Introduction
Since the global outbreak of coronavirus disease 2019 (COVID-19), this viral pneumonia has attracted intense attention worldwide. The usual symptoms of COVID-19 include fever, cough, shortness of breath, and muscle ache, which are similar to those of severe acute respiratory system (SARS) and Middle Eastern respiratory syndrome (MERS) (1). As of March 20th, 2020, confirmed cases of severe acute respiratory system coronavirus 2 (SARS-CoV-2) infection have been reported in more than 180 countries and regions, with Asia, Europe and North America being the most heavily affected. SARS-CoV-2 was discovered via unbiased sequencing in samples from patients with pneumonia, and the virus showed the typical coronavirus morphology via transmission electron microscopy (TEM). The high resolving power of TEM permits studies at nanometer scale, providing direct images of viruses for diagnosis and research (2). TEM plays an important role in virus detection and is particularly useful for identifying unknown agents involved in particular outbreaks or transmission clusters; it has contributed to the discovery of many types of viruses and served as a diagnostic tool for direct identification of viruses in biological samples (3). Therefore, we used TEM to compare the morphology of the HCoV-229E and SARS-CoV-2 coronaviruses in ultrathin sections of sensitive infected cells and to further investigate the morphology of cells in bronchoalveolar lavage fluid (BALF) from two ICU patients with confirmed SARS-CoV-2 infection, so as to providing morphological bases for study of SARS-CoV-2
Methods
Huh7 cells infected with HCoV-229E and Vero E6 cells infected with SARS-CoV-2 were cultured at 37 °C for 48–72 h, and cells exhibiting cytopathic effects (CPEs) were obtained. BALF was obtained from ICU patients with confirmed COVID-19. Cells were centrifuged at 800–1,200 rpm for 5 minutes, and the supernatant was removed, inactivated by fixation and processed for standard TEM as follows: the sample was fixed in a mixed fixative solution containing 2.5% glutaraldehyde and 4% paraformaldehyde for 2 h at 4 °C, fixed in 1% osmium tetroxide, dehydrated through a graded series of ethanol (30%, 50%, 70%, 90% and 100%) and embedded in pure Epon 812 resin. Sections (70 nm) from samples were stained with uranyl acetate and lead citrate. Imaging was performed at 100 kV using a Hitachi JEM-1400 PLUS transmission electron microscope (Japan Electron Optics Laboratory Co., Ltd). Digital images of the specimens were acquired using an EMSIS VELETA G3 CCD camera and analysed by experienced electron microscopists.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was reviewed and approved by the Dongguan People’s Hospital Medical Ethics Board (approval #KYKT2020-005). All enrolled patients completed the informed consent form.
Results
Ultrastructural characteristics of SARS-CoV-2-infected cultured cells
The human coronaviruses OC43/229E and HKU1 have been confirmed to replicate steadily and induce CPEs in Huh7 cells (a human hepatoma cell line) (4-6). Coronavirus-infected cells usually show an irregular morphology with spreading from the edge to the centre of the cell layer and a gradual appearance as small aggregates; the number of interstitial particles increases, and the diseased cells become rounded, condensed, detached or fused, although the cytopathy usually does not extend throughout the cell monolayer (7). The same CPEs were observed in Huh7 cells infected with HCoV-229E in our cell culture model; most of the cells became rounded and aggregated, and some cells began to detach or fuse (Figure 1). TEM showed that HCoV-229E replicated and assembled in the cytoplasm of Huh7 cells, matured on the inner membrane of the rough endoplasmic reticulum, and filled in significantly expanded vesicles (Figure 2A). The virions were spherical or polymorphic with diameters ranging from 80 to 120 nm and were coated with elongated coronal fibrils arranged on the envelope surface. The nucleocapsid of the virions was helically symmetric, with a diameter ranging from 9 to 13 nm. Viral particles were released into the extracellular environment through exocytosis and often adhered to the cell membrane surface as aggregates (Figure 2B). Vero E6 cells, or rhesus monkey embryonic kidney cells, were the first cells in which SARS-CoV was isolated (8-10). We tried to inoculate Vero E6 cells with a clinically isolated SARS-CoV-2 virus (GenBank accession no. MT123290.1). Obvious CPEs were observed after culture: SARS-CoV-2-infected cells became rounded or irregular in shape and gradually aggregated, with relatively few cell lesions (Figure 1). In addition, TEM of Vero E6 cells infected with SARS-CoV-2 showed mature virions attached to the inner walls of vesicles (Figure 3A), and large amounts of viral particles were adhered to the cytoplasmic and extracellular membrane surfaces as well as the cellular processes of the Vero E6 cells (Figure 3B). Moreover, many infected cells showed typical ultrastructural changes consistent with apoptosis, suggesting that target cell apoptosis induced by SARS-CoV-2 may play an important role in the pathogenesis of COVID-19.
