Abstract
Background
With a high incidence rate and mortality rate, tuberculosis (TB) is a major public health concern worldwide. There is an urgent need to develop the rapid, reliable and affordable diagnostic test for the detection of Mycobacterium tuberculosis (Mtb). Metagenomic next-generation sequencing (mNGS) showed promising values in the rapid diagnosis of clinical TB, while studies on the application of mNGS in pulmonary and extrapulmonary TB remain scarce.
Methods
In this study, we collected the results of both mNGS and culture of 70 specimens from suspected TB patients at the Shanghai Pulmonary Hospital of Tongji University. Results of GeneXpert MTB/RIF assay (Xpert) were obtained from 19 patients. We also assessed the diagnostic performance, relationship and consistency between mNGS, traditional culture method, and Xpert.
Results
Overall, 36 of 70 suspected patients were finally diagnosed with TB. The sensitivity, specificity, positive predictive value, negative predictive value and the Youden index of mNGS in all clinical TB specimens were calculated as 66.7% (95% CI: 0.489–0.809), 97.1% (95% CI: 0.829–0.998), 96.0% (95% CI: 0.777–0.998), 73.3% (95% CI: 0.578–0.849), 63.8%, respectively. The sensitivity of mNGS was much superior to that of conventional culture method (66.7%, 95% CI: 0.489–0.809 vs. 36.1%, 95% CI: 0.213–0.538) and Xpert (76.9%, 95% CI: 0.460–0.938 vs. 61.5%, 95% CI: 0.323–0.849). In pulmonary TB cases, mNGS, culture, and Xpert all demonstrated better diagnostic capacity for TB when compared with extrapulmonary TB cases. Among all methods and sample classifications, the Youden index of parallel diagnostic test in pulmonary samples was outstanding (mNGS/culture 88.2%; mNGS/Xpert 100%). Furthermore, the correlation and concordance between mNGS and culture method in all types of specimens was statistically significant (both P<0.001).
Conclusions
With high sensitivity and specificity, mNGS showed remarkable diagnostic performance in various samples from suspected TB patients. Combining mNGS and culture or Xpert method had the potential for clinical application in TB.
Keywords: Metagenomic next-generation sequencing (mNGS) technology, culture method, GeneXpert MTB/RIF assay (Xpert), tuberculosis (TB), Mycobacterium tuberculosis (Mtb)
Introduction
Tuberculosis (TB), as one of infectious diseases, is caused by the Mycobacterium tuberculosis (Mtb). According to the report of Word Health Organization (WHO), about 25% of worldwide people are infected with Mtb and hence be in danger of developing TB disease (1). With a high incidence rate, TB is the major reason of mortality in infectious cases, and ranks as one of the top ten killers in all diseases (1). The difficulty of detecting Mtb quickly and accurately is considered as one of the leading causes of high mortality rate in TB (2).
At present, microbiological tests, such as traditional culture methods and multiple molecular approaches, are the dominating choices for the diagnosis of TB. Solid or liquid culture is still recommended as the “gold-standard” method for TB diagnosis, which could also be used for drug susceptibility testing (3,4). Nonetheless, conventional Mtb culture method is time-consuming, prone to be polluted, and requires further biochemical tests (5,6). Sputum smear microscopy, as an economic and convenient Mtb detection tool, is short of high sensitivity and reproducibility (7). Ninet et al. (8) illustrated that Acid-fast bacilli (AFB) microscopy using Ziehl-Neelsen (ZN) staining was a simple and rapid method, while its low sensitivity and specificity was unsatisfying. GeneXpert MTB/RIF assay (Xpert) (9,10) and updated ultra (11,12), as the rapid diagnostic technology involving nucleic acid amplification (NAAT), have high sensitivity and specificity for the identification of Mtb and drug-resistance Mtb. Kurtoglu et al. (13) also found the clinical value of combing Xpert with culture in the diagnosis of Mtb infection. However, an obvious limitation of Xpert is its poor performance in extrapulmonary TB cases (14,15).
Metagenomics next-generation sequencing (mNGS) technology, as one of rapid diagnostic tools, is also known as the whole genome sequencing (WGS) approach. In DNA sequencing platforms, mNGS could reveal the possible causative pathogens by multiple steps, such as identifying the nucleotides of clinical samples, constructing reference databases of organisms, and strictly sequence read mapping (16-18). In the areas of medical microbiology, mNGS showed its outstanding advantages like high-throughput capabilities, unbiased detection, and relatively short turnaround time (19,20). While for TB, a certain number of researches revealed the clinical value of mNGS in the diagnosis of Mtb infection (21,22). Several countries and the WHO also employed mNGS for the surveillance of drug-resistance Mtb, and the instruction of choosing antitubercular agents (23-25). To fully evaluate the performance of mNGS in pulmonary and extrapulmonary TB samples, we collected the results of mNGS and culture of 70 specimens from suspected TB patients, and Xpert results from 19 patients. We also calculated the diagnostic performance of mNGS, traditional culture method, Xpert, parallel diagnostic test, and serial diagnostic test in the identification of Mtb. Further, this study assessed the correlation and consistency between mNGS and traditional culture or Xpert method in TB.
We present the following article in accordance with the STARD reporting checklist (available at http://dx.doi.org/10.21037/jtd-20-1232).
Methods
Patients
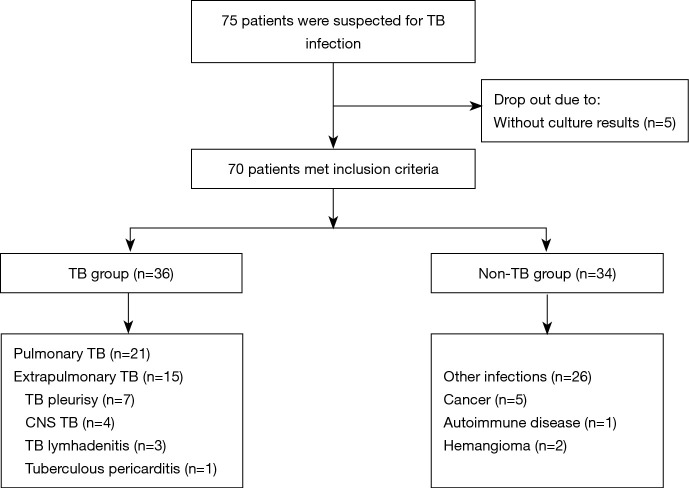
This retrospective study incorporated a total of 70 patients at the Shanghai Pulmonary Hospital of Tongji University between 1 January and 30 September 2019 (Figure 1). Among all enrolled patients, 36 patients (51.4%) were finally diagnosed with pulmonary or extrapulmonary TB according to the criterion of China Clinical Treatment Guide for Tuberculosis (The 2017 Version) and other clinical guidelines. Thirty-four patients (49.6%) were clinically suspected of having TB at the beginning, but finally diagnosed with diseases other than TB, including bacterial or fungal infection, cancer, and autoimmune disease. The results of conventional culture, mNGS, and Xpert of all patients were collected. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and approved by Shanghai Pulmonary Hospital (ethical number K19-158). All patients signed the written informed consent before the collection of samples.
Figure 1.
Study flowchart showing patient enrollment.
