Abstract
Cytosolic acetyl-CoA is an intermediate of the synthesis of most secondary metabolites and the source of acetyl for protein acetylation. The formation of cytosolic acetyl-CoA from citrate is catalysed by ATP-citrate lyase (ACL). However, the function of ACL in global metabolite synthesis and global protein acetylation is not well known. Here, four genes, PaACLA1, PaACLA2, PaACLB1, and PaACLB2, which encode the ACLA and ACLB subunits of ACL in Petunia axillaris, were identified as the same sequences in Petunia hybrida ‘Ultra’. Silencing of PaACLA1-A2 and PaACLB1-B2 led to abnormal leaf and flower development, reduced total anthocyanin content, and accelerated flower senescence in petunia ‘Ultra’. Metabolome and acetylome analysis revealed that PaACLB1-B2 silencing increased the content of many downstream metabolites of acetyl-CoA metabolism and the levels of acetylation of many proteins in petunia corollas. Mechanistically, the metabolic stress induced by reduction of acetyl-CoA in PaACL-silenced petunia corollas caused global and specific changes in the transcriptome, the proteome, and the acetylome, with the effect of maintaining metabolic homeostasis. In addition, the global proteome and acetylome were negatively correlated under acetyl-CoA deficiency. Together, our results suggest that ACL acts as an important metabolic regulator that maintains metabolic homeostasis by promoting changes in the transcriptome, proteome. and acetylome.
Keywords: Acetyl-CoA, acetylome, ATP-citrate lyase, metabolic homeostasis, petunia, proteome
PaACL silencing reduced anthocyanin contents, accelerated flower senescence, and changed the proteome and acetylome to maintain metabolic homeostasis in petunia
Introduction
Acetyl-coenzyme A (CoA) is the intermediate precursor for the biosynthesis of a wide variety of phytochemicals, including carbohydrates, malonyl-CoA, organic acids, fatty acids, isoprenoids, flavonoids, and stilbenoids (Fatland et al., 2002; Oliver et al., 2009; Finkemeier et al., 2011; Wu et al., 2011; Chypre et al., 2012; Xing and Poirier, 2012). Acetyl-CoA is found in the cytosol and in organelles, and is membrane impermeable. Therefore, acetyl-CoA biogenesis is thought to occur in each subcellular compartment where it is required, except for the nucleus, into which cytosolic acetyl-CoA most likely diffuses via the nuclear pore (Brooks and Stumpf, 1966; Fatland et al., 2002; Chen et al., 2017a). Cytosolic acetyl-CoA is also the source of acetyl for protein acetylation, including histone acetylation (Chen et al., 2017a). In yeast and mammalian cells, the levels of acetylation of histone H3 and H4 were affected by cellular acetyl-CoA levels (Takahashi et al., 2006; Wellen et al., 2009; Cai et al., 2011; Galdieri and Vancura, 2012; Lee et al., 2014). In the plant Arabidopsis thaliana, the increase in H3K27 acetylation (H3K27ac) is dependent on cytoplasmic ATP-citrate lyase (ACL), and H3K27ac is an important link between the cytosolic level of acetyl-CoA and gene expression in response to the dynamic metabolic environments in the plant (Chen et al., 2017a).
The formation of cytosolic acetyl-CoA from citrate is catalysed by ACL (Aoshima, 2007). ACL has been characterized in plants at the genomic level; it is a heteropolymer composed of ACLA and ACLB subunits located in the cytoplasm, implying that it generates a cytosolic pool of acetyl-CoA (Fatland et al., 2002). In A. thaliana, the ACLA subunit is encoded by three genes and the ACLB subunit is encoded by two genes (Fatland et al., 2002). ACL is involved in plant growth and development, secondary metabolism, and stress. In A. thaliana, moderately reduced ACL activity resulted in miniaturized organs, smaller cells, aberrant plastid morphology, and reduced cuticular wax deposition in vegetative tissue (Fatland et al., 2002). Overexpression of AtACL can promote the synthesis of wax, cutin, and rubber, revealing the important position of ACL in controlling the direction of carbon flow through cytoplasmic acetyl-CoA (Xing et al., 2014). Overexpression of sugarcane (Saccharum officinarum) SoACLA-1 in tobacco enhanced its drought resistance (Phan et al., 2016). In canola (Brassica napus), unchanged ACL protein might maintain the activity of the tricarboxylic acid cycle for adaptation to osmotic stress in leaves of drought-tolerant canola inoculated with Enterobacter sp. S16-3 as plant growth-promoting bacteria (Kazemi et al., 2017). The expression of ACL in Raphanus sativus roots was up-regulated under heavy metal stress (Wang et al., 2016). After treatment with low light, strong light, water stress, and abscisic acid, the expression level of ACL increased over a certain period (Tong, 2009). However, the function of ACL in global metabolite synthesis and global protein acetylation in plants is not known.
Petunia (Petunia hybrida) is a good system for research on plant growth and development (Gerats and Vandenbussche, 2005). In this study, we found that both the ACLA and ACLB subunits were encoded by two genes in petunia. Silencing of any single PaACL gene did not result in a visible phenotypic change, whereas silencing of both PaACLA1 and PaACLA2 (PaACLA1-A2) or both PaACLB1 and PaACLB2 (PaACLB1-B2) resulted in a similar phenotype, comprising abnormal development of leaves and flowers, decreased total anthocyanin content, increased chlorophyll content, acceleration of flower senescence, and increased ethylene production. Metabolomic analysis of petunia revealed that PaACLB1-B2 silencing changed the metabolite profile and that most metabolites, including flavonoids, were up-regulated. Mechanistically, the metabolic stress induced by the reduction of acetyl-CoA in PaACL-silenced petunia corollas resulted in global and specific changes in the transcriptome, the proteome, and the acetylome to maintain metabolic homeostasis.
Materials and methods
Plant materials and growth conditions
Petunia hybrida ‘Ultra’ plants were grown under greenhouse conditions (23±2 °C, 14 h light/10 h dark) (Yang et al., 2017). The roots, stems, and leaves were collected from plants in the vegetative stage when the height of the plants was approximately 25 cm. The flowers were harvested at anthesis (corollas 90° reflexed) and immediately placed in tap water. All tissues were frozen in liquid nitrogen and stored at –80 °C until use. All experiments were conducted at least three times with independently collected and extracted tissues unless otherwise noted.
RNA extraction, RT–PCR, and cloning of the petunia ACL genes
Total RNA was isolated according to the methods of Liu et al. (2011). A commercially available kit (TSK314s, Tsingke Guangzhou, China) was used for reverse transcription of petunia mRNA. Full-length PaACLA1 (Peaxi162Scf00357g00428.1), PaACLA2 (Peaxi162Scf00158g00011.1), PaACLB1 (Peaxi162Scf00228g00327.1), and PaACLB2 (Peaxi162Scf01160g00016.1) cDNA was isolated from P. hybrida ‘Ultra’ using specific primers (see Supplementary Table S1 at JXB online) based on their sequences in the Petunia axillaris genome (https://solgenomics.net/organism/Petunia_axillaris/genome).
Sequence analysis
Alignments were conducted, and a phylogenetic tree was generated using DNAMAN software. An identity search for nucleotides and translated amino acids was conducted using the National Center for Biotechnology Information (NCBI) BLAST network server (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Quantitative real-time PCR assays
Quantitative real-time PCR (qPCR) assays were performed according to previous methods (Liu et al., 2011). Analyses were conducted following the Minimum Information for Publication of Quantitative Real-Time PCR Experiments guidelines (Bustin et al., 2009; Tan et al., 2014). Cyclophilin (CYP) (accession no. EST883944) and Actin (accession no. FN014209) were used as the internal reference genes to quantify cDNA abundance (Mallona et al., 2010; Yang et al., 2017). Similar results were obtained with both reference genes. The data reported in the Results represent relative expression values calculated by the 2−ΔΔCt approximation method (Livak and Schmittgen, 2001). The sequences of all primers used for qPCR analysis are listed in Supplementary Table S2. Three biological replicates, each including three technical repeats, were analysed for each treatment.
Agroinoculation of pTRV vectors
pTRV2-PaACLA1, pTRV2-PaACLA2, pTRV2-PaACLB1, pTRV2-PaACLB2, pTRV2-PaACLA1-PaACLA2, and pTRV2-PaACLB1-PaACLB2 vectors were constructed by amplifying the gene sequences of approximately 250 bp of the 3′ untranslated region of the four PaACL genes using the specific primers listed in Supplementary Table S3 and cloning them into the pTRV2 vector. Agrobacterium tumefaciens (strain GV3101) transformed with pTRV1 and pTRV2 derivatives were prepared as previously described (Spitzer-Rimon et al., 2012; Tan et al., 2014). Approximately 30 petunia plants were inoculated with each vector for virus-induced gene silencing (VIGS) experiments. The inoculated plants were grown under greenhouse conditions.
Determination of ACL activity
Crude protein extracts were desalted by chromatography through Sephadex G-25 (Sigma-Aldrich, St. Louis, MO, USA), and ACL activity was determined using a coupled spectrophotometric assay (Fatland et al., 2002).
Pigment measurement
Four or five mature petunia leaves were collected from each plant and freeze-dried, and three replicate methanol extractions were prepared for each type of plant using the method of Wellburn (1994). The absorbance of the solution was read with a spectrophotometer at 646.8 nm, 663.2 nm, and 470.0 nm against the solvent (acetone) blank. The individual concentrations of total chlorophyll and total carotenoids were calculated from the spectrophotometric measurements (Lichtenthaler, 1987).
Anthocyanin extraction and analysis were performed as previously described (Chen et al., 2017b). Petunia flowers were harvested at anthesis and the corolla limbs were collected. Three biological replicates were analysed for each treatment.
Acetyl-CoA measurement
Approximately 1 g of corollas was ground in the presence of liquid nitrogen, and 9 ml of phosphate-buffered saline was then added. The sample was thoroughly vortexed and incubated for 20 min at room temperature. The mixture was centrifuged at 16 873 g for 15 min to separate the layers, and the supernatant was collected. The content of acetyl-CoA in the supernatant was determined using a kit (JN709212, Ji-Ning Bio-tech, Shanghai, China) according to the manufacturer’s instructions.
Measurement of ethylene
Ethylene measurements were performed according to the method of Tan et al. (2014). Flowers were collected and placed individually in 150 ml airtight containers. The containers were capped and incubated at 25 °C for 1 h. Then, 2 µl of head-space gas was withdrawn using a gas-tight hypodermic syringe and injected into a gas chromatograph (GC 17A, Shimadzu, Kyoto, Japan) for measurement of ethylene concentration. The gas chromatograph was equipped with a flame ionization detector and an activated alumina column. All measurements were performed with five replicates.
Scanning electron microscopy
The mature leaves and corolla limbs from petunia plants were cut into 3–5 mm2 pieces. The samples were fixed, dehydrated, and coated with gold by using ion sputtering equipment, according to our previously described protocol (Yang et al., 2017). Samples were observed with a scanning electron microscope (XL-30-ESEM, FEI, The Netherlands) at 10 kV acceleration and photographed.
Analysis of widely targeted metabolites
The collected petunia corollas were freeze-dried and ground to powder, and 0.1 g of powder was placed in 1.0 ml of 70% aqueous methanol at 4 °C and stored overnight. Then, the extract was centrifuged at 10 000 g for 10 min, and the supernatant was filtered through a microporous membrane (0.22 μm pore size) for analysis.
The metabolites were analysed by ultra-performance liquid chromatography (UPLC) (Shim-pack UFLC SHIMADZU CBM30A, http://www.shimadzu.com.cn/) and tandem mass spectrometry (MS/MS) (AB SCIEX 6500 QTRAP) under the conditions described by Li and Song (2019). In the triple quadrupole, each ion pair was scanned for detection based on the optimized decompression potential and collision energy (Chen et al., 2013). The qualitative and quantitative MS analysis of metabolites in the samples was performed on metabolites based on the self-built Metware database (MWDB) (Metware, Wuhan, China), and multiple reaction monitoring (MRM) (Chen et al., 2013).
