Abstract
Background
Lung cancer is a complex disease that diagnosed the most common cancer and led cause of cancer death. MDM2 (MDM2 proto-oncogene) encodes a nuclear-localized E3 ubiquitin ligase. The encoded protein can promote tumor formation by targeting tumor suppressor proteins, such as TP53, for proteasomal degradation. Epidemiology studies have investigated the association of MDM2 single nucleotide polymorphisms (SNP) and interaction between genetic and environmental factors with lung cancer.
Methods
This Chinese case-control study comprised 627 cases and 633 controls explored the role of MDM2 five htSNPs (rs1690924, rs1846402, rs2291857, rs3730581 and rs3730635, haplotype-tagging SNP) tagging 95% of the common haplotypes across the gene and the interactions of MDM2, PPP1R13L, CD3EAP and TP53 in the same pathological pathway on lung cancer risk, together with smoking-duration.
Results
None of the htSNPs in MDM2 were associated with lung cancer risk in co-dominant, dominant, recessive, and log-additive models (adjusted for smoking-duration). Haplotype analysis showed that global haplotype association was statistically significant (P=0.0036, adjusted for smoking-duration) and haplotype5 (rs1690924A-rs1846402G-rs2291857C-rs3730581G-rs3730635A) was associated with reduced risk of lung cancer [OR (95%) =0.52 (0.33–0.82), P=0.0053, adjusted for smoking-duration]. MDR interaction analysis showed that two the best significant models and strong synergy between MDM2 and TP53.
Conclusions
MDM2 five-htSNPs haplotype exhibited association with lung cancer susceptibility, interaction of MDM2 and TP53 htSNPs and smoking-duration contributed to lung cancer risk and strong synergy between MDM2 and TP53 htSNPs influenced lung cancer predisposition. Our results suggest that MDM2, TP53 and smoking-duration interact in relation to lung carcinogenesis.
Keywords: MDM2 and PPP1R13L and CD3EAP and TP53, genetic variants, smoking duration, interaction, lung cancer
Introduction
Cancer incidence and mortality are rapidly growing worldwide. Lung cancer is the most commonly diagnosed cancer (11.6% of the total cases) and the leading cause of cancer death (18.4% of the total cancer deaths) (1). Lung cancer is complex disease affected by many genetic factors and environmental exposures. Nicotine and carbon monoxide caused by cigarette smoking have been considered as causative environmental factors for the development of lung cancer. Another possible mechanism may involve interactions between smoking and various susceptibility genes in relation to lung cancer (2).
MDM2 (MDM2 proto-oncogene) (Gene ID: 4193) is located on chromosome 12q15. The gene consists of 13 exons and encodes a nuclear-localized E3 ubiquitin ligase. The encoded protein promotes tumor formation by targeting tumor suppressor proteins, such as TP53, for proteasomal degradation. This gene is itself transcriptionally-regulated by TP53. Over-expression or amplification of MDM2 is detected in many human malignancies, including lung cancer (https://www.ncbi.nlm.nih.gov/gene/4193) (3). The effects of single nucleotide polymorphisms (SNP) at MDM2 have been investigated in relation to lung cancer with inconsistent results (4-15).
The two genes PPP1R13L [protein phosphatase 1, regulatory (inhibitor) subunit 13 like)] (Gene ID: 10848) and CD3EAP (CD3e molecule, epsilon-associated protein) (Gene ID: 10849) located on chromosome 19q13.3 relate to DNA repair and cell survival and cell proliferation, respectively. The gene TP53 (tumor protein p53) located on chromosome 17p13.1 encodes the tumor suppressor p53, which in response to diverse types of cellular stress regulates expression of target genes. We previously reported that PPP1R13L rs1970764, CD3EAP rs967591 and rs735482, and TP53 htNP2 were associated with lung cancer or interacted in relation to lung cancer risk among both Caucasian Danes and Chinese [(16-20), Yin et al. submitted and revised].
MDM2, TP53, PPP1R13L, and CD3EAP all belong to the pathway of gene expression. MDM2, TP53 and PPP1R13L belong to the pathways of gene expression and p53 pathway. Both MDM2 and TP53 share the pathways of gene expression, p53 pathway and TP53 Network (https://www.ncbi.nlm.nih.gov/gene/4193).
Previous epidemiology studies concerning MDM2 SNPs and lung cancer risk were mainly focused on single SNP and interactions (4-15). No systematical investigations have been reported on the associations between MDM2 htSNPs (Haplotype-tagging SNP) and lung cancer risk. In this Chinese case-control study we explored the role of htSNPs tagging 95% common haplotypes across the MDM2 gene and assess gene-gene and gene-gene-environment interactions in the same pathological pathway related to lung cancer risk, including the interaction between MDM2 htSNPs, TP53 htSNPs, and PPP1R13L and CD3EAP risk SNPs. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-19-4784).
Methods
Ethics permission
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study protocol was approved by Chinese Administration Office of Human Genetic Resources {no. [2001]015}. Subjects were informed about the study and written or oral informed consent was achieved from all study participants.
Study population
A total of 1,260 subjects were enrolled in this hospital-based case-control study, comprising 627 cases and 633 controls. The patients with lung cancer were diagnosed using standard clinical and histological criteria. Qualified cases were previously untreated (no chemotherapy or radiotherapy for cancer prior to recruitment). Cancer-free controls were selected from the orthopedics wards in the same area. All subjects were unrelated ethnic Han Chinese. Demographic and covariate data were acquired from medical records and questionnaires through personal interview with a professional physician. Stratification criteria were determined as follows age (10 years an interval), gender, family history and smoking duration (20 years an interval).
