Abstract
Background
RACGAP1 has significant involvement in tumorigenesis of cancers, including liver cancer, stomach cancer, and colon cancer. However, the role and the exact mechanism of RACGAP1 in esophageal squamous cell carcinoma (ESCC) has not been explored.
Methods
QPCR and Western blots analysis was performed to analyze the expression of RACGAP1 in ESCC. MTT assays and colony formation assays were performed to explore the functional role of RACGAP1 in ESCC. Cell cycle analysis and immunofluorescence assays were used to investigate the function of RACGAP1 involvement in mitotic catastrophe. At last, we conducted the public datasets mining to explore the expression status and prognosis value of RACGAP1 as well as the correlation between RACGAP1 and E2F3 in various cancers.
Results
The high abnormal expression of RACGAP1 is observed in ESCC and associated with worse clinical outcomes of patients with ESCC. RACGAP1, a novel cell cycle associated gene regulated by E2F3, acts as an oncogenic driver in ESCC cell lines. Notably, for the first time, RACGAP1 depletion induced severe mitotic catastrophe, followed by massive cell death.
Conclusions
Our findings showed the essential role of RACGAP1 in ESCC cancer cell survival and the therapeutic potential of RACGAP1 as a molecular target for ESCC.
Keywords: Esophageal squamous cell carcinoma (ESCC), RACGAP1, E2F3, mitotic catastrophe
Introduction
Esophageal cancer (EC) is the seventh most common cancer, and the sixth cancer-related cause of death throughout the world (1). In contrast to adenocarcinoma as the primary histological subtype in Western countries, squamous cell carcinoma accounts for about 90% of EC and ranks as the sixth most common cancer and the fourth leading cause of cancer-related deaths in China (2). Although advances in surgical techniques and therapeutic options during the past decade, ESCC is still characterized by its poor outcome, with a 5-year survival rate of around 25% in China (3,4). Notably, esophageal adenocarcinoma (EAC) exhibits a durable clinical response to therapies targeting VEGF signaling or receptor tyrosine kinases such as EGFR, ERBB2/HER2, and MET (5-7). However, although the molecular feature of ESCC has been described in depth (8-10), effective targeted therapies for ESCC patients are minimal. Recently, early-phase immunotherapy trials showed response rates of approximately 20–30% for advanced ESCC patients, which are encouraging but inadequate (11,12). Therefore, it is urgent to identify ideal targets and establish novel therapeutic approaches based on an improved understanding of the malignant biological properties of ESCC.
RACGAP1 is an evolutionarily conserved GTPase activating protein (GAP) highly related to the Rho family of GTPases (13,14). RACGAP1 was initially identified in testis and male germ cells and has been demonstrated to be a key regulator in various malignancies, such as colorectal cancer, ovarian cancer, meningioma, uterine cancer, liver cancer as well as breast cancer. It is associated with aggressive clinical behavior (15-22). However, the clinical value of RACGAP1 expression, its functional role and regulation in ESCC oncogenesis, and, more important, the therapeutic target potential of RACGAP1 have not been shown previously.
In this study, we found that RACGAP1 serves as an independent predictive biomarker for poor prognosis in ESCC patients. We also proved that the oncogenic function of RACGAP1 in vitro by loss-of-function and gain-of-function experiments in ESCC cell lines. Notably, RACGAP1 was a cell cycle associated gene and was transcriptionally regulated by E2F3. By multiple approaches, we demonstrated for the first time that RACGAP1 depletion induced severe mitotic catastrophe followed by massive cell death, suggesting RACGAP1 might be a potential drug target for the development of novel anti-cancer therapies.
Methods
Sample collection
Fresh frozen tissues for QPCR and WB analysis were surgically resected from histopathologically and clinically diagnosed ESCC patients with detailed long-term follow-up clinical data. All samples were obtained from the tumor resource bank of Guangzhou Medical University Affiliated Cancer Hospital and Institute. All patients supplied informed consent. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by Ethics, the Committee of Affiliated Cancer Hospital (No. 20170930b0601231), and the Institute of Guangzhou Medical University.
Cell culture
The human ESCC cancer cell lines and standard cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 5% FBS at 37 °C and 5% CO2 in the cell incubator.
MTT assay
The ESCC cancer cells were seeded in a 96-well culture plate (Corning) at a density of 1,000 cells every well and incubated at 37 °C for 24 h. Every day after seeding, the cells in each well were stained with MTT (Sigma, M2003) for 4 h, the media was discarded, and 200 µL of dimethyl sulfoxide (DMSO) was added to each well and incubated and shake at room temperature for 15 mins. The absorbance was measured at 560 nm via a microplate reader. All experiments were repeated three times.
Colony assay
The cells were seeded in a 6-well culture plate (Corning) at a density of 200 cells/well followed by incubation at 37 °C. After 15 days of seeding, the colonies were washed with PBS, fixed with 4% paraformaldehyde for 10 min at room temperature, and stained with 1% crystal violet for 20 min. The colony was counted. All experiments were repeated three times.
