Abstract
Background
The prognostic roles of granulocyte-/granulocyte-macrophage colony-stimulating factor (G-/GM-CSF) and its receptors (CSFRs) from the genomic perspective remain controversial. The aim of our study was to evaluate their prognostic value in multiple cancer types by analyzing omics data.
Methods
The omics data of G-/GM-CSF and receptors were obtained from the cBioportal database. Cutoff values were determined by X-tile. Overall survival (OS) was assessed by Kaplan–Meier curves. Differentially expressed genes (DEGs) and common regulated genes were analyzed using R software and Venny 2.1.0, while enrichment pathway analyses were performed by Metascape.
Results
A comprehensive mRNA analysis was performed in 8,565 patients across 24 cancer types. The combination subgroup of CSF2 and its receptors with high expression and favorable prognosis was associated with the activation of immune-related pathways, while the subgroup with unfavorable prognosis was associated with the activation of inflammatory and cellular pathways. As for the combination subgroup of CSF3 and its receptor, the high expression and poor prognosis subgroup was accompanied by the activation of inflammation and signaling transduction pathways.
Conclusions
The prognostic value of CSFs and CSFRs are cancer-type dependent. Therefore, personalized risk stratification based on CSF and CSFR pathway should be considered for cancer patients.
Keywords: Colony-stimulating factors (CSFs), colony-stimulating factor receptors (CSFRs), survival analysis, differentially expressed genes, enrichment pathway
Introduction
Granulocyte-macrophage colony-stimulating factor (GM-CSF, also called CSF2) and granulocyte-colony stimulating factor (G-CSF, also called CSF3) are members of the colony-stimulating factor (CSF) superfamily which are defined as growth factors capable of stimulating the proliferation, differentiation, survival, and function of macrophages, dendritic cells (DCs), monocytes, neutrophils, and eosinophils. They have established themselves as effective antitumor drugs in components of cellular vaccines for adjuvant radiotherapy and chemotherapy to stimulate the maturation, proliferation, and differentiation of a variety of blood cells and alleviate bone marrow suppression (1-3).
GM-CSF (or CSF2) and G-CSF (or CSF3) are secreted as monomeric glycoproteins (4). As widely expressed cytokines, CSF’s and its receptors’ effect in different tumors is considerably complex and remains controversial. Recent studies have shown that GM-CSF can impair antitumor immune responses and has an immunosuppressive effect by expanding myeloid-derived suppressor cells (MDSCs) and stimulating FoxP3+ regulatory T cells (Tregs) in the blood and tumor microenvironment (5-7). GM-CSF has antitumor potential by inducing the differentiation of macrophages and monocytes, enhancing the natural cytotoxicity, and augmenting antibody-dependent cellular cytotoxicity. It can enhance antitumor efficacy by modulating T-cell infiltration in the tumor microenvironment and facilitating antigen recognition (8-10). Cancer vaccinations using GM-CSF as an immune adjuvant have been applied in clinical trials (11,12). G-CSF can promote tumor proliferation, migration, and angiogenesis, and is frequently associated with a poor clinical outcome in multiple cancer types (13-20).
CSFs and colony-stimulating factor receptors (CSFRs) play important roles in cancer biology; however, the expression and prognostic value of CSFs and CSFRs remain inconsistent across different solid tumors, and the molecular mechanisms behind these potential effects have not been clarified. Therefore, we retrospectively reviewed a large cohort of 8565 patients across 24 cancer types using omics data in order to facilitate personalized risk stratification and provide a theoretical basis for the effect of CSFs and CSFRs on different cancers. We present the following article in accordance with the Materials Design Analysis Reporting (MDAR) reporting checklist (available at http://dx.doi.org/10.21037/atm-20-5363).
Methods
Data collection and preprocessing
Genomic data and clinicopathologic data including age, gender, race, and American Joint Committee on Cancer (AJCC) stage for 24 The Cancer Genome Atlas (TCGA) cancer types were downloaded from the cBioportal data database (http://www.cBioportalportal.org/). We classified the single ligand/receptor group into two (high and low) subgroups and divided the ligand-receptor binding group into three subgroups (low, low ligand and low receptor expression; high, high ligand and high receptor expression; moderate, the remaining cohort) according to CSF and CSFR expression using X-tile to calculate the optimal cutoff values.
Functional pathway enrichment analysis
Differentially expressed genes (DEGs) of each subgroup were identified by R software using “edgeR” package (R Foundation for Statistical Computing, Vienna, Austria). The upregulated and downregulated genes of each cancer type were obtained. The Venn diagrams of common DEGs of each cancer type that had a consistent prognostic value were then generated using Venny 2.1.0 (http://bioinfogp.cnb.csic.es/tools/venny/index.html). Pathway enrichment was then assessed with input of common DEGs using Metascape (http://metascape.org/gp/index.html#/main/step1) to complete the pathway enrichment and biological process annotation. A heatmap and network of enriched terms across input gene lists were generated. For each input gene list, Metascape referred to the following data resources when conducting pathway and biological process enrichment analysis: Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway; Gene Ontology (GO) Biological Processes; Reactome Gene Sets; Canonical Pathways; Classical Pathways Canonical Pathways; the CORUM mammalian protein complex database. A gene list analysis report was created and visualized.
Statistical analysis
All statistical analyses and data plotting were performed using IBM SPSS version 22 (IBM Corporation, Armonk, NY, USA) and R software version 3.5.0 with the “edgeR” package (R Foundation for Statistical Computing, Vienna, Austria). Crosstab with Chi-square test was used to summarize the correlation between CSF and CSFR expression and clinicopathological parameters. Any association of CSF and CSFR expression with overall survival (OS) was determined using the Kaplan-Meier analysis after grouping. The Cox proportional hazards regression model was applied to perform univariate and univariate analyses. A statistically significant difference was defined as a P value <0.05.
Ethical statement
This study was approved by The First Affiliated Hospital of Shandong First Medical University review board (No. S033). Informed consent from patients was waived due to the anonymity of individual patient data. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Results
Patient characteristics
The following solid tumor types were selected: adrenocortical carcinoma (ACC, N=79), cholangiocarcinoma (CHOL, N=36), bladder urothelial carcinoma (BLCA, N=407), colorectal adenocarcinoma (COAD, N=376), breast cancer (BRCA, N=1,093), lower grade glioma (LGG, N=515), glioblastoma multiforme (GBM, N=137), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC, N=304), esophageal carcinoma (ESCA, N=184), stomach adenocarcinoma (STAD, N=415), head and neck squamous cell carcinoma (HNSC, N=520), kidney renal clear cell carcinoma (KIRC, N=533), liver hepatocellular carcinoma (LIHC, N=371), lung adenocarcinoma (LUAD, N=514), lung squamous cell carcinoma (LUSC, N=501), ovarian serous cystadenocarcinoma (OV, N=303), pancreatic adenocarcinoma (PAAD, N=178), prostate adenocarcinoma (PRAD, N=497), sarcoma (SARC, N=258), skin cutaneous melanoma (SKCM, N=467), thyroid carcinoma (THCA, N=501), thymoma (THYM, N=120), uterine corpus endometrial carcinoma (UCEC, N=176), and uterine carcinosarcoma (UCS, N=80). Altogether, data on 24 cancer type samples (N=8,565) were downloaded from the cBioportal database (Table 1).