Determined MDM2 htSNPs
Using the TagSNPs software online and approaches of the algorithm-Tagger-pairwiseTagging, htSNPs of MDM2 gene from the International HapMap Project were determined in the relevant region of chromosome 12 (http://www.hapmap.org, HapMap Data Rel 27 PhaseII+III, Feb09, on NCBI B36 assembly, dbSNP b26). Qualified criteria were: r2-cut off of 0.8 and MAF (minor allele frequency)-cut off of 0.05 in HCB (Han Chinese in Beijing) samples. Five htSNPs (rs1690924, rs1846402, rs2291857, rs3730581 and rs3730635) were selected, tagging 95% of the common haplotype diversity across the MDM2 gene. Table 1 shows the information of MDM2 five htSNPs, three risk SNPs of PPP1R13L and CD3EAP and TP53 five htSNPs. Three risk SNPs of PPP1R13L and CD3EAP were previously reported (20,21) while we increased the number of included samples in present study. The genotype data of PPP1R13L and CD3EAP three risk SNPs and TP53 five htSNPs were employed for interaction analyses of gene-gene and gene-gene-environment in this study.
Table 1. Characteristics for MDM2 htSNPs selected and SNPs in PPP1R13L and CD3EAP and htSNPs in TP53a.
| dbSNP ID | Position | Location | Base change | Allele frequency in HapMap HCBb | MAFc in controls for current study |
|---|---|---|---|---|---|
| Chr12q15 | |||||
| MDM2 | |||||
| rs1690924 | 68811541 | Intron | A/G | A: 0.814/G: 0.186 | G: 0.24 |
| rs1846402 | 68814798 | Intron | G/T | G: 0.826/T: 0.174 | T: 0.16 |
| rs2291857 | 68824258 | Intron | C/A | C: 0.700/A: 0.300 | A: 0.32 |
| rs3730581 | 68825712 | Intron | A/G | A: 0.581/G: 0.419 | G: 0.49 |
| rs3730635 | 68835343 | Intron | A/G | A: 0.946/G: 0.054 | G: 0.02 |
| Chr19q13.3 | |||||
| PPP1R13La | |||||
| rs1970764 | 45387615 | Intron | A/G | No | G: 0.48 |
| CD3EAPa | |||||
| rs967591 | 45406676 | 5’UTR | G/A | G: 0.525/A: 0.475d | A: 0.42 |
| rs735482 | 45408744 | Exon3 | A/C | A: 0.558/C: 0.442 | C: 0.45 |
| Codon 261 (K [Lys] [AAA] ⇒ T [Thr] [ACA]) (missense) | |||||
| Chr17p13.1 | |||||
| TP53 | |||||
| rs12951053 | 7674089 | Intron | A/C | A: 0.667/C: 0.333 | C: 0.34e |
| rs1042522 | 7676154 | Exon4 | G/C | G: 0.511/C: 0.489 | C: 0.45e |
| Codon 72 (R [Arg] [CGC]) ⇒ P [Pro] [CCC] (missense) | |||||
| rs8079544 | 7676734 | Intron | C/T | C: 0.878/T: 0.122 | T: 0.08e |
| rs12602273 | 7679695 | Intron | C/G | C: 0.678/G: 0.322 | G: 0.28e |
| rs8064946 | 7685993 | Intron | G/C | G: 0.622/C: 0.378 | C: 0.32 e |
a, information from NCBI SNP database (GRCh38.p7) and HapMap database; b, Han Chinese in Beijing; c, minor allele frequency; d, CHB+JPT (Han Chinese in Beijing + Japanese from 1000 GENOMES); e, from previous result, here this is employed for interaction analysis.
DNA isolation and genotyping
A volume of 5 mL of peripheral blood was taken from each volunteer. Genomic DNA of peripheral blood samples was extracted with the Puregene DNA Isolation Kit or FlexiGene DNA kit 250 (Gentra Systems, Minneapolis, MN, USA or Qiagen, Germany) following the manufacturer’s instructions. Genotyping of rs1690924 (A > G), rs1846402 (G > T), rs2291857 (C > A), rs3730581 (A > G), and rs3730635 (A > G) of the MDM2 gene was performed using ligase detection reaction coupled with polymerase chain reaction (LDR-PCR) as previously published (22) in Shanghai Generay Biotechnology Co. Ltd. (China). Genotypes of PPP1R13L rs1970764 (A > G) and CD3EAP rs967591(G > A) and rs735482 (A > C) have been previously reported (20,21). This study only genotyped the loci for the increased samples. The sequences (5'-3') of primers and probes of MDM2 five htSNPs are listed in Table 2. Each group of LDR probes consisted of 1 common probe and 2 discriminating probes for the 2 alleles. The steps for genotyping were in short: performed PCR reactions, completed LDR reactions and sequenced LDR products. The genotyping call-rate was 96% on average for the MDM2 five htSNPs. As a quality control, some samples were genotyped in duplicate. Repeated genotyping yielded 100% identity.
Table 2. The sequences (5'-3') of primers and probes for MDM2 5 htSNPs examined.
