Abstract
Background
Proline levels are significantly increased in tumor specimens and urine samples from gastric cancer (GC) patients, and we previously showed that intracellular proline levels significantly differ between human GC cell lines and normal gastric epithelial cells. Pyrroline-5-carboxylate reductase 1 (PYCR1) is the key enzyme in intracellular proline synthesis, but its role in GC remains largely unknown.
Methods
Bioinformatic analysis and immunohistochemical (IHC) staining with a tissue microarray were conducted to assess the association between PYCR1 expression and clinical parameters. PYCR1 downregulation and overexpression were then established in two GC cell lines (AGS and MKN28 cells) to determine whether PYCR1 promotes malignant behavior in GC. Gene set enrichment analysis (GSEA) was further performed to investigate the pathway regulating PYCR1 in GC.
Results
PYCR1 expression was up-regulated in different GC cohorts. High PYCR1 protein expression was correlated with advanced tumor stage, aggressive histological type and high Ki-67 index. High PYCR1 expression in GC tissues was an indicator of poor outcome in GC patients. In vitro, PYCR1 knockdown markedly attenuated GC cells growth and promoted apoptosis, while overexpression produced the opposite effects. GSEA analysis indicated PI3K/Akt axis was strongly correlated with PYCR1 expression and that PIK3CB and AKT1 mRNA expression was positively associated with PYCR1 in GC tissues. PI3K inhibition further significantly reduced PYCR1 mRNA and protein expression. Moreover, as PYCR1 is a mitochondrial endomembrane protein, nutrient stress induced by glucose deprivation also regulated PYCR1 expression.
Conclusions
PYCR1 is highly expressed in GC and acts as a mitochondrial oncogene to induce cancer progression by enhancing tumor proliferation and responding to metabolic stress. PYCR1 is a novel prognostic marker and a potential therapeutic target in GC.
Keywords: Gastric cancer (GC), proline metabolism, Pyrroline-5-carboxylate reductase 1 (PYCR1), PI3K/Akt, metabolic stress
Introduction
Gastric cancer (GC) is the third leading cause of cancer mortality worldwide and remains a major health threat in Asia-Pacific regions (1). GC patients with advanced-stage disease often have a poor prognosis due to tumor metastasis and recurrence; therefore, it is of clinical significance to identify new biomarkers that control the severity of GC and exhibit predictive value for prognosis.
Recently, there has been intense interest in understanding metabolic reprogramming in cancer aiming to develop a metabolism-based anticancer therapy. Accordingly, several studies aimed at identifying metabolic changes in macroenvironment (blood or urine) and microenvironment (carcinoma tissue and gastric juice) have been performed to map global metabolic profiles and interpret the possible underlying mechanism in GC (2). Among several metabolites, proline was identified to exhibit significantly increased levels in tumor specimens (3) and urine samples (4,5) from GC patients and to be a possible predictive factor for patient outcome (5) and tumor metastasis (6). More importantly, our recent study using gas chromatograph-mass spectrometer found that human GC cell lines and normal gastric epithelial cell exhibited a significant difference in intracellular proline among 278 metabolites (7). These findings suggest that proline metabolism, including the metabolites and metabolic enzymes in this pathway, might play an important role in GC, and more intensive research is warranted.
Pyrroline-5-carboxylate reductase 1 (PYCR1), a mitochondrial inner membrane protein, catalyzes the rate-limiting step in proline synthesis from pyrroline-5-carboxylate (P5C). PYCR1 has been shown to dramatically affects cellular energetic, physiological and pathological processes (8). Additionally, the ability of proline to regulate redox equivalent shuttling, resulting in the protection of cells from oxidative stress, has been demonstrated (9-11). Despite recent efforts to understand the activities of PYCR1 in cancer metabolic reprogramming, little is known about the role of PYCR1 in GC, and it remains unclear whether PYCR1 expression may correlate with any clinicopathological features of GC.
In this present study, we therefore combined in-silico analyses of three independent cohorts and tumor microarray to investigate the significance of PYCR1 in GC and performed further in vitro studies to investigate both its potential effects on GC cell phenotype and its associated regulatory pathway.
Methods
Bioinformatic analyses
Profile of basic PYCR1 expression in multiple human tissues was evaluated by GTEx (https://gtexportal.org). The datasets GSE13861(Cho dataset) and GSE13911(DErrico dataset) were downloaded from the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/). The Cancer Genome Atlas (TCGA), which has both sequencing and pathological data on 30 different cancers including GC, was selected for further analyses of PYCR1 using GEPIA (http://gepia.cancer-pku.cn/) (12). The mutation counts and fraction of copy number altered genome data for TCGA GC were directly downloaded from the cBioPortal for Cancer Genomics (http://cbioportal.org) (13). Prognostic values of PYCR1 mRNA were investigated using an online database Kaplan-Meier plotter (http://kmplot.com/analysis/) (14). Gene Set Enrichment Analysis (GSEA), which is designed to detect whether a predefined gene set shows concordant differences between two biological states, was performed to investigate the PYCR1-associated genes and pathways (15).
Tissue microarray
Tissue microarray (HStm-Ade180-Sur-08) containing 72 paired GC/para-cancerous tissues and 18 GC tissues were purchased from Shanghai Outdo Biotech (Shanghai, China). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Sample collection was approved by the Ethics Committees of National Engineering Center for Biochip. Written informed consent was obtained from the patient for publication of this study and any accompanying images. Patients who underwent gastric surgical resection were followed up, and database of clinical variables (sex, age, diagnosis, stage, Ki-67 index, survival status, follow-up period) was established.
Immunohistochemical (IHC) staining and scoring
The expression of PYCR1 protein was examined using IHC staining with primary antibody (PYCR1, 1:2000, Proteintech). The slide was then photographed under a microscope and analyzed by two observers who were blinded to the clinical and pathological data. PYCR1 expression was scored semi-quantitatively by multiplying the staining intensity (0, no staining; 1, weak staining; 2, moderate staining; 3, strong staining) and the staining rate (graded on the percentage of positive tumor cells; 0, none; 1, 1–25%; 2, 26–50%; 3, 51–75%; 4, >75%). Protein expression level was then classified into two categories: low expression (grades 0–3) and high expression (grades 4–12).