Transcriptomic analysis of corollas by RNA sequencing
Corollas of PaACLB1-B2 silenced petunia lines and controls were collected for mRNA library construction and sequencing as described before (Guo et al., 2017). Corolla RNA was isolated using TRIzol reagent (Promega, USA) according to the manufacturer’s protocol. The extracted RNA was quantified and its integrity was measured on a 1.0% denaturing agarose gel. The poly(A)-containing mRNA molecules were purified using Sera-mag Magnetic Oligo(dT) Beads (Thermo Scientific) and were fragmented by incubation in RNA fragmentation reagent (Ambion). The fragmented mRNA was then converted into double-stranded cDNA using a SuperScript double-stranded cDNA synthesis kit (Invitrogen) by priming with random hexamers. Strand non-specific transcriptome libraries were prepared using the NEBNext mRNA Library Prep Reagent Set (New England Biolabs). These cDNA libraries were amplified and sequenced on an Illumina HiSeq platform (Metware, Wuhan, China). Raw reads, including the adaptor sequences, low-quality sequences, and unknown nucleotides, were filtered into clean reads using the standard quality control technique. Clean reads were aligned to petunia reference genome sequences (https://solgenomics.net/organism/Petunia_axillaris/genome) by hierarchical indexing for spliced alignment of transcripts in the HISAT2 application. The fragments per kilobase of transcript per million reads mapped (FPKM) method was used to calculate normalized expression levels. Differentially expressed genes (DEGs) between PaACLB1-B2 silenced samples and controls were identified by the NOISeq method according to the default criterion of a 2-fold change (P-value <0.05).
Protein extraction and trypsin digestion
Protein extraction and trypsin digestion were performed according our previous protocol (Guo et al., 2017). The corollas (~2.5 g for each sample) were ground to a powder in liquid nitrogen and then transferred to a 5 ml centrifuge tube. A volume of lysis buffer (8 M urea, 1% Triton-100, 10 mM dithiothreitol, and 1% protease inhibitor cocktail) four times the volume of the sample powder was added, followed by sonication three times on ice using a high-intensity ultrasonic processor (Scientz). The remaining debris was removed by centrifugation at 20 000 g at 4 °C for 10 min. Finally, the protein was precipitated with cold 20% trichloroacetic acid for 2 h at –20 °C. After centrifugation at 12 000 g at 4 °C for 10 min, the supernatant was discarded. The remaining precipitate was washed with cold acetone three times. The protein was redissolved in 8 M urea, and the protein concentration was determined with a BCA kit according to the manufacturer’s instructions (P0011, Beyotime Biotechnology, Shanghai, China).
For digestion, the protein solution was reduced with 5 mM dithiothreitol for 30 min at 56 °C and alkylated with 11 mM iodoacetamide for 15 min at room temperature in darkness. The protein sample was then diluted by adding 100 mM triethylammonium bicarbonate (TEAB) to a urea concentration less than 2 M. Finally, trypsin was added at a 1:50 trypsin-to-protein mass ratio for the first digestion overnight, and at a 1:100 trypsin-to-protein mass ratio for a second 4 h digestion.
TMT labelling
After trypsin digestion, the peptide was desalted in a Strata X C18 SPE column (Phenomenex) and vacuum-dried. Peptide was reconstituted in 0.5 M TEAB and processed according to the manufacturer’s protocol for the tandem mass tag (TMT) kit (No. 90068, Thermo Scientific, Waltham, MA, USA). Briefly, one unit of TMT reagent was thawed and reconstituted in acetonitrile. The peptide mixtures were incubated for 2 h at room temperature and then pooled, desalted, and dried by vacuum centrifugation.
HPLC fractionation
The tryptic peptides were fractionated by high pH reverse-phase HPLC using a Thermo Betasil C18 column (5 μm particles, 10 mm internal diameter, 250 mm length). Briefly, peptides were first separated with a gradient of 8% to 32% acetonitrile (pH 9.0) over 60 min into 60 fractions. Then, the peptides were combined into acetylation fractions and dried by vacuum centrifuging.
Drawing of protein–metabolite co-expression networks
Protein–metabolite co-expression networks were constructed based on the Spearman correlation coefficient (Kovarik, 2013). The Spearman correlation coefficients between the quantitative values of differential proteins and differential metabolites in samples were calculated by using the cor function in R. Data with Spearman correlation coefficients greater than 0.95 or less than –0.95 between metabolites and proteins were screened out to construct the co-expression network. The network was visualized and analyzed by using cycloscape v3.7.2 software (Smoot et al., 2011).
Antibody-based acetylation peptide enrichment
To enrich acetylation peptides, tryptic peptides dissolved in NETN buffer (100 mM NaCl, 1 mM EDTA, 50 mM Tris–HCl, 0.5% NP-40, pH 8.0) were incubated with pre-washed antibody beads (lot number PTM-104, PTM Bio) at 4 °C overnight with gentle shaking. Then, the beads were washed four times with NETN buffer and twice with water. The bound peptides were eluted from the beads with 0.1% trifluoroacetic acid. Finally, the eluted fractions were combined and vacuum-dried. For LC-MS/MS analysis, the resulting peptides were desalted with C18 ZipTips (Millipore) according to the manufacturer’s instructions.
LC-MS/MS analysis
LC-MS/MS analysis was performed according to a previously described protocol (Guo et al., 2017). The tryptic peptides were dissolved in 0.1% formic acid (solvent A) and directly loaded on to a homemade reversed-phase analytical column (15 cm length, 75 μm internal diameter). The gradient consisted of an increase from 6% to 23% of solvent B (0.1% formic acid in 98% acetonitrile) over 26 min, followed by an increase from 23% to 35% in 8 min, then climbing to 80% in 3 min and then holding at 80% for the last 3 min, all at a constant flow rate of 400 nl min–1, on an EASY-nLC 1000 UPLC system.
The peptides were subjected to nanospray ionization followed by MS/MS in Q ExactiveTM Plus (Thermo Fisher Scientific) coupled online to the UPLC. The electrospray voltage applied was 2.0 kV. The m/z scan range was 350 to 1800 for full scan, and intact peptides were detected in the Orbitrap (Thermo Fisher Scientific) at a resolution of 70 000. Peptides were then selected for MS/MS using the normalized collision energy setting at 28, and the fragments were detected in the Orbitrap at a resolution of 17 500. A data-dependent procedure was used that alternated between one MS scan and 20 MS/MS scans with 15.0 s dynamic exclusion. Automatic gain control was set at 5E4. The fixed first mass was set at 100 m/z.
Database search
The MS/MS data were processed using a Maxquant search engine (v.1.5.2.8). Tandem mass spectra were searched against the petunia database (https://solgenomics.net/organism/Petunia_axillaris/genome) concatenated with a reverse decoy database. Trypsin/P was specified as a cleavage enzyme allowing up to four missing cleavages. The mass tolerance for precursor ions was set at 20 ppm in the first search and 5 ppm in the main search, and the mass tolerance for fragment ions was set at 0.02 Da. Carbamidomethyl on Cys was specified as a fixed modification, and acetylation modification and oxidation on Met were specified as variable modifications. The false discovery rate (FDR) was adjusted to <1%, and the minimum score for modified peptides was set to >40.
Subcellular localization analysis
We used WoLF PSORT (v0.2, http://www.genscript.com/psort/wolf_psort.html), a subcellular localization predication software program, to predict the subcellular localization of proteins (Horton et al., 2007). WoLF PSORT is an updated version of PSORT/PSORT II for the prediction of eukaryotic sequences.
Gene ontology enrichment and pathway analysis
Proteins were classified by gene ontology (GO) annotation into three categories: biological process, cellular compartment, and molecular function. For each category, a two-tailed Fisher’s exact test was used to test the enrichment of the differentially modified protein against all identified proteins. GO annotations with a corrected P-value <0.05 were considered significant.
The KEGG database (v.2.5, http://www.kegg.jp/kegg/mapper.html) was used to identify enriched pathways by a two-tailed Fisher’s exact test to test the enrichment of the differentially modified protein against all identified proteins (Kanehisa and Goto, 2000). Pathways with a corrected P-value <0.05 were considered significant. These pathways were classified into hierarchical categories according to the KEGG website.
Motif analysis
The software Motif-x was used to analyse the model of sequences constituted with amino acids in specific positions of modify-21-mers (10 amino acids upstream and downstream of the site) in all protein sequences. All of the database protein sequences were used as background database parameters and other default parameters.
Preparation of specific antibodies and western blot analysis
Western blot analyses were performed according to the methods of Liu et al. (2019). The peptides (Supplementary Table S4) expressed in Escherichia coli of three proteins, PamCS (Peaxi162Scf00402g00612.1), PaPDC2 (pyruvate decarboxylase-2, Peaxi162Scf00014g00030.1) and PaGELP (GDSL, esterase/lipase, Peaxi162Scf00038g01135.1), and the synthetic acetylation peptides (QQFNKacKMNS) of the protein PaGELP255Kac, were used as antigens for antibody production in rabbits from Diaan (http://www.dia-an.cn/). These antibodies were used for blotting analysis (Liu et al. 2019).
Statistical analysis
Statistical analysis was performed using one-way analysis of variance followed by Duncan’s multiple range test with at least three replicates. P-values ≤0.05 were considered significant.
Results
Identification and expression analysis of the ACL gene family in Petunia hybrida
To identify petunia ACL genes, we used the cDNA sequences of AtACLA1 (AY113979) and AtACLB1 (AY050858) from A. thaliana (Fatland et al., 2002) as a query in BLAST and searched the P. axillaris draft genome sequence v1.6.2. Sequences from two ACLAs, PaACLA1 and PaACLA2, and two ACLBs, PaACLB1 and PaACLB2, of P. axillaris were recovered. Using the special primers of these gene sequences, we obtained the same full-length sequences of these genes from P. hybrida ‘Ultra’ by RT–PCR. The results of multiple sequence alignment showed that the four PaACLs had high identity to their homologs in A. thaliana and Solanum lycopersicum (Supplementary Fig. S1).
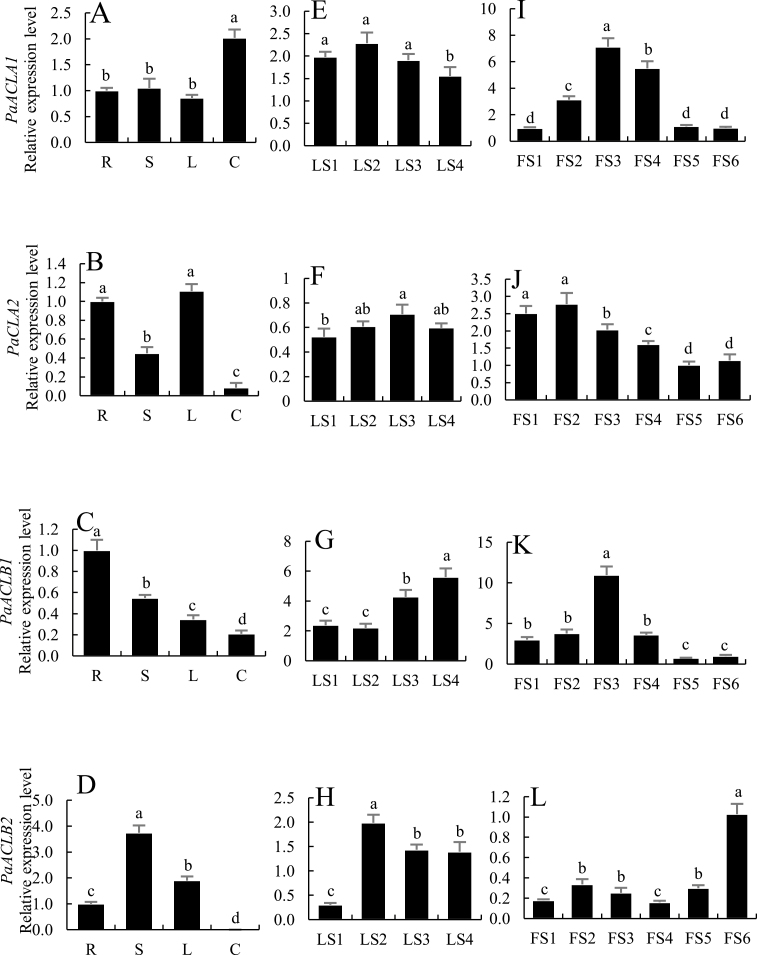
Four PaACL transcriptional levels were examined in different plant organs and during leaf and bud development by qPCR using CYP and Actin as the internal reference genes. PaACLA1, PaACLA2, PaACLB1, and PaACLB2 transcripts were all detected in roots, stems, leaves, and corollas. The expression levels of PaACLA1, PaACLA2, PaACLB1 and PaACLB2 were strongest in the corollas, leaves, roots, and stems, respectively. The lowest expression of PaACLA1 was in leaves, and the expression of the other three genes was lowest in flowers (Fig. 1A–D; Supplementary Fig. S2A–D).