| rs number | Primers and probes |
|---|---|
| rs1690924 | Forward primer: TGTAATGGAAAGCCATCAGTAT |
| Reverse primer: TCTCCTGTCCCAAGATCTTGC | |
| Common probe:-P-GCTATAAAAGATAATAGCATTTGTA-FAM- | |
| Discriminating probe G: TTTAAAGACATGTATTAATGAGAAAACG | |
| Discriminating probe A: AAAGACATGTATTAATGAGAAAACA | |
| rs1846402 | Forward primer: TAAGTGGGAGAGACAGAGAAC |
| Reverse primer: CCAGGTTAAGAACTTCTGCAC | |
| Common probe:-P-GCTCAATCTGTCACTGAAAATCATGTTT-FAM- | |
| Discriminating probe T: TTTTTTTTCACACTGAAATTCTGCCTAAGGTT | |
| Discriminating probe G: TTTTTCACACTGAAATTCTGCCTAAGGTG | |
| rs2291857 | Forward primer: CTACTCAGTAGATATGCTATCAG |
| Reverse primer: CAACATTTAGTATGAGGATGC | |
| Common probe:-P-ACCTTTCAATAAACATTAAACATGT-HEX- | |
| Discriminating probe A: CTAACACAAACCCTTATGCAATTTA | |
| Discriminating probe C: TTTCTAACACAAACCCTTATGCAATTTC | |
| rs3730581 | Forward primer: AGAAAATAGTTGACAGAGAGAA |
| Reverse primer: GCATGTACGAGATTCTGGTCT | |
| Common probe:-P-TAGTAGACGAGAAGGCTGTTGCCTGTTT-HEX- | |
| Discriminating probe G: TTTTTTTAATAGTTGAGAACAGTTAGTAGACG | |
| Discriminating probe A: TTTTAATAGTTGAGAACAGTTAGTAGACA | |
| rs3730635 | Forward primer: AAGGTGGAAGAGCCTTTTCAG |
| Reverse primer: CGAAAGTACCTACAGTGTGAC | |
| Common probe:-P-GTTAGAGGGGGAAAGTGTGGAAGTT-FAM- | |
| Discriminating probe G: TTTGGATTTTGAAACTGAAATTATTCTG | |
| Discriminating probe A: GGATTTTGAAACTGAAATTATTCTA |
Statistical analysis
Characteristics of cases and controls, allele frequencies, genotype frequencies, Hardy-Weinberg equilibrium, co-dominant model; dominant model; recessive model and log-additive model for case-control association of each single-locus, haplotype associations, and pair-wise linkage disequilibrium (LD), unconditional logistic regression for measurement of odd ratio, 95% confidence interval (OR, 95% CI) after adjustment for smoking-duration were explored employing SPSS© v16.0 (SPSS Inc., Chicago, IL, USA) or SNPStats program (23) or Haploview software 4.2 (24). Haplotypes with frequency <0.01 among both cases and controls were excluded from the analysis. The interaction analyses of gene-gene and gene-gene-smoking duration in relation to lung cancer risk were implemented employing platform of multifactor dimensionality reduction (MDR). This software (3.0.3. dev. Jar) (25) is an updated version where permutation testing has been added into the main MDR program. The MDR method is nonparametric and free model. MDR has rational power to recognize interactions between two or more loci in relatively small samples. MDR has excellent power for identifying high-order gene-gene interactions. MDR is directly usable to case-control and discordant-sib-pair studies (25). If P value was less than 0.05, the difference was considered to be statistically significant. Power test was determined employing online statistical software: Unmatched Case/Control Studies (https://www.stat.ubc.ca/~rollin/stats/ssize/caco.html).
Results
Study population
The MDM2 five htSNPs were genotyped in a Chinese hospital-based case-control study of 627 lung cancer patients and 633 control subjects. There were no statistically significant differences for the distribution of age and gender between cases and controls. However, more cases had a family history of cancer and cases had longer smoking history than controls (>20 years) (both P<0.0001) (Table 3).
Table 3. Distribution of selected characteristics in the case-control study population.
| Characteristics | Cases, n (%) | Controls, n (%) | P value |
|---|---|---|---|
| Overall | 627 | 633 | |
| Age (years) | |||
| Mean (±SD) | 58 (±10.4) | 58 (±10.5) | 0.9a |
| ≤40 | 29 (4.6) | 29 (4.6) | |
| 41–50 | 109 (17.4) | 125 (19.7) | |
| 51–60 | 222 (35.4) | 214 (33.8) | 0.748b |
| >60 | 267 (42.6) | 265 (41.9) | |
| Gender | |||
| Female | 185 (29.5) | 184 (29.1) | |
| Male | 442 (70.5) | 449 (70.9) | 0.86b |
| Family historyc | |||
| No | 536 (85.5) | 628 (99.2) | |
| Yes | 91 (14.5) | 5 (0.8) | <0.0001b,d |
| Smoking duration | |||
| Never | 241 (38.4) | 333 (52.6) | |
| ≤20 (years) | 104 (16.6) | 98 (15.5) | <0.0001b,d |
| >20 (years) | 282 (45.0) | 202 (31.9) |
a, for t-test; b, for χ2 test (two-sided); c, family history of cancer; d, statistical significance.
Allele frequencies of MDM2 five htSNPs
The minor-allele frequencies (MAF) among the controls (G =0.24, T =0.16, A =0.32, G =0.49 and G =0.02 for rs1690924, rs1846402, rs2291857, rs3730581 and rs3730635, respectively), were similar to the MAF of HapMap-HCB reported by NCBI SNP database (https://www.ncbi.nlm.nih.gov/snp) (P=0.154, 0.552, 0.523, 0.078, respectively) except for rs3730635 (P=0.017) (Table 1). The genotype distribution in control population was in Hardy-Weinberg equilibrium for rs1690924 (P=0.29), rs1846402 (P=0.54), rs2291857(P=0.93), rs3730581 (P=0.23) and rs3730635 (P=1).
Association between MDM2 five htSNPs and lung cancer risk
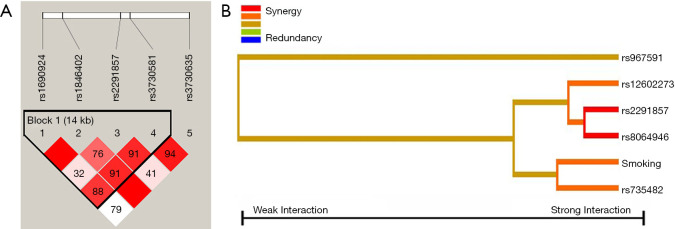
No significant associations were found between genotype distributions and lung cancer risk for MDM2 five htSNPs in co-dominant, dominant, recessive, and log-additive models after adjustment for smoking status (Table 4). Next, LD analysis was implemented. Strong LD was found between the five htSNPs (D’ values from 0.768 to 0.9984 for pair-wise LD) except for rs1690924 and rs2291857 (D’=0.3291) and rs2291857 and rs3730635 (D’=0.4139) in middle LD (Table 5, Figure 1A). Five-locus haplotype analysis was performed (Table 6). Among 19 possible haplotypes, 9 commonly occurring haplotypes were identified (frequency: about or above 1%), capturing 97.28% (cumulative frequency) of all possible haplotypes. The haplotype analysis revealed that a statistically significant global haplotype association after adjustment for smoking duration (P=0.0036) and that haplotype5 (rs1690924A-rs1846402G-rs2291857C-rs3730581G-rs3730635A) was associated with reduced risk of lung cancer after adjustment for smoking duration (0.0279 for cases and 0.0552 for controls) [OR (95% CI) =0.52 (0.33–0.82), P=0.0053]. Of minor importance, the combined group of 10 rare haplotypes was associated with increased risk of lung cancer after adjustment for smoking duration [OR (95% CI) =2.23 (1.21–4.09), P=0.0099]. No evidence was found for individual htSNPs of MDM2 in smoking-subgroup analyses (data not shown).