Table 1. Correlations between CSFs and CSFRs and clinicopathological features across 24 cancer types.
| Variables | All (N=8,565) | CSF2, N (%) | CSF3, N (%) | |||||
|---|---|---|---|---|---|---|---|---|
| Low | High | P value | Low | High | P value | |||
| Sex | 0.980 | <0.001 | ||||||
| Female | 4,354 (50.8) | 2,798 (50.8) | 1,556 (50.8) | 2,716 (53.2) | 1,638 (47.3) | |||
| Male | 4,211 (49.2) | 2,705 (49.2) | 1,506 (49.2) | 2,386 (46.8) | 1,825 (52.7) | |||
| Age | <0.001 | 0.009 | ||||||
| <65 | 5,248 (61.3) | 3,476 (63.2) | 1,772 (57.9) | 3,068 (60.1) | 2,180 (63.0) | |||
| ≥65 | 3,317 (38.7) | 2,027 (36.8) | 1,290 (42.1) | 2,034 (39.9) | 1,283 (37.0) | |||
| Race | 0.871 | 0.002 | ||||||
| Asian | 535 (6.2) | 342 (6.2) | 193 (6.3) | 353 (6.9) | 182 (5.3) | |||
| Others | 8,030 (93.8) | 2,869 (93.7) | 5,161 (93.8) | 4,749 (93.1) | 3,281 (94.7) | |||
| AJCC stage | <0.001 | <0.001 | ||||||
| I–II | 3,527 (41.2) | 2,161 (39.3) | 1,366 (44.6) | 2,409 (47.2) | 1,118 (32.3) | |||
| III–IV | 2,065 (24.1) | 1,176 (21.4) | 889 (29.0) | 1,313 (25.7) | 752 (21.7) | |||
| NA | 2,973 (34.7) | 2,166 (39.4) | 807 (26.4) | 1,380 (27.0) | 1,593 (46.0) | |||
CSF, colony-stimulating factor; CSFR, colony-stimulating factor receptor; CSF2, colony-stimulating factor 2; CSF2RA, colony-stimulating factor 2 receptor A; CSF2RB, colony-stimulating factor 2 receptor B; CSF3, colony-stimulating factor 3; CSF3R, granulocyte-colony stimulating factor 3 receptor; NA, not available; AJCC, American Joint Committee on Cancer, AJCC is a classification system developed by the American Joint Committee on Cancer for describing the extent of disease progression in cancer patients.
Evaluating the prognostic value of CSFs and CSFRs in 24 types of cancer based on the cBioportal database
To evaluate the prognostic value of CSFs and CSFRs in the 24 types of cancer, Kaplan-Meier analyses were performed. The distributions of expression level showed large variations across different samples from most of the 24 TCGA cancer types (Table 2). High CSF2 indicated poor prognosis in 6 (43%) cancer types including CESC (HR: 2.096; 95% CI: 1.312–3.349; P=0.002), ESCA (HR: 2.477; 95% CI: 1.321–4.645; P=0.005), HNSC (HR: 1.582; 95% CI: 1.179–2.122; P=0.002), etc. However, high CSF2 expression predicted favorable survival in 8 (57%) cancer types including BRCA (HR: 0.408; 95% CI: 0.208–0.801; P=0.009), BLCA (HR: 0.681; 95% CI: 0.467–0.995; P=0.047), LUAD (HR: 0.529; 95% CI: 0.36–0.776; P=0.001), etc. High expression of CSF2RA was an unfavorable prognostic factor of OS in 5 (45%) cancer types including COAD (HR: 1.676; 95% CI: 1.027–2.734; P=0.039), ESCA (HR: 2.217; 95% CI: 1.252–3.928; P=0.006), LUSC (HR: 1.375; 95% CI: 1.026–1.842; P=0.033), etc. In contrast, it was identified as a favorable prognostic factor in 6 (55%) cancer types including CESC (HR: 0.388; 95% CI: 0.238–0.633; P<0.001), HNSC (HR: 0.554; 95% CI: 0.413–0.745; P<0.001), SKCM (HR: 0.585; 95% CI: 0.448–0.764; P<0.001), etc. High CSF2RB expression was associated with poor prognosis in 8 (47%) cancer types, including BLCA (HR: 1.567; 95% CI: 1.026–2.393; P=0.038), ESCA (HR: 3.132; 95% CI: 1.357–7.226; P=0.007), GBM (HR: 1.699; 95% CI: 1.016–2.839; P=0.043), etc.; meanwhile, it was associated with favorable prognosis in 9 (53%) cancer types including BRCA (HR: 0.687; 95% CI: 0.489–0.964; P=0.030), CESC (HR: 0.266; 95% CI: 0.107–0.66; P=0.004), COAD (HR: 0.619; 95% CI: 0.401–0.954; P=0.030), etc.
Table 2. The effects of CSFs and CSFRs on overall survival across 24 cancer types by multivariate Cox regression model.