Sample processing and DNA extraction
For the TB group, sample sources for mNGS and traditional culture were as follows: 14 bronchoalveolar lavage fluid (BALF) samples, 14 pleural fluid samples, 4 cerebrospinal fluid (CSF) samples, 3 sputum samples, and one pericardial fluid sample. For the non-TB group, sample sources for mNGS and traditional culture were as follows: 14 BALF samples, 11 pleural fluid samples, 3 sputum samples, 3 lung tissue, 2 pus samples, and one CSF sample. In all 19 samples which were further tested by Xpert, 8 were BALF samples, 7 were pleural fluid samples, 2 were sputum samples, and 2 were pus samples.
Volumes of 200–300 µL of BALF, pleural fluid, CSF, pericardial fluid and pus were collected by standard procedures from enrolled patients, and placed in sterile tubes for further analysis. The lung tissue samples were gathered by fine needle aspiration, and applied to next steps after the procedure of homogenate. The sputum samples were used for DNA extraction following by liquefaction. According to the manufacturer’s operational guidebook, TIANamp Micro DNA Kit (DP316, TIANGEN BIOTECH, Beijing, China) was employed to the process of DNA extraction.
Construction of DNA libraries
Firstly, for the purpose of generate 200- to 300-bp fragments, we used Bioruptor Pico instrument to fragment DNA that extracted from different samples. Secondly, an end-repair method was carried out for the DNA fragments. Subsequently, after adding bubble adapters overnight, we amplified the DNA fragments by polymerase chain reaction (PCR). Then, Ion Torrent Proton Sequencer (Life Technologies, Carlsbad, CA, USA) was applied. Go through in sequence, DNA libraries were eventually constructed. The quality control of the DNA libraries was performed by an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), which could evaluate the DNA concentration and fragment size. Then, quality qualified libraries were sequenced by BGISEQ-50 platform (MGI BGISEQ-50, China).
Bioinformatic analysis
High-quality sequencing data were obtained through getting rid of low quality, low complexity, and short reads that less than 35 bp in length. Then, the human host sequences mapped to the human reference genome (hg19) and Yanhuang genome sequence were excluded by Burrows-Wheeler Alignment (BWA, Version: 0.7.10) (26,27). The next step in the process of advanced data analysis is to match the remaining reads with four microbial genome databases, including bacterial, virus, fungal, and parasite databases. Subsequently, the parameters of different species including Mtb were classified and recorded, such as the read counts and the genomic coverage. The SOAPCoverage software (https://github.com/aquaskyline/SOAPcoverage) was used for assessing the coverage. The National Center for Biotechnology Information (NCBI, ftp://ftp.ncbi.nlm.nih.gov/genomes/), as one of public databases, was used for downloading the integrated classification reference databases. So far, more than 4,000 viral genomes, nearly 3,000 bacterial genomes, over 200 clinically prevalent fungi, as well as 140 disease related parasites were contained in the reference databases. Concretely, the genome DNA of Mtb is NC_00962.3. Pathogens that identified in suspected background databases and negative control samples were filtered once they reached the threshold level (26,27).
Determination of pathogens
Normal clinical samples were sequenced in the study. We determined the detection value of normalized 20 million clean reads of each pathogen at the beginning and assessed the reference range by a boxplot algorithm. For the reference range, the peak value was the result of upper quartile plus threefold interquartile range, while the minimum value was lower quartile minus threefold interquartile range. By means of BWA alignment, we collected the percentage of pathogenic sequences mapped to the entire genome and the number of pathogenic reads with over 10 mapping quality in healthy individuals. The procedures of scientifically estimating the existence of pathogenic organisms was as follows: first, the percentage of sequences of all pathogens that detected in single sample were ranked; then, among top ten pathogens, the final selected pathogens must simultaneously met the requirements of both over three strictly mapped reads and all the parameters transcending the upper limit of the reference range.
Mtb culture
In the process of Mtb culture, 10 mL clinical samples were firstly mixed with the same volume of 2% NALC-NaOH in the 50 mL sterile plastic centrifuge tube for 20 minutes. After centrifuging for 15 minutes, sediment were retained and washed with saline solution for one time. Then, 2 mL saline solution was used for floating and sinking process. For liquid culture, 0.5–1 mL suspension of the pathogen was inoculated in Mycobacteria Growth Indicator Tubes (MGIT). Tubes were then putted into the BACTEC MGIT 960 System at 37 °C and monitored for detecting the growth of Mtb. For solid culture, 0.2–0.5 mL suspension of each sample was inoculated in media at 37 °C. Kinyoun staining and light microscope were utilized to detect AFB from smears of positive tubes. With the help of the MGIT TB ID method (MPT 64: Becton Dickinson, Sparks, Maryland, USA), Mtb could be assessed.
Statistical analysis
The sensitivity, specificity, positive predictive value, negative predictive value, and Youden index of mNGS, traditional culture method, and Xpert were calculated. The Cohen’s Kappa test and McNemar’s test for a paired fourfold table were conducted for comparing the results of mNGS and traditional culture method or Xpert. We also carried out the parallel and serial diagnostic test. All P values were two-sided, and statistical significance was defined as P<0.05. The Statistical Package for the Social Sciences (SPSS) 22.0 statistical software (IBM SPSS, Chicago, IL, USA) for Windows version 19.0 was employed.
Results
mNGS data of clinical samples
More than one billion raw reads of all clinical samples were collected by BGISEQ-50 instrument. After filtering over 50 million short and low-quality reads, the clean reads ultimately accounted for nearly 50% of all detected reads. By analyzing and assessing the data, we found that a large proportion of clean reads were mapped to the human genome and Yanhuang genome. And only less than 1% of whole clean reads were mapped to the microbial genomes, including viruses, bacteria, fungi and parasites. For different kinds of clinical specimens, the detected sequences were also varied.
Patient characteristics
All clinical samples that finally diagnosis of TB was validated through specific PCR or pathological test. Among a total of enrolled 36 TB patients, 27 (75.0%) were male and 9 (25.0%) were female. In the non-TB group, there were more males than females (73.5% and 26.5% respectively). Samples were divided into two categories, including pulmonary (52.9%, 37/70) and extrapulmonary samples (47.1%, 33/70). In the whole cohort, BALF made up the largest proportion of clinical samples (40%, 28/70; Figure 1, Tables 1,S1). For 19 samples which were tested by mNGS, culture, and Xpert, the ratio of pulmonary (52.7%, 10/19) and extrapulmonary samples (47.4%, 9/19) was close (Table S2).
Table 1. Clinical characteristics of all patients (n=70).
| Characteristics | TB group (n=36) | Non-TB group (n=34) |
|---|---|---|
| Gender, n (%) | ||
| Female | 9 (25.0) | 9 (26.5) |
| Male | 27 (75.0) | 25 (73.5) |
| Pulmonary samples, n | ||
| BALF | 14 | 14 |
| Sputum | 3 | 3 |
| Lung tissue | – | 3 |
| Extrapulmonary samples, n | ||
| Pleural fluid | 14 | 11 |
| CSF | 4 | 1 |
| Pericardial fluid | 1 | – |
| Pus | – | 2 |
BALF, bronchoalveolar lavage fluid; CSF, cerebrospinal fluid; TB, tuberculosis.
