Abstract
A large body of scientific evidence corroborated by clinical and animal model experiments indicates that tumor-associated macrophages (TAMs) play a crucial role in tumor development and progression. TAMs are a key immune cell type present in tumor microenvironment (TME) and associated with poor prognosis, drug resistance, enhanced angiogenesis and metastasis in cancer. TAMs are a phenotypically diverse population of myeloid cells which display tremendous plasticity and dynamic metabolic nature. A complete interpretation of pro-tumoral and anti-tumoral metabolic switch in TAMs is essential to understand immune evasion mechanisms in cancer. Recent studies have also implicated epigenetic mechanisms as significantly regulators of TAM functions. In this review we provide an overview of metabolic circuitry in TAMs, its impact on immune effector cells and interventions aimed at rewiring the metabolic circuits in TAMs. Mechanisms responsible for TAM polarization in cancer are also discussed.
Keywords: Tumor-associated macrophages (TAMs), metabolic reprogramming, immunotherapy, immune-metabolic crosstalk, tumor microenvironment (TME)
Introduction
Development and progression of tumors is characterized by selective survival of immune resistant tumor variants in an immunosuppressive tumor microenvironment (TME) (1). Tumor-associated macrophages (TAMs) are a major component of immune cell types present in TME. TAMs have a key role in inducing evolution of TME and supporting tumor growth (2). A fairly large body of scientific evidence has implicated TAMs to have a major role in tumor progression, angiogenesis, metastasis, evasion of immune response and unfavorable response to therapy. TAMs like other immune cells respond to environmental signals by acquiring a wide spectrum of phenotypic and functional states. Emerging evidence indicates that ‘immunosuppressive and protumoral’ behavior of TAMs results from a rewired metabolic program which affects disease progression and outcome in cancer.
TAMs account for a majority of myeloid cell population in almost all solid and hematologic malignancies and proposed as potential diagnostic and prognostic biomarkers for cancer (3). Studies from various human cancers have indicated an important role for TAMs in disease progression (Table 1). TAMs belong to the class of immune myeloid cells along with myeloid-derived suppressor cells (MDSCs) and tumor-associated neutrophils (TANs) (36). Macrophages exhibit polarization into distinct phenotypic and functional subsets known as M1 and M2 macrophages in response to various activation stimuli. The dichotomy in M1 and M2 macrophages is also evident at the metabolic level; M1 macrophages exhibit preponderance of glycolysis, fatty acid synthesis (FAS) and pentose phosphate pathway (PPP) pathways while M2 macrophages largely use oxidative phosphorylation (OXPHOS) (37). A comparison of immunologic and metabolic features of M1 and M2 macrophages is presented as Table 2. This classification of M1 and M2 macrophages as originally proposed by Mills (42) has served as a useful platform for studying macrophage polarization, however, this classification has been based on in vitro conditions and does not take into account the tissue microenvironment. Macrophages have been shown to exist as a highly diverse population; transcriptome based network analysis of macrophage activation suggests a activation stimulus specific activation gene expression in macrophages which may mean that the classification into M1 and M2 macrophages may not suffice to account for the heterogeneity and plasticity in macrophage biology (43,44).
Table 1. TAM phenotype and signaling pathway in various solid tumors and blood cancers.
| Cancer type (solid and blood malignancies) | Macrophage phenotype | Signaling pathway | References |
|---|---|---|---|
| Breast cancer | IL-4/IL-13 stimulated macrophages; M2a derived CCL18 and VEGF promote metastasis in breast cancer; M1-like macrophages helps in the infiltration of CTLs, CD8 T cells and undergoes the inflammatory cytokine cascade to eliminate the tumor; CD68+ macrophages; COX2+; CD163+ | Signaling pathway involved—ROCK signaling; COX2+ TAMs activate PI3K-AKT pathway in cancer cells | (4-7) |
| Prostate cancer | CD206; TNF-α and IL-1β | TAMs promote prostate cancer progression through the activation of the CCL2–CCR2 axis, followed by the activation of the CCL17/CCL22–CCR4 axis; prostate cancer derived CCN3 mediates polarization to protumoral M2-like macrophages; VEGF mediated angiogenesis | (8-10) |
| Lung cancer (NSCLC) | CD204+ M2 and pan-CD68+ TAMs found in both stromal and intratumoral component | M2-like macrophages promote migration and angiogenesis in NSCLC via associated factors like MMP and VEGF | (11,12) |
| Ovarian cancer | M2-like macrophages with phenotype IL-10, CD163, CD204; CD206(MR) overexpressing CCL18 and CCL22 | M2-like macrophages enhance cancer cell proliferation via MMP9/HB-EGF axis. This EGF leads to αMβ2 integrin upregulation which further increases EGFR, ICAM-1 expression and then activates the VEGF/VEGFR pathway promoting angiogenesis and thus metastasis; the main TF involved is IRF4 | (13-19) |
| Glioblastoma | M2 marker arginase 1 was upregulated (10-fold) and also IL-1β upregulated (5-fold) | ERK1/2 signaling regulates macrophage recruitment in glioblastoma | (20-22) |
| Blood cancer | CD163+ M2 TAMs (mouse) and CD163+CD206+ M2 TAMs in multiple myeloma and classical Hodgkin Lymphoma; CLL: Markers expressed were: CD11b, CD163, CD206, HLA-DR, HGF, IDO—all resembling M2 phenotype. Burger et al. first found nurse like cells helping CLL microenvironment to be tumor supporting and later Tsukada et al. demonstrated that these were component of the TME similar to the TAMs in solid tumors; AML: splenic leukemia-associated macrophages turned out to have M2 characteristics while bone marrow leukemia-associated macrophages turned out M1 characterized | ALL: proliferation of T-ALL cell lines showed significant increase after being co-cultured with M2 macrophage subset due to secretion of TNFα, growth related oncogeneα, C5a, IL-6, CCL1 | (23-31) |
| Pancreatic cancer (PC) | REDD–; CD163+ M2-polarized macrophages were significantly more abundant in primary PDA samples | M2 TAMs found in the TME is associated to YAP1 signaling which correlates with tumorigenesis in many cancer types. YAP1/HIF-α pathway has been recently found responsible for promoting cancer stem cells in PC. REDD1 deficient TAMs outplay normal cells and form more vascular junctions favoring angiogenesis; notch signaling plays a role in macrophage polarization within the PDA (pancreatic ductal adenocarcinoma) microenvironment; pancreatic cancer-educated macrophages induced the upregulation of CD59 in pancreatic cancer cells via the IL-6R/STAT3 pathway; YAP1/HIF-α pathway | (32-35) |
TF, transcription factor.
Table 2. Major differences between M1 and M2 macrophages with respect to antigen presentation, activation stimuli, phenotypes expressed in various disease conditions, signaling pathway and molecules involved, biomarkers, metabolism, immune function and genes present.
| Functional aspect | M1 macrophage | M2 macrophage |
|---|---|---|
| Antigen presentation | Yes | No |
| Activation stimuli | IFN-γ, TNF-α, and lipopolysaccharide (classical activation) | IL-4, IL-10, IL-13, TGF-β, glucocorticoids (alternate activation) |
| Metabolism | Glycolysis, HIF-1α, iNOS/NO, PKF2, mTOR, Induction of pro-inflammatory cytokines like IL-1β | OXPHOS, Arginase 1,2, AMPK, PFKFB1, Th2 type response induction, cholesterol efflux (38), efferocytosis (39) |
| Nature of Immune function | Pro-inflammatory killer mediated by ROS, RNS, TNFα, IL-6, IL-12, IL-23 | Immunomodulation mediated by IL-10, TGFβ, PDGF, VEGF, EGF, Arginase, α-KG |
| Biomarkers—used in combination or isolated markers to identify macrophage subset (40) | CCL2, CCL3, CCL5; CXCL8, CXCL9, CXCL10, CXCL11, CXCL16; IL-12, TNFα, IL-6, IL-1, IL-23; CD80, CD86, NOS, ROS, MHCII, TLR2/TLR4 | CCL17, CCL18, CCL22, CCL24; CXCR1, CXCR2; IL-10, IL-2RA; CD23, CD163, CD36, CD86, Mannose receptor (CD68+ MR+/CD206+MR+), scavenger receptor class A (SR-A), lectin-like oxidized LDL, Arginase, MHCII (39) |
| Genes/enzymes | NOS2, Ciita, IL-12 (41) | Arg1, Ym1, Fizz1, MMP12, MMP7, MERTK, Mcr1, IL-10, CD81 (41) |
| Signaling pathways and molecules present (41) | PI3K, p65, IRF5, STAT1, STAT2 | PI3K, p50, IRF4, STAT3, STAT6 |
| Phenotype in various disease condition | Bacterial and viral infections, auto-immune diseases (ex; Rheumatoid arthritis), obesity, diabetes, cardiovascular diseases, atherosclerosis, steatosis/fatty liver | Fibrosis, wound healing, sepsis, allergy, asthma, parasitic diseases (e.g., helminths), several tumors/cancers |
α-KG, α-ketoglutarate.
Experimental data suggests TAMs to be largely biased towards M2 phenotype (2,45). Molecular profiling of macrophages and the transcriptome data reveals that TAMs encompass a diverse macrophage population that shares features of both the M1 and M2 phenotype with a greater expression of genes involved in embryonic and tissue development and is not a distinct subset (46-48). Metabolic plasticity and intimate crosstalk with tumor cells is an important characteristic feature of TAMs. TAMs respond to altered metabolic profile of TME by polarizing to a cellular state which utilizes glycolysis, alterations in tricarboxylic acid (TCA) cycle, FAS and altered nitrogen cycle metabolism (49). These functionally reprogrammed TAMs contribute to production of altered cytokines and angiogenic factors which support tumor growth and survival (50,51). The bidirectional immune-metabolic communication between cancer cells and TAMs is orchestrated by multitude of metabolites, chemokines, cytokines, growth factors etc. helps to maintain the immunosuppressive nature of TME (52). The various cytokines involved in metabolic programming include IL-6, TNF, CCL5, CCL18. IL-6 which favor glycolysis by converting phosphoinositide dependent protein kinase 1 (PDPK1) to phosphoglycerate kinase (PGK), whereas TNF, CCL5 and CCL18 help in synthesis of metabolic enzymes like lactate dehydrogenase A (LDHA), pyruvate kinase M1/2 (PKM1/2), pyruvate dehydrogenase kinase 1 (PDK1), G6PD, PDH, glucose transporter 1 (GLUT1) and adhesion molecule VCAM1.