Table 4. Associations of single htSNP in MDM2 and PPP1R13L and CD3EAP with lung cancer riska,b.
| Gene/rsCa/Co | Co–dominant(AB vs. AA)/(BB vs. AA)/P | Dominant (AB + BB vs. AA)/P | Recessive (BB vs. AA + AB)/P | Log–additive– –/P |
|---|---|---|---|---|
| MDM2 | ||||
| rs1690924(A>G) | ||||
| 571/568 | 1.24 (0.96–1.59)/1.24 (0.77–2.00)/0.22 | 1.24 (0.97–1.57)/0.08 | 1.14 (0.71–1.83)/0.58 | 1.17 (0.97–1.42)/0.11 |
| rs1846402(G>T) | ||||
| 608/603 | 1.20 (0.93–1.56)/0.74 (0.35–1.59)/0.25 | 1.16 (0.90–1.48)/0.25 | 0.70 (0.33–1.50)/0.36 | 1.09 (0.87–1.36)/0.46 |
| rs2291857(C>A) | ||||
| 611/613 | 1.17(0.92–1.49)/1.02 (0.69–1.51)/0.42 | 1.14 (0.91–1.43)/0.26 | 0.94 (0.65–1.37)/0.76 | 1.06 (0.90–1.27)/0.47 |
| rs3730581(A>G) | ||||
| 613/621 | 1.18 (0.90–1.55)/0.99 (0.72–1.36)/0.35 | 1.11 (0.86–1.44)/0.42 | 0.89 (0.68–1.16)/0.39 | 1.00 (0.85–1.17)/0.99 |
| rs3730635(A>G) | ||||
| 612/625 | 1.24 (0.74–2.10)/NA (0.00–NA)/0.17 | 1.32 (0.79–2.21)/0.29 | NA (0.00–NA)/0.092 | 1.37 (0.84–2.26)/0.21 |
| PPP1R13L | ||||
| rs1970764(A>G) | ||||
| 594/590 | 1.06 (0.79–1.40)/1.39(0.99–1.94)/0.11 | 1.15 (0.88–1.50)/0.32 | 1.34 (1.02–1.76)/0.037c | 1.18 (0.99–1.39)/0.057 |
| CD3EAP | ||||
| rs967591(G>A) | ||||
| 594/590 | 1.31 (1.01–1.70)c/1.28 (0.92–1.78)/0.11 | 1.30 (1.01–1.67)/0.038c | 1.09 (0.81–1.45)/0.58 | 1.15 (0.98–1.35)/0.092 |
| rs735482(A>C) | ||||
| 603/595 | 1.13 (0.87–1.48)/1.14 (0.82–1.57)/0.62 | 1.13 (0.88–1.46)/0.32 | 1.05 (0.79–1.39)/0.73 | 1.07 (0.91–1.26)/0.4 |
a, Dominant model: AB (Heterozygote) + BB (Homozygous variant-type) versus AA (Homozygous wild-type), Recessive model: BB versus AA + AB, Co-dominant model: AB versus AA and BB versus AA, Log-additive model: analysis of trend where AA is ‘0’, AB is ‘1’ and BB is ‘2’; b, OR (95% CI), adjusted for smoking duration; c, statistical significance.
Table 5. D’ statistics of linkage disequilibrium analysis for MDM2 htSNPsa.
| rs number | rs1690924 | rs1846402 | rs2291857 | rs3730581 | rs3730635 |
|---|---|---|---|---|---|
| rs1690924 | – | 0.9984 | 0.3291 | 0.8816 | 0.7944 |
| rs1846402 | <2e-16 | – | 0.768 | 0.9125 | 0.9903 |
| rs2291857 | <2e-16 | <2e-16 | – | 0.9121 | 0.4139 |
| rs3730581 | <2e-16 | <2e-16 | <2e-16 | – | 0.9452 |
| rs3730635 | 0.000339 | 0.000501 | 1.92e-06 | 5.55e-15 | – |
a, above is the D’ value, below is the P value.
Figure 1.
(A) D’ LD map and LD plot of MDM2 five htSNPs generated by Haploview 4.2. One block was detected. The criteria of block partition were based on solid spine LD. The digit in the boxes represents D’ value (e.g., 99 means 0.99; 1 means 0.01; empty boxes means 1.0). Deep red boxes designate strong evidence of LD. Light red boxes designate uninformative. White boxes designate strong evidence of recombination. (B) Interaction dendrogram resulting from MDR analysis of 14 attributors in MDM2, PPP1R13L, CD3EAP, TP53 and smoking-duration [entropy-based IG (the value of information gain) for the SNP pairs]. Red bar and orange bar indicate the high-level synergies on the phenotype, while the brown indicate a medium-level interaction, green and blue connections indicate redundancy or lack of synergistic interactions between the markers.