Fig. 1.
Expression patterns of PaACLAs and PaACLBs determined using quantitative real-time PCR. (A–D) Expression of PaACLA1 (A), PaACLA2 (B), PaACLB1 (C), and PaACLB2 (D) in different organs. R, roots; S, stems; L, leaves; C, corollas. (E–H) Expression of PaACLA1 (E), PaACLA2 (F), PaACLB1 (G), and PaACLB2 (H) during leaf development. LS1, young leaves (1 cm); LS2, growing leaves (3 cm); LS3, mature leaves (4 cm); LS4, old leaves. (I–L) Expression of PaACLA1 (I), PaACLA2 (J), PaACLB1 (K), and PaACLB2 (L) during flower development. Flower development stages: FS1 (0.5 cm in length), FS2 (1.0 cm), FS3 (2.0 cm), FS4 (3.0 cm), FS5 (4.0 cm), and FS6 (anthesis). Cyclophilin (accession no. EST883944) was used as the internal reference gene to quantify cDNA abundance. Data are presented as means ±SD (n=3). Different letters indicate significant differences (P≤0.05).
To further assess the expression of PaACLAs and PaACLBs during leaf development, leaf development was divided into four stages: LS1 (young leaves, 1.0 cm in length), LS2 (growing leaves, 3.0 cm), LS3 (mature leaves, 4.0 cm), and LS4 (old leaves). As shown in Fig. 1E–H and Supplementary Fig. S2E–H, the expression of PaACLA1 did not change significantly from stage LS1 to LS3, whereas the expression of PaACLA2 increased significantly from LS1 to LS3. The expression of PaACLB1 increased gradually from stage LS2 to LS4. The expression of PaACLB2 significantly increased from stage LS1 to LS2 and then decreased.
Flower development was divided into six stages: FS1 (0.5 cm in length), FS2 (1.0 cm), FS3 (2.0 cm), FS4 (3.0 cm), FS5 (4.0 cm), and FS6 (anthesis). As shown in Fig. 1I–L and Supplementary Fig. S2I–L, the expression of PaACLA1 significantly increased from stage FS1 to FS3 and then decreased, the expression of PaACLA2 decreased gradually from FS1 to FS5, the expression of PaACLB1 significantly increased from FS2 to FS3 and then decreased, and the expression of PaACLB2 remained fairly stable from FS1 to FS5 and then increased.
VIGS-mediated PaACLA1-A2 and PaACLB1-B2 silencing leads to abnormal leaf and flower development and reduces total anthocyanin content
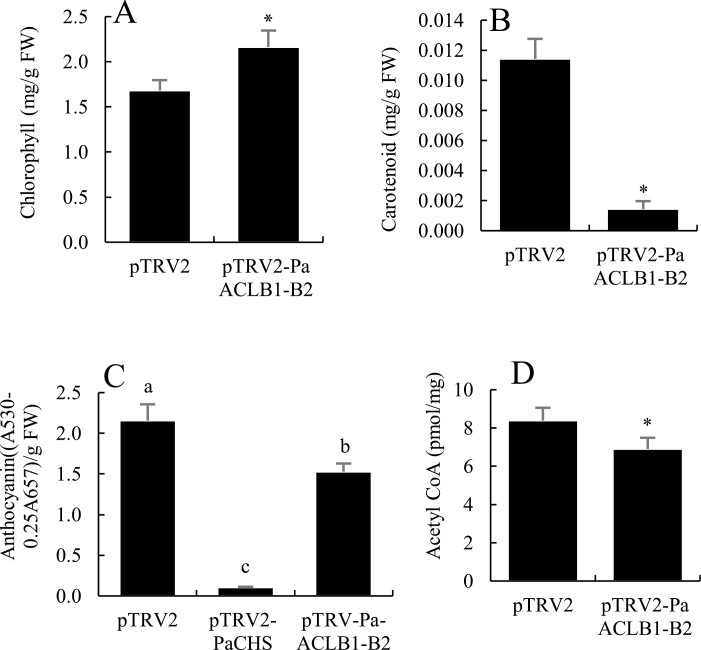
To identify the function of the PaACLA1-A2 and PaACLB1-B2 genes, vectors were constructed to suppress the expression of these genes because we had already established a highly efficient VIGS system in petunia ‘Ultra’ (Tan et al., 2014; Liu et al., 2017). Approximately 30 days after petunia seedlings were infected, no visible change was observed in any single-gene-silenced plants compared with the controls. By contrast, pTRV2-PaACLA1-A2- and pTRV2-PaACLB1-B2-infected plants had similar abnormal phenotypes, with decreased plant height, internodes, and crown width (Supplementary Table S5; Fig. 2A–C; Supplementary Fig. S3A–C). The plants were small, and the leaves were dark green, small, and uneven in shape (Fig. 2A–D; Supplementary Fig. S3A–D). The chlorophyll content was significantly higher and the carotenoid content was significantly lower in leaves, compared with controls (Fig. 3A). The flowers were smaller and the floral colour was lighter than controls, and there were some cracked and necrotic tissues at the edge of the corollas in the PaACLA1-A2- and PaACLB1-B2-silenced plants (Fig. 2H; Supplementary Fig. S3E–H). The pedicels, filaments, and styles of PaACLB1-B2-silenced plants were short, and the stigmas were small (Supplementary Table S5; Fig. 2E). The sepals were smaller and had a deeper colour than the controls (Fig. 2F, G). The colour of the anthers, the top of the pedicels, and the base of the sepals was lighter than that of the control (Fig. 2I, J). Little or no pollen was found in the anthers of PaACLB1-B2-silenced plants (Supplementary Fig. S3I). Self-pollination yielded hardly any seeds, while pollination with normal pollen yielded more seeds than self-pollination but fewer seeds than the controls (168.80±11.84 for PaACLB1-B2-silenced plants pollinated with normal pollen versus 221.80±15.83 for controls; P<0.05). The seeds produced by cross-pollinated PaACLB1-B2-silenced plants were lighter in colour than control seeds (Supplementary Fig. S3J). The total content of anthocyanins in PaACLB1-B2-silenced corollas was significantly lower compared than that of control corollas, but higher than that of PaCHS-silenced flowers, used as a positive control (Fig. 3B). In addition, the content of acetyl-CoA in PaACLB1-B2-silenced corollas was significantly lower compared with the control (Fig. 3C).
Fig. 2.
Phenotypic alterations caused by VIGS-mediated silencing of PaACLAs and PaACLBs in petunia plants. (A–G) Left to right: 5-week-old pTRV2-treated (control) and PaACLA1-, PaACLA2-, PaACLB1-, PaACLB2-, PaACLA1-A2-, and PaACLB1-B2-silenced plants: side view (A), top view (B), stems (C), leaves (D), pedicels (E), abaxial surface of sepals (F), and adaxial surface of sepals (H). (H) Left to right: flowers of the control and PaACLA1-, PaACLA2-, PaACLB1-, PaACLB2-, PaACLA1-A2-, and PaACLB1-B2-silenced plants. (I–K) Left to right: control and PaACLA1-, PaACLA2-, PaACLB1-, PaACLB2-, PaACLA1-A2- and PaACLB1-B2-silenced stamens (I), pistils (J), and stigmas (K) from 5-week-old plants. Bars=4 cm in (A) and (B); 1 cm in (C), (E), and (H); 0.5 cm in (D), (F), (G), (I), and (J); 0.15 cm in (K).
Fig. 3.
Effects of PaACLB1-B2 silencing on the chlorophyll, carotenoid, anthocyanin, and acetyl-CoA content in petunia. (A) Chlorophyll and (B) carotenoid content in leaves. (C) Anthocyanin content in corollas. (D) Acetyl-CoA content in corollas. Data are presented as means ±SD (n=3). *P≤0.05.
We examined the expression of PaACLAs and PaACLBs in corollas of different silenced plants by qPCR using CYP and Actin as internal reference genes (Supplementary Figs S4 and S5). As expected, treatment with pTRV2-PaACLA1, pTRV2-PaACLA2, pTRV2-PaACLB1, and pTRV2-PaACLB2 significantly reduced the expression of PaACLA1, PaACLA2, PaACLB1, and PaACLB2, respectively. pTRV2-PaACLA1-A2 and pTRV2-PaACLB1-B2 treatment significantly reduced the expression of the two PaACLAs and the two PaACLBs, respectively, but did not significantly change the expression of PaACLBs and PaACLAs, respectively (Supplementary Figs S4 and S5).
The ACL activities in leaves and corollas treated with pTRV2-PaACLB1-B2 and pTRV2 were determined using the purified proteins and a coupled spectrophotometric assay (Fatland et al., 2002). The results showed that PaACLB1-B2 silencing significantly reduced ACL activities in corollas compared with the control (Supplementary Fig. S6).
PaACLB1-B2 silencing changes the shape and size of cells
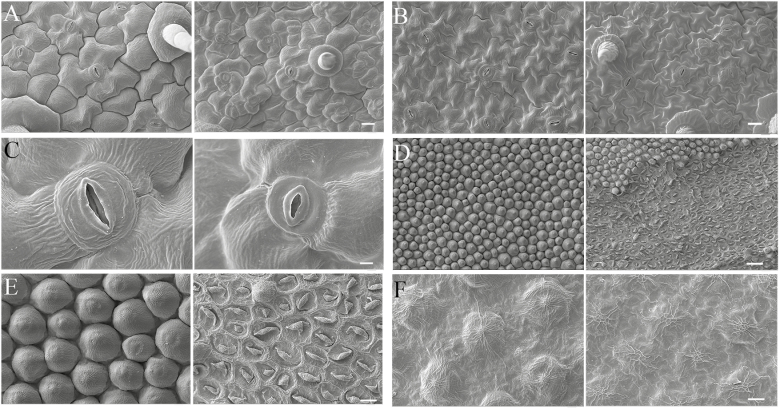
Scanning electron micrographs showed that the size of leaf and corolla abaxial and adaxial epidermis cells was smaller in the PaACLB1-B2-silenced plants than in controls (Supplementary Table S5; Fig. 4). In addition, there were a large number of dead cells in necrotic corolla tissue (Supplementary Table S5; Fig. 4D, E).
Fig. 4.
Scanning electron micrographs of the leaves of PaACLB1-B2-silenced plants (left panels) compared with those of control plants (right panels). (A) Leaf abaxial epidermal cells. (B) Leaf adaxial epidermal cells. (C) Guard cells of leaf abaxial surface. (D, E) Petal abaxial epidermal cells. (F) Petal adaxial epidermal cells. Bars=20 µm in (A) and (B); 4 µm in (C); 40 µm in (D); 10 µm in (E) and (F).
PaACLB1-B2 silencing accelerates flower senescence and increases ethylene production
We found that the longevity of PaACLB1-B2-silenced flowers was shorter than that of control flowers (5.17±0.26 versus 7.25±0.42 days) (Fig. 5A). Trypan blue staining showed that there were more dead cells in the corollas of PaACLB1-B2-silenced plants than in control flowers (Fig. 5B–D). In addition, the flowers of PaACLB1-B2-silenced plants produced more ethylene than control flowers after being open for 3 days (Fig. 5E).
Fig. 5.
PaACLB1-B2 silencing accelerates flower senescence in petunia. (A) Flowers of PaACLB1-B2-silenced and control plants. (B) Corollas for trypan blue dyeing. (C, D) Corolla tissues after trypan blue dyeing. (E) Ethylene production in PaACLB1-B2-silenced and control corollas. *P≤0.05. Bars=1 cm in (A) and (B); 0.1 cm in (C); 500 μm in (D).