Metabolic pathways in TAMs
As discussed earlier, M2 phenotype of TAMs is immunosuppressive and protumoral in function (53). TME has the ability to recruit and polarize macrophages into M2 type or TAMs. Transcription factors (TFs) such as NF-κB, STAT-3 and HIF-1 act as key factors in initiating a transcriptional program in TAMs that defines their protumoral function (54). Polarization of TAMs is also regulated by a variety of cytokines, chemokines and growth factors produced by tumor cells and TME. Colony stimulating factor-1 (CSF-1) and CCL2 have been clinically correlated to be associated with increased recruitment of M2-like macrophages at tumor site and disease severity (55). Vascular endothelial growth factor-1 (VEGF-1) produced in tumors as a result of angiogenic switch also has been shown to induce recruitment of TAMs (56). Tumor hypoxia also correlates with preferential localization of TAMs in areas of low oxygen tension. Tumor hypoxia induces transcription of gene involved in glucose and nitrogen metabolism. Studies in mouse mammary tumors indicate that TAMs are a mixed population of both M1 and M2-like macrophages (57). M1-like macrophages are localized in normoxic region of the tumor, however, M2-like macrophages with greater angiogenic potential are more concentrated in the hypoxic zones of tumor and their number increased as the disease progressed (58). Hypoxia has been shown to promote protumoral activity of TAMs by at least two mechanisms: (I) inducing an iron donor phenotype in TAMs and increased proliferation of tumor cells (II) upregulation of DNA damage inducible transcript 4 (DDT4 or REDD1) an endogenous inhibitor of mechanistic target of rapamycin (mTOR) pathway in humans which promotes OXPHOS and reduced glucose intake in TAMs. Hypoxia has also been shown to regulate exosome cargo from tumor cells (59). Studies in ovarian cancer have shown that hypoxia induces the expression of miR-940 in tumor exosomes which stimulated M2 phenotype polarization (60). Exosomes derived from tumor cells could influence macrophage differentiation by altering the miRNA profiles of TAMs (61). It also suggests that metabolic programming in TAMs is a combined effect of hypoxia and cytokines in microenvironment. Hypoxia in TME also increases arginase-1 and Mannose receptor (CD206) levels on TAMs (62). TAMs present in these hypoxic regions also induce expression of HIF-1α which induces a switch to glycolytic fermentation. Furthermore, cancer cell derived lactic acid in the TME stabilizes the expression of HIF-1α under hypoxic and normoxic conditions. These conditions cause the polarization of M1 phenotype to the M2 phenotype aided by lactic acid and characterized by increased arginase-1 level, CD206 and VEGF levels (63).
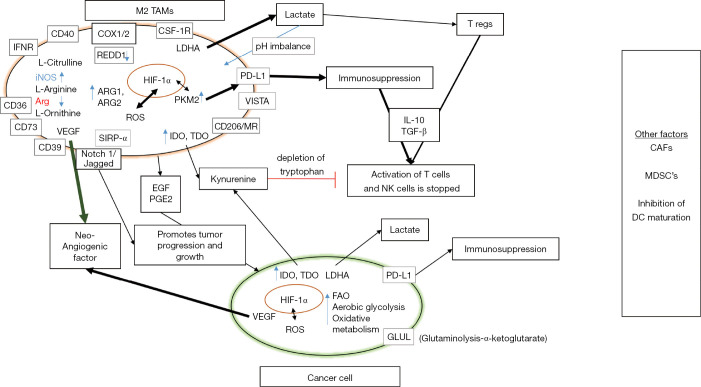
Seth et al. have shown that the deletion of LDHA and depletion or deletion of lactate levels in immune regulatory myeloid cells leads to the lung cancer regression and is supported by strong anti-tumor immune responses (64). Cancer cells maintain this vicious cycle by the constant production of lactic acid, maintaining an acidic pH and activating hypoxic response in TAMs. TAMs also secrete immunosuppressive and anti-inflammatory cytokines like IL-10 in response to the increased lactate levels which induces immunosuppression (65). There are also other metabolic genes such as tumor PKM which inhibits lipopolysaccharide (LPS) induced IL-1β, and is involved in M1–M2 polarization and controlling the phenotypic profile of macrophages (66). Recent work has implicated IL-4-induced signaling through AKT and mTOR complex 1 (mTORC1) in the regulation of glucose metabolism for M2 activation (67). Similarly, after stimulation with IL-4, mTORC2 is activated by PI3K which then activates AKT and this pathway is important for the changes in metabolism that are essential to M2 activation (67). Goossens et al. have shown that extracellular matrix component hyaluronic acid released by ovarian cancer cells induces a TAM phenotype by enhancing cholesterol efflux (38). Halbrook et al., using a metabolomics approach have identified a range of metabolites, including many pyrimidines released by TAMs into their culture medium (68). The TAM niche exhibits highly heterogeneous and dynamic cell populations as experimentally documented in many patient studies. This diversity signifies the ability of TAMs to polarize in functional and metabolic terms in accordance with the habitat they are present in (69,70). The metabolic phenotype of TAMs is highly diverse and keeps evolving with metabolic changes accompanying tumor progression (Figure 1). The dynamic adaptation of TAMs in response to malignant cells has a profound effect on not only on survival of TAMs and tumor cells but also other immune cells at the tumor site. Analysis of immune-metabolic crosstalk between TAMs, tumor cells and TME holds promise for deciphering novel targets in cancer therapy.
Figure 1.
Immune-metabolic crosstalk between TAMs and cancer cell. This representation shows various TAM mediated pathways involved in immunosuppression and tumor progression. Effector functions of other immune cells are inhibited in the process allowing immune escape of tumor variants. The bold arrows and highlighted words show the dominant pathways in TAM mediated immunosuppression. Other factors which regulate TAM mediated immunosuppression and TME directly/indirectly include: CAFs actively promoting the recruitment of monocytes to the TME and their differentiation toward M2 macrophages (71). In particular, the secretion of CXCL12/SDF1, macrophage colony-stimulating factor (M-CSF also known as CSF-1), IL-6, and CCL2/MCP-1 by CAFs actively promotes the recruitment of monocytes to the TME and their differentiation into a M2 immunosuppressive phenotype (72-74). TAM dependent recruitment of MDSCs, another group of potent immunosuppressive cells of myeloid origin can further suppress innate and adaptive immune responses to cancer. MDSCs are a heterogeneous group of cells consisting of immature precursors of monocytes and granulocytes. However, tumor-infiltrating monocytic MDSCs can differentiate into TAMs by CSF-1 and HIF-1α (75). TAMs, like M2-polarized macrophages, abundantly produce CCL2 which promotes CCR2+ monocytic MDSCs trafficking from bone marrow to tumor (76). Soluble factors secreted like IL-6 and M-CSF have been shown to block DC differentiation from CD34+ progenitors and promote lineage commitment toward CD14+ monocytes that express little to no MHC and costimulatory molecules (77,78). TAM, tumor-associated macrophage; TME, tumor microenvironment; CAF, cancer-associated fibroblast; HF, hypoxia inducible factor; SIRP, signaling regulatory protein; IDO, indolamine-2,3-dioxygenase; TDO, tryptophan-2,3-dioxygenase; EGF, epidermal growth factor; VEGF, vascular endothelial growth factor; pEGF, precursor epidermal growth factor; PGE2, prostaglandin E2; ROS, reactive oxygen species; NOS, nitric oxide synthase; Arg, arginine; IFN, interferon; TGF, tumor growth factor; FAO, fatty acid oxidation; CSF, colony stimulating factor; PD-L1, programed cell death ligand 1; PKM, protein kinase M; MR, mannose receptor; LDHA, lactate dehydrogenase A; REDD1, protein regulated in development and DNA damage response 1; COX, cyclooxygenase; DC, dendritic cells; MDSC, myeloid derived suppressor cell.
Glucose metabolism reprogramming in TAMs
TAMs are metabolically distinct from conventional M2 polarized subset in prioritizing usage of glycolysis as a key metabolic pathway. In aerobic glycolysis (the Warburg effect), pyruvate is converted to lactic acid by LDHA (79,80). Lactic acid activates VEGF, transforming growth factor β (TGF-β), and HIF-1α in oxidative tumor cells (81). Simultaneously, lactic acid released by glycolytic cancer cells into the TME also upregulates HIF-1α expression in bone marrow-derived macrophages (BMDMs) (81). Tumor cells are highly glycolytic in nature as they adapt to survive in nutrient limited microenvironment. TAMs compete with TME for nutrients such as glucose and undergo changes in glucose metabolism in a manner similar to tumor cells. Studies from murine models and human cells stimulated with tumor extract solution show upregulation of enzymes hexokinase-2 (HK2), phosphofructokinase and enolase 1 (ENO1) (82,83). Over expression of HIF-1α in TAMs upregulates genes responsible for glycolysis pathway such as PDK1, PGK1, GLUT1, glucokinase (GCK) and PKM2 (84). High amounts of lactate are released into TME as a result of enhanced glycolysis in TAMs, increased levels of lactate receptor have also been observed in vivo in thyroid cancer patients. Increased lactic acid levels induce TF HIF-1α and mTOR dependent aerobic glycolysis (85).
Overall, aerobic glycolysis is a characteristic feature of TAMs due to over-expression of GLUT1, over expression of enzyme HK2, lactic acid fermentation and presence of mTOR-AKT signaling pathway in TAMs. Co-culture of normal monocytes with conditioned medium from tumor cell line upregulates genes involved in glycolysis leading to angiogenesis, metastasis and epithelial-mesenchymal transition (EMT) (86,87). Aerobic glycolysis has been shown to be essential for tumor progression, angiogenesis and EMT in cancer cells. Lactate activated human TAMs stimulated the secretion of CCL5 via Notch signaling in macrophages (88). CCL5 increased cell migration, induced cancer cell EMT, and promoted aerobic glycolysis in breast cancer cells, by a positive metabolic feedback loop in the co-culture system. Inhibition of aerobic glycolysis significantly reduced breast cancer cell EMT (89). Recent data from de-Brito et al. indicates that TAMs show high glycolytic activity, with high lactate secretion similar to the M1/M(LPS + IFN-γ) phenotype. This activity seems to be essential for the M2 profile of TAMs, since the inhibition of glycolysis, but not the impairment of the OXPHOS or PPP, diminished the expression of M2/M(IL-4) markers (90). These novel data indicate that TAMs, although are usually phenotyped as M2/M(IL-4)-like macrophages, they are metabolically distinct from these cells, being rather similar to M1/M(LPS + IFN-γ) macrophages, depending on the glycolytic metabolism to support their profile and functions.
Lipid metabolism reprogramming in TAMs
Alternatively, activated macrophages prefer to use fatty acid (FA) metabolism. Since TME is also a FA rich environment, it is plausible that unsaturated FAs present in TME may play a role in polarizing tissue macrophages to an M2 phenotype. Anti-inflammatory cytokines like IL-10 are also associated with lipid metabolism. Whether lipid metabolism is a consequence of polarization of macrophages or the TME drives lipid metabolism in TAMs is not known. Ip et al. have shown that IL-10 inhibits lipopolysaccharide-induced glucose uptake and glycolysis and promotes OXPHOS. Upon LPS activation, IL-10-deficient macrophages had further reduced OXPHOS as compared with the already reduced OXPHOS in control macrophages (91). For a cell to grow and proliferate lipids are required. If the lipid levels in the cell are not sufficient then the FAS pathway can be initiated in the cytoplasm to allow cells to generate lipids from precursors derived from other cell intrinsic metabolic pathways including the TCA cycle, glycolysis and the pentose-phosphate pathway (92,93). mTOR signaling promotes FAS through the induction of sterol regulatory element binding protein (SREBP) a TF which in turn induces fatty acid synthase (FASN) and acetyl CoA carboxylase (ACC) (94). Recent data indicates that macrophages from both murine tumor models and human tumors show enhanced lipid uptake and lipid metabolism was critical in inducing TAM polarization (95). Increased lipid uptake in tumor cells by upregulation of FA receptor CD36 has been shown to be responsible for metastasis (96,97), and CD36 has also been proposed as a potential biomarker for cancer. CD36 deficiency is representative of the tumor stroma and high cancer risk: the lower the CD36 level in the stroma, the more aggressive the tumor, levels of CD 36 expression and mammographic density have also been proposed as potential areas of therapeutic intervention (98). Lipid metabolism also has been shown to play an important role in EMT, cholesterol lowering drug simvastatin was able to reverse EMT in A549T cells and repolarize M2 to M1 phenotype in macrophages (99). This suggests lipid metabolism to be a promising therapeutic target in cancer. Caspase-1 mediated lipid accumulation in TAMs and metabolic reprogramming has also been demonstrated in THP-1 macrophages co-cultured with MCF-7 tumor cells (100). Long chain fatty acid oxidation (FAO) in TAMs can help in OXPHOS pathway. In an in vitro study using TAMs and hepatocellular carcinoma cells (HCCs), it has been shown that M2 monocyte-derived macrophages (MDMs) enhanced the proliferation, migration, and invasion of HCC cells via the FAO pathway and that FAO played a key role in protumoral function of macrophages (101).