| Cancer types | CSF2 | CSF2RA | CSF2RB | CSF3 | CSF3R | CSF2 + CSF2RA | CSF2 + CSF2RB | CSF3 + CSF3R | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | ||||||||
| BRCA | 0.408 (0.208–0.801) | 0.009 | 0.713 (0.486–1.045) | 0.083 | 0.687 (0.489–0.964) | 0.03 | 0.702 (0.509–0.968) | 0.031 | 0.653 (0.469–0.909) | 0.010 | 0.313 (0.145–0.679) | 0.003 | 0.323 (0.142–0.739) | 0.007 | 0.488 (0.311–0.767) | 0.002 | |||||||
| BLCA | 0.681 (0.467–0.995) | 0.047 | 1.378 (0.959–1.982) | 0.073 | 1.567 (1.026–2.393) | 0.038 | 1.714 (1.216–2.414) | 0.002 | 1.591 (1.040–2.434) | 0.032 | 0.948 (0.572–1.569) | 0.834 | 1.702 (0.794–3.644) | 0.171 | 2.857 (1.537–5.310) | 0.001 | |||||||
| CESC | 2.096 (1.312–3.349) | 0.002 | 0.388 (0.238–0.633) | <0.001 | 0.266 (0.107–0.66) | 0.004 | 1.782 (1.053–3.017) | 0.031 | 0.536 (0.320–0.899) | 0.018 | 0.877 (0.436–1.764) | 0.714 | 1.494 (0.152–1.603) | 0.241 | 0.948 (0.445–2.020) | 0.889 | |||||||
| COAD | 1.411 (0.913–2.181) | 0.121 | 1.676 (1.027–2.734) | 0.039 | 0.619 (0.401–0.954) | 0.03 | 1.549 (1.002–2.394) | 0.049 | 0.658 (0.380–1.136) | 0.133 | 2.151 (1.128–4.101) | 0.020 | 0.930 (0.521–1.661) | 0.807 | 1.152 (0.554–2.392) | 0.705 | |||||||
| ESCA | 2.477 (1.321–4.645) | 0.005 | 2.217 (1.252–3.928) | 0.006 | 3.132 (1.357–7.226) | 0.007 | 0.607 (0.289–1.275) | 0.187 | 1.876 (1.078–3.266) | 0.026 | 6.362 (2.768–14.624) | <0.001 | 8.016 (2.936–21.885) | <0.001 | 1.212 (0.514–2.858) | 0.661 | |||||||
| HNSC | 1.582 (1.179–2.122) | 0.002 | 0.554 (0.413–0.745) | <0.001 | 0.605 (0.463–0.790) | <0.001 | 1.395 (1.060–1.829) | 0.016 | 1.675 (1.144–2.453) | 0.008 | 1.072 (0.623–1.844) | 0.801 | 1.040 (0.673–1.605) | 0.861 | 2.033 (1.316–3.139) | 0.001 | |||||||
| LUAD | 0.529 (0.360–0.776) | 0.001 | 0.762 (0.538–1.079) | 0.126 | 0.530 (0.367–0.767) | 0.001 | 0.707 (0.459–1.087) | 0.114 | 0.589 (0.439–0.790) | <0.001 | 0.514 (0.295–0.895) | 0.019 | 0.342 (0.208–0.562) | <0.001 | 0.462 (0.278–0.766) | 0.003 | |||||||
| LUSC | 2.066 (1.393–3.064) | <0.001 | 1.375 (1.026–1.842) | 0.033 | 1.349 (0.937–1.942) | 0.108 | 1.441 (1.009–2.059) | 0.044 | 1.375 (1.041–1.815) | 0.025 | 3.175 (1.817–5.547) | <0.001 | 4.572 (2.128–9.822) | <0.001 | 1.761 (1.139–2.724) | 0.011 | |||||||
| PRAD | 0.294 (0.036–2.398) | 0.253 | 3.994 (0.504–31.626) | 0.19 | 0.490 (0.142–1.693) | 0.259 | 0.151 (0.039–0.588) | 0.006 | 0.593 (0.167–2.112) | 0.420 | 0.232 (0.024–2.237) | 0.206 | 0.290 (0.030–2.794) | 0.284 | 0.690 (0.077–6.181) | 0.740 | |||||||
| SKCM | 0.641 (0.492–0.835) | 0.001 | 0.585 (0.448–0.764) | <0.001 | 0.568 (0.435–0.742) | <0.001 | 1.194 (0.906–1.573) | 0.209 | 0.587 (0.449–0.767) | <0.001 | 0.458 (0.323–0.649) | <0.001 | 0.488 (0.352–0.680) | <0.001 | 1.761 (1.139–2.724) | 0.011 | |||||||
| STAD | 0.793 (0.564–1.115) | 0.182 | 0.825 (0.633–1.077) | 0.157 | 0.866 (0.672–1.117) | 0.268 | 0.846 (0.611–1.170) | 0.311 | 0.886 (0.687–1.143) | 0.350 | 0.693 (0.453–1.060) | 0.091 | 0.780 (0.511–1.191) | 0.250 | 0.753 (0.487–1.163) | 0.201 | |||||||
| UCEC | 0.633 (0.312–1.284) | 0.205 | 0.500 (0.247–1.013) | 0.054 | 0.278 (0.123–0.631) | 0.002 | 0.634 (0.304–1.318) | 0.222 | 2.456 (1.133–5.325) | 0.023 | 0.346 (0.139–0.859) | 0.022 | 0.296 (0.107–0.814) | 0.018 | 1.471 (0.545–3.968) | 0.446 | |||||||
| ACC | 0.543 (0.206–1.430) | 0.216 | 0.347 (0.131–0.923) | 0.034 | 0.275 (0.104–0.729) | 0.009 | 2.318 (1.096–4.905) | 0.028 | 0.445 (0.188–1.052) | 0.065 | 0.300 (0.070–1.288) | 0.105 | 0.329 (0.098–1.106) | 0.072 | 1.002 (0.220–4.567) | 0.998 | |||||||
| CHOL | 0.238 (0.064–0.888) | 0.033 | 0.723 (0.161–3.251) | 0.672 | 1.629 (0.591–4.492) | 0.345 | 0.506 (0.173–1.481) | 0.214 | 0.758 (0.283–2.028) | 0.581 | 0.273 (0.043–1.722) | 0.167 | 0.544 (0.134–2.204) | 0.393 | 0.507 (0.146–1.757) | 0.284 | |||||||
| GBM | 0.865 (0.538–1.390) | 0.549 | 0.527 (0.286–0.972) | 0.04 | 1.699 (1.016–2.839) | 0.043 | 1.335 (0.881–2.023) | 0.174 | 1.735 (1.035–2.910) | 0.037 | 0.629 (0.253–1.961) | 0.317 | 1.779 (0.849–3.122) | 0.127 | 1.810 (0.967–3.388) | 0.064 | |||||||
| KIRC | 2,836 (2.019–3.983) | <0.001 | 1.538 (0.891–2.657) | 0.122 | 1.828 (1.225–2.727) | 0.003 | 1.592 (1.175–2.156) | 0.003 | 2.323 (1.573–3.431) | <0.001 | 4.153 (2.137–8.072) | <0.001 | 3.090 (1.737–5.497) | <0.001 | 3.526 (2.066–6.019) | <0.001 | |||||||
| LGG | 1.338 (0.545–3.284) | 0.526 | 1.572 (1.074–2.300) | 0.02 | 2.207 (1.440–3.383) | <0.001 | 0.543 (0.376–0.784) | 0.001 | 2.220 (1.552–3.177) | <0.001 | 1.645 (0.584–4.631) | 0.346 | 1.224 (0.300–4.994) | 0.778 | 1.203 (0.724–1.997) | 0.476 | |||||||
| LIHC | 0.651 (0.433–0.980) | 0.04 | 1.533 (0.984–2.389) | 0.059 | 0.691 (0.439–1.086) | 0.109 | 0.704 (0.463–1.070) | 0.100 | 1.698 (1.081–2.668) | 0.022 | 1.002 (0.579–1.732) | 0.995 | 0.510 (0.289–0.899) | 0.020 | 1.156 (0.502–2.660) | 0.734 | |||||||
| OV | 0.558 (0.323–0.964) | 0.036 | 1.308 (0.916–1.868) | 0.14 | 1.484 (0.939–2.345) | 0.091 | 1.216 (0.816–1.812) | 0.337 | 1.035 (0.729–1.471) | 0.846 | 0.805 (0.355–1.826) | 0.603 | 0.592 (0.219–1.600) | 0.301 | 2.209 (0.964–5.060) | 0.061 | |||||||