Cell lines and cell culture
Human GC cell lines BGC823, AGS, MKN45, SGC7901 and MKN28 were purchased from National Infrastructure of Cell Line Resource (Beijing, China). Cells were cultured in RPMI 1640 medium (Hyclone) with 10% fetal bovine serum (Gibco) and 1% Penicillin-Streptomycin antibiotics (Hyclone). The cell culture was maintained in an incubator at 37 °C with 5% CO2.
PYCR1 knockdown and overexpression
For PYCR1 knockdown, PYCR1 specific siRNA (siPYCR1) and negative control siRNA (siNEG) were chemically synthesized by Sangon Biotech (Shanghai, China). Transfection of cells with siRNA was performed using Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific) according to the manufacturer’s recommendation. The sequence of the siRNAs used as follows: siNEG: sense, 5'-UUCUCCGAACGUGUCACGUTT-3'; antisense, 5'-CGUGACACGUUCGGAGAATT-3'; siPYCR1sense, 5'-GCUGUGAAGCCACACAUCATT-3'; antisense, 5'-UGAUGUGUGGCUUCACAGCTT-3'. For PYCR1 overexpression, cell lines were transfected with PYCR1 overexpression plasmid (pENTER-PYCR1-Flag) or blank plasmid (purchasing from Vigene Biosciences) using Lipofectamine 2000. Then western blot and real time PCR was performed to determine the efficiency of reduction or overexpression.
Cell proliferation and colony formation assay
Cell proliferation was measured using CCK-8 (Dojindo, Japan) following the manufacturer’s protocol. Simply, transfected cells were seeded on 96-well plates at a density of around 2,000 cells/well, then 10 µL of CCK-8 from kit was added to each well containing 100 µL fresh media for 2 h incubation at 37 °C at the test timepoint. Finally, absorbance values were determined at 450 nm by a plate reader (Varioskan Flash, Thermo Fisher Scientific). For colony formation assay, transfected cells were seeded into 6-well plat at a density of 500 cells/well, and were maintained in culture medium for 10 days. Then, cells were fixed (absolute alcohol) and stained (crystal violet) for colony count.
Apoptosis detection
Apoptosis was determined by Hoechst 33342 staining and flow cytometric analysis respectively. Nuclear Hoechst 33342 staining was performed to visualized the changes of nuclear morphology of transfected cells. Briefly, transfected cells were washed twice with PBS, and then incubated with 10 µg/mL Hoechst 33342 (Solarbio, China) in the dark for 20 minutes, washed twice with PBS and observed using a fluorescence microscopy. For flow cytometric analysis, transfected cells were trypsinized and washed twice with PBS, and resuspended in binding buffer. Cell suspensions were then stained with Annexin V-FITC and 7-AAD (KeyGen BioTECH, China) for 10 minutes in the dark at room temperature before flow cytometric analysis.
RNA extraction and quantitative real time PCR
Total RNA from different treatment groups was harvested using TRIzol reagent (Invitrogen). The cDNA was synthesized using the FastKing RT Kit (TIANGEN BIOTECH, China), quantitative real-time PCR using Talent qPCR PreMix Kit(TIANGEN BIOTECH, China) was then performed with the following procedures: 95 °C for 3min, followed by 95 °C for 5 s and 60 °C for 15 s for 40 cycles (QuantStudio 5 Real-Time PCR System). The relative expression of the mRNA of PYCR1were detected, and the β-actin was used as an internal control. Primers are as follows: β-actin: Forward 5'-CCTGGCACCCAGCACAAT-3', Reverse 5'-GGGCCGGAC TCGTCATAC-3'; PYCR1: Forward 5'-CCTACGGGGCTGCCAAGAT-3', Reverse 5'-ACTCT CCAGCACATGCAAGG-3'. The fold change in gene expression was calculated as: Fold change =2-△△Ct.
Western blot
Cells were harvested and lysed in RIPA containing proteinase and phosphatase inhibitors. The concentration of proteins in cell lysates were quantified by BCA kit (Applygen Technologies, China) following the instruction. Equal amounts of cell lysates (20 µg protein/lane) were electrophoresed on SDS-polyacrylamide gels and transferred to nitrocellulose membrane. Membranes were blocked and then probed with the appropriate primary antibody [PYCR1, 1:5,000 (Proteintech); PARP, 1:1,000 (Proteintech); Caspase-3, 1:1,000 (CST); cleaved-Caspase 3, 1:1000 (CST); p-PI3K, 1:2,000 (Proteintech); p-Akt, 1:4,000 (Proteintech); Akt, 1:1,000 (Proteintech); β-actin, 1:8,000 (Applygen)] in blocking buffer overnight at 4 °C. The bound antibodies were detected with IRDye 800CW-conjugated goat anti-rabbit IgG or anti-mouse IgG (at 1:10,000 dilution; LI-COR Bioscience, Lincoln, NE, USA) and visualized with Odyssey 290 infrared imaging system (LI-COR Bioscience).
Statistical analyses
Normally distributed data were presented as mean ± SEM, data with skewed distribution were presented as median with interquartile. Difference between groups was determined by two-tailed Student’s t-test or one-way ANOVA for normally distributed data, while comparisons were performed by nonparametric tests for skewed distribution. Enumeration data were determined by chi square test or Fisher exact test. The correlation between Ki-67 index and PYCR1 score was examined by Spearman correlation test. Overall survival in relation to PYCR1 expression was evaluated by the Kaplan-Meier survival curve and Log-rank test. Risk factors of prognosis in patients with GC were evaluated using univariate and multivariate analyses. All statistical analyses were carried out using SPSS for Windows 21.0 software, the value of P<0.05 was considered statistically significant.