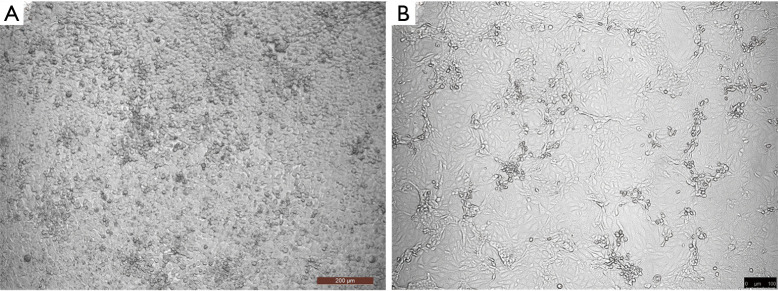
Figure 1.
CPEs of HCoV-229E in cultured Huh7 cells and SARS-CoV-2 in cultured Vero E6 cells, 100×. (A) CPEs of HCoV-229E in cultured Huh7 cells after 72 h. The cells became rounded and aggregated, and some cells began to detach or fuse. (B) CPEs of SARS-CoV-2 in cultured Vero E6 cells after 48 h. There were few cytopathic changes, and the cells became rounded or irregular in shape and aggregated. CPEs, cytopathic effects.
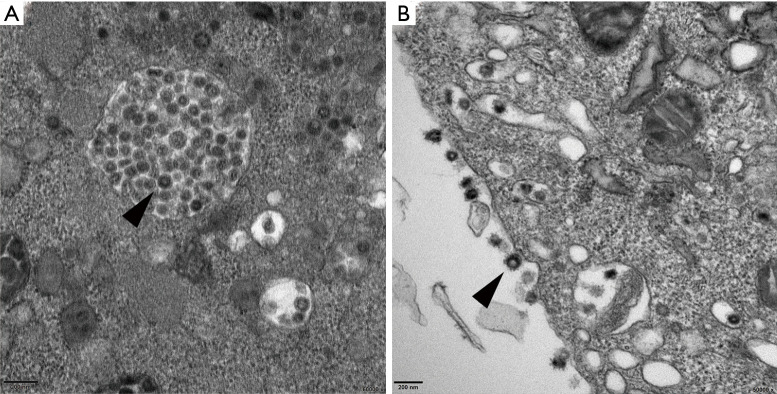
Figure 2.
Thin-section electron micrographs of HCoV-229E-infected Huh7 cells. (A) The vesicles in infected cells were filled with coronavirus virions of various sizes and spherical or multiform morphology (arrowhead) (Bar =200 nm, 50,000×); (B) The typical coronavirus virions were attached to the cell membrane via coronal spikes (arrowhead) (Bar =200 nm, 50,000×).
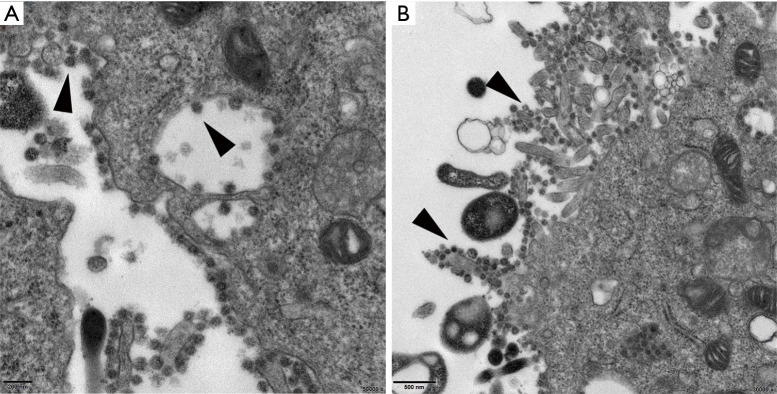
Figure 3.
Thin-section electron micrographs of Vero E6 cells infected with a SARS-CoV-2-positive throat swab sample. (A) Most viral particles were in the cytoplasm and outside the cell membrane and between the cellular processes; the mature virus particles were also attached to the inner wall of the vesicles (arrowhead) (Bar =200 nm, 50,000×); (B) Most mature virions were attached around the cytoplasm and between the cellular processes (arrowhead) (Bar =500 nm, 30,000×).
Ultrastructural characteristics of SARS-CoV-2-infected BALF cells
We explored the morphology of cells in BALF from two ICU patients with confirmed COVID-19.
Patient 1: A 69-year-old female developed symptoms of cough and production of sputum with unknown properties after returning to her hometown in Hubei province on January 25th, 2020 and developed fever two days later with an unknown maximum body temperature, and no dizziness or headache. Chest high-resolution computed tomography (HRCT) showed right middle lobular pneumonia. A throat swab tested positive for SARS-CoV-2 by real-time reverse transcription-polymerase chain reaction (rRT-PCR) on Jan 19th.
