Abstract
The centromere is a chromosome locus that directs equal segregation of chromosomes during cell division. A nucleosome containing the histone H3 variant CENP-A epigenetically defines the centromere. Here, we summarize findings from recent structural biology studies, including several CryoEM structures, that contributed to elucidate specific features of the CENP-A nucleosome and molecular determinants of its interactions with CENP-C and CENP-N, the only two centromere proteins that directly bind to it. Based on those findings, we propose a role of the CENP-A nucleosome in the organization of centromeric chromatin beyond binding centromeric proteins.
Keywords: CENP-A, centromere, chromatin
Introduction
During cell division, centromeres play a fundamental role in directing the accurate segregation of sister chromatids. They mediate the recruitment of multiprotein megastructures, ‘kinetochores’. Kinetochores attach to microtubulebased spindle fibers that subsequently pull condensed chromosomes to opposite poles of the dividing cell [1]. Centromere defects have been reported to result in aneuploidy, cell death, and cancer [2]. The centromeric locus has specific DNA and protein composition. In complex eukaryotes, centromeric DNA is usually repetitive (termed as α-satellites in humans). Bioinformatics analysis has shown that centromeric DNA evolves very fast and shows little conservation between species [3,4]. Furthermore, the discovery of neocentromeres [5–7], new functional centromeres formed at ectopic loci (often as a consequence of the disruption of the original centromere), has reinforced the idea that a specific DNA sequence is not essential for centromere function. In contrast, the protein component of the centromere (in complex eukaryotes called constitutive centromere associated network (of proteins) - CCAN) is a hallmark of all functional centromeres, although its composition varies between species [8]. Interestingly, only one of the proteins in this complex, CENP-B, recognizes a particular DNA sequence (17-bp CENP-B box) [9,10] but its presence is neither conserved nor necessary for centromere function [5,8,11]. This information led to the conclusion that centromeres are defined epigenetically. In particular, numerous experiments have helped to establish the histone H3 variant CENP-A as an epigenetic mark of the centromere. CENP-A is sufficient to seed and propagate a functional centromere/kinetochore [12–20].
What the specific features of CENP-A are, and how they help to establish and maintain the centromere, have been critical questions in the field for the last couple of decades. Intensive work on the reconstitution of chromatin and centromeric complexes from purified components, together with advances in cryo-electron microscopy, have paved the way toward understanding the molecular determinants of centromeric chromatin. Here, we review the atomic structures of the CENP-A nucleosome (in isolation and in complex with other centromeric proteins) obtained to date, and we speculate on how the presence of CENP-A nucleosomes can modify chromatin architecture.
Structure of the CENP-A nucleosome
The presence of the centromere-specific histone H3 variant CENP-A is a conserved feature in most eukaryotic centromeres [21], although its sequence is changing fast across species [22–25]. Rapid evolution of CENP-A is connected with co-evolution of other centromere and kinetochore proteins [8,21] and with the evolution of underlining DNA [26]. Thus, although mediating the essential and evolutionally conserved process of chromosome segregation, the centromeres have a different architecture and compositions that are species specific. In this review, we will focus mainly on human and yeast CENP-A nucleosome analysis as most of the structural work has been done with proteins from these species.
CENP-A is the most divergent histone H3 variant in human (Figure 1A) [27]. In its histone-fold domain (HFD) and associated αN helix, it shares 62% sequence identity with the canonical H3, while the histone tails are highly dissimilar between the two. As a consequence, human CENP-A is subject to a unique, and limited, set of post-translational modifications (reviewed in [28]) (Figure 1A). Post-translational modifications are implicated in the CENP-A deposition at centromeres, its stability and maintenance, recruitment of CCAN proteins and in the specific organization of centromeric chromatin [28].
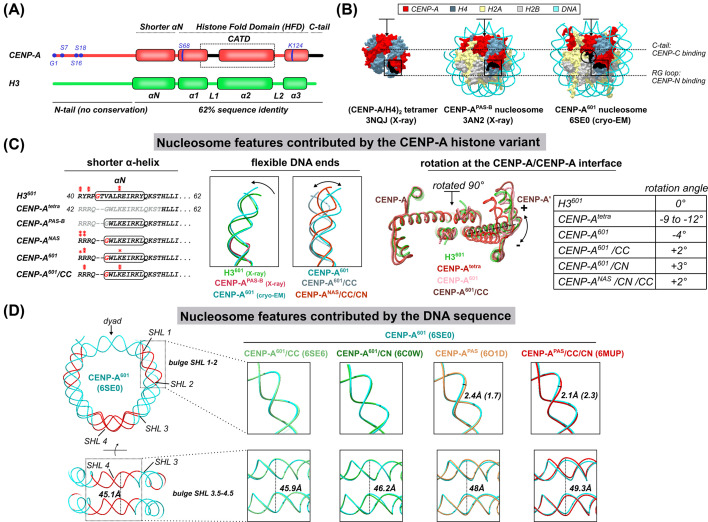
Figure 1. Structural features of CENP-A nucleosome.
(A) Schematic representation of H3 and CENP-A. The post-translational sites on CENP-A are colored in blue and the L1 loop and C-terminal tail, which specifically bind CENP-N and CENP-C respectively, are colored in black. (B) Surface representation of CENP-A-containing (sub)nucleosomal complexes, solved by X-ray crystallography and cryo-EM. CENP-A is in red, H4 is in blue, H2A is in yellow, H2B is in gray and the DNA is in cyan. CENP-A-specific features recognized by CENP-C (C-terminal tail) and CENP-N (RG-loop) are shown in black and boxed on each of the structures. The arrow is showing the position of the four-helix bundle between two CENP-A molecules (rotated 90° in (C), right). PDB ID and the experimental method used are indicated below each structure. (C) Nucleosome features contributed by CENP-A. Left: Multiple sequence alignment, illustrating a shorter α-N helix (boxed sequence) of CENP-A, observed in various structures. Residues interacting with DNA are labeled with one star if only one side of the nucleosome is involved in interactions, and with two stars if residues on both sides are interacting. Middle: overlay of terminal DNA in H3 nucleosome (3LZ0) and CENP-A nucleosome structures obtained by X-ray crystallography (3AN2) or cryo-EM (6SE0). The flexibility of terminal DNA is modulated by the binding of CCAN components (6SE0, 6MUP, 6SEE). Right: Rotation at the four-helix bundle formed between two CENP-A or two H3 molecules (indicated with the arrow in (B) and rotated 90° away from the viewer). The angle between α2 helices (amino acids 86–113) in the canonical nucleosome is taken as a reference. The rotation is most pronounced in the (CENP-A/H4)2 tetramer (3NQJ) and is greatly reduced in the CENP-A nucleosome (6SE0), while it virtually disappears when CENP-C and CENP-N bind the CENP-A nucleosome (6MUP). A table summarizing the measured rotation angles is shown on the right. (D) Nucleosome features contributed by the DNA sequence. Left: the DNA of the CENP-A601 nucleosome (6SE0) is shown in cyan and regions where important DNA path deviation is observed, in comparison with nucleosome wrapped in the α-satellite DNA, are highlighted in red. The distance between DNA gyres is measured between I-36 phosphate and I-41 phosphate. In order to highlight differences in the DNA path around the nucleosome in budge SHL 1–2 and budge SHL 3.5–4.5, on the right, are shown as zoomed views of different overlays. The first four panels show that there are no significant differences in the DNA path between the CENP-A601 nucleosome (cyan; 6SE0) and the CENP-A601 nucleosome, in complex with CENP-C (light green, 6SE6), or with CENP-N (dark green, 6C0W). The overlay of the CENP-A601 nucleosome (cyan; 6SE0) with the CENP-APAS nucleosome (orange; 6O1D) reveals a 2.4-Å deviation in the SHL 1–2 (1.7 Å at the opposite side of the nucleosome) and 2 Å widening of DNA gyres. When the CENP-A601 nucleosome (cyan; 6SE0) is overlaid with the CENP-APAS nucleosome with two CENP-C and two CENP-N molecules (orange; 6O1D), the difference at the SHL 1–2 is 2.4 Å (2.3 Å on the other side of the nucleosome) and widening between DNA gyres increases to 49.3 Å. All nucleosomes were overlaid by aligning the histone cores. Abbreviation: CENP-A601, CENP-A nucleosome on super-positioning 601 sequence. CENP-APAS, CENP-A nucleosome on palindromic α-satellite DNA sequence.
The indispensable role of CENP-A in epigenetic specification of centromeres and the relatively high sequence divergence, compared with other H3 variants, have long been motivators for understanding its structure within the chromatin. Over the years, studies done with proteins from different organisms, using both natural and artificial DNA sequences and employing different biophysical techniques, have led to various models for the CENP-A containing nucleosomes, evoking a different histone stoichiometry and composition along with a different DNA path (reviewed in [29]). However, to date all atomic resolution structures of CENP-A nucleosomes (Table 1) are octameric in nature with negatively supercoiled DNA, highly resembling canonical nucleosomes. Rather than altering the composition or the overall shape of the nucleosome, CENP-A is inferring unique dynamic properties to the otherwise expected nucleosome shape.
Table 1. High-resolution structures of CENP-A (sub)nucleosome.
| PDB | DNA | Species | Method | Stabilizing reagents | CCANs | Res. (Å) | Reference |
|---|---|---|---|---|---|---|---|
| CENP-A subnucleosomal complex: CENP-A/H4 tetramer | |||||||
| 3NQJ | - | Homo sapiens | X-ray | - | - | 2.1 | [31] |
| 3NQU | - | Homo sapiens | X-ray | - | - | 2.5 | [31] |
| CENP-A nucleosome | |||||||
| 3AN2 | Palindromic α-Satellite with CENP-B boxes (PAS-B) | Homo sapiens | X-ray | - | - | 3.6 | [35] |
| 6O1D | α-Satellite (NAS) | Homo sapiens | Cryo-EM | - | - | 3.4 | [40] |
| 6E0P | α-Satellite (NAS) | Homo sapiens | Cryo-EM | scFv* | - | 2.6 | |
| 6E0C | 601 | Homo sapiens | Cryo-EM | scFv* | - | 2.6 | |
| 6SE0 | 601 | Homo sapiens | Cryo-EM | - | - | 3.8 | [37] |
| 6UPH | 601 | Saccharomyces cerevisiae | Cryo-EM | Tween 20 | - | 2.7 | [44] |
| 6TEM | 601 | Homo sapiens and Xenopus laevis | Cryo-EM | - | - | 3.9 | [41] |
| CENP-A/H3.3 hybrid nucleosome | |||||||
| 3WTP | Palindromic α-Satellite (PAS) | Homo sapiens | X-ray | - | - | 2.7 | [45] |
| CENP-A nucleosome in complex with CCAN component(s) | |||||||
| 6BUZ | 601 | Homo sapiens | Cryo-EM | - | 1x CENP-N | 3.9 | [88] |
| 6C0W | 601 | Homo sapiens | Cryo-EM | - | 1x CENP-N | 4 | [89] |
| 6MUP | α-Satellite (NAS) | Homo sapiens | Cryo-EM | Glutaraldehyde | 2x CENP-C 2x CENP-N |
3.5 | [48] |
| 6MUO | α-Satellite (NAS) | Homo sapiens | Cryo-EM | 2x CENP-C 1x CENP-N |
3.6 | ||
| 6SE6 | 601 | Homo sapiens | Cryo-EM | - | 2x CENP-C | 3.5 | [37] |
| 6SEG | 601 | Homo sapiens | Cryo-EM | - | 2x CENP-C | 3.1 | |
| 6QLD | 601 | Saccharomyces cerevisiae | Cryo-EM | BS3† | CCAN | 4.1 | [46] |
| H3-CENP-A-H3 tri-nucleosome | |||||||
| 6L49 | 601 | Homo sapiens | Cryo-EM | Mg2+ | - | 18.9 | [43] |
Abbreviations: NAS, natural (non-palindromic) α-satellite DNA sequence; PAS, palindromic (engineered) α-satellite DNA sequence; PAS-B, PAS that has CENP-B box at the ends.
scFv - engineered a single-chain fragment from the PL2-6 antibody, which includes the variable heavy and light chains connected by a flexible linker.