Table S1. Detailed results of mNGS and culture method.
| Patient No. | Sample type | Group | Clinical diagnosis | Culture | mNGS | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mtb | NTM | Virus | Mtb | Bacteria and fungi | Virus | |||||
| 1 | Pleural fluid | TB | Pulmonary TB | Mtb | N | N | Mtb | Massilia timonae | CMV | |
| 2 | BALF | TB | Pulmonary TB | N | N | N | Mtb | N | N | |
| 3 | BALF | TB | Pulmonary TB | Mtb | N | N | Mtb | Haemophilus influenzae, Haemophilus aegyptius | N | |
| 4 | CSF | TB | CNS TB | N | N | N | Mtb | N | N | |
| 5 | Pleural fluid | TB | TB pleurisy | N | N | N | N | N | N | |
| 6 | Pleural fluid | TB | TB pleurisy | N | N | N | N | N | N | |
| 7 | Pleural fluid | TB | Pulmonary TB | Mtb | N | N | Mtb | N | CMV, EBV | |
| 8 | BALF | TB | Pulmonary TB | N | N | N | Mtb | Prevotella melaninogenicus, rothia mucilaginosa, actinomycetes | N | |
| 9 | BALF | TB | Pulmonary TB | Mtb | N | N | Mtb | Veillonella atypica | EBV | |
| 10 | BALF | TB | Pulmonary TB | Mtb | N | N | Mtb | Haemophilus influenzae | N | |
| 11 | BALF | TB | Pulmonary TB | N | N | N | N | pseudomonas fluorescens, Veillonella dispar, Actinomycesgraevenitzii, Streptococcus parasanguis, Talaromyces marneffei | HHV-7 | |
| 12 | Pericardial fluid | TB | Tuberculous pericarditis | N | N | N | N | N | N | |
| 13 | Pleural fluid | TB | TB pleurisy | N | N | N | Mtb | N | EBV | |
| 14 | Pleural fluid | TB | TB pleurisy | N | N | N | N | Staphylococcus aureus | N | |
| 15 | BALF | TB | Pulmonary TB, lymphoma | N | N | N | N | Haemophilus parainfluenzae, Prevotella salivae, Actinomycetes, Streptococcus parasanguis, Veillonella dispar | EBV | |
| 16 | Pleural fluid | TB | Pulmonary TB | N | N | N | N | Porphyromonas endodontalis, Fusobacterium nucleatum, Prevotella buccae, Slackia, Bifidobacterium adolescentis, Campylobacter rectus, Streptococcus constellatus | N | |
| 17 | Sputum | TB | Pulmonary TB | N | N | N | Mtb | Aspergillus niger, Aspergillus brasiliensis, Neisseria sicca, Neisseria macaque, Streptococcus parasanguis, Porphyromonas sordellii, rothia mucilaginosa, Prevotella salivae, Prevotella species, Veillonella dispar, Veillonella parvula | N | |
| 18 | BALF | TB | Pulmonary TB | Mtb | N | N | Mtb | Staphylococcus aureus | N | |
| 19 | BALF | TB | Pulmonary TB, lung cancer | N | N | N | Mtb | Prevotella melaninogenicus, Streptococcus oralis, Fusobacterium nucleatum, Porphyromonas sordellii | N | |
| 20 | Sputum | TB | Pulmonary TB | Mtb | NTM (rapidly growing mycobacteria) | NTB | Mtb | Candida albicans, Actinomyces odontolyticus, rothia mucilaginosa, Neisseria flavescens, Veillonella atypica, Prevotella salivae | EBV | |
| 21 | BALF | TB | Pulmonary TB | Mtb | N | N | Mtb | Acinetobacter baumanni, Pseudomonas aeruginosa, Porphyromonas sordellii, Candida tropicalis, Streptococcus mitis, rothia mucilaginosa, Prevotella melaninogenicus | N | |
| 22 | Pleural fluid | TB | TB Pleurisy | N | N | N | N | N | N | |
| 23 | CSF | TB | CNS TB | Mtb | N | N | Mtb | N | N | |
| 24 | BALF | TB | Pulmonary TB | Mtb | N | N | Mtb | N | N | |
| 25 | Pleural fluid | TB | Pulmonary TB, lung cancer | Mtb | N | N | N | Streptococcus pneumoniae | EBV | |
| 26 | Pleural fluid | TB | TB Pleurisy | N | N | N | Mtb | N | N | |
| 27 | CSF | TB | CNS TB | Mtb | N | N | Mtb | – | N | |
| 28 | BALF | TB | Pulmonary TB, lung cancer | Mtb | N | N | N | Staphylococcus epidermidis, Prevotella histicola, Ochrobactrum anthropic, Bradyrhizobium elkanii, Kocuria palustris, Veillonella dispar, Veillonella atypica, Dangerous Ralstonia, Megasphaera elsdenii | N | |
| 29 | Sputum | TB | Pulmonary TB | N | N | N | Mtb | Lactobacillus salivarius, Lactobacillus crispatusC, Veillonella parvula, Streptococcus gordonii, rothia mucilaginosa | N | |
| 30 | BALF | TB | Pulmonary TB | N | N | N | Mtb | Bacteria | N | |
| 31 | CSF | TB | CNS TB | N | N | N | Mtb | N | N | |
| 32 | Pleural fluid | TB | TB lymhadenitis | N | N | N | Mtb | N | N | |
| 33 | BALF | TB | Pulmonary TB | N | N | N | Mtb | Bacteria | N | |
| 34 | Pleural fluid | TB | TB lymhadenitis | N | N | N | N | N | N | |
| 35 | Pleural fluid | TB | TB pleurisy | N | N | N | N | N | N | |
| 36 | Pleural fluid | TB | TB lymhadenitis | N | N | N | Mtb | N | N | |
| 37 | BALF | Non-TB | Lymphoma | N | N | N | N | Porphyromonas endodontalis, Neisseria elongate, Neisseria mucosa, Streptococcus salivarius, Veillonella dispar, Veillonella parvula, Mycoplasma orale | EBV | |
| 38 | Sputum | Non-TB | Bacterial pneumonia | N | N | N | N | Haemophilus parainfluenzae, Streptococcus sanguinis, Actinomyces odontolyticus, Veillonella parvula, Prevotella | N | |
| 39 | Sputum | Non-TB | Bacterial pneumonia | N | N | N | N | Veillonella parvula, Streptococcus infantarius, Rothia mucilaginosa, Enterococcus faecium, Candida albicans, Candida glabrata | HHV-1, EBV | |
| 40 | Sputum | Non-TB | Bacterial pneumonia | N | NTM (Mycobacterium intracellulare) | N | Prevotella, Veillonella atypica, Veillonella dispar, Neisseria macaque, Neisseria sicca, Streptococcus parasanguis, Streptococcus salivarius, Actinomyces odontolyticus | N | ||
| 41 | BALF | Non-TB | Granulomatous pneumonitis | N | N | N | Prevotella melaninogenicus, Streptococcus parasanguis, Veillonella atypica, Staphylococcus epidermidis, Gemella sanguinis | N | ||
| 42 | Pleural fluid | Non-TB | Hemangioendothelioma | N | N | N | N | N | N | |
| 43 | BALF | Non-TB | Lung cancer, fungal pneumonia | N | N | N | N | Actinomyces odontolyticus, Aspergillus fumigatus | N | |
| 44 | BALF | Non-TB | Tongue cancer | N | N | N | Actinomyces odontolyticus, Neisseria mucosa, Prevotella, Haemophilus parainfluenzae, Veillonella atypica, Rothia dentocariosa, Campylobacter rectus | N | ||