Glutamine metabolism in TAMs
Apart from the glycolytic switch, dependence on glutamine-glutamate pathway (i.e.,) amino acid switch is an important metabolic characteristic of TAMs. Cancer cells show an enhanced level of intracellular glutamine and experiments conducted in in vivo settings have emphasized the contribution of glutamine metabolism in cancer (102,103). Glutamine synthesis is upregulated in most human cancers and levels of enzyme glutamine synthetase (GS) (GLUL). Transcripts of another key enzyme glutaminase (GLS) are upregulated in malignancies of colon, esophagus, liver, stomach, thyroid and head and neck cancer. However, distinct patterns of glutamine metabolism have also been noted even in different cancer subtypes from the same tissue (104). Quantitative PCR analysis showed increased expression of GRIA2 (GluA receptor), SLC1A2, SLC1A3, increased expression of GS (GLUL) and a decreased expression of cysteine glutamate antiporter in glioblastoma model (105). Glutamine metabolism assists macrophage activation and elicits desired immune responses, but the underlying mechanisms remain uncertain. Glutaminolysis is an important activation signature for the alternative activation (M2) of macrophages accompanied by FAO and JMJD3 dependent epigenetic reprogramming of M2 genes (106). Glutaminolysis results in production of α-ketoglutarate (α-KG) which along with the levels of succinate determines the M1–M2 polarization of macrophages. High α-KG/succinate ratio represents M2 phenotype while low α-KG/succinate ratio represents M1 phenotype (106,107). Glutamine is also the most abundant amino acid in plasma, is an intermediate for TCA cycle and acts as nitrogen donor for synthesis of purines, pyrimidines, NAD, asparagine etc. via its terminal amide group. Glutamine also acts as an activator of mTOR pathway and uptake of essential amino acids. Enzyme GS is also expressed by human macrophages and BMDMs derived from GS –/– mice display a unique metabolic feature after IL-10 stimulation, similar to that observed in blood-derived human macrophages treated with IL-10 in the presence of methionine sulfoximine, a GS inhibitor (108). GS deletion in TAMs strongly favored a decrease in tumor metastasis and angiogenesis with an increase in number of anti-tumor CD8 T cells (108). Extracellular supraphysiological glutamine supplementation also induced polarization of macrophages to M2 phenotype. All these data point towards a fundamental role for TAM glutamine metabolism in shaping tumor development. Interleukin 10 has been shown to be a principle inducer of GS expression in blood derived macrophages. IL-10 signaling has been shown to inhibit glycolysis and promote OXPHOS (91). IL-10 induced anti-inflammatory activity of macrophages is accompanied by mTORC1 inhibition, by means of a specific activation of REDD1. According to the hypothesis proposed by Mazzone et al., increased intracellular levels of glutamine due to IL-10-mediated GS expression may promote the anti-inflammatory events typical of M2-like macrophages in synergy with REDD1 expression through Sp1 TF which inhibits mTOR (49). TAM specific REDD1 deletion promotes tumor vessel normalization and metastasis inhibition. Therefore, GS and REDD1 may represent key molecules which help to polarize TAMs to M2 like phenotype (49).
TAM mediated immunosuppression
Immune cells involved in immune surveillance of cancer include CD4+ T helper cells, cytotoxic CD8+ T cells, natural killer (NK) cells. Enhanced glucose consumption by tumors creates a nutritional competition between cells in TME. These effector cells exhibit high glycolytic metabolism to meet the escalating energy demands of developing tumor cell. Immunosuppressive T reg cells depend on OXPHOS for increased bioenergetics (109). Immune cells and the cancer cells in the TME compete for the availability of glucose and the M2 TAMs strategically employ OXPHOS to avoid competition with T lymphocytes. TAMs also limit glycolytic flux in many effector cells by expressing and upregulating CD274, also known as PDL1 (110-112). The effector cells secrete IFN-γ and the malignant cancer cells, endothelial cells and the TAMs in response to IFN-γ, upregulate the expression of PD-L1 (113). Zhang et al. showed that TAMs promote and upregulate PD-L1 expression in cancer cells by secreting EGF (114). The PD-L1 and PD-1 interaction produces a reverse signal which elicits pro-glycolytic effects and limits glycolysis and proliferation in TAMs (110). Cancer cells and TAMs interact in such a way that they direct the metabolic competition to favor tumor progression. The expression level of PD-L1 in TAMs is controlled by PKM2, HIF-1α and as well as the signaling cascade initiated by PGE2 (115-117). PGE2 is present in TME is due to arachidonic acid metabolism and apoptosis activation in cancer cells in response to therapy (118,119). Apart from recruiting TAMs into TME and favoring repolarization of M1 to M2 phenotype, PGE2 has pro-oncogenic functions which help in cancer growth, survival and proliferation (120,121). The M2 TAMs deprive the TME of amino acids. TAMs have high expression of arginase-1, arginase-2, indoleamine-2,3-dioxygenase (IDO) which converts tryptophan to kynurenine in TME. Kynurenine induces functional impairment of T cells and NK cells by limiting tryptophan availability. Kynurenine also favors the growth and proliferation of T reg cells (122,123).
M2 TAMs are known to express high levels of CD38 which initiates adenosine synthesis from NAD+, CD39 or the ectonucleoside triphosphate diphosphohydrolase 1 (ENTDP1) and CD73 or the 5’-nucleotidase ecto (NT5E) which hydrolyses ATP to adenosine. These enzymes starve the immune-effector cells of the essential nutrients and induce immunosuppression. Montalbán del Barrio et al. showed that migration of monocytes was restricted after blocking CD73 or CD39 activity on ovarian cancer cells (124). In-vitro and ex-situ TAMs upregulated the ectonucleotidases CD39 and CD73 expression which convert extracellular ATP to biologically active adenosine. This resulted in suppressed CD4 T cell proliferation and tumor immune evasion. TAMs overexpress CD39 and inhibitors against these can improve immune responses (124,125). Studies also show that IL-27 mediated CD39 induction on TAMs produced similar amount of adenosine as produced by cancer cells and thus mediated anti-proliferative effect on T cells (124,126,127). These mechanisms reduce the extracellular availability of nucleotides like adenosine and glutamate which limits the recruitment, activation and presentation of APCs. The over production of these nucleotides by ectonucleotidases present on cancer cells and T regs induces immunosuppression as the adenosine produced directly inhibits T cell mechanisms. Hence these are promising targets for novel and strategic therapeutic interventions (128). Lactate is another important immunosuppressive metabolite shown to induce anergy in T cells and NK cells (129). High lactate levels also alter chemokine receptor signaling thereby limiting T cell motility and migration (130-132). T cells under the influence of high lactic acid levels also tend to polarize to suppressive T regs phenotype (133). PKM acts as a determinant of the Warburg effect in LPS activated macrophages (i.e.,) the M1 phenotype (66,134). PKM and HIF-1α promotes PDL-1 expression on TAMs, T cells and cancer cells by binding to the PDL-1 promoter. O Neill showed that blocking PKM downregulated and suppressed the PDL-1 expression on the immune cells and cancer cells (116). Seth et al. showed that LDHA also promoted PDL-1 expression in a similar fashion to PKM and HIF-1α, but the overall mechanism remains unknown. It is possible that it may be similar or may even involve HIF-1α (64,115).
Hashimoto et al. studied the interaction of TAMs with other components in the TME and reported that TAMs may influence the cancer-associated fibroblasts (CAFs). They also showed that TAMs increase the expression of CXCL2 and enhance invasion of neuroblastoma cells via CXCL2/CXCR2 signaling (135). Other studies showed that inhibition of CXCL2/CXCR2 signaling resulted in the suppression of cell proliferation and metastasis in hepatocellular carcinoma and breast cancer (136,137). Kortlever et al. 2017 reported that oncogenes like KRAS and MYC seems to influence the recruitment of MDMs and mediate their polarization to the M2 phenotype which in turn supports tumor progression (138). The following graphical representation shows how the TAMs play a role in TME via their reciprocal crosstalk and helps in tumor initiation, progression, growth and survival (Figure 2).
Figure 2.
Graphical representation of how TAMs contribute to TME. The primary and secondary components present in TME which support tumor growth and progression are also indicated in the figure. TAM, tumor-associated macrophage; TME, tumor microenvironment; CAF, cancer-associated fibroblast; VEGF, vascular endothelial growth factor; pEGF, precursor epidermal growth factor; CSF, colony stimulating factor; EMT, epithelial-mesenchymal transition; IDO, indolamine-2,3-dioxygenase; TDO, tryptophan-2,3-dioxygenase; FAO, fatty acid oxidation; MDSC, myeloid derived suppressor cell; PGE2, prostaglandin E2; TAN, tumor-associated neutrophil.
Epigenetic regulation and effect of tissue environment on macrophage metabolism
Epigenetic modifications have recently emerged as key drivers for immune cell phenotypes. Epigenetic enzymes that induce processes such as DNA methylation, histone modifications etc. have been implicated in shaping the functional response of immune cells to various stimuli (139). Epigenetic phenomenon has been so far implicated for disease pathology in situations such as diabetes, atherosclerosis, and obesity. In cancer as well, epigenetic modifications are being investigated as a novel paradigm for regulating polarization and metabolic rewiring of TAMs. Epigenetic dysregulation has also been implicated as a prominent cause for malignant transformation in cancer cells through non-coding RNAs, DNA methylation and histone modifications (140). Recent findings on epigenetic regulation of functions of tumor infiltrating immune cells, macrophages in particular, has provided novel targets for tumor immunotherapy. However, it is apparent that the relationship between metabolism and epigenetics is bidirectional and highly intertwined event in cancer, i.e., metabolic plasticity of immune cells in TME can induce epigenetic changes via production of certain metabolites. Conversely, epigenetic changes can regulate expression of metabolic genes and influence cellular differentiation. Understanding the crosstalk between epigenetics and metabolism is critical for identifying novel therapeutic targets in cancer.
Epigenetic changes can be classified into three main categories: (I) DNA methylation; (II) posttranslational histone modifications; and (III) noncoding RNA including lncRNA and miRNA. DNA methylation refers to transcriptional repression and is characterized by transfer of a methyl group to the cytosine ring of DNA by DNA methyltransferases (DNMTs) (To form 5-methylcytosine). DNA methylation is removed by another set of enzymes known as ten-eleven translocation (TET) proteins. Histone modifications include histone acetylation and deacetylation and are orchestrated by enzymes histone acetyltransferases and histone deacetylases (HDACs), respectively. Histone acetylation is linked to transcriptional activity, whereas histone deacetylation is associated with transcriptional repression (141). Similarly, methylation and demethylation of histones are achieved by histone methyltransferases and histone demethylases, respectively. Noncoding RNAs like lncRNA and miRNAs play an essential role in post transcriptional control of gene expression (142).