Results
PYCR1 is commonly upregulated in various cancer types and GC tissues
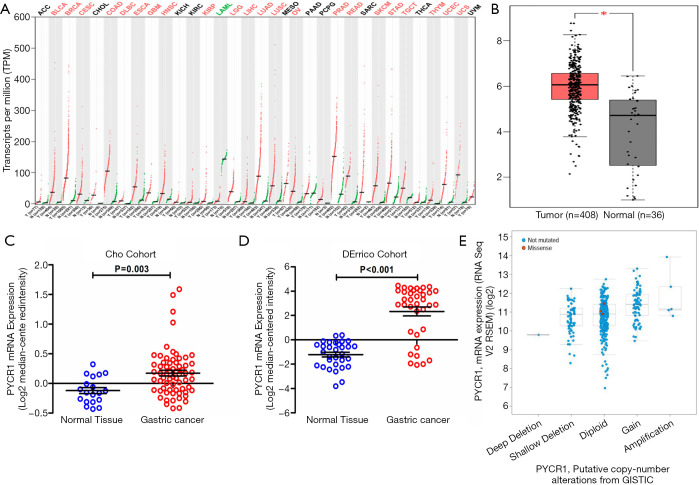
Genes that are consistently expressed across different cancers, such as the well-known oncogenes Ras and Myc, might characterize the fundamental process by which normal cells undergo malignant transformation. We found that PYCR1 was widely overexpressed across 22 cancer types in a dataset from The Cancer Genome Atlas (TCGA) (Figure 1A). Additionally, the basal expression of PYCR1 mRNA was relatively higher in the stomach, pancreas and prostate compared with that in other tissues, indicating that PYCR1 mRNA might be specific to these organs (available online: http://gtexportal.org/gene/PYCR1). Thus, up-regulation of PYCR1 might be cancer-specific and might play an important role in stomach-related diseases such as GC.
Figure 1.
Expression of PYCR1 mRNA in pan-cancer and GC. (A) PYCR1 mRNA expression in 33 pan-cancers. Of them, PYCR1 mRNA is significantly upregulated in 22 cancer types including GC. (B) Comparison of PYCR1 mRNA level between normal (n=36) and GC tissues (n=408) in the TCGA cohort. (C,D) Statistic analysis of PYCR1 mRNA expression in normal and GC tissues in two independent datasets-Cho dataset (GSE13861) & DErrico dataset (GSE13911) (both P<0.05, by Student t test). (E) PYCR1 mRNA levels were higher in samples with PYCR1 gained copy number variance (CNV) compared with the samples without CNV in the TCGA dataset (cBioPortal). *, P<0.05.
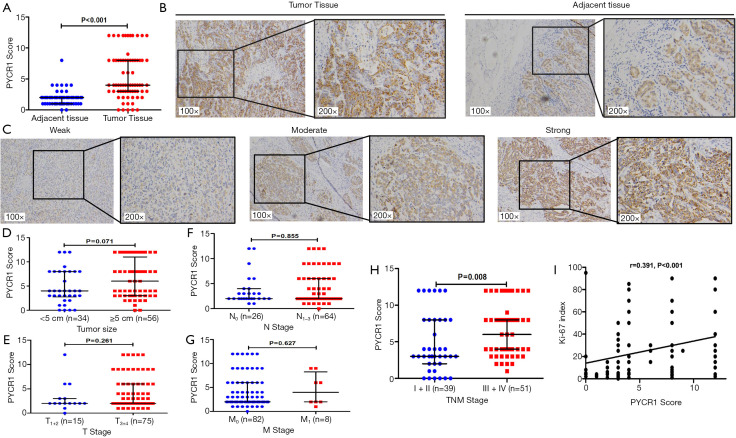
To further evaluate the PYCR1 expression in GC and normal gastric tissues, changes in the PYCR1 transcriptional level were first validated using microarray values from Gene Expression Omnibus (GEO) and TCGA datasets. PYCR1 mRNA was increased in GC tissues in the TCGA cohort (Figure 1B) and GEO datasets (Cho cohort and DErrico cohort) (Figure 1C,D). In addition, we determined that the increase in the PYCR1 mRNA abundance was consistent with the enhancement of its DNA level seen in TCGA dataset (Figure 1E). To ensure that conclusions derived from in silico analysis were reliable, we then assessed the PYCR1 protein expression using immunostaining in 72 paired GC and adjacent tissues. IHC staining indicated that the PYCR1 staining score was higher in tumor tissues than in para-cancer tissues (Figure 2A). PYCR1 staining was specifically localized in the cytoplasm; representative IHC results for PYCR1 in GC tissues and adjacent tissues, along with examples of different staining intensity, are shown in Figure 2B,C, respectively. The PYCR1 score did not show significant difference in cancer tissues regarding the tumor size (Figure 2D), T stage (Figure 2E), N stage (Figure 2F) and M stage (Figure 2G), but it was higher in patients with advanced TNM stage (Stage III + IV) disease (Figure 2H). Moreover, PYCR1 immunostaining was positively associated with the Ki-67 index (r=0.391, P<0.001 by Spearman’s correlation coefficient) (Figure 2I). These findings suggest that PYCR1 may play an important role in GC progression.
Figure 2.
Overexpression of PYCR1 protein in GC. (A) Statistical analysis of PYCR1 score in GC tissues and para-cancerous tissues (P<0.001, by Wilcoxon test). (B) Expression of PYCR1 protein in paired tumorous and peri-tumorous tissues (n=72) was determined by immunohistochemical staining (magnification, 100× and 200×). (C) Representative images of PYCR1 staining intensity (weak, moderate and strong) in GC tissues (magnification, 100× and 200×). Statistical analyses of PYCR1 score in cancer tissues (n=90) with regard to tumor size (D), T stage (E), N stage (F), M stage (G) and TNM stage (H) (P value by Mann-Whitney test). (I) Correlation between Ki-67 index and PYCR1 score in tumor tissues (n=90) was analyzed by Spearmen test (r=0.319, 95% CI, 0.194–0.557; P<0.001).
Associations between PYCR1 expression and clinicopathological characteristics in GC
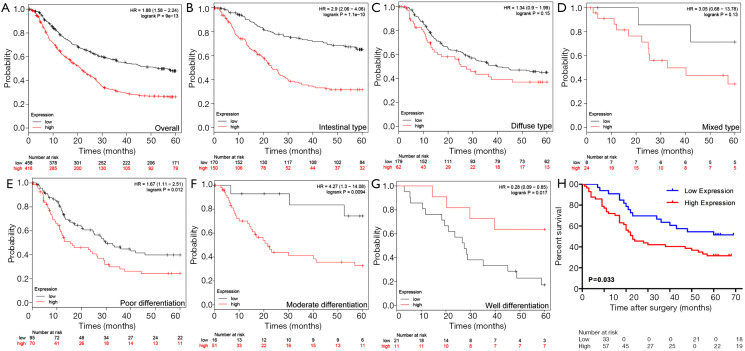
We then studied whether PYCR1 expression was associated with clinicopathological features and had prognostic value in GC patients. The prognostic value of PYCR1 mRNA expression in GC was first assessed using the online Kaplan-Meier Plotter tool. Lower levels of PYCR1 mRNA were associated with longer overall survival (OS) times in the entire cohorts of GC patients (HR =1.88, 95% CI, 1.58–2.24, P<0.001) (Figure 3A). Assessment of the prognostic value based on the Lauren classification of GC indicated that high expression of PYCR1 in intestinal-type gastric adenocarcinoma predicated an adverse outcome (HR =2.9, 95% CI, 2.06–4.06, P<0.001) (Figure 3B,C,D). Subgroup analyses based on stratification by the degree of differentiation identified that high PYCR1 mRNA expression was also associated with poor prognosis in patients with poorly and moderately differentiated GC (HR =1.67, 95% CI, 1.11–2.51, P=0.012; HR =4.27, 95% CI, 1.3–14.08, P=0.0094; respectively), while high PYCR1 expression was associated improved OS in patients with well-differentiated GC (HR =0.28, 95% CI, 0.09–0.85, P=0.017) (Figure 3E,F,G).