Quantitative PCR
Total RNA was extracted using the TRIzol Reagent (Invitrogen) following the manufacturer’s user guide. Reverse transcription using a cDNA Synthesis Kit (Thermo, K1622). Quantitative real-time polymerase chain reaction using ChamQ SYBR Color qPCR Master Mix (Vazyme). ACTB was used as the internal control for comparison and normalization. The sequences of the amplification QPCR primers were as follows:
E2F1: 5'-GACGGCTTGAGGGGTTGAC-3' 5'-ATGCTACGAAGGTCCTGACAC-3'
E2F2: 5'- TGGGTAGGCAGGGGAATGTTT-3 5'- TGGTAAGTCACATAGGCCAGC-3'
E2F3: 5'- TCCCTAAACCCGCTTCCAAAG-3' 5'-AGTCTGCTGTAAGAGGTTGGC-3'
RACGAP1: 5'-GAAAGCAGAGACTGAGCGAAG-3' 5'-GTTGAATGCTGCCAGATGTGT-3'
ACTB: 5'- CGCGAGAAGATGACCCAGAT-3' 5'- GGGCATACCCCTCGTAGATG-3'
siRNA transfection
The specific siRNAs and control siRNA were obtained from RiboBio Co., Ltd. Cancer cells were counted and seeded in plates. After 24 h, the cells were 40% confluent and were transfected with siRNAs using RNAiMAX transfection reagents (Thermo Fisher) according to the manufacturer’s directions. After 48 h, cells with treatment were harvested for further experiments.
Western blotting
Protein lysates were electrophoresed by SDS/PAGE (10% gel) followed by iBlot transfer to PVDF (Thermo Fisher). Membranes were blocked in 5% skim milk powder in TBS with 0.1% Tween-20, and then incubated overnight with a primary antibody, The primary antibodies were used: anti-RACGAP1 (PTG, 13739-1-AP, RRID:AB_2176276), anti-ACTB (PTG, 20536-1-AP, RRID:AB_10700003), anti-GAPDH (PTG, 10494-1-AP, RRID:AB_2263076), anti-mouse and anti-rabbit peroxidase conjugated secondary antibodies (R&D Systems, HAF007, RRID:AB_357234, and HAF008, RRID:AB_357235).
Cell cycle analysis
ESCC were transfected with a control siRNA and RACGAP1 siRNAs. After 48 h, cells were detached using trypsin, washed twice with 1X PBS, and fixed overnight in cold 70% ethanol at −20 °C. Then cells were washed twice in cold PBS and stained with PI (propidium iodide, Sigma-Aldrich). The DNA content in the cells was analyzed by flow cytometry using a Cytomics FC 500 (Beckman Coulter, Fullerton) in duplicate and FlowJo 7.6 software (Beckman). All experiments were repeated three times.
Dual-luciferase reporter assay
Luciferase activity was measured using a Dual-Luciferase Reporter Assay System (Promega). Cells at 30% confluence in 24-well dishes were transfected with reporter plasmid using Lipofectamine 3000 (Invitrogen). After 24 h, siRNA was transfected with Lipofectamine RNAiMAX. Firefly and Renilla luciferase activities were measured 48 h after transfection following the manufacturer’s instructions. All reporter gene assays were performed in duplicate and repeated three times.
Data acquisition
The full set of data of differentially expressed RACGAP1 mRNA in ES tumor tissue and normal tissue was downloaded from the NCBI Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA), the Series accession numbers: GSE23400, GSE20347, GSE45670 and TCGA.
Statistical analysis
All data are expressed as mean ± SEM. Statistical analyses were performed using GraphPad Prism 8 (GraphPad Software, Inc., La Jolla, CA, USA). Differences between two groups or multiple group differences were analyzed by Student’s t-test or one-way ANOVA, respectively. P value <0.05 was considered statistically significant.
