Abstract
Storage of meristematic tissue at ultra-low temperatures offers a mean to maintain valuable genetic resources from vegetatively reproduced plants. To reveal the biology underlying cryo-stress, shoot tips of the model plant Arabidopsis thaliana were subjected to a standard preservation procedure. A transcriptomic approach was taken to describe the subsequent cellular events which occurred. The cryoprotectant treatment induced the changes in the transcript levels of genes associated with RNA processing and primary metabolism. Explants of a mutant lacking a functional copy of the transcription factor WRKY22 were compromised for recovery. A number of putative downstream targets of WRKY22 were identified, some related to phytohormone-mediated defense, to the osmotic stress response, and to development. There were also alterations in the abundance of transcript produced by genes encoding photosynthesis-related proteins. The wrky22 mutant plants developed an open stomata phenotype in response to their exposure to the cryoprotectant solution. WRKY22 probably regulates a transcriptional network during cryo-stress, linking the explant’s defense and osmotic stress responses to changes in its primary metabolism. A model is proposed linking WRKY53 and WRKY70 downstream of the action of WRKY22.
Keywords: Abiotic stress, cryoprotectant, shoot tip, stomatal closure, transcription factor, transcriptomics, ultra-low temperature
The transcription factor WRKY22 links the explant’s defense and osmotic stress response during Arabidopsis shoot tip cryo-stress.
Introduction
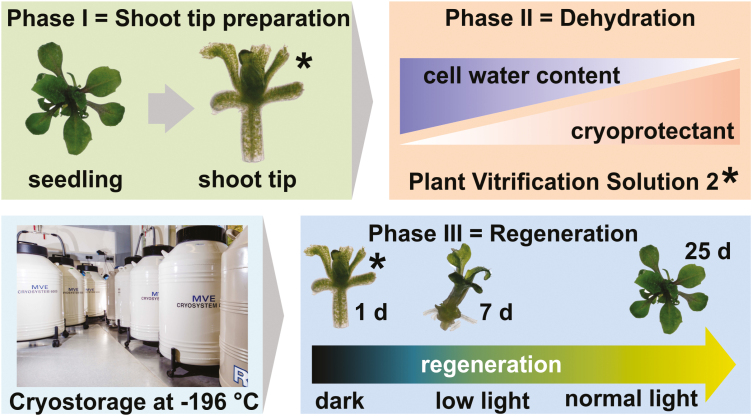
Plant biodiversity disappears rapidly as a direct impact of humankind’s use of plant natural resources. It is of tremendous importance to preserve genetic resources, which are otherwise irretrievably lost. In plant gene banks, several ex situ plant conservation and propagation techniques, starting from seed storage, in field maintenance, tissue culture, or long-term storage at ultra-low temperatures, so-called cryopreservation, were established (Engelmann and Takagi, 2000). Plant storage at ultra-low temperatures, between –140 °C and –196 °C, is frequently used in the context of maintaining plants which can only be reproduced vegetatively or to preserve selected clonal material/varieties of heterozygous plant species (Panis et al., 1996; Wu et al., 2003; Keller, 2005). The storage procedure imposes a spectrum of abiotic stresses, including wounding following the preparation of the explant from its mother plant, osmotic stress occurring as a result of the dehydration, and the chemical toxicity of the cryoprotectant itself. During cryo-storage, the explant also suffers from a rapid and large variation in temperature. Studies focusing on the regulation of the acclimation to cryo-associated abiotic stress have begun to reveal that oxidative stress is also of relevance (Basu, 2008; El-Banna et al., 2010; Ren et al., 2013, 2015; Gross et al., 2017). A range of tissues/organs are used as explants, including non-differentiated callus, cell cultures, buds, and shoot tips (Benson et al., 2007; Reed, 2008). Various protocols were optimized for a number of key gene bank species, seeking to avoid the formation of ice crystals in the tissue, as the expansion associated with freezing can result in severe mechanical damage. This goal is usually supported by treating the explant with cryoprotectants, such as Plant Vitrification Solution 2 (PVS2), before being ultra-rapidly cooled (Fig. 1) (Sakai et al., 1990; Engelmann, 2004).
Fig. 1.
Schematic overview of the long-term storage protocol. Phase I, excision of shoot tips; phase II, gradual reduction in explant hydration by treatment with cryoprotectant; phase III, recovery from cryopreservation over 25 d. PVS2, Plant Vitrification Solution 2. *Sampled for RNA-seq analysis.
Currently, there is no rational design of a protocol for cryopreservation, as the molecular framework underlying successful long-term storage of plant meristems is not understood. Shoot tips of the model plant Arabidopsis thaliana can be successfully cryopreserved and regenerated, which allows us to take advantage of the extensive genomic and genetic resources developed for this species (Stock et al., 2019). Several genes are known to be differentially transcribed during the cryo-stress of A. thaliana shoot tips: these include SQD1 (At4g33030), MBF1C (At3g24500), a gene encoding a putative aspartyl protease (At1g66180), PR5 (At1g75040), and the gene encoding the WRKY22 (At4g01250) transcription factor (TF) (Gross et al., 2017). However, their functional necessity for cryoprotection was only addressed for WRKY22 in this study.
WRKY22 itself is known to regulate upstream processes of low temperature acclimation (Chawade et al., 2007; Park et al., 2015), hypoxia-induced immunity (Hsu et al., 2013), pathogen-triggered immunity (Dong et al., 2003; Göhre et al., 2012), and leaf senescence (Zhou et al., 2011). Therefore, we hypothesize that WRKY22 has a substantial role when combinations of different abiotic stressors, such as during cryopreservation, appear during plant development.
In our study, we demonstrated that the absence of a functional copy of the WRK22 TF results in a compromised level of post-cryogenic recovery of Arabidopsis and, hence, aimed to model the functional role of WRKY22 when different cryogenic stressors were applied.
The first objective of the present research was to evaluate the molecular events occurring during the cryo-storage of wild-type (WT) A. thaliana shoot tips. This was achieved by determining its transcriptome during the preparation stage of the shoot tip explants (phase I), during their cryoprotectant treatment (phase II), and during their rewarming and recovery (phase III) (Fig. 1). The second objective was to characterize the differences between the shoot tip transcriptomes of a wrky22 knockout mutant (KO) and the WT, with the intention of detecting the downstream transcriptional effects (particularly during phase II) associated with the presence of WRKY22. A complex gene regulatory network dependent on the TF WRKY22 was unraveled. Our study demonstrates that Arabidopsis shoot tips are an excellent model to elucidate the molecular crosstalk necessary to cope with cryo-stress.
Materials and methods
Plant material and growing conditions
Seeds of the A. thaliana ecotype Columbia-0 (WT) and relevant T-DNA insertion lines (see Supplementary Table S1a at JXB online) were obtained from The Nottingham Arabidopsis Stock Centre (NASC) (http://arabidopsis.info). The T-DNA insertion lines were validated by a genomic PCR (primer sequences shown in Supplementary Table S1b). Surface-sterilized seeds were plated on solidified Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) containing 3% (w/v) sucrose, and held for 21 d under an 8 h photoperiod (light intensity 150 µmol m–2 s–1) with a day/night temperature regime of 22/20 °C. After this, the seedlings were shifted to a 22/8 °C regime for 3 weeks to provide the explants used for cryo-storage. The full-length coding sequence of WRKY22 was amplified from cDNA and cloned into the binary vector pB2GW7.0 (Karimi et al., 2002) under the control of the Cauliflower mosaic virus (CaMV) 35S promoter (35S:comp) using the Gateway system (www.thermofisher.com), according to the supplier’s protocol. The vector was introduced into the wrky22.1 mutant using the Agrobacterium-mediated floral dip method (Bechtold et al., 1993). Successfully transformed plants were identified using Basta selection, supported by a reverse transcription real-time PCR (RT–PCR) assay directed at WRKY22. The relevant primer sequences are given in Supplementary Table S1b.
Transmission electron microscopy
For comparative ultrastructural analysis, Arabidopsis apical shoot meristem tissue of 2–3 mm length of the WT and the wrky22.1 mutant was dissected out and used for microwave-assisted sample preparation. Therefore 3–5 explants of preparation stages I, II, and III have been used for aldehyde/osmium tetroxide fixation, substitution in acetone, embedding in Spurr resin, and sample polymerization as described in Supplementary Table S3. Ultrathin sectioning and ultrastructure analysis were performed as described (Daghma et al., 2011).
Cryo-storage and regeneration
The chosen protocol was previously described in detail by Stock et al. (2017). For experiments using whole seedlings instead of shoot tips, seedlings were treated as conducted for shoot tips. In brief, after an overnight immersion of the excised shoot tips in liquid MS medium (pH 5.8) containing 0.1 M sucrose, the material was partially desiccated for 20 min by immersion in MS medium (pH 5.8) containing 2 M glycerol and 0.4 M sucrose. This solution was then replaced by PVS2 [30% (w/v) glycerol, 15% (w/v) ethylene glycol, 15% (w/v) DMSO, 0.4 M sucrose in MS, pH 5.8] for 1 h at 4 °C in the dark. After treatment with liquid nitrogen, shoot tips were rewarmed and placed onto recovery medium. For regeneration, the explants were maintained in the dark for 3 d at 22 °C, then for 4 d under low-light, long-day conditions (16 h photoperiod, irradiance 20–30 µmol m–2 s–1, 22/20 °C), and finally under a normal light regime (16 h photoperiod, irradiance 150 µmol m–2 s–1, 22/20 °C) for an additional 18 d. Visual assessments were made after a recovery period of 25 d: explants showing no sign of any development were considered as ‘dead’; those which regenerated incomplete shoot/root/leaf structures or callus were classed as ‘surviving’; and the third category represented those which developed into normal plants (‘recovered’). Only recovered plantlets were included in the statistical analyses, which were based on the Win Fisher test. The values reported here represent the mean of three replicates, each of which comprised a group of 30 shoot tips.
RNA extraction, cDNA synthesis, and quantitative RT–PCR
RNA extraction from 3–5 shoot tips at each of the three cryo-stress phases as defined in Fig. 1 (at the end of phases I, II, and III), was performed using an RNeasy Plus Micro Kit (Qiagen GmbH, Hilden, Germany). A 0.5 µg aliquot of DNase I-treated RNA was used as the template for synthesis of the first cDNA strand, using Maxima Reverse Transcriptase, primed by oligo(dT)18 (Thermo Fisher Scientific, Waltham, MA, USA). The resulting cDNAs were subjected to qRT–PCRs driven by a variety of gene-specific primers (sequences given in Supplementary Table S1b) in reactions based on SsoAdvanced™ Universal SYBR® Green Supermix (BioRad Laboratories, Hercules, CA, USA). The amplifications were run on a LightCycler® 480 Real Time PCR System (Roche Diagnostics GmbH, Mannheim, Germany), with three technical replicates.