Figure 3.
High PYCR1 expression predicts poor prognosis in GC patients. (A) Kaplan-Meier plotter analysis of GC patients’ survival stratified by PYCR1 mRNA expression level. (B,C,D) Survival analyses of PYCR1 in GC patients based on Lauren classification (Intestinal, diffuse and mixed type respectively) through Kaplan-Meier plotter. (E,F,G) Survival analyses of PYCR1 in GC patients according to tumor differentiation status (poor, moderate and well differentiation respectively) using Kaplan-Meier plotter. (H) Kaplan-Meier survival analysis of GC patients stratified by PYCR1 protein expression level (P=0.033, by Log-rank test).
Further evaluation of PYCR1 protein expression in 90 GC patients with different clinicopathologic features revealed that high PYCR1 expression was positively correlated with patients’ AJCC stage (P=0.003), histologic type (P=0.006) and tumor Ki-67 index (P<0.001) (Table 1). The cumulative survival rate within 5 years after surgery was significantly increased in those with low PYCR1 expression (P<0.05) (Figure 3H), but low PYCR1 expression was not an independent risk factor in multivariate analysis (HR =1.436, 95% CI, 0.758–2.723, P=0.267) (Table 2).
Table 1. Clinicopathological characteristics of GC patients with high and low PYCR1 protein expression.
| Clinical parameters | Total (n=90) | PYCR1 expression | χ2 | P | |
|---|---|---|---|---|---|
| Low (n=33) | High (n=57) | ||||
| Sex (male/female) | 61/29 | 21/12 | 40/17 | 0.409 | 0.522 |
| Age (<60/≥60 years) | 30/60 | 15/18 | 15/42 | 3.445 | 0.063 |
| Tumor size (<5/≥5 cm) | 34/56 | 15/18 | 19/38 | 1.306 | 0.253 |
| Histologic type | 0.006§ | ||||
| Tubular/papillary | 68 | 19 | 49 | – | |
| Mucinous | 9 | 7 | 2 | ||
| Signet-ring-cell and others‡ | 13 | 7 | 6 | ||
| Differentiation (well + moderate/poor) | 14/76 | 4/29 | 10/47 | 0.468 | 0.494 |
| Ki-67 Index (low/high)¶ | 52/38 | 26/7 | 26/31 | 9.428 | 0.002 |
| Invasion depth (T1 + 2/T3 + 4) | 15/75 | 8/25 | 7/50 | 2.153 | 0.142 |
| Lymph node metastasis (N0/N1–3) | 26/64 | 12/21 | 14/43 | 1.417 | 0.234 |
| Distant metastasis (M0/M1) | 82/8 | 31/2 | 51/6 | 0.111 | 0.739† |
| Stage (I + II/III + IV) | 39/41 | 21/12 | 18/39 | 8.747 | 0.003 |
Statistical analyses was performed by chi square test. †, continuity adjusted value for chi square test; ‡, other refers to one case of non-differentiated cancer; §, Fisher’s exact test; ¶, split by median value.
Table 2. Univariate and multivariate analysis of risk factors for overall survival of GC patients.
| Factors | Univariate analysis | Multivariate analysis† | |||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| Sex (female/male) | 0.990 (0.563–1.739) | 0.972 | 0.792 (0.442–1.417) | 0.432 | |
| Age (<60/≥60 years) | 1.121 (0.638–1.970) | 0.69 | 1.338 (0.749–2.391) | 0.326 | |
| Tumor size (<5/≥5 cm) | 2.257 (1.245–4.093) | 0.007 | 1.683 (0.894–3.167) | 0.107 | |
| Histologic type | |||||
| Others/tubular + papillary | 0.804 (0.449–1.441) | 0.464 | 0.867 (0.462–1.627) | 0.656 | |
| Differentiation | |||||
| Well + moderate/poor | 1.890 (0.808–4.418) | 0.142 | 1.421 (0.568–3.553) | 0.453 | |
| Stage (I + II/III + IV) | 2.992 (1.663–5.384) | <0.001 | 2.360 (1.245–4.476) | 0.009 | |
| PYCR1 level (low/high) | 1.857 (1.037–3.326) | 0.037 | 1.436 (0.758–2.723) | 0.267 | |
†, multivariate analysis was performed by Enter method.