| 45 | Pleural fluid | Non-TB | Fungal pneumonia | N | N | N | N | N | EBV | |
| 46 | BALF | Non-TB | Autoimmune disease | N | N | N | N | Corynebacterium striatum | N | |
| 47 | Pleural fluid | Non-TB | Atypical pneumonia) | N | N | N | N | N | EBV | |
| 48 | BALF | Non-TB | Fungal pneumonia | N | N | N | N | Aspergillus fumigatus | N | |
| 49 | BALF | Non-TB | Bacterial and fungal pneumonia | N | N | N | N | Hemophilus | N | |
| 50 | BALF | Non-TB | Lung cancer | N | N | N | N | Prevotella melaninogenicus, Porphyromonas sordellii | N | |
| 51 | Lung tissue | Non-TB | Fungal pneumonia | N | N | N | N | Talaromyces marneffei, Echinococcus canadensis (parasite) | HHV-7 | |
| 52 | Lung tissue | Non-TB | HIV, fungal pneumonia | N | N | N | N | Talaromyces marneffei | EBV | |
| 53 | Lung tissue | Non-TB | Cryptococcal pneumonia | N | N | N | N | N | N | |
| 54 | Pleural fluid | Non-TB | Hemangioma | N | N | N | N | N | N | |
| 55 | BALF | Non-TB | Pneumonoconiosis | N | N | N | N | Pyramidobacter pisco-lens, Prevotella intermedia, Porphyromonas endodontalis, Prevotella, Micromonas micros, Campylobacter rectus, Olsenella uli, Treponema denticola | EBV | |
| 56 | Pleural fluid | Non-TB | Lung cancer | N | N | N | N | Klebsiella pneumoniae | N | |
| 57 | BALF | Non-TB | Lung cancer | N | N | N | N | N | N | |
| 58 | Pleural fluid | Non-TB | Bacterial pneumonia | N | N | N | Mtb | N | N | |
| 59 | Pleural fluid | Non-TB | Lymphoma | N | N | N | N | N | EBV | |
| 60 | BALF | Non-TB | Bacterial pneumonia | N | N | N | N | Prevotella melaninogenicus | N | |
| 61 | BALF | Non-TB | Bacterial pneumonia | N | N | N | N | Prevotella melaninogenicus, Prevotella salivae, Rothia mucilaginosa, Rothia dentocariosa, Streptococcus parasanguis, Streptococcus oralis, Fusobacterium periodonticum, Fusobacterium nucleatum, Veillonella atypica, Veillonella dispar | HHV-7 | |
| 62 | Pleural fluid | Non-TB | Lung abscess | N | N | N | N | N | N | |
| 63 | Pleural fluid | Non-TB | pleural effusion | N | N | N | N | N | N | |
| 64 | BALF | Non-TB | Bacterial pneumonia | N | N | N | N | Prevotella melaninogenicus, Ochrobactrum anthropic, Veillonella dispar, Veillonella atypica, Streptococcus parasanguis, Streptococcus mitis, Porphyromonas sordellii, Porphyromonas endodontalis, Campylobacter concisus, Campylobacter showae | N | |
| 65 | CSF | Non-TB | Fungal meningitis | N | N | N | N | Candida parapsilosis | N | |
| 66 | BALF | Non-TB | Bacterial pneumonia | N | N | N | N | Gram-negative bacteria | N | |
| 67 | Pus | Non-TB | Soft tissue abscesses | N | N | N | N | Bacteria | N | |
| 68 | Pus | Non-TB | Pleural effusion | N | N | N | N | Bacteria | N | |
| 69 | Pleural fluid | Non-TB | Lung abscess | N | N | N | N | N | N | |
| 70 | Pleural fluid | Non-TB | Disseminated NTM infection | N | N | N | NTM | N | N | |
BALF, bronchoalveolar lavage fluid; CSF, cerebrospinal fluid; CMV, cytomegalovirus; EBV, Epstein-Barr virus; HHV, human herpesvirus; Mtb, mycobacterium tuberculosis; mNGS, metagenomic next- generation sequencing; NTM, non-tuberculous mycobacterium; N, none; TB, tuberculosis.
Table S2. Clinical characteristics of 19 patients received Xpert test (n=19).
| Characteristics | TB group (n=13) | Non-TB group (n=6) |
|---|---|---|
| Gender, n (%) | ||
| Female | 4 (30.8) | 1 (16.7) |
| Male | 9 (69.2) | 5 (83.3) |
| Pulmonary samples, n | ||
| BALF | 6 | 2 |
| Sputum | 2 | – |
| Extrapulmonary samples, n | ||
| Pleural fluid | 5 | 2 |
| Pus | – | 2 |
BALF, bronchoalveolar lavage fluid; CSF, cerebrospinal fluid; TB, tuberculosis; Xpert, GeneXpert MTB/RIF assay.
Diagnostic performance of mNGS, culture, and Xpert in TB
On the basis of the classification of different specimens, the diagnostic performance of traditional culture, mNGS technology, Xpert, parallel diagnostic test, and serial diagnostic test were estimated (Tables 2,S3).
Table 2. Diagnostic performance of mNGS and traditional culture in TB.
| Samples | Sensitivity (95% CI) | Specificity (95% CI) | +PV (95% CI) | −PV (95% CI) | Youden index |
|---|---|---|---|---|---|
| All samples | |||||
| Traditional culture | 36.1% (0.213–0.538) | 100% (0.874–1.000) | 100% (0.717–1.000) | 59.6% (0.458–0.722) | 36.1% |
| mNGS | 66.7% (0.489–0.809) | 97.1% (0.829–0.998) | 96.0% (0.777–0.998) | 73.3% (0.578–0.849) | 63.8% |
| Parallel diagnostic test | 72.2% (0.546–0.852) | 97.1% (0.829–0.998) | 96.3% (0.791–0.998) | 76.7% (0.610–0.877) | 69.3% |
| Serial diagnostic test | 69.4% (0.517–0.831) | 100% (0.834–1.000) | 96.3% (0.791–0.998) | 76.7% (0.610–0.877) | 69.4% |
| Pulmonary samples | |||||
| Traditional culture | 47.1% (0.239–0.715) | 100% (0.800–1.000) | 100% (0.598–1.000) | 69.0% (0.490–0.840) | 47.1% |
| mNGS | 82.4% (0.558–0.953) | 100% (0.800–1.000) | 100% (0.732–1.000) | 87.0% (0.653–0.966) | 82.4% |
| Parallel diagnostic test | 88.2% (0.623–0.979) | 100% (0.800–1.000) | 100% (0.747–1.000) | 90.9% (0.694–0.984) | 88.2% |
| Serial diagnostic test | 41.2% (0.194–0.665) | 100% (0.800–1.000) | 100% (0.561–1.000) | 66.7% (0.471–0.821) | 41.2% |
| Extrapulmonary samples | |||||
| Traditional culture | 26.3% (0.101–0.514) | 100% (0.732–1.000) | 100% (0.463–1.000) | 50.0% (0.311–0.689) | 26.3% |
| mNGS | 47.4% (0.252–0.705) | 92.9% (0.642–0.996) | 90.0% (0.541–0.995) | 56.5% (0.349–0.761) | 40.3% |
| Parallel diagnostic test | 57.9% (0.340–0.789) | 92.9% (0.642–0.996) | 91.7% (0.598–0.996) | 61.9% (0.387–0.810) | 50.8% |
| Serial diagnostic test | 21.1% (0.070–0.461) | 100% (0.732–1.000) | 100% (0.396–1.000) | 48.3% (0.299–0.671) | 21.1% |
TB, tuberculosis; mNGS, metagenomic next-generation sequencing; 95% CI, 95% confidence interval; +PV, positive predictive value; −PV, negative predictive value.
