignification and their expression profiles in loquat fruit with different lignin accumulation. Planta 227, 1243–1254. [DOI] [PubMed] [Google Scholar]
- Shi R, Sun YH, Li Q, Heber S, Sederoff R, Chiang VL. 2010. Towards a systems approach for lignin biosynthesis in Populus trichocarpa: transcript abundance and specificity of the monolignol biosynthetic genes. Plant & Cell Physiology 51, 144–163. [DOI] [PubMed] [Google Scholar]
- Shoji T, Kajikawa M, Hashimoto T. 2010. Clustered transcription factor genes regulate nicotine biosynthesis in tobacco. The Plant Cell 22, 3390–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF. 1997. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proceedings of the National Academy of Sciences, USA 94, 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo JT, Li H, Ban QY, Han Y, Meng K, Jin MJ, Zhang ZK, Rao JP. 2018. Characteristics of chilling injury-induced lignification in kiwifruit with different sensitivities to low temperatures. Postharvest Biology and Technology 135, 8–18. [Google Scholar]
- Thomashow MF. 1999. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annual Review of Plant Physiology and Plant Molecular Biology 50, 571–599. [DOI] [PubMed] [Google Scholar]
- Tucker G, Yin XR, Zhang AD, Wang MM, Zhu QG, Liu XF, Xie XL, Chen KS, Grierson D. 2017. Ethylene and fruit softening. Food Quality and Safety 1, 253–267. [Google Scholar]
- Vágújfalvi A, Galiba G, Cattivelli L, Dubcovsky J. 2003. The cold-regulated transcriptional activator Cbf3 is linked to the frost-tolerance locus Fr-A2 on wheat chromosome 5A. Molecular Genetics and Genomics 269, 60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Fits L, Memelink J. 2000. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289, 295–297. [DOI] [PubMed] [Google Scholar]
- van der Fits L, Memelink J. 2001. The jasmonate-inducible AP2/ERF-domain transcription factor ORCA3 activates gene expression via interaction with a jasmonate-responsive promoter element. The Plant Journal 25, 43–53. [DOI] [PubMed] [Google Scholar]
- Vélez-Bermúdez IC, Salazar-Henao JE, Fornalé S, et al. . 2015. A MYB/ZML complex regulates wound-induced lignin genes in maize. The Plant Cell 27, 3245–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Yin XR, Zhang B, Grierson D, Xu CJ, Chen KS. 2017. Transcriptomic and metabolic analyses provide new insights into chilling injury in peach fruit. Plant, Cell & Environment 40, 1531–1551. [DOI] [PubMed] [Google Scholar]
- Wang P, Zhang B, Li X, Xu C, Yin X, Shan L, Ferguson I, Chen K. 2010. Ethylene signal transduction elements involved in chilling injury in non-climacteric loquat fruit. Journal of Experimental Botany 61, 179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WQ, Zhang J, Ge H, Li SJ, Li X, Yin XR, Grierson D, Chen KS. 2016. EjMYB8 transcriptionally regulates flesh lignification in loquat fruit. PLoS One 11, e0154399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Yin XR, Zeng JK, Ge H, Song M, Xu CJ, Li X, Ferguson IB, Chen KS. 2014. Activator- and repressor-type MYB transcription factors are involved in chilling injury induced flesh lignification in loquat via their interactions with the phenylpropanoid pathway. Journal of Experimental Botany 65, 4349–4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao G, Ming M, Allan AC, et al. . 2017. Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. The Plant Journal 92, 437–451. [DOI] [PubMed] [Google Scholar]
- Yin XR, Allan AC, Chen KS, Ferguson IB. 2010. Kiwifruit EIL and ERF genes involved in regulating fruit ripening. Plant Physiology 153, 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu ZX, Li JX, Yang CQ, Hu WL, Wang LJ, Chen XY. 2012. The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Molecular Plant 5, 353–365. [DOI] [PubMed] [Google Scholar]
- Zeng JK, Li X, Xu Q, Chen JY, Yin XR, Ferguson IB, Chen KS. 2015. EjAP2-1, an AP2/ERF gene, is a novel regulator of fruit lignification induced by chilling injury, via interaction with EjMYB transcription factors. Plant Biotechnology Journal 13, 1325–1334. [DOI] [PubMed] [Google Scholar]
- Zeng JK, Li X, Zhang J, Ge H, Yin XR, Chen KS. 2016. Regulation of loquat fruit low temperature response and lignification involves interaction of heat shock factors and genes associated with lignin biosynthesis. Plant, Cell & Environment 39, 1780–1789. [DOI] [PubMed] [Google Scholar]
- Zhang X, Fowler SG, Cheng H, Lou Y, Rhee SY, Stockinger EJ, Thomashow MF. 2004. Freezing-sensitive tomato has a functional CBF cold response pathway, but a CBF regulon that differs from that of freezing-tolerant Arabidopsis. The Plant Journal 39, 905–919. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yin X, Xiao Y, et al. . 2018. An ETHYLENE RESPONSE FACTOR-MYB transcription complex regulates furaneol biosynthesis by activating QUINONE OXIDOREDUCTASE expression in strawberry. Plant Physiology 178, 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhu Q, Hu M, Gao Z, An F, Li M, Jiang Y. 2017. Low-temperature conditioning induces chilling tolerance in stored mango fruit. Food Chemistry 219, 76–84. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.