Abstract
This article contains data on the bacterial communities of lagoon sediments with fish potential in the Central Andes of Peru. The surface sediment samples were collected from four lagoons destined for continental water fish farming. DNA extraction was performed from 0.5 g of sample through the Presto™ Soil DNA Extraction Kit. Bacterial sequencing of the 16S rRNA amplicon was performed on the DNA extracted from the sediment. At least 36 Phyla bacteria were detected, the bacterial communities being dominated by Proteobacteria, Cyanobacteria, Actinobacteria, Firmicutes, Chloroflexi. These data can be used for predictive analysis to gain a better understanding of the dynamics of bacterial communities in environments under pressure from fish farming.
Keywords: Gen 16S rRNA, Bacterial composition, Sediment, Gaps, Fish farming
Specifications Table
| Subject | Biology |
| Specific subject area | Microbial ecology |
| Type of data | Tables, figures, FASTQ |
| How data was acquired | High performance sequencing data of the 16S rRNA gene amplicon using Illumina MiSeq sequencing [1]. |
| Data format | Raw and analyzed |
| Parameters for data collection | Identification of ponds with fish activity and sediment collection. |
| Description of data collection | Extraction and amplification of bacterial DNA by PCR and sequencing of 16S bacterial rRNA amplicon [2]. |
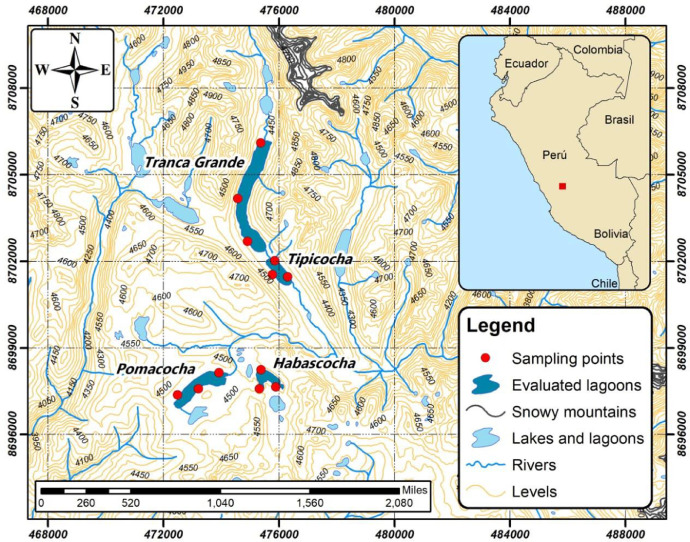
| Data source location | Lagoons with fish potential located in the Central Andes of Peru, between latitude −11.7808°, longitude −75.2454° and latitude −11.7198, longitude −75.2311 (Fig. 1). |
| Data accessibility | Data is available in the article. |
Value of the Data
-
•
These data are the first generated using 16S rRNA genes from bacterial communities in lake environments pressured by fish farming in the Peruvian Andes.
-
•
These metagenomic data may be useful to other researchers to expand molecular studies and compare the composition of bacterial communities under different environmental and anthropogenic factors.
-
•
These data can be used for predictive analysis to gain a better understanding of the dynamics of bacterial communities in environments under pressure from fish farming.
1. Data Description
1.1. Study area
The study was conducted in the Pomacocha, Habascocha, Tipicocha and Tranca Grande lagoons of glacial origin located in the Central Andes of Peru, in the upper basin of the Perene River, at an altitude between 4310 and 4330 m.a.s.l. [3]. The four lagoons are used for intensive farming of Oncorhynchus mykiss (rainbow trout) in large floating cages (Fig. 1).
Fig. 1.
Location map of the study area in the Perene river watershed, Peru.