Patient 2: A 72-year-old female developed fever without an obvious cause on January 28th, 2020, the fever was accompanied by cough and expectoration of white, sticky sputum. A throat swab tested positive for SARS-CoV-2 by rRT-PCR on Jan 30th. Chest HRCT showed a small amount of chronic inflammation in the medial segment of the middle right lung lobe and local nodular thickening in the oblique fissure of the left lung on Jan 31st. She reported a travel history to Wuhan on Jan 18th–19th, 2020. The clinical diagnosis was consistent with COVID-19, and the patient was admitted to the hospital with essential hypertension and type 2 diabetes. BALF (4.5 mL) from patients was prepared for electron microscopy as described above. Type II alveolar epithelial cells with characteristic hemilamellar bodies were observed among BALF cells under electron microscopy; these organelles were filled with unequal numbers of mature virions in the markedly expanded rough endoplasmic reticulum and vesicles. The virions were spherical or pleomorphic, 80–120 nm in diameter, and composed of an envelope with coronal fibrils and a helical nucleocapsid (Figure 4A). Nucleic acid isolated from BALF of patient 1 tested positive for SARS-CoV-2 [the cycle thresholds (CT) value was approximately 21], consistent with the TEM results. Moreover, the viral particles existed mainly in type II alveolar epithelial cells, consistent with clinicopathological observations indicating that the virus destroyed alveolar epithelial cells and produced virus-induced cytopathic changes. In addition, we observed circular objects in the vesicles, which may have been polyvacuolar bodies or autophagic vesicles that engulfed particles (Figure 4A). Virions were observed in BALF from patient 2 under TEM (Figure 4B). However, nucleic acid isolated from BALF tested negative for SARS- CoV-2 (CT >37), inconsistent with the TEM results. We speculated that this discrepancy may have resulted from sampling error; in addition, the viral load of the patients did not decrease completely, possibly explaining why some discharged patients again have positive nucleic acid tests later. In addition, some cells exhibited deteriorated, autolysed, and disintegrated organelles resulting from a failure to fix BALF cells in time to prevent these events (data not shown). Therefore, timely fixation of electron microscopy specimens is extremely important for the interpretation of results.
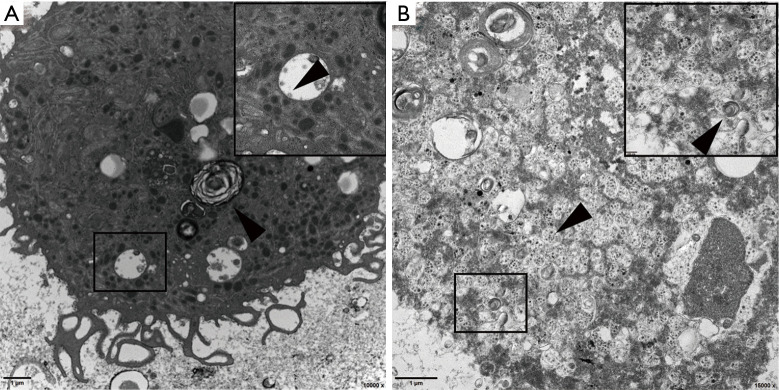
Figure 4.
Electron micrographs of (BALF from a patient infected with SARS-CoV-2. The top right corner shows an enlargement of the selected part of the image. (A) Electron micrographs of BALF from Patient 1showing lamellar bodies in type II alveolar epithelial cells (arrowhead). The virions attached to the smooth vesicles in the cytoplasm (arrowhead). Low magnification (Bar =1 µm, 10,000×), high magnification (Bar =200 nm, 40,000×). (B) Electron micrographs of BALF from Patient 2. The cells were fragmented and the organelles disintegrated; however, the cells, which should be type II alveolar epithelial cells, contained lamellar bodies (arrowhead), and virions were attached to the vesicles (arrowhead). Low magnification (Bar =1 µm, 15,000×), high magnification (Bar =200 nm, 50,000×).
Discussion
Our study shows that the morphology of SARS-CoV-2 did not differ significantly from that of HCoV-229E, as determined by electron microscopy. The SARS coronavirus has been reported to enter cells through membrane fusion instead of endocytosis, and the nucleocapsids then assemble in the endoplasmic reticulum and mature by budding into smooth vesicles derived from the Golgi apparatus. These smooth vesicles fuse with the cell membrane, and the mature virions are released (11,12). In our study, TEM also showed that SARS-CoV-2 entered cells through membrane fusion and matured in vesicles. This phenomenon suggests that the life cycle of SARS-CoV-2 might be similar to that of SARS-CoV. This finding may provide a morphological basis for further studying the mechanism of SARS-CoV-2. However, the clinical characteristics and pathological manifestations of COVID-19 caused by SARS-CoV-2 are different from those of SARS (13). Fu-Sheng Wang reported that the pathological features of a COVID-19 patient showed evident pneumocyte desquamation and hyaline membrane formation, and a large amount of mucus can be seen in the bronchial lumen, which may cause dyspnea and hypoxemia in clinical patients. These findings suggested that SARS-CoV-2 may have invaded bronchial epithelial cells and mucus cells (14). These findings suggest that SARS-CoV-2 may invade bronchial mucosal epithelial cells and mucus-secreting cells. Similarly, we also observed ultrastructural deterioration of the ciliated columnar epithelium in BALF from COVID-19 patients via TEM, which is consistent with pathological changes of COVID-19. So the establishment of animal models of SARS-CoV-2 infection is necessary for further study of the infection, transmission and pathogenesis of this disease.