Specific epigenetic modifications relevant to tumor macrophages have indicated prominent roles for epigenetic enzymes in inducing M2 polarization. Amongst DNMTs, DNMT3B is the only DNMT having role in M2 polarization. Knockdown of DNMT3B in RAW264.7 macrophages and mouse BMDMs has been shown to induce M2 polarization and prevent M1 marker expression (143). As for demethylation, TET2 loss of function mutations are implicated in myeloid malignancies, however a direct role for TET proteins in macrophage polarization is still to be established. Another HMT which is a H3K4 methyltransferase, SMYD3, is speculated to positively regulate M2 polarization. Its expression levels in human monocyte-derived macrophages (HMDMs) decrease with exposure to LPS + IFN-γ and increases with exposure to the combination of M-CSF, IL-4, and IL-13 (M-CSF + IL-4 + IL-13) (144). JMJD3 (KDM6B) which is an H3K27 demethylase, has been recognized as an essential regulator of M2 polarization through its induction of IRF4, Arg1, CD206, and other M2 markers in IL4- stimulated (15) and IL-4 + IL-13-stimulated (145) mouse BMDMs. In histone acetylation, HDAC9 is another negative regulator of M2 polarization as peritoneal macrophages from HDAC9-deficient mice expressed lower levels of M1 genes and higher levels of M2 genes compared to wild type mice (146).
Metabolic intermediates can also induce epigenetic changes in immune cell types. TAM polarization is governed by the interplay between metabolic intermediates, TFs and enhancers leading to specific gene expression. Classical activation of macrophages with TLR4 ligand and LPS results in signal dependent activation of TF’s leading to impairment of mitochondrial respiration and a glycolytic shift (147). This glycolytic shift is accompanied by an increase of lactate which serves as an inhibitor of histone de-acetylases (class II—HDAC’s) (148). TLR signaling also leads to shift in NAD+-NADH ratio which further influences class III HDAC’s (SIRT1 and SIRT6) activities causing de-acetylation of non-histone and histone substrates (149). Cancer cells use ATP-citrate lyase, an enzyme which converts citrate to Acetyl-Co-A, as a substrate for histone acetylation (150). Increase in Acetyl-CoA (required for histone acetylation) and reduction in 2-oxoglutarate levels [required for TET proteins and JHDM (Jumonji domain-containing histone demethylase)] may be two crucial metabolites responsible for determining the metabolic switch between M1 and M2 macrophage. Immune-responsive gene 1 (IRG1) is responsible for itaconate production and IRG1 deficient murine BMDMs show significant reduction in succinate levels and inflammatory cytokines (151). Itaconate, a metabolite found highly induced in classically activated macrophages has modulatory effect on succinate levels (151). It inhibits succinate dehydrogenase-mediated succinate oxidation leading to shift in macrophage metabolism and effector functions. In case of alternatively activated macrophages (IL-4 stimulation) STAT6-PPARγ-PGC1α signaling axis plays a key role in oxidative metabolism and in regulation of mitochondrial function (152,153). The other signaling axis found in addition to the former is mTORC2-IRF4 signaling axis (154). Alternative activation of macrophages has been shown to be influenced by FAO, polyamine synthesis and uridine diphosphate N-acetylglucosamine (UDP-GlcNAc) synthesis (153). Increased polyamine synthesis leads to activation of HAT enzymes, DNA hypomethylation and induction of M2 genes (155).
Impact of tissue microenvironment on macrophage polarization
Apart from epigenetic regulation of TAM functions, tissue microenvironment is another important factor that shapes the phenotype and functional characteristics of macrophages. This indicates that macrophage functions may vary according to the surrounding tissue and local imprinting endows macrophage with particular tissue specific functions. Tissue specificity may account for high plasticity and versatility of TAMs and their dynamic evolution within TME during cancer progression. Experiments to unearth the ontogeny of TAMs in mouse breast cancer and lung cancer models have suggested two possible developmental routes for TAMs in a given tissue: (I) tissue-resident macrophages of either embryonic or monocytic origin that may undergo a change in phenotype/function during tumor development [tissue-resident TAMs, (trTAMs)], or (II) monocytes that undergo a distinct differentiation step to become macrophages in response to tumor growth [tumor-induced TAMs, (tiTAMs)]. These two populations may both be present simultaneously in a particular tumor, or alternatively, trTAMs may dominate at early stages of tumor growth, while tiTAMs become prominent at later stages. In addition, monocytes that enter tumor tissues may undergo phenotypic changes in response to the TME without terminal differentiation into macrophages. To determine the relative contribution of these two-macrophage population, experiments were conducted in genetic models on the PyMT tumor background to delete either the MTM (trTAM) or TAM (tiTAM) populations. Inhibition of tiTAM differentiation resulted in decreased tumor growth, trTAM depletion however did not have any impact on tumor growth (3). These data suggest that in some tissues, the trTAM population may have a relatively lesser role as compared to tiTAM in tumor development.
Experiments using both transplantation of fluorescently-labeled bone marrow (156,157) and tracking of microsphere-labeled monocytes (158) suggested a monocyte origin for TAMs. Recent studies have confirmed that TAM of different origins accumulate within the TME in mouse cancer models. Using parabiotic mice and bone marrow transfer, it was shown that the pool of TAM was composed of both newly recruited MDMs and resident macrophages in a model of pancreatic ductal adenocarcinoma (159). Studies in brain have indicated conflicting results, some studies indicate microglial cells as a predominant population while some suggest monocyte derived TAMs major population in glioblastoma models (21). Therefore, studies on ontogeny of TAMs, although still in nascent stage may be an important factor for understanding TAM diversity. Establishing a specific phenotypic and functional profile for each subset of TAM may therefore be essential to promote anti-tumor immune response.
Therapeutics targeting metabolic pathways in TAMs
There has been an increasing interest in developing therapeutic targets that can modulate TAM metabolism and reverse metabolic reprogramming (70,160). Tumor tissue of origin, genetic factors and TME have merged as major drivers for determining the metabolic phenotype of tumor cells. Comprehending the metabolic equilibrium between the pro and anti-tumoral and inflammatory characteristics of TAMs can help design specific inhibition of certain molecules. Repolarizing myeloid cells to perform anti-tumoral function seems to be the best approach for cancer therapy. M2 macrophages may be repolarized to M1 macrophages by altering specific cytokines in tumor. For example, inhibition of CSF-CSF1R axis using small molecules or monoclonal antibodies has been considered for repolarization of M2 like macrophages to M1 like macrophages. CSF1R inhibitors are currently undergoing clinical trials as monotherapy or combination drugs in cancer (161). Apart from cytokines, chemokines also have important role in TAM biology. PF-04136309, a CCR2 antagonist, was shown to completely block the mobility of CCR2+ monocytes into the tumor in a pancreatic cancer mouse model (162). Likewise, an anti-CCL2 monoclonal antibody, carlumab (CNTO88) also showed to be efficient in preventing the development and progression of several tumors in mouse models (163).
Since glycolysis is essential for tumor promoting functions of TAMS, inhibiting glycolysis pathway has been explored as a method to induce repolarization of TAMs. Glycolytic inhibitors such as 3-bromopyruvate (3-BP) (164), MJE3 (165), 3-(3-pyridinyl)-1-(4-pyridinyl)-2-propen-1-one (3PO) (166), 3-dihydroxy-6-methyl-7-(phenylmethyl)-4-propylnaphthalene-1-carboxylic acid (FX11) (167), and dichloroacetate (DCA) (168) target HK2, PFKFB3, PGAM1, LDHA and PDH, respectively in cancer cells and have been effective in suppressing tumor growth. However, specific targeting of glycolytic inhibitors only in tumor cells is a potential caveat as glycolysis is also an important metabolic pathway for normal cells. Other similar approaches aimed at interfering with glycolysis pathway that have been explored are mTORC1 targeting (169), blockage of VEGFA (170), inhibition of hypoxia (171,172) or modulation of iron metabolism. Myeloid cells in TME also utilize glutamine metabolism as a predominant pathway, experimental evidence from 4T-1 breast cancer model indicates that blocking glutamine metabolism produced the dual benefit of retarding recruitment of myeloid cells in TME and TAM repolarization as well (173). Immune checkpoint blockade (monoclonal antibodies targeting PD-L1 or PD-1) has also been shown to influence TAM metabolism and induce partial restoration of M1 like functions in murine models of B16 melanoma (174). Another dimension in TAM targeted therapeutics is aimed at disrupting the crosstalk between TAMs and other malignant cells; these include blocking TAM derived cytokines/chemokines (175,176), inhibition of adhesion molecule such as VCAM1 on malignant cells or inhibition of NF-κB mediated transcriptional program in TAMs (177,178). All these strategies seem to be promising interventions according to the results obtained so far in rodent tumor models.
Strategies like usage of liposomal clodronate which helps in TAM depletion have shown improved survival in few pre-clinical models (179). Clo-Lipo-DOTAP (clodronate containing liposomes) developed by Piaggio et al. showed significant reduction in primary tumor volume mediated by macrophage clearance in melanoma mouse model (180). Enhancement of phagocytic ability of TAMS has been suggested as a method to repolarize TAM. Two approaches to upregulate phagocytic activity (which is a characteristic of M1 macrophages) are facilitating antibody-dependent cellular phagocytosis or inhibition of CD47-signal regulatory protein alpha (CD47-SIRPα) signaling. Antibody dependent cellular phagocytosis is dependent on the interaction between Fc domain of antibody and Fc receptor on cancer cells. Examples include rituximab which promotes phagocytic activity of macrophages thus inhibiting tumor development (181), and trastuzumab used for HER2-overexpressing breast cancer therapy which triggers the phagocytic activity of macrophages both in-vitro and in-vivo (182). CD47 on interaction with SIRPα protein on macrophages transmits the ‘don’t eat me’ signal thus blocking phagocytosis. Many therapeutic antibodies have been designed against CD47 and SIRPα (183,184). Weiskopf et al. has shown that Hu5F9-G4, the CD47 antibody, promotes phagocytic activity of macrophages, thus, eradicating tumor cells and may be used as an immunotherapeutic drug for human small cell lung carcinoma (SCLC) (185). Similarly Petrova et al. have shown that TTI-621, a CD47 antibody (SIRPαFc), blocks the CD47-SIRPα axis and enhances macrophage phagocytosis in both hematological malignancies and solid tumors (186). Non-antibody approaches to repolarize M2 TAMs to M1-like type such as hydroxychloroquine or iron oxide nanoparticles have also been reported (187,188). Autophagy may also be key role in macrophage polarization. Shan et al. have demonstrated that isoprenaline induced M2 polarization was suppressed by autophagy via the mTOR and ROS/ERK pathway (189).
Conclusions
Enormous heterogeneity and plasticity in macrophages present a complex landscape in tumor biology. The metabolic by-products from TAMs also modulate the neighboring cells by acting as signaling mediators. Existence of tissue residence and recruited TAMs further adds to the complexity of metabolic pathways in various macrophage subsets. However, there is broad consensus on existence of multiple metabolic pathways rather than preference of any one pathway for TAMs. Furthermore, many of these pathways may have bypass or compensatory mechanisms; alternative pathways may be triggered after inhibition of primary pathway. No indicators are currently available which can define which metabolic pathway will be preferred in a given tumor scenario in metastatic, dormant or primary tumors. An added pitfall of targeting metabolic pathways in TAMs is that most of these pathways are also shared by normal cells, therefore the impact of sustained metabolic inhibition may have drastic and unpredictable effects on non-malignant cells. Perhaps, immunostimulatory strategies may have to be coupled with suppressing TAM functions to obtain maximum clinical benefit. A better understanding of these gaps in TAM metabolism may unravel novel avenues for combination tumor immunotherapy.