BS3, bis(sulfosuccinimidyl)suberate.
In solution, CENP-A forms stable dimers with histone H4 that further dimerizes to form dimer-of-dimers or (CENP-A/H4)2 tetramers (similar to the canonical (H3/H4)2 tetramer). Hydrogen-deuterium exchange studies of the (CENP-A/H4)2 tetramer, indicated special solution dynamics imposed by CENP-A [30], inferring a more compact and rigid complex compared with the canonical counterpart. The first atomic resolution X-ray crystal structure of the human (CENP-A/H4)2 tetramer [31] clearly identified stronger hydrophobic interactions at the CENP-A/H4 interface, versus the H3/H4 interface and 9–12° rotation at the dimer/dimer interface, providing an explanation for solution studies (Figure 1B,C). The region conferring CENP-A rigidity in solution was mapped to loop 1 (L1) and the α2 helix within HFD and named CENP-A targeting domain (CATD) for its ability to drive centromeric localization when swapped in the H3 histone sequence. Later studies [32–34] showed that CATD is being recognized by the specific CENP-A chaperone HJURP, which ensures deposition at the centromere. The first high-resolution structure of the CENP-containing nucleosome came from crystallographic studies on human nucleosomes [35] (Figure 1B). The structure revealed an H3-like nucleosome with two copies of H2A, H2B, H4 and CENP-A and negatively supercoiled DNA, albeit disordered at the ends. Despite this, the high structural similarity with the canonical nucleosome could have been enforced by crystal contacts. Several high-resolution cryo-EM structures of the CENP-A nucleosome were recently published, which largely confirmed the findings observed in the X-ray structure. The cryo-EM structures also revealed fine details contributed by species-specific histone variants, DNA sequence and the binding of other proteins to the CENP-A nucleosome. Here, we summarize what we have so far learned about the unique features of the CENP-A nucleosome, contributed by the histone variant itself and structural features emphasized by α-satellite DNA.
Nucleosome features contributed by histone variant CENP-A
Shorter αN-helix and flexible DNA ends
The human CENP-A, just like H3, has a disordered N-terminal tail followed by an αN helix and a typical histone fold, consisting of three α-helices (Figure 1A). While the helices in the histone fold are stable, the αN helix of CENP-A is dynamic. In the absence of DNA, in (CENP-A/H4)2 tetramer, this helix is completely disordered [30,31] (Figure 1C, left). When the CENP-A packs in the nucleosome, the DNA stabilizes the αN helix, but to a lesser extent than the analogous helix of H3 in the canonical nucleosome. The αN helix in CENP-A is half a turn shorter than in H3 due to the position of the amino acids 46GW47 in CENP-A, which interfere with helix formation (glycine is a helix breaker) (Figure 1C). Furthermore, αN helix in H3 interacts with DNA through bi-dentate H3R49 interaction, which is replaced by the weaker CENP-AK49. The shorter αN helix, together with looser DNA binding, contribute to increased dynamics of the DNA ends in CENP-A nucleosome, clearly observed in solution during MNase digestion [31,35–38]. Shorter CENP-A nucleosomal DNA is in agreement with CENP-A ChIP-seq data, indicating that this feature is retained in cellular context [39]. The flexible DNA is observed in almost all structures of the CENP-A nucleosome, independently of the species, DNA sequence, or methods used [35,37,40–44]. Indeed, in the heterotypic human nucleosome with one copy of H3 and one copy of CENP-A [45], the flexibility of the DNA is clearly associated with the CENP-A part of the nucleosome, while the DNA on the H3 side is stably wrapped. The most extreme flexibility of the DNA ends in human nucleosomes are observed in the X-ray structure of the human CENP-A nucleosome [35] where the terminal 13-bp pairs on each DNA end are disordered. The use of an engineered palindromic DNA sequence with a CENP-B binding motif at the ends, and tight packaging in the crystal, might have contributed to pronounced DNA dynamics. Interestingly, the cryo-EM structure of the budding yeast CENP-A nucleosome assembled on the super-positioning 601 sequence, reveals almost the same extent of DNA disorder [44], while the binding of CCAN [46] further unwraps DNA, leaving only 105 bp to interact with the histone core. In contrast, the cryo-EM structures of the human CENP-A nucleosome in isolation [37,40] show traceable density for almost the entire 145–147 bp sequence of the nucleosomal DNA, albeit further from the nucleosome core than in the case of H3 nucleosomes, and with less intensity [47] indicating some degree of flexibility. A recent cryo-EM study on CENP-A nucleosome [41] exploited phase plate technology to decipher differential 601 DNA unwrapping from the two sides of the CENP-A nucleosome. However, while the nucleosomal DNA sequence generally had a minor effect on the terminal DNA flexibility in the reported CENP-A nucleosome in solution [37], the binding of the CCAN proteins showed an impact [37,48]. The binding of CENP-C potentiates DNA dynamics, indirectly through the destabilization of the C-terminal tail of H2A [37], but upon subsequent CENP-N binding, DNA tails become more rigid [48] (Figure 1C, middle). Although the structure of the CENP-A nucleosome in complex with part of CENP-C and CENP-N [48] demonstrates that both proteins can concomitantly bind the nucleosome, some studies are suggesting a dynamic nature of these associations through the cell cycle [49–51]. Thus, it is possible that the extent of terminal DNA dynamics on the CENP-A nucleosome also changes through the cell cycle as a consequence of modulations by different binding partners. One might also wonder what would be the functional implication of altered nucleosome DNA dynamics. Studies on nucleosomes assembled on longer DNA [42,43] show that the flexible nature of the DNA at the entry/exit sites results in an alternative path of the linker DNA, which does not cross above the dyad and thus cannot accommodate the binding of the histone H1. In this context, flexible DNA ends on the CENP-A nucleosome would translate into a unique chromatin structure specific for the centromeric region. Additionally, flexible DNA ends might help accommodate binding of other CCAN components, that was impaired when the CENP-A nucleosome was deprived of its terminal DNA flexibility [42,46]. In summary, the shorter αN helix and the looser DNA binding are intrinsically well-conserved features of the CENP-A nucleosome that are not heavily influenced by the DNA sequence itself, but are modulated by CCAN binding.
Rotation at the CENP-A/CENP-A interface
Another highly unique part of the CENP-A nucleosome is the tetramerization interface between two CENP-A/H4 dimers (Figure 1C). This interface is mediated exclusively with the CENP-A residues which make a four-helix bundle just below the dyad (Figure 1B, arrow). SAXS experiments in solution showed the (CENP-A/H4)2 tetramer to be much more compact than the (H3/H4)2 tetramer [31] and the crystal structure of the (CENP-A/H4)2 tetramer shows substantial (9–12°) rotation at the dimer–dimer interface. This rotation is significantly reduced when CENP-A forms nucleosomes (Figure 1C, right). Interestingly, the binding of CENP-C and/or CENP-N to the CENP-A nucleosome induces further rotation, resulting in a canonically shaped nucleosome. It is possible that the forces at the CENP-A/CENP-A interface mediate nucleosome dynamics or stability, but more research is required to establish functional relevance of these observations.
Nucleosome features contributed by DNA sequence
In organisms with a point centromere, like S. cerevisiae, the DNA sequence of the centromeric DNA is absolutely essential, as it is recognized by sequence-specific binding proteins. However, its importance in more complex eukaryotes is still controversial. In many organisms, natural centromeres are embedded in repetitive DNA, but at the same time, they can form and function elsewhere on the chromosome [5,52–54]. In humans, the centromeric, α-satellite, DNA, is a repetitive AT-rich sequence with the monomer length of 171 bp [3,4]. The 17-bp CENP-B box within this sequence is the only DNA sequence specifically recognized by the CCAN, and in particular CENP-B. However, since CENP-B knockout mice are viable [11,55] and neocentromeres [5,6] and human chromosome Y [56] are deprived of CENP-B, it seems like CENP-B function at the centromere is redundant.