PaACLB1-B2 silencing changes the metabolomic profile of petunia corollas
To further analyse the effects of PaACL silencing on the content of the metabolites, widely targeted metabolites were detected with UPLC and MS/MS in corollas from PaACLB1-B2-silenced plants and controls. To ensure that the sample collected was the PaACLB1- and PaACLB2-silenced corollas, the lighter part of the corollas of plants treated with pTRV2-PaACLB1-B2 was sampled, and silencing of both PaACLB1 and PaACLB2 in the samples was confirmed by qPCR. The qualitative and quantitative MS analysis of metabolites in the samples was performed based on the KEGG database, MWDB database, and MRM. We identified 715 metabolites, including 182 flavonoids (Supplementary Dataset S1).
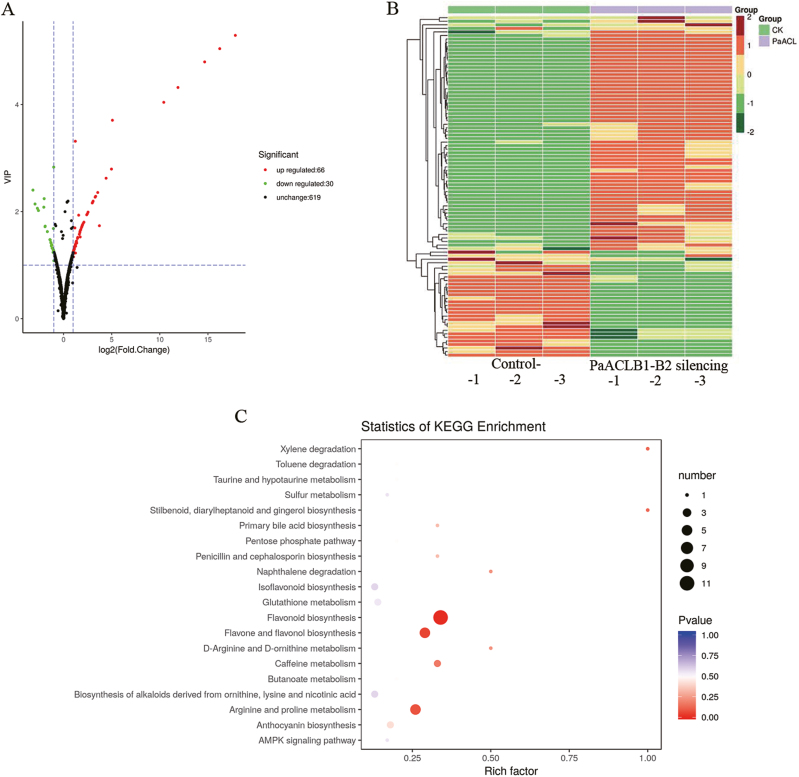
Differential metabolites were screened: the criteria for screening included a fold change value of ≥2 or ≤0.5 and a variable importance in projection value of ≥1. Thirty metabolites were down-regulated and 66 were up-regulated with a high degree of repeatability (Supplementary Dataset S2; Fig. 6A, B, Supplementary Fig. S7A), and all differential metabolites were organized based on the KEGG database using FoldChange (P<0.05). The differential metabolites were enriched in flavonoid biosynthesis, flavone and flavonol biosynthesis, and arginine and proline metabolism in PaACLB1-B2-silenced corollas (Supplementary Dataset S3; Fig. 6C). Metabolites with antioxidant activity and stress-related activity, such as 17 flavonoid metabolites [including hesperetin 7-rutinoside, 3,7-Di-O-methylquercetin, and hesperetin 7-O-neohesperidoside (>30-fold)], 9 phenolamide metabolites [including N-caffeoyl putrescine, N-feruloyl tyramine, and N′-feruloyl putrescine (>8-fold) (Supplementary Dataset S2)], and 7 coumarin metabolites [including esculin, 6,7-dimethoxy-4-methylcoumarin, and O-feruloyl 3-hydroxylcoumarin (2-3-fold)], accumulated in PaACLB1-B2-silenced plants. In addition, two anthocyanin metabolites were significantly increased, and two anthocyanin metabolites were significantly reduced. These results are not inconsistent with the reduction of total anthocyanin content in corollas, since different anthocyanin metabolites have different content in corollas (Ando et al., 1999). One terpenoid metabolite was up-regulated, and one terpenoid metabolite was down-regulated (Supplementary Dataset S2). Among the differential metabolites, all carbohydrates, coumarins, organic acids, and most differential phenolamides (9 out of 10) and lipid fatty acids (5 out of 7) were up-regulated in PaACLB1-B2-silenced corollas. In addition, the content of proline in PaACLB1-B2-silenced corollas was higher than in the control.
Fig. 6.
Analysis of the differential metabolites in PaACLB1-B2-silenced petunia corollas compared with controls. (A) Volcano plot. The green dots in the figure represent the differentially expressed metabolites that were decreased, the red dots represent the differentially expressed metabolites that were increased, and the black dots indicates metabolites for which there was no significant difference. (B) Heat map. Green indicates the differentially expressed metabolites that were decreased and red indicates the differentially expressed metabolites that were increased. The columns labelled control 1–3 and PaACLB1-B2 silencing 1–3 refer to corolla samples from independent plants. The tree on the left side of the map indicates the relationship among the metabolites according to the normalized contents of the metabolites in different samples. The smaller the differences between metabolite contents in different samples, the closer the two metabolites are placed in the tree. (C) KEGG enrichment analysis.
It is noteworthy that PaACLB1-B2 silencing up-regulated many downstream metabolites, including 17 flavonoids, 4 organic acids, 4 lipid fatty acids, 3 lipid glycerophospholipids, and 1 terpenoid of acetyl-CoA metabolism, indicating that PaACLB1-B2 silencing maintained metabolic homeostasis in petunia corollas.
Transcriptome analysis reveals that PaACLB1-B2 silencing increases the mRNA levels of genes encoding the key enzymes of biosynthesis of secondary metabolites
To explore the mechanism of PaACL silencing-induced up-regulation of many metabolites, we performed transcriptome analysis of corollas from PaACLB1-B2-silenced plants and controls. Three biological replicates were analysed for each treatment. In total, ~29.34 million clean reads were generated (Supplementary Dataset S4). PaACLB1-B2 silencing resulted in the differential expression of 932 genes, with 733 up-regulated and 199 down-regulated genes (Supplementary Dataset S5), with a high degree of repeatability (Supplementary Fig. S7B).
To investigate the influence of the DEGs on pathways, statistical pathway enrichment analysis of PaACLB1-B2-silenced corollas and controls was performed based on the KEGG database using FoldChange and FDR. The DEGs were enriched in 13 KEGG metabolic pathways (Supplementary Dataset S6). The top 10 significant (P<0.05) metabolic pathways of the DEGs were phenylpropanoid biosynthesis; ubiquinone and other terpenoid-quinone biosynthesis; biosynthesis of secondary metabolites; metabolism of xenobiotics by cytochrome P450; glutathione metabolism; cutin, suberine, and wax biosynthesis; amino sugar and nucleotide sugar metabolism; phenylalanine metabolism; ascorbate and aldarate metabolism; and pentose and glucuronate interconversions (Supplementary Fig. S8). These results supported that acetyl-CoA is widely involved in plant secondary metabolism.
To elucidate the functional differences between the DEGs, they were analysed for GO enrichment based on clustering analysis (Supplementary Dataset S7). In the cellular component category, many of the DEGs were enriched in extracellular region, cell wall, external encapsulating structure, apoplast, intrinsic component of plasma membrane, plant-type cell wall, extracellular space, extracellular region part, plasma membrane part, and anchored component of plasma membrane. These results suggest that acetyl-CoA could be associated with the intracellular environment.
In terms of molecular functions, a large proportion of the DEGs were highly enriched in haem binding, oxidoreductase activity, monooxygenase activity, tetrapyrrole binding, iron ion binding, cofactor binding, transition metal ion binding, carbohydrate transmembrane transporter activity, hydrolase activity, and peroxidase activity. These results suggested that the reduction of acetyl-CoA could affect the activities of many enzymes, including the enzymes of stress-response and catabolic processes (Supplementary Dataset S7). The analysis of biological processes showed that many DEGs were highly enriched for the following: hormone biosynthetic process, phytosteroid metabolic process, hormone metabolic process, carbohydrate transport, regulation of hormone levels, steroid metabolic process, cellular response to chemical stimulus, hormone-mediated signalling pathway, chemical homeostasis, and steroid biosynthetic process (Supplementary Dataset S7). Significant pathway enrichment analysis showed that plant hormone biosynthesis was the most important pathway in PaACLB1-B2 silencing and control, and plant hormone biosynthesis was the key biological event. Plant hormone biosynthesis is very important for plant growth and development, including flower senescence.
As expected, the mRNA levels of both PaACLB1 and PaACLB2 were down-regulated (4.5-fold) and those of both PaACLA1 and PaACLA2 were not significantly changed. We found that the mRNA levels of the genes encoding two key enzymes of ethylene synthesis, 1-aminocyclopropane-1-carboxylate synthase (ACS), PaACS9 (Peaxi162Scf00096g01846, >100-fold) and PaACS11 (Peaxi162Scf00498g00034, 29.5-fold), and 1-aminocyclopropane-1-carboxylate oxidase (ACO), PaACO4 (Peaxi162Scf01333g10016, 10.2-fold) and PaACO1 (Peaxi162Scf00047g01927, 3.5-fold), as well as ethylene receptor 2 (PaETR2, Peaxi162Scf00024g00157, 10-fold), were up-regulated. In addition, the expression of histone genes (Peaxi162Scf00037g00927 and Peaxi162Scf00697g00054) was up-regulated (Supplementary Dataset S6). Furthermore, we performed qPCR of PaACS9, PaACS11, PaACO1, PaACO4, PaETR2, and a senescence-related transcription factor gene, PaWRKY23 (Peaxi162Scf00770g00114) (Ruan et al., 2019). The results of qPCR assays (Supplementary Figs S9 and S10) for these genes were in agreement with the DEG results.
PaACLB1-B2 silencing-induced changes in the proteome profile of petunia corollas
We investigated the proteome in petunia corollas treated with pTRV2-PaACLB and pTRV2. Three biological replicates were analysed for each treatment. In total, 6200 protein groups were identified from petunia, among which 5343 proteins were quantified (Supplementary Dataset S8). A total of 345 proteins were up-regulated and 182 proteins were down-regulated (with a threshold of 1.2-fold) in PaACLB1-B2-silenced plants compared with the control (P<0.05), with a high degree of repeatability (Supplementary Dataset S9; Supplementary Fig. S11).
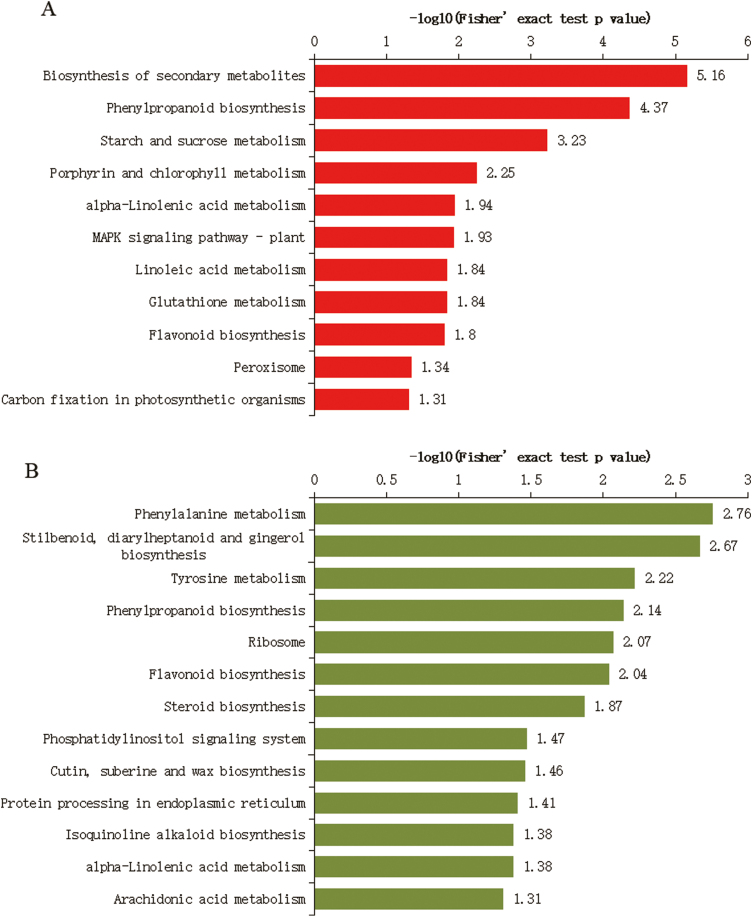
The up- and down-regulated proteins were enriched in 11 and 13 KEGG metabolic pathways, respectively (Supplementary Dataset S10). Many of the up-regulated proteins were enriched in the biosynthesis of secondary metabolites, phenylpropanoid biosynthesis, starch and sucrose metabolism, porphyrin and chlorophyll metabolism, alpha-linolenic acid metabolism, MAPK signalling pathway, linoleic acid metabolism, glutathione metabolism, flavonoid biosynthesis, peroxisome, and carbon fixation in photosynthetic organisms (Fig. 7A). Many of the down-regulated proteins were enriched in phenylalanine metabolism, stilbenoid, diarylheptanoid and gingerol biosynthesis, tyrosine metabolism, phenylpropanoid biosynthesis, ribosome, flavonoid biosynthesis, steroid biosynthesis, phosphatidylinositol signalling system, cutin, suberine and wax biosynthesis, and protein processing in endoplasmic reticulum (Fig. 7B). These results further showed that acetyl-CoA is widely involved in secondary metabolism, including flavonoid biosynthesis and the stress response.