Supplementary
The article’s supplementary files as
Acknowledgments
Infrastructure support from Amity University Uttar Pradesh is acknowledged.
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Provenance and Peer Review: This article was commissioned by the Guest Editor (Gaurav Pandey) for the series “Tumor Associated Macrophages in Solid Tumor: Friend or Foe” published in Annals of Translational Medicine. The article was sent for external peer review organized by the Guest Editor and the editorial office.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-2037). The series “Tumor Associated Macrophages in Solid Tumor: Friend or Foe” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
References
- 1.Shankaran V, Ikeda H, Bruce AT, et al. IFNγ, and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature 2001;410:1107-11. 10.1038/35074122 [DOI] [PubMed] [Google Scholar]
- 2.Chen Y, Song Y, Du W, et al. Tumor-associated macrophages: an accomplice in solid tumor progression. J Biomed Sci 2019;26:78. 10.1186/s12929-019-0568-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franklin RA, Liao W, Sarkar A, et al. The cellular and molecular origin of tumor-associated macrophages. Science 2014;344:921-5. 10.1126/science.1252510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Little AC, Pathanjeli P, Wu Z, et al. IL-4/IL-13 stimulated macrophages enhance breast cancer invasion via rho-GTPase regulation of synergistic VEGF/CCL-18 signaling. Front Oncol 2019;9:456. 10.3389/fonc.2019.00456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laoui D, Movahedi K, van Overmeire E, et al. Tumor-associated macrophages in breast cancer: Distinct subsets, distinct functions. Int J Dev Biol 2011;55:861-7. 10.1387/ijdb.113371dl [DOI] [PubMed] [Google Scholar]
- 6.Qiu SQ, Waaijer SJH, Zwager MC, et al. Tumor-associated macrophages in breast cancer: Innocent bystander or important player? Cancer Treat Rev 2018;70:178-89. 10.1016/j.ctrv.2018.08.010 [DOI] [PubMed] [Google Scholar]
- 7.Ramos RN, Rodriguez C, Hubert M, et al. CD163+ tumor-associated macrophage accumulation in breast cancer patients reflects both local differentiation signals and systemic skewing of monocytes. Clin Transl Immunology 2020;9:e1108. 10.1002/cti2.1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen PC, Cheng HC, Wang J, et al. Prostate cancer-derived CCN3 induces M2 macrophage infiltration and contributes to angiogenesis in prostate cancer microenvironment. Oncotarget 2014;5:1595-608. 10.18632/oncotarget.1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riabov V, Kim D, Chhina S, et al. Immunostimulatory early phenotype of tumor-associated macrophages does not predict tumor growth outcome in an HLA-DR mouse model of prostate cancer. Cancer Immunol Immunother 2015;64:873-83. 10.1007/s00262-015-1697-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maolake A, Izumi K, Shigehara K, et al. Tumor-associated macrophages promote prostate cancer migration through activation of the CCL22-CCR4 axis. Oncotarget 2017;8:9739-51. 10.18632/oncotarget.14185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rakaee M, Busund LTR, Jamaly S, et al. Prognostic value of macrophage phenotypes in resectable non-small cell lung cancer assessed by multiplex immunohistochemistry. Neoplasia 2019;21:282-93. 10.1016/j.neo.2019.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Huang N, Zhu W, et al. Modulation the crosstalk between tumor-associated macrophages and non-small cell lung cancer to inhibit tumor migration and invasion by ginsenoside Rh2. BMC Cancer 2018;18:579. 10.1186/s12885-018-4299-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carroll MJ, Kapur A, Felder M, et al. M2 macrophages induce ovarian cancer cell proliferation via a heparin binding epidermal growth factor/matrix metalloproteinase 9 intercellular feedback loop. Oncotarget 2016;7:86608-20. 10.18632/oncotarget.13474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta V, Yull F, Khabele D. Bipolar tumor-associated macrophages in ovarian cancer as targets for therapy. Cancers 2018;10:366. 10.3390/cancers10100366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Satoh T, Takeuchi O, Vandenbon A, et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol 2010;11:936-44. 10.1038/ni.1920 [DOI] [PubMed] [Google Scholar]
- 16.Kawamura K, Komohara Y, Takaishi K, et al. Detection of M2 macrophages and colony-stimulating factor 1 expression in serous and mucinous ovarian epithelial tumors. Pathol Int 2009;59:300-5. 10.1111/j.1440-1827.2009.02369.x [DOI] [PubMed] [Google Scholar]
- 17.Allavena P, Chieppa M, Bianchi G, et al. Engagement of the Mannose receptor by tumoral mucins activates an immune suppressive phenotype in human tumor-associated macrophages. Clin Dev Immunol 2010;2010:547179. [DOI] [PMC free article] [PubMed]
- 18.Schutyser E, Struyf S, Proost P, et al. Identification of biologically active chemokine isoforms from ascitic fluid and elevated levels of CCL18/pulmonary and activation-regulated chemokine in ovarian carcinoma. J Biol Chem 2002;277:24584-93. 10.1074/jbc.M112275200 [DOI] [PubMed] [Google Scholar]
- 19.Colvin EK. Tumor-associated macrophages contribute to tumor progression in ovarian cancer. Front Oncol 2014;4:137. 10.3389/fonc.2014.00137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: Cancer as a paradigm. Nat Immunol 2010;11:889-96. 10.1038/ni.1937 [DOI] [PubMed] [Google Scholar]
- 21.Chen Z, Hambardzumyan D. Immune microenvironment in glioblastoma subtypes. Front Immunol. 2018;9:1004. 10.3389/fimmu.2018.01004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lailler C, Louandre C, Morisse MC, et al. ERK1/2 signaling regulates the immune microenvironment and macrophage recruitment in glioblastoma. Biosci Rep 2019;39:BSR20191433. 10.1042/BSR20191433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beider K, Bitner H, Leiba M, et al. Multiple myeloma cells recruit tumor-supportive macrophages through the CXCR4/CXCL12 axis and promote their polarization toward the M2 phenotype. Oncotarget 2014;5:11283-96. 10.18632/oncotarget.2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris JA, Jain S, Ren Q, et al. CD163 versus CD68 in tumor associated macrophages of classical hodgkin lymphoma. Diagn Pathol 2012;7:12. 10.1186/1746-1596-7-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al-Matary YS, Botezatu L, Opalka B, et al. Acute myeloid leukemia cells polarize macrophages towards a leukemia supporting state in a growth factor independence 1 dependent manner. Haematologica 2016;101:1216-27. 10.3324/haematol.2016.143180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Komohara Y, Niino D, Saito Y, et al. Clinical significance of CD163+ tumor-associated macrophages in patients with adult T-cell leukemia/lymphoma. Cancer Sci 2013;104:945-51. 10.1111/cas.12167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang X, Feng W, Wang R, et al. Repolarizing heterogeneous leukemia-associated macrophages with more M1 characteristics eliminates their pro-leukemic effects. Oncoimmunology 2017;7:e1412910. 10.1080/2162402X.2017.1412910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burger JA, Tsukada N, Burger M, et al. Blood-derived nurse-like cells protect chronic lymphocytic leukemia B cells from spontaneous apoptosis through stromal cell-derived factor-1. Blood 2000;96:2655-63. 10.1182/blood.V96.8.2655 [DOI] [PubMed] [Google Scholar]
- 29.Tsukada N, Burger JA, Zvaifler NJ, et al. Distinctive features of “nurselike” cells that differentiate in the context of chronic lymphocytic leukemia. Blood 2002;99:1030-7. 10.1182/blood.V99.3.1030 [DOI] [PubMed] [Google Scholar]
- 30.Boissard F, Fournié JJ, Laurent C, et al. Nurse like cells: chronic lymphocytic leukemia associated macrophages. Leuk Lymphoma 2015;56:1570-2. 10.3109/10428194.2014.991731 [DOI] [PubMed] [Google Scholar]
- 31.Giannoni P, Pietra G, Travaini G, et al. Chronic lymphocytic leukemia nurse-like cells express hepatocyte growth factor receptor (c-MET) and indoleamine 2,3-dioxygenase and display features of immunosuppressive type 2 skewed macrophages. Haematologica 2014;99:1078-87. 10.3324/haematol.2013.091405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan B, Jiang Z, Cheng L, et al. Paracrine HGF/c-MET enhances the stem cell-like potential and glycolysis of pancreatic cancer cells via activation of YAP/HIF-1α. Exp Cell Res 2018;371:63-71. 10.1016/j.yexcr.2018.07.041 [DOI] [PubMed] [Google Scholar]
- 33.Lankadasari MB, Mukhopadhyay P, Mohammed S, et al. TAMing pancreatic cancer: Combat with a double edged sword. Mol Cancer 2019;18:48. 10.1186/s12943-019-0966-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang R, Liu Q, Peng J, et al. Pancreatic cancer-educated macrophages protect cancer cells from complement-dependent cytotoxicity by up-regulation of CD59. Cell Death Dis 2019;10:836. 10.1038/s41419-019-2065-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu H, Hang JJ, Han T, et al. The M2 phenotype of tumor-associated macrophages in the stroma confers a poor prognosis in pancreatic cancer. Tumour Biol 2016;37:8657-64. 10.1007/s13277-015-4741-z [DOI] [PubMed] [Google Scholar]
- 36.Lindau D, Gielen P, Kroesen M, et al. The immunosuppressive tumour network: myeloid-derived suppressor cells, regulatory T cells and natural killer T cells. Immunology 2013;138:105-15. 10.1111/imm.12036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koo SJ, Garg NJ. Metabolic programming of macrophage functions and pathogens control. Redox Biol 2019;24:101198. 10.1016/j.redox.2019.101198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goossens P, Rodriguez-Vita J, Etzerodt A, et al. Membrane cholesterol efflux drives tumor-associated macrophage reprogramming and tumor progression. Cell Metab 2019;29:1376-89.e4. 10.1016/j.cmet.2019.02.016 [DOI] [PubMed] [Google Scholar]
- 39.Bi Y, Chen J, Hu F, et al. M2 macrophages as a potential target for antiatherosclerosis treatment. Neural Plast 2019;2019:6724903. [DOI] [PMC free article] [PubMed]
- 40.Ka MB, Daumas A, Textoris J, et al. Phenotypic diversity and emerging new tools to study macrophage activation in bacterial infectious diseases. Front Immunol 2014;5:500. 10.3389/fimmu.2014.00500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chávez-Galán L, Olleros ML, Vesin D, et al. Much more than M1 and M2 macrophages, there are also CD169+ and TCR+ macrophages. Front Immunol 2015;6:263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mills CD, Kincaid K, Alt JM, et al. M-1/M-2 macrophages and the Th1/Th2 paradigm. J Immunol 2000;164:6166-73. 10.4049/jimmunol.164.12.6166 [DOI] [PubMed] [Google Scholar]
- 43.Xue J, Schmidt SV, Sander J, et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 2014;40:274-88. 10.1016/j.immuni.2014.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ginhoux F, Schultze JL, Murray PJ, et al. New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat Immunol 2016;17:34-40. 10.1038/ni.3324 [DOI] [PubMed] [Google Scholar]