Table 6. Haplotype association of MDM2 htSNPs with lung cancer riska.
| Haplotypeb | Constructionc | Case frequency | Control frequency | OR (95% CI) | P value |
|---|---|---|---|---|---|
| 1 | AGCAA | 0.476 | 0.4844 | 1.00 (Reference) | – |
| 2 | ATAGA | 0.1303 | 0.1403 | 1.00 (0.77–1.28) | 0.98 |
| 3 | GGAGA | 0.1456 | 0.1219 | 1.17 (0.91–1.51) | 0.23 |
| 4 | GGCGA | 0.1021 | 0.1073 | 0.97 (0.73–1.29) | 0.82 |
| 5 | AGCGA | 0.0279 | 0.0552 | 0.52 (0.33–0.82)d | 0.0053d |
| 6 | AGAGA | 0.0284 | 0.0311 | 1.07 (0.64–1.79) | 0.79 |
| 7 | ATCGA | 0.0242 | 0.0142 | 1.64 (0.81–3.31) | 0.17 |
| 8 | AGAGG | 0.0196 | 0.0121 | 1.41 (0.72–2.76) | 0.32 |
| 9 | AGAAA | 0.009 | 0.0164 | 0.54 (0.23–1.29) | 0.17 |
| Rare | – | NA | NA | 2.23 (1.21–4.09)d | 0.0099d |
a, global haplotype association P value: 0.0036, adjusted for smoking duration; b, five-locus order: rs1690924-rs1846402-rs2291857-rs3730581-rs3730635; c, underlined indicates minor allele; d, statistical significance.
Association of PPP1R13L and CD3EAP SNPs and lung cancer
PPP1R13L rs1970764(A > G) in recessive model [OR (95% CI) =1.34 (1.02–1.76), P=0.037] and CD3EAP rs967591(G > A) in dominant model [OR (95% CI) =1.30 (1.01–1.67), P=0.038] were associated with increased risk of lung cancer after controlling for smoking duration in the current expanded study (Table 4).
Interactions of gene-gene-smoking
Table 7 summarizes the best significant candidate models of gene-gene-smoking duration interactions for combinations of htSNPs in MDM2, PPP1R13L, CD3EAP and smoking-duration, for combinations of htSNPs in MDM2, PPP1R13L, CD3EAP, TP53, and smoking-duration and for combinations of htSNPs in MDM2, TP53, and smoking-duration from MDR analysis. In the analysis for MDM2, TP53, and smoking-duration, one three-locus model and one four-locus model had relative higher values of balanced accuracy overall (0.6073 or 0.6446) and cross-validation consistency (8/10) that were significant at the 0.0030-0.0040 or 0.0250-0.0260 level. The three-locus model and four-locus model consisted of MDM2, TP53 and smoking duration. One two-locus model including MDM2 rs1846402 was significant. In the analysis for MDM2, PPP1R13L, CD3EAP, and smoking-duration or MDM2, PPP1R13L, CD3EAP, TP53, and smoking-duration, one two-locus (CD3EAP rs735482 and smoking duration) had values of balanced accuracy overall of 0.5831 and cross-validation consistency of 8/10 that was significant at the 0.0040-0.0050 or 0.0130-0.0140 level. No significant interaction model was found when smoking-duration was excluded.
Table 7. The best candidate models for interactions of gene-gene-smoking duration from MDR analysisa,b.
| Model | Attributes | Bal. Acc.c overall | Bal. Acc. CVd training | Bal. Acc. CV testing | CV consistency | P valuee |
|---|---|---|---|---|---|---|
| MDM2 + PPP1R13L + CD3EAP + smoking-duration | ||||||
| One-locus | Smoking | 0.5708 | 0.5708 | 0.5708 | 10/10 | 0.0020–0.0030 |
| Two-locus | Smoking | |||||
| rs735482 | 0.5831 | 0.5833 | 0.5657 | 8/10 | 0.0040–0.0050 | |
| MDM2 + PPP1R13L + CD3EAP + TP53 + smoking-duration | ||||||
| One-locus | Smoking | 0.5708 | 0.5708 | 0.5708 | 10/10 | 0.0080–0.0090 |
| Two-locus | Smoking | |||||
| rs735482 | 0.5831 | 0.5833 | 0.5657 | 8/10 | 0.0130–0.0140 | |
| MDM2 + TP53 + smoking-duration | ||||||
| One-locus | Smoking | 0.5708 | 0.5708 | 0.5708 | 10/10 | 0.0030–0.0040 |
| Two-locus | Smoking | |||||
| rs1846402 | 0.579 | 0.5807 | 0.557 | 7/10 | 0.0170–0.0180 | |
| Three-locus | Smoking | |||||
| rs2291857 | ||||||
| rs8064946 | 0.6073 | 0.609 | 0.57 | 8/10 | 0.0030–0.0040 | |
| Fourth-locus | Smoking | |||||
| rs1690924 | ||||||
| rs2291857 | ||||||
| rs8064946 | 0.6446 | 0.6486 | 0.5535 | 8/10 | 0.0250–0.0260 | |
a, analyzed by MDR 3.0.3. dev. Jar, data of TP53 from previous study (Yin et al. submitted and revised); b, only list statistical significant models; C, balanced accuracy; d, cross-validation; e, P value based on 1,000 permutation test, statistical significant P value.
Figure 1B shows the dendrogram of 14 attributes in the interaction analysis of MDM2, PPP1R13L, CD3EAP, TP53, and smoking-duration built using the MDR software. The entropy-based dendrogram indicated that a high degree of synergy interaction exists between MDM2 rs2291857 and TP53 rs8064946. There was a lesser degree of synergy interaction between CD3EAP rs735482 and smoking duration.
Discussion
We evaluated five htSNPs of MDM2 gene in relation to lung cancer risk in current study. Only one of these SNPs, rs1690924, has previously been assessed in a study of the chemotherapy outcome in lung cancer patients (26-29). To the best of our knowledge, none of the remaining four htSNPs have been assessed previously in relation to lung cancer risk. The majority of the MDM2 SNP studies have focused on MDM2 rs2279744 (SNP T309G) in relation to lung cancer risk with inconsistent results (4-15).