Fig. 7.
KEGG pathway enrichment analysis of differentially expressed proteins in PaACLB1-B2-silenced petunia corollas compared with controls. (A) Up-regulated proteins; (B) down-regulated proteins. The significance level was set at P<0.05 (Fisher’s exact test). Data are from Supplementary Dataset S10.
As expected, both PaACLB1 (3.0-fold) and PaACLB2 (>4.0-fold) were significantly down-regulated. Unexpectedly, the abundance of both PaACLA1 (1.4-fold) and PaACLA2 (6.2-fold) was significantly reduced. The abundance of PaACO1, PaCHSA (Peaxi162Scf00047g01225, 2.0-fold), four acyl transferases (Peaxi162Scf00003g00487, 1.2-fold; Peaxi162Scf00047 g00128, 1.2-fold; Peaxi162Scf00160g01037.1, 1.2-fold; Peaxi162Scf00075g01225, 1.3-fold) and two histone proteins (H2A.11, Peaxi162Scf71358g00001, and another histone protein, Peaxi162Scf00836g00036) was significantly increased (Supplementary Dataset S9). The abundance of all six differentially expressed proteins enriched in porphyrin and chlorophyll metabolism were up-regulated in PaACLB1-B2-silenced plants compared with controls (Supplementary Dataset S9).
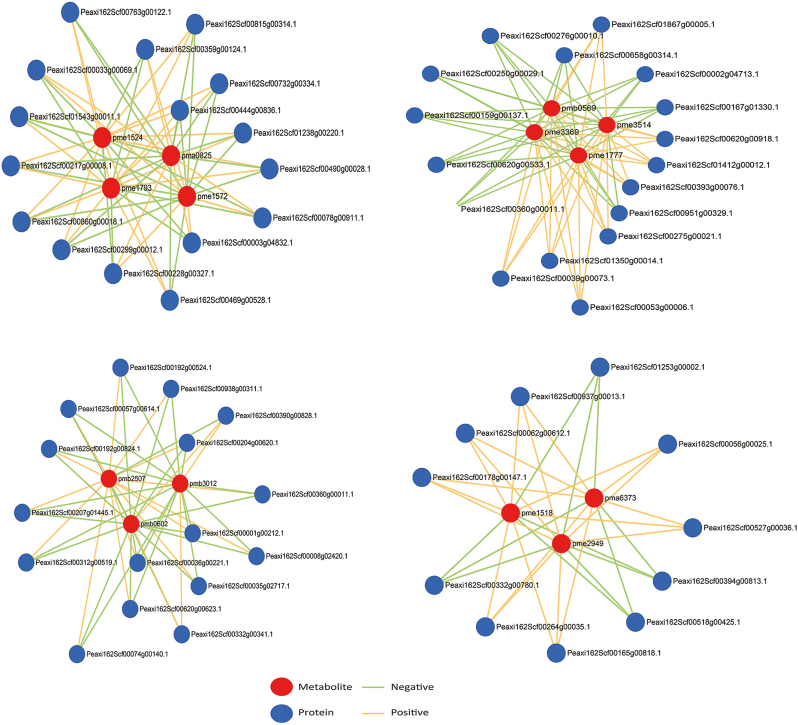
To study the potential relationship between differential proteins and differential metabolites, we analysed the KEGG pathway annotation of differential proteins and differential metabolites. Twenty pathways with both differential proteins and differential metabolites, which were enriched in the flavonoid biosynthesis pathway, were screened out (Supplementary Figs S12 and S13). The metabolite–protein co-expression network of 14 differential flavonoid metabolites and 58 differential proteins in the corollas of PaACLB1-B2 silenced plants and controls is shown in Fig. 8.
Fig. 8.
Metabolite–protein co-expression network of the significant differential flavonoid metabolites and differential proteins in the corollas of PaACLB1-B2 silenced petunia plants and controls. The node size correlates with the ratio between responders and non-responders.
Effects of PaACLB1-B2 silencing on lysine acetylation
Since acetyl-CoA is the substrate of lysine acetylation, the acetylated proteins and their modification sites were identified using TMT labelling and lysine acetylated (Kac) affinity enrichment followed by high-resolution LC-MS/MS in petunia corollas treated with pTRV2-PaACLB1-B2 or pTRV2. A total of 2210 lysine acetylation sites in 1148 protein groups were identified, among which 1744 sites in 921 proteins were accurately quantified (Supplementary Dataset S11). In the identification, the mass errors were lower than 10 ppm, confirming that the sample preparation reached the standard (Supplementary Fig. S14A). The length of most peptides was between 8 and 20 amino acids, consistent with the properties of tryptic peptides (Supplementary Fig. S14B). We subsequently used the quantification results of the global proteome to normalize the acetylome quantification data. Unexpectedly, 68 sites in 54 lysine acetylation proteins were quantified as up-regulated targets, and 40 sites in 38 lysine acetylation proteins were quantified as down-regulated targets at a threshold of 1.2 (P<0.05) in PaACLB1-B2-silenced plants compared with the control, with a high degree of repeatability (Supplementary Dataset S12; Supplementary Fig. S15). The acetylated proteins contained from one to 21 acetylation sites, and there were 678 acetylated proteins containing only one acetylation site, accounting for 59.0% of the total acetylated proteins. The proportions of proteins with two, three, four, or more modification sites were 19.2, 10.0, 5.1, and 6.6%, respectively. Several MS/MS spectra corresponding to sites from proteins that undergo acetylation are presented in Supplementary Fig. S16. These results are expected to supply valuable resources for post-translational modification studies in the future.
We performed enrichment pathway analysis based on the KEGG database using FoldChange (P<0.05). The proteins with up-regulated acetylation levels were enriched in one KEGG metabolic pathway, tyrosine metabolism (Supplementary Dataset S13; Supplementary Fig. S17A). The proteins with down-regulated acetylation levels were enriched in five KEGG metabolic pathways: oxidative phosphorylation, glycolysis/gluconeogenesis, pentose phosphate pathway, fructose and mannose metabolism, and purine metabolism (Supplementary Dataset S13; Supplementary Fig. S17B). These results further showed that the reduction of acetyl-CoA is associated with primary metabolism.
We further performed GO analysis of the proteins with differential acetylation levels (Supplementary Dataset S14). With regard to molecular function, many proteins with up-regulated acetylation levels were enriched in phosphoglycerate kinase activity, transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer, protein dimerization activity, carbon–nitrogen lyase activity, phosphotransferase activity, and carboxyl group as acceptor. Many proteins with down-regulated acetylation levels were enriched in fructose-bisphosphate aldolase activity, aldehyde-lyase activity, carbon–carbon lyase activity, lipid binding, lyase activity, nucleobase-containing compound kinase activity, metal cluster binding, and iron–sulfur cluster binding. These results showed that acetyl-CoA is involved in many post-translational modifications of proteins or transferring acyl or carboxyl groups of metabolites, and affects enzyme activities.
In terms of biological processes, a large proportion of proteins with up-regulated acetylation levels were enriched in nucleosome organization, DNA packaging, nucleosome assembly, chromatin assembly, protein–DNA complex subunit organization, chromatin assembly or disassembly, protein–DNA complex assembly, cellular component assembly, macromolecular complex assembly, and DNA conformation change. Many proteins with down-regulated acetylation levels were enriched in purine nucleoside metabolic process, purine ribonucleoside metabolic process, ribonucleoside metabolic process, nucleobase-containing small molecule metabolic process, purine-containing compound metabolic process, purine ribonucleoside triphosphate metabolic process, purine nucleoside triphosphate metabolic process, nucleoside metabolic process, glycosyl compound metabolic process, and ribonucleoside triphosphate metabolic process. These results showed that the reduction of acetyl-CoA resulted in the occurrence of a large number of nuclear events, including transcription regulation and DNA synthesis.
Unexpectedly, the levels of acetylation were down-regulated in 15 and up-regulated in 17 proteins located in the cytoplasm, and the levels of acetylation of many proteins located in the chloroplast, nucleus, and plasma membrane were significantly changed because acetyl-CoA usually does not penetrate cell membranes. These results indicated that acetyl-CoA deficiency, as caused by reduced PaACL activity, affects crosstalk between different intracellular organelles and compartments, which is a response to metabolic stress and causes significant metabolic adaptation.
The acetylation of PaACLA1 (six sites), PaACLB1 (one site), and PaACLB2 (two sites) was detected, and interestingly, the differential acetylation levels of two, one, and two sites in PaACLA1, PaACLB1, and PaACLB2, respectively, were all up-regulated in PaACLB1-B2-silenced plants compared with the control. Thus, a negatively regulated loop was formed, such that PaACL negatively regulated the acetylation level of PaACL itself by regulating the level of acetyl-CoA (Fig. 9). Whether the acetylation of PaACL enhances enzyme activity requires further study. The acetylation levels of eight sites of two histones, including histone H2A.11, were up-regulated (Supplementary Dataset S12).
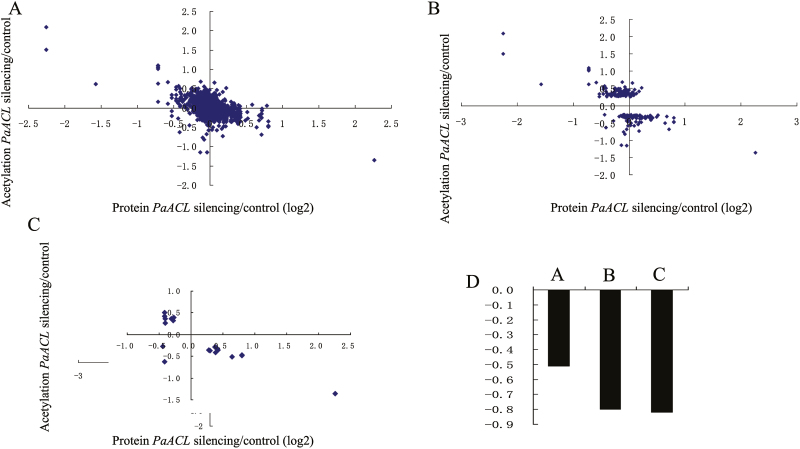
Fig. 9.
Concordance between changes in proteins and their acetylation. (A–C) Correlation between protein and acetylation fold changes in petunia corollas treated with pTRV2-PaACLB1-B2 compared with those treated with pTRV2 (control) for all acetylation/protein pairs (A), significantly changed proteins (B), and significantly changed acetylation sites (C). (D) Pearson correlations of the comparisons shown in (A–C).
In addition, we identified a total of 12 conserved motifs of acetylated sites (Supplementary Fig. S18A) and analysed the frequencies of neighbouring amino acid residues for acetylated lysines using iceLogo (Supplementary Fig. S18B) (Colaert et al., 2009). To study the evolutionary conservation of acetylated lysine and non-acetylated lysine in plants, we aligned petunia proteins with their respective orthologues from three other species. The results unexpectedly showed that acetylated lysines are significantly more conserved than non-acetylated lysines, suggesting that acetylated lysines maintain a stronger selective pressure than non-acetylated lysines in plants (Supplementary Fig. S19).