Functional implication of PYCR1 in GC in vitro
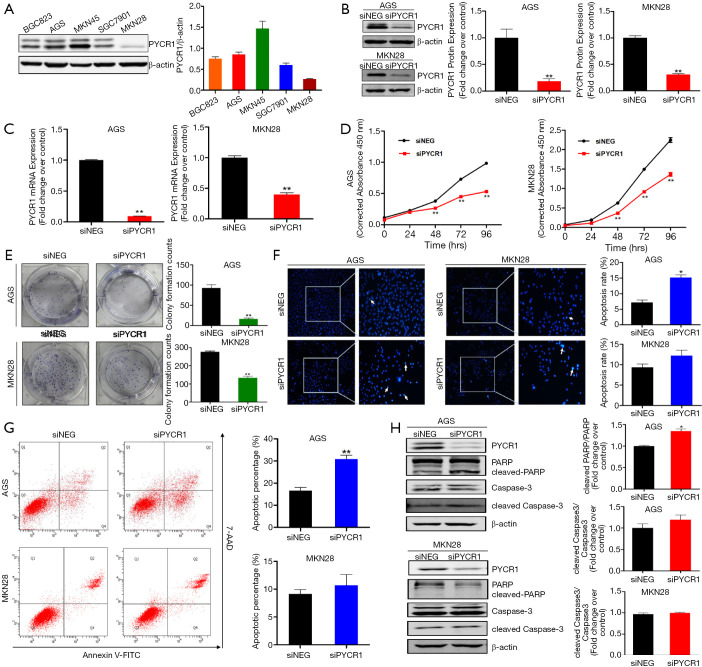
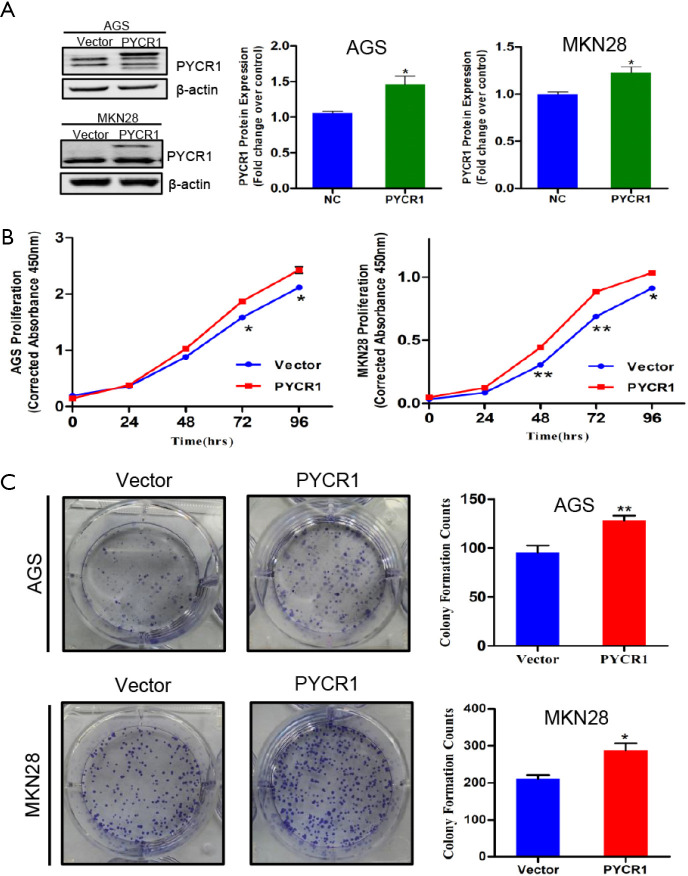
To elucidate the biological function of PYCR1 in GC, we performed a further in vitro experiment. First, the protein levels of PYCR1 were detected in five GC cell lines by western blotting, the results demonstrated that PYCR1 expression was variable in GC cell lines with different differentiation (Figure 4A). Based on these results, AGS (high PYCR1 expression) and MKN28 (low PYCR1 expression) were selected to perform further in vitro assays. To determine whether PYCR1 is required for GC cells proliferation, RNA interference (Figure 4B,C) and plasmid overexpression (Figure S1A) techniques were respectively applied to knockdown and upregulate PYCR1 in GC cells. Quantitative analysis of the CCK-8 assay results indicated that AGS and MKN28 cells exhibited a significantly reduction in the cell viability following PYCR1 knockdown (Figure 4D). The colony formation assay results also showed that PYCR1 downregulation reduced the number of colonies (Figure 4E). In contrast, cell viability and colony formation were promoted in both cell lines by exogenous PYCR1 overexpression (Figure S1).
Figure 4.
Biological function of PYCR1 in GC cells in vitro. (A) Expression profile of PYCR1 protein in 5 GC cell lines was analyzed by western blotting. (B,C) Efficiency of PYCR1 knockdown in GC cells transfected with siNEG and siPYCR1, which was detected by western blotting and real time RT-PCR respectively (n=3). (D) PYCR1 knockdown inhibited cell proliferation via CCK-8 assay (n=3). (E) Colony formation assay showed clonogenicity of indicated cells (n=3). (F) Hoechst 33342 staining showed deeply stained nuclei and chromatin condensation in cells transfected with siPYCR1 (Magnification, 100×, 200×) (n=3). (G) Cell apoptosis after PYCR1 downregulation in AGS and MKN28 GC cells was assessed by flow cytometry (n=3). (H) Representative blots of apoptosis-associated markers (PARP, cleaved PARP, Caspase-3, cleaved Caspase-3) determined by immunoblot test and the quantitative results of gray intensity (n=3). (Note: Histogram of cleaved PARP/PARP in MKN28 cell was not presented, because the blot of cleaved PARP cannot be detected in MKN28 cells in three independent assays). *, P<0.05; **, P<0.001, by Student t test.
Figure S1.
PYCR1 overexpression promotes GC cells growth. (A) PYCR1 was overexpressed in AGS and MKN28 cells (n=3). (B) PYCR1 overexpression increased cell proliferation via CCK-8 assay (n=3). (C) Colony formation assay showed PYCR1 overexpression improved clonogenicity of indicated cells (n=3). *, P<0.05; **, P<0.001, by Student t-test.
To investigate the mechanism behind regulating cell growth, we further examined the effect of PYCR1 on apoptosis. At 48 hours after transfection, PYCR1-downregulated cells displayed deeply stained nuclei and chromatin condensation, as detected by nuclear staining with Hoechst 33342 (Figure 4F). Flow cytometry analysis further identified a significant increase in apoptotic AGS GC cells after PYCR1 downregulation (AGS, siNEG vs. siPYCR1, 16.60±1.46 vs. 30.80±1.76), but such an increase was not evident in MKN28 cells (MKN28, siNEG vs. siPYCR1, 9.133±0.80 vs. 10.7±1.929) (Figure 4G). In addition, apoptosis-associated markers (cleaved PARP and cleaved Caspase-3) were upregulated in AGS cells after PYCR1 knockdown (Figure 4H). These in vitro results suggest that PYCR1 promote GC progression mainly through the induction of tumor cell proliferation.