Nevertheless, it is well established that the DNA sequence plays an important role in the formation and stability of the nucleosome itself [57,58]. Actually, the majority of the structural studies are done on nucleosomes wrapped with DNA sequences engineered with the aim to increase the nucleosome symmetry (palindromic sequences [59,60]) or nucleosome stability (super-positioning [61]). While engineered sequences make nucleosomes more amenable for biophysical studies, providing homogeneity and symmetry, the trade-off is the loss of valuable information about the nucleosome in its natural DNA environment. The research involving CENP-A nucleosomes is also affected by the need to artificially increase nucleosome stability in order to study them at high resolution. The crystal structure of the human CENP-A nucleosome was obtained using palindromic α-satellite DNA with CENP-B boxes at the ends (potentially increasing terminal DNA flexibility), while the use of the super-positioning 601 sequence [37,40], antibody fragments [40], cross-linking reagents [48] or detergents [44] were used to stabilize the CENP-A nucleosome for cryo-EM studies (Table 1). Interestingly, although the overall nucleosome shape is the same on different DNA sequences, a careful overlay points out clear deviations in the DNA path that is sequence specific. This indicates possible role of the DNA sequence in the centromere structure. While the tightly wrapped 601 sequence has the same path on both canonical and CENP-A nucleosome, with no or very slight deviations upon the binding of CENP-C or CENP-N, the natural centromeric α-satellite DNA deviates from that path in two places on the CENP-A nucleosome (Figure 1D). First, a 2-Å bulge between SHL1–SHL2 (Figure 1D, top) is present on all CENP-A structures with natural α-satellite DNA [40,48]. Interestingly, chromatin remodeling enzymes [62,63] and pioneering transcription factors [64,65] interact with the nucleosome at this site, while inducing nucleosome sliding or unwrapping. Other distinctive bulges of natural α-satellite DNA are detected between SHL 3.5–4.5 and −3.5 to −4.5, resulting in 2.9-Å widening of DNA gyres opposite from the dyad if CENP-A nucleosome on super-positioning 601 sequence (CENP-A601) and CENP-A nucleosome on natural α-satellite sequence (CENP-ANAS) are compared. However, in contrast with the SHL 1–2 location, which is basically insensitive to ligands binding to the nucleosome, this bulge decreases upon binding of the nucleosome-specific antibody and increases upon binding of CENP-C and CENP-N, resulting in total 4.2 Å gyre widening (Figure 1D, bottom). The binding of CENP-N accommodates a ∼1.5 Å shift of the DNA regions (SHL −3.5 and SHL 3.5) toward CENP-N binding site, which generates two symmetric CENP-N binding sites [48].
It is clear, ever since Lowary and Widom identified super-positioning 601 sequence for canonical nucleosome [58] that the DNA sequence affects the nucleosome stability. However, although computational methods are starting to address DNA sequence–nucleosome structure relationship [66] it is still extremely difficult to study experimentally at high resolution. Until recently, the majority of high-resolution nucleosome structures were obtained by X-ray crystallography, which imposes the constraints of crystal packing which, in turn, interferes with the high-quality crystal formation of nucleosomes wrapped with array of different sequences. Still, the impact of the DNA sequence on nucleosome structure was clearly observed in X-ray structures of nucleosome [57,67,68]. We hope the fast development of cryo-EM will enable a more thorough structural analysis of nucleosomes on different DNA sequences and advance our understanding of the DNA role in nucleosome structure. Present research has identified complex physico-chemical properties (like bendability and stretching) of a given DNA sequence (rather than the invariable order of nucleotides) to be responsible for DNA sequence stabilization of nucleosomes [57,69]. It is very possible that the CENP-A nucleosome has a particular and potentially slightly different preference for the DNA sequence in comparison with canonical nucleosome. This fine DNA sequence preference could be instrumental in positioning CENP-A nucleosome within monomers of centromere repeats. This in turn would lead to specific nucleosome phasing at the centromere, resulting in a unique centromeric chromatin structure. In fact, nucleosome phasing has been observed on human and mice α-satellite DNA monomers [39,70]. Nucleosomal DNA could also promote association of the CENP-A nucleosome with CCAN components and in that way assure stable centromeric chromatin. If indeed a specific DNA sequence emphasizes important dynamic features of the CENP-A nucleosome, one would expect a clear connection in the evolution of centromeric sequence and the CENP-A histone, but this study is still not available. Further understanding of the interplay between the DNA sequence and the histone octamer is necessary to resolve the role of the DNA sequence in centromere formation and functioning.
CENP-A nucleosomes are specifically recognized by CENP-C and CENP-N
The human CENP-A nucleosome is essential in establishing and maintaining a group of 16 proteins defining a centromere—the CCAN [71–73]. The complex is present at the chromosomes in all phases of the cell cycle, although it is proposed that the interaction between subunits changes through the cell cycle [50]. It is a platform for the formation of the kinetochore, which attaches the mitotic chromosome to the microtubules. The CCAN proteins can be divided into five subgroups based on biochemical and recruitment studies: CENP-C, CENP-L-N, CENP-H-I-K-M, CENP-T-W-S-X and CENP-O-P-Q-U-R [74–83]. CENP-C and CENP-N are the only two CCANs that bind directly to CENP-A nucleosomes [19,76,84–87]. The role of CENP-A in epigenetically marking the centromere directly depends on its ability to interact with CENP-C and CENP-N, but both interactions might vary through the cell cycle. Recently, the binding of both of these proteins to the CENP-A nucleosome has been elucidated at the atomic level [37,46,48,88–90], and findings have revealed that both CENP-C and CENP-N not only interact with CENP-A (specific interactions) but also make contacts elsewhere at the nucleosome (nonspecific interactions) (Figures 2 and 3). These studies have also revealed structural changes in the CENP-A nucleosome that are associated with the binding.
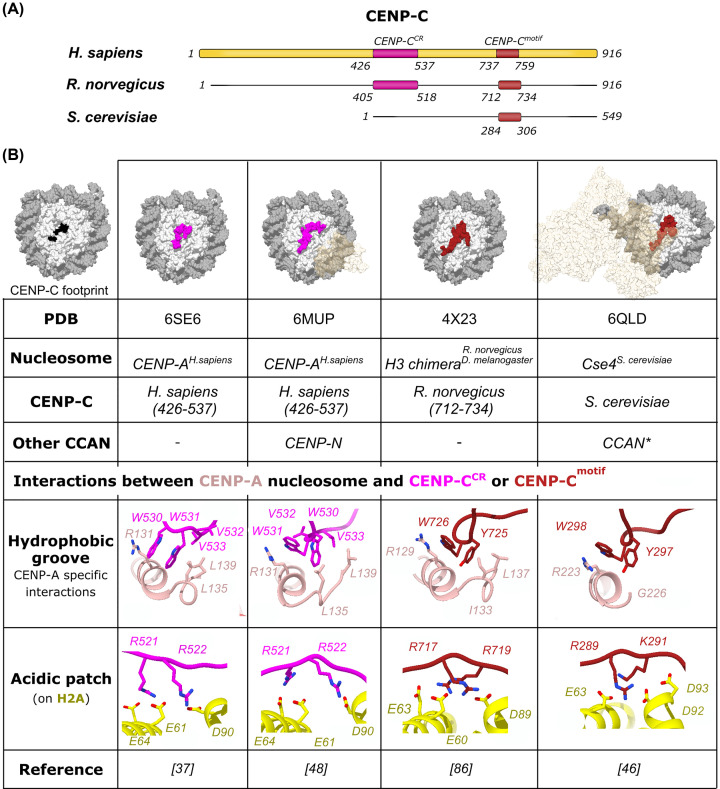
Figure 2. CENP-A nucleosome in complex with CENP-C.
(A) A schematic representation of CENP-C from Homo sapiens, Rattus norvegicus and Saccharomyces cerevisiae, indicating nucleosome-binding regions, CENP-CCR and CENP-Cmotif, used in structural studies. (B) A table summarizing information revealed from published 3D-structures of nucleosomes in complex with CENP-C. A surface representation of the CENP-A nucleosome with mapped CENP-C footprint in black is shown on the far left. In the surface representation, histones are shown in light gray, DNA is colored dark gray, CENP-CCR is magenta, the CENP-Cmotif is dark red and other CCAN components shown in transparent beige. The CENP-C residues involved in the interaction with the acidic patch and the CENP-A C-terminal tail are shown as sticks.
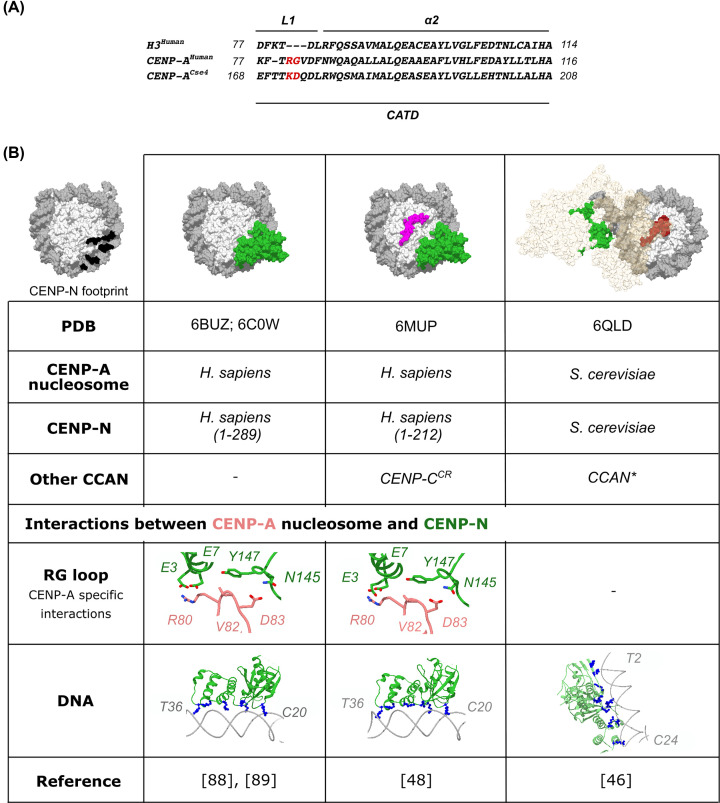
Figure 3. CENP-A nucleosome in complex with CENP-N.
(A) A multiple sequence alignment of the CATD region of CENP-A (H. sapiens) and CENP-ACse4 (S. cerevisiae) with the corresponding region of H3 (H. sapiens). Insertions in the L1 loop are colored red. (B) A table summarizing information revealed from published 3D-structures of nucleosomes in complex with CENP-N. A surface representation of the CENP-A nucleosome with the mapped CENP-N footprint in black is shown on the far left. In the surface representation, histones are light gray, DNA is dark gray, CENP-N is green, CENP-CCR is magenta, the CENP-Cmotif is dark red, and the other CCAN components are shown in transparent beige. The CENP-N residues interacting with the CENP-A RG loop are shown as sticks. Positively charged residues (arginines and lysines), interacting with the nucleosomal DNA, are shown in blue.