We selected three proteins, PaACO1 (Peaxi162Scf00047g01927), PaCHSA, and PamCS (mitochondrial citrate synthase, Peaxi162Scf00402g00612), and one additional protein, Argonaute (PaARG717Kac, Peaxi162Scf00050g00169), to examine their protein and acetylation abundance by western blotting. The results of western blotting using antibodies raised against these proteins were consistent with the results of the proteome and acetylome analyses (Supplementary Fig. S20).
Correlation between the global proteome and acetylome
Our data on the proteome and acetylome of pTRV2- and pTRV2-PaACLB1-B2-treated corollas allowed us to examine the correlation between the global proteome and acetylome. There were 1744 quantified Kac sites in 920 quantified proteins in this study (Supplementary Dataset S15). Of the 920 quantified proteins, 25 proteins were down-regulated and 58 were up-regulated. Pearson’s correlation coefficient was calculated as –0.51 when all quantified proteins were considered in terms of their quantified acetylation (Fig. 9A, D) and increased to –0.80 when all significantly altered proteins were considered in terms of their quantified acetylation (Fig. 9B, D). For acetylation/protein pairs with significantly altered proteins and acetylation, a larger negative correlation was observed (r=–0.82; Fig. 9C, D). Therefore, the global proteome and acetylome were negatively correlated.
Interestingly, in acetylation/protein pairs with significantly altered proteins and acetylation, the changed direction of all acetylation sites in one protein was the same, which allowed us to perform KEGG pathway analysis of the differential proteins with acetylation sites in the opposite direction. These proteins were enriched in two KEGG pathways, fatty acid degradation and alpha-linolenic acid metabolism (Supplementary Dataset S16, Supplementary Fig. S21).
Discussion
Acetyl-CoA is a central metabolite and the acetyl source for acetylation of many metabolites and proteins (Oliver et al., 2009; Chen et al., 2017a). In this study, we investigated the effects of the reduction of ACL-derived cytosolic acetyl-CoA on growth, development, and the global metabolome, and performed transcriptome, proteome, and acetylome analyses of petunia corollas.
In this study, we first analysed the spatial and temporal expression of four genes encoding two subunits of PaACL during plant growth and development. The mRNAs of four PaACLs were detected in all organs (roots, stems, leaves, and corollas) examined, but were present at different levels in different organs and stages of development (Fig. 1Supplementary Fig. S2), indicating the importance of PaACL in the growth and development of plants. Although the expression patterns of the four genes were not completely consistent, there was functional redundancy in the different genes encoding each subunit.
We produced VIGS-mediated PaACLA- and PaACLB-silenced plants. Silencing of both PaACLB1 and PaACLB2 (PaACLB1-B2) or both PaACLA1 and PaACLA2 (PaACLA1-A2) resulted in similar abnormal growth and development of plants, while silencing of any single PaACL gene did not result in any visible phenotype change; this demonstrated that both ACL subunits were required to form a functional ATP-citrate lyase, and that there was functional redundancy between PaACLB1 and PaACLB2, and between PaACLA1 and PaACLA2. Similarly, both acl1 and acl2 subunits are required to form a functional ATP-citrate lyase in Aspergillus niger (Chen et al., 2013).
Silencing of PaACLB1-B2 reduced the content of acetyl-CoA and resulted in abnormal leaf and flower development (Fig. 2D–JSupplementary Fig. S3E). Small flower sizes, necrotic tissues in the corolla edge, and short filaments, styles, and pedicels were observed in PaACLB1-B2-silenced plants. Consistently, the adaxial and abaxial epidermal cells of corollas in the PaACLB1-B2-silenced plants were small compared with the control, as observed by scanning electron microscopy (Fig. 4). In A. thaliana, plants with reduced ACL activity showed a similar phenotype (Fatland et al., 2005). In addition, PaACLB1-B2 silencing reduced the fertility of both stamens and pistils, accelerated flower senescence, and increased ethylene production, features that were not reported in A. thaliana. These results indicate the important roles of PaACL in plant growth and development.
Consistent with A. thaliana with antisense ACLA (Fatland et al., 2005), PaACLB1-B2 silencing changed the chlorophyll content and anthocyanin accumulation in both corollas and seeds (Fig. 3A, C; Supplementary Fig. S3J). In PaACLB1-B2-silenced plants, the amino acid content in leaves changed greatly, especially the accumulation of proline (Supplementary Dataset S2). Proline accumulation has been reported during conditions of drought, oxidative stress, and in response to biotic stresses (Fabro et al., 2004; Choudhary et al., 2005; Haudecoeur et al., 2009; Yang et al., 2009). Fatland et al. (2005) suggested that the shortage of acetyl-CoA in A. thaliana resulted in physiological stress, termed ‘metabolic stress’. In this study, the shortage of acetyl-CoA induced by PaACLB1-B2 silencing resulted in abnormal leaf and flower development, necrotic tissues in the corolla edge, acceleration of flower senescence, and higher proline content, which further supports the hypothesis that metabolic stress is induced by ACL silencing.
Metabolome analysis of petunia corollas showed that PaACLB1-B2 silencing changed the metabolome profile (Supplementary Dataset S2). The accumulation of primary metabolites (e.g. soluble sugars, amino acids, organic acids, lipids) exerts resistance to metabolic stress and maintains the metabolism of cells. Secondary metabolites may play an important role as antioxidants; these include flavonoids, phenolamides, and coumarins, and other metabolites that were significantly induced or inhibited by metabolic stress in PaACLB1-B2-silenced plants. The involvement of phenolamides in plant defence against biotic and abiotic stresses has also been proposed (Buanafina, 2009; Park et al., 2009; Bassard et al., 2010; Demkura et al., 2010; Kaur et al., 2010; Onkokesung et al., 2012). Some coumarin compounds have been identified as phytotoxic metabolites and function in the defence response of plants against biotic and abiotic stresses (Baillieul et al., 2003; Shimizu et al., 2005; Prats et al., 2007; Gnonlonfin et al., 2012; Sun et al., 2014; Liu et al., 2017b; Duan et al., 2019). Although acetyl-CoA is the precursor of many metabolites, including flavonoids, PaACLB1-B2 silencing up-regulated the content of 66 metabolites, including 29 downstream metabolites of acetyl-CoA metabolism, while only 30 metabolites were down-regulated. In A. thaliana plants in which ACLA was suppressed, the concentration of anthocyanins, a group of flavonoids, in rosettes and inflorescence stems increased. These results demonstrated the reconstitution of metabolic homeostasis in the case of a shortage of acetyl-CoA in PaACLB1-B2-silenced petunia corollas.
The transcriptome analysis in this study showed that PaACLB1-B2 silencing activated the transcription of many genes, especially genes involved in the biosynthesis of secondary metabolites, phenylpropanoid biosynthesis, and arginine and proline metabolism (Supplementary Dataset S4). In line with these results, in the metabolome analysis, the contents of many secondary metabolites, including phenylpropanoids and proline, increased. These results showed that PaACLB1-B2 silencing changed the transcriptome profile to some extent to maintain metabolic homeostasis.
The proteome analysis showed that 345 proteins were up-regulated and only 182 proteins were down-regulated in PaACLB-B2-silenced plants compared with the control. Most differentially expressed proteins were enriched in KEGG pathways including biosynthesis of secondary metabolites, phenylpropanoid biosynthesis pathways, and phenylalanine metabolism. In the proteome and metabolome analyses, both the differential proteins and the differential metabolites were enriched in the flavonoid biosynthesis pathway. In addition, many up-regulated proteins were enriched in porphyrin and chlorophyll metabolism and starch and sucrose metabolism, and the contents of chlorophyll and all differential carbohydrate metabolites were up-regulated. Moreover, there were 20 pathways in which both differential proteins and differential metabolites were distributed, some of which were enriched in the flavonoid biosynthesis pathway. These results show that PaACLB1-B2 silencing changed the proteome profile to maintain metabolic homeostasis. The down-regulated proteins were enriched in the KEGG pathways stilbenoid, diarylheptanoid and gingerol biosynthesis, tyrosine metabolism, flavonoid biosynthesis, and cutin, suberine and wax biosynthesis, most likely indicating the feedback regulation of metabolites on the abundance of synthetic enzymes.
Analysis of qPCR and transcriptome data showed that the expression of PaACS9, PaACS11, PaACO1, and PaACO4, which encode the key enzymes in ethylene biosynthesis, was up-regulated, and proteome data showed that PaACO1 abundance was up-regulated compared with the control. A recent study in rice (Oryza sativa) showed that a reduction in OsACL-A2 abundance resulted in evident phenotypes, with small lesion-mimic leaves and enhanced immunity to bacterial blight, and an increase in the expression of senescence-related genes (Ruan et al., 2019). It is possible that the metabolic stress induced by the reduction of acetyl-CoA in PaACLB1-B2-silenced plants promoted flower senescence by increasing the expression of cell death- and senescence-related genes, including PaACO and PaACS (Supplementary Fig. S22). These results indicate that the changes in transcriptome and proteome profiles are associated with abnormal growth and development caused by PaACLB1-B2 silencing.
Acetyl-CoA is the source of acetyl for protein acetylation, including histone acetylation, which promotes gene expression. Unexpectedly, protein lysine acetylome analysis showed that the reduction of acetyl-CoA up-regulated the acetylation level of more proteins of different localization. Consistently, the abundance of four acyl-transferases was up-regulated (Supplementary Dataset S6). Only a minority of differentially Lys-acetylated proteins (~30%) was localized in the cytosol of ACL-depleted plants. Since other compartments have their own acetyl-CoA sources, the major fraction of affected proteins appears to be perturbed due to pleiotropic effects. This may also explain why more proteins were more frequently Lys-acetylated in the mutants. The proteins with different acetylation levels were enriched in several KEGG metabolic pathways associated with primary metabolism (Supplementary Dataset S13). The results of GO analysis showed that the proteins with different levels of acetylation were enriched in many post-translational modifications of proteins or transferring acyl or carboxyl groups of metabolites, and affected the enzyme activities. It is possible that acetyl-CoA may be used effectively under conditions of a shortage of acetyl-CoA. These results showed that PaACLB1-B2 silencing changed the acetylome profile to maintain metabolic homeostasis.
Two categories of enzymes, CHS and CHI, play critical roles in flavonoid synthesis. Interestingly, CHS and CHI usually play key roles in the plant response to stress (Koes et al., 2005; Falcone Ferreyra et al., 2012; Ishida et al., 2016). AtCHS expression was significantly increased concomitantly with the accumulation of anthocyanins in A. thaliana plants subjected to salt stress (Piao et al., 2001). In this study, the abundance of PaCHSA and the acetylation level of PaCHI increased, which explained, at least partially, the cause of the increase in many flavonoids in the corollas of PaACLB1-B2 silenced plants. Correspondingly, the increase in the abundance of PaANS could be connected with the increase in some anthocyanin metabolites (Supplementary Fig. S23). The abundance of the biosynthetic enzyme of proline, PaP5CS1 (Peaxi162Scf00658g00314), whose transcript level did not change, was significantly up-regulated, and could be connected to the observed increase of proline contents. These results further support that metabolic homeostasis is maintained in PaACLB1-B2 silenced plants by changing the proteome and acetylome.
In mammalian cells, ACL is required for increases in histone acetylation in response to growth factor stimulation and during differentiation, and fluctuation of cellular acetyl-CoA levels affects histone acetylation in yeast and mammalian cells at multiple lysine residues of histone (H3 and H4) tails (Wellen et al., 2009). In A. thaliana, the increase of H3K27 acetylation (H3K27ac) is dependent on cytoplasmic ACL (Chen et al., 2017a). In this study, we did not detect a change in the acetylation level of H3K27, but we did detect up-regulation of the levels of acetylation of histones H2A.11 and another histone superfamily protein. Obviously, these were not direct consequences of the reduction of acetyl-CoA, which could be attributed to PaACLB1-B2 silencing-induced metabolic stress.