Correlation of PYCR1 and PI3K/Akt pathway
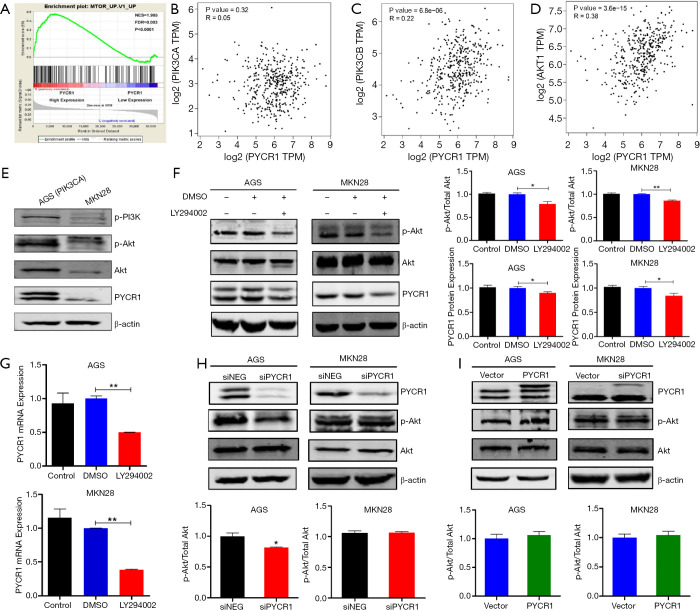
To probe the relevant pathways regulating PYCR1 expression on an unbiased basis, we performed GSEA using the RNA sequencing data from the TCGA GC cohort. Among the 189 predefined ‘oncogenic signature’ gene sets, the MTOR pathway was identified to be strongly associated with PYCR1 expression in this dataset (Figure 5A), suggesting that PYCR1 is involved in the PI3K/Akt/mTOR axis. In addition, correlation analysis with the GEPIA tool reconfirmed that PYCR1 mRNA expression was positively associated with PIK3CB and AKT1 mRNA expression in the same cohort but was not associated with PIK3CA mRNA (Figure 5B,C,D). Additionally, the protein expression level of PYCR1 was higher in GC cells with PI3KCA mutation (AGS) than in MKN28 cells, possibly validating the regulatory effect of this pathway on PYCR1 (Figure 5E). Accordingly, the PI3K inhibitor LY294002 was applied to confirm this correlation from the upstream direction. As shown in Figure 5F,G, LY294002 treatment obviously inhibited both protein and mRNA expression of PYCR1 in AGS (40 µM) and MKN28 (100 µM) cells, demonstrating that PI3K/Akt axis is an upstream regulator of PYCR1. We also evaluated the changes in the levels of phosphorylated Akt (p-Akt) following PYCR1 knockdown and overexpression. It was found that the phosphorylation level of p-Akt was not changed in either cell lines following PYCR1 downregulation (Figure 5H) or upregulation (Figure 5I). These results indicate that PI3K/Akt pathway affects proline metabolism via PYCR1 in GC.
Figure 5.
PYCR1 and PI3K/Akt signaling pathway. (A) Gene Set Enrichment Analysis (GSEA) identified significant association between PYCR1 and mTOR signaling pathway in the TCGA GC dataset. (B,C,D) Correlation between PYCR1 mRNA and PIK3CA mRNA(r=0.05, P=0.32, by Spearman test), PIK3CB Mrna (r=0.22 P=6.8×10−6, by Spearman test) or AKT1 mRNA(r=0.38, P=3.6×10−15; by Spearman test) in TCGA GC tissues. (E) Western blot showing the baseline levels of p-PI3K, p-Akt, Akt and PYCR1 in AGS and MKN28 cells. PYCR1 expression is upregulated in GC cell with PIK3CA mutation. (F,G) The effects of PI3K phosphorylation inhibitor LY294002 on the expression of PYCR1 protein and mRNA (n=3). The effects of PYCR1 knockdown (H) or overexpression (I) on the protein expression of p-Akt and Akt in AGS and MKN28 (n=3). *, P<0.05; **, P<0.001, by Student t test.
PYCR1 responses to nutrient stress in tumor microenvironment
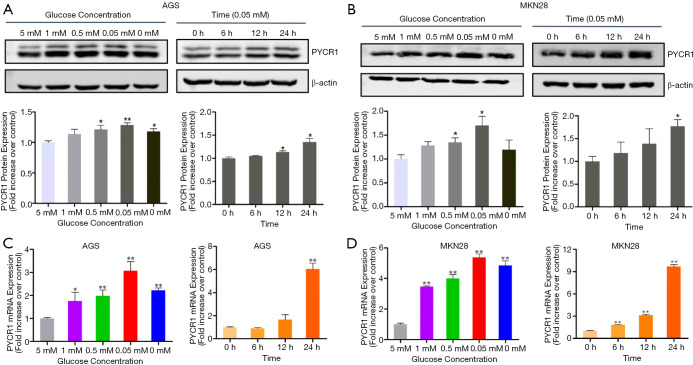
Nutrient deficiency is an important feature of tumor microenvironment, and it affects tumor progression and drives changes in cancer metabolism (16). PYCR1 is localized in the mitochondrial membrane, and we reasoned that it might respond to nutrient stress in the tumor microenvironment due to this special localization. Thus, we investigated the glucose concentration- and time-dependent response of PYCR1 mRNA and protein levels. As the glucose concentration in the medium decreased (from 5 to 0 mM), an increase in PYCR1 protein expression was observed along with an increase in PYCR1 mRNA after 24 hours of treatment (Figure 6). A time-dependent increase in PYCR1 mRNA and protein expression was also seen in the presence of 0.05mM glucose concentration (Figure 6). These data show that PYCR1 can act as a stress-induced metabolic gene to support cell growth.
Figure 6.
Metabolic stress induced by glucose withdrawal upregulates PYCR1. (A,B) GC cells were exposure for 24 hours to medium with decreasing glucose concentration from 5 mM (control) to 0 mM (left panel), cells were treated with medium containing 0.05mM glucose and collected at indicated time points (0 h as control) (right panel). Levels of PYCR1 protein was detected by western blotting, densitometry analyses show the band density ratios of PYCR1 to β-actin in indicated cells (n=3). (C,D) PYCR1 mRNA level was measured by real-time RT-PCR using β-actin as an internal control. (n=3, *, P<0.05; **, P<0.001, by Student t test).
Discussion
Proline and its related metabolites and associated metabolic pathways have been reported to be central to cancer growth and metastasis (17). PYCR1, as the critical enzyme in the last step of the proline synthesis pathway in mitochondria, has recently received much attention in cancer research. Our data based on TCGA mining revealed that PYCR1 was commonly upregulated across 22 cancer types, consistent with Nilsson’s result (18) which indicated that PYCR1 was the most frequently overexpressed metabolic gene across 1981 tumor samples consisting of 19 cancer types (18). In addition, some studies applying metabolomics have found increased de novo proline synthesis in metastatic breast cancer cells (17), melanoma cell lines (19) and ovarian cancer stem cells (20). Thus, upregulation of PYCR1 in tumors might explain the metabolic shift in proline synthesis in cancer. Collectively, this evidence from different studies robustly supports the hypothesis that PYCR1 is the fundamental gene for tumor transformation, and ultimately results in metabolic adaptation in cancer cells.