1.2. Analytical data
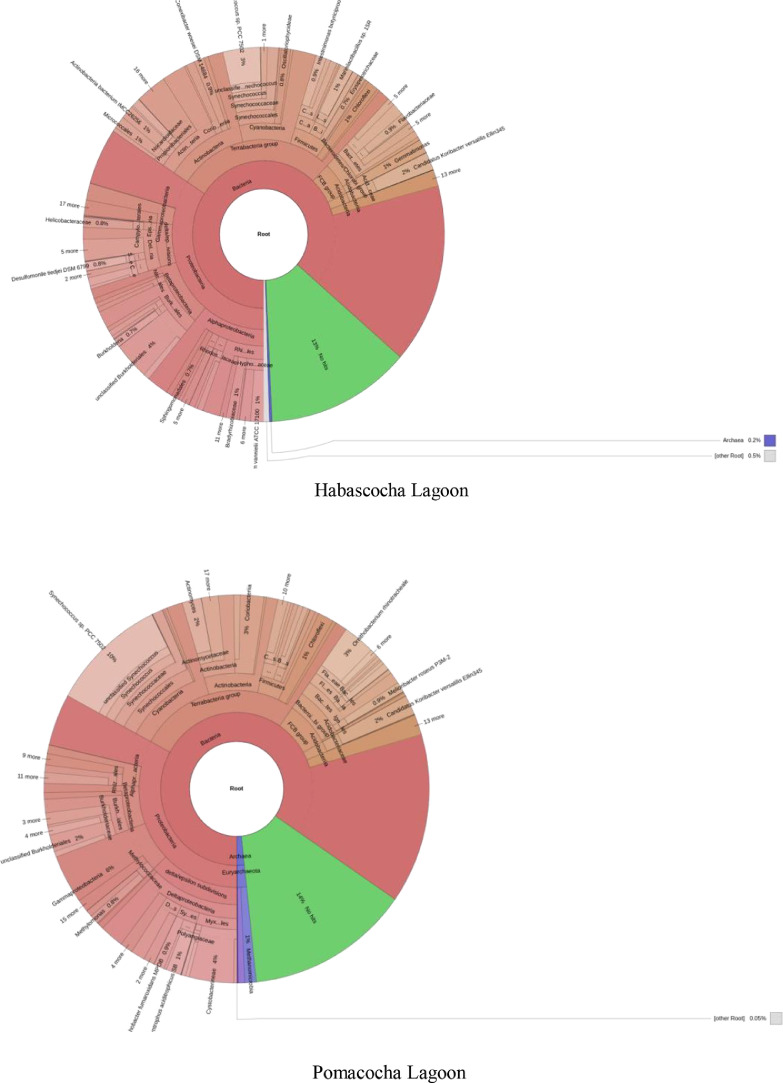
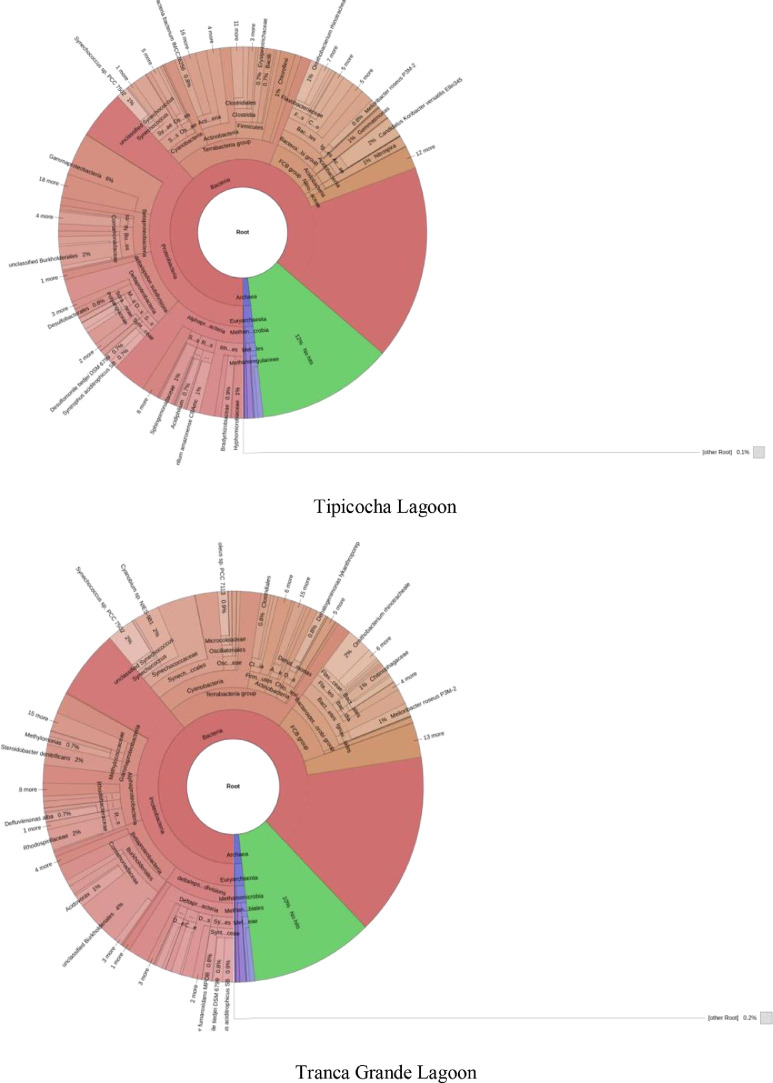
The metagenomic data presented in this manuscript provide information on the bacterial communities of lagoon sediments intended for the cultivation of Oncorhynchus mykiss in the Central Andes of Peru. The bacterial taxonomic composition generated through sequencing of the 16S rRNA amplicon using the standard next-generation Illumina MiSeq protocol is shown in Fig. 2. Analysis of the final readings revealed the Bacteria and Archaea domains. In the Habascocha lagoon the readings revealed 33 phyla, 64 classes and 127 orders, in the Pomacocha lagoon 30 phyla, 61 classes and 120 orders, in the Tipicocha lagoon 34 phyla, 61 classes and 130 orders and, in the Tranca Grande lagoon 31 phyla, 55 classes and 127 orders. The readings also revealed 276 bacterial families in the four lakes. However, between 10% and 14% of the total readings were not classified.
Fig. 2.