An Asian-Korean study reported that MDM2 rs2279744 was associated with increased risk of lung adenocarcinoma [GG versus TT, adjusted OR (95% CI) =1.91 (1.16–3.14), P=0.01] and the risk of lung adenocarcinoma increased as the number of rs2279744 G alleles increased [P (trend) =0.01] (4). An Asian-Chinese study reported that MDM2 rs2279744 was associated with an increased lung cancer risk [GG versus TT, OR (95% CI) =1.83 (1.45–2.32)] and [TG versus TT, OR (95% CI) =1.33 (1.09–1.63)] and that the interactions between MDM2 and TP53 polymorphisms increased lung cancer risk [for the presence of both MDM2 rs2279744 GG and TP53 rs1042522 CC, OR (95% CI) =4.56 (2.76–7.54)] and interactions of the polymorphisms (respectively and jointly) and smoking [smokers with both the MDM2 rs2279744 GG and TP53 rs1042522 CC, OR (95% CI) =10.41 (5.26–20.58)] (5). A Caucasian-Norwegian study reported that the MDM2 rs2279744 GG genotype was associated with risk of non-small cell lung cancer (NSCLC) [OR (95% CI) =1.62 (1.06–2.50)] and the GG genotype was associated with higher age at diagnosis in individuals with TP53 mutations (P=0.037) (6). A Caucasian-Norwegian study found a slightly reduced risk for lung cancer among individuals harboring the MDM2 rs2279744 G allele [TG/GG versus TT, OR (95% CI) =0.86 (0.67–0.98)] (7). An Asian-Singaporean study reported that MDM2 rs2279744 TT genotype was associated with increased risk of lung cancer [TT versus GG, OR (95% CI) =2.1 (1.01–4.36)] and carriers of this genotype with the TP53 rs1042522 C allele had a 2.5-fold increased risk [OR (95% CI) =2.5 (1.2–5.0)] among non-smoking Chinese women (8). An Asian-Chinese study reported increased risk for carriers of the MDM2 rs2279744 GG genotype in relation to lung adenocarcinoma risk [GG versus TT, adjusted OR (95% CI) =1.68 (1.27–2.21)]. The combination of TP53 rs1042522 CC and MDM2 rs2279744 GG genotypes [adjusted OR (95% CI) =2.66 (1.54–4.60)] interacted in relation to lung adenocarcinoma risk (9). An Asian-Chinese study reported that the P73 rs2273953 and rs1801173 AT/AT [AT/AT versus GC/GC, OR (95% CI) =0.46 (0.22–0.97)] and MDM2 rs2279744 TT [TT versus GG, OR (95% CI) =0.48 (0.26–0.86)] genotypes were associated with a decreased risk of developing NSCLC, and interaction between the P73 and MDM2 polymorphisms such that carriers of both the P73 AT/AT and MDM2 TT genotypes were at reduced risk of developing NSCLC [OR (95% CI) =0.13 (0.03–0.59)] (10).
A meta-analysis including 7 studies encompassing in total 4,276 cases and 5,318 controls revealed that MDM2 rs2279744 (SNP T309G) was associated with increased risk of lung cancer for homozygous G-allele carriers [GG versus TT, OR (95% CI) =1.27 (1.12–1.44)] (11). Recently, a meta-analysis including 11 articles with a total 6,470 NSCLC patients and 8,027 controls concluded that the MDM2 rs2279744 (SNP T309G) polymorphism may contribute to NSCLC susceptibility, especially for Asians and women (12).
A Caucasian-Canada study found no overall association between the MDM2 rs2279744 genotypes and NSCLC risk [T/G versus TT, adjusted OR (95% CI) =0.82 (0.6–1.1)] and [G/G versus TT, adjusted OR (95% CI) =1.32 (0.9–2.0)] and but reported interaction (P=0.01) between smoking and MDM2 rs2279744 genotypes (13). A study on a population consisting of Caucasians in the United States and African-Americans reported that MDM2 rs2279744 (SNP T309G) and MDM2 rs769412 (SNP A354G) were not associated with lung cancer risk (14). An Asian-Japanese study reported no association between MDM2 rs2279744 (SNP T309G) and lung cancer risk (15).
An Asian-Japanese study reported reduced overall survival of carriers of the MDM2 rs2279744 TT genotype as compared to carriers of the TG or GG genotypes (P=0.02) for patients with stage I lung adenocarcinoma (26). An Asian-Chinese study reported that carriers of MDM2 rs1690924 AG genotype were more sensitive to gastrointestinal toxicity than carriers of the wild-type homozygote GG [OR (95% CI) =2.32 (1.30–4.14), P=0.004)], suggesting MDM2 rs1690924 could be used to predict the toxicities of platinum-based chemotherapy in patients with advanced NSCLC (27). Recently, a Caucasian-Spanish study reported that MDM2 rs1690924 GG genotype presented higher risk of death [HR (95% CI) =1.99 (1.05–3.80), P=0.0345], suggesting one may significantly act as predictive factors of survival among NSCLC patients treated with platinum-based chemotherapy (28). A Caucasian-Spanish study did not find the influence of MDM2 rs1690924 on platinum-based chemotherapy toxicity for NSCLC patients (29).
To the best of our knowledge, the current study is the first systematically assess the association and interaction of MDM2 common variants in relation to lung cancer risk. In this study, the role of five htSNPs (rs1690924, rs1846402, rs2291857, rs3730581 and rs3730635) across MDM2 gene tagging 95% common haplotype diversity was evaluated. The five htSNPs are all located in intron regions. The five htSNPs selected did not include MDM2 rs2279744 (SNP T309G) SNP which is the most extensively studied polymorphism in previous epidemiological studies because the htSNP choice wads random. The htSNPs were selected solely based on their linkage with other SNPs in MDM2.