Results
High RACGAP1 expression in ESCC
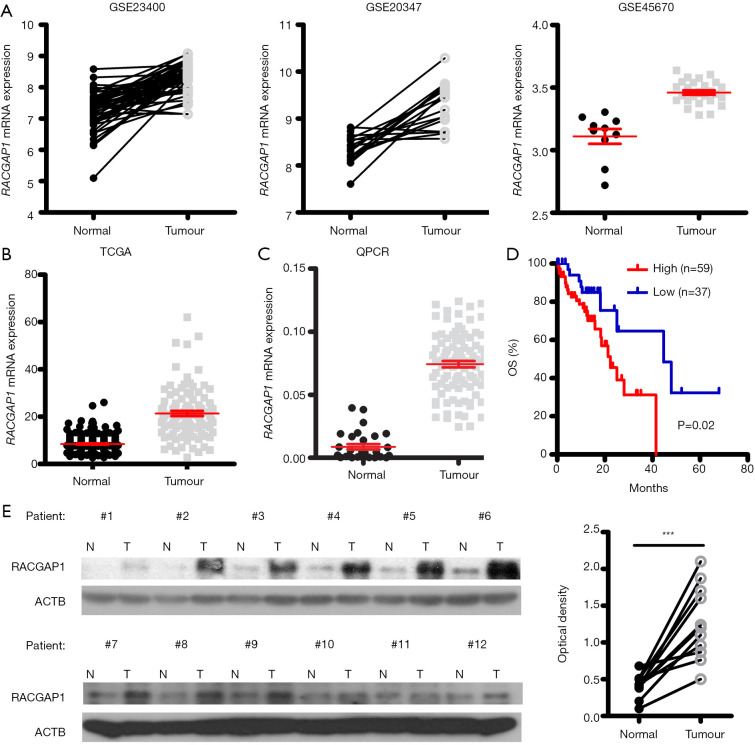
In order to identify whether RACGAP1 gene is vital for ESCC tumor growth, we retrieved 3 ESCC microarray datasets [GSE23400 (23), GSE20347 (24), GSE45670 (25)] and found that RACGAP1 displayed overexpression in the tumors when compared with the adjacent healthy tissues (Figure 1A). Also, RNA-seq data from TCGA and GTEx showed that RACGAP1 is significantly upregulated in ESCC tissues compared to esophageal epithelial tissues (Figure 1B). Consistent with the publicly available datasets, RACGAP1 mRNA level in our ESCC cohort was dramatically higher than that of regular counterparts (Figure 1C). Importantly, the Kaplan-Meier method was used to analyze the correlation between RACGAP1 expression and overall survival (OS) of patients and found that higher RACGAP1 level was positively associated with more inferior OS time (Figure 1D). Moreover, the WB result showed that the protein level of RACGAP1 is much higher in ESCC tissues than matched adjacent healthy tissues (Figure 1E).
Figure 1.
RACGAP1 is highly upregulated in ESCC. (A,B) mRNA expression of RACGAP1 is significantly upregulated in ESCC tissues compared to healthy tissues, indicated by three microarray datasets from GEO (A) as well as RNA-seq data from TCGA data (B). (C) QPCR assay was conducted to validate the upregulation of RACGAP1 mRNA level in ESCC tissues (n=96) compared to adjacent healthy tissues (n=20). (D) The Kaplan-Meier curves showed that the overall survival (OS) time was significantly shorter in the high RACGAP1 expression group. The P value was determined by the log-rank test. (E) Left panel: Western blotting assay shown that the RACGAP1 protein level in ESCC tissues is higher than in matched adjacent healthy tissues. Right panel: statistical results of the grayscale analysis. The P value was determined with a paired t-test. ***, P<0.001.
Oncogenic role of RACGAP1 in ESCC
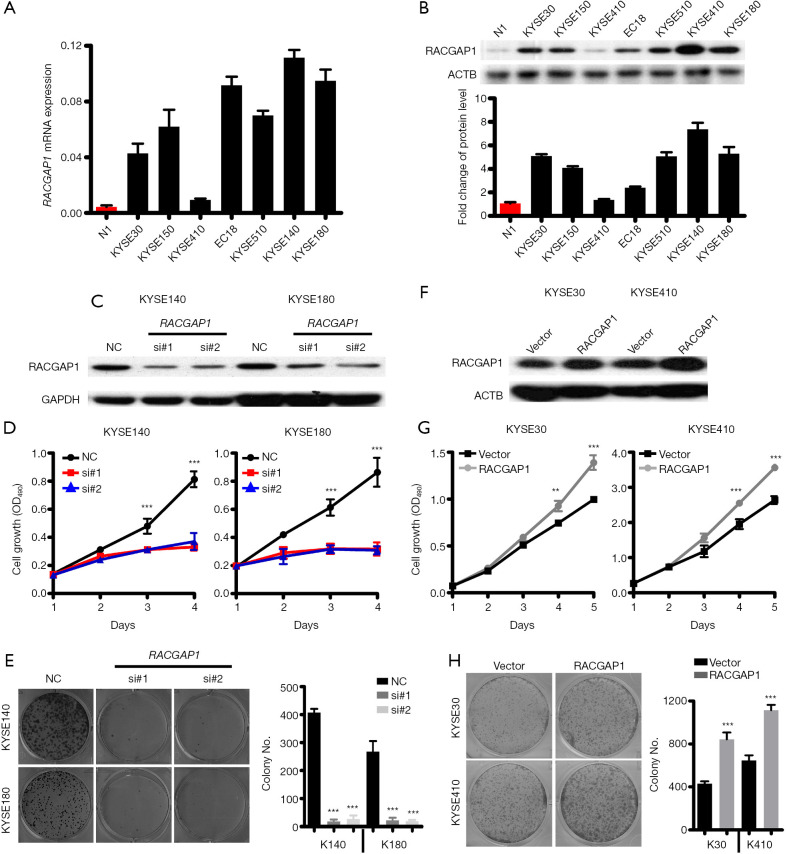
Further, we investigated RACGAP1 expression levels in diverse human ESCC cell lines, including KYSE30, KYSE150, KYSE410, EC18, KYSE510, KYSE140, and KYSE180 cells as well as a normal cell line N1 which was originated from normal esophageal squamous mucosa. Expression profiles via QPCR and WB revealed that RACGAP1 expression was hardly detectable in the standard esophageal epithelial cell lines but was much higher in almost all (6/7) ESCC cell lines (Figure 2A,B). Based on RACGAP1 expression profiles, we selected KYSE140 and KYSE180 cell lines, which expressed higher RACGAP1 for further loss-of-function experiments. Both MTT assay and colony formation assay were performed after RACGAP1 knockdown by siRNAs (Figure 2C) and demonstrated that RACGAP1 knockdown tremendously reduced the growth and clonogenicity of ESCC cancer cells (Figure 2D,E), indicating that RACGAP1 was crucial for the survival of the ESCC cells that are addicted to its expression. Next, KYSE30 and KYSE410, which expressed lower RACGAP1, were chosen to conduct gain-of-function experiments. As expected, RACGAP1 overexpression (Figure 2F) markedly promoted cell proliferation and increased the colony number of ESCC cells (Figure 2G,H). Collectively, our findings showed that RACGAP1 plays a vital role in maintaining the malignant phenotypes of ESCC Cells.