Composition of bacterial communities in lake sediments with fish potential in the Central Andes of Peru.
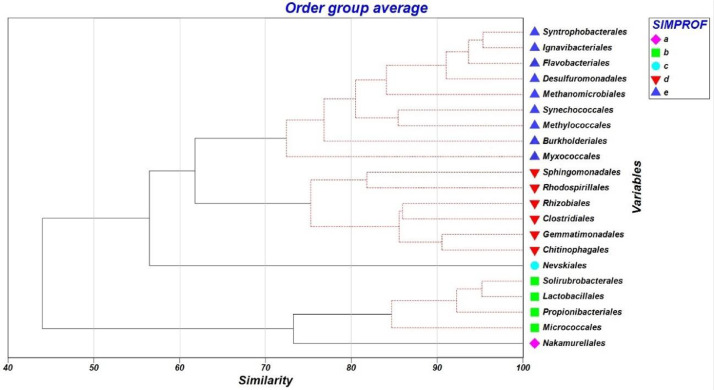
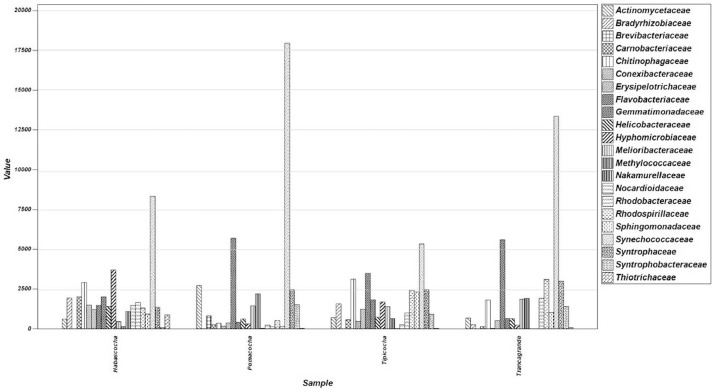
Table 1 shows the abundance of bacteria in surface sediments of lagoons with fish potential in the Central Andes of Peru, according to phylum, obtained through high performance sequencing. Table 2 shows the mean abundance and percentage contribution of phyla bacteria to the differentiation or similarity between groups, according to the SIMPER analysis. Phylum Actinobacteria presented the highest percentage of contribution to the bacterial communities (29.20%), followed by Cyanobacteria (16.11%) and Proteobacteria (14.66%). The grouping of bacterial orders by SIMPROF analysis, reported five statistically different groups in relation to the number and site of sampling (Fig. 3). The distribution of bacterial families in surface sediments of ponds with fish potential at 70% contribution by SIMPER analysis is shown in Fig. 4.
Table 1.