Table S3. Diagnostic performance of mNGS, traditional culture, and Xpert in TB.
| Samples | Sensitivity (95% CI) | Specificity (95% CI) | +PV (95% CI) | −PV (95% CI) | Youden index |
|---|---|---|---|---|---|
| All samples | |||||
| Traditional culture | 46.2% (0.204–0.739) | 100% (0.517–1.000) | 100% (0.517–1.000) | 46.2% (0.204–0.739) | 46.2% |
| mNGS | 76.9% (0.460–0.938) | 100% (0.517–1.000) | 100% (0.655–1.000) | 66.7% (0.309–0.910) | 76.9% |
| Xpert | 61.5% (0.323–0.849) | 100% (0.517–1.000) | 100% (0.598–1.000) | 54.5% (0.246–0.819) | 61.5% |
| mNGS + Xpert | 84.6% (0.537–0.973) | 100% (0.517–1.000) | 100% (0.679–1.000) | 75.0% (0.356–0.955) | 84.6% |
| mNGS + Xpert + traditional culture | 92.3% (0.621–0.996) | 100% (0.517–1.000) | 100% (0.699–1.000) | 85.7% (0.420–0.992) | 92.3% |
| Pulmonary samples | |||||
| Traditional culture | 62.5% (0.259–0.898) | 100% (0.198–1.000) | 100% (0.463–1.000) | 40.0% (0.073–0.830) | 62.5% |
| mNGS | 87.5% (0.467–0.993) | 100% (0.198–1.000) | 100% (0.560–1.000) | 66.7% (0.125–0.982) | 87.5% |
| Xpert | 75.0% (0.356–0.955) | 100% (0.198–1.000) | 100% (0.517–1.000) | 50.0% (0.092–0.908) | 75.0% |
| mNGS + Xpert | 100.0% (0.598–1.00) | 100% (0.198–1.000) | 100% (0.598–1.000) | 100% (0.198–1.00) | 100% |
| mNGS + Xpert + traditional culture | 100.0% (0.598–1.00) | 100% (0.198–1.000) | 100% (0.598–1.000) | 100% (0.198–1.00) | 100% |
| Extrapulmonary samples | |||||
| Traditional culture | 20.0% (0.011–0.701) | 100% (0.396–1.000) | 100% (0.055–1.000) | 50.0% (0.174–0.826) | 20.0% |
| mNGS | 60.0% (0.170–0.927) | 100% (0.396–1.000) | 100% (0.310–1.000) | 66.7% (0.241–0.940) | 60.0% |
| Xpert | 40.0% (0.073–0.830) | 100% (0.396–1.000) | 100% (0.198–1.000) | 57.1% (0.202–0.882) | 40.0% |
| mNGS + Xpert | 60.0% (0.170–0.927) | 100% (0.396–1.000) | 100% (0.310–1.000) | 66.7% (0.241–0.940) | 66.7% |
| mNGS + Xpert + traditional culture | 80.0% (0.396–1.00) | 100% (0.396–1.000) | 100% (0.396–1.000) | 80.0% (0.299–0.989) | 80.0% |
TB, tuberculosis; mNGS, metagenomic next-generation sequencing; Xpert, GeneXpert MTB/RIF assay; 95% CI, 95% confidence interval; +PV, positive predictive value; −PV, negative predictive value.
Regarding all clinical TB specimens, mNGS reached a sensitivity of 66.7% (95% CI: 0.489–0.809), which was much superior to traditional culture method (36.1%, 95% CI: 0.213–0.538). Rather, the specificity of applying mNGS to detecting Mtb was 97.1% (95% CI: 0.829–0.998), which was slightly lower than that of traditional culture (100%, 95% CI: 0.874–1.000). Taking clinical final diagnosis as the standard, the positive predictive value and negative predictive value of mNGS in the analysis of Mtb were calculated as 96.0%, 73.3%, respectively. In comparison with the traditional culture method, the results also showed that the Youden index of all these clinical samples by mNGS was higher (36.1% vs. 63.8%).
The diagnostic performance of mNGS and culture in TB varied from samples to samples. In 17 pulmonary TB cases, mNGS identified fourteen cases with the best sensitivity of 82.4% (95% CI: 0.558–0.953), which was still outperformed conventional culture (47.1%, 95% CI: 0.239–0.715). In 19 extrapulmonary TB cases, mNGS produced a sensitivity of 47.4% (95% CI: 0.252–0.705), which was better than traditional culture (26.3%, 95% CI: 0.101–0.514). The same specificity of mNGS and culture in detecting Mtb in 20 pulmonary samples of non-TB patients had been achieved (both 100%, 95% CI: 0.800–1.000). However, in 14 extrapulmonary samples of non-TB participants, the specificity of mNGS dropped slightly to 92.9% (95% CI: 0.642–0.996), whereas traditional culture did not (100%, 95% CI: 0.732–1.000). For pulmonary or extrapulmonary samples, the Youden index of mNGS was calculated as 82.4%,73.3%, respectively. And traditional culture had the Youden index with 47.1%, 26.3%, respectively.
In comparison with Xpert, mNGS also showed better sensitivity in all clinical (76.9%, 95% CI: 0.460–0.938 vs. 61.5%, 95% CI: 0.323–0.849), pulmonary (87.5%, 95% CI: 0.467–0.993 vs. 75.0%, 95% CI: 0.356–0.955), and extrapulmonary samples (60.0%, 95% CI: 0.170–0.927 vs. 40.0%, 95% CI: 0.073–0.830). The sensitivity and positive predictive value of mNGS, culture, and Xpert all reached 100% in patients who received all three tests. In terms of detecting Mtb by single method, the Youden index of mNGS ranked first, Xpert second, and culture third (Table S3).
For the whole cohort, the only false-positive result was produced by mNGS (Patient nos. 58, Table S1). mNGS reported the infection of Mtb in the sample of pleural fluid, while the negative results were demonstrated by Mtb culture and PCR in the same sample. The hospitalized patient, with clinical final diagnosis of bacterial pneumonia, was discharged after receiving anti-infection therapy.
Diagnostic performance of mNGS in combination with culture or Xpert in TB
We combined NGS with traditional culture through two ways and measured the diagnostic efficacy of combined diagnostic test in all enrolled patients. When a total of 36 TB cases were taken into consideration, the sensitivity of parallel diagnostic test was improved to 72.2% (95% CI: 0.546–0.852), which was higher than mNGS alone, traditional culture method alone, and serial diagnostic test (69.4%, 95% CI: 0.517–0.831). Similar elevated results were obtained when all TB cases were divided into two groups, including pulmonary and extrapulmonary TB group. However, the specificity of parallel and serial diagnostic test in detecting Mtb were not heightened remarkably Among all methods and sample classifications, the Youden index of parallel diagnostic test in pulmonary samples was the highest (mNGS/culture 88.2%; Table 2).
The improvement of diagnostic performance of NGS in combination with Xpert in TB were also found. The sensitivity, specificity, positive predictive value, negative predictive value, and Youden index of mNGS plus Xpert in all clinical samples was 84.6% (95% CI: 0.537–0.973), 100% (95% CI: 0.517–1.000), 100% (95% CI: 0.679–1.000), 75.0% (95% CI: 0.356–0.955), 84.6%, respectively. The Youden index of mNGS/Xpert in pulmonary samples was outstanding (100%; Table 2). When combined mNGS with culture and Xpert, the diagnostic performance in TB was further enhanced in extrapulmonary samples (Youden index: mNGS/Xpert 66.7% vs. mNGS/Xpert/culture, 80.0%; Table S3).