Notably, the present study is the first to highlight the clinical implication and biological function of PYCR1 in GC. We found that PYCR1 was highly expressed in tumor tissues and was positively associated with advanced TNM stage in GC patients from different cohorts. However, whether PYCR1 expression differs during the progression from normal gastric epithelium to precancerous lesions (intestinal metaplasia and dysplasia) to GC is not known. Therefore, further study is warranted to uncover whether proline metabolism reprogramming via PYCR1 may contribute to gastric carcinogenesis. GC patients with low PYCR1 expression had favorable prognoses during the five-years postsurgical period. Similarly, PYCR1 also displays prognostic value in breast cancer (21), lung cancer (22) and melanoma (23). However, high expression of PYCR1 was not an independent risk factor for GC prognosis in our present research, this might be attributed to the limited sample in the tissue microarray. Thus, a large sample size might be required to allow for adjusting all those variables of interest to determine its role in prognostic prediction. We further demonstrated that PYCR1 regulated GC cell proliferation and apoptosis in cultured GC cells. GSEA performed by Ding et al. demonstrated that PYCR1 overexpression lead to marked enrichment of the gene signatures related to cell proliferation (21). We identified that PYCR1 protein expression was positively associated with the Ki-67 index, a cellular biomarker for proliferation, in tumor samples. These results in cells and clinical specimens strongly indicate that PYCR1 is critical for tumor growth. The induction of apoptosis by PYCR1 reduction might be attributed to the dysfunction of PYCR1 in maintaining the mitochondria membrane potential (8) and mitochondrial redox homeostasis (24). In addition, downregulation of PYCR1 leads to decreased intracellular proline synthesis, which might cause accumulation of the proapoptotic product P5C to elicit apoptotic events. However, a result from Liu’s group showed that a reduction in the PYCR1 level did not increase apoptosis in P493 lymphoma cells (25), suggesting the heterogeneity of different tumor cells. Therefore, the role of PYCR1 in regulating apoptosis is warranted to be further clarified.
Furthermore, our results based on GSEA and in-vitro assays confirmed that the PI3K/Akt pathway is an upstream regulator of PYCR1. The PI3K/Akt pathway has been recognized as an important signaling cascade in the pathogenesis of GC and as a possible therapeutic target for GC (26). Genetic alteration (mutation or amplification) of the genes encoding crucial components of this pathway is the main mechanism of PI3K/Akt pathway dysregulation in GC. Inactivation of 4E-BP1 resulting from PI3K/Akt activation can lead to release of the transcription factor eIF4E, which triggers the transcription of multiple oncogenes, including c-MYC, CCND1 and others (27). Previous studies have identified that PYCR1 is regulated by c-MYC (28), and that its amplification is the main mechanism of MYC dysregulation in GC (29). Collectively, these lines of evidence show that activation of the PI3K/Akt/c-MYC axis might explain PYCR1 upregulation in GC tissues, which is a promising target for precision treatment of GC. On the other hand, this signaling pathway is also critical in regulating cell fate decisions, including proliferation, apoptosis or autophagy (30). Thus, PYCR1 knockdown-mediated inactivation of target proteins (e.g., p-Akt) in the PI3K/Akt signaling pathway may contribute to the induction of apoptosis and inhibition of growth in the GC cell lines, especially in cells with PIK3CA mutation, while overexpression results in the opposite effect. However, this pattern was not observed in MCF-7 and MDA-MB-231 breast cancer cells in Ding’s study (21); thus, additional evidence in other cancer types is needed.
The tumor microenvironment is another determining factor for tumor biological behavior. Cancer cells are frequently exposed to a nutrient-limited microenvironment, and interest in understanding the metabolic plasticity that supports cancer cell survival and proliferation under such conditions in increasing. The glucose concentration in GC tissue is relatively lower than that in normal tissue (2,3), suggesting that metabolic stress is a usual state in GC. However, the mechanism by which GC cells overcome nutrient stress in the tumor microenvironment to continue growing in known. In this study, we discovered that PYCR1 was upregulated in response to glucose deprivation in a concentration- and time-dependent manner. Some research has identified that cancer cells rely on high levels of PYCR1 to confer resistance to oxidative stress for cell survival (9-11). Additionally, Pandhare et al. found that as the levels of glucose in the culture medium decreased, the increased intracellular proline level improved colorectal cell survival (31). Thus, PYCR1 upregulation under glucose starvation conditions could result in an increase in the intracellular proline level to help cancer cells overcome nutrient deprivation and keep growing. Collectively, these finding indicate that PYCR1 might function as a stress response oncogene enabling GC cells to resist metabolic stress in the tumor microenvironment.
There are some limitations in this study. First, we only showed phenomenological findings in vitro, so detailed mechanism behind the regulation of PYCR1 in gastric carcinogenesis is needed to be investigated in vivo and in vitro in our future studies. Second, as an enzyme located in inner membrane of mitochondria, we only studied its role in response to metabolic stress, but its associations with mitochondria metabolism was not investigated at present work.
Conclusions
In summary, the present work revealed for the first time that PYCR1 overexpression can promote GC progression by enhancing tumor proliferation, and responding to metabolic stress and that it could serve as a marker for prognosis of GC patients. We also provide evidence that the PI3K/Akt pathway can affect proline metabolism via PYCR1 in GC. Together with the results in our previous report (7), these findings highlight the potential therapeutic value of targeting PYCR1 as a modulator of proline metabolism in GC.
Supplementary
The article’s supplementary files as
Acknowledgement
We thank the whole staff in the Medical Research Center of Peking University Third Hospital for their excellent technical assistance on experiments. This work has been selected as a poster presentation (Su1054) in Digestive Disease Week (San Diego 2019) and published as a meeting abstract in Gastroenterology. 156(6):S496-S496.
Funding: This work was supported by National Natural Science Foundation of China [81672410].
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Sample collection was approved by the Ethics Committees of National Engineering Center for Biochip. Written informed consent was obtained from the patient for publication of this study and any accompanying images.