Abundance of bacteria in surface sediment of lagoons with fish potential in the Central Andes of Peru, according to phylum.
| Phylum | Habascocha | Pomacocha | Tipicocha | Trancagrande |
|---|---|---|---|---|
| Acidobacteria | 2668 | 2901 | 3284 | 1023 |
| Actinobacteria | 20,234 | 13,410 | 8668 | 3407 |
| Aquificae | 83 | 1 | 19 | 7 |
| Armatimonadetes | 23 | 15 | 66 | 18 |
| Bacteroidetes | 7001 | 10,302 | 10,980 | 12,591 |
| Caldiserica | 20 | 76 | 87 | 74 |
| Candidatus Cloacimonetes | 34 | 22 | 70 | 53 |
| Candidatus Korarchaeota | 0 | 0 | 4 | 6 |
| Candidatus Saccharibacteria | 473 | 306 | 578 | 278 |
| Chlamydiae | 15 | 110 | 81 | 61 |
| Chlorobi | 14 | 69 | 142 | 51 |
| Chloroflexi | 1740 | 2234 | 2260 | 2964 |
| Chrysiogenetes | 1 | 0 | 0 | 0 |
| Crenarchaeota | 0 | 0 | 0 | 1 |
| Cyanobacteria | 13,986 | 20,855 | 10,245 | 22,762 |
| Deferribacteres | 0 | 4 | 4 | 0 |
| Deinococcus Thermus | 190 | 335 | 653 | 250 |
| Dictyoglomi | 112 | 758 | 733 | 1097 |
| Elusimicrobia | 2 | 1 | 2 | 2 |
| Euryarchaeota | 319 | 2534 | 2837 | 2668 |
| Fibrobacteres | 12 | 32 | 25 | 41 |
| Firmicutes | 8616 | 5975 | 7613 | 4841 |
| Fusobacteria | 35 | 43 | 65 | 55 |
| Gemmatimonadetes | 2008 | 407 | 1818 | 654 |
| Ignavibacteriae | 442 | 1440 | 1418 | 1876 |
| Kiritimatiellaeota | 2 | 3 | 2 | 2 |
| Nitrospirae | 653 | 116 | 1761 | 82 |
| Planctomycetes | 45 | 59 | 55 | 148 |
| Proteobacteria | 58,539 | 55,169 | 66,426 | 64,971 |
| Spirochaetes | 108 | 447 | 469 | 612 |
| Synergistetes | 16 | 38 | 52 | 74 |
| Tenericutes | 147 | 116 | 198 | 134 |
| Thaumarchaeota | 7 | 2 | 66 | 3 |
| Thermodesulfobacteria | 173 | 329 | 353 | 459 |
| Thermotogae | 14 | 15 | 13 | 19 |
| Verrucomicrobia | 636 | 516 | 969 | 814 |
Table 2.
Mean abundance and percentage contribution of bacterial phyla in lagoon sediment with fish potential in the Central Andes of Peru, according to SIMPER analysis.
| Phylum | Av. dissim | Contrib.% | Cumulative% | Mean A | Mean B |
|---|---|---|---|---|---|
| Actinobacteria | 4.90 | 29.20 | 29.20 | 20,200 | 8500 |
| Cyanobacteria | 2.70 | 16.11 | 45.30 | 14,000 | 18,000 |
| Proteobacteria | 2.46 | 14.66 | 59.97 | 58,500 | 62,200 |
| Bacteroidetes | 1.79 | 10.68 | 70.65 | 7000 | 11,300 |
| Firmicutes | 1.03 | 6.17 | 76.81 | 8620 | 6140 |
| Euryarchaeota | 0.99 | 5.88 | 82.69 | 319 | 2680 |
| Ignavibacteriae | 0.47 | 2.83 | 85.52 | 442 | 1580 |
| Gemmatimonadetes | 0.44 | 2.62 | 88.14 | 2010 | 960 |
| Acidobacteria | 0.35 | 2.06 | 90.21 | 2670 | 2400 |
| Dictyoglomi | 0.31 | 1.87 | 92.07 | 112 | 863 |
| Chloroflexi | 0.31 | 1.86 | 93.93 | 1740 | 2490 |
| Nitrospirae | 0.31 | 1.84 | 95.77 | 653 | 653 |
| Spirochaetes | 0.17 | 1.00 | 96.77 | 108 | 509 |
| Deinococcus Thermus | 0.09 | 0.55 | 97.32 | 190 | 413 |
| Verrucomicrobia | 0.09 | 0.52 | 97.85 | 636 | 766 |
| Thermodesulfobacteria | 0.09 | 0.52 | 98.36 | 173 | 380 |
| Candidatus Saccharibacteria | 0.07 | 0.39 | 98.75 | 473 | 387 |
| Aquificae | 0.03 | 0.18 | 98.93 | 83 | 9 |
| Chlorobi | 0.03 | 0.18 | 99.12 | 14 | 87.3 |
| Chlamydiae | 0.03 | 0.17 | 99.29 | 15 | 84 |
| Caldiserica | 0.02 | 0.15 | 99.44 | 20 | 79 |
| Planctomycetes | 0.02 | 0.11 | 99.54 | 45 | 87.3 |
| Synergistetes | 0.02 | 0.10 | 99.64 | 16 | 54.7 |
| Tenericutes | 0.01 | 0.08 | 99.72 | 147 | 149 |