CENP-A nucleosome in complex with CENP-C
CENP-C exists in many species across the animal, plant and fungi kingdom, but its sequence is evolutionally very dynamic [26,91,92]. It is a key CCAN component that plays a central role in the organization of the centromere and kinetochore recruitment [93]. Human CENP-C is a 934-amino acid long protein with many disordered regions that, besides binding to the CENP-A nucleosome, directly interacts with CENP-N-L, CENP-H-I-K-M and the DNA-binding protein, CENP-B [86,93–97]. The ability of CENP-C to bind several CCAN complexes, while at the same time directly connecting the CENP-A nucleosome through its C-terminus and the outer kinetochore components (KMN complex) through its N-terminus, together with having a dimerization domain that duplicates all of these interactions, have led to the proposal that CENP-C is a major organizer of the centromere [93]. The human CENP-C has two domains that specifically bind the CENP-A nucleosome, CENP-C central region 426–537 (CENP-CCR) and CENP-C motif 736–758 (CENP-Cmotif) (Figure 2A) [72,76,84,98]. The CENP-Cmotif is one of the most evolutionarily conserved parts of the CENP-C protein [26,99], but it is neither necessary nor sufficient for kinetochore targeting in human cells [100]. On the other hand, CENP-CCR is required for centromeric retention of the newly incorporated CENP-A [100]. Based on studies in chicken cells, the Fukagawa lab has proposed the interaction between the CENP-A nucleosome and CENP-C to be dependent on the cell cycle [49–51]. In the interphase cells, CENP-C is not interacting with the CENP-A nucleosome, while during mitosis CDK1-mediated phosphorylation of the Thr651 residue adjacent to CENP-Cmotif (the only known CENP-C binding module in chicken cells), induces CENP-A nucleosome binding [49]. Until recently, the only available molecular insight into CENP-C binding was the X-ray crystal structure of a chimeric nucleosome bound with rat CENP-Cmotif [86]. In the last 2 years, our understanding of the CENP-A nucleosome/CENP-C interactions was complemented with cryo-EM structures of human CENP-A nucleosomes, in complex with CENP-CCR [37], and in complex with both CENP-CCR and CENP-N [48], as well as a cryo-EM structure of S. cerevisiae CENP-A homolog (Cse4) in complex with 14 CCAN subunits, named Ctf19 complex in S. cerevisiae [46] (Cse4Ctf19). The CENP-A nucleosome has two binding sites for CENP-C, one on each face of the nucleosome, and makes a 1:2 complex with CENP-C. However, one copy of CENP-C is more disordered in almost all structures [37,46,48,86], which could be a consequence of instability of the complex during sample preparation (crystal formation or freezing in vitreous ice). The structural comparison reveals that CENP-CCR and CENP-Cmotif have similar modes of nucleosome binding, exploiting the hydrophobic interaction with the C-terminal tail of CENP-A, and electrostatic interaction with the acidic patch formed by H2A/H2B on the surface of the nucleosome (Figure 2B). The acidic patch is a hotspot for canonical nucleosome binding, and a similar interaction has been described for several chromatin complexes [101–104]. The CENP-A specific interaction is mediated by a hydrophobic stretch of amino acids that are longer in CENP-CCR, resulting in a stronger binding affinity toward the CENP-A nucleosome in comparison with CENP-Cmotif [37]. Interestingly, despite evidence that CENP-C interacts with multiple modules within CCANs, those interactions are not visible in the Cse4Ctf19 complex, probably due to the dynamic nature of the protein parts involved [46]. Together, solution studies and high-resolution structures have indicated several structural changes that take place in the CENP-A nucleosome upon CENP-C binding. First, CENP-CCR binding increases flexibility of the terminal DNA that is mediated through destabilization of the H2A C-terminal tail [37]. Interestingly, while the binding of the CENP-CCR to CENP-A nucleosome destabilizes the DNA wrap, subsequent binding of CENP-N reverts this effect [40,48] (Figure 1C). Second, HDX, fluorescence and AFM studies detected rigidification and compaction of the CENP-A core upon CENP-C binding [85,105], and structural studies are in agreement with this [37,46,48]. Third, the CENP‐C binding rigidifies the N‐terminal tail of H4 in the conformation favoring H4K20 mono-methylation, required for the establishment and maintenance of functional centromeres [37,38,48]. Together, structural studies have started to reveal the effects and the complexity of the interaction between the essential CCAN protein, CENP-C, and the specialized centromeric nucleosome. However, a full understanding of how CENP-C organizes the centromere is awaiting more complete complexes.
CENP-A nucleosome in complex with CENP-N
In humans, CENP-N is a 339-aa globular protein that binds the CENP-A nucleosome through its N-terminal region (1–286), and CENP-L through its C-terminal region (287-339). CENP-L in turn directly interacts with the CENP-H-I-K-M [93,96,100,106,107]. Although CENP-N directly binds the CENP-A nucleosome, its recruitment to the centromere is mediated by CENP-C during mitosis [89,96]. Initial biochemical studies have identified the exposed L1 loop within the CATD of CENP-A (Figure 3A) and the DNA, as binding determinants for CENP-N [76,84,100,108], and recent cryo-EM structures have confirmed the same binding elements [48,88–90].
The L1 loop is the only surface-exposed part of the CATD on the CENP-A nucleosome. It has two residues’ insertion, in comparison with H3—bulky charged Arg80 and small flexible Gly81 (usually referred as RG-loop). CENP-N engages with the RG-loop using both electrostatic and hydrophobic interactions (Figure 3B). The CENP-AR80 forms hydrogen bonds with both CENP-NE3 and CENP-NE7, while the CENP-NY147 creates hydrophobic interaction with CENP-AV82. In comparison with the CENP-A nucleosome/CENP-C interaction, which has a relatively small footprint on the nucleosome (Figure 2B), the CENP-A nucleosome/CENP-N complex has a large interaction interface (2400 Å2), half of which is engaged in the CENP-N/DNA interaction (Figure 3B). However, the CENP-N/DNA interaction is not specific in nature, but rather several positively charged residues of CENP-N (Lys15, Arg42, Lys45, Lys81 and Arg194) interact with the backbone phosphates of DNA at the super-helical location SHL −2 to −3. The structure of the budding yeast CENP-N, in complex with the CENP-A (Cse4) nucleosome, which was solved in the presence of the other 13 Ctf19 components (Cse4Ctf19) [46], reveals striking differences from the structure of the human complex. Neither the interaction of CENP-N with L1 loop of CENP-A (Cse4) (171KD172 in CENP-ACse4), nor the binding to the DNA at the SHL −2 to −3 are conserved in budding yeast. Instead, the CENP-N-L complex is reported to tightly bind the nucleosomal DNA, which is unwrapped from the nucleosome, using several positively charged Arginine and Lysine residues. However, no direct interaction is reported with the CENP-A (Cse4) nucleosome. Mutational analysis showed that, in the absence of CENP-C, the integrity of the CCAN complex depends on CENP-N-L/DNA interaction [46]. Pronounced differences in the mode of CENP-N binding to the CENP-A (Cse4) nucleosome, in comparison with the vertebrate CENP-A nucleosome, could reflect true differences between species or it could be a consequence of different in vitro sample preparation or a lack of other CCAN components in human complexes. Notably, the structures of Cse4Ctf19 and CENP-A/CENP-N [46,88–90] are asymmetric, with only one copy of the Ctf19 or CENP-N, respectively per nucleosome (two copies are expected based on solution studies), probably due to dissociation during freezing. Symmetrical occupancy of CENP-C and CENP-N, reported by Allu at al., was facilitated with mild cross-linking conditions (see Table 1) [48].
From the structure of the human CENP-A nucleosome, in complex with both CENP-CCR and CENP-N [48], it is clear that these two modules can coexist on the same nucleosome without affecting each other’s binding. However, they both interact with the N-terminal tail of histone H4 and drive it in a specific orientation along the surface of the nucleosome, orienting it in a position that probably facilitates the centromere-specific H4K20 monomethylation essential for the epigenetic establishment of the kinetochore [109]. Furthermore, the concomitant binding of CENP-N and CENP-CCR induces distinctive bulges of the DNA between SHLs 3.5/4.5 and SHLs −3.5/−4.5, which locally widen the DNA gyre distance by 3–4 Å (nucleosome gapping) accommodating a ∼1.5 Å DNA slide toward each of the CENP-N binding sites. This rearrangement is facilitated with natural α-satellite DNA, and it was not observed in the CENP-A nucleosome/CENP-N complexes on 601 DNA [48,88,89]. The same study reported two different stoichiometries of CENP-A/CENP-C/CENP-N complexes. One that has two copies of each, CENP-C and CENP-N, and the other one that has two copies of CENP-C and only one copy of CENP-N. The authors propose a model, based on cell experiments, where the chromosome condensation in mitosis results in the loss of one CENP-N molecule, generating asymmetrical arrangement at the CENP-A nucleosome that favors the formation of kinetochore.
In summary, the vertebrate CENP-N binds the RG-loop and the DNA between SHL2 and SHL3. The binding is facilitated on natural α-satellite DNA [48,100], where it further increases the sequence-specific DNA gyre gap opposite of the dyad. Also, the CENP-A nucleosome structures, in complex with CENP-C and CENP-N, have highlighted not only the importance of cumulative structural changes (for example, DNA unwrapping induced by CENP-C but stabilized again by CENP-N binding), but also the complexity of the stoichiometry within the CCAN that might be changing during the cell cycle [48,50,108].
CENP-A nucleosome—a chromatin-embedded pedestal for the centromere
The structure of the whole CCAN (Ctf19) complex from the budding yeast, in isolation [106] and in complex with the centromeric nucleosome [46], is a huge step toward understanding the molecular determinants of the centromere and kinetochore architecture. Although interactions between CCAN subunits and CENP-A nucleosome might be different in the more complex eukaryotes, the structure of the CseCtf19 gives a perception of the spatial organization around the CENP-A nucleosome, justifying the requirement for unwrapped DNA ends and accessible nucleosome faces that can accommodate CCAN binding. It is truly impressive to see a single nucleosome, ∼50 × 100 Å in size, supporting the megastructure of ∼200 × 300 Å (assuming 2 CCAN:1 nucleosome stoichiometry) (see model in Figure 4A). However, although this impressive structural work gives an idea of how the CCAN components are organized on the CENP-A (or Cse4) nucleosome, it, at the same time, opens other questions. How is this bulky structure embedded in chromatin? How does it re-organize with chromatin changes during the cell cycle, and how does it become available for kinetochore formation in mitosis?
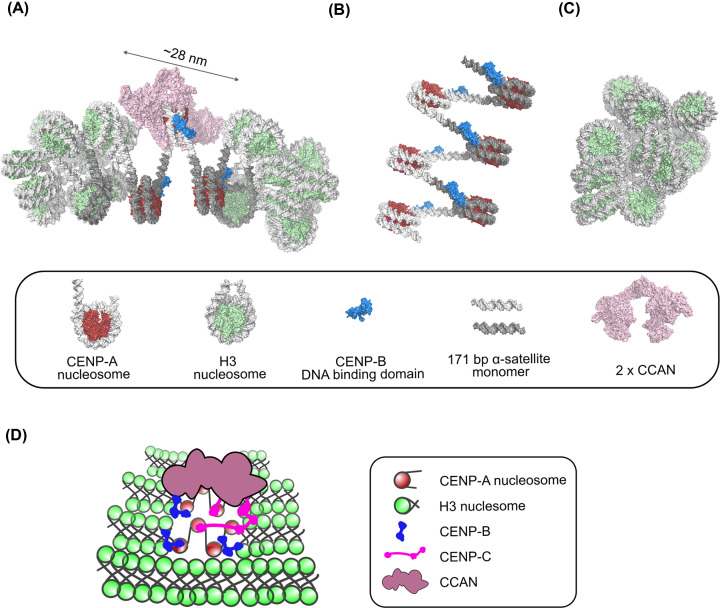
Figure 4. A model of the human CCAN complex in centromeric chromatin.