| PAAD | 3.078 (1.416–6.690) | 0.005 | 1.796 (0.898–3.690) | 0.098 | 0.736 (0.448–1.211) | 0.228 | 0.744 (0.489–1.130) | 0.166 | 1.773 (0.914–3.443) | 0.090 | 5.317 (1.297–21.790) | 0.020 | 2.019 (0.846–4.815) | 0.113 | 5.317 (1.297–21.790) | 0.020 | |||||||
| SARC | 0.748 (0.493–1.136) | 0.173 | 0.646 (0.430–0.970) | 0.035 | 0.434 (0.284–0.664) | <0.001 | 0.658 (0.415–1.043) | 0.075 | 0.331 (0.134–0.814) | 0.016 | 0.594 (0.358–0.984) | 0.043 | 0.431 (0.255–0.729) | 0.002 | 0.164 (0.023–1.184) | 0.073 | |||||||
| THCA | 0.185 (0.064–0.540) | 0.002 | 0.636 (0.221–1.835) | 0.403 | 3.545 (1.231–10.206) | 0.019 | 2.824 (1.015–7.858) | 0.047 | 1.800 (0.652–4.971) | 0.257 | 0.184 (0.057–0.595) | 0.005 | 0.682 (0.196–2.375) | 0.547 | 4.189 (1.035–16.951) | 0.045 | |||||||
| THYM | 1.429 (0.863–2.365) | 0.165 | 1.002 (0.999–1.005) | 0.172 | 1.001 (1.001–1.002) | 0.001 | 1.002 (1.001–1.004) | 0.007 | 1.004 (1.002–1.006) | 0.001 | 0.219 (0.045–1.079) | 0.062 | 6,964.802 (0.001–1.307E+122) | 0.949 | 1,122.132 (0.001–1.706E+177) | 0.973 | |||||||
| UCS | 4.879 (0.971–24.526) | 0.054 | 6.694 (1.769–25.332) | 0.005 | 1.013 (1.003–1.023) | 0.009 | 5.420 (1.550–18.943) | 0.008 | 1.016 (1.005–1.027) | 0.004 | 7.750 (0.771–77.880) | 0.082 | 5.770 (1.155–28.830) | 0.033 | 17.561 (2.754–111.976) | 0.002 | |||||||
All the P values were calculated by multivariate Cox regression model. Multivariate analysis showing HRs and P values for 24 cancer types. CSF, colony-stimulating factor; CSFR, colony-stimulating factor receptor; HR, hazard ratio; CI, confidence interval.
High CSF3 predicted bad outcome in 10 (77%) cancer types, including BLCA (HR: 1.714; 95% CI: 1.216–2.414; P=0.002), CESC (HR: 1.782 95% CI: 1.503–3.017; P=0.031), COAD (HR: 1.549; 95% CI: 1.002–2.394; P=0.049), etc.; meanwhile, it was associated with favorable outcome in 3 (23%) cancer types including BRCA (HR: 0.702; 95% CI: 0.509–0.968; P=0.031), PRAD (HR: 0.151; 95% CI: 0.039–0.588; P=0.006), and LGG (HR: 0.543; 95% CI: 0.376–0.784). High CSF3R expression indicated poor prognosis in 11 (69%) cancer types including BLCA (HR: 1.591; 95% CI: 1.040–2.434; P=0.032), CESC (HR: 1.536; 95% CI: 0.320–0.899; P=0.018), ESCA (HR: 1.876; 95% CI: 1.078–3.266; P=0.026), etc. Conversely, high CSF3R expression indicated good prognosis in 5 (31%) cancer types including BRCA (HR: 0.653; 95% CI: 0.469–0.909; P=0.010), LUAD (HR: 0.589; 95% CI: 0.439–0.790; P<0.001), CESC (HR: 0.536; 95% CI: 0.320–0.899), etc. (Table 2).
We further analyzed the prognostic value of the combination subgroups of CSFs and CSFRs. High expression of CSF2 and CSF2RA indicated a favorable prognosis for BRCA (HR: 0.313; 95% CI: 0.145–0.679; P=0.003), SKCM (HR: 0.458; 95% CI: 0.323–0.649; P<0.001), UCEC (HR: 0.346; 95% CI: 0.139–0.859; P=0.022), SARC (HR: 0.594; 95% CI: 0.358–0.984; P=0.043), and THCA (HR: 0.184; 95% CI: 0.057–0.595; P=0.005), whereas it indicated an unfavorable prognosis for COAD (HR:2.151; 95% CI: 1.128–4.101; P=0.020), ESCA (HR: 6.362; 95% CI: 2.768–14.624; P<0.001), LUSC (HR: 3.175; 95% CI: 1.817–5.547; P<0.001), KIRC (HR: 4.153; 95% CI: 2.137–8.072; P<0.001), and PAAD (HR: 5.317; 95% CI: 1.297–21.790; P=0.020). Tumors with overexpressed CSF2 and CSF2RB were associated with increased OS in BRCA (HR: 0.323; 95% CI: 0.142–0.739; P=0.007), LUAD (HR: 0.342; 95% CI: 0.208–0.562; P<0.001), SKCM (HR: 0.488; 95% CI: 0.352–0.680; P<0.001), UCEC (HR: 0.296; 95% CI: 0.107–0.814; P=0.018), LIHC (HR: 0.510; 95% CI: 0.289–0.899; P=0.020), and SARC (HR: 0.431; 95% CI: 0.255–0.729; P=0.002), and were associated with decreased OS in ESCA (HR: 8.016; 95% CI: 2.936–21.885; P<0.001), LUSC (HR: 4.572; 95% CI: 2.128–9.822; P<0.001), KIRC (HR: 3.090; 95% CI: 1.737–5.491; P<0.001), and UCS (HR: 5.770; 95% CI: 1.155–28.830; P=0.033). In addition, the binding subgroup of CSF3 and CSF3R was characterized by high expression indicating improved OS in BRCA (HR: 0.488; 95% CI: 0.311–0.767; P=0.002) and LUAD (HR: 0.462; 95% CI: 0.278–0.766; P=0.003) in contrast to decreased OS in BLCA (HR: 2.857; 95% CI: 1.537–5.310; P=0.001), HNSC (HR: 2.033; 95% CI: 1.316–3.139; P=0.001), LUSC (HR: 1.761; 95% CI: 1.139–2.724; P=0.011), KIRC (HR: 3.526; 95% CI: 2.066–6.019; P<0.001), THCA (HR: 4.189; 95% CI: 1.035–16.951; P=0.045), and UCS (HR: 17.561; 95% CI: 2.754–111.976 ; P=0.002) (Table 2).
Functional enrichment analysis of CSFs and CSFRs and the related common DEGs
To identify the enriched biological processes affecting the prognostic roles of CSFs and CSFRs pathway in different cancer types, we divided the cancer types with consistent prognosis into three pan-cancer categories based on molecular similarities among histologically or anatomically related cancer types, including pan-gastrointestinal, pan-gynecological, and pan-squamous cancers (21). Then, we performed Metascape network analysis using the common DEGs of cancer types with consistent prognosis and pan-cancer categories.