Relationship and concordance between mNGS, culture, and Xpert
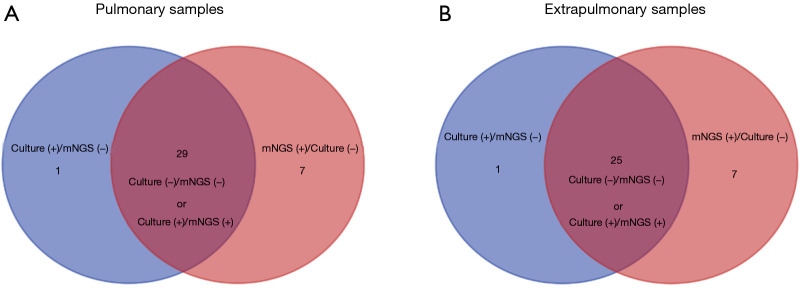
The same Mtb outcomes of both mNGS and culture method were observed in 54 of 70 cases (Figure 2). On the contrary, 16 of 70 samples showed the opposite Mtb results by mNGS and culture. The infection of Mtb was detected in patient nos.25 and nos.28 by culture, but not by mNGS (Table S1). In order to better understand the relationship between mNGS and traditional culture or Xpert method, we carried out the McNemar’s test and the consistency test based on the different classifications of clinical specimens (Tables 3,S4).
Figure 2.
Venn diagrams in different classification of clinical samples. (A) The same Mtb outcomes of both mNGS and culture method were observed in 29 of 37 cases with pulmonary samples; (B) the same Mtb outcomes of both mNGS and culture method were observed in 25 of 33 cases with extrapulmonary samples.
Table 3. Relationship between mNGS and traditional culture.
| mNGS of samples | Culture | P1 | Kappa value (P2) | |
|---|---|---|---|---|
| Negative | Positive | |||
| All samples, n (%) | <0.001 | 0.433 (<0.001) | ||
| Negative | 43 (75.4) | 2 (15.4) | ||
| Positive | 14 (24.6) | 11 (84.6) | ||
| Pulmonary samples, n (%) | 0.004 | 0.498 (0.001) | ||
| Negative | 22 (75.9) | 1 (12.5) | ||
| Positive | 7 (24.1) | 7 (87.5) | ||
| Extrapulmonary samples, n (%) | 0.033 | 0.368 (0.016) | ||
| Negative | 21 (75.0) | 1 (20.0) | ||
| Positive | 7 (25.0) | 4 (80.0) | ||
Statistically significant data were marked with bold and underline. TB, tuberculosis; mNGS, metagenomic next-generation sequencing; P1, P value of McNemar’s test; P2, P value of Cohen’s Kappa test.
Table S4. Relationship between mNGS and Xpert.
| mNGS of samples | Xpert | P1 | Kappa value (P2) | ||
|---|---|---|---|---|---|
| Negative | Positive | ||||
| All samples, n (%) | |||||
| Negative | 8 (88.9) | 3 (30.0) | 0.020 | 0.582 (0.009) | |
| Positive | 1 (11.1) | 7 (70.0) | |||
| Pulmonary samples, n (%) | |||||
| Negative | 2 (66.7) | 2 (28.6) | 0.500 | 0.348 (0.260) | |
| Positive | 1 (33.3) | 2 (71.4) | |||
| Extrapulmonary samples, n (%) | |||||
| Negative | 6 (100.0) | 1 (33.3) | 0.083 | 0.727 (0.023) | |
| Positive | 0 (0.0) | 2 (66.7) | |||
Statistically significant data were marked with bold and underline. TB, tuberculosis; mNGS, metagenomic next-generation sequencing; Xpert, GeneXpert MTB/RIF assay; P1, P value of McNemar’s test; P2, P value of Cohen’s Kappa test.
The correlation between mNGS and culture in all 70 specimens was statistically significant (P<0.001). With a kappa statistic of 0.443 (P<0.001), a certain degree of concordance between two methods was demonstrated. Similarly, the significant relationship and modest consistency between NGS and culture were also found in 37 pulmonary samples (P=0.004; kappa =0.498, P=0.001), and 33 extrapulmonary samples (P=0.033; kappa =0.368, P=0.016; Table 3). The significant correlation between mNGS and Xpert was observed only when all 19 samples were taken into account (P=0.020). The kappa value of mNGS and Xpert in all, pulmonary, and extrapulmonary samples was 0.582 (P=0.009), 0.348 (P=0.260), 0.727 (P=0.023), respectively. The kappa statistic illustrated satisfying concordance between mNGS and Xpert in all and extrapulmonary samples (Table S4).
The classification results of different pathogens
The single patient diagnosed with TB could carry a number of pathogens rather than only Mtb, such as Haemophilus influenzae, Candida albicans, Aspergillus, etc. Apart from Mtb, 143 bacteria, 12 fungus, and 19 viruses were detected in total of 70 clinical samples by mNGS technology. By comparison, traditional culture method only revealed the results of mycobacteria, including Mtb and non-tuberculous mycobacterium (NTM). For Patient nos. 20, who was diagnosed with Mtb infection by both these two means, the Candida albicans infection was detected only by means of mNGS. It is noteworthy that mNGS has great advantages in the detection of viruses, whereas traditional culture method do not. For example, Patient nos. 7 was found to contain Epstein-Barr virus (EBV) and cytomegalovirus (CMV) infection by mNGS (Table S1).
Discussion
The whole cohort of clinically suspected for TB were measured by mNGS or traditional culture method, among whom 19 patients also received Xpert test. The aim of this research was to assess the diagnostic performance of mNGS in TB, and the relationship between mNGS and traditional culture or Xpert method. The results underlined that the sensitivity of mNGS was better than traditional culture and Xpert, especially in pulmonary specimens. In pulmonary TB cases, all mNGS, culture, and Xpert methods demonstrated better diagnostic capacity for TB when compared with extrapulmonary TB cases. The remarkable diagnostic efficacy of parallel diagnostic test showed the potential of combining mNGS with culture or Xpert in TB clinical application. Meanwhile, the correlation and concordance between mNGS and culture method in all types of specimens was statistically significant (both P<0.001).
Multiple studies demonstrated the clinical value of mNGS in multiple types of diseases, including TB, cancer, and other infection disease. In metastatic melanoma (28), lung cancer (29) and breast cancer (30), mNGS showed its distinct advantages in guiding therapeutic options by detecting multiple mutations simultaneously. For intensive care unit (ICU) patients with sepsis, mNGS technology was employed to identify various pathogens, which could help select the appropriate antibiotic therapy quickly and accurately (31,32). Fang et al. (33) reported that a HIV-infected patient without classical rash was firstly diagnosed with Varicella-zoster virus (VZV) infection by mNGS and further verified by other tests, which showed the clinical benefits of mNGS. When it comes to respiratory samples in TB, Votintseva et al. (21) confirmed the ability of mNGS in providing outcomes of TB infection and antitubercular susceptibility. For sputum samples in TB, with the help of mNGS, the drug-resistant Mtb was identified faster than traditional culture (34-36). However, there is limited information about the diagnostic performance of mNGS in different samples in TB. Our study indicated that the clinical potential of applying mNGS to a variety of clinical specimens, including pulmonary and extrapulmonary specimens. Meanwhile, mNGS also showed its advantages in improving the sensitivity of detecting Mtb and providing more comprehensive profiles of pathogen constitution when compared with traditional culture and Xpert method. The finding was in accord with a recent study of Zhou et al. (22), further enhancing high credibility of these results. Moreover, we also found the potential of combining mNGS with culture or Xpert parallelly in identifying Mtb infection.