- 45.Biswas SK, Allavena P, Mantovani A. Tumor-associated macrophages: functional diversity, clinical significance, and open questions. Semin Immunopathol 2013;35:585-600. 10.1007/s00281-013-0367-7 [DOI] [PubMed] [Google Scholar]
- 46.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest 2012;122:787-95. 10.1172/JCI59643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol 2012;12:253-68. 10.1038/nri3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chittezhath M, Dhillon MK, Lim JY, et al. Molecular profiling reveals a tumor-promoting phenotype of monocytes and macrophages in human cancer progression. Immunity 2014;41:815-29. 10.1016/j.immuni.2014.09.014 [DOI] [PubMed] [Google Scholar]
- 49.Mazzone M, Menga A, Castegna A. Metabolism and TAM functions—it takes two to tango. FEBS J 2018;285:700-16. 10.1111/febs.14295 [DOI] [PubMed] [Google Scholar]
- 50.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell 2010;141:39-51. 10.1016/j.cell.2010.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell 2006;124:263-6. 10.1016/j.cell.2006.01.007 [DOI] [PubMed] [Google Scholar]
- 52.Andrejeva G, Rathmell JC. Similarities and distinctions of cancer and immune metabolism in inflammation and tumors. Cell Metab 2017;26:49-70. 10.1016/j.cmet.2017.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sica A, Allavena P, Mantovani A. Cancer related inflammation: the macrophage connection. Cancer Lett 2008;267:204-15. 10.1016/j.canlet.2008.03.028 [DOI] [PubMed] [Google Scholar]
- 54.Quatromoni JG, Eruslanov E. Tumor-associated macrophages: function, phenotype, and link to prognosis in human lung cancer. Am J Transl Res 2012;4:376-89. [PMC free article] [PubMed] [Google Scholar]
- 55.Poh AR, Ernst M. Targeting macrophages in cancer: from bench to bedside. Front Oncol 2018;8:49. 10.3389/fonc.2018.00049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Riabov V, Gudima A, Wang N, et al. Role of tumor associated macrophages in tumor angiogenesis and lymphangiogenesis. Front Physiol 2014;5:75. 10.3389/fphys.2014.00075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ojalvo LS, King W, Cox D, et al. High-density gene expression analysis of tumor-associated macrophages from mouse mammary tumors. Am J Pathol 2009;174:1048-64. 10.2353/ajpath.2009.080676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tripathi C, Tewari BN, Kanchan RK, et al. Macrophages are recruited to hypoxic tumor areas and acquire a Pro-Angiogenic M2-Polarized phenotype via hypoxic cancer cell derived cytokines Oncostatin M and Eotaxin. Oncotarget 2014;5:5350-68. 10.18632/oncotarget.2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shao C, Yang F, Miao S, et al. Role of hypoxia-induced exosomes in tumor biology. Mol Cancer 2018;17:120. 10.1186/s12943-018-0869-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen X, Zhou J, Li X, et al. Exosomes derived from hypoxic epithelial ovarian cancer cells deliver microRNAs to macrophages and elicit a tumor-promoted phenotype. Cancer Lett 2018;435:80-91. 10.1016/j.canlet.2018.08.001 [DOI] [PubMed] [Google Scholar]
- 61.Meng W, Hao Y, He C, et al. Exosome-orchestrated hypoxic tumor microenvironment. Mol Cancer 2019;18:57. 10.1186/s12943-019-0982-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Laoui D, Van Overmeire E, Conza G, Di, et al. Tumor hypoxia does not drive differentiation of tumor-associated macrophages but rather fine-tunes the M2-like macrophage population. Cancer Res 2014;74:24-30. 10.1158/0008-5472.CAN-13-1196 [DOI] [PubMed] [Google Scholar]
- 63.Colegio OR, Chu NQ, Szabo AL, et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 2014;513:559-63. 10.1038/nature13490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seth P, Csizmadia E, Hedblom A, et al. Deletion of lactate dehydrogenase-A in myeloid cells triggers antitumor immunity. Cancer Res 2017;77:3632-43. 10.1158/0008-5472.CAN-16-2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petty AJ, Yang Y. Tumor-associated macrophages: implications in cancer immunotherapy. Immunotherapy 2017;9:289-302. 10.2217/imt-2016-0135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Palsson-McDermott EM, Curtis AM, Goel G, et al. Pyruvate kinase M2 regulates Hif-1α activity and IL-1β induction and is a critical determinant of the Warburg effect in LPS-activated macrophages. Cell Metab 2015;21:347. 10.1016/j.cmet.2015.01.017 [DOI] [PubMed] [Google Scholar]
- 67.Linke M, Fritsch SD, Sukhbaatar N, et al. mTORC1 and mTORC2 as regulators of cell metabolism in immunity. FEBS Lett 2017;591:3089-103. 10.1002/1873-3468.12711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Halbrook CJ, Pontious C, Kovalenko I, et al. Macrophage-released pyrimidines inhibit gemcitabine therapy in pancreatic cancer. Cell Metab 2019;29:1390-9.e6. 10.1016/j.cmet.2019.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cuccarese MF, Dubach JM, Pfirschke C, et al. Heterogeneity of macrophage infiltration and therapeutic response in lung carcinoma revealed by 3D organ imaging. Nat Commun 2017;8:14293. 10.1038/ncomms14293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cassetta L, Pollard JW. Targeting macrophages: therapeutic approaches in cancer. Nat Rev Drug Discov 2018;17:887-904. 10.1038/nrd.2018.169 [DOI] [PubMed] [Google Scholar]
- 71.Kuen J, Darowski D, Kluge T, et al. Pancreatic cancer cell/fibroblast co-culture induces M2 like macrophages that influence therapeutic response in a 3D model. PLoS One 2017;12:e0182039. 10.1371/journal.pone.0182039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Comito G, Giannoni E, Segura CP, et al. Cancer-associated fibroblasts and M2-polarized macrophages synergize during prostate carcinoma progression. Oncogene 2014;33:2423-31. 10.1038/onc.2013.191 [DOI] [PubMed] [Google Scholar]
- 73.Zhang J, Chen L, Xiao M, et al. FSP1+ fibroblasts promote skin carcinogenesis by maintaining MCP-1-mediated macrophage infiltration and chronic inflammation. Am J Pathol 2011;178:382-90. 10.1016/j.ajpath.2010.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Takahashi H, Sakakura K, Kudo T, et al. Cancer-associated fibroblasts promote an immunosuppressive microenvironment through the induction and accumulation of protumoral macrophages. Oncotarget 2017;8:8633-47. 10.18632/oncotarget.14374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Corzo CA, Condamine T, Lu L, et al. HIF-1α regulates function and differentiation of myeloid-derived suppressor cells in the tumor microenvironment. J Exp Med 2010;207:2439-53. 10.1084/jem.20100587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lesokhin AM, Hohl TM, Kitano S, et al. Monocytic CCR2 + myeloid-derived suppressor cells promote immune escape by limiting activated CD8 T-cell infiltration into the tumor microenvironment. Cancer Res 2012;72:876-86. 10.1158/0008-5472.CAN-11-1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Menetrier-Caux C, Montmain G, Dieu MC, et al. Inhibition of the differentiation of dendritic cells from CD34+ progenitors by tumor cells: role of interleukin-6 and macrophage colony- stimulating factor. Blood 1998;92:4778-91. 10.1182/blood.V92.12.4778 [DOI] [PubMed] [Google Scholar]
- 78.Bharadwaj U, Li M, Zhang R, et al. Elevated interleukin-6 and G-CSF in human pancreatic cancer cell conditioned medium suppress dendritic cell differentiation and activation. Cancer Res 2007;67:5479-88. 10.1158/0008-5472.CAN-06-3963 [DOI] [PubMed] [Google Scholar]
- 79.Wallace DC. Mitochondria and cancer: Warburg addressed. Cold Spring Harb Symp Quant Biol 2005;70:363-74. 10.1101/sqb.2005.70.035 [DOI] [PubMed] [Google Scholar]
- 80.Liberti MV, Locasale JW. The Warburg effect: how does it benefit cancer cells? Trends Biochem Sci 2016;41:287. 10.1016/j.tibs.2016.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dietl K, Renner K, Dettmer K, et al. Lactic acid and acidification inhibit TNF secretion and glycolysis of human monocytes. J Immunol 2010;184:1200-9. 10.4049/jimmunol.0902584 [DOI] [PubMed] [Google Scholar]
- 82.Liu D, Chang C, Lu N, et al. Comprehensive proteomics analysis reveals metabolic reprogramming of tumor-associated macrophages stimulated by the tumor microenvironment. J Proteome Res 2017;16:288-97. 10.1021/acs.jproteome.6b00604 [DOI] [PubMed] [Google Scholar]
- 83.Rabold K, Netea MG, Adema GJ, et al. Cellular metabolism of tumor-associated macrophages – functional impact and consequences. FEBS Lett 2017;591:3022-41. 10.1002/1873-3468.12771 [DOI] [PubMed] [Google Scholar]
- 84.Wang T, Liu H, Lian G, et al. HIF1α-induced glycolysis metabolism is essential to the activation of inflammatory macrophages. Mediators Inflamm 2017;2017:9029327. [DOI] [PMC free article] [PubMed]
- 85.Cheng SC, Quintin J, Cramer RA, et al. mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science 2014;345:1250684. 10.1126/science.1250684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ye H, Zhou Q, Zheng S, et al. Tumor-associated macrophages promote progression and the Warburg effect via CCL18/NF-kB/VCAM-1 pathway in pancreatic ductal adenocarcinoma. Cell Death Dis 2018;9:453. 10.1038/s41419-018-0486-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Penny HL, Sieow JL, Adriani G, et al. Warburg metabolism in tumor-conditioned macrophages promotes metastasis in human pancreatic ductal adenocarcinoma. Oncoimmunology 2016;5:e1191731. 10.1080/2162402X.2016.1191731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hirschhaeuser F, Sattler UGA, Mueller-Klieser W. Lactate: a metabolic key player in cancer. Cancer Res 2011;71:6921-5. 10.1158/0008-5472.CAN-11-1457 [DOI] [PubMed] [Google Scholar]
- 89.Lin S, Sun L, Lyu X, et al. Lactate-activated macrophages induced aerobic glycolysis and epithelial-mesenchymal transition in breast cancer by regulation of CCL5-CCR5 axis: a positive metabolic feedback loop. Oncotarget 2017;8:110426-43. 10.18632/oncotarget.22786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.de-Brito NM, Duncan-Moretti J, da-Costa HC, et al. Aerobic glycolysis is a metabolic requirement to maintain the M2-like polarization of tumor-associated macrophages. Biochim Biophys Acta Mol Cell Res 2020;1867:118604. [DOI] [PubMed]
- 91.Ip WKE, Hoshi N, Shouval DS, et al. Anti-inflammatory effect of IL-10 mediated by metabolic reprogramming of macrophages. Science 2017;356:513-9. 10.1126/science.aal3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.O’Neill LAJ, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nat Rev Immunol 2016;16:553-65. 10.1038/nri.2016.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mehta MM, Weinberg SE, Chandel NS. Mitochondrial control of immunity: Beyond ATP. Nat Rev Immunol 2017;17:608-20. 10.1038/nri.2017.66 [DOI] [PubMed] [Google Scholar]
- 94.Jones RG, Pearce EJ. MenTORing immunity: mTOR signaling in the development and function of tissue-resident immune cells. Immunity 2017;46:730-42. 10.1016/j.immuni.2017.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Baenke F, Peck B, Miess H, et al. Hooked on fat: the role of lipid synthesis in cancer metabolism and tumour development. Dis Model Mech 2013;6:1353-63. 10.1242/dmm.011338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ladanyi A, Mukherjee A, Kenny HA, et al. Adipocyte-induced CD36 expression drives ovarian cancer progression and metastasis. Oncogene 2018;37:2285-301. 10.1038/s41388-017-0093-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pascual G, Avgustinova A, Mejetta S, et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature 2017;541:41-5. 10.1038/nature20791 [DOI] [PubMed] [Google Scholar]