For experiments using whole Arabidopsis seedlings, ~30 mg of Arabidopsis plant material was harvested, immediately frozen in liquid nitrogen, and ground by vortexing four times with four steel beads. Total RNA was extracted using NucleoSpin RNA Plant Kits and DNase I treatment according to the manufacturer’s instructions (Macherey-Nagel, Germany). cDNA was synthesized from 1 µg of total RNA using MuLV-Reverse Transcriptase (Fermentas, Germany) in a total volume of 20 µl and diluted to 1:20 with nuclease-free water. qRT–PCR was performed in a 384-well thermocycler (CFX384 Touch™ Real-Time PCR Detection System, Bio-Rad) using the GoTaq qPCR Mastermix (Promega, USA). Seven identically treated biological replicates were analyzed.
The resulting data were analyzed using QBASEPLUS v2.3 software (Biogazelle, Ghent, Belgium),employing Clath (At5g46630) and TIP41 (At4g34270) (Czechowski et al., 2005) as reference genes (primer sequences are given in Supplementary Table S1b). Primer specificity was assessed by inspection of a melting curve derived after 40 amplification cycles. For the generation of the standard curves, aliquots of all cDNAs per time point were combined into a mixture. This mixture was serially diluted by 2-fold dilutions down to 1:64 with nuclease-free water to generate standard curve templates and to determine PCR efficiencies for each primer pair.
RNA-seq analysis
For the purpose of the RNA-seq analysis, RNA was extracted using an RNeasy Plant Mini Kit (Qiagen) from a bulk of 100 shoot tips per replicate (three) per sampling point from both WT and wrky22.1 mutant shoot tips and treated with DNase. For mRNA purification and poly(A) selection, the Illumina TruSeq RNA Sample Preparation v2 Kit (Illumina, San Diego, CA, USA) was used. Library preparation was performed using the ScriptSeq™ v2 RNA-Seq Library Preparation Kit (Epicentre, Madison, WI, USA) following the manufacturer’s protocol. Quality assessment of the libraries was done using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Cluster generation of the prepared libraries was performed using the cBot (Illumina) and TruSeq SR Cluster Kit v3-cBot-HS (Illumina) following the manufacturer’s instructions. The concentration of libraries loaded in the flowcells was 12 pM, followed by sequencing on a HiSeq 2500 instrument with the TruSeq SBS Kit v3-HS (Illumina) for 50 cycles. Image analysis and base calling were performed using the Illumina pipeline v 1.8.
Accession numbers
The RNA-seq data from this article have been deposited in the European Nucleotide Archive (http://www.ebi.ac.uk/ena) with the accession number PRJEB22967.
Read mapping and gene expression profiling, GO term enrichment analysis, and MapMan functional annotation
Single end reads of triplicated WT and KO samples were mapped onto representative A. thaliana transcripts (TAIR v10; https://www.arabidopsis.org/) with kallisto (-l 190, -s 20) (Bray et al., 2016). Transcript counts and normalized transcripts per million reads (tpm) were combined and analyzed using R software (www.r-project.org). Differentially expressed genes (DEGs) were determined using the Bioconductor packager edgeR (www.bioconductor.org/packages/release/bioc/html/edgeR.html) (Robinson et al., 2010) including multiple hypothesis testing correction (Bonferroni, 1936) to avoid false positives at the possible expense of power. Gene annotations and ontology were retrieved from TAIR v10 and functional annotations from the Mapman repository (http://mapman.gabipd.org/) (see Supplementary Dataset 3 at JXB online). Gene Ontology (GO) term enrichment analyses were conducted with the Bioconductor package topGO (www.bioconductor.org/packages/devel/bioc/html/topGO.html) (Alexa and Rahnenfuhrer, 2010) based on Fisher’s exact test (Fisher, 1922).
Estimation of stomatal density and aperture
A method adapted from Li et al. (2013) was used to estimate stomatal density and aperture from leaves detached from 4-week-old WT, wrky22.1, and wrky22.2 mutant plants. Plants of each of the WT and the wrky22.1 and wrky22.2 mutants were soil grown under an 8 h photoperiod (light intensity 150 µmol m–2 s–1) with a day/night temperature regime of 22/20 °C for 4 weeks. Estimates of stomatal closure were based on observations taken from three biological replicates. The leaves were floated in 30 mM KCl, 10 mM MES-KOH (pH 6.1) under 150 µmol m–2 s–1 light for 2 h at room temperature, then in the same buffer containing either 10 µM abscisic acid (ABA) for 2 h at room temperature, or PVS2 at 4 °C for 1 h in the dark. Stomatal aperture was represented by the ratio between the stomatal width and length, obtained from a sample of 80 stomata. Stomatal density and aperture were recorded with a Keyence Digital Microscope VHX-5000 (KEYENCE GmbH, Neu-Isenburg, Germany).
Drought stress experiment
A set of 40 plants of each of the WT or the wrky22.1 or wrky22.2 mutants was soil grown under an 8 h photoperiod (light intensity 150 µmol m–2 s–1) with a day/night temperature regime of 22/20 °C for 4 weeks, then transferred into a cabinet delivering an 8 h photoperiod (light intensity 120 µmol m–2 s–1), a temperature regime of 22/20 °C, and a relative humidity of 40%. Each pot was initially well watered, after which water was withheld for 0–21 d. At each sampling point, the FW of three plants per genotype was obtained and their rosette diameter measured. Soil moisture was determined using a HH2 Moisture Meter (DELTA-T DEVICES, Cambridge, UK).
Promoter in silico analysis
Prediction of the promoter region and putative cis-binding elements for target genes of WRKY22 was conducted using http://PlantPAN2.itps.ncku.edu.tw.
Results
The inactivation of WRKY22 compromises the regrowth of cryopreserved explants
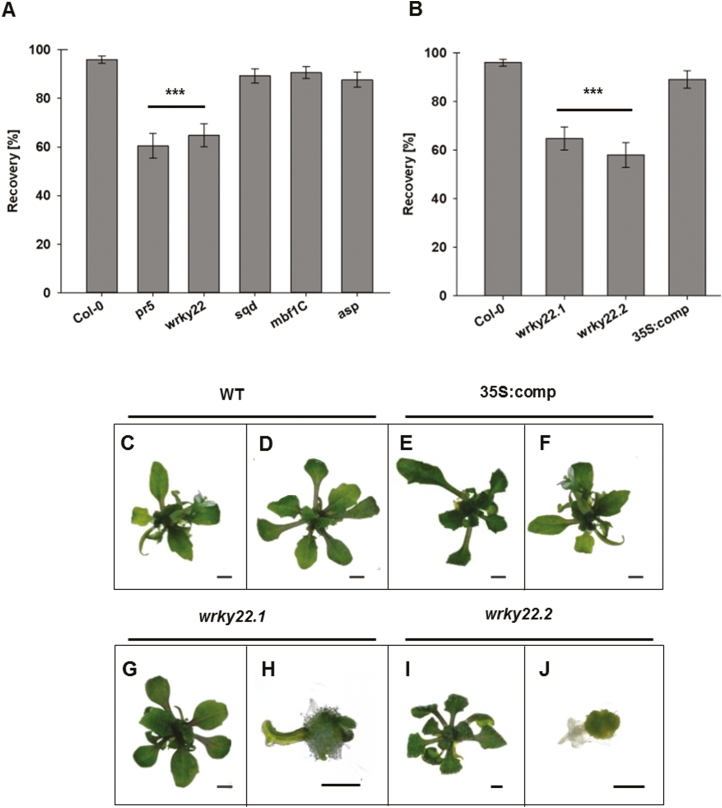
The effect of knocking out the genes At4g33030 (SQD1), At3g24500 (MBF1C), At1g66180 (ASP), At1g75040 (PR5), and At4g01250 (WRKY22) on the performance of cryopreserved explants was explored by exposing T-DNA KO lines for each gene to the cryopreservation protocol. The regeneration capacity of A. thaliana shoot tips was analyzed after a cryo-storage of 30 min. As at –196 °C metabolic activity ceases, preservation of the shoot tips will be similar between a short-term (30 min) and a long-term (>2 years) storage. The vast majority (98%) of WT shoot tips regenerated into viable plantlets after cryogenic treatment, as did explants from the mutant lines involving At4g33030, At3g24500, and At1g66180. In contrast, the lack of a fully functional copy of either WRKY22 or PR5 resulted in a significantly impaired level of regeneration (Fig. 2A). As little is known about the molecular mechanism relevant for successful cryopreservation, we focused on the role of the TF WRKY22. By testing two independent T-DNA insertion mutants (wrky22.1 and wrky22.2), it was shown that the loss of function of WRKY22 was responsible for the observed loss in regeneration. In both cases, the genes’ highly conserved WRKY domain sequence was disrupted by a T-DNA sequence, resulting in a highly reduced abundance of WRKY22 transcripts (Supplementary Fig. S1). The regeneration rate was reduced from 98% for WT explants to 60% for those derived from each of the mutants (Fig. 2B). Introducing a copy of WT WRKY22 driven by the CaMV 35S promoter (35S:comp) into the wrky22.1 mutant resulted in a WRKY22 transcript level similar to that measured in the WT explant and restored the WT phenotype (Fig. 2B; Supplementary Fig. S1). Regenerated plantlets from the WT (Fig. 2C, D) and 35S:comp (Fig. 2E, F) resembled one another with respect to their rosette leaves, roots, and shoots, while the wrky22 plantlets exhibited a distinct phenotype: 60% of the plantlets retained a WT phenotype (Fig. 2G, I), while 40% developed incomplete leaves and roots, produced some callus material, or stayed green without any further development (Fig. 2H,J).
Fig. 2.
Regrowth of Arabidopsis T-DNA mutant explants. The proportion of recovered plantlets after a 25 d recovery period of (A) the WT and mutants pr5, wrky22, sqd1, mbf1C, and asp, and (B) the wrky22.1 and wrky22.2 mutants and a transgenic wrky22.1 mutant plant harboring the transgene 35S:comp. The bars represent the mean % of successful regeneration, with its associated SD. The performance of the mutants was compared with that of the WT using the Win Fisher test (***P≤0.001, n≥90). (C–J) The appearance of recovered plantlets derived from shoot tips of (C, D) WT, (E, F) transgenic wrky22.1 mutant plant harboring the 35S:comp, (G, H) wrky22.1 mutant, and (I, J) wrky22.2 mutant showing (C–G, I) regenerated and (H, J) surviving but non-recovering shoot tips. Scale bar=1 mm.
PVS2 treatment induces WRKY22 abundance and ultrastructural changes
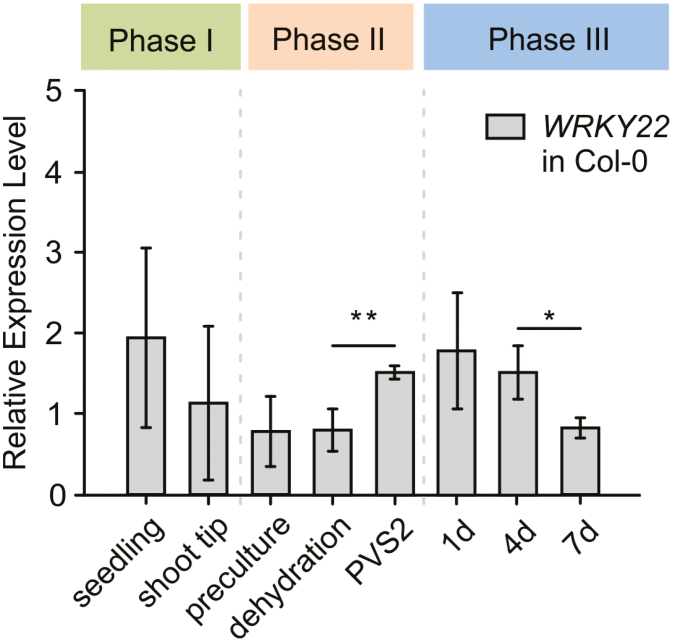
qRT–PCR of WT explants revealed that WRKY22 transcription increased significantly over phase II, and decreased after the fourth day of phase III (Fig. 3), indicating that the TF is likely to be involved in the response to PVS2-induced stress and the early phase of regeneration.