Silencing of PaACLA1-A2 or PaACLB1-B2 did not result in a change in the mRNA levels of the PaACLBs and PaACLAs (Supplementary Figs S4 and S5), respectively, but resulted in not only a significant reduction in PaACLB1 and PaACLB2 abundance (as expected) but also a significant reduction in PaACLA1 and PaACLA2 abundance. Similarly, in A. thaliana, the abundance of both ACLA and ACLB is reduced in antisense ACLA plants (Fatland et al., 2005). There is no sequence similarity between any of the ACLA and ACLB genes; thus, this coordination in expression is not a co-suppression mechanism. We speculate that coordination between ACLA and ACLB expression occurs via a post-transcriptional mechanism and that excess ACLA subunits may be turned over in the absence of ACLB. In addition, the levels of acetylation of PaACLA1, PaACLB1, and PaACLB2 were up-regulated in PaACLB1-B2-silenced plants, and we could not rule out the possibility that increasing the acetylation of PaACLs increased their activities.
Previous studies suggested that the proteome and ubiquitylome were negatively correlated in petunia corollas and human cells, since ubiquitination plays important roles in protein degradation (Guo et al., 2017; Jiang et al., 2018). In this study, surprisingly, the global proteome and acetylome were negatively correlated in petunia corollas treated with pTRV2-PaACLB1-B2 and pTRV2 (Supplementary Dataset S15), implying that proteome expression levels were negatively regulated by acetylation in PaACLB1-B2-silenced plants compared with the control. It is possible that under conditions of acetyl-CoA deficiency, the abundance of some proteins with high acetylation levels was reduced to save the acetyl-CoA source. In eukaryotes, acetylation has been shown to affect the rate of turnover of certain proteins (Hershko et al., 1984). The negative correlation between the proteome and acetylome might occur only under particular conditions, such as acetyl-CoA deficiency.
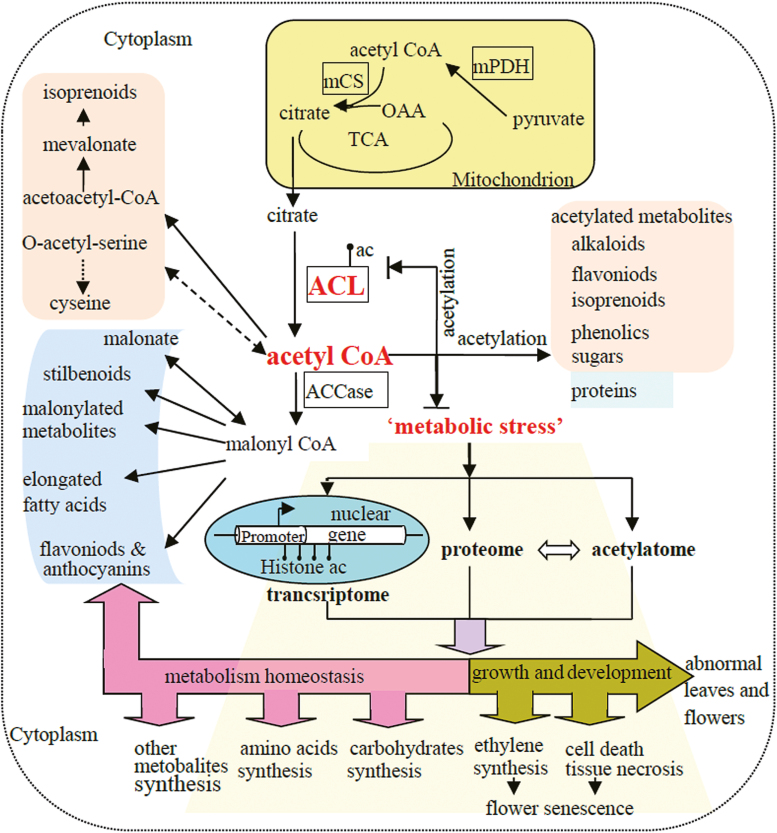
In conclusion, our results show that petunia PaACL plays important roles in the synthesis of most secondary metabolites and in plant growth and development, including flower senescence. Moreover, the reduction in cytosolic acetyl-CoA increased the content of many secondary metabolites and produced a new metabolic homeostasis by metabolic stress-induced changes in the transcriptome, proteome, and acetylome profiles of petunia corollas (Fig. 10). In addition, the negative correlation between the proteome and acetylome, which has not been reported previously, might occur only under specific conditions, such as metabolic stress induced by PaACL silencing.
Fig. 10.
Acetyl-CoA metabolism and the regulation of PaACLB1-B2 silencing-induced metabolic stress in petunia. ACL, ATP-citrate lyase; ac, acetylation; ACCase, acetyl-CoA carboxylase; mCS, mitochondrial citrate synthase; OAA, oxaloacetate; TCA, tricarboxylic acid cycle; mPDH, mitochondrial pyruvate dehydrogenase.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Specific primer sequences used for full-length ACL gene isolation from Petunia hybrida ‘Ultra’.
Table S2. Primer sequences used in quantitative real-time PCR.
Table S3. Specific primer sequences used for VIGS vector construction.
Table S4. Peptides expressed in Escherichia coli of three proteins, PamCS, PaPDC2, and PaGELP.
Table S5. Effects of PaACLB1-B2 silencing on the plants and cell size.
Fig. S1. Predicted amino acid sequence alignments and neighbour-joining trees of ACLAs and ACLBs.
Fig. S2. Expression patterns of PaACLAs and PaACLBs determined by quantitative real-time PCR using Actin as the internal reference gene.
Fig. S3. Phenotypic alteration of VIGS-mediated silencing of PaACLB1 and PaACLB2 in plants.
Fig. S4. Expression of PaACLAs and PaACLBs determined using quantitative real-time PCR.
Fig. S5. Expression of PaACLAs and PaACLBs determined by quantitative real-time PCR using Actin as the internal reference gene.
Fig. S6. ACL activities of PaACLB1-B2 silenced flowers and controls.
Fig. S7. Repeatability analysis of metabolome and transcriptome.
Fig. S8. KEGG pathway enrichment analysis of differentially expressed genes in petunia corollas of PaACLB1-B2-silenced plants and controls.
Fig. S9. Confirmation of expression data for six senescence-related genes by quantitative real-time PCR using Cyclophilin as the internal reference gene.
Fig. S10. Confirmation of expression data for six senescence-related genes by quantitative real-time PCR using Actin as the internal reference gene.
Fig. S11. Repeatability test between samples in the proteome of petunia corollas of PaACLB1-B2-silenced plants and controls.
Fig. S12. Distribution of KEGG pathways of differential proteins and differential metabolites.
Fig. S13. Bubble chart of KEGG enrichment analysis.
Fig. S14. Quality control of mass spectrometry data.
Fig. S15. Repeatability test between samples in protein lysine acetylome.
Fig. S16. MS/MS spectra of lysine acetylation of several proteins.
Fig. S17. KEGG pathway enrichment analysis of proteins with up-regulated and down-regulated Kac sites.
Fig. S18. Motif analysis of all the identified Kac sites in petunia.
Fig. S19. Evolutionary conservation of acetylated and non-acetylated lysines on protein orthologues in selected species.
Fig. S20. Confirmation of proteome and acetylome data.
Fig. S21. Differential proteins with acetylation sites in the opposite direction were enriched in the fatty acid degradation and alpha-linolenic acid metabolism KEGG pathways.
Fig. S22. Effects of PaACLB1-B2 silencing on the proteins engaged in ethylene biosynthesis in petunia.
Fig. S23. Effects of PaACLB1-B2 silencing on anthocyanin and other flavonoid biosynthesis and the proteins engaged in their biosynthesis pathway in petunia.
Dataset S1. Total metabolites identified in petunia corollas.
Dataset S2. Differential metabolites in petunia corollas of PaACLB1-B2 silenced plants and controls.
Dataset S3. KEGG pathway analysis of differential metabolites in petunia corollas of PaACLB1-B2-silenced plants and controls.
Dataset S4. Transcriptome analysis of corollas from the PaACLB1-B2-silenced plants and controls.
Dataset S5. Differential mRNAs in corollas from the PaACLB1-B2-silenced plants and controls.
Dataset S6. KEGG pathway enrichment of differential mRNAs in corollas from the PaACLB1-B2-silenced plants and controls.
Dataset S7. GO enrichment of differential mRNAs in corollas from the PaACLB1-B2-silenced plants and controls.
Dataset S8. Proteome analysis of corollas from the PaACLB1-B2-silenced plants and controls.
Dataset S9. Differential proteins in corollas from the PaACLB1-B2-silenced plants and controls.
Dataset S10. KEGG pathway enrichment of differential proteins in corollas from the PaACLB1-B2-silenced plants and controls.
Dataset S11. Acetylome analysis of the corollas from the PaACLB1-B2-silenced plants and controls.
Dataset S12. Differential acetylation sites in proteins in corollas from the PaACLB1-B2-silenced plants and controls.
Dataset S13. KEGG pathway enrichment of proteins with differential acetylation sites in corollas from the PaACLB1-B2-silenced plants and controls.
Dataset S14. GO enrichment of proteins with up- and down-regulated acetylation sites in corollas from the PaACLB1-B2-silenced plants and controls.
Dataset S15. Connection of proteome and acetylome
Dataset S16. Differential proteins with opposite direction differential acetylation sites.
Acknowledgements
We thank Jingjie PTM Biolab Co. Ltd (Hangzhou, China) and Metware Ltd Co. (Wuhan, China) for the multi-omics service. This study was supported by the National Natural Science Foundation of China (31870692, 31770737, 31701953, and 31661143047) and the National Key Research and Development Plan (2018YFD1000407).
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (https://www.ebi.ac.uk/pride/; Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD015480. The raw RNA sequence data have been submitted to NCBI with accession number PRJNA577189 (https://dataview.ncbi.nlm.nih.gov/object/PRJNA577189).