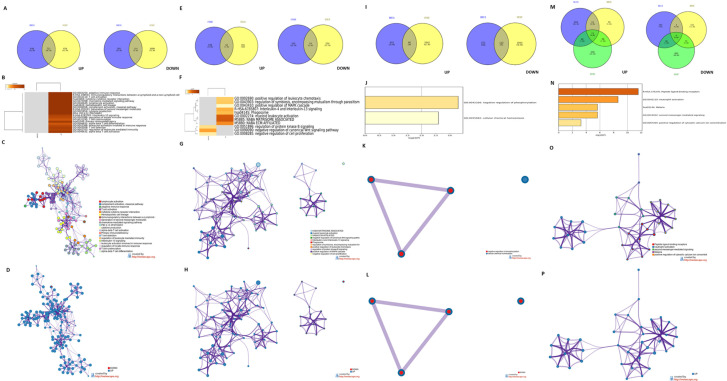
In the binding subgroup of CSF2 and CSF2RA, high expression predicted favorable outcome in BRCA, LUAD, UCEC, SARC, THCA, and SKCM, and unfavorable outcome in COAD, ESCA, LUSC, KIRC, and PAAD (P<0.05, Table 2). According to our previous classification, BRCA and UCEC belonged to pan-gynecological cancers, while COAD and ESCA belonged to pan-gastrointestinal cancers. The Venn diagram showed 721 (36.1%) common upregulated genes and 114 (5.1%) common downregulated genes between BRCA and UCEC (Figure S1A). As shown in Figure S1B,C,D, almost all of the upregulated genes regulated the following immunological enrichment pathways: lymphocyte activation, immunoregulatory interactions between a lymphoid cell and a non-lymphoid cell, regulation of lymphocyte mediated immunity, adaptive immune response, T cell activation, T cell costimulation, alpha-beta T cell proliferation, cytokine–cytokine receptor interaction, primary immunodeficiency, interleukin-10 signaling, and others. Moreover, 52 (4.4%) common upregulated genes and 19 (2.7%) common downregulated genes between COAD and ESCA were also found in the binding group of CSF2 and CSF2RA (Figure S1E). As illustrated in the Figure S1F,G,H, biological pathways associated with inflammatory response were regulated by almost all common upregulated genes which included positive regulation of leukocyte chemotaxis, phagosome, myeloid leukocyte activation, negative regulation of cell proliferation, and positive regulation of MAPK cascade.
In the binding subgroup of CSF2 and CSF2RB, high expression was associated with increased OS in BRCA, LUAD, UCEC, LIHC, SARC, and SKCM, and decreased OS in ESCA, LUSC, KIRC, and UCS (P<0.05, Table 2). Based on the pan-cancer analysis, BRCA and UCEC belonged to pan-gynecological cancers. However, ESCA, LUSC, KIRC, and UCS, in which high expression indicated poor prognosis, did not belong to any pan-cancer types. In BRCA and UCEC, a total of 80 (3%) common upregulated genes and 122 (7%) common downregulated genes were detected (Figure S1I). As we can see from Figure S1J,K,L, pathways correlated with cellular response were regulated by all common downregulated genes which included negative regulation of phosphorylation and cellular chemical homeostasis.
In the binding subgroup of CSF3 and CSF3R, high expression was associated with good prognosis in BRCA and LUAD and poor prognosis in BLCA, HNSC, LUSC, KIRC, and THCA (P<0.05) (Table 2). According to our categories, no common pan-cancer types were found in BRCA and LUAD which had high expression implying good prognosis, whereas BLCA, HNSC, and LUSC were classified as pan-squamous cancer groups. Our results showed that 13 (0.9%) common upregulated genes and 8 (0.6%) common downregulated genes were found among BLCA, LUSC, and HNSC (Figure S1M). All common upregulated genes regulated the following pathways related to inflammatory and signaling transduction: neutrophil activation, peptide ligand binding receptors, second-messenger–mediated signaling, and positive regulation of cytosolic calcium ion concentration (Figure S1N,O,P).
Discussion
GM-CSF (CSF2) and G-CSF (CSF3) are hematopoietic growth factors that regulate the growth, proliferation, and differentiation of cells of hematopoietic lineages (1,2). Numerous experiments have shown that G-/GM-CSF can be used as tumour-derived factors to promote tumor growth and progression, and the excessive secretion of CSFs and CSFRs have been observed in multiple cancer types (22-27).
In our results, we found that a high CSF2 expression was an independent predictor of diminished OS for CESC, ESCA, HNSC, LUSC, KIRC, and PAAD patients, while a high CSF3 expression was an independent predictor of diminished OS for BLCA, CESC, COAD, HNSC, LUSC, ACC, KIRC, THCA, THYM, and UCS patients. As previously reported, GM-CSF has been found to promote tumor growth, proliferation, and migration in a variety of cancers including lung cancer (28), skin carcinoma (29,30), glioma (31), bladder cancer (32), prostate cancer (33), colorectal cancer, and head and neck squamous cell carcinoma (34) via an autocrine or paracrine loop. G-CSF has also been found to promote cancer development and invasion in various cancers, including gastric and colorectal carcinomas (16), pancreatic adenocarcinoma (35), breast cancer (20), head and neck squamous cell carcinoma (17), skin carcinoma (30), and glioma (31), which is consistent with our results. In addition, some studies further confirmed that high G/GM-CSF expression was associated with poor prognosis by immunohistochemistry (IHC) (15,35-38).
However, some research has also suggested that G-/GM-CSF can exert an antitumor effects when used as adjuvant and tumor vaccine in immunotherapy in an immune-dependent manner (12,39-41) while exerting antiproliferative effects on tumors in a non-immune-dependent manner (42,43). In our study, highly expressed CSF2 indicating favorable prognosis was found in BRCA, BLCA, LUAD, SKCM, CHOL, LIHC, OV, and THCA, while highly expressed CSF3 indicating good prognosis was found in BRCA, PRAD, and LGG. In previous studies confirmed by IHC, the production of GM-CSF in colorectal cancer cells had a favorable prognostic effect (8). Furthermore, the increased survival in prostate cancer was associated with recruitment of CD8+ T cells and T-cell infiltration modulation in the tumor microenvironment (9). Another IHC finding showed that GM-CSF suppresses growth and enhances the immunogenicity of glioma (44). Overall, our research provides real-world data to confirm that CSFs and CSFRs serve as independent prognostic factors in multiple cancer types, and that the prognostic value of CSFs and CSFRs is cancer-type dependent.