Figure 2.
RACGAP1 as an oncogenic driver in ESCC. (A,B) QPCR assay (A) and Western blotting assay (B) shown that the RACGAP1 mRNA level is highly upregulated in 6/7 ESCC cell lines compared with healthy esophageal epithelial cells. (C) Western blotting assay shown RACGAP1 was knockdown in KYSE140 and KYSE180 by two specific siRNAs. (D) MTT assay showed that RACGAP1 depletion strongly inhibits the cell growth of KYSE140 and KYSE180. (E) Colony formation assay showed that RACGAP1 depletion suppressed the proliferation of KYSE140 and KYSE180. (F) The western blotting assay showed stable overexpression of RACGAP1 in KYSE30 and KYSE410 cell lines. (G) MTT assay showed that RACGAP1 overexpression enhances the cell growth of KYSE30 and KYSE410. (H) Colony formation assay showed that RACGAP1 overexpression promoted the proliferation of KYSE30 and KYSE410. The data are presented as means ± SEMs. The P values were determined with Student’s t-test (two groups) or one-way ANOVA (three groups). **, P<0.01; ***, P<0.001. ESCC, esophageal squamous cell carcinoma.
RACGAP1 is a cell cycle gene and its depletion induced mitotic catastrophe
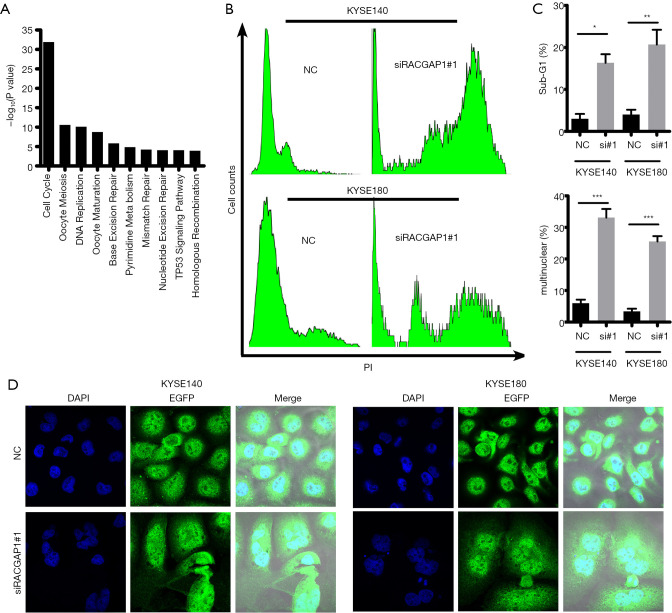
Co-expression network analysis is a method that constructs networks of genes which were co-activated or repressed across a group of samples and subsequently investigated this network (26). It is a powerful tool to infer gene function as well as the associations between gene and disease from genome-wide gene expression. Thus, we investigated the functional status of RACGAP1 from a systematic perspective via analyzing the RNA sequencing (RNA-seq) data from the TCGA ESCC cohort. Indeed, 368 genes which were positively correlated with RACGAP1 (r≥0.5, P<0.05) were identified and GSEA analysis (27) shown that these genes were mainly clustered into DNA repair pathways and cell cycle modules (Figure 3A), indicating that RACGAP1 involves in cell cycle dysregulation of ESCC cancer cells. Therefore, we conducted cell cycle analysis after RACGAP1 repression by siRNAs and found that there is a dramatic alteration of cell cycle profiles in RACGAP1 knockdown cells compared with control cells (Figure 3B). In detail, a higher proportion of sub-G1 phase and multiple nuclear cells indicated by abnormal DNA content was observed in RACGAP1 repression cells than control cells (Figure 3C). Immunostaining in KYSE140 and KYSE180 cells showed that RACGAP1 depletion resulted in about 60% of abnormal mitosis, as evidenced by forming giant cells with binucleation and multinucleation (Figure 3D), suggesting that cells with RACGAP1 knockdown would eventually undergo mitotic catastrophe. Thus, our data suggest that RACGAP1 is a cell cycle associated gene, whose loss causes severe mitotic mistakes and triggers mitotic catastrophe in cancer cells, leading to the ensuing extensive cell death.