The results showed that mNGS failed to identify12 of 36 TB cases. The extrapulmonary cases accounted for the majority of 12 false-negative cases (75.0%, 9/12). The feature of releasing rather fewer extracellular nucleic acids by Mtb, which is one of the intracellular bacteria, may finally add difficulty in detecting Mtb by mNGS. In addition, the empirical treatments of anti-tuberculosis drugs may lead to the death of Mtb, thus resulting in the decreased sensitivity of both mNGS and traditional culture method (22,37). In our view, it is of great significance to improve the ability of mNGS in grasping meaningful reads of Mtb genomes, and collect samples for diagnostic tests before antibiotic therapy.
The only false-positive TB case of the study happened in Patient nos. 58. Both culture and PCR demonstrated the negative results of Mtb in Patient nos. 58, which was contrary to the mNGS result. In addition, the detection of non-pathogenic microbe also limited the diagnostic performance of mNGS. The sequences of non-pathogens, such as Staphylococcus epidermidis and Neisseria mucosa, was detected in our study. One of the reasons for false-positive errors is the common existence of trace quantities of DNA contaminants in laboratory reagents and centrifuge tubes (38,39). The false-positive errors also occur in the process of mNGS, such as the pollution in one of the multiple steps and misannotated species. To fully utilize the power of mNGS, it is worth further improving the sterile process of collecting and processing clinical specimens, establishing the negative control libraries of high quality, and applying scientific calibration procedures. We hold the opinion that using mNGS in TB should be considered upon a fair balance of its sensitivity and specificity.
When compared with the positive culture result, the difficulty of releasing sufficient genomic DNA by Mtb may also lead to the false-negative result of mNGS. Besides, due to the insufficient number of Mtb in clinical samples which failed to meet the analytical concentration of mNGS, the opposite result between mNGS and culture in patient nos.25 and nos.c28 was found (40). When compared with Xpert which was recommended for Mtb detection by WHO, mNGS shown better performance in pulmonary and extrapulmonary samples. In addition, the combination of mNGS and Xpert could make up the defect of Xpert in detecting Mtb in extrapulmonary samples. By means of mNGS, the diagnostic speed can be nearly 5 times faster than it of traditional culture method in Mtb detection (3 days vs. over 14 days). However, the testing time of mNGS has not surpass Xpert (3 days vs. 2–3 hours). In consideration of the influences of samples types in the comparison of mNGS, culture and Xpert results, we applied the same batch of samples in these methods. Another limitation is that specimen preservation for few days may affect the accuracy of mNGS, culture and Xpert. Given these, we should appraise the mNGS report dialectically and make clinical diagnosis on the basis of various testing data. Moreover, in order to the better clinical application, the turnaround time and cost of mNGS can’t be neglected as well. Automation, standardization process, and optimization of the connection between hospital departments are all good choices to advance the development of mNGS. In addition, the lower price of reagent and the parallel processing technique which refers to test different types of samples simultaneously, also do good to the turnaround time and the cost of mNGS.
There are some limitations in our study. Firstly, the results and hypothesis of the study were presented on the basis of a rather small and heterogeneous cohort. Secondly, as a retrospective and single-centered study, some clinical information like treatment data before samples collection was insufficient. For further investigation and stronger credibility, a prospective and multi-centered study with larger sample size is necessary.
Conclusions
In this study, mNGS showed better diagnostic performance than traditional culture and Xpert method in patients who were clinically suspected of TB infection. With high sensitivity and specificity, we also found that combining mNGS with culture or Xpert showed outstanding advantages in clinical diagnosis of TB. Furthermore, the significant correlation and concordance between mNGS and culture method in all types of specimens was elucidated. More researches need to be conducted to explore better clinical application of mNGS in the future.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This study was supported in part by a grant from National Natural Science Foundation of China (81802255), Shanghai Pujiang Program (17PJD036) and a grant from Shanghai Municipal Commission of Health and Family Planning Program (20174Y0131), National Key Research & Development Project (2016YFC0902300), Major disease clinical skills enhancement program of three year action plan for promoting clinical skills and clinical innovation in municipal hospitals, Shanghai Shen Kang Hospital Development Center Clinical Research Plan of SHDC (16CR1001A), “Dream Tutor” Outstanding Young Talents Program (fkyq1901), Key Disciplines of Shanghai Pulmonary Hospital (2017ZZ02012), Grant of Shanghai Science and Technology Commission (16JC1405900).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and approved by Shanghai Pulmonary Hospital (ethical number K19-158). All patients signed the written informed consent before the collection of samples.
Footnotes
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at http://dx.doi.org/10.21037/jtd-20-1232
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jtd-20-1232
Peer Review File: Available at http://dx.doi.org/10.21037/jtd-20-1232
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-20-1232). The authors have no conflicts of interest to declare.