Footnotes
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-19-4402
Peer Review File: Available at http://dx.doi.org/10.21037/atm-19-4402
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-19-4402). The authors have no conflicts of interest to declare.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Xiao S, Zhou L. Gastric cancer: Metabolic and metabolomics perspectives (Review). Int J Oncol 2017;51:5-17. 10.3892/ijo.2017.4000 [DOI] [PubMed] [Google Scholar]
- 3.Hirayama A, Kami K, Sugimoto M, et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res 2009;69:4918-25. 10.1158/0008-5472.CAN-08-4806 [DOI] [PubMed] [Google Scholar]
- 4.Wu H, Xue R, Tang Z, et al. Metabolomic investigation of gastric cancer tissue using gas chromatography/mass spectrometry. Anal Bioanal Chem 2010;396:1385-95. 10.1007/s00216-009-3317-4 [DOI] [PubMed] [Google Scholar]
- 5.Chen Y, Zhang J, Guo L, et al. A characteristic biosignature for discrimination of gastric cancer from healthy population by high throughput GC-MS analysis. Oncotarget 2016;7:87496-510. 10.18632/oncotarget.11754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen JL, Tang HQ, Hu JD, et al. Metabolomics of gastric cancer metastasis detected by gas chromatography and mass spectrometry. World J Gastroenterol 2010;16:5874-80. 10.3748/wjg.v16.i46.5874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang W, Zhou L, Lin S, et al. Metabolic profiles of gastric cancer cell lines with different degrees of differentiation. International Journal of Clinical and Experimental Pathology 2018;11:869-75. [PMC free article] [PubMed] [Google Scholar]
- 8.Reversade B, Escande-Beillard N, Dimopoulou A, et al. Mutations in PYCR1 cause cutis laxa with progeroid features. Nat Genet 2009;41:1016-21. 10.1038/ng.413 [DOI] [PubMed] [Google Scholar]
- 9.Krishnan N, Dickman MB, Becker DF. Proline modulates the intracellular redox environment and protects mammalian cells against oxidative stress. Free Radic Biol Med 2008;44:671-81. 10.1016/j.freeradbiomed.2007.10.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yasuda T, Kaji Y, Agatsuma T, et al. DJ-1 cooperates with PYCR1 in cell protection against oxidative stress. Biochem Biophys Res Commun 2013;436:289-94. 10.1016/j.bbrc.2013.05.095 [DOI] [PubMed] [Google Scholar]
- 11.Kuo ML, Lee MB, Tang M, et al. PYCR1 and PYCR2 Interact and Collaborate with RRM2B to Protect Cells from Overt Oxidative Stress. Sci Rep 2016;6:18846. 10.1038/srep18846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szász AM, Lanczky A, Nagy A, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget 2016;7:49322-33. 10.18632/oncotarget.10337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Subramanian A, Kuehn H, Gould J, et al. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics 2007;23:3251-3. 10.1093/bioinformatics/btm369 [DOI] [PubMed] [Google Scholar]
- 16.Schulze A, Harris AL. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature 2012;491:364-73. 10.1038/nature11706 [DOI] [PubMed] [Google Scholar]
- 17.Elia I, Broekaert D, Christen S, et al. Proline metabolism supports metastasis formation and could be inhibited to selectively target metastasizing cancer cells. Nat Commun 2017;8:15267. 10.1038/ncomms15267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nilsson R, Jain M, Madhusudhan N, et al. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat Commun 2014;5:3128. 10.1038/ncomms4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Ingeniis J, Ratnikov B, Richardson AD, et al. Functional specialization in proline biosynthesis of melanoma. PLoS One 2012;7:e45190. 10.1371/journal.pone.0045190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vermeersch KA, Wang L, Mezencev R, et al. OVCAR-3 spheroid-derived cells display distinct metabolic profiles. PLoS One 2015;10:e0118262. 10.1371/journal.pone.0118262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding J, Kuo ML, Su L, et al. Human mitochondrial pyrroline-5-carboxylate reductase 1 promotes invasiveness and impacts survival in breast cancers. Carcinogenesis 2017;38:519-31. 10.1093/carcin/bgx022 [DOI] [PubMed] [Google Scholar]
- 22.Cai F, Miao Y, Liu C, et al. Pyrroline-5-carboxylate reductase 1 promotes proliferation and inhibits apoptosis in non-small cell lung cancer. Oncol Lett 2018;15:731-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye Y, Wu Y, Wang J. Pyrroline-5-carboxylate reductase 1 promotes cell proliferation via inhibiting apoptosis in human malignant melanoma. Cancer Manag Res 2018;10:6399-407. 10.2147/CMAR.S166711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hollinshead KER, Munford H, Eales KL, et al. Oncogenic IDH1 Mutations Promote Enhanced Proline Synthesis through PYCR1 to Support the Maintenance of Mitochondrial Redox Homeostasis. Cell Rep 2018;22:3107-14. 10.1016/j.celrep.2018.02.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu W, Hancock CN, Fischer JW, et al. Proline biosynthesis augments tumor cell growth and aerobic glycolysis: involvement of pyridine nucleotides. Sci Rep 2015;5:17206. 10.1038/srep17206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuoka T, Yashiro M. The Role of PI3K/Akt/mTOR Signaling in Gastric Carcinoma. Cancers (Basel) 2014;6:1441-63. 10.3390/cancers6031441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roux PP, Topisirovic I. Signaling Pathways Involved in the Regulation of mRNA Translation. Mol Cell Biol 2018;38:e00070-18. 10.1128/MCB.00070-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu W, Le A, Hancock C, et al. Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc Natl Acad Sci U S A 2012;109:8983-8. 10.1073/pnas.1203244109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calcagno DQ, Leal MF, Assumpcao PP, et al. MYC and gastric adenocarcinoma carcinogenesis. World J Gastroenterol 2008;14:5962-8. 10.3748/wjg.14.5962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martini M, De Santis MC, Braccini L, et al. PI3K/AKT signaling pathway and cancer: an updated review. Ann Med 2014;46:372-83. 10.3109/07853890.2014.912836 [DOI] [PubMed] [Google Scholar]
- 31.Pandhare J, Donald SP, Cooper SK, et al. Regulation and function of proline oxidase under nutrient stress. J Cell Biochem 2009;107:759-68. 10.1002/jcb.22174 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as