The deeper molecular mechanisms of GM-CSF and G-CSF in multiple cancers have yet to be fully elucidated and require further investigation. Recently, enrichment pathways of different tumors have begun to provide additional ways to predict and thus improve patient outcome. In the conventional sense, the classification of tumor patients in many previous studies has been based on pathological classification. Recent studies have found that subgroups of tumor patients based on the similarity of anatomical and histological structures have unique and significantly different enrichment pathway disorders in different pan-cancer classifications (21). Results in our study clarified the molecular network induced by the common DEGs of CSFs and CSFRs, evidencing important biological enrichment pathways. DEGs contribute to the complex outcomes of CSFs and CSFRs, and so we used an enrichment analysis tool to identify the important enrichment pathways. In our results, the highly expressed combination subgroup of CSF2 and CSF2RA indicating favorable prognosis was found to be coregulated by almost all common upregulated genes associated with the activation of immune response, while subgroups with unfavorable prognosis coregulated by the majority of common upregulated genes were often correlated with inflammatory response. In a similar way, the highly expressed combination subgroup of CSF2 and CSF2RB predicting good prognosis coregulated by all common downregulated genes correlated with the activation of cellular response, whereas the highly expressed binding subgroup of CSF3 and CSF3R indicating unfavorable prognosis coregulated by all common upregulated genes correlated with inflammatory and signaling transduction. In clinical oncology, the immune response to inflammation and cancer cells can be activated by GM-CSF, which acts as an immunostimulant to activate the activity of various immune cells. The cells secreting GM-CSF can promote the phagocytosis and apoptosis of tumor cells by promoting the maturation and aggregation of DCs and antitumor immunity, and promote the secretion of multiple co-stimulators (45). It is important to mention that GM-CSF can enhance T-cell activation through different potential mechanisms and can stimulate the recruitment, maturation, and function of circulating antigen presenting cells, including DCs (46-49). Alternatively, GM-CSF may increase the numbers of Fc receptor-bearing antigen-presenting cells as has been postulated in a clinical trial (50). In addition, previous studies reported that GM-CSF influenced inflammation by recruiting, priming, and activating inflammatory cells such as neutrophils, macrophages, and eosinophils. At the same time, GM-CSF promotes the production of pro-inflammatory cytokines, antigen presentation, phagocytosis, and leukocyte chemotaxis and adhesion (51,52), and greatly enhances the production of neutrophils and monocytes in bone marrow. During inflammation, GM-CSF is significantly increased in circulation and plays a hormone-like function on bone cells. Overproduction of GM-CSF has been shown to play a key role in the pathological inflammation of animal models and human diseases. GM-CSF deficiency is associated with chronic inflammatory responses (2,52). A major role for G-CSF is to stimulate the differentiation, growth, and effector functions of neutrophils and macrophages and regulate the activity and function of developing and mature neutrophils. However, there are numerous reports of G-CSF exacerbating the inflammation of sterile tissue and inducing the proliferation pathway to serve as a pro-inflammatory cytokine (14,53), which was further confirmed in our study. Overall, the common DEGs in the binding subgroups with consistent prognosis were upregulated or downregulated. Accordingly, the enrichment pathways were activated or inhibited, and this may serve as a complement to explain the preceding prognostic results.
This study systematically documented the expression level of CSFs and CSFRs for TCGA samples and interactively analyzed the survival time with different cancer profiles. However, the major limitation of this study was its retrospective nature. Additionally, the amount of data of certain cancer types was insufficient, so significant outcomes were difficult to obtain. Future studies, including cell or animal study, will be necessary to determine the regulatory effects of CSFs and CSFRs in different tumors. Another limitation was that the specific biological mechanisms of CSFs and CSFRs affecting prognosis were not further investigated. It is difficult to describe the specific mechanism as no detailed explanation is yet available. Therefore, more in vitro and in vivo experiments to decipher the therapeutic values and mechanisms of action of CSFs and CSFRs are needed.
Conclusions
In conclusion, we completed genomics analysis of CSF and CSFR expression based on the cBioportal data of 24 cancer types. Our findings indicate that CSFs and CSFRs may be independent prognostic factors for OS, and that the prognostic value of CSFs and CSFRs is cancer-type dependent. The CSF2 and CSF2RA binding subgroups with favorable prognosis are often associated with the activation of immune response, while subgroups with unfavorable prognosis are often correlated with inflammatory response. Similarly, CSF2 and CSF2RB binding subgroups with increased OS are often associated with the activation of cellular response. Meanwhile, the CSF3 and CSF3R binding subgroups associated with decreased OS are often accompanied by the inflammation and signaling transduction pathways, and thus may be novel therapeutic strategies for personalized treatment. Therefore, personalized risk stratification based on CSFs and CSFRs pathway should be considered for cancer patients.
Figure S1.
Enrichment network analysis for the binding of CSFs and CSFRs. High expression subgroup of CSF2 and CSF2RA with favorable and unfavorable prognosis (A,B,C,D); (E,F,G,H) High expression subgroup of CSF2 and CSF2RB with favorable prognosis. (I,J,K) High expression subgroup of CSF3 and CSF3R with unfavorable prognosis (L,N,O). (A,E,I,M) The Venn diagrams of common upregulated and downregulated genes. (B,F,J,N) Heatmaps of enriched terms across input common genes, colored by P values. (C,G,K,O) Network of enriched terms colored by cluster ID, where nodes that share the same cluster ID are typically close to each other. (D,H,L,P) Network of enriched terms represented as pie charts, where pies are color-coded based on the identities of the gene lists. CSF, colony-stimulating factor; CSFR, colony-stimulating factor receptor; UP, common upregulated genes; DOWN, common downregulated genes.
Supplementary
The article’s supplementary files as
Acknowledgments
We gratefully acknowledge The Cancer Genome Atlas (TCGA) database for providing the primary data.
Funding: This study was funded by the National Natural Science Foundation of China (no. 81672974 and 81602719).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by The First Affiliated Hospital of Shandong First Medical University review board (no. S033). Informed consent from patients was waived due to the anonymity of individual patient data. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-5363
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-5363). The authors have no conflicts of interest to declare.