Fig. 3.
Relative abundance of WRKY22 transcripts present in Arabidopsis WT explants. Six-week-old Columbia seedlings were sampled at each stages of the cryopreservation/acclimation process. After shoot tip preparation (phase I), stepwise cellular dehydration and cryoprotection using Plant Vitrification Solution 2 (PVS2, phase II), and post-cryogenic recovery (phase III), transcript levels were detected by qRT–PCR using specific primers. The data represent means ±SD from four independent biological replicates (n=4). ** and *: means differ at P≤0.01 and ≤0.05, respectively, using one-way ANOVA followed by Holm–Sidak post-hoc test.
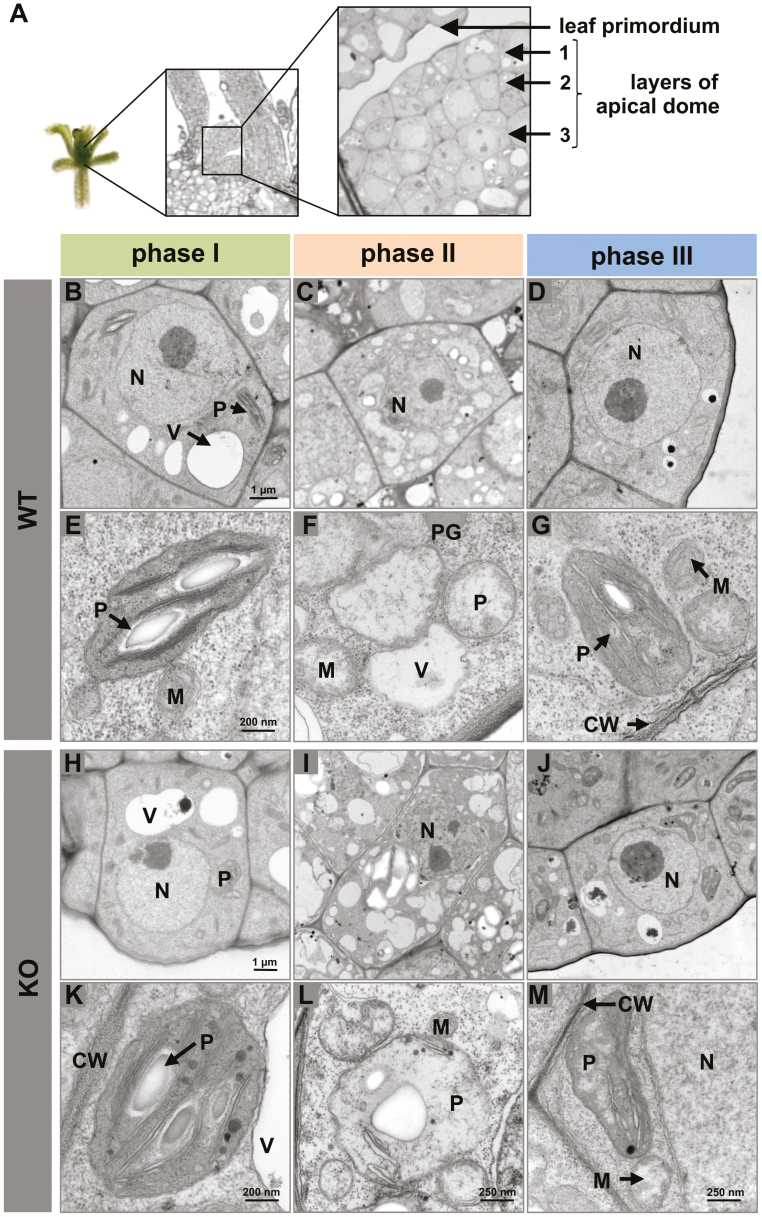
Ultrastructure analysis of meristematic (Fig. 4) cells revealed that cell size and vacuoles appear to be reduced as an effect of PVS2 treatment (phase II) (Fig. 4C). Cell organelles, most pronounced for plastids and mitochondria, either increased in size or began to degrade, characterized by formation of plastoglobuli (Fig. 4F). In comparison, WT cells of phase I (Fig. 4B, E) and phase III (Fig. 4D, G) look similar. Nuclei and vacuoles of the cells appeared prominent and the cytoplasm of cells was homogeneous and even structured. During the recovery period (phase III), all symptoms associated with dehydration and organelle degradation disappeared (Fig. 4D, G). The ultrastructure of the wrky22.1 mutant tissue in all three phases was indistinguishable from that of the WT cells (Fig. 4H–M).
Fig. 4.
The effect of cryo-stress on the cellular ultrastructure of the shoot tip of the WT and the wrky22.1 mutant. (A) Schematic view of the shoot tip, showing the first three layers of the apical dome. (B–M) Transmission electron micrographs of meristematic cells visualized after phases I–III. (B–G) WT, (H–M) wrky22.1. CW, cell wall; M, mitochondrion; N, nucleus; P, plastid; PG, plastoglobulus; V, vacuole.
RNA processing acts as a key regulator during cryoprotectant treatment
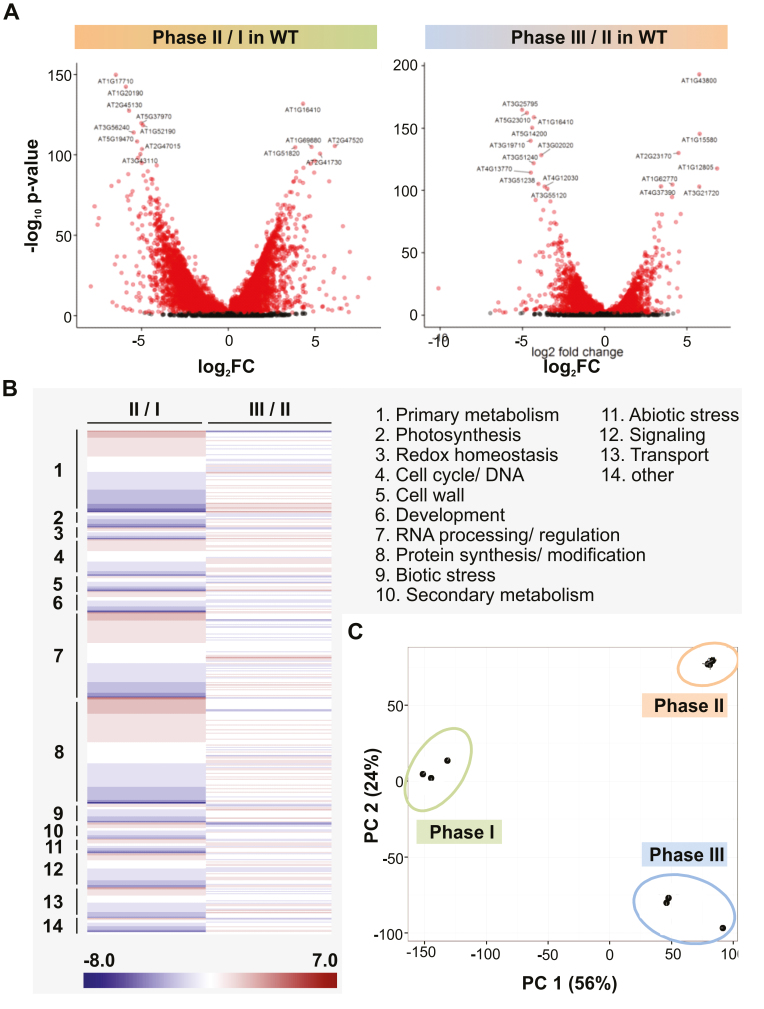
The comparison between the transcriptomes of the WT explants sampled in phases I and II identified 12 067 DEGs, and that between phases II and III 6349 DEGs (Fig. 5A). The two sets of DEGs were assigned to MapMan bins, as depicted in Fig. 5B. A principal component analysis showed a clear separation between the three phases (Fig. 5C). The GO enrichment analysis established that the PVS2 treatment had a major positive impact on the abundance of transcript generated by genes encoding proteins involved in RNA processing and methylation, mitochondrial processes, DNA modification, and nuclear targeting, while the major classes of genes negatively impacted were related to photosynthesis (in particular the light response and chlorophyll synthesis) as well as the metabolism of saccharides, lipids, fatty acids, and amino acids (Fig. 6A; Supplementary Dataset 1). The MapMan analysis confirmed the conclusions drawn from the GO term enrichment analysis: transcripts encoding proteins involved in ribosomal protein synthesis became notably more abundant in phase II than in phase I, but this difference was not apparent between phases II and III (Fig. 6C; Supplementary Table S2). Genes encoding components of RNA processing and ribosomal protein synthesis, as well as metabolism and photosynthesis, were represented in the set of the most highly up- and down-regulated genes (Tables 1, 2; Supplementary Dataset 2). The transcriptomic data-based conclusions were validated for the three selected RNA processing genes NOP56 (At1g56110), NOP58 (At3g05060), and EBP2 (At3g22660) using qRT–PCR: in each case, transcript abundance was boosted by the PVS2 treatment (Supplementary Fig. S2).
Fig. 5.
The transcriptome of WT shoot tip explants at the end of phases I–III. (A) The analysis identified 12 067 genes as changed with respect to their transcript abundance between phases I and II, and 6349 between phases II and III (P-value <0.01 after multiple hypothesis correction). Genes associated with a P value <10–100 are labeled with their AGI code. (B) The MapMan bins of the DEGs identified in the contrasts phases II versus I and III versus II. Red indicates increased abundance and blue decreased abundance, with the color intensity reflecting the fold of differential gene expression. (C) A principal component analysis confirms the difference between the three phases.
Fig. 6.
GO term enrichment and MapMan functional assignment of the WT transcriptome. Enriched GO terms among the regulated genes in the contrast (A) phase II versus I and (B) phase III versus II; enriched GO terms among genes with higher abundance are shown in red and among lower abundance in blue. (C) MapMan mapping of RNA–protein synthesis. Each square represents the transcription of a single gene within a given pathway. Hochberg-corrected transcripts with higher abundance are shown in red, and lower abundance in blue. The color intensity reflects the fold of differential gene expression.
Table 1.