Figure 3.
RACGAP1 depletion induced mitotic catastrophe. (A) KEGG pathway enrichment analysis shown RACGAP1 co-expression genes involve in cell cycle and cell cycle-associated pathways. (B) Cell cycle analysis showed that RACGAP1 depletion results in Severe cell cycle abnormality. (C) Statistical analysis of the percentage of tumor cells in sub-G1 and tumor cells with multinuclear in (B). The data are presented as means ± SEMs. The P values were determined with Student’s t-test (two groups) or one-way ANOVA (three groups). *, P<0.05; **, P<0.01; ***, P<0.001. (D) IF analysis of RACGAP1-knockdown cells and control cells (magnification, 180×). Green: GFP, Blue: DAPI.
RACGAP1 was a transcription target of E2F3
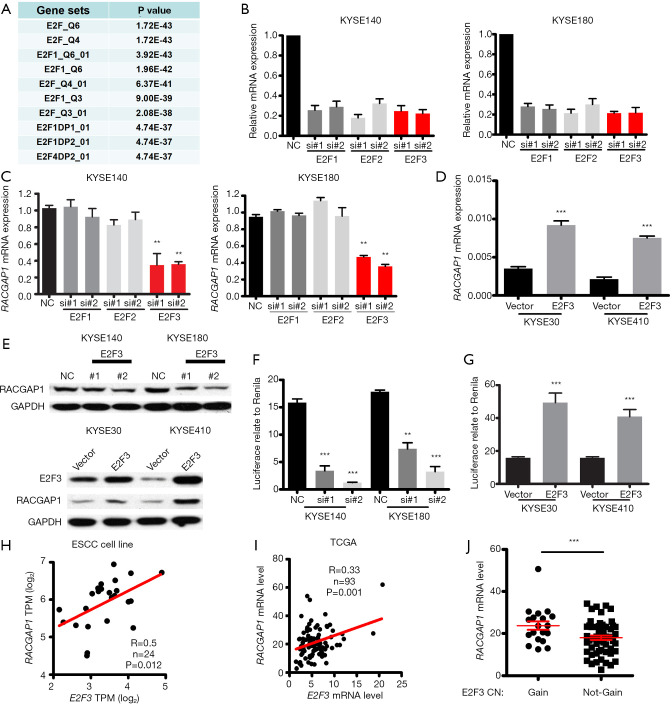
Cell cycle genes were coordinated by different transcription factors during cell cycle progression (28). Thus far, the transcription factor which controls the transcription of RACGAP1 is still to be named. Gene co-expression networks can also be used for the identification of regulatory genes. Thus, GSEA analysis was performed to explore which transcription factor regulated the expression of RACGAP1 and its co-expressed genes and revealed the most significant enrichment of transcription factor is the E2F family proteins, including E2F1, E2F2, and E2F3 (Figure 4A). Thus, we supposed that RACGAP1 is one of the targets of E2F family proteins during the cell cycle progression. Then, we use siRNAs to knockdown these three genes (Figure 4B), respectively, and found that only E2F3 inhibition could downregulate the mRNA expression of RACGAP1 (Figure 4C). Besides, consistent with the result of the loss-of-function experiment, exogenous expression of E2F3 upregulated the mRNA expression of RACGAP1 (Figure 4D). In parallel, immunoblotting assays were perform to confirm the regulation of RACGAP1 mediated by E2F3 (Figure 4E).
Figure 4.
RACGAP1 is regulated by E2F3. (A) Transcription factor analysis showed that RACGAP1 co-expression genes are regulated by E2F family members. (B) QPCR assay confirmed the efficiency of siRNAs. (C) QPCR assay showed that Only E2F3 knockdown could repress the mRNA expression of RACGAP1 in KYSE140 and KYSE180. (D) QPCR assay showed that E2F3 overexpression induced the mRNA expression of RACGAP1. (E) WB assay confirmed that RACGAP1 is regulated by E2F3. (F,G) Luciferase reporter assay showed that E2F3 knockdown decreases the promoter activity of RACGAP1 (F), and E2F3 overexpression increase the promoter activity of RACGAP1 (G). (H,I) E2F3 mRNA level is positively correlated with the RACGAP1 mRNA level in ESCC cell line RNA-seq data (H) and TCGA ESCC data (I). (J) RACGAP1 mRNA level is significantly higher in E2F3 copy number gain ESCC samples than E2F3 copy number not-gain ESCC samples. The data are presented as means ± SEMs. The P values were determined with Student’s t-test (two groups) or one-way ANOVA (three groups). **, P<0.01; ***, P<0.001. ESCC, esophageal squamous cell carcinoma.
Moreover, illustrated by luciferase reporter assay, E2F3 knockdown actively suppresses the promoter activity of RACGAP1 (Figure 4F), and vice versa, E2F3 overexpression enhanced the promoter activity of RACGAP1 (Figure 4G). Finally, correlation analysis demonstrated that the mRNA level of E2F3 is positively correlated with that of RACGAP1 significantly in both our ESCC cohort (Figure 4H) and TCGA cohort (Figure 4I). Notably, in the TCGA cohort, samples with E2F3 copy number gain or amplification displayed a higher mRNA level of RACGAP1 (Figure 4J). Taken together, RACGAP1 is a bona fide transcription target of E2F3.