(English Language Editor: J. Gray)
References
- 1.Liu J, Qian C, Cao X. Post-Translational Modification Control of Innate Immunity. Immunity 2016;45:15-30. 10.1016/j.immuni.2016.06.020 [DOI] [PubMed] [Google Scholar]
- 2.Wang T, Wu B, Hu X, et al. A randomized multicenter phase II trial of mecapegfilgrastim single administration versus granulocyte colony-stimulating growth factor on treating chemotherapy-induced neutropenia in breast cancer patients. Ann Transl Med 2019;7:196. 10.21037/atm.2019.04.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Demetri GD, Griffin JD. Granulocyte colony-stimulating factor and its receptor. Blood 1991;78:2791-808. 10.1182/blood.V78.11.2791.bloodjournal78112791 [DOI] [PubMed] [Google Scholar]
- 4.Touw IP, van de Geijn GJ. Granulocyte colony-stimulating factor and its receptor in normal myeloid cell development, leukemia and related blood cell disorders. Front Biosci 2007;12:800-15. 10.2741/2103 [DOI] [PubMed] [Google Scholar]
- 5.Filipazzi P, Valenti R, Huber V, et al. Identification of a new subset of myeloid suppressor cells in peripheral blood of melanoma patients with modulation by a granulocyte-macrophage colony-stimulation factor-based antitumor vaccine. J Clin Oncol 2007;25:2546-53. 10.1200/JCO.2006.08.5829 [DOI] [PubMed] [Google Scholar]
- 6.Jinushi M, Nakazaki Y, Dougan M, et al. MFG-E8-mediated uptake of apoptotic cells by APCs links the pro- and antiinflammatory activities of GM-CSF. J Clin Invest 2007;117:1902-13. 10.1172/JCI30966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Serafini P, Carbley R, Noonan KA, et al. High-dose granulocyte-macrophage colony-stimulating factor-producing vaccines impair the immune response through the recruitment of myeloid suppressor cells. Cancer Res 2004;64:6337-43. 10.1158/0008-5472.CAN-04-0757 [DOI] [PubMed] [Google Scholar]
- 8.Nebiker CA, Han JY, Eppenberger-Castori S, et al. GM-CSF Production by Tumor Cells Is Associated with Improved Survival in Colorectal Cancer. Clinical Cancer Research 2014;20:3094-106. 10.1158/1078-0432.CCR-13-2774 [DOI] [PubMed] [Google Scholar]
- 9.Wei XX, Chan S, Kwek S, et al. Systemic GM-CSF Recruits Effector T Cells into the Tumor Microenvironment in Localized Prostate Cancer. Cancer Immunology Research 2016;4:948-58. 10.1158/2326-6066.CIR-16-0042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van den Eertwegh AJ, Versluis J, van den Berg HP, et al. Combined immunotherapy with granulocyte-macrophage colony-stimulating factor-transduced allogeneic prostate cancer cells and ipilimumab in patients with metastatic castration-resistant prostate cancer: a phase 1 dose-escalation trial. Lancet Oncol 2012;13:509-17. 10.1016/S1470-2045(12)70007-4 [DOI] [PubMed] [Google Scholar]
- 11.Parmiani G, Castelli C, Pilla L, et al. Opposite immune functions of GM-CSF administered as vaccine adjuvant in cancer patients. Ann Oncol 2007;18:226-32. 10.1093/annonc/mdl158 [DOI] [PubMed] [Google Scholar]
- 12.Creelan BC, Antonia S, Noyes D, et al. Phase II trial of a GM-CSF-producing and CD40L-expressing bystander cell line combined with an allogeneic tumor cell-based vaccine for refractory lung adenocarcinoma. J Immunother 2013;36:442-50. 10.1097/CJI.0b013e3182a80237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kitade H, Yanagida H, Yamada M, et al. Granulocyte-colony stimulating factor producing anaplastic carcinoma of the pancreas treated by distal pancreatectomy and chemotherapy: report of a case. Surg Case Rep 2015;1:46. 10.1186/s40792-015-0048-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirai K, Kumakiri M, Fujieda S, et al. Expression of granulocyte colony-stimulating factor and its receptor in epithelial skin tumors. J Dermatol Sci 2001;25:179-88. 10.1016/S0923-1811(00)00131-6 [DOI] [PubMed] [Google Scholar]
- 15.Fan Z, Li Y, Zhao Q, et al. Highly Expressed Granulocyte Colony-Stimulating Factor (G-CSF) and Granulocyte Colony-Stimulating Factor Receptor (G-CSFR) in Human Gastric Cancer Leads to Poor Survival. Med Sci Monit 2018;24:1701-11. 10.12659/MSM.909128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morris KT, Khan H, Ahmad A, et al. G-CSF and G-CSFR are highly expressed in human gastric and colon cancers and promote carcinoma cell proliferation and migration. Br J Cancer 2014;110:1211-20. 10.1038/bjc.2013.822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutschalk CM, Herold-Mende CC, Fusenig NE, et al. Granulocyte colony-stimulating factor and granulocyte-macrophage colony-stimulating factor promote malignant growth of cells from head and neck squamous cell carcinomas in vivo. Cancer Res 2006;66:8026-36. 10.1158/0008-5472.CAN-06-0158 [DOI] [PubMed] [Google Scholar]
- 18.Yokoyama T, Hyodo M, Hosoya Y, et al. Aggressive G-CSF-producing gastric cancer complicated by lung and brain abscesses, mimicking metastases. Gastric Cancer 2005;8:198-201. 10.1007/s10120-005-0335-6 [DOI] [PubMed] [Google Scholar]
- 19.Tachibana M, Murai M. G-CSF production in human bladder cancer and its ability to promote autocrine growth: a review. Cytokines Cell Mol Ther 1998;4:113-20. [PubMed] [Google Scholar]
- 20.Hollmén M, Karaman S, Schwager S, et al. G-CSF regulates macrophage phenotype and associates with poor overall survival in human triple-negative breast cancer. Oncoimmunology 2015;5:e1115177. 10.1080/2162402X.2015.1115177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoadley KA, Yau C, Hinoue T, et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell 2018;173:291-304.e6. 10.1016/j.cell.2018.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lammel V, Stoeckle C, Padberg B, et al. Hypereosinophilia driven by GM-CSF in large-cell carcinoma of the lung. Lung Cancer 2012;76:493-5. 10.1016/j.lungcan.2012.02.014 [DOI] [PubMed] [Google Scholar]
- 23.Perez FA, Fligner CL, Yu EY. Rapid clinical deterioration and leukemoid reaction after treatment of urothelial carcinoma of the bladder: possible effect of granulocyte colony-stimulating factor. J Clin Oncol 2009;27:e215-7. 10.1200/JCO.2009.22.4931 [DOI] [PubMed] [Google Scholar]
- 24.Revoltella RP, Menicagli M, Campani D. Granulocyte-macrophage colony-stimulating factor as an autocrine survival-growth factor in human gliomas. Cytokine 2012;57:347-59. 10.1016/j.cyto.2011.11.016 [DOI] [PubMed] [Google Scholar]
- 25.Taghipour Fard Ardekani M, Malekzadeh M, Hosseini SV, et al. Evaluation of Pre-Treatment Serum Levels of IL-7 and GM-CSF in Colorectal Cancer Patients. Int J Mol Cell Med 2014;3:27-34. [PMC free article] [PubMed] [Google Scholar]
- 26.Urdinguio RG, Fernandez AF, Moncada-Pazos A, et al. Immune-dependent and independent antitumor activity of GM-CSF aberrantly expressed by mouse and human colorectal tumors. Cancer Res 2013;73:395-405. 10.1158/0008-5472.CAN-12-0806 [DOI] [PubMed] [Google Scholar]