(A) 3D-model of the CCAN/centromeric chromatin complex. The nucleosome modeled harboring CCAN is built using structural information from the H3-CENP-A-H3 tri-nucleosome structure (6L49). CENP-C and CENP-N are modeled on the central nucleosome as in the human CENP-A/CENP-C/CENP-N structure (6MUP). The yeast CCAN complex structure (pink; extracted from 6QLD) deprived of the CENP-ACse4 nucleosome, is docked on the central CENP-A nucleosome by aligning one copy of the CCAN complex on each CENP-N molecule. The DNA binding domain of CENP-B (DBD) (blue; 1HLV) is docked on a degenerated CENP-B box in the α-satellite sequence, as positioned in the CENP-A/CENP-C/CENP-N structure (6MUP). The DNA path was slightly adjusted to accommodate a kink observed in the CENP-B/DNA structure. The 171-bp monomers in the α-satellite sequence are colored light and dark gray (the nucleosome position is based on the CENP-A/CENP-C/CENP-N structure (6MUP)). It is obvious that nucleosomes adjacent to the CCAN loaded CENP-A cannot adopt a H3-H3-H3 packing (6L4A), so two CENP-A nucleosomes with open DNA ends are modelled on each side of the CENP-A, loaded with CCAN to avoid steric clashes. The H3 containing histone core is colored light green, and the CENP-A histone core is colored red. The size of the 2xCCAN:1CENP-A nucleosome complex is indicated. (B) A 3D-model of 6x CENP-A nucleosomes/CENP-B (DBD), based on the open DNA conformation of the CENP-A nucleosome in the H3-CENP-A-H3 tri-nucleosome structure (6L49). Notice the bigger spacing between nucleosomes. (C) A 3D-model of 18x H3 nucleosomes array built based on the H3-H3-H3 tri-nucleosome structure (6L4A). Notice the tight nucleosome packing imposed with DNA crossing over the nucleosome dyad. (D) A schematic diagram of human centromeric chromatin. H3 loaded chromatin is more compact than the CENP-A loaded chromatin. CENP-B binding is enforcing the CENP-A-like (looser) chromatin by introducing a kink close to the nucleosome entry/exit site. Because CENP-B has a dimerization domain, it is also bridging two nucleosomes. CENP-C is structuring the chromatin by binding four CENP-A nucleosomes (dimer with two nucleosome-binding regions) and thus increasing their local concentration. It also interacts with the CCAN.
CENP-A nucleosomes are present in the chromatin at fairly low levels. Indeed, according to Bodor at al. [110] even at the centromere, H3 nucleosomes are 25-times more abundant than the CENP-A nucleosomes; raising a question of how the CENP-A nucleosomes are organized within the centromere. Immunofluorescence and super-resolution microscopy studies [111–113] revealed that the CENP-A nucleosomes are interspersed between regions containing H3 nucleosomes, and cluster in G1. It is still not clear how the CENP-A nucleosomes organize in the higher order chromatin. We propose that distinct dynamic features of the CENP-A nucleosomes (flexible DNA ends and plastic histone core) generate chromatin with unique properties [36,43,108,114,115] which is more relaxed and can accommodate the bulky CCAN complex (Figure 4B). The tightly packed nucleosome arrays [36,108,114], where DNA crosses after it exits the nucleosome, are not compatible with the CCAN presence (Figure 4C). Instead, a flexible chromatin organization generated with untwisted linker DNA between nucleosomes [43] is needed in the neighborhood of the CCAN loaded CENP-A nucleosome (Figure 4A). Thus, the function of the vertebrate CENP-A nucleosome at the centromere might be dual. One fraction serves as a pedestal for the CCAN complex, while the other fraction participates in generating special chromatin environment, allowing space for the CENP-ACCAN complex. In the point centromeres, like S. cerevisiae, the two functions are separated and both are genetically encoded [116,117]. The CENP-A (Cse4) assembled on 78–88 bp AT-rich CDEII is carrying Ctf19 complex, and surrounding CDEI and CDEIII DNA are binding Cfb1 and CBF3 complex, respectively, that are modifying surrounding chromatin.
Actually, recent efforts to visualize chromatin in cells using cryo-electron tomography (cryo-ET) and related techniques [118,119] are supporting a dynamic fluid-like organization of chromatin [120] rather than the rigid 30-nm chromatin fiber model based on in vitro chromatin studies that was widely accepted for decades. Comprehensive chromatin models based on an array of biophysical studies propose chromatin to be organized in distinct domains that have irregular fluid-like nucleosome organization [121]. Different cellular events like DNA replication, DNA repair/recombination and RNA transcription are all altering chromatin distribution between chromatin domains, and a phase separation is emerging as the mechanistic drive behind those processes [122]. Furthermore, the chromatin structure and nucleosome organization is dynamically changing during the cell cycle [119,123–125]. In the light of these observations, we propose that special dynamic properties of the chromatin loaded with CENP-A nucleosomes, distinguishes the centromere from the rest of the chromosome, but further studies are needed to confirm this model.
The intrinsic properties of the underlying DNA could also contribute toward the formation of the functional centromere. The repetitive nature of centromeric DNA could have a function in nucleosome spacing and positioning [39], and the DNA sequence properties are assuring the nucleosome ‘fit’ that promotes productive binding of CCAN components [48]. The distinct human CENP-A nucleosomes loaded with CCANs are further cross-linked by the CENP-C, a major organizer of the centromere, through its two nucleosome-binding sites and dimerization domain [37,93]. The DNA-binding CCAN components, CENP-N [76,84,88,89], CENP-T-W-S-X [81,126] and CENP-B [127] might help in further adjusting the centromeric chromatin structure and compaction. Intriguingly, CENP-B, the only CCAN component with DNA sequence-specific binding, has a redundant role in the established centromeric chromatin, but it becomes essential in de novo centromere formation on human artificial chromosomes [128]. One explanation could be that CENP-B conditions the chromatin for centromere formation, in the absence of pre-existing CENP-A, by binding close to the entry/exit site of the nucleosomes [129,130], which helps with nucleosome phasing [39,131] and DNA unwrapping (it induces a 59° kink [10]) and its dimerization domain [132] could play a role in specific chromatin cross-linking. Neocentromeres, which are natural centromeres that arise at the naïve chromatin (deprived from CENP-A and CENP-B) might exploit different mechanisms to establish functional centromere. A recent study by Kasinathan and Henikoff [133] used a bioinformatics approach to identify dyad symmetries in centromeres where CENP-B is absent and in neocentromeres, suggesting that DNA secondary structures could also play a role in centromere formation.
Finally, it is becoming more and more clear that strong epigenetic and weaker, but important, genetic traits both guide the centromere formation and function; the dynamic nature of which is yet to be understood.
Conclusions and future perspectives
Amazing advances in the use of cryo-electron microscopy for single particle analysis cryo-ET and related techniques in the last decade [118,134,135] has revolutionized structural biology and introduced a much needed tool for studying complex molecular machines like the centromere and kinetochore. In the last 2 years, we have witnessed a large number of impressive high-resolution cryo-EM structures of the centromere and kinetochore complexes [37,46,48,88,89,106,136–138]. Yet, we are just starting to chip the tip of the iceberg when it comes to fully understanding these complex megastructures. Fortunately, other powerful and complementary techniques are developing, like structural mass-spectroscopy, computational modeling, chromosome capture techniques and super-resolution microscopy. An integrative approach, combining information from high-resolution structures with other complementary techniques that can assess fluctuations in dynamics, stoichiometry or precise location in the cell, will be a next step toward a holistic understanding of centromeres and kinetochores.
Summary
Centromeres are epigenetically specified by the histone H3 variant, CENP-A.
CENP-A specific and DNA sequence-specific features are identified in high-resolution structures of the CENP-A nucleosome.
CENP-N and CENP-C directly and specifically bind the CENP-A nucleosome and induce conformational changes.
A specialized chromatin environment, contributed by both genetic and epigenetic elements, is defining the centromere.
Abbreviations
- aa
amino acid
- AFM
atomic force microscopy
- CATD
CENP-A targeting domain
- CCAN
constitutive centromere associated network
- cryo-EM
cryo-electron microscopy
- cryo-ET
cryo-electron tomography
- HDX
hydrogen-deuterium exchange
- HFD
histone-fold domain
- MNase
micrococcal nuclease
- SAXS
small angle X-ray scattering
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
AAA and NS are funded by Centre for Molecular Medicine Norway (NCMM) and Norwegian Research Council [grant #261395]. NS is funded by Department of Chemistry, University of Oslo.