Pan-cancer analysis of RACGAP1
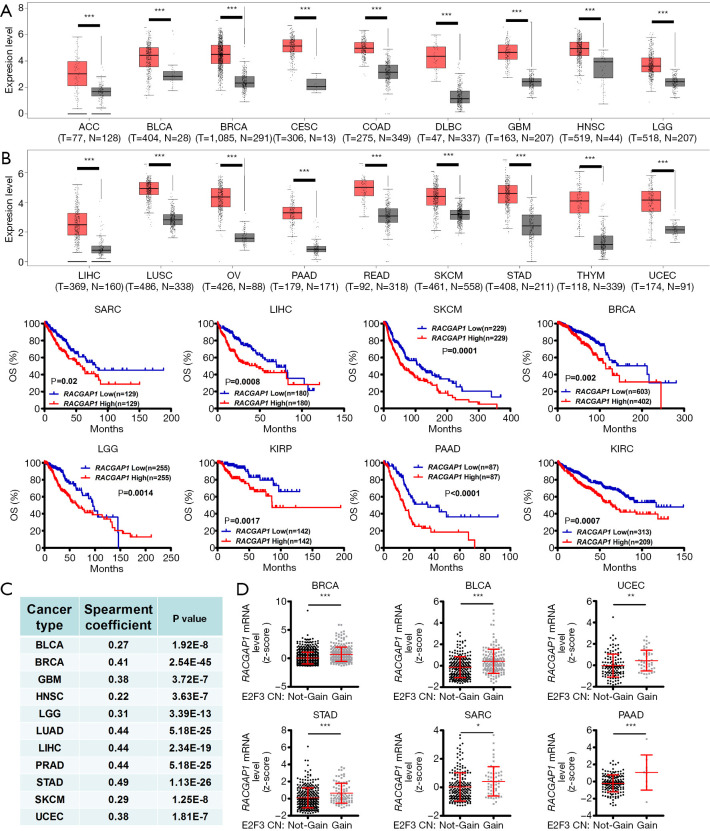
Importantly, more than ESCC, the mRNA expression of RACGAP1 was significantly increased in various human cancer types compared with the healthy tissues, indicated by the GEPIA database (29) (Figure 5A). Strikingly, in public datasets of the cBioPortal (http://cbioportal.org/), high RACGAP1 expression was statistically significantly associated with more reduced survival of cancer patients (Figure 5B). The analysis of data from TCGA was conducted and displayed the high positive correlation between the mRNA level of E2F3 and the mRNA level of RACGAP1 in different cancers to explore whether the association between E2F3 and RACGAP1 is ESCC specific or universal in pan-cancers (Figure 5C). Notably, the mRNA level of RACGAP1 is significantly higher in tumor samples with E2F3 copy number gain than that without E2F3 copy number gain (Figure 5D). Thus, the high RACGAP1 level in human cancers significantly correlated with worse outcomes.
Figure 5.
Pan-cancer analysis of RACGAP1. (A) The mRNA Level of RACGAP1 is significantly upregulated in 18 types of human cancer. (B) High expression of RACGAP1 was related to worse overall survival (OS) in 8 types of cancer. (C) RACGAP1 mRNA level is positively correlated with the E2F3 mRNA level in 11 types of cancer. (D) RACGAP1 mRNA level is significantly higher in E2F3 copy number gain tumor samples than E2F3 copy number not-gain tumor samples in 6 types of cancer. Error bars represent means ± SD. *, P<0.05; **, P<0.01; ***, P<0.001, the P values were calculated by Student’s t-test or long-rank test. ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; COAD, colon adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B-cell lymphoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; LGG, low grade glioma; LIHC, liver hepatocellular carcinoma; LUSC, lung squamous cell carcinoma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma.
Discussion
Because the use of targeted therapy is minimal, the prognosis of ESCC patients remains poor. However, this could be improved by understanding the disease mechanism, which might also result in identifying molecular targets for developing novel therapies. Although RACGAP1 as an oncogenic driver has recently been revealed in various kinds of cancer, its regulation factor, and the potential as a drug target in ESCC needs to be clarified. In this study, we determined that RACGAP1 was expressed at significantly higher levels in ESCC cells compared to adjacent normal mucosae. Second, the high expression of RACGAP1 was significantly correlated with dismal OS in these patients, indicating that RACGAP1 could serve as a prognostic biomarker and might even play an essential role in disease progression. Finally, a series of in vitro experiments verified the tumor-promoting effect of RACGAP1 in ESCC.