The 50 most highly up-regulated genes in the WT explants identified in the contrast phase II versus I
| Locus | FC | Gene name | Mapman functional description | |
|---|---|---|---|---|
| 1* | AT2G47520 | 6.1 | HYPOXIA RESPONSIVE (ERF) 2 (HRE2) | RNA.regulation of transcription |
| 2 | AT4G12490 | 6.1 | AZI3 | misc.protease inhibitor |
| 3 | AT3G46280 | 5.0 | kinase-like protein | signalling.receptor kinases |
| 4 | AT2G26150 | 4.9 | HEAT SHOCK TRANSCRIPTION FACTOR A2 | stress.abiotic.heat |
| 5 | AT1G10585 | 4.9 | basic helix-loop-helix DNA-binding superfamily protein | RNA.regulation of transcription |
| 6 | AT1G69880 | 4.8 | THIOREDOXIN H-TYPE 8 | redox.thioredoxin |
| 7 | AT5G59240 | 4.7 | Ribosomal protein S8e family protein | protein.synthesis.ribosomal protein |
| 8 | AT3G17609 | 4.7 | HY5-HOMOLOG | RNA.regulation of transcription |
| 9 | AT4G22470 | 4.6 | Protease inhibitor/lipid-transfer protein | misc.protease inhibitor |
| 10 | AT5G51440 | 4.5 | HSP20-like chaperones superfamily protein | stress.abiotic.heat |
| 11 | AT1G05680 | 4.4 | UGT74E2 | hormone metabolism.salicylic acid |
| 12* | AT4G06746 | 4.4 | RELATED TO AP2 9 (RAP2.9) | RNA.regulation of transcription |
| 13** | AT3G51240 | 4.4 | FLAVANONE 3-HYDROXYLASE | secondary metabolism.flavonoids |
| 14 | AT1G64220 | 4.4 | TRANSLOCASE OF OUTER MEMBRANE 7-2 | transport mitochondrial membrane |
| 15 | AT1G17180 | 4.4 | GLUTATHIONE S-TRANSFERASE TAU 25 | misc.glutathione S transferases |
| 16 | AT3G09680 | 4.3 | Ribosomal protein S12/S23 family protein | protein.synthesis.ribosomal protein |
| 17** | AT1G16410 | 4.3 | CYTOCHROME P450 79F | secondary metabolism.sulfur-containing |
| 18 | AT2G16060 | 4.1 | HEMOGLOBIN 1 | redox.heme |
| 19** | AT5G13930 | 4.1 | CHALCONE SYNTHASE | secondary metabolism.flavonoids |
| 20 | AT5G14200 | 4.0 | ISOPROPYLMALATE DEHYDROGENASE 1 | amino acid metabolism.synthesis |
| 21 | AT5G40040 | 4.0 | 60S acidic ribosomal protein family | protein.synthesis.ribosomal protein |
| 22 | AT5G39580 | 4.0 | Peroxidase superfamily protein | misc.peroxidases |
| 23 | AT2G15620 | 3.9 | NITRITE REDUCTASE 1 | N-metabolism.nitrate metabolism |
| 24 | AT3G19710 | 3.9 | BRANCHED-CHAIN AMINOTRANSFERASE4 | amino acid metabolism.synthesis |
| 25 | AT3G12860 | 3.9 | NOP56-like pre RNA processing ribonucleoprotein | protein.synthesis.ribosome biogenesis |
| 26 | AT5G41670 | 3.9 | 6-phosphogluconate dehydrogenase family protein | OPP.oxidative |
| 27 | AT3G46230 | 3.9 | HSP17.4 | stress.abiotic.heat |
| 28** | AT5G07990 | 3.9 | CYTOCHROME P450 75B1 | secondary metabolism.flavonoids |
| 29 | AT1G32880 | 3.9 | ARM repeat superfamily protein | protein.targeting.nucleus |
| 30 | AT1G51820 | 3.9 | Leucine-rich repeat protein kinase family protein | signalling.receptor kinases.misc |
| 31 | AT1G58684 | 3.8 | Ribosomal protein S5 family protein | protein.synthesis.ribosomal protein |
| 32 | AT1G58983 | 3.8 | Ribosomal protein S5 family protein | protein.synthesis.ribosomal protein |
| 33 | AT1G02820 | 3.8 | LEA3 | development |
| 34 | AT3G06900 | 3.8 | U4 SMALL NUCLEOLAR RNA2 | RNA.processing |
| 35 | AT5G40850 | 3.8 | UROPHORPHYRIN METHYLASE 1 | tetrapyrrole synthesis |
| 36 | AT1G51850 | 3.8 | Leucine-rich repeat protein kinase family protein | signalling.receptor kinases.misc |
| 37 | AT4G33070 | 3.7 | ATPDC1 | fermentation.PDC |
| 38 | AT1G24280 | 3.7 | G6PD3 | OPP.oxidative |
| 39** | AT5G23010 | 3.7 | 2-ISOPROPYLMALATE SYNTHASE 3 | secondary metabolism.sulfur-containing |
| 40 | AT4G12480 | 3.7 | PEARLI 1 | misc.protease inhibitor |
| 41 | AT1G23410 | 3.7 | Ribosomal protein S27a | protein.synthesis.ribosomal protein |
| 42 | AT4G12500 | 3.6 | Bifunctional inhibitor/lipid-transfer protein | misc.protease inhibitor |
| 43 | AT1G78050 | 3.6 | PGM | glycolysis.unclear |
| 44 | AT1G14120 | 3.6 | AUXIN OXIDASE | misc.oxidases |
| 45 | AT5G13490 | 3.6 | ADP/ATP CARRIER 2 | transport.unspecified cations |
| 46 | AT3G02020 | 3.5 | ASPARTATE KINASE 3 | amino acid metabolism |
| 47 | AT2G03230 | 3.5 | GCK domain-containing protein | not assigned.unknown |
| 48 | AT4G25630 | 3.5 | FIBRILLARIN 2 | protein.synthesis.ribosome biogenesis |
| 49 | AT5G27120 | 3.5 | NOP58-like pre RNA processing ribonucleoprotein | RNA.regulation of transcription |
| 50 | AT5G53290 | 3.5 | CYTOKININ RESPONSE FACTOR 3 | RNA.regulation of transcription |
MapMan bins consistent with GO term enrichment are shown in bold. Genes labeled with an asterisk have been associated in the literature with either the drought stress response (*) or products of secondary metabolism (**).
Table 2.
The 50 most highly down-regulated genes in the WT explants identified in the contrast phase II versus I
| Locus | FC | Gene Name | Mapman Functional Description | |
|---|---|---|---|---|
| 1 | AT2G33830 | –7.7 | DORMANCY ASSOCIATED GENE 2 | hormone metabolism |
| 2 | AT1G31580 | –7.6 | ECS1 | stress.biotic |
| 3 | AT1G56600 | –7.5 | GALACTINOL SYNTHASE 2 | minor CHO metabolism |
| 4* | AT1G20440 | –6.5 | COLD-REGULATED 47 | stress.abiotic.unspecified |
| 5 | AT1G17710 | –6.5 | Pyridoxal phosphate phosphatase-related | misc.acid and other phosphatases |
| 6 | AT1G26945 | –6.4 | PACLOBUTRAZOL RESISTANCE 6 | not assigned.unknown |
| 7 | AT5G45890 | –6.2 | SENESCENCE-ASSOCIATED GENE 12 | protein.degradation |
| 8 | AT1G20190 | –5.9 | EXPANSIN 11 | cell wall.modification |
| 9* | AT1G29395 | –5.9 | COLD REGULATED 314 INNER MEMBRANE 1 | not assigned.no ontology |
| 10 | AT1G52690 | –5.9 | LATE EMBRYOGENESIS ABUNDANT | development |
| 11 | AT3G09922 | –5.8 | INDUCED BY PHOSPHATE STARVATION1 | not assigned.unknown |
| 12 | AT1G56220 | –5.8 | Dormancy/auxin associated family protein | development.unspecified |
| 13 | AT2G45130 | –5.7 | SPX DOMAIN GENE 3 | stress.abiotic |
| 14* | AT1G73330 | –5.7 | DROUGHT-REPRESSED 4 | stress.biotic |
| 15 | AT3G27690 | –5.6 | LHCB2.3 | PS.lightreaction.photosystem II |
| 16 | AT5G24490 | –5.6 | 30S ribosomal protein | protein.synthesis.ribosomal protein |
| 17 | AT3G01500 | –5.5 | SALICYLIC ACID-BINDING PROTEIN 3 | TCA/org transformation |
| 18 | AT3G15450 | –5.5 | Aluminium induced protein | hormone metabolism |
| 19 | AT3G56240 | –5.5 | COPPER CHAPERONE | metal handling |
| 20 | AT5G14565 | –5.3 | MICRORNA398C | micro RNA, natural antisense |
| 21 | AT1G09350 | –5.3 | GALACTINOL SYNTHASE 3 | minor CHO metabolism |
| 22 | AT5G19470 | –5.3 | NUDIX HYDROLASE HOMOLOG 24 | nucleotide metabolism |
| 23 | AT3G26180 | –5.2 | CYP71B20 | misc.cytochrome P450 |
| 24 | AT3G02040 | –5.2 | SENESCENCE-RELATED GENE 3 | lipid metabolism |
| 25* | AT1G20450 | –5.2 | EARLY RESPONSIVE TO DEHYDRATION 10 | stress.abiotic.unspecified |
| 26 | AT2G41870 | –5.2 | Remorin family protein | RNA.regulation of transcription |
| 27 | AT3G16670 | –5.2 | Pollen Ole e 1 allergen | not assigned.unknown |
| 28 | AT3G26740 | –5.2 | CCR-LIKE | signalling.light |
| 29 | AT1G80920 | –5.2 | TOC12 | stress.abiotic.heat |
| 30 | AT5G06760 | –5.1 | LATE EMBRYOGENESIS ABUNDANT 4–5 | development |
| 31 | AT3G62550 | –5.1 | Adenine nucleotide alpha hydrolases-like protein | hormone metabolism |
| 32 | AT3G55240 | –5.1 | Protein coding | not assigned.unknown |
| 33 | AT5G37970 | –5.0 | S-adenosyl-L-methionine-dependent methyltransferases superfamily protein | hormone metabolism.salicylic acid |
| 34* | AT4G25490 | –5.0 | DRE BINDING PROTEIN 1B (CBF1) | RNA.regulation of transcription |
| 35 | AT3G63210 | –5.0 | MEDIATOR OF ABA-REGULATED DORMANCY 1 | hormone metabolism |
| 36 | AT2G47015 | –5.0 | MICRORNA408 | micro RNA, natural antisense |
| 37 | AT1G75380 | –5.0 | BIFUNCTIONAL NUCLEASE IN BASAL DEFENSE RESPONSE 1 | stress.abiotic.touch/wounding |
| 38 | AT1G67265 | –4.9 | ROTUNDIFOLIA LIKE 21 | development.unspecified |
| 39 | AT1G52190 | –4.9 | NITRATE TRANSPORTER 1.11 | transport.peptides and oligopeptides |
| 40 | AT5G49360 | –4.9 | BETA-XYLOSIDASE 1 | cell wall.degradation |
| 41 | AT1G01470 | –4.9 | LATE EMBRYOGENESIS ABUNDANT 14 | development |
| 42 | AT1G23730 | –4.9 | BETA CARBONIC ANHYDRASE 3 | TCA/org transformation |
| 43 | AT1G79040 | –4.9 | PHOTOSYSTEM II SUBUNIT R | PS.lightreaction.photosystem II |
| 44 | AT1G18870 | –4.9 | ISOCHORISMATE SYNTHASE 2 | Co-factor and vitamine metabolism |
| 45 | AT2G17040 | –4.9 | NAC DOMAIN CONTAINING PROTEIN 36 | development.unspecified |
| 46 | AT1G20620 | –4.8 | CATALASE 3 | redox.dismutases and catalases |
| 47 | AT1G73540 | –4.8 | NUDIX HYDROLASE HOMOLOG 21 | nucleotide metabolism |
| 48 | AT5G39520 | –4.8 | hypothetical protein | not assigned.unknown |
| 49 | AT1G11530 | –4.7 | ATCXXS1 | redox.thioredoxin |
| 50 | AT1G28330 | –4.7 | DORMANCY-ASSOCIATED PROTEIN 1 | development.unspecified |
MapMan bins consistent with GO term enrichment are shown in bold. Genes labeled with an asterisk have been associated in the literature with either the drought stress response (*) or products of secondary metabolism (**)
The DEGs identified included five genes known to be inducible by drought stress [ERF2 (At4g06746), RAP2.9 (At2g47520), DR4 (At1g73330), ERD10 (At1g20450), and CBF1 (At4g25490)], along with two low temperature stress-inducible genes [COR47 (At1g20440) and COR413IM1 (At1g29395)]: these are marked by a single asterisk in Tables 1 and 2. The set of genes with higher abundance included a number associated with the defense response, in particular related to products of secondary metabolism (marked with a double asterisk in Table 1).