- 98.DeFilippis RA, Chang H, Dumont N, et al. CD36 repression activates a multicellular stromal program shared by high mammographic density and tumor tissues. Cancer Discov 2012;2:826-39. 10.1158/2159-8290.CD-12-0107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jin H, He Y, Zhao P, et al. Targeting lipid metabolism to overcome EMT-associated drug resistance via integrin β3/FAK pathway and tumor-associated macrophage repolarization using legumain-activatable delivery. Theranostics 2019;9:265-78. 10.7150/thno.27246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Niu Z, Shi Q, Zhang W, et al. Caspase-1 cleaves PPARγ for potentiating the pro-tumor action of TAMs. Nat Commun 2017;8:766. 10.1038/s41467-017-00523-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang Q, Wang H, Mao C, et al. Fatty acid oxidation contributes to IL-1β secretion in M2 macrophages and promotes macrophage-mediated tumor cell migration. Mol Immunol 2018;94:27-35. 10.1016/j.molimm.2017.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gross MI, Demo SD, Dennison JB, et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol Cancer Ther 2014;13;890-901. 10.1158/1535-7163.MCT-13-0870 [DOI] [PubMed] [Google Scholar]
- 103.Shroff EH, Eberlin LS, Dang VM, et al. MYC oncogene overexpression drives renal cell carcinoma in a mouse model through glutamine metabolism. Proc Natl Acad Sci U S A 2015;112:6539-44. 10.1073/pnas.1507228112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kung HN, Marks JR, Chi JT. Glutamine synthetase is a genetic determinant of cell type-specific glutamine independence in breast epithelia. PLoS Genet 2011;7:e1002229. 10.1371/journal.pgen.1002229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Choi J, Stradmann-Bellinghausen B, Yakubov E, et al. Glioblastoma cells induce differential glutamatergic gene expressions in human tumor-associated microglia/macrophages and monocyte-derived macrophages. Cancer Biol Ther 2015;16:1205-13. 10.1080/15384047.2015.1056406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu PS, Wang H, Li X, et al. α-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat Immunol 2017;18:985-94. 10.1038/ni.3796 [DOI] [PubMed] [Google Scholar]
- 107.Newsholme P, Curi R, Gordon S, et al. Metabolism of glucose, glutamine, long-chain fatty acids and ketone bodies by murine macrophages. Biochem J 1986;239:121-5. 10.1042/bj2390121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Palmieri EM, Menga A, Martín-Pérez R, et al. Pharmacologic or genetic targeting of glutamine synthetase skews macrophages toward an M1-like phenotype and inhibits tumor metastasis. Cell Rep 2017;20:1654-66. 10.1016/j.celrep.2017.07.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bantug GR, Galluzzi L, Kroemer G, et al. The spectrum of T cell metabolism in health and disease. Nat Rev Immunol 2018;18:19-34. 10.1038/nri.2017.99 [DOI] [PubMed] [Google Scholar]
- 110.Hartley GP, Chow L, Ammons DT, et al. Programmed cell death ligand 1 (PD-L1) signaling regulates macrophage proliferation and activation. Cancer Immunol Res 2018;6:1260-73. 10.1158/2326-6066.CIR-17-0537 [DOI] [PubMed] [Google Scholar]
- 111.Wagner J, Rapsomaniki MA, Chevrier S, et al. A single-cell atlas of the tumor and immune ecosystem of human breast cancer. Cell 2019;177:1330-45.e18. 10.1016/j.cell.2019.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lin H, Wei S, Hurt EM, et al. Host expression of PD-L1 determines efficacy of PD-L1 pathway blockade-mediated tumor regression. J Clin Invest 2018;128:805-15. 10.1172/JCI96113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lane RS, Femel J, Breazeale AP, et al. IFNγ-activated dermal lymphatic vessels inhibit cytotoxic T cells in melanoma and inflamed skin. J Exp Med 2018;215:3057-74. 10.1084/jem.20180654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang Y, Velez-Delgado A, Mathew E, et al. Myeloid cells are required for PD-1/PD-L1 checkpoint activation and the establishment of an immunosuppressive environment in pancreatic cancer. Gut 2017;66:124-36. 10.1136/gutjnl-2016-312078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Noman MZ, Desantis G, Janji B, et al. PD-L1 is a novel direct target of HIF-1α, and its blockade under hypoxia enhanced: MDSC-mediated T cell activation. J Exp Med 2014;211:781-90. 10.1084/jem.20131916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Palsson-McDermott EM, Dyck L, Zaslona Z, et al. Pyruvate kinase M2 is required for the expression of the immune checkpoint PD-L1 in immune cells and tumors. Front Immunol 2017;8:1300. 10.3389/fimmu.2017.01300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Prima V, Kaliberova LN, Kaliberov S, et al. COX2/mPGES1/PGE2 pathway regulates PD-L1 expression in tumor-associated macrophages and myeloid-derived suppressor cells. Proc Natl Acad Sci U S A 2017;114:1117-22. 10.1073/pnas.1612920114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Daurkin I, Eruslanov E, Stoffs T, et al. Tumor-associated macrophages mediate immunosuppression in the renal cancer microenvironment by activating the 15-lipoxygenase-2 pathway. Cancer Res 2011;71:6400-9. 10.1158/0008-5472.CAN-11-1261 [DOI] [PubMed] [Google Scholar]
- 119.Huang Q, Li F, Liu X, et al. Caspase 3-mediated stimulation of tumor cell repopulation during cancer radiotherapy. Nat Med 2011;17:860-6. 10.1038/nm.2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wen Z, Liu H, Li M, et al. Increased metabolites of 5-lipoxygenase from hypoxic ovarian cancer cells promote tumor-associated macrophage infiltration. Oncogene 2015;34:1241-52. 10.1038/onc.2014.85 [DOI] [PubMed] [Google Scholar]
- 121.Pennock ND, Martinson HA, Guo Q, et al. Ibuprofen supports macrophage differentiation, T cell recruitment, and tumor suppression in a model of postpartum breast cancer. J Immunother Cancer 2018;6:98. 10.1186/s40425-018-0406-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Labadie BW, Bao R, Luke JJ. Reimagining IDO pathway inhibition in cancer immunotherapy via downstream focus on the tryptophan-kynurenine-aryl hydrocarbon axis. Clin Cancer Res 2019;25:1462-71. 10.1158/1078-0432.CCR-18-2882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Geiger R, Rieckmann JC, Wolf T, et al. L-arginine modulates T cell metabolism and enhances survival and anti-tumor activity. Cell 2016;167:829-42.e13. 10.1016/j.cell.2016.09.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Montalbán Del Barrio I, Penski C, Schlahsa L, et al. Adenosine-generating ovarian cancer cells attract myeloid cells which differentiate into adenosine-generating tumor associated macrophages - a self-amplifying, CD39- and CD73-dependent mechanism for tumor immune escape. J Immunother Cancer 2016;4:49. 10.1186/s40425-016-0154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Häusler SFM, Montalbán Del Barrio I, Strohschein J, et al. Ectonucleotidases CD39 and CD73 on OvCA cells are potent adenosine-generating enzymes responsible for adenosine receptor 2A-dependent suppression of T cell function and NK cell cytotoxicity. Cancer Immunol Immunother 2011;60:1405-18. 10.1007/s00262-011-1040-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.d’Almeida SM, Kauffenstein G, Roy C, et al. The ecto-ATPDase CD39 is involved in the acquisition of the immunoregulatory phenotype by M-CSF-macrophages and ovarian cancer tumor-associated macrophages: regulatory role of IL-27. Oncoimmunology 2016;5:e1178025. 10.1080/2162402X.2016.1178025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Karakasheva TA, Waldron TJ, Eruslanov E, et al. CD38-expressing myeloid-derived suppressor cells promote tumor growth in a murine model of esophageal cancer. Cancer Res 2015;75:4074-85. 10.1158/0008-5472.CAN-14-3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kepp O, Loos F, Liu P, et al. Extracellular nucleosides and nucleotides as immunomodulators. Immunol Rev 2017;280:83-92. 10.1111/imr.12571 [DOI] [PubMed] [Google Scholar]
- 129.de la Cruz-López KG, Castro-Muñoz LJ, Reyes-Hernández DO, et al. Lactate in the regulation of tumor microenvironment and therapeutic approaches. Front Oncol 2019;9:1143. 10.3389/fonc.2019.01143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Haas R, Smith J, Rocher-Ros V, et al. Lactate regulates metabolic and proinflammatory circuits in control of T cell migration and effector functions. PLoS Biol 2015;13:e1002202. 10.1371/journal.pbio.1002202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Brand A, Singer K, Koehl GE, et al. LDHA-associated lactic acid production blunts tumor immunosurveillance by T and NK cells. Cell Metab 2016;24:657-71. 10.1016/j.cmet.2016.08.011 [DOI] [PubMed] [Google Scholar]
- 132.Harmon C, Robinson MW, Hand F, et al. Lactate-mediated acidification of tumor microenvironment induces apoptosis of liver-resident NK cells in colorectal liver metastasis. Cancer Immunol Res 2019;7:335-46. 10.1158/2326-6066.CIR-18-0481 [DOI] [PubMed] [Google Scholar]
- 133.Angelin A, Gil-de-Gómez L, Dahiya S, et al. Foxp3 reprograms T cell metabolism to function in low-glucose, high-lactate environments. Cell Metab 2017;25:1282-93.e7. 10.1016/j.cmet.2016.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell 2008;13:472-82. 10.1016/j.ccr.2008.05.005 [DOI] [PubMed] [Google Scholar]
- 135.Hashimoto O, Yoshida M, Koma Y, et al. Collaboration of cancer-associated fibroblasts and tumour-associated macrophages for neuroblastoma development. J Pathol 2016;240:211-23. 10.1002/path.4769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Erin N, Nizam E, Tanrıöver G, et al. Autocrine control of MIP-2 secretion from metastatic breast cancer cells is mediated by CXCR2: a mechanism for possible resistance to CXCR2 antagonists. Breast Cancer Res Treat 2015;150:57-69. 10.1007/s10549-015-3297-3 [DOI] [PubMed] [Google Scholar]
- 137.Song X, Wang Z, Jin Y, et al. Loss of miR-532-5p in vitro promotes cell proliferation and metastasis by influencing CXCL2 expression in HCC. Am J Transl Res 2015;7:2254-61. [PMC free article] [PubMed] [Google Scholar]