Uncontrolled cancer cell division after oncogenic transformation requires continuous high-efficiency coordination of various cell cycle-associated genes. The analysis of comprehensive mutational catalogs using high-throughput sequencing technologies has underscored cell cycle pathway dysregulation as a common characteristic in ESCC (8-10). Hence, active research efforts are exploring cell cycle-related genes as biomarkers and therapeutic targets. In the present work, we found RACGAP1 as a novel cell cycle associated gene, and it is under the control of E2F3, an E2F family protein. Indeed, we proved that knockdown of E2F3 reduced RACGAP1 expression and while forced overexpression of E2F3 was able to induce RACGAP1 expression. Additionally, via luciferase reporter assay, RACGAP1 promoter activity is associated with the expression of E2F3. These findings prove that RACGAP1 is a direct downstream transcriptional target of E2F3.
Since mitotic catastrophe represents a mode of cell death, it acts as a tumor-suppressive mechanism, which might be used for cancer therapy (30,31). Three reasons are suggesting that the stimulation of mitotic catastrophe might be an attractive approach to the development of new therapies. First, since cancer cells are typical genomic instability represents by aneuploidy, this gives them more sensitivity to mitotic aberrations. Second, different from cell death induced by multiple chemotherapeutic agents at relatively high doses, mitotic catastrophe is activated in response to significantly lower doses of cytotoxic agents (32). Third, escape from mitotic catastrophe is a crucial mechanism of chemo-resistance in various cancers. It is reported that RACGAP1 participates in cytokinesis as a part of the centralspindlin complex. Surprisingly, RACGAP1 knockdown induced massive apoptosis in ESCC cancer lines due to severe mitotic catastrophe. Therefore, RACGAP1 activation is essential for the survival and growth of ESCC cancer cells and is also their “Achilles heel.”
Our work altogether represents information about the clinical significance and biological function of RACGAP1 in ESCC and, more important, provides new insight into the regulation and drug target probability of RACGAP1 in ESCC. Our results showed that a clinical evaluation of RACGAP1 expression in primary tumors could afford the prognostic information for patients with ESCC and that targeting RACGAP1 might be a novel treatment approach.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work. The authors have ensured that questions related to the accuracy or integrity of any part of the work have been appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by Ethics, the Committee of Affiliated Cancer Hospital (No. 20170930b0601231), and the Institute of Guangzhou Medical University. Informed consent was provided by all participants.
Footnotes
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-2901
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-2901). The authors have no conflicts of interest to declare.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Shang QX, Yang YS, Hu WP, et al. Prognostic significance and role of thoracic lymph node metastasis based on Chinese expert consensus in esophageal cancer. Ann Transl Med 2019;7:381. 10.21037/atm.2019.07.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeng H, Chen W, Zheng R, et al. Changing cancer survival in China during 2003-15: a pooled analysis of 17 population-based cancer registries. Lancet Glob Health 2018;6:e555-67. 10.1016/S2214-109X(18)30127-X [DOI] [PubMed] [Google Scholar]
- 4.Guo LW, Huang HY, Shi JF, et al. Medical expenditure for esophageal cancer in China: a 10-year multicenter retrospective survey (2002-2011). Chin J Cancer 2017;36:73. 10.1186/s40880-017-0242-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilke H, Muro K, Van Cutsem E, et al. Ramucirumab plus paclitaxel versus placebo plus paclitaxel in patients with previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (RAINBOW): a double-blind, randomised phase 3 trial. Lancet Oncol 2014;15:1224-35. 10.1016/S1470-2045(14)70420-6 [DOI] [PubMed] [Google Scholar]
- 6.Fuchs CS, Tomasek J, Yong CJ, et al. Ramucirumab monotherapy for previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (REGARD): an international, randomised, multicentre, placebo-controlled, phase 3 trial. Lancet 2014;383:31-9. 10.1016/S0140-6736(13)61719-5 [DOI] [PubMed] [Google Scholar]
- 7.Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 2010;376:687. 10.1016/S0140-6736(10)61121-X [DOI] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research Network. Analysis Working Group : Asan University; BC Cancer Agency; Integrated genomic characterization of oesophageal carcinoma. Nature 2017;541:169-75. 10.1038/nature20805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lau KW, Zeng H, Liang H, et al. Bioinformatics-based identification of differentiated expressed microRNA in esophageal squamous cell carcinoma. Transl Cancer Res 2018;7:1366-75. 10.21037/tcr.2018.10.15 [DOI] [Google Scholar]