A GO term enrichment analysis of the set of DEGs in phase III indicated that genes with higher abundance could be assigned to terms of development, cell cycling, protein modification/ubiquitination, and DNA modification/replication, while apoptosis and defense were suppressed (Fig. 6B; Supplementary Dataset S1). Consistent with MapMan analysis, genes showing higher abundance were prominently related to auxin-mediated cell growth, and those with lower abundance to products of secondary metabolism (Supplementary Fig. S3).
Cryoprotectant treatment affects genes relevant for photosynthesis in the wrky22 mutant shoot tip explants
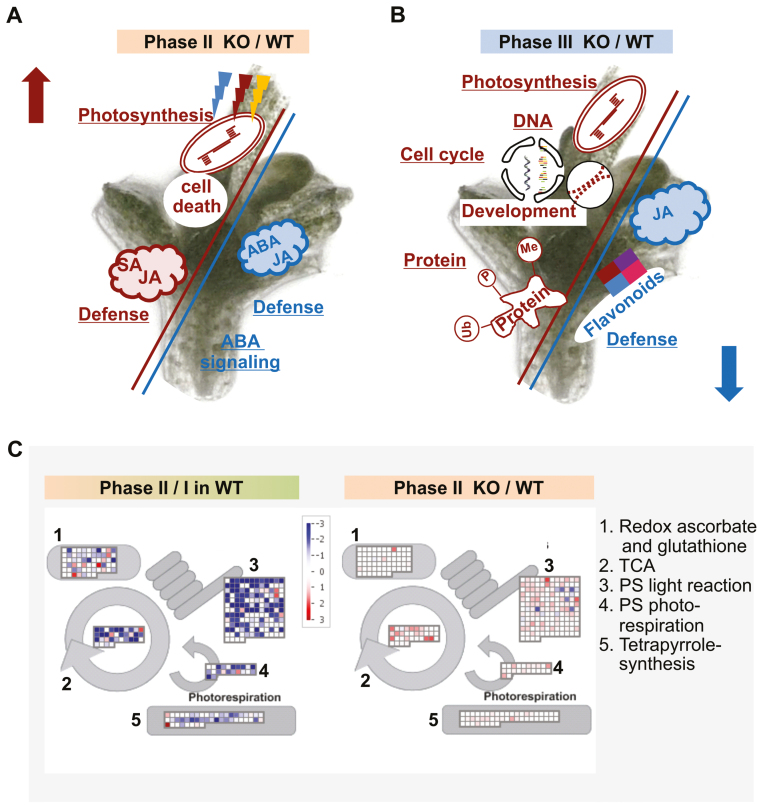
In all, 124 genes were differentially transcribed between the WT and the wrky22 mutant when the explants were sampled at the end of phase I, as were 2599 at the end of phase II and 1119 at the end of phase III. A principal component analysis clearly distinguished the three phases, and highlighted the genotypic difference between the WT and the wrky22 mutant in phases II and III (Supplementary Fig. S4). Effects of WRKY22 were marginal during shoot tip preparation. The most over-represented category of DEGs was associated with secondary cell wall synthesis, in particular genes encoding PROLINE-RICH EXTENSIN-LIKE FAMILY PROTEINS (Supplementary Datasets S1, S2). These findings suggested that the wrky22 mutant shoot tips were compromised with respect to the strength of their secondary cell walls. Transcriptome changes associated with cryoprotectant treatment in phase II of the wrky22 mutant explants showed the high importance of photosynthesis and subsequently an adapted response to light, scavenging of reactive oxygen species (ROS), and apoptosis. The MapMan analysis confirmed the importance of photosynthesis during phase II. Transcripts relevant for PSI and PSII and the Calvin cycle (Fig. 7C; Supplementary Fig. S5) seemed to be key aspects in the wrky22 mutant acclimation response. These findings were validated for the three selected genes RCA (At2g39730), PSAN (At5g64040), and RBCS3B (At5g38410) using qRT–PCR. The analysis showed reduced transcript abundance during phase II compared with phase I in both the wrky22 mutant and the WT explants, and higher transcript expression in the wrky22 mutant than in the WT during phase II (Supplementary Fig. S2).
Fig. 7.
GO term enrichment and MapMan functional assignment of the wrky22 mutant transcriptome. Enriched GO terms among the regulated genes in the contrast (A) phase II and (B) phase III in the wrky22.1 mutant over the WT. Enriched GO terms among genes with higher abundance are shown in red and with lower abundance in blue. (C) MapMan mapping of primary metabolism. Each square represents the transcription of a single gene within a given pathway. Hochberg-corrected transcripts with higher abundance are shown in red, and lower abundance in blue. The color intensity reflects the fold of differential gene expression. TCA, tricarboxylic acid cycle.
Genes encoding the salicylic acid (SA)- and jasmonic acid (JA)-regulated defense response, along with products of secondary metabolism, were over-represented in both the high- and low-abundance categories, while processes relying on ABA signaling were suppressed (Fig. 7A; Supplementary Dataset S1). During phase III, development, DNA/RNA modification, protein modification, and photosynthesis were all promoted in the wrky22 mutant, while the JA-mediated defense response and flavonoid synthesis were suppressed (Fig. 7B; Supplementary Dataset S1).
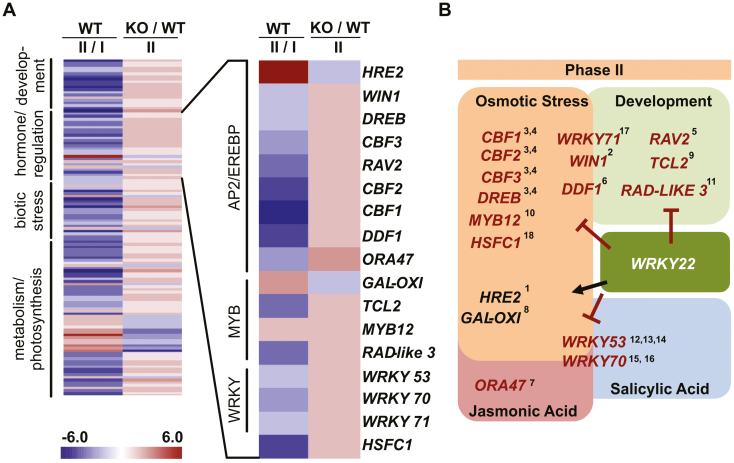
Changes in the mutant transcriptome indicated that WRKY22 is involved in phytohormone-mediated drought and defense acclimation through crosstalk with assorted transcription factors
The set of DEGs (selected on the basis of a log2 fold change threshold in transcript abundance of 1.5) between phases I and II for the WT, and between the wrky22 mutant and the WT during phase II, was assembled to identify potential targets of WRKY22. Of these, 145 were assigned to MapMan bins associated with development, hormone and transcript regulation, biotic stress, and photosynthesis (Fig. 8A). The group of DEGs assigned to the hormone and regulation category included a number of members of the AP2-EREBP, MYB, and WRKY TF families. Four of these [WRKY71 (At1g29860), WIN1 (At5g11190), WRKY53 (At4g23810), and WRKY70 (At3g56400) are known to be inducible by more than one stress agent. The products of certain dehydration-responsive binding protein/C-repeat binding factor (CBF)-encoding genes, as well as those of DDF 1 (At1g12610), HRE2 (At2g47520), and GAL-OXI (At3g27220) are known to be involved in the regulation of the osmotic stress response, while those of RAV2 (At1g68840), TCL2 (At2g30424), and RAD-LIKE3 (At4g36570) control two or more developmental processes; finally, the product of ORA47 (At1g74930) was identified as acting in the JA-regulated defense response (see references in Fig. 8B). The presence of WRKY22 resulted in the suppression of most of these genes (the exceptions were GAL-OXI and HRE2), which supported the existence of crosstalk during osmotic stress acclimation between WRKY22 and members of the AP2-EREBP, MYB, and WRKY families. Overall, 25 putative interaction partners of WRKY22 in response to osmotic stress were identified. All of the identified targets, except RAB18 (AT5G66400), contain several WRKY22-binding motifs (W-boxes) in their promoter region, confirming a crosstalk between the TFs and target genes (Table 3). In silico analysis of the WRKY70 promotor sequence showed a W box (C/T)TGAC(T/C) motif, within the region 1000 bp upstream from the WRKY70 genomic DNA sequence known to interact with WRKY22 (Supplementary Fig. S6A). We were able to validate the transcriptomic data-based conclusions for the WRKY70 (At3g56400) TF, using qRT–PCR on Arabidopsis seedlings, which have been exposed to a cryoprotectant and a cooling treatment. In each case, WRKY70 transcript abundance was increased in the wrky22.1 and wrky22.2 T-DNA insertion lines compared with WT seedlings. After PVS2 treatment, a significant increase of WRKY70 transcript was observed in mutant seedlings (Supplementary Fig. S6B). In contrast to the expression of WRKY70, WRKY53 showed no generally increased transcript abundance in different samples. This may be explained by the fact that whole Arabidopsis seedlings and not only meristem-including shoot tips have been used in the validation experiment.
Fig. 8.