Author contributions
YY planned and designed the research. HZ, SZ, LS, XZ, ZC, QW, GC, and JL performed experiments, conducted fieldwork, and analysed the data. YY and HZ wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Ando T, Saito N, Tatsuzawa F, et al. 1999. Floral anthocyanins in wild taxa of Petunia (Solanaceae). Biochemical Systematics and Ecology 27, 623–650. [Google Scholar]
- Aoshima M. 2007. Novel enzyme reactions related to the tricarboxylic acid cycle: phylogenetic/functional implications and biotechnological applications. Applied Microbiology and Biotechnology 75, 249–255. [DOI] [PubMed] [Google Scholar]
- Baillieul F, de Ruffray P, Kauffmann S. 2003. Molecular cloning and biological activity of α-, β-, and γ-megaspermin, three elicitins secreted by Phytophthora megasperma H20. Plant Physiology 131, 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassard JE, Ullmann P, Bernier F, Werck-Reichhart D. 2010. Phenolamides: bridging polyamines to the phenolic metabolism. Phytochemistry 71, 1808–1824. [DOI] [PubMed] [Google Scholar]
- Brooks JL, Stumpf PK. 1966. Fat metabolism in higher plants. XXXIX. Properties of a soluble fatty acid synthesizing system from lettuce chloroplasts. Archives of Biochemistry and Biophysics 116, 108–116. [DOI] [PubMed] [Google Scholar]
- Buanafina M. 2009. Feruloylation in grasses: current and future perspectives. Molecular Plant 2, 861–872. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, et al. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry 55, 611–622. [DOI] [PubMed] [Google Scholar]
- Cai L, Sutter BM, Li B, Tu BP. 2011. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Molecular Cell 42, 426–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Li C, Wang Y, et al. 2017a. Cytosolic acetyl-CoA promotes histone acetylation predominantly at H3K27 in Arabidopsis. Nature Plants 3, 814–824. [DOI] [PubMed] [Google Scholar]
- Chen G, Liu H, Wei Q, Zhao H, Liu J, Yu Y. 2017b. The acyl-activating enzyme PhAAE13 is an alternative enzymatic source of precursors for anthocyanin biosynthesis in petunia flowers. Journal of Experimental Botany 68, 457–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Gong L, Guo Z, Wang W, Zhang H, Liu X, Yu S, Xiong L, Luo J. 2013. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Molecular Plant 6, 1769–1780. [DOI] [PubMed] [Google Scholar]
- Choudhary NL, Sairam RK, Tyagi A. 2005. Expression of Δ 1-pyrroline-5-carboxylate synthetase gene during drought in rice (Oryza sativa L.). Indian Journal of Biochemistry & Biophysics 42, 366–370. [PubMed] [Google Scholar]
- Chypre M, Zaidi N, Smans K. 2012. ATP-citrate lyase: a mini-review. Biochemical and Biophysical Research Communications 422, 1–4. [DOI] [PubMed] [Google Scholar]
- Colaert N, Helsens K, Martens L, Vandekerckhove J, Gevaert K. 2009. Improved visualization of protein consensus sequences by iceLogo. Nature Methods 6, 786–787. [DOI] [PubMed] [Google Scholar]
- Demkura PV, Abdala G, Baldwin IT, Ballaré CL. 2010. Jasmonate-dependent and -independent pathways mediate specific effects of solar ultraviolet B radiation on leaf phenolics and antiherbivore defense. Plant Physiology 152, 1084–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan C, Mao T, Sun S, et al. 2019. Constitutive expression of GmF6′H1 from soybean improves salt tolerance in transgenic Arabidopsis. Plant Physiology and Biochemistry 141, 446–455. [DOI] [PubMed] [Google Scholar]
- Fabro G, Kovács I, Pavet V, Szabados L, Alvarez ME. 2004. Proline accumulation and AtP5CS2 gene activation are induced by plant-pathogen incompatible interactions in Arabidopsis. Molecular Plant-Microbe Interactions 17, 343–350. [DOI] [PubMed] [Google Scholar]
- Falcone Ferreyra ML, Rius SP, Casati P. 2012. Flavonoids: biosynthesis, biological functions, and biotechnological applications. Frontiers in Plant Science 3, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatland BL, Ke J, Anderson MD, Mentzen WI, Cui LW, Allred CC, Johnston JL, Nikolau BJ, Wurtele ES. 2002. Molecular characterization of a heteromeric ATP-citrate lyase that generates cytosolic acetyl-coenzyme A in Arabidopsis. Plant Physiology 130, 740–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatland BL, Nikolau BJ, Wurtele ES. 2005. Reverse genetic characterization of cytosolic acetyl-CoA generation by ATP-citrate lyase in Arabidopsis. The Plant Cell 17, 182–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkemeier I, Laxa M, Miguet L, Howden AJ, Sweetlove LJ. 2011. Proteins of diverse function and subcellular location are lysine acetylated in Arabidopsis. Plant Physiology 155, 1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdieri L, Vancura A. 2012. Acetyl-CoA carboxylase regulates global histone acetylation. Journal of Biological Chemistry 287, 23865–23876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerats T, Vandenbussche M. 2005. A model system for comparative research: Petunia. Trends in Plant Science 10, 251–256. [DOI] [PubMed] [Google Scholar]
- Gnonlonfin G, Sanni A, Brimer L. 2012. Review Scopoletin – a coumarin phytoalexin with medicinal properties. Critical Reviews in Plant Sciences 31, 47–56. [Google Scholar]
- Guo J, Liu J, Wei Q, Wang R, Yang W, Ma Y, Chen G, Yu Y. 2017. Proteomes and ubiquitylomes analysis reveals the involvement of ubiquitination in protein degradation in petunias. Plant Physiology 173, 668–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haudecoeur E, Planamente S, Cirou A, Tannières M, Shelp BJ, Moréra S, Faure D. 2009. Proline antagonizes GABA-induced quenching of quorum-sensing in Agrobacterium tumefaciens. Proceedings of the National Academy of Sciences, USA 106, 14587–14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Heller H, Eytan E, Kaklij G, Rose IA. 1984. Role of the α-amino group of protein in ubiquitin-mediated protein breakdown. Proceedings of the National Academy of Sciences, USA 81, 7021–7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. 2007. WoLF PSORT: protein localization predictor. Nucleic Acids Research 35, W585–W587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida JK, Wakatake T, Yoshida S, Takebayashi Y, Kasahara H, Wafula E, dePamphilis CW, Namba S, Shirasu K. 2016. Local auxin biosynthesis mediated by a YUCCA flavin monooxygenase regulates haustorium development in the parasitic plant Phtheirospermum japonicum. The Plant Cell 28, 1795–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Hua Z, Dong Y, Liu Z, Thiele CJ, Li Z. 2018. Quantitative ubiquitylome analysis and crosstalk with proteome/acetylome analysis identified novel pathways and targets of perifosine treatment in neuroblastoma. Translational Cancer Research 7, 1548–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H, Heinzel N, Schöttner M, Baldwin IT, Gális I. 2010. R2R3-NaMYB8 regulates the accumulation of phenylpropanoid-polyamine conjugates, which are essential for local and systemic defense against insect herbivores in Nicotiana attenuata. Plant Physiology 152, 1731–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazemi B, Bandehagh A, Sarikhani M, Komatsu S. 2017. Protein profiles underlying the effect of plant growth-promoting rhizobacteria on canola under osmotic stress. Journal of Plant Growth Regulation 37, 560–574. [Google Scholar]
- Koes R, Verweij W, Quattrocchio F. 2005. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends in Plant Science 10, 236–242. [DOI] [PubMed] [Google Scholar]
- Kovarik J. 2013. From immunosuppression to immunomodulation: current principles and future strategies. Pathobiology 80, 275–281. [DOI] [PubMed] [Google Scholar]
- Lee JV, Carrer A, Shah S, et al. 2014. Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metabolism 20, 306–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Song J. 2019. Analysis of widely targeted metabolites of the euhalophyte Suaeda salsa under saline conditions provides new insights into salt tolerance and nutritional value in halophytic species. BMC Plant Biology 19, 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK. 1987. Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods in Enzymology 148, 350–382. [Google Scholar]
- Liu F, Xiao Z, Yang L, Chen Q, Shao L, Liu J, Yu Y. 2017a. PhERF6, interacting with EOBI, negatively regulates fragrance biosynthesis in petunia flowers. New Phytologist 215, 1490–1502. [DOI] [PubMed] [Google Scholar]
- Liu J, Chang X, Ding B, Zhong S, Peng L, Wei Q, Meng J, Yu Y. 2019. PhDHS is involved in chloroplast development in petunia. Frontiers in Plant Science 10, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Li J, Wang H, Fu Z, Liu J, Yu Y. 2011. Identification and expression analysis of ERF transcription factor genes in petunia during flower senescence and in response to hormone treatments. Journal of Experimental Botany 62, 825–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Zainuddin IM, Vanderschuren H, Doughty J, Beeching JR. 2017b. RNAi inhibition of feruloyl CoA 6′-hydroxylase reduces scopoletin biosynthesis and post-harvest physiological deterioration in cassava (Manihot esculenta Crantz) storage roots. Plant Molecular Biology 94, 185–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Mallona I, Lischewski S, Weiss J, Hause B, Egea-Cortines M. 2010. Validation of reference genes for quantitative real-time PCR during leaf and flower development in Petunia hybrida. BMC Plant Biology 10, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver D, Nikolau B, Wurtele E. 2009. Acetyl-CoA—life at the metabolic nexus. Plant Science 176, 597–601. [Google Scholar]
- Onkokesung N, Gaquerel E, Kotkar H, Kaur H, Baldwin IT, Galis I. 2012. MYB8 controls inducible phenolamide levels by activating three novel hydroxycinnamoyl-coenzyme A: polyamine transferases in Nicotiana attenuata. Plant Physiology 158, 389–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Kang K, Lee K, Choi D, Kim YS, Back K. 2009. Induction of serotonin biosynthesis is uncoupled from the coordinated induction of tryptophan biosynthesis in pepper fruits (Capsicum annuum) upon pathogen infection. Planta 230, 1197–1206. [DOI] [PubMed] [Google Scholar]
- Perez-Riverol Y, Csordas A, Bai J, et al. 2019. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Research 47, D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T, Li J, Sun B, Liu J, Zhao W, Huang C, Yang L, Li Y. 2016. ATP-citrate lyase gene (SoACLA-1), a novel ACLA gene in sugarcane, and its overexpression enhance drought tolerance of transgenic tobacco. Sugar Technology 19, 258–269. [Google Scholar]
- Piao HL, Lim JH, Kim SJ, Cheong GW, Hwang I. 2001. Constitutive over-expression of AtGSK1 induces NaCl stress responses in the absence of NaCl stress and results in enhanced NaCl tolerance in Arabidopsis. The Plant Journal 27, 305–314. [DOI] [PubMed] [Google Scholar]
- Prats E, Llamas M, Jorrin J, Rubiales D. 2007. Constitutive coumarin accumulation on sunflower leaf surface prevents rust germ tube growth and appressorium differentiation. Crop Science 47, 1119–1124. [Google Scholar]
- Ruan B, Hua Z, Zhao J, et al. 2019. OsACL-A2 negatively regulates cell death and disease resistance in rice. Plant Biotechnology Journal 17, 1344–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu B, Miyagawa H, Ueno T, Sakata K, Watanabe K, Ogawa K. 2005. Morning glory systemically accumulates scopoletin and scopolin after interaction with Fusarium oxysporum. Zeitschrift für Naturforschung C 60, 83–90. [DOI] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. 2011. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer-Rimon B, Farhi M, Albo B, et al. 2012. The R2R3-MYB-like regulatory factor EOBI, acting downstream of EOBII, regulates scent production by activating ODO1 and structural scent-related genes in petunia. The Plant Cell 24, 5089–5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Wang L, Zhang B, Ma J, Hettenhausen C, Cao G, Sun G, Wu J, Wu J. 2014. Scopoletin is a phytoalexin against Alternaria alternata in wild tobacco dependent on jasmonate signalling. Journal of Experimental Botany 65, 4305–4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, McCaffery JM, Irizarry RA, Boeke JD. 2006. Nucleocytosolic acetyl-coenzyme A synthetase is required for histone acetylation and global transcription. Molecular Cell 23, 207–217. [DOI] [PubMed] [Google Scholar]
- Tan Y, Liu J, Huang F, Guan J, Zhong S, Tang N, Zhao J, Yang W, Yu Y. 2014. PhGRL2 protein, interacting with PhACO1, is involved in flower senescence in the petunia. Molecular Plant 7, 1384–1387. [DOI] [PubMed] [Google Scholar]
- Tong J. 2009. Cloning and functional research of citrate synthase and ATP-citrate lyase of Brassica napus L. PhD thesis, Chinese Academy of Agricultural Sciences.
- Wang Y, Xu L, Tang M, Jiang H, Chen W, Zhang W, Wang R, Liu LW. 2016. Functional and integrative analysis of the proteomic profile of radish root under Pb exposure. Frontiers in Plant Science 7, 1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellburn AR. 1994.The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. Journal of Plant Physiology 144, 307–313. [Google Scholar]
- Wellen K, Hatzivassiliou G, Sachdeva U, Bui T, Cross J, Thompson C. 2009. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Oh M, Schwarz E, Larue C, Sivaguru M, Imai B, Yau P, Ort D, Huber S. 2011. Lysine acetylation is a widespread protein modification for diverse proteins in Arabidopsis. Plant Physiology 155, 1769–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S, Poirier Y. 2012. The protein acetylome and the regulation of metabolism. Trends in Plant Science 17, 423–430. [DOI] [PubMed] [Google Scholar]
- Xing S, van Deenen N, Magliano P, Frahm L, Forestier E, Nawrath C, Schaller H, Gronover C, Prufer D, Poirier Y. 2014. ATP citrate lyase activity is post-translationally regulated by sink strength and impacts the wax, cutin and rubber biosynthetic pathways. The Plant Journal 79, 270–284. [DOI] [PubMed] [Google Scholar]
- Yang S, Lan S, Gong M. 2009. Hydrogen peroxide-induced proline and metabolic pathway of its accumulation in maize seedlings. Journal of Plant Physiology 166, 1694–1699. [DOI] [PubMed] [Google Scholar]
- Yang W, Cai Y, Hu L, Wei Q, Chen G, Bai M, Wu H, Liu J, Yu Y. 2017. PhCESA3 silencing inhibits elongation and stimulates radial expansion in petunia. Scientific Reports 7, 41471. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (https://www.ebi.ac.uk/pride/; Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD015480. The raw RNA sequence data have been submitted to NCBI with accession number PRJNA577189 (https://dataview.ncbi.nlm.nih.gov/object/PRJNA577189).