| Thaumarchaeota | 0.01 | 0.06 | 99.77 | 7 | 23.7 |
| Candidatus Cloacimonetes | 0.01 | 0.06 | 99.83 | 34 | 48.3 |
| Fibrobacteres | 0.01 | 0.05 | 99.88 | 12 | 32.7 |
| Fusobacteria | 0.01 | 0.05 | 99.93 | 35 | 54.3 |
| Armatimonadetes | 0.01 | 0.05 | 99.97 | 23 | 33 |
| Candidatus Korarchaeota | 0.00 | 0.01 | 99.98 | 0 | 3.33 |
| Deferribacteres | 0.00 | 0.01 | 99.99 | 0 | 2.67 |
| Thermotogae | 0.00 | 0.01 | 100.00 | 14 | 15.7 |
| Chrysiogenetes | 0.00 | 0.00 | 100.00 | 1 | 0 |
| Kiritimatiellaeota | 0.00 | 0.00 | 100.00 | 2 | 2.33 |
| Elusimicrobia | 0.00 | 0.00 | 100.00 | 2 | 1.67 |
| Crenarchaeota | 0.00 | 0.00 | 100.00 | 0 | 0.333 |
Fig. 3.
Dendrogram of similarity of bacterial orders in surface sediment of lagoons with fish potential at 70% accumulated contribution, according to SIMPROF analysis.
Fig. 4.
Distribution of bacterial families in surface sediment of ponds with fish potential at 70% contribution.
2. Experimental design, materials and methods
2.1. Sediment sampling
Surface sediment samples (10 cm) were collected from four inland water fish (Oncorhynchus mykiss) culture ponds in November 2019. Sediment samples from each lagoon were conditioned in airtight plastic bags and transported on ice to the Universidad Nacional de Tumbes laboratory for analysis [4].
2.2. DNA extraction, 16S rRNA genes PCR amplification and sequencing
DNA extraction was performed from 0.5 g sample using the PrestoTM Soil DNA Extraction Kit, in accordance with the manufacturer's instructions and standard protocols. DNA concentration and quality were determined using a NanodropTM ONe quantification spectrophotometer (Thermo Fisher Scientific, Massachusetts, USA) obtaining ranges from 0.3 to 88.5 ng/µl.
PCR amplification was performed using the Gene One and GE Healthcare Life Sciences kits by mixing 1 µl of the 16S rRNA F universal primer, 1 µl of the 16S rRNA R universal primer, 22 µl of the PCR mix (containing premix buffer, MgCl2, dNTPs and taqPolymerase) and 1 µl DNA sample obtaining a total reaction volume of 25 µl. Primers 27 F (5′-AGAGTTGATCCTGGCTCAG-3′) and 1392R (5′-GGTACCTTGTACGACTT-3′) were used and amplified for a product of about 1365 bp. Bacterial sequencing of the 16S rRNA amplicon was performed using the standard next-generation Illumina MiSeq [5], [6], [7], [8]. The construction of the library was carried out commercially (ADMERA HEALTH LLC, USA).
2.3. Bioinformatic analysis of sequence readings
The FASTQ files generated by the program FASTQC v0.11.9 were processed to know the length of the readings, the quality of the bases and the percentage of nucleotide bases. Subsequently, quality filtering and removal of regions of the primer and adapters present in the readings was performed using the Trimmomatic v0.39 program [9] with minimum trimming values of Q30 and trimming of readings below 30 bp. All individual reads were greater than 150,000 per isolate with a read length of 251 nucleotides and a quality value of each sequenced base greater than 30. The taxonomic analysis was performed using the program [10], based on the database minikraken_20,171,019_4GB. This program also handles multiple scripts for circular representation. Finally, operational taxonomic units were identified and abundances calculated [11,12].