Crosstalk between transcription factors associated with WRKY22 activity. (A) A heatmap identifying putative interaction partners with WRKY22 during phase II belonging to the AP2-EREBP, MYB, and WRKY transcription factor family. Genes differentially expressed between phases II and I in WT explants were chosen on the basis of a log2 fold change of >1.5, and in the wrky22.1 mutant explants on the basis of a log2 fold change >1.3. Red indicates increased abundance and blue decreased abundance, with the color intensity reflecting the fold of differential gene expression. Genes repressed by WRK22 are indicated by a red arrow, and those promoted by it by a black arrow. (B) Assignment of function in the context of the biotic and/or abiotic stress response: 1(Park et al., 2011), 2(Al-Abdallat et al., 2014), 3 (Novillo et al., 2004), 4(Sakuma et al., 2002), 5(Matías-Hernández et al., 2016), 6(Kang et al., 2011), 7(Chen et al., 2016), 8(Loreti et al., 2005), 9(Tominaga-Wada et al., 2013), 10(Wang et al., 2016), 11(Baxter et al., 2007), 12(Sun et al., 2003), 13(Sun and Yu, 2015), 14(Miao and Zentgraf, 2007), 15(Li et al., 2013), 16(Chen et al., 2017), 17(Guo and Qin, 2016), 18(Rizhsky et al., 2004).
Table 3.
DEGs identified in explants from the contrast phase II versus I in the WT, on the basis of a log2 fold change threshold of 1.5, and in the phase II wrky22.1 mutant versus the WT, on the basis of a log2 fold change threshold of 1.3
| Locus | WT II/I | KO II | Description | Mapman functional description | Reference | |
|---|---|---|---|---|---|---|
| 1 | AT1G01470 | –4.9 | 1.4 | LATE EMBRYOGENESIS ABUNDANT 14 (LEA14) | development.late embryogenesis abundant | Jia et al. 2014 |
| 2 | AT1G12610 | –3.9 | 1.7 | DWARF AND DELAYED FLOWERING 1 (DDF1) | RNA.regulation of transcription.AP2/EREBP | Magome et al., 2008; Kang et al., 2011 |
| 3 | AT4G25490 | –5.0 | 1.7 | C-repeat/DRE binding factor 1 (CBF1) | RNA.regulation of transcription.AP2/EREBP | Liu et al., 1998; Sakuma et al., 2002 |
| 4 | AT4G25470 | –4.3 | 1.7 | C-repeat/DRE binding factor 2 (CBF2) | RNA.regulation of transcription.AP2/EREBP | Liu et al., 1998; Sakuma et al., 2002 |
| 5 | AT4G25480 | –2.1 | 1.4 | C-repeat/DRE binding factor 2 (CBF3) | RNA.regulation of transcription.AP2/EREBP | Liu et al., 1998; Sakuma et al., 2002 |
| 6 | AT5G21960 | –1.6 | 1.4 | DREB | RNA.regulation of transcription.AP2/EREBP | Liu et al., 1998; Sakuma et al., 2002 |
| 7 | AT2G47520 | 6.1 | –1.9 | HYPOXIA RESPONSIVE ETHYLENE RESPONSE FACTOR 2 (HRE2) | RNA.regulation of transcription.AP2/EREBP | Park et al., 2011 |
| 8 | AT2G47460 | 1.8 | 1.8 | MYB12 | RNA.regulation of transcription.MYB | Wang et al., 2016 |
| 9 | AT3G27220 | 2.4 | –1.8 | Galactose oxidase/kelch repeat superfamily (GAL-OXI) | RNA.regulation of transcription.MYB | Loreti et al 2005 |
| 10 | AT3G56400 | –2.5 | 1.3 | WRKY70 | RNA.regulation of transcription.WRKY | Li et al., 2013; Chen et al., 2017 |
| 11 | AT4G23810 | –2.0 | 1.3 | WRKY53 | RNA.regulation of transcription.WRKY | Sun et al., 2003; Sun and Yu, 2015; |
| 12 | AT1G29860 | –1.9 | 1.6 | WRKY71 | RNA.regulation of transcription.WRKY | Guo and Quin, 2016 |
| 13 | AT4G11650 | –4.2 | 1.3 | OSMOTIN 34 (OSM34) | stress.abiotic | Sharma et al., 2013 |
| 14 | AT3G24520 | –3.8 | 1.4 | Heat shock transcription factor C1 (HSFC1) | RNA.regulation.transcription.HSF | Rizhsky et al 2004 |
| 15 | AT1G20440 | –6.5 | 2.2 | COLD REGULATED 47 (COR47) | stress.abiotic.unspecified | Wu et al., 2017 |
| 16 | AT1G20450 | –5.1 | 1.6 | EARLY RESPONSIVE TO DEHYDRATION 10 (ERD10) | stress.abiotic.unspecified | Wu et al., 2017 |
| 17 | AT5G66400 | –4,5 | 1.6 | RESPONSIVE TO ABA 18 (RAB18) | stress.abiotic.unspecified | Wu et al., 2017 |
| 18 | AT1G73330 | –5.6 | 2.8 | DROUGHT-REPRESSED 4 (DR4) | stress.biotic.PR-proteins.proteinase inhibitors | Boyce et al., 2003 |
| 19 | AT3G62410 | –4.0 | 1.5 | CP12 domain-containing protein 2 (CP12-2) | PS.calvin cycle | López-Calcagno et al., 2017 |
| 20 | AT3G54050 | –3.7 | 1.5 | HIGH CYCLIC ELECTRON FLOW 1 (HCEF1) | PS.calvin cycle.FBPase | Soto-Suárez et al., 2016 |
| 21 | AT1G32060 | –4.0 | 1.5 | PHOSPHORIBULOKINASE (PRK) | PS.calvin cycle.PRK | López-Calcagno et al., 2017 |
| 22 | AT2G39730 | –4.0 | 1.6 | RUBISCO ACTIVASE (RCA) | PS.calvin cycle.rubisco interacting | Zhang et al., 2015 |
| 23 | AT1G29395 | –5.8 | 1.6 | COLD REGULATED 314 INNER MEMBRANE 1 (COR413IM1) | not assigned.no ontology | Magome et al., 2008 |
| 24 | AT1G62480 | –4.0 | 1.3 | Vacuolar calcium-binding protein-related | signalling.calcium | Boyce et al., 2003 |
| 25 | AT4G17340 | –4.3 | 1.4 | tonoplast intrinsic protein 2;2 (TIP2;2) | transport.Major Intrinsic Proteins.TIP | Zhu et al., 2014 |
Gene functions are assigned either by MapMan analysis or from the literature in the context of the regulation of osmotic stress.
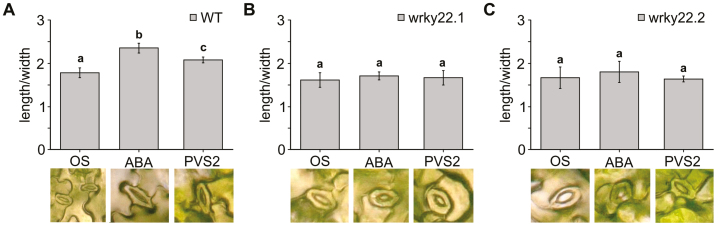
Stomatal closure induced by PVS2 treatment differed between the WT and the wrky22 mutants
To reveal the function of WRKY22 in stomatal movement, leaves of both the WT and the two independent wrky22 mutants were treated with either ABA or PVS2. Both treatments promoted the closure of guard cells in WT leaves (Fig. 9A) but not in those of either mutant (Fig. 9B,C), which implied that the loss of WRKY22 function induced a greater level of sensitivity to osmotic stress. A follow-up experiment, in which water was withheld from both WT and wrky22 mutant plants for 18 d, confirmed that the loss of WRKY22 function significantly reduced the plants’ FW and the rosette size (Supplementary Fig. S7).
Fig. 9.
Changes in stomatal aperture induced by WRKY22 in the presence of ABA and PVS2. (A) WT, (B) wrky22.1 mutant, (C) wrky22.2 mutant. The stomatal aperture ratio (length/width) was calculated from 80 stomata in three biological replicates; SD (n=3). Statistical significance was calculated using one-way ANOVA followed by Holm–Sidak post-hoc test. Mean values marked by the same letter did not differ significantly from one another (P≤0.001).
Discussion
Achieving a high level of post-cryogenic viability is important to preserve currently endangered plant species and maintain biodiversity ex situ. This requires explants to have the capacity to properly respond to a variety of stresses, which include wounding and the exposure to osmotic, chemical, and low temperature stress.
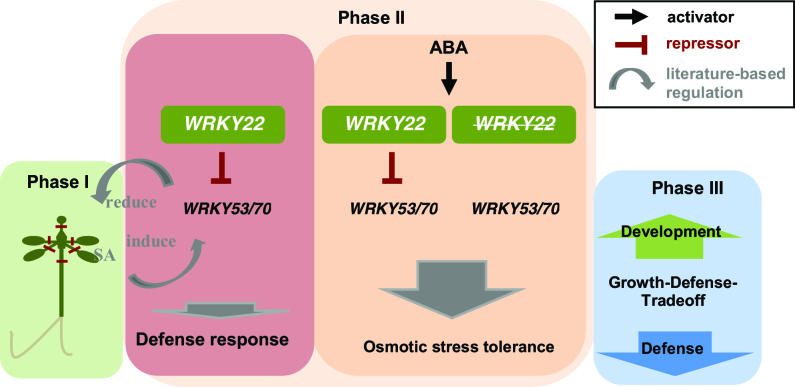
This study showed that Arabidopsis WT shoot tips could overcome cryo-induced stress response accompanied by high post-cryogenic recovery. A transcriptomic and modeling (Fig. 10) approach and further molecular characterization of WT and T-DNA insertion plants unraveled the molecular mechanisms underlying cryopreservation after shoot tip preparation (phase I), cryoprotectant treatment (phase II), and first day of recovery (phase III).
Fig. 10.
A proposed function for WRKY22 during the cryopreservation process. WRKY22 suppresses the transcription of WRKY53 and WRKY70, resulting in an altered salicylic acid (SA)-mediated wounding response and an altered osmotic stress response as suggested by the changed stomatal opening behavior. The open stomata phenotype exhibited by the wrky22.1 mutant results in a greater volume of H2O loss and CO2 fixation, and a change to the chloroplasts’ capacity. A higher energy demand may limit the trade-off between growth and defense, resulting in the mutant explants suffering a compromised level of post-cryopreservation recovery.