References
- 1.McKinley K.L. and Cheeseman I.M. (2016) The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol. 17, 16–29 10.1038/nrm.2015.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Potapova T. and Gorbsky G.J. (2017) The consequences of chromosome segregation errors in mitosis and meiosis. Biology 6, 12 10.3390/biology6010012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henikoff S., Ahmad K. and Malik H.S. (2001) The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293, 1098–1102 10.1126/science.1062939 [DOI] [PubMed] [Google Scholar]
- 4.Melters D.P., Bradnam K.R., Young H.A., Telis N., May M.R., Ruby J.G. et al. (2013) Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 14, R10 10.1186/gb-2013-14-1-r10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Voullaire L.E., Slater H.R., Petrovic V. and Choo K.H. (1993) A functional marker centromere with no detectable alpha-satellite, satellite III, or CENP-B protein: activation of a latent centromere? Am. J. Hum. Genet. 52, 1153–1163 [PMC free article] [PubMed] [Google Scholar]
- 6.Marshall O.J., Chueh A.C., Wong L.H. and Choo K.H.A. (2008) Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am. J. Hum. Genet. 82, 261–282 10.1016/j.ajhg.2007.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott K.C. and Sullivan B.A. (2014) Neocentromeres: a place for everything and everything in its place. Trends Genet. 30, 66–74 10.1016/j.tig.2013.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Hooff J.J., Tromer E., van Wijk L.M., Snel B. and Kops G.J. (2017) Evolutionary dynamics of the kinetochore network in eukaryotes as revealed by comparative genomics. EMBO Rep. 18, 1559–1571 10.15252/embr.201744102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masumoto H., Masukata H., Muro Y., Nozaki N. and Okazaki T. (1989) A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J. Cell Biol. 109, 1963–1973 10.1083/jcb.109.5.1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka Y., Nureki O., Kurumizaka H., Fukai S., Kawaguchi S., Ikuta M. et al. (2001) Crystal structure of the CENP-B protein-DNA complex: the DNA-binding domains of CENP-B induce kinks in the CENP-B box DNA. EMBO J. 20, 6612–6618 10.1093/emboj/20.23.6612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perez-Castro A.V., Shamanski F.L., Meneses J.J., Lovato T.L., Vogel K.G., Moyzis R.K. et al. (1998) Centromeric protein B null mice are viable with no apparent abnormalities. Dev. Biol. 201, 135–143 10.1006/dbio.1998.9005 [DOI] [PubMed] [Google Scholar]
- 12.Sekulic N. and Black B.E. (2012) Molecular underpinnings of centromere identity and maintenance. Trends Biochem. Sci. 37, 220–229 10.1016/j.tibs.2012.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roure V., Medina-Pritchard B., Lazou V., Rago L., Anselm E., Venegas D. et al. (2019) Reconstituting Drosophila centromere identity in human cells. Cell Rep. 29, 464.e5–479.e5 10.1016/j.celrep.2019.08.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hori T., Shang W.-H., Takeuchi K. and Fukagawa T. (2013) The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J. Cell Biol. 200, 45–60 10.1083/jcb.201210106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Logsdon G.A., Barrey E.J., Bassett E.A., DeNizio J.E., Guo L.Y., Panchenko T. et al. (2015) Both tails and the centromere targeting domain of CENP-A are required for centromere establishment. J. Cell Biol. 208, 521–531 10.1083/jcb.201412011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heun P., Erhardt S., Blower M.D., Weiss S., Skora A.D. and Karpen G.H. (2006) Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev. Cell 10, 303–315 10.1016/j.devcel.2006.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olszak A.M., van Essen D., Pereira A.J., Diehl S., Manke T., Maiato H. et al. (2011) Heterochromatin boundaries are hotspots for de novo kinetochore formation. Nat. Cell Biol. 13, 799–808 10.1038/ncb2272 [DOI] [PubMed] [Google Scholar]
- 18.Mendiburo M.J., Padeken J., Fülöp S., Schepers A. and Heun P. (2011) Drosophila CENH3 is sufficient for centromere formation. Science 334, 686–690 10.1126/science.1206880 [DOI] [PubMed] [Google Scholar]
- 19.Guse A., Carroll C.W., Moree B., Fuller C.J. and Straight A.F. (2011) In vitro centromere and kinetochore assembly on defined chromatin templates. Nature 477, 354–358 10.1038/nature10379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furuyama S. and Biggins S. (2007) Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc. Natl. Acad. Sci. U.S.A. 104, 14706–14711 10.1073/pnas.0706985104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tromer E.C., van Hooff J.J.E., Kops G.J.P.L. and Snel B. (2019) Mosaic origin of the eukaryotic kinetochore. Proc. Natl. Acad. Sci. U.S.A. 116, 12873–12882 10.1073/pnas.1821945116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cooper J.L. and Henikoff S. (2004) Adaptive evolution of the histone fold domain in centromeric histones. Mol. Biol. Evol. 21, 1712–1718 10.1093/molbev/msh179 [DOI] [PubMed] [Google Scholar]
- 23.Rosin L. and Mellone B.G. (2016) Co-evolving CENP-A and CAL1 domains mediate centromeric CENP-A deposition across Drosophila species. Dev. Cell 37, 136–147 10.1016/j.devcel.2016.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malik H.S. and Henikoff S. (2003) Phylogenomics of the nucleosome. Nat. Struct. Mol. Biol. 10, 882–891 10.1038/nsb996 [DOI] [PubMed] [Google Scholar]
- 25.Drinnenberg I.A., Henikoff S. and Malik H.S. (2016) Evolutionary turnover of kinetochore proteins: a ship of Theseus? Trends Cell Biol. 26, 498–510 10.1016/j.tcb.2016.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Talbert P.B., Bryson T.D. and Henikoff S. (2004) Adaptive evolution of centromere proteins in plants and animals. J. Biol. 3, 18 10.1186/jbiol11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Filipescu D., Müller S. and Almouzni G. (2014) Histone H3 variants and their chaperones during development and disease: contributing to epigenetic control. Annu. Rev. Cell Dev. Biol. 30, 615–646 10.1146/annurev-cellbio-100913-013311 [DOI] [PubMed] [Google Scholar]
- 28.Srivastava S. and Foltz D.R. (2018) Posttranslational modifications of CENP-A: marks of distinction. Chromosoma 127, 279–290 10.1007/s00412-018-0665-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Black B.E. and Cleveland D.W. (2011) Epigenetic centromere propagation and the nature of CENP-A nucleosomes. Cell 144, 471–479 10.1016/j.cell.2011.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Black B.E., Foltz D.R., Chakravarthy S., Luger K., Woods V.L. and Cleveland D.W. (2004) Structural determinants for generating centromeric chromatin. Nature 430, 578–582 10.1038/nature02766 [DOI] [PubMed] [Google Scholar]
- 31.Sekulic N., Bassett E.A., Rogers D.J. and Black B.E. (2010) The structure of (CENP-A-H4)2 reveals physical features that mark centromeres. Nature 467, 347–351 10.1038/nature09323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu W., Yuan B., Flygare J. and Lodish H.F. (2011) Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev. 25, 2573–2578 10.1101/gad.178780.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho U.-S. and Harrison S.C. (2011) Recognition of the centromere-specific histone Cse4 by the chaperone Scm3. Proc. Natl. Acad. Sci. U.S.A. 108, 9367–9371 10.1073/pnas.1106389108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Z., Feng H., Zhou B.-R., Ghirlando R., Hu K., Zwolak A. et al. (2011) Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature 472, 234–237 10.1038/nature09854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tachiwana H., Kagawa W., Shiga T., Osakabe A., Miya Y., Saito K. et al. (2011) Crystal structure of the human centromeric nucleosome containing CENP-A. Nature 476, 232–235 10.1038/nature10258 [DOI] [PubMed] [Google Scholar]
- 36.Panchenko T., Sorensen T.C., Woodcock C.L., Kan Z.-Y., Wood S., Resch M.G. et al. (2011) Replacement of histone H3 with CENP-A directs global nucleosome array condensation and loosening of nucleosome superhelical termini. Proc. Natl. Acad. Sci. U.S.A. 108, 16588–16593 10.1073/pnas.1113621108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ali-Ahmad A., Bilokapić S., Schäfer I.B., Halić M. and Sekulić N. (2019) CENP-C unwraps the human CENP-A nucleosome through the H2A C-terminal tail. EMBO Rep. 20, e48913 10.15252/embr.201948913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arimura Y., Tachiwana H., Takagi H., Hori T., Kimura H., Fukagawa T. et al. (2019) The CENP-A centromere targeting domain facilitates H4K20 monomethylation in the nucleosome by structural polymorphism. Nat. Commun. 10, 1–10 10.1038/s41467-019-08314-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hasson D., Panchenko T., Salimian K.J., Salman M.U., Sekulic N., Alonso A. et al. (2013) The octamer is the major form of CENP-A nucleosomes at human centromeres. Nat. Struct. Mol. Biol. 20, 687–695 10.1038/nsmb.2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou B.-R., Yadav K.N.S., Borgnia M., Hong J., Cao B., Olins A.L. et al. (2019) Atomic resolution cryo-EM structure of a native-like CENP-A nucleosome aided by an antibody fragment. Nat. Commun. 10, 2301 10.1038/s41467-019-10247-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boopathi R., Danev R., Khoshouei M., Kale S., Nahata S., Ramos L. et al. (2020) Phase-plate cryo-EM structure of the Widom 601 CENP-A nucleosome core particle reveals differential flexibility of the DNA ends. Nucleic Acids Res. 40, 5735–5748 10.1093/nar/gkaa246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roulland Y., Ouararhni K., Naidenov M., Ramos L., Shuaib M., Syed S.H. et al. (2016) The flexible ends of CENP-A nucleosome are required for mitotic fidelity. Mol. Cell 63, 674–685 10.1016/j.molcel.2016.06.023 [DOI] [PubMed] [Google Scholar]
- 43.Takizawa Y., Ho C.-H., Tachiwana H., Matsunami H., Kobayashi W., Suzuki M. et al. (2020) Cryo-EM structures of centromeric tri-nucleosomes containing a central CENP-A nucleosome. Structure 28, 44.e4–53.e4 10.1016/j.str.2019.10.016 [DOI] [PubMed] [Google Scholar]