In the present study, we documented positive association of global five-locus haplotype and negative association of haplotype5 (rs1690924A-rs1846402G-rs2291857C-rs3730581G-rs3730635A) containing one variant-allele of rs3730581 with risk of lung cancer after adjusting smoking-duration. This finding is close to findings reported by others that carriers at least one variant-allele of MDM2 SNP [rs2279744 (SNP T309G)] were at reduced risk of lung cancer in a study of Caucasian-Norwegians (7).
We also documented that smoking-duration was the most important risk factor for lung cancer (one-locus model) and found interactions between MDM2 rs2291857 and TP53 rs8064946 (three-locus model), or MDM2 rs1690924, rs2291857 and TP53 rs8064946 (four-locus model), and or MDM2 rs1846402 (two-locus model) with smoking-duration in relation to lung cancer risk. Furthermore, we documented strong synergy interaction between MDM2 rs2291857 and TP53 rs8064946 in relation to smoking-induced lung cancer. These findings are in agreement with previous Chinese studies showing interactions between MDM2 and TP53 polymorphisms and smoking in relation to lung cancer risk (5,9). Smoking-duration has been considered a more important risk factor for lung cancer developing than other smoking-related factors such as cumulative smoking (17). Tobacco carcinogens may induce various types of DNA damage and increase genomic instability during long-term smoking. The TP53 tumor suppressor gene, cellular gatekeeper and guardian of genome, coordinates protective cellular responses to oncogenic stressors, such as DNA damage (30). TP53 activity is tightly controlled by MDM2. MDM2 is a primary negative regulatory factor for TP53 (3). The TP53-MDM2 negative feedback loop constitutes the core module of a network of regulatory interactions activated under cellular stress. In normal cells, the level of TP53 proteins is kept low through negative regulation by MDM2 (31). Through its N-terminal domain, MDM2 binds to TP53 and forms the MDM2–TP53 complex. The binding process obscures the TP53 transcription activation region and reduces TP53 transcription activity. MDM2 exerts the inhibitory effect not only through blocking its transcriptional activity, but also through directly eliminating it from the cell for down-regulating TP53 (3). Overall, the observed interactions support the notion that smoking-duration interact with genetically determined variation in MDM2 activity to modulate the individual’s predisposition towards smoking-induced lung cancer.
In this expanded study group including 83 new cases and 78 new controls, we were also able to reproduce our previous findings that PPP1R13L rs1970764 and CD3EAP rs967591 were associated with lung cancer risk and a previously reported interaction between CD3EAP rs735482 and smoking-duration in relation to lung cancer risk (19-21).
The present study included 1,260 participants and power analyses showed that we had 89%, 92%, 94%, 86% and 34% chance for rs1690924, rs1846402, rs2291857, rs3730581 and rs3730635, respectively, to detect OR =1.5 at the 0.05 significant level using two-sided tests under the dominant model. The low statistical power for rs3730635 is due to the low MAF of 0.02 in present controls population, which is significantly lower than reported HCB frequency in NCBI dbSNP (https://www.ncbi.nlm.nih.gov/snp). Thus, further larger population-based studies are warranted to confirm present findings.
Conclusions
In conclusion, MDM2 five-htSNPs haplotype exhibited association with lung cancer susceptibility, interaction of MDM2 and TP53 htSNPs and smoking-duration contributed to lung cancer risk and strong synergy between MDM2 and TP53 htSNPs influenced lung cancer predisposition. Our results suggest that MDM2, TP53 and smoking-duration interact in relation to lung carcinogenesis.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This study was supported by the National Natural Science Foundation of China (Grant No. 30571016 and No. 81072384).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study protocol was approved by the Chinese Administration Office of Human Genetic Resources {no. [2001]015} and informed consent was taken from all the participants.
Footnotes
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-19-4784
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-19-4784
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-19-4784). The authors have no conflicts of interest to declare.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Yang D, Liu Y, Bai C, et al. Epidemiology of lung cancer and lung cancer screening program in China and the United States. Cancer Lett 2020;468:82-7. 10.1016/j.canlet.2019.10.009 [DOI] [PubMed] [Google Scholar]
- 3.Hou H, Sun D, Zhang X. The role of MDM2 amplification and overexpression in therapeutic resistance of malignant tumors. Cancer Cell Int 2019;19:216. 10.1186/s12935-019-0937-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park SH, Choi JE, Kim EJ, et al. MDM2 309T>G polymorphism and risk of lung cancer in a Korean population. Lung Cancer 2006;54:19-24. 10.1016/j.lungcan.2006.06.008 [DOI] [PubMed] [Google Scholar]
- 5.Zhang X, Miao X, Guo Y, et al. Genetic polymorphisms in cell cycle regulatory genes MDM2 and TP53 are associated with susceptibility to lung cancer. Hum Mutat 2006;27:110-7. 10.1002/humu.20277 [DOI] [PubMed] [Google Scholar]
- 6.Lind H, Zienolddiny S, Ekstrøm PO, et al. Association of a functional polymorphism in the promoter of the MDM2 gene with risk of nonsmall cell lung cancer. Int J Cancer 2006;119:718-21. 10.1002/ijc.21872 [DOI] [PubMed] [Google Scholar]