In brief, transmission electron microscopy is a valuable technique. The species of virus can be distinguished by electron microscopy directly, which provides morphology information and counts of all viral particles, whether or not they are infectious. But there is no advantage in distinguishing virus subtypes, because of its limitation. However, TEM has a wider range of applications. It could be also used to investigate antiviral drugs. The process of antiviral drug suppressed viral invasion of cells could be observed by TEM, and this approach may be helpful for identifying the cellular target of viral invasion and further investigating viral pathogenesis.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This work was supported by National Key Research and Development Program of China (Grant No. 2018YFC1200503.), National Key Research and Development Program of China (Grant No. 2020YFC0842400), National Key Research and Development Program of China (Grant No. 2020YFC0841500), Guangdong Province (Grant No. 2020B111110001), Science and Technology Development Fund in Macao Special Administrative Region (Grant No. 0042/2020/A), The Key Projects of Dongguan City Social Science and Technology Development Plan (Grant No. 2018507150011645) and The Scientific Research and Development Fund Project of Dongguan people’s Hospital (Grant No. K201909).
Ethics Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was reviewed and approved by the Dongguan People’s Hospital Medical Ethics Board (approval #KYKT2020-005). All enrolled patients completed the informed consent form.
Footnotes
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jtd-20-1368
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-20-1368). The authors have no conflicts of interest to declare.
References
- 1.Du Y, Tu L, Zhu P, et al. Clinical Features of 85 Fatal Cases of COVID-19 from Wuhan. A Retrospective Observational Study. Am J Respir Crit Care Med 2020;201:1372-9. 10.1164/rccm.202003-0543OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richert-Pggeler KR, Franzke K, Hipp K, et al. Electron Microscopy Methods for Virus Diagnosis and High Resolution Analysis of Viruses. Front Microbiol 2019;9:3255. 10.3389/fmicb.2018.03255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roingeard P, Raynal PI, Eymieux S, et al. Virus detection by transmission electron microscopy: Still useful for diagnosis and a plus for biosafety. Rev Med Virol 2019;29:e2019. 10.1002/rmv.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freymuth F, Vabret A, Rozenberg F, et al. Replication of respiratory viruses, particularly influenza virus, rhinovirus, and coronavirus in HuH7 hepatocarcinoma cell line. J Med Virol 2005;77:295-301. 10.1002/jmv.20449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerna G, Campanini G, Rovida F, et al. Genetic variability of human coronavirus OC43-, 229E-, and NL63-like strains and their association with lower respiratory tract infections of hospitalized infants and immunocompromised patients. J Med Virol 2006;78:938-49. 10.1002/jmv.20645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vabret A, Dina J, Gouarin S, et al. Detection of the new human coronavirus HKU1: a report of 6 cases. Clin Infect Dis 2006;42:634-9. 10.1086/500136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou ZYR, editor. Manual of clinical common respiratory virus isolation and culture. 2015. [Google Scholar]
- 8.Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 2003;348:1953-66. 10.1056/NEJMoa030781 [DOI] [PubMed] [Google Scholar]
- 9.Drosten C, Gunther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 2003;348:1967-76. 10.1056/NEJMoa030747 [DOI] [PubMed] [Google Scholar]
- 10.Peiris JSM, Lai ST, Poon LLM, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 2003;361:1319-25. 10.1016/S0140-6736(03)13077-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qinfen Z, Jinming C, Xiaojun H, et al. The life cycle of SARS coronavirus in Vero. E6 cells. J Med Virol 2004;73:332-7. 10.1002/jmv.20095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ng ML, Tan SH, See EE, et al. Early events of SARS coronavirus infection in vero cells. J Med Virol 2003;71:323-31. 10.1002/jmv.10499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicholls JM, Poon LLM, Lee KC, et al. Lung pathology of fatal severe acute respiratory syndrome. Lancet 2003;361:1773-8. 10.1016/S0140-6736(03)13413-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Z, Shi L, Wang Y, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med 2020;8:420-2. 10.1016/S2213-2600(20)30076-X [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as