- 44.Migl D., Kschonsak M., Arthur C.P., Khin Y., Harrison S.C., Ciferri C. et al. (2020) Cryoelectron microscopy structure of a yeast centromeric nucleosome at 2.7 Å resolution. Structure 28, 363.e3–370.e3 10.1016/j.str.2019.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arimura Y., Shirayama K., Horikoshi N., Fujita R., Taguchi H., Kagawa W. et al. (2014) Crystal structure and stable property of the cancer-associated heterotypic nucleosome containing CENP-A and H3.3. Sci. Rep. 4, 1–7, 10.1038/srep07115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan K., Yang J., Zhang Z., McLaughlin S.H., Chang L., Fasci D. et al. (2019) Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome. Nature 574, 278–282 10.1038/s41586-019-1609-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bilokapic S., Strauss M. and Halic M. (2018) Histone octamer rearranges to adapt to DNA unwrapping. Nat. Struct. Mol. Biol. 25, 101 10.1038/s41594-017-0005-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Allu P.K., Dawicki-McKenna J.M., Eeuwen T.V., Slavin M., Braitbard M., Xu C. et al. (2019) Structure of the human core centromeric nucleosome complex. Curr. Biol. 29, 2625.e5–2639.e5 10.1016/j.cub.2019.06.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watanabe R., Hara M., Okumura E., Hervé S., Fachinetti D., Ariyoshi M. et al. (2019) CDK1-mediated CENP-C phosphorylation modulates CENP-A binding and mitotic kinetochore localization. J. Cell Biol. 218, 4042–4062 10.1083/jcb.201907006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nagpal H. and Fukagawa T. (2016) Kinetochore assembly and function through the cell cycle. Chromosoma 125, 645–659 10.1007/s00412-016-0608-3 [DOI] [PubMed] [Google Scholar]
- 51.Nagpal H., Hori T., Furukawa A., Sugase K., Osakabe A., Kurumizaka H. et al. (2015) Dynamic changes in CCAN organization through CENP-C during cell-cycle progression. Mol. Biol. Cell 26, 3768–3776 10.1091/mbc.E15-07-0531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nergadze S.G., Piras F.M., Gamba R., Corbo M., Cerutti F., McCarter J.G.W. et al. (2018) Birth, evolution, and transmission of satellite-free mammalian centromeric domains. Genome Res. 6, 789–799 10.1101/gr.231159.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Logsdon G.A., Gambogi C.W., Liskovykh M.A., Barrey E.J., Larionov V., Miga K.H. et al. (2019) Human artificial chromosomes that bypass centromeric DNA. Cell 178, 624.e19–639.e19 10.1016/j.cell.2019.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murillo-Pineda M. and Jansen L.E.T. (2020) Genetics, epigenetics and back again: Lessons learned from neocentromeres. Exp. Cell Res. 389, 111909 10.1016/j.yexcr.2020.111909 [DOI] [PubMed] [Google Scholar]
- 55.Hudson D.F., Fowler K.J., Earle E., Saffery R., Kalitsis P., Trowell H. et al. (1998) Centromere protein B null mice are mitotically and meiotically normal but have lower body and testis weights. J. Cell Biol. 141, 309–319 10.1083/jcb.141.2.309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolfe J., Darling S.M., Erickson R.P., Craig I.W., Buckle V.J., Rigby P.W.J. et al. (1985) Isolation and characterization of an alphoid centromeric repeat family from the human Y chromosome. J. Mol. Biol. 182, 477–485 10.1016/0022-2836(85)90234-7 [DOI] [PubMed] [Google Scholar]
- 57.Chua E.Y.D., Vasudevan D., Davey G.E., Wu B. and Davey C.A. (2012) The mechanics behind DNA sequence-dependent properties of the nucleosome. Nucleic Acids Res. 40, 6338–6352 10.1093/nar/gks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lowary P.T. and Widom J. (1998) New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 10.1006/jmbi.1997.1494 [DOI] [PubMed] [Google Scholar]
- 59.Luger K., Mäder A.W., Richmond R.K., Sargent D.F. and Richmond T.J. (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 10.1038/38444 [DOI] [PubMed] [Google Scholar]
- 60.Davey C.A., Sargent D.F., Luger K., Maeder A.W. and Richmond T.J. (2002) Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 319, 1097–1113 10.1016/S0022-2836(02)00386-8 [DOI] [PubMed] [Google Scholar]
- 61.Vasudevan D., Chua E.Y.D. and Davey C.A. (2010) Crystal structures of nucleosome core particles containing the “601” strong positioning sequence. J. Mol. Biol. 403, 1–10 10.1016/j.jmb.2010.08.039 [DOI] [PubMed] [Google Scholar]
- 62.Willhoft O., Ghoneim M., Lin C.-L., Chua E.Y.D., Wilkinson M., Chaban Y. et al. (2018) Structure and dynamics of the yeast SWR1-nucleosome complex. Science 362, eaat7716 10.1126/science.aat7716 [DOI] [PubMed] [Google Scholar]
- 63.Liu X., Li M., Xia X., Li X. and Chen Z. (2017) Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure. Nature 544, 440–445 10.1038/nature22036 [DOI] [PubMed] [Google Scholar]
- 64.Dodonova S.O., Zhu F., Dienemann C., Taipale J. and Cramer P. (2020) Nucleosome-bound SOX2 and SOX11 structures elucidate pioneer factor function. Nature 580, 669–672 10.1038/s41586-020-2195-y [DOI] [PubMed] [Google Scholar]
- 65.Habig M., Bahena‐Garrido S.M., Barkmann F., Haueisen J. and Stukenbrock E.H. (2020) The transcription factor Zt107320 affects the dimorphic switch, growth and virulence of the fungal wheat pathogen Zymoseptoria tritici. Mol. Plant Pathol. 21, 124–138 10.1111/mpp.12886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Öztürk M.A., De M., Cojocaru V. and Wade R.C. (2020) Chromatosome structure and dynamics from molecular simulations. Annu. Rev. Phys. Chem. 71, 101–119 10.1146/annurev-physchem-071119-040043 [DOI] [PubMed] [Google Scholar]
- 67.Frouws T.D., Duda S.C. and Richmond T.J. (2016) X-ray structure of the MMTV-A nucleosome core. Proc. Natl. Acad. Sci. U.S.A. 113, 1214–1219 10.1073/pnas.1524607113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Makde R.D., England J.R., Yennawar H.P. and Tan S. (2010) Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature 467, 562–566 10.1038/nature09321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Segal E., Fondufe-Mittendorf Y., Chen L., Thåström A., Field Y., Moore I.K. et al. (2006) A genomic code for nucleosome positioning. Nature 442, 772–778 10.1038/nature04979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Iwata-Otsubo A., Dawicki-McKenna J.M., Akera T., Falk S.J., Chmátal L., Yang K. et al. (2017) Expanded satellite repeats amplify a discrete CENP-a nucleosome assembly site on chromosomes that drive in female meiosis. Curr. Biol. 27, 2365.e8–2373.e8 10.1016/j.cub.2017.06.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheeseman I.M. and Desai A. (2008) Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9, 33–46 10.1038/nrm2310 [DOI] [PubMed] [Google Scholar]
- 72.Musacchio A. and Desai A. (2017) A molecular view of kinetochore assembly and function. Biology 6, 5 10.3390/biology6010005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kixmoeller K., Allu P.K. and Black B.E. (2020) The centromere comes into focus: from CENP-A nucleosomes to kinetochore connections with the spindle. Open Biol. 10, 200051 10.1098/rsob.200051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Amano T., Sagai T., Tanabe H., Mizushina Y., Nakazawa H. and Shiroishi T. (2009) Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Dev. Cell 16, 47–57 10.1016/j.devcel.2008.11.011 [DOI] [PubMed] [Google Scholar]
- 75.Basilico F., Maffini S., Weir J.R., Prumbaum D., Rojas A.M., Zimniak T. et al. (2014) The pseudo GTPase CENP-M drives human kinetochore assembly. eLife 3, e02978 10.7554/eLife.02978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Carroll C.W., Silva M.C.C., Godek K.M., Jansen L.E.T. and Straight A.F. (2009) Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat. Cell Biol. 11, 896–902 10.1038/ncb1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Earnshaw W.C. and Rothfield N. (1985) Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91, 313–321 10.1007/BF00328227 [DOI] [PubMed] [Google Scholar]
- 78.Foltz D.R., Jansen L.E.T., Black B.E., Bailey A.O., Yates J.R. and Cleveland D.W. (2006) The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8, 458–469 10.1038/ncb1397 [DOI] [PubMed] [Google Scholar]
- 79.Hori T., Amano M., Suzuki A., Backer C.B., Welburn J.P., Dong Y. et al. (2008) CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell 135, 1039–1052 10.1016/j.cell.2008.10.019 [DOI] [PubMed] [Google Scholar]
- 80.Izuta H., Ikeno M., Suzuki N., Tomonaga T., Nozaki N., Obuse C. et al. (2006) Comprehensive analysis of the ICEN (Interphase Centromere Complex) components enriched in the CENP-A chromatin of human cells. Genes Cells 11, 673–684 10.1111/j.1365-2443.2006.00969.x [DOI] [PubMed] [Google Scholar]
- 81.Nishino T., Takeuchi K., Gascoigne K.E., Suzuki A., Hori T., Oyama T. et al. (2012) CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell 148, 487–501 10.1016/j.cell.2011.11.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Okada M., Cheeseman I.M., Hori T., Okawa K., McLeod I.X., Yates J.R. et al. (2006) The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8, 446–457 10.1038/ncb1396 [DOI] [PubMed] [Google Scholar]
- 83.Saitoh H., Tomkiel J., Cooke C.A., Ratrie H., Maurer M., Rothfield N.F. et al. (1992) CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell 70, 115–125 10.1016/0092-8674(92)90538-N [DOI] [PubMed] [Google Scholar]
- 84.Carroll C.W., Milks K.J. and Straight A.F. (2010) Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 189, 1143–1155 10.1083/jcb.201001013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Falk S.J., Guo L.Y., Sekulic N., Smoak E.M., Mani T., Logsdon G.A. et al. (2015) CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere. Science 348, 699–703 10.1126/science.1259308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kato H., Jiang J., Zhou B.-R., Rozendaal M., Feng H., Ghirlando R. et al. (2013) A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science 340, 1110–1113 10.1126/science.1235532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cao S., Zhou K., Zhang Z., Luger K. and Straight A.F. (2018) Constitutive centromere-associated network contacts confer differential stability on CENP-A nucleosomes in vitro and in the cell. Mol. Biol. Cell 29, 751–762 10.1091/mbc.E17-10-0596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chittori S., Hong J., Saunders H., Feng H., Ghirlando R., Kelly A.E. et al. (2018) Structural mechanisms of centromeric nucleosome recognition by the kinetochore protein CENP-N. Science 359, 339–343 10.1126/science.aar2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pentakota S., Zhou K., Smith C., Maffini S., Petrovic A., Morgan G.P. et al. (2017) Decoding the centromeric nucleosome through CENP-N. eLife 6, e33442 10.7554/eLife.33442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tian T., Li X., Liu Y., Wang C., Liu X., Bi G. et al. (2018) Molecular basis for CENP-N recognition of CENP-A nucleosome on the human kinetochore. Cell Res. 28, 374–378 10.1038/cr.2018.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ogura Y., Shibata F., Sato H. and Murata M. (2004) Characterization of a CENP-C homolog in Arabidopsis thaliana. Genes Genet. Syst. 79, 139–144 10.1266/ggs.79.139 [DOI] [PubMed] [Google Scholar]