- 7.Gansmo LB, Knappskog S, Romundstad P, et al. Influence of MDM2 SNP309 and SNP285 status on the risk of cancer in the breast, prostate, lung and colon. Int J Cancer 2015;137:96-103. 10.1002/ijc.29358 [DOI] [PubMed] [Google Scholar]
- 8.Chua HW, Ng D, Choo S, et al. Effect of MDM2 SNP309 and p53 codon 72 polymorphisms on lung cancer risk and survival among non-smoking Chinese women in Singapore. BMC Cancer 2010;10:88. 10.1186/1471-2407-10-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ren YW, Yin ZH, Wan Y, et al. P53 Arg72Pro and MDM2 SNP309 polymorphisms cooperate to increase lung adenocarcinoma risk in Chinese female non-smokers: a case control study. Asian Pac J Cancer Prev 2013;14:5415-20. 10.7314/APJCP.2013.14.9.5415 [DOI] [PubMed] [Google Scholar]
- 10.Li W, Wang SS, Deng J, et al. Association of p73 gene G4C14-A4T14 polymorphism and MDM2 gene SNP309 with non-small cell lung cancer risk in a Chinese population. Oncol Lett 2017;14:1817-22. 10.3892/ol.2017.6327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilkening S, Bermejo JL, Hemminki K. MDM2 SNP309 and cancer risk: a combined analysis. Carcinogenesis 2007;28:2262-7. 10.1093/carcin/bgm191 [DOI] [PubMed] [Google Scholar]
- 12.Luan L, Wang H, Zhao B, et al. Association of MDM2 gene SNP 309 polymorphism and human non-small cell lung cancer susceptibility: A meta-analysis. Pathol Res Pract 2019;215:152538. 10.1016/j.prp.2019.152538 [DOI] [PubMed] [Google Scholar]
- 13.Liu G, Wheatley-Price P, Zhou W, et al. Genetic polymorphisms of MDM2, cumulative cigarette smoking and nonsmall cell lung cancer risk. Int J Cancer 2008;122:915-8. 10.1002/ijc.23178 [DOI] [PubMed] [Google Scholar]
- 14.Pine SR, Mechanic LE, Bowman ED, et al. MDM2 SNP309 and SNP354 are not associated with lung cancer risk. Cancer Epidemiol Biomarkers Prev 2006;15:1559-61. 10.1158/1055-9965.EPI-06-0217 [DOI] [PubMed] [Google Scholar]
- 15.Enokida Y, Shimizu K, Kakegawa S, et al. Single-nucleotide polymorphism (c.309T>G) in the MDM2 gene and lung cancer risk. Biomed Rep 2014;2:719-24. 10.3892/br.2014.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin JY, Ma YG, Vogel U, et al. GLTSCR1, ATM, PPP1R13L and CD3EAP Genetic Variants and Lung Cancer Risk in a Chinese Population. Curr Med Sci 2018;38:734-40. 10.1007/s11596-018-1938-6 [DOI] [PubMed] [Google Scholar]
- 17.Vogel U, Laros I, Jacobsen NR, et al. Two regions in chromosome 19q13.2-3 are associated with risk of lung cancer. Mutat Res 2004; 546:65-74. 10.1016/j.mrfmmm.2003.11.001 [DOI] [PubMed] [Google Scholar]
- 18.Yin J, Vogel U, Ma Y, et al. A specific diplotype defined by PPP1R13L rs1970764, CD3EAP rs967591 and ERCC1 rs11615 and lung cancer risk in a Chinese population. Lung Cancer 2012;76:286-91. 10.1016/j.lungcan.2012.01.001 [DOI] [PubMed] [Google Scholar]
- 19.Yin J, Wang H, Vogel U, et al. Association and interaction of NFKB1 rs28362491 insertion/deletion ATTG polymorphism and PPP1R13L and CD3EAP related to lung cancer risk in a Chinese population. Tumour Biol 2016;37:5467-73. 10.1007/s13277-015-4373-3 [DOI] [PubMed] [Google Scholar]
- 20.Yin J, Wang H, Vogel U, et al. Fine-mapping markers of lung cancer susceptibility in a sub-region of chromosome19q13.3 among Chinese. Oncotarget 2016;7:60929-39. 10.18632/oncotarget.9279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou W, Yin J, Vogel U, et al. 19p13.3-GADD45B common variants and 19q13.3-PPP1R13L and 19q13.3-CD3EAP in lung cancer risk among Chinese. Chem Biol Interact 2017;277:74-8. 10.1016/j.cbi.2017.08.018 [DOI] [PubMed] [Google Scholar]
- 22.Xiao Z, Xiao J, Jiang Y, et al. A novel method based on ligase detection reaction for low abundant YIDD mutants detection in hepatitis B virus. Hepatol Res 2006;34:150-5. 10.1016/j.hepres.2005.12.007 [DOI] [PubMed] [Google Scholar]
- 23.Solé X, Guinó E, Valls J, et al. SNPStats: a web tool for the analysis of association studies. Bioinformatics 2006;22:1928-9. 10.1093/bioinformatics/btl268 [DOI] [PubMed] [Google Scholar]
- 24.Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263-5. 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- 25.Ritchie MD, Hahn LW, Roodi N, et al. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet 2001;69:138-47. 10.1086/321276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enokida Y, Shimizu K, Atsumi J, et al. Prognostic potential of the MDM2 309T>G polymorphism in stage I lung adenocarcinoma. Cancer Med 2016;5:1791-801. 10.1002/cam4.750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qian J, Liu H, Gu S, et al. Genetic variants of the MDM2 gene are predictive of treatment-related toxicities and overall survival in patients with advanced NSCLC. Clin Lung Cancer 2015;16:e37-53. 10.1016/j.cllc.2015.02.001 [DOI] [PubMed] [Google Scholar]
- 28.Pérez-Ramírez C, Cañadas-Garre M, Alnatsha A, et al. Pharmacogenetics of platinum-based chemotherapy: impact of DNA repair and folate metabolism gene polymorphisms on prognosis of non-small cell lung cancer patients. Pharmacogenomics J 2019;19:164-77. 10.1038/s41397-018-0014-8 [DOI] [PubMed] [Google Scholar]
- 29.Pérez-Ramírez C, Cañadas-Garre M, Alnatsha A, et al. Pharmacogenetic predictors of toxicity to platinum based chemotherapy in non-small cell lung cancer patients. Pharmacol Res 2016;111:877-84. 10.1016/j.phrs.2016.08.002 [DOI] [PubMed] [Google Scholar]
- 30.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature 2000;408:307-10. 10.1038/35042675 [DOI] [PubMed] [Google Scholar]
- 31.Bose I, Ghosh B.The p53-MDM2 network: from oscillations to apoptosis. J Biosci 2007; 32:991-7. 10.1007/s12038-007-0103-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as