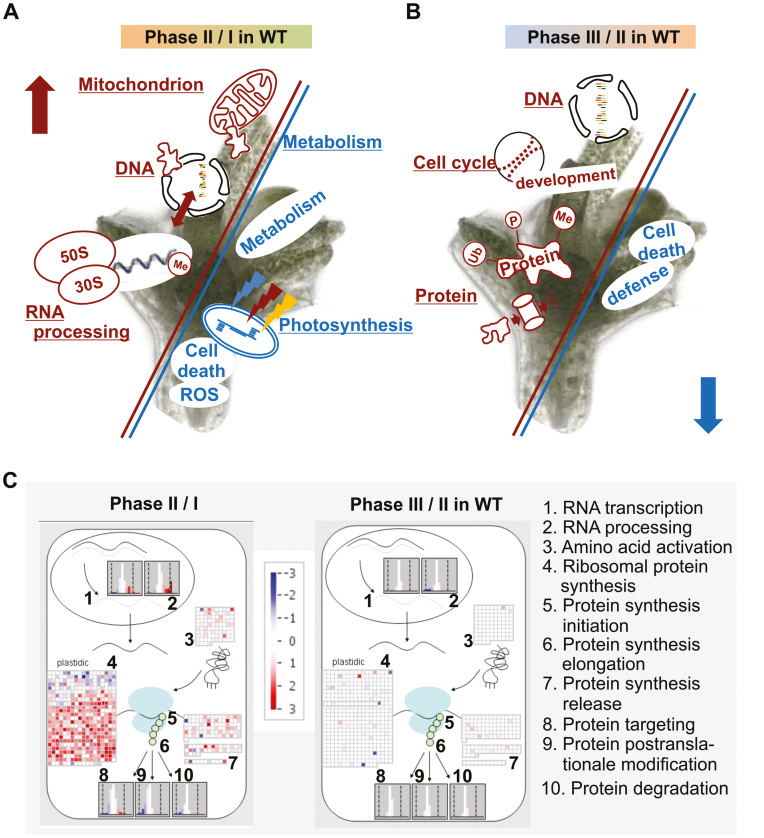
Cryoprotectant treatment initiated a number of changes to A. thaliana WT shoot tips, including the induction of a more de-differentiated status (Fig. 4) and an increase in protein synthesis (Fig. 6). At the same time, a limited response to apoptosis, ROS production, and photosynthesis promoted processes related to development, cell cycling, and protein turnover in phase III.
Moreover, this study provided significant insight into the basic function of WRKY22 and into processes in which multiple abiotic stressors act simultaneously. The response of the wrky22 mutant was rather different compared with the WT, comprising a less organized stress response, specifically related to phytohormone-mediated processes and photosynthesis. This response was more permissive of damage, resulting in a significant proportion of the rewarmed explants being able to form only non-differentiated callus rather than new shoot material. The loss of the functional WRKY22 transcript limited the responsiveness to phytohormone-mediated defense (SA and JA) and drought stress (ABA) responses in mutant shoot tips in phase I and phase II.
The WT explant’s transcriptome response to cryo-stressors
It has been proposed that recovery post-cryopreservation is largely compromised by a build-up of ROS, together with a reduced capacity to produce the detoxifying antioxidant enzymes (Uchendu et al., 2010; Chen et al., 2015; Ren et al., 2015; Gross et al., 2017). The transcript abundance of the redox stress marker genes ATH8 (At1g69880), AHB1 (At2g16060), APX2 (At3g09640), and GPX7 (At4g31870) was increased by the cryoprotectant treatment, but many other known ROS marker genes were down-regulated in the WT explants, and only marginally altered in the wrky22 mutant. To further address the role of ROS during cryopreservation, additional biochemical investigations will be needed.
The PVS2 reagent combines a number of different cryoprotectant substances, some of which are potentially toxic for meristematic cells (Volk et al., 2006). One of the PVS2-intrinsic cryoprotectants is DMSO; given that this compound inhibits electron transport in the chloroplast (Reeves and Hall, 1977), it can be expected to affect the transcription of genes associated with photosynthesis. To reduce its chemotoxicity, the cryoprotectant treatment is typically conducted in the dark at a low temperature, conditions which suppress photosynthesis. Osmotic stress, resulting from the partial dehydration of the explant, is required to avoid the formation of ice crystals during the cooling step. The ultrastructural analysis showed typical cellular stress indicators such as less dense appearing cytoplasm, plasmolysis, formation of plastoglobuli, and an increasing number of small vacuoles. These effects agree with those which have been seen in potato (Kaczmarczyk et al., 2008). It is known that salinity treatment, which imposes osmotic stress, can induce stomatal closure, the inhibition of CO2 fixation, and a reduced flux of electrons through PSII (Kilian et al., 2007; Stepien and Johnson, 2009); here, the intensity of transcription in the treated WT explants was reduced with respect to both of the PSII-associated genes PSBR (At1g79040) and LHCB2.3 (At3g27690) (Table 3). Thus, the suggestion is that chemotoxicity and osmotic stress represent significant components of the overall stress imposed by the cryo-stressors.
The regulatory role of WRKY22 during cryopreservation
Consistent with what has been reported in the literature (Kloth et al., 2016), changes in WRKY22 expression initiated transcriptional changes in genes related to the synthesis of the cell wall (Supplementary Dataset S1) and the SA-mediated stress response (Fig. 7A). Most of the changes induced in the transcriptome took place during phase II, possibly reflecting a delayed wounding response. The altered nature of the SA-mediated defense response may have arisen through crosstalk with WRKY53 and WRKY70, both of which are known to act as regulators of SA-mediated gene transcription (Li et al., 2004; Wang et al., 2006; Miao et al., 2007; Kloth et al., 2016). Such crosstalk is supported by the observation that WRKY70 gene expression is increased in both independent wrky22 KO mutant lines compared with the WT when the seedlings were exposed to a cold treatment. Since both WRKY53 and WRKY70 have been identified as repressors of stomatal opening (Li et al., 2013; Sun and Yu, 2015), it is tempting to speculate that WRKY22 cooperates with these two TFs in the context of the explants’ acclimation to osmotic stress. WRKY22 might participate in the explants’ acclimation to osmotic stress by impacting on stomata opening–closing control and thereby on the plants’ osmotic stress behavior (Fig. 9). During recovery, a number of genes involved in auxin-driven growth or in histone modification showed higher abundance in the wrky22 mutant explants, while certain defense response genes were expressed at a lower level (Figs 6B, 7B; Supplementary Dataset S1; Supplementary Fig. S3): this represents a strategy whereby a choice is made between growth and defense (Huot et al., 2014).
The schematic model presented in Fig. 10 summarizes key aspects of the WRKY22-mediated regulation of both the osmotic stress and defense responses. The PVS2 treatment and excision of the explant trigger stomatal closure, probably involving WRKY53 and WRKY70, while at the same time the wounding response is orchestrated by genes responding to an SA signal. The putative wrky22 mutant’s open stomata phenotype would enhance the volume of CO2 fixation, driving changes in the transcription of genes encoding PSII at the expense of defense responses necessary during regeneration.
In summary, Arabidopsis represents a suitable model for identifying the mechanistic basis of the response to the combined abiotic stresses imposed by the cryopreservation process. Successful recovery requires a balance between ensuring cellular survival during low temperature storage through de-differentiation and the ability to regenerate a viable plant upon rewarming. Elucidation of the underlying processes is informative to better understand combinatorial stress defense mechanisms in plants.
Supplementary data
Supplementary data are available at JXB online, and access to the transcriptional data set is provided via doi: 10.5447/ipk/2020/6.
Fig. S1. Verification of the inactivated WRKY22 transcript.
Fig. S2. qRT–PCR-selected genes.
Fig. S3. MapMan_Auxin_Secondary Metabolites.
Fig. S4. Principal component analysis of the wrky22.1 mutant.
Fig. S5. MapMan_Photosynthesis_Wilcoxon Sum Rank.
Fig. S6. qRT–PCR of WRKY70 dependent on Arabidopsis genotype.
Fig. S7. Drought stress experiment.
Dataset S1. GO term enrichment.
Dataset S2. Hitlist 50 UP_DOWN.
Dataset S3. Mothertable RNA-seq reads.
Table S1. Primer list and Arabidopsis genotypes.
Table S2. MapMan_Protein Synthesis_Wilcoxon Sum Rank.
Table S3. Sample preparation for electron microscopy.
Acknowledgements
We acknowledge the excellent technical assistance of Petra Linow, Jacqueline Fuge, and Marion Benecke, and thank Marion Grübe, Doris Büchner, and Dr Angelika Senula for the introduction to cryopreservation techniques. The authors wish to thank Dr Armin Meister for support in statistical analyses, and Robert Köbner for language editing. This work was supported by the Leibniz Gemeinschaft within the Leibniz Competition Funding line 3 (Networking) (grant no. SAW-2013-DSMZ-3).