- 10.Sawada G, Niida A, Uchi R, et al. Genomic Landscape of Esophageal Squamous Cell Carcinoma in a Japanese Population. Gastroenterology 2016;150:1171-82. 10.1053/j.gastro.2016.01.035 [DOI] [PubMed] [Google Scholar]
- 11.Shah MA, Kojima T, Hochhauser D, et al. Efficacy and Safety of Pembrolizumab for Heavily Pretreated Patients With Advanced, Metastatic Adenocarcinoma or Squamous Cell Carcinoma of the Esophagus: The Phase 2 KEYNOTE-180 Study. JAMA Oncol 2019;5:546-50. 10.1001/jamaoncol.2018.5441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doi T, Piha-Paul SA, Jalal SI, et al. Safety and Antitumor Activity of the Anti-Programmed Death-1 Antibody Pembrolizumab in Patients With Advanced Esophageal Carcinoma. J Clin Oncol 2018;36:61-7. 10.1200/JCO.2017.74.9846 [DOI] [PubMed] [Google Scholar]
- 13.Hirose K, Kawashima T, Iwamoto I, et al. MgcRacGAP is involved in cytokinesis through associating with mitotic spindle and midbody. J Biol Chem 2001;276:5821-8. 10.1074/jbc.M007252200 [DOI] [PubMed] [Google Scholar]
- 14.Zhao WM, Fang G. MgcRacGAP controls the assembly of the contractile ring and the initiation of cytokinesis. Proc Natl Acad Sci U S A 2005;102:13158-63. 10.1073/pnas.0504145102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mi S, Lin M, Brouwer-Visser J, et al. RNA-seq Identification of RACGAP1 as a Metastatic Driver in Uterine Carcinosarcoma. Clin Cancer Res 2016;22:4676-86. 10.1158/1078-0432.CCR-15-2116 [DOI] [PubMed] [Google Scholar]
- 16.Ke HL, Ke RH, Li ST, et al. Expression of RACGAP1 in high grade meningiomas: a potential role in cancer progression. J Neurooncol 2013;113:327-32. 10.1007/s11060-013-1121-7 [DOI] [PubMed] [Google Scholar]
- 17.Imaoka H, Toiyama Y, Saigusa S, et al. RacGAP1 expression, increasing tumor malignant potential, as a predictive biomarker for lymph node metastasis and poor prognosis in colorectal cancer. Carcinogenesis 2015;36:346-54. 10.1093/carcin/bgu327 [DOI] [PubMed] [Google Scholar]
- 18.Wang SM, Ooi LL, Hui KM. Upregulation of Rac GTPase-activating protein 1 is significantly associated with the early recurrence of human hepatocellular carcinoma. Clin Cancer Res 2011;17:6040-51. 10.1158/1078-0432.CCR-11-0557 [DOI] [PubMed] [Google Scholar]
- 19.Wang C, Wang W, Liu Y, et al. Rac GTPase activating protein 1 promotes oncogenic progression of epithelial ovarian cancer. Cancer Sci 2018;109:84-93. 10.1111/cas.13434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawson CD, Der CJ. Filling GAPs in our knowledge: ARHGAP11A and RACGAP1 act as oncogenes in basal-like breast cancers. Small GTPases 2018;9:290-6. 10.1080/21541248.2016.1220350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pliarchopoulou K, Kalogeras KT, Kronenwett R, et al. Prognostic significance of RACGAP1 mRNA expression in high-risk early breast cancer: a study in primary tumors of breast cancer patients participating in a randomized Hellenic Cooperative Oncology Group trial. Cancer Chemother Pharmacol 2013;71:245-55. 10.1007/s00280-012-2002-z [DOI] [PubMed] [Google Scholar]
- 22.Saigusa S, Tanaka K, Mohri Y, et al. Clinical significance of RacGAP1 expression at the invasive front of gastric cancer. Gastric Cancer 2015;18:84-92. 10.1007/s10120-014-0355-1 [DOI] [PubMed] [Google Scholar]
- 23.Hu N, Clifford RJ, Yang HH, et al. Genome wide analysis of DNA copy number neutral loss of heterozygosity (CNNLOH) and its relation to gene expression in esophageal squamous cell carcinoma. BMC Genomics 2010;11:576. 10.1186/1471-2164-11-576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su H, Hu N, Yang HH, et al. Global gene expression profiling and validation in esophageal squamous cell carcinoma and its association with clinical phenotypes. Clin Cancer Res 2011;17:2955-66. 10.1158/1078-0432.CCR-10-2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wen J, Yang H, Liu MZ, et al. Gene expression analysis of pretreatment biopsies predicts the pathological response of esophageal squamous cell carcinomas to neo-chemoradiotherapy. Ann Oncol 2014;25:1769-74. 10.1093/annonc/mdu201 [DOI] [PubMed] [Google Scholar]
- 26.Galán-Vásquez E, Perez-Rueda E. Identification of Modules With Similar Gene Regulation and Metabolic Functions Based on Co-expression Data. Front Mol Biosci 2019;6:139. 10.3389/fmolb.2019.00139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sadasivam S, DeCaprio JA. The DREAM complex: master coordinator of cell cycle-dependent gene expression. Nat Rev Cancer 2013;13:585-95. 10.1038/nrc3556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Denisenko TV, Sorokina IV, Gogvadze V, et al. Mitotic catastrophe and cancer drug resistance: A link that must to be broken. Drug Resist Updat 2016;24:1-12. 10.1016/j.drup.2015.11.002 [DOI] [PubMed] [Google Scholar]
- 31.Vitale I, Galluzzi L, Castedo M, et al. Mitotic catastrophe: a mechanism for avoiding genomic instability. Nat Rev Mol Cell Biol 2011;12:385-92. 10.1038/nrm3115 [DOI] [PubMed] [Google Scholar]
- 32.Huertas D, Soler M, Moreto J, et al. Antitumor activity of a small-molecule inhibitor of the histone kinase Haspin. Oncogene 2012;31:1408-18. 10.1038/onc.2011.335 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as