- 138.Kortlever RM, Sodir NM, Wilson CH, et al. Myc cooperates with ras by programming inflammation and immune suppression. Cell 2017;171:1301-15.e14. 10.1016/j.cell.2017.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chen S, Yang J, Wei Y, et al. Epigenetic regulation of macrophages: from homeostasis maintenance to host defense. Cell Mol Immunol 2020;17:36-49. 10.1038/s41423-019-0315-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Phan AT, Goldrath AW, Glass CK. Metabolic and epigenetic coordination of t cell and macrophage immunity. Immunity 2017;46:714-29. 10.1016/j.immuni.2017.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Deckert J, Struhl K. Histone acetylation at promoters is differentially affected by specific activators and repressors. Mol Cell Biol 2001;21:2726-35. 10.1128/MCB.21.8.2726-2735.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov 2013;12:847-65. 10.1038/nrd4140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yang X, Wang X, Liu D, et al. Epigenetic regulation of macrophage polarization by DNA methyltransferase 3b. Mol Endocrinol 2014;28:565-74. 10.1210/me.2013-1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kittan NA, Allen RM, Dhaliwal A, et al. Cytokine induced phenotypic and epigenetic signatures are key to establishing specific macrophage phenotypes. PLoS One 2013;8:e78045. 10.1371/journal.pone.0078045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ishii M, Wen H, Corsa CAS, et al. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood 2009;114:3244-54. 10.1182/blood-2009-04-217620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cao Q, Rong S, Repa JJ, et al. Histone deacetylase 9 represses cholesterol efflux and alternatively activated macrophages in atherosclerosis development. Arterioscler Thromb Vasc Biol 2014;34:1871-9. 10.1161/ATVBAHA.114.303393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rodríguez-Prados JC, Través PG, Cuenca J, et al. Substrate fate in activated macrophages: a comparison between innate, classic, and alternative activation. J Immunol 2010;185:605-14. 10.4049/jimmunol.0901698 [DOI] [PubMed] [Google Scholar]
- 148.Latham T, MacKay L, Sproul D, et al. Lactate, a product of glycolytic metabolism, inhibits histone deacetylase activity and promotes changes in gene expression. Nucleic Acids Res 2012;40:4794-803. 10.1093/nar/gks066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Liu TF, Vachharajani VT, Yoza BK, et al. NAD+ -dependent sirtuin 1 and 6 proteins coordinate a switch from glucose to fatty acid oxidation during the acute inflammatory response. J Biol Chem 2012;287:25758-69. 10.1074/jbc.M112.362343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wellen KE, Hatzivassiliou G, Sachdeva UM, et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 2009;324:1076-80. 10.1126/science.1164097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lampropoulou V, Sergushichev A, Bambouskova M, et al. Itaconate links inhibition of succinate dehydrogenase with macrophage metabolic remodeling and regulation of inflammation. Cell Metab 2016;24:158-66. 10.1016/j.cmet.2016.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Vats D, Mukundan L, Odegaard JI, et al. Oxidative metabolism and PGC-1β attenuate macrophage-mediated inflammation. Cell Metab 2006;4:13-24. 10.1016/j.cmet.2006.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jha AK, Huang SCC, Sergushichev A, et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 2015;42:419-30. 10.1016/j.immuni.2015.02.005 [DOI] [PubMed] [Google Scholar]
- 154.Huang SCC, Smith AM, Everts B, et al. Metabolic reprogramming mediated by the mTORC2-IRF4 signaling axis is essential for macrophage alternative activation. Immunity 2016;45:817-30. 10.1016/j.immuni.2016.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pasini A, Caldarera CM, Giordano E. Chromatin remodeling by polyamines and polyamine analogs. Amino Acids 2014;46:595-603. 10.1007/s00726-013-1550-9 [DOI] [PubMed] [Google Scholar]
- 156.De Palma M, Venneri MA, Galli R, et al. Tie2 identifies a hematopoietic lineage of proangiogenic monocytes required for tumor vessel formation and a mesenchymal population of pericyte progenitors. Cancer Cell 2005;8:211-26. 10.1016/j.ccr.2005.08.002 [DOI] [PubMed] [Google Scholar]
- 157.De Palma M, Venneri MA, Roca C, et al. Targeting exogenous genes to tumor angiogenesis by transplantation of genetically modified hematopoietic stem cells. Nat Med 2003;9:789-95. 10.1038/nm871 [DOI] [PubMed] [Google Scholar]
- 158.Movahedi K, Laoui D, Gysemans C, et al. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes. Cancer Res 2010;70:5728-39. 10.1158/0008-5472.CAN-09-4672 [DOI] [PubMed] [Google Scholar]
- 159.Laviron M, Boissonnas A. Ontogeny of tumor-associated macrophages. Front Immunol 2019;10:1799. 10.3389/fimmu.2019.01799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.DeNardo DG, Ruffell B. Macrophages as regulators of tumour immunity and immunotherapy. Nat Rev Immunol 2019;19:369-82. 10.1038/s41577-019-0127-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ruffell B, Coussens LM. Macrophages and therapeutic resistance in cancer. Cancer Cell 2015;27:462-72. 10.1016/j.ccell.2015.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sanford DE, Belt BA, Panni RZ, et al. Inflammatory monocyte mobilization decreases patient survival in pancreatic cancer: a role for targeting the CCL2/CCR2 axis. Clin Cancer Res 2013;19:3404-15. 10.1158/1078-0432.CCR-13-0525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mantovani A, Marchesi F, Malesci A, et al. Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol 2017;14:399-416. 10.1038/nrclinonc.2016.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Yun J, Rago C, Cheong I, et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science 2009;325:1555-9. 10.1126/science.1174229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Evans MJ, Saghatelian A, Sorensen EJ, et al. Target discovery in small-molecule cell-based screens by in situ proteome reactivity profiling. Nat Biotechnol 2005;23:1303-7. 10.1038/nbt1149 [DOI] [PubMed] [Google Scholar]
- 166.Clem B, Telang S, Clem A, et al. Small-molecule inhibition of 6-phosphofructo-2-kinase activity suppresses glycolytic flux and tumor growth. Mol Cancer Ther 2008;7:110-20. 10.1158/1535-7163.MCT-07-0482 [DOI] [PubMed] [Google Scholar]
- 167.Le A, Cooper CR, Gouw AM, et al. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci U S A 2010;107:2037-42. 10.1073/pnas.0914433107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sutendra G, Michelakis ED. Pyruvate dehydrogenase kinase as a novel therapeutic target in oncology. Front Oncol 2013;3:38. 10.3389/fonc.2013.00038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhao X, Jiang P, Deng X, et al. Inhibition of mTORC1 signaling sensitizes hepatocellular carcinoma cells to glycolytic stress. Am J Cancer Res 2016;6:2289-98. [PMC free article] [PubMed] [Google Scholar]
- 170.Schoors S, De Bock K, Cantelmo AR, et al. Partial and transient reduction of glycolysis by PFKFB3 blockade reduces pathological angiogenesis. Cell Metab 2014;19:37-48. 10.1016/j.cmet.2013.11.008 [DOI] [PubMed] [Google Scholar]
- 171.Del Rey MJ, Valín Á, Usategui A, et al. Hif-1α knockdown reduces glycolytic metabolism and induces cell death of human synovial fibroblasts under normoxic conditions. Sci Rep 2017;7:3644. 10.1038/s41598-017-03921-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Singh D, Arora R, Kaur P, et al. Overexpression of hypoxia-inducible factor and metabolic pathways: Possible targets of cancer. Cell Biosci 2017;7:62. 10.1186/s13578-017-0190-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Brown JM, Recht L, Strober S. The promise of targeting macrophages in cancer therapy. Clin Cancer Res 2017;23:3241-50. 10.1158/1078-0432.CCR-16-3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Curran MA, Montalvo W, Yagita H, et al. PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proc Natl Acad Sci U S A 2010;107:4275-80. 10.1073/pnas.0915174107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ngambenjawong C, Gustafson HH, Pun SH. Progress in tumor-associated macrophage (TAM)-targeted therapeutics. Adv Drug Deliv Rev 2017;114:206-21. 10.1016/j.addr.2017.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lin Y, Xu J, Lan H. Tumor-associated macrophages in tumor metastasis: Biological roles and clinical therapeutic applications. J Hematol Oncol 2019;12:76. 10.1186/s13045-019-0760-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kühnemuth B, Mühlberg L, Schipper M, et al. CUX1 modulates polarization of tumor-associated macrophages by antagonizing NF-κB signaling. Oncogene 2015;34:177-87. 10.1038/onc.2013.530 [DOI] [PubMed] [Google Scholar]
- 178.Hagemann T, Lawrence T, McNeish I, et al. “Re-educating” tumor-associated macrophages by targeting NF-κB. J Exp Med 2008;205:1261-8. 10.1084/jem.20080108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Goulielmaki E, Bermudez-Brito M, Andreou M, et al. Pharmacological inactivation of the PI3K p110δ prevents breast tumour progression by targeting cancer cells and macrophages article. Cell Death Dis 2018;9:678. 10.1038/s41419-018-0717-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Piaggio F, Kondylis V, Pastorino F, et al. A novel liposomal Clodronate depletes tumor-associated macrophages in primary and metastatic melanoma: anti-angiogenic and anti-tumor effects. J Control Release 2016;223:165-77. 10.1016/j.jconrel.2015.12.037 [DOI] [PubMed] [Google Scholar]
- 181.Chao MP, Alizadeh AA, Tang C, et al. Anti-CD47 antibody synergizes with rituximab to promote phagocytosis and eradicate non-Hodgkin lymphoma. Cell 2010;142:699-713. 10.1016/j.cell.2010.07.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Shi Y, Fan X, Deng H, et al. Trastuzumab triggers phagocytic killing of high HER2 cancer cells in vitro and in vivo by interaction with Fcγ receptors on macrophages. J Immunol 2015;194:4379-86. 10.4049/jimmunol.1402891 [DOI] [PubMed] [Google Scholar]
- 183.Zhang W, Huang Q, Xiao W, et al. Advances in anti-tumor treatments targeting the CD47/SIRPα axis. Front Immunol 2020;11:18. 10.3389/fimmu.2020.00018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Zhang X, Fan J, Ju D. Insights into CD47/SIRPα axis-targeting tumor immunotherapy. Antib Ther 2018;1:27-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Weiskopf K, Jahchan NS, Schnorr PJ, et al. CD47-blocking immunotherapies stimulate macrophage-mediated destruction of small-cell lung cancer. J Clin Invest 2016;126:2610-20. 10.1172/JCI81603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Petrova PS, Viller NN, Wong M, et al. TTI-621 (SIRPαFc): a CD47-blocking innate immune checkpoint inhibitor with broad antitumor activity and minimal erythrocyte binding. Clin Cancer Res 2017;23:1068-79. 10.1158/1078-0432.CCR-16-1700 [DOI] [PubMed] [Google Scholar]
- 187.Zanganeh S, Hutter G, Spitler R, et al. Iron oxide nanoparticles inhibit tumour growth by inducing pro-inflammatory macrophage polarization in tumour tissues. Nat Nanotechnol 2016;11:986-94. 10.1038/nnano.2016.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Li Y, Cao F, Li M, et al. Hydroxychloroquine induced lung cancer suppression by enhancing chemo-sensitization and promoting the transition of M2-TAMs to M1-like macrophages. J Exp Clin Cancer Res 2018;37:259. 10.1186/s13046-018-0938-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Shan M, Qin J, Jin F, et al. Autophagy suppresses isoprenaline-induced M2 macrophage polarization via the ROS/ERK and mTOR signaling pathway. Free Radic Biol Med 2017;110:432-43. 10.1016/j.freeradbiomed.2017.05.021 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as