References
- Al-Abdallat AM, Al-Debei HS, Ayad JY, Hasan S. 2014. Over-expression of SlSHN1 gene improves drought tolerance by increasing cuticular wax accumulation in tomato. International Journal of Molecular Sciences 15, 19499–19515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexa A, Rahnenfuhrer J. 2010. topGO: enrichment analysis for gene ontology. R package version 2. https://bioconductor.org/packages/release/bioc/html/topGO.html [Google Scholar]
- Basu C. 2008. Gene amplification from cryopreserved Arabidopsis thaliana shoot tips. Current Issues in Molecular Biology 10, 55–60. [PubMed] [Google Scholar]
- Baxter CE, Costa MM, Coen ES. 2007. Diversification and co-option of RAD-like genes in the evolution of floral asymmetry. The Plant Journal 52, 105–113. [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. 1993. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. Comptes Rendus de l’Académie des Sciences. Série 3, Sciences de la vie 316, 1194–1199. [Google Scholar]
- Benson EE, Harding K, Johnston JW. 2007. Cryopreservation of shoot tips and meristems. Methods in Molecular Biology 368, 163–183. [DOI] [PubMed] [Google Scholar]
- Bonferroni CE. 1936. Teoria statistica delle classi e calcolo delle probabilita. Libreria internazionale Seeber. [Google Scholar]
- Boyce JM, Knight H, Deyholos M, Openshaw MR, Galbraith DW, Warren G, Knight MR. 2003. The sfr6 mutant of Arabidopsis is defective in transcriptional activation via CBF/DREB1 and DREB2 and shows sensitivity to osmotic stress. The Plant Journal 34, 395–406. [DOI] [PubMed] [Google Scholar]
- Bray N, Pimentel H, Melsted P, Pachter L. 2016. Near-optimal RNA-Seq quantification. Nature Biotechnology 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Chawade A, Bräutigam M, Lindlöf A, Olsson O, Olsson B. 2007. Putative cold acclimation pathways in Arabidopsis thaliana identified by a combined analysis of mRNA co-expression patterns, promoter motifs and transcription factors. BMC Genomics 8, 304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GQ, Ren L, Zhang J, Reed BM, Zhang D, Shen XH. 2015. Cryopreservation affects ROS-induced oxidative stress and antioxidant response in Arabidopsis seedlings. Cryobiology 70, 38–47. [DOI] [PubMed] [Google Scholar]
- Chen HY, Hsieh EJ, Cheng MC, Chen CY, Hwang SY, Lin TP. 2016. ORA47 (octadecanoid-responsive AP2/ERF-domain transcription factor 47) regulates jasmonic acid and abscisic acid biosynthesis and signaling through binding to a novel cis-element. New Phytologist 211, 599–613. [DOI] [PubMed] [Google Scholar]
- Chen J, Nolan TM, Ye H, Zhang M, Tong H, Xin P, Chu J, Chu C, Li Z, Yin Y. 2017. Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses. The Plant Cell 29, 1425–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. 2005. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology 139, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daghma DS, Kumlehn J, Melzer M. 2011. The use of cyanobacteria as filler in nitrocellulose capillaries improves ultrastructural preservation of immature barley pollen upon high pressure freezing. Journal of Microscopy 244, 79–84. [DOI] [PubMed] [Google Scholar]
- Dong J, Chen C, Chen Z. 2003. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Molecular Biology 51, 21–37. [DOI] [PubMed] [Google Scholar]
- El-Banna A, Hajirezaei MR, Wissing J, Ali Z, Vaas L, Heine-Dobbernack E, Jacobsen HJ, Schumacher HM, Kiesecker H. 2010. Over-expression of PR-10a leads to increased salt and osmotic tolerance in potato cell cultures. Journal of Biotechnology 150, 277–287. [DOI] [PubMed] [Google Scholar]
- Engelmann F. 2004. Plant cryopreservation: progress and prospects. In Vitro Cellular & Developmental Biology - Plant 40, 427–433. [Google Scholar]
- Engelmann F, Takagi H. 2000. Cryopreservation of tropical plant germplasm—current research progress and applications. Tsukuba: JIRCAS. [Google Scholar]
- Fisher RA. 1922. On the interpretation of χ2 from contingency tables, and the calculation of P. Journal of the Royal Statistical Society 85, 87–94. [Google Scholar]
- Göhre V, Jones AM, Sklenář J, Robatzek S, Weber AP. 2012. Molecular crosstalk between PAMP-triggered immunity and photosynthesis. Molecular Plant-Microbe Interactions 25, 1083–1092. [DOI] [PubMed] [Google Scholar]
- Gross BL, Henk AD, Bonnart R, Volk GM. 2017. Changes in transcript expression patterns as a result of cryoprotectant treatment and liquid nitrogen exposure in Arabidopsis shoot tips. Plant Cell Reports 36, 1–12. [DOI] [PubMed] [Google Scholar]
- Guo D, Qin G. 2016. EXB1/WRKY71 transcription factor regulates both shoot branching and responses to abiotic stresses. Plant Signaling & Behavior 11, e1150404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu FC, Chou MY, Chou SJ, Li YR, Peng HP, Shih MC. 2013. Submergence confers immunity mediated by the WRKY22 transcription factor in Arabidopsis. The Plant Cell 25, 2699–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huot B, Yao J, Montgomery BL, He SY. 2014. Growth–defense tradeoffs in plants: a balancing act to optimize fitness. Molecular Plant 7, 1267–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia F, Qi S, Li H, Liu P, Li P, Wu C, Zheng C, Huang J.Overexpression of Late Embryogenesis Abundant 14 enhances Arabidopsis salt stress tolerance. 2014. [DOI] [PubMed]
- Kaczmarczyk A, Rutten T, Melzer M, Keller ER. 2008. Ultrastructural changes associated with cryopreservation of potato (Solanum tuberosum L.) shoot tips. Cryo Letters 29, 145–156. [PubMed] [Google Scholar]
- Kang H-G, Kim J, Kim B, Jeong H, Choi SH, Kim EK, Lee H-Y, Lim PO. 2011. Overexpression of FTL1/DDF1, an AP2 transcription factor, enhances tolerance to cold, drought, and heat stresses in Arabidopsis thaliana. Plant Science 180, 634–641. [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. 2002. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Keller ER. 2005. Improvement of cryopreservation results in garlic using low temperature preculture and high-quality in vitro plantlets. Cryo Letters 26, 357–366. [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K. 2007. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. The Plant Journal 50, 347–363. [DOI] [PubMed] [Google Scholar]
- Kloth KJ, Wiegers GL, Busscher-Lange J, van Haarst JC, Kruijer W, Bouwmeester HJ, Dicke M, Jongsma MA. 2016. AtWRKY22 promotes susceptibility to aphids and modulates salicylic acid and jasmonic acid signalling. Journal of Experimental Botany 67, 3383–3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Besseau S, Törönen P, Sipari N, Kollist H, Holm L, Palva ET. 2013. Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytologist 200, 457–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Brader G, Palva ET. 2004. The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. The Plant Cell 16, 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. 1998. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. The Plant Cell 10, 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Calcagno EP, Abuzaid OA, Lawson T, Raines AC. 2017. Arabidopsis CP12 mutants have reduced levels of phosphoribulokinase and impaired function of the Calvin–Benson cycle. Journal of Experimental Botany 68, 2285–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E, Poggi A, Novi G, Alpi A, Perata P. 2005. A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiology 137, 1130–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magome H, Yamaguchi S, Hanada A, Kamiya Y, Oda K. 2008. The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis. The Plant Journal 56, 613–626. [DOI] [PubMed] [Google Scholar]
- Matías-Hernández L, Aguilar-Jaramillo AE, Osnato M, Weinstain R, Shani E, Suárez-López P, Pelaz S. 2016. TEMPRANILLO reveals the mesophyll as crucial for epidermal trichome formation. Plant Physiology 170, 1624–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Laun TM, Smykowski A, Zentgraf U. 2007. Arabidopsis MEKK1 can take a short cut: it can directly interact with senescence-related WRKY53 transcription factor on the protein level and can bind to its promoter. Plant Molecular Biology 65, 63–76. [DOI] [PubMed] [Google Scholar]
- Miao Y, Zentgraf U. 2007. The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. The Plant Cell 19, 819–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. 1962. A revised medium for rapid growth and bio assay with tobacco tissue cultures. Physiologia Plantarum 15, 473–497. [Google Scholar]
- Novillo F, Alonso JM, Ecker JR, Salinas J. 2004. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proceedings of the National Academy of Sciences, USA 101, 3985–3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panis B, Totte N, Van Nimmen K, Withers L, Swennen R. 1996. Cryopreservation of banana (Musa spp.) meristem cultures after preculture on sucrose. Plant Science 121, 95–106. [Google Scholar]
- Park HY, Seok HY, Woo DH, Lee SY, Tarte VN, Lee EH, Lee CH, Moon YH. 2011. AtERF71/HRE2 transcription factor mediates osmotic stress response as well as hypoxia response in Arabidopsis. Biochemical and Biophysical Research Communications 414, 135–141. [DOI] [PubMed] [Google Scholar]
- Park S, Lee CM, Doherty CJ, Gilmour SJ, Kim Y, Thomashow MF. 2015. Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. The Plant Journal 82, 193–207. [DOI] [PubMed] [Google Scholar]
- Reed BM. 2008. Plant cryopreservation: a practical guide. Springer. [Google Scholar]
- Reeves SG, Hall DO. 1977. The effect of dimethyl sulphoxide on electron transport in chloroplasts. Cell Biology International Reports 1, 353–361. [DOI] [PubMed] [Google Scholar]
- Ren L, Zhang D, Chen GQ, Reed BM, Shen XH, Chen HY. 2015. Transcriptomic profiling revealed the regulatory mechanism of Arabidopsis seedlings response to oxidative stress from cryopreservation. Plant Cell Reports 34, 2161–2178. [DOI] [PubMed] [Google Scholar]
- Ren L, Zhang D, Jiang XN, Gai Y, Wang WM, Reed BM, Shen XH. 2013. Peroxidation due to cryoprotectant treatment is a vital factor for cell survival in Arabidopsis cryopreservation. Plant Science 212, 37–47. [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Shuman J, Shulaev V, Davletova S, Mittler R. 2004. When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiology 134, 1683–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A, Kobayashi S, Oiyama I. 1990. Cryopreservation of nucellar cells of navel orange (Citrus sinensis Osb. var. brasiliensis Tanaka) by vitrification. Plant Cell Reports 9, 30–33. [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. 2002. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochemical and Biophysical Research Communications 290, 998–1009. [DOI] [PubMed] [Google Scholar]
- Sharma S, Lin W, Villamor JG, Verslues PE. 2013. Divergent low water potential response in Arabidopsis thaliana accessions Landsberg erecta and Shahdara. Plant, Cell & Environment 36, 994–1008. [DOI] [PubMed] [Google Scholar]
- Soto-Suárez M, Serrato AJ, Rojas-González JA, Bautista R, Sahrawy M. 2016. Transcriptomic and proteomic approach to identify differentially expressed genes and proteins in Arabidopsis thaliana mutants lacking chloroplastic 1 and cytosolic FBPases reveals several levels of metabolic regulation. BMC Plant Biology 16, 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepien P, Johnson GN. 2009. Contrasting responses of photosynthesis to salt stress in the glycophyte Arabidopsis and the halophyte Thellungiella: role of the plastid terminal oxidase as an alternative electron sink. Plant Physiology 149, 1154–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J, Senula A, Nagel M, Mock H-P, Keller ERJ. 2017. A simple method for cryopreservation of shoot tips of Arabidopsis genotypes. Cryo Letters 38, 364–371. [PubMed] [Google Scholar]
- Sun C, Palmqvist S, Olsson H, Borén M, Ahlandsberg S, Jansson C. 2003. A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. The Plant Cell 15, 2076–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Yu D. 2015. Activated expression of AtWRKY53 negatively regulates drought tolerance by mediating stomatal movement. Plant Cell Reports 34, 1295–1306. [DOI] [PubMed] [Google Scholar]
- Tominaga-Wada R, Nukumizu Y, Sato S, Wada T. 2013. Control of plant trichome and root-hair development by a tomato (Solanum lycopersicum) R3 MYB transcription factor. PLoS One 8, e54019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchendu EE, Leonard SW, Traber MG, Reed BM. 2010. Vitamins C and E improve regrowth and reduce lipid peroxidation of blackberry shoot tips following cryopreservation. Plant Cell Reports 29, 25–35. [DOI] [PubMed] [Google Scholar]
- Volk GM, Harris JL, Rotindo KE. 2006. Survival of mint shoot tips after exposure to cryoprotectant solution components. Cryobiology 52, 305–308. [DOI] [PubMed] [Google Scholar]
- Wang D, Amornsiripanitch N, Dong X. 2006. A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathogens 2, e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Kong W, Wong G, Fu L, Peng R, Li Z, Yao Q. 2016. AtMYB12 regulates flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis thaliana. Molecular Genetics and Genomics 291, 1545–1559. [DOI] [PubMed] [Google Scholar]
- Wu X, Qiao Z, Liu H, Acharya BR, Li C, Zhang W. 2017. CML20, an Arabidopsis calmodulin-like protein, negatively regulates guard cell ABA signaling and drought stress tolerance. Frontiers in Plant Science 8, 824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YJ, Huang XL, Xiao JN, Li XJ, Zhou MD, Engelmann F. 2003. Cryopreservation of mango (Mangifera indica L.) embryogenic cultures. Cryo Letters 24, 303–314. [PubMed] [Google Scholar]
- Zhang M, Li X, Yang Y, Luo Z, Liu C, Gong M, Zou Z. 2015. An acidified thermostabilizing mini-peptide derived from the carboxyl extension of the larger isoform of the plant Rubisco activase. Journal of Biotechnology 212, 116–124. [DOI] [PubMed] [Google Scholar]
- Zhou X, Jiang Y, Yu D. 2011. WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Molecules and Cells 31, 303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Wu Z, Cao G, Li J, Wei J, Tsuge T, Gu H, Aoyama T, Qu LJ. 2014. TRANSLUCENT GREEN, an ERF family transcription factor, controls water balance in Arabidopsis by activating the expression of aquaporin genes. Molecular Plant 7, 601–615. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.