References
- 1.Harding E. WHO global progress report on tuberculosis elimination. Lancet Respir Med 2020;8:19. 10.1016/S2213-2600(19)30418-7 [DOI] [PubMed] [Google Scholar]
- 2.Lytras T, Kalkouni O. The global tuberculosis epidemic: turning political will into concrete action. J Thorac Dis 2018;10:S3149-52. 10.21037/jtd.2018.08.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reisner BS, Gatson AM, Woods GL. Evaluation of mycobacteria growth indicator tubes for susceptibility testing of Mycobacterium tuberculosis to isoniazid and rifampin. Diagn Microbiol Infect Dis 1995;22:325-9. 10.1016/0732-8893(95)00147-7 [DOI] [PubMed] [Google Scholar]
- 4.Diacon AH, Pym A, Grobusch MP, et al. Multidrug-resistant tuberculosis and culture conversion with bedaquiline. N Engl J Med 2014;371:723-32. 10.1056/NEJMoa1313865 [DOI] [PubMed] [Google Scholar]
- 5.Hanna BA, Ebrahimzadeh A, Elliott LB, et al. Multicenter evaluation of the BACTEC MGIT 960 system for recovery of mycobacteria. J Clin Microbiol 1999;37:748-52. 10.1128/JCM.37.3.748-752.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krajewski W, Kasielski M, Król M, et al. The Abbott LCx-MTB assay is more sensitive than culture for detection of Mycobacterium tuberculosis in respiratory specimens. Arch Med Sci 2005;1:80-8. [Google Scholar]
- 7.Steingart KR, Henry M, Ng V, et al. Fluorescence versus conventional sputum smear microscopy for tuberculosis: a systematic review. Lancet Infect Dis 2006;6:570-81. 10.1016/S1473-3099(06)70578-3 [DOI] [PubMed] [Google Scholar]
- 8.Ninet B, Rohner P, Metral C, et al. Assessment of use of the COBAS AMPLICOR system with BACTEC 12B cultures for rapid detection of frequently identified mycobacteria. J Clin Microbiol 1999;37:782-4. 10.1128/JCM.37.3.782-784.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boehme CC, Nicol MP, Nabeta P, et al. Feasibility, diagnostic accuracy, and effectiveness of decentralised use of the Xpert MTB/RIF test for diagnosis of tuberculosis and multidrug resistance: a multicentre implementation study. Lancet 2011;377:1495-505. 10.1016/S0140-6736(11)60438-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zignol M, Dean AS, Alikhanova N, et al. Population-based resistance of Mycobacterium tuberculosis isolates to pyrazinamide and fluoroquinolones: results from a multicountry surveillance project. Lancet Infect Dis 2016;16:1185-92. 10.1016/S1473-3099(16)30190-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chakravorty S, Simmons AM, Rowneki M, et al. The New Xpert MTB/RIF Ultra: Improving Detection of Mycobacterium tuberculosis and Resistance to Rifampin in an Assay Suitable for Point-of-Care Testing. mBio 2017;8:e00812-17. 10.1128/mBio.00812-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ng KCS, van Deun A, Meehan CJ, et al. Xpert Ultra Can Unambiguously Identify Specific Rifampin Resistance-Conferring Mutations. J Clin Microbiol 2018;56:e00686-18. 10.1128/JCM.00686-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurtoglu MG, Ozdemir M, Kesli R, et al. Comparison of the GenoType((R)) MTBC Molecular Genetic Assay with culture methods in the diagnosis of tuberculosis. Arch Med Sci 2014;10:315-8. 10.5114/aoms.2012.29216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen YZ, Sun LC, Wen YH, et al. Pooled analysis of the Xpert MTB/RIF assay for diagnosing tuberculous meningitis. Biosci Rep 2020;40:BSR20191312. 10.1042/BSR20191312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allahyartorkaman M, Mirsaeidi M, Hamzehloo G, et al. Low diagnostic accuracy of Xpert MTB/RIF assay for extrapulmonary tuberculosis: A multicenter surveillance. Sci Rep 2019;9:18515. 10.1038/s41598-019-55112-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spitaleri A, Ghodousi A, Miotto P, et al. Whole genome sequencing in Mycobacterium tuberculosis. Ann Transl Med 2019;7:S197. 10.21037/atm.2019.07.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cole ST, Brosch R, Parkhill J, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 1998;393:537-44. 10.1038/31159 [DOI] [PubMed] [Google Scholar]
- 18.Wang J, Han Y, Feng J. Metagenomic next-generation sequencing for mixed pulmonary infection diagnosis. BMC Pulm Med 2019;19:252. 10.1186/s12890-019-1022-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan S, Qiao X, Liu L, et al. Next-Generation Sequencing of Cerebrospinal Fluid for the Diagnosis of Neurocysticercosis. Front Neurol 2018;9:471. 10.3389/fneur.2018.00471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson MR, Naccache SN, Samayoa E, et al. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med 2014;370:2408-17. 10.1056/NEJMoa1401268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Votintseva AA, Bradley P, Pankhurst L, et al. Same-Day Diagnostic and Surveillance Data for Tuberculosis via Whole-Genome Sequencing of Direct Respiratory Samples. J Clin Microbiol 2017;55:1285-98. 10.1128/JCM.02483-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou X, Wu H, Ruan Q, et al. Clinical Evaluation of Diagnosis Efficacy of Active Mycobacterium tuberculosis Complex Infection via Metagenomic Next-Generation Sequencing of Direct Clinical Samples. Front Cell Infect Microbiol 2019;9:351. 10.3389/fcimb.2019.00351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tagliani E, Cirillo DM, Kodmon C, et al. EUSeqMyTB to set standards and build capacity for whole genome sequencing for tuberculosis in the EU. Lancet Infect Dis 2018;18:377. 10.1016/S1473-3099(18)30132-4 [DOI] [PubMed] [Google Scholar]
- 24.Zignol M, Cabibbe AM, Dean AS, et al. Genetic sequencing for surveillance of drug resistance in tuberculosis in highly endemic countries: a multi-country population-based surveillance study. Lancet Infect Dis 2018;18:675-83. 10.1016/S1473-3099(18)30073-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gröschel MI, Walker TM, van der Werf TS, et al. Pathogen-based precision medicine for drug-resistant tuberculosis. PLoS Pathog 2018;14:e1007297. 10.1371/journal.ppat.1007297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589-95. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754-60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu ML, Zhou L, Sadri N. Comparison of targeted next generation sequencing (NGS) versus isolated BRAF V600E analysis in patients with metastatic melanoma. Virchows Arch 2018;473:371-7. 10.1007/s00428-018-2393-2 [DOI] [PubMed] [Google Scholar]
- 29.Sumimoto H, Takano A, Teramoto K, et al. Identification of candidate lung cancer neoantignes by next generation sequencing towards precision medicine. Ann Oncol 2017;28 Suppl 9:ix84-ix5. 10.1093/annonc/mdx697.046 [DOI] [Google Scholar]
- 30.Tan K, Yen YJ, Chen CJ, et al. Comprehensive next-generation sequencing to identify targetable genetic alterations in advanced breast cancer patients. Ann Oncol 2017;28 Suppl 9:ix79. 10.1093/annonc/mdx697.021 [DOI] [Google Scholar]
- 31.Long Y, Zhang Y, Gong Y, et al. Diagnosis of Sepsis with Cell-free DNA by Next-Generation Sequencing Technology in ICU Patients. Arch Med Res 2016;47:365-71. 10.1016/j.arcmed.2016.08.004 [DOI] [PubMed] [Google Scholar]
- 32.Burillo A, Bouza E. Use of rapid diagnostic techniques in ICU patients with infections. BMC Infect Dis 2014;14:593. 10.1186/s12879-014-0593-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang M, Weng X, Chen L, et al. Fulminant central nervous system varicella-zoster virus infection unexpectedly diagnosed by metagenomic next-generation sequencing in an HIV-infected patient: a case report. BMC Infect Dis 2020;20:159. 10.1186/s12879-020-4872-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doyle RM, Burgess C, Williams R, et al. Direct Whole-Genome Sequencing of Sputum Accurately Identifies Drug-Resistant Mycobacterium tuberculosis Faster than MGIT Culture Sequencing. J Clin Microbiol 2018;56:e00666-18. 10.1128/JCM.00666-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown AC, Bryant JM. Rapid Whole-Genome Sequencing of Mycobacterium tuberculosis Isolates Directly from Clinical Samples. J Clin Microbiol 2015;53:2230-7. 10.1128/JCM.00486-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nimmo C, Doyle R, Burgess C, et al. Rapid identification of a Mycobacterium tuberculosis full genetic drug resistance profile through whole genome sequencing directly from sputum. Int J Infect Dis 2017;62:44-6. 10.1016/j.ijid.2017.07.007 [DOI] [PubMed] [Google Scholar]
- 37.Causse M, Ruiz P, Gutierrez-Aroca JB, et al. Comparison of two molecular methods for rapid diagnosis of extrapulmonary tuberculosis. J Clin Microbiol 2011;49:3065-7. 10.1128/JCM.00491-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lusk RW. Diverse and widespread contamination evident in the unmapped depths of high throughput sequencing data. PLoS One 2014;9:e110808. 10.1371/journal.pone.0110808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salter SJ, Cox MJ, Turek EM, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biology 2014;12:87. 10.1186/s12915-014-0087-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gu W, Miller S, Chiu CY. Clinical Metagenomic Next-Generation Sequencing for Pathogen Detection. Annu Rev Pathol 2019;14:319-38. 10.1146/annurev-pathmechdis-012418-012751 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as