- 27.Murayama T, Imoto S, Natazuka T, et al. Proliferative reaction of myelogenous leukemia cells with cytokines G-CSF, GM-CSF, M-CSF, SCF and TPO. Leuk Res 1998;22:557-60. 10.1016/S0145-2126(98)00042-3 [DOI] [PubMed] [Google Scholar]
- 28.Uemura Y, Kobayashi M, Nakata H, et al. Effects of GM-CSF and M-CSF on tumor progression of lung cancer: roles of MEK1/ERK and AKT/PKB pathways. Int J Mol Med 2006;18:365-73. [PubMed] [Google Scholar]
- 29.Obermueller E, Vosseler S, Fusenig NE, et al. Cooperative autocrine and paracrine functions of granulocyte colony-stimulating factor and granulocyte-macrophage colony-stimulating factor in the progression of skin carcinoma cells. Cancer Res 2004;64:7801-12. 10.1158/0008-5472.CAN-03-3301 [DOI] [PubMed] [Google Scholar]
- 30.Mueller MM, Fusenig NE. Constitutive expression of G-CSF and GM-CSF in human skin carcinoma cells with functional consequence for tumor progression. Int J Cancer 1999;83:780-9. [DOI] [PubMed] [Google Scholar]
- 31.Mueller MM, Herold-Mende CC, Riede D, et al. Autocrine growth regulation by granulocyte colony-stimulating factor and granulocyte macrophage colony-stimulating factor in human gliomas with tumor progression. Am J Pathol 1999;155:1557-67. 10.1016/S0002-9440(10)65472-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perez FA, Fligner CL, Yu EY. Rapid clinical deterioration and leukemoid reaction after treatment of urothelial carcinoma of the bladder: possible effect of granulocyte colony-stimulating factor. J Clin Oncol 2009;27:e215-7. 10.1200/JCO.2009.22.4931 [DOI] [PubMed] [Google Scholar]
- 33.Lang SH, Miller WR, Duncan W, et al. Production and response of human prostate cancer cell lines to granulocyte macrophage-colony stimulating factor. Int J Cancer 1994;59:235-41. 10.1002/ijc.2910590216 [DOI] [PubMed] [Google Scholar]
- 34.Ninck S, Reisser C, Dyckhoff G, et al. Expression profiles of angiogenic growth factors in squamous cell carcinomas of the head and neck. Int J Cancer 2003;106:34-44. 10.1002/ijc.11188 [DOI] [PubMed] [Google Scholar]
- 35.Zhang L, Tao L, Guo L, et al. G-CSF associates with neurogenesis and predicts prognosis and sensitivity to chemotherapy in pancreatic ductal adenocarcinoma. Cancer Manag Res 2018;10:2767-75. 10.2147/CMAR.S165226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, Jia R, Zhao T, et al. HIF-1α mediates tumor-nerve interactions through the up-regulation of GM-CSF in pancreatic ductal adenocarcinoma. Cancer Lett 2019;453:10-20. 10.1016/j.canlet.2019.03.036 [DOI] [PubMed] [Google Scholar]
- 37.Hollmén M, Karaman S, Schwager S, et al. G-CSF regulates macrophage phenotype and associates with poor overall survival in human triple-negative breast cancer. Oncoimmunology 2015;5:e1115177. 10.1080/2162402X.2015.1115177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tavakkoli M, Wilkins CR, Mones JV, et al. A Novel Paradigm Between Leukocytosis, G-CSF Secretion, Neutrophil-to-Lymphocyte Ratio, Myeloid-Derived Suppressor Cells, and Prognosis in Non-small Cell Lung Cancer. Front Oncol 2019;9:295. 10.3389/fonc.2019.00295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harada T, Hamada A, Shimokawa M, et al. A phase I/II trial of irinotecan plus amrubicin supported with G-CSF for extended small-cell lung cancer. Jpn J Clin Oncol 2014;44:127-33. 10.1093/jjco/hyt198 [DOI] [PubMed] [Google Scholar]
- 40.Bauhofer A, Plaul U, Torossian A, et al. Perioperative prophylaxis with granulocyte colony-stimulating factor (G-CSF) in high-risk colorectal cancer patients for an improved recovery: A randomized, controlled trial. Surgery 2007;141:501-10. 10.1016/j.surg.2006.09.004 [DOI] [PubMed] [Google Scholar]
- 41.Xavier L, Cunha M, Goncalves C, et al. Hematological remission and long term hematological control of acute myeloblastic leukemia induced and maintained by granulocyte-colony stimulating factor (G-CSF) therapy. Leuk Lymphoma 2003;44:2137-42. 10.1080/1042819031000111053 [DOI] [PubMed] [Google Scholar]
- 42.Ruff MR, Farrar WL, Pert CB. Interferon gamma and granulocyte/macrophage colony-stimulating factor inhibit growth and induce antigens characteristic of myeloid differentiation in small-cell lung cancer cell lines. Proc Natl Acad Sci U S A 1986;83:6613-7. 10.1073/pnas.83.17.6613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamashita Y, Nara N, Aoki N. Antiproliferative and differentiative effect of granulocyte-macrophage colony-stimulating factor on a variant human small cell lung cancer cell line. Cancer Res 1989;49:5334-8. [PubMed] [Google Scholar]
- 44.Xu H, Wang Q, Lin C, et al. Synergism between cryoablation and GM-CSF: enhanced immune function of splenic dendritic cells in mice with glioma. Neuroreport 2015;26:346-53. 10.1097/WNR.0000000000000351 [DOI] [PubMed] [Google Scholar]
- 45.Zarei S, Schwenter F, Luy P, et al. Role of GM-CSF signaling in cell-based tumor immunization. Blood 2009;113:6658-68. 10.1182/blood-2008-06-161075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dranoff G. GM-CSF-secreting melanoma vaccines. Oncogene 2003;22:3188-92. 10.1038/sj.onc.1206459 [DOI] [PubMed] [Google Scholar]
- 47.Li B, Simmons A, Du T, et al. Allogeneic GM-CSF-secreting tumor cell immunotherapies generate potent anti-tumor responses comparable to autologous tumor cell immunotherapies. Clin Immunol 2009;133:184-97. 10.1016/j.clim.2009.07.008 [DOI] [PubMed] [Google Scholar]
- 48.Mach N, Gillessen S, Wilson SB, et al. Differences in dendritic cells stimulated in vivo by tumors engineered to secrete granulocyte-macrophage colony-stimulating factor or Flt3-ligand. Cancer Res 2000;60:3239-46. [PubMed] [Google Scholar]
- 49.Dranoff G. Cytokines in cancer pathogenesis and cancer therapy. Nat Rev Cancer 2004;4:11-22. 10.1038/nrc1252 [DOI] [PubMed] [Google Scholar]
- 50.Cartron G, Zhao-Yang L, Baudard M, et al. Granulocyte-macrophage colony-stimulating factor potentiates rituximab in patients with relapsed follicular lymphoma: results of a phase II study. J Clin Oncol 2008;26:2725-31. 10.1200/JCO.2007.13.7729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shiomi A, Usui T. Pivotal roles of GM-CSF in autoimmunity and inflammation. Mediators Inflamm 2015;2015:568543. [DOI] [PMC free article] [PubMed]
- 52.Dougan M, Dranoff G, Dougan SK. GM-CSF, IL-3, and IL-5 Family of Cytokines: Regulators of Inflammation. Immunity 2019;50:796-811. 10.1016/j.immuni.2019.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bo L, Wang F, Zhu J, et al. Granulocyte-colony stimulating factor (G-CSF) and granulocyte-macrophage colony stimulating factor (GM-CSF) for sepsis: a meta-analysis. Crit Care 2011;15:R58. 10.1186/cc10031 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as