- 92.Dawe R.K., Reed L.M., Yu H.-G., Muszynski M.G. and Hiatt E.N. (1999) A maize homolog of mammalian CENPC is a constitutive component of the inner kinetochore. Plant Cell 11, 1227–1238 10.1105/tpc.11.7.1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Klare K., Weir J.R., Basilico F., Zimniak T., Massimiliano L., Ludwigs N. et al. (2015) CENP-C is a blueprint for constitutive centromere-associated network assembly within human kinetochores. J. Cell Biol. 210, 11–22 10.1083/jcb.201412028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gascoigne K.E., Takeuchi K., Suzuki A., Hori T., Fukagawa T. and Cheeseman I.M. (2011) Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell 145, 410–422 10.1016/j.cell.2011.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Screpanti E., De Antoni A., Alushin G.M., Petrovic A., Melis T., Nogales E. et al. (2011) Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr. Biol. 21, 391–398 10.1016/j.cub.2010.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.McKinley K.L., Sekulic N., Guo L.Y., Tsinman T., Black B.E. and Cheeseman I.M. (2015) The CENP-L-N complex forms a critical node in an integrated meshwork of interactions at the centromere-kinetochore interface. Mol. Cell 60, 886–898 10.1016/j.molcel.2015.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Przewloka M.R., Venkei Z., Bolanos-Garcia V.M., Debski J., Dadlez M. and Glover D.M. (2011) CENP-C is a structural platform for kinetochore assembly. Curr. Biol. 21, 399–405 10.1016/j.cub.2011.02.005 [DOI] [PubMed] [Google Scholar]
- 98.Milks K.J., Moree B. and Straight A.F. (2009) Dissection of CENP-C-directed centromere and kinetochore assembly. Mol. Biol. Cell 20, 4246–4255 10.1091/mbc.e09-05-0378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brown M.T. (1995) Sequence similarities between the yeast chromosome segregation protein Mif2 and the mammalian centromere protein CENP-C. Gene 160, 111–116 10.1016/0378-1119(95)00163-Z [DOI] [PubMed] [Google Scholar]
- 100.Guo L.Y., Allu P.K., Zandarashvili L., McKinley K.L., Sekulic N., Dawicki-McKenna J.M. et al. (2017) Centromeres are maintained by fastening CENP-A to DNA and directing an arginine anchor-dependent nucleosome transition. Nat. Commun. 8, 15775 10.1038/ncomms15775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wilson M.D., Benlekbir S., Fradet-Turcotte A., Sherker A., Julien J.-P., McEwan A. et al. (2016) The structural basis of modified nucleosome recognition by 53BP1. Nature 536, 100–103 10.1038/nature18951 [DOI] [PubMed] [Google Scholar]
- 102.Lesbats P., Serrao E., Maskell D.P., Pye V.E., O’Reilly N., Lindemann D. et al. (2017) Structural basis for spumavirus GAG tethering to chromatin. Proc. Natl. Acad. Sci. U.S.A. 114, 5509–5514 10.1073/pnas.1621159114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fang Q., Chen P., Wang M., Fang J., Yang N., Li G. et al. (2016) Human cytomegalovirus IE1 protein alters the higher-order chromatin structure by targeting the acidic patch of the nucleosome. eLife 5, e11911 10.7554/eLife.11911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Barbera A.J., Chodaparambil J.V., Kelley-Clarke B., Joukov V., Walter J.C., Luger K. et al. (2006) The nucleosomal surface as a docking station for Kaposi’s Sarcoma Herpesvirus LANA. Science 311, 856–861 10.1126/science.1120541 [DOI] [PubMed] [Google Scholar]
- 105.Melters D.P., Pitman M., Rakshit T., Dimitriadis E.K., Bui M., Papoian G.A. et al. (2019) Intrinsic elasticity of nucleosomes is encoded by histone variants and calibrated by their binding partners. Proc. Natl. Acad. Sci. U.S.A. 116, 24066–24074 10.1073/pnas.1911880116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hinshaw S.M. and Harrison S.C. (2019) The structure of the Ctf19c/CCAN from budding yeast. eLife 8, e44239 10.7554/eLife.44239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Weir J.R., Faesen A.C., Klare K., Petrovic A., Basilico F., Fischböck J. et al. (2016) Insights from biochemical reconstitution into the architecture of human kinetochores. Nature 537, 249–253 10.1038/nature19333 [DOI] [PubMed] [Google Scholar]
- 108.Fang J., Liu Y., Wei Y., Deng W., Yu Z., Huang L. et al. (2015) Structural transitions of centromeric chromatin regulate the cell cycle-dependent recruitment of CENP-N. Genes Dev. 29, 1058–1073 10.1101/gad.259432.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hori T., Shang W.-H., Toyoda A., Misu S., Monma N., Ikeo K. et al. (2014) Histone H4 Lys 20 monomethylation of the CENP-A nucleosome is essential for kinetochore assembly. Dev. Cell 29, 740–749 10.1016/j.devcel.2014.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bodor D.L., Mata J.F., Sergeev M., David A.F., Salimian K.J., Panchenko T. et al. (2014) The quantitative architecture of centromeric chromatin. eLife 3, e02137 10.7554/eLife.02137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Blower M.D., Sullivan B.A. and Karpen G.H. (2002) Conserved organization of centromeric chromatin in flies and humans. Dev. Cell 2, 319–330 10.1016/S1534-5807(02)00135-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ribeiro S.A., Vagnarelli P., Dong Y., Hori T., McEwen B.F., Fukagawa T. et al. (2010) A super-resolution map of the vertebrate kinetochore. Proc. Natl. Acad. Sci. U.S.A. 107, 10484–10489 10.1073/pnas.1002325107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Andronov L., Ouararhni K., Stoll I., Klaholz B.P. and Hamiche A. (2019) CENP-A nucleosome clusters form rosette-like structures around HJURP during G1. Nat. Commun. 10, 1–8 10.1038/s41467-019-12383-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Geiss C.P., Keramisanou D., Sekulic N., Scheffer M.P., Black B.E. and Frangakis A.S. (2014) CENP-A arrays are more condensed than canonical arrays at low ionic strength. Biophys. J. 106, 875–882 10.1016/j.bpj.2014.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Naughton C. and Gilbert N. (2020) Centromere chromatin structure - lessons from neocentromeres. Exp. Cell Res. 389, 111899 10.1016/j.yexcr.2020.111899 [DOI] [PubMed] [Google Scholar]
- 116.Clarke L. and Carbon J. (1980) Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature 287, 504–509 10.1038/287504a0 [DOI] [PubMed] [Google Scholar]
- 117.Clarke L. and Carbon J. (1983) Genomic substitutions of centromeres in Saccharomyces cerevisiae. Nature 305, 23–28 10.1038/305023a0 [DOI] [PubMed] [Google Scholar]
- 118.Ou H.D., Phan S., Deerinck T.J., Thor A., Ellisman M.H. and O’Shea C.C. (2017) ChromEMT: visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science 357, eaag0025 10.1126/science.aag0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cai S., Böck D., Pilhofer M. and Gan L. (2018) The in situ structures of mono-, di-, and trinucleosomes in human heterochromatin. Mol. Biol. Cell 29, 2450–2457 10.1091/mbc.E18-05-0331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ashwin S.S., Maeshima K. and Sasai M. (2020) Heterogeneous fluid-like movements of chromatin and their implications to transcription. Biophys. Rev. 12, 461–468 10.1007/s12551-020-00675-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Maeshima K., Tamura S., Hansen J.C. and Itoh Y. (2020) Fluid-like chromatin: Toward understanding the real chromatin organization present in the cell. Curr. Opin. Cell Biol. 64, 77–89 10.1016/j.ceb.2020.02.016 [DOI] [PubMed] [Google Scholar]
- 122.Hildebrand E.M. and Dekker J. (2020) Mechanisms and functions of chromosome compartmentalization. Trends Biochem. Sci. 45, 385–396 10.1016/j.tibs.2020.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ma H., Tu L.-C., Chung Y.-C., Naseri A., Grunwald D., Zhang S. et al. (2019) Cell cycle- and genomic distance-dependent dynamics of a discrete chromosomal region. J. Cell Biol. 218, 1467–1477 10.1083/jcb.201807162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kakui Y., Rabinowitz A., Barry D.J. and Uhlmann F. (2017) Condensin-mediated remodeling of the mitotic chromatin landscape in fission yeast. Nat. Genet. 49, 1553–1557 10.1038/ng.3938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tanizawa H., Kim K.-D., Iwasaki O. and Noma K.-I. (2017) Architectural alterations of the fission yeast genome during the cell cycle. Nat. Struct. Mol. Biol. 24, 965–976 10.1038/nsmb.3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Takeuchi K., Nishino T., Mayanagi K., Horikoshi N., Osakabe A., Tachiwana H. et al. (2014) The centromeric nucleosome-like CENP-T-W-S-X complex induces positive supercoils into DNA. Nucleic Acids Res. 42, 1644–1655 10.1093/nar/gkt1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gamba R. and Fachinetti D. (2020) From evolution to function: two sides of the same CENP-B coin? Exp. Cell Res. 111959 10.1016/j.yexcr.2020.111959 [DOI] [PubMed] [Google Scholar]
- 128.Ohzeki J., Nakano M., Okada T. and Masumoto H. (2002) CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J. Cell Biol. 159, 765–775 10.1083/jcb.200207112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fujita R., Otake K., Arimura Y., Horikoshi N., Miya Y., Shiga T. et al. (2015) Stable complex formation of CENP-B with the CENP-A nucleosome. Nucleic Acids Res. 43, 4909–4922 10.1093/nar/gkv405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fachinetti D., Han J.S., McMahon M.A., Ly P., Abdullah A., Wong A.J. et al. (2015) DNA sequence-specific binding of CENP-B enhances the fidelity of human centromere function. Dev. Cell 33, 314–327 10.1016/j.devcel.2015.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Henikoff S. and Smith M.M. (2015) Histone variants and epigenetics. Cold Spring Harb. Perspect. Biol. 7, a019364 10.1101/cshperspect.a019364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kitagawa K., Masumoto H., Ikeda M. and Okazaki T. (1995) Analysis of protein-DNA and protein-protein interactions of centromere protein B (CENP-B) and properties of the DNA-CENP-B complex in the cell cycle. Mol. Cell. Biol. 15, 1602–1612 10.1128/MCB.15.3.1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kasinathan S. and Henikoff S. (2018) Non-B-Form DNA is enriched at centromeres. Mol. Biol. Evol. 35, 949–962 10.1093/molbev/msy010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bai X., McMullan G. and Scheres S.H.W. (2015) How cryo-EM is revolutionizing structural biology. Trends Biochem. Sci. 40, 49–57 10.1016/j.tibs.2014.10.005 [DOI] [PubMed] [Google Scholar]
- 135.Merk A., Bartesaghi A., Banerjee S., Falconieri V., Rao P., Davis M.I. et al. (2016) Breaking Cryo-EM resolution barriers to facilitate drug discovery. Cell 165, 1698–1707 10.1016/j.cell.2016.05.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jenni S. and Harrison S.C. (2018) Structure of the DASH/Dam1 complex shows its role at the yeast kinetochore-microtubule interface. Science 360, 552–558 10.1126/science.aar6436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yan K., Zhang Z., Yang J., McLaughlin S.H. and Barford D. (2018) Architecture of the CBF3-centromere complex of the budding yeast kinetochore. Nat. Struct. Mol. Biol. 25, 1103–1110 10.1038/s41594-018-0154-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Higashi T.L., Eickhoff P., Simoes J.S., Locke J., Nans A., Flynn H.R. et al. (2020) A structure-based mechanism for DNA entry into the cohesin ring. bioRxiv 10.1101/2020.04.21.052944 [DOI] [PMC free article] [PubMed] [Google Scholar]