2.4. Statistical analysis
Similarity percentage analysis (SIMPER) was performed to calculate the relative contribution of each taxon to the overall average dissimilarity observed between two or more groups of taxonomic assemblages. The groups were defined on the basis of a preliminary similarity profile clustering analysis (SIMPROF) of the same taxonomic occurrence data set [13]. The SIMPROF analysis allowed to test the multivariate structure within groups of samples. Square-root transformed abundances were used to calculate Bray Curtis similarities [14], showing patterns between samples determined by significant similarity measurements (p < 0.05), using clustering and ordering [15]. These analyses were performed in the Primer V7.
2.5. Nucleotide sequence access numbers
The 16S rRNA gene sequences reported in this study were sent to the GenBank database with the access number PRJNA657251 (https://www.ncbi.nlm.nih.gov/sra/PRJNA657251).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was funded by CONCYTEC-FONDECYT under the call E041–01 [contract number 76–2018- FONDECYT-BM-IADT-MU].
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.dib.2020.106228.
Appendix. Supplementary materials
References
- 1.Caporaso J.G., Lauber C.L., Walters W.A., Berg-lyons D., Huntley J., Fierer N., Owens S.M., Betley J., Fraser L., Bauer M., Gormley N., Gilbert J.A., Smith G., Knight R. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuhn R., Böllmann J., Krahl K., Bryant I.M., Martienssen M. Data on DNA gel sample load, gel electrophoresis, PCR and cost analysis. Data Br. 2018;16:732–751. doi: 10.1016/j.dib.2017.11.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mariano M., Huaman P., Mayta E., Montoya H., Chanco M.C. Contamination produced by intensive fish farming in Andean lagoons of Junin, Peru. Rev. Peru. Biol. 2011;17:137–140. [Google Scholar]
- 4.Miller D.N., Bryant J.E., Madsen E.L., Ghiorse W.C. Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl. Environ. Microbiol. 1999;65:4715–4724. doi: 10.1128/aem.65.11.4715-4724.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fadrosh D.W., Bing Ma P.G., Sengamalay N., Ott S., Brotman R.M., Ravel J. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. 2014;2:1–7. doi: 10.1186/2049-2618-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K., Schloss P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Appl. Environ. Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salipante S.J., Kawashima T., Rosenthal C., Hoogestraat D.R., Cummings L.A., Sengupta D.J., Harkins T.T., Cookson B.T., Hoffman N.G. Performance comparison of Illumina and Ion Torrent next-generation sequencing platforms for 16S rRNA-based bacterial community profiling. Appl. Environ. Microbiol. 2014;80:7583–7591. doi: 10.1128/AEM.02206-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fouhy F., Clooney A.G., Stanton C., Claesson M.J., Cotter P.D. 16S rRNA gene sequencing of mock microbial populations-impact of DNA extraction method, primer choice and sequencing platform. BMC Microbiol. 2016;16:1–13. doi: 10.1186/s12866-016-0738-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wood D.E., Salzberg S.L. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:1–12. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edgar R.C. UPARSE : highly accurate OTU sequences from microbial amplicon reads. Br. Commun. 2013;10:996–1000. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 12.Kabeer F.A., Jabir T., Krishnan K.P., Abdulla M.H. Metagenomic data of fungal community in Kongsfjorden, Arctic using Illumina next generation sequencing. Data Br. 2019;22:195–198. doi: 10.1016/j.dib.2018.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibert C., Escarguel G. PER-SIMPER—A new tool for inferring community assembly processes from taxon occurrences. Glob. Ecol. Biogeogr. 2019;28:374–385. [Google Scholar]
- 14.Somerfield P.J., Burton M., Sanderson W.G. Analyses of sublittoral macrobenthic community change in a marine nature reserve using similarity profiles (SIMPROF) Mar. Environ. Res. 2014;102:51–58. doi: 10.1016/j.marenvres.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 15.Izegaegbe J.I., Vivier L., Mzimela H.M. Spatial and temporal distribution of macrobenthic fauna of subtropical Richards Bay Harbour, South Africa. Reg. Stud. Mar. Sci. 2020;36 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.