Abstract
Background
The BAP1 mutation is commonly found kidney renal clear cell carcinoma (KIRC) and a potential biomarker of individualized therapy. We evaluated the clinical significance of BAP1 mutation in the prognosis and treatment therapies for KIRC. Potential key pathways and related genes associated with these mechanisms were also identified in this investigation.
Methods
We identified the relevant data of patients BAP1 mutated on the cBioPortal and the compounds with significant selectivity to BAP1 mutations on the Genomics of Drug Sensitivity in Cancer (GDSC). And then, we identified the differences in mRNA expression levels of biological function annotation and pathways between mutated and wild type BAP1 patients by GSEA analysis. Furthermore, we screened the differentially expressed genes (DEGs) between BAP1 mutated and wild typed in KIRC patients and performed the GO and KEGG analysis. Finally, we conducted a protein-protein interaction (PPI) network to investigate the interaction between proteins encoded by candidate DEGs.
Results
Review of the TCGA data revealed 41 patients (10%) with KIRC displayed the BAP1 mutation. Further analysis led to the identification of 730 DEGs, while 617 genes were shown to be down-regulated, with 113 genes displaying upregulation. GO and KEGG pathway analysis indicated DEGs as enriched in metabolism, drug metabolism-cytochrome P450, and Drug-metabolizing enzymes. Subsequently, the top 10 hub genes, ranked by the degree in the PPI network were identified. Furthermore, our findings verify that the BAP1 mutation was associated with the deterioration of prognosis in patients with KIRC. Additionally, analysis of the GDSC database revealed that KIRC patients with BPP1 mutation are more prone to responding to Linsitinib.
Conclusions
Our investigation identified the main pathways and relevant genes related to the BAP1 mutation in KIRC, which can contribute to the development of targeted treatment strategies for enhanced prognostic predictions of KIRC.
Keywords: Kidney renal clear cell carcinoma (KIRC), BAP1 mutation, TCGA, RNA sequencing, bioinformatics analysis
Introduction
Kidney cancer [renal cell carcinoma (RCC)] is the most common prevailing urogenital tumor and comprises approximately 3% of all adult malignant diseases (1). More specifically, kidney renal clear cell carcinoma (KIRC) is established as highly treatment-resistant, with an incidence rate that is steadily increasing around the world (2). It has also been consistently associated with genetic mutations (3), with VHL, PBRM1/BAF180, SETD2, and BAP1 identified as four of the most commonly mutated genes in KIRC (4). Moreover, treatment resistance has been shown to result from genetic mutations, further highlighting the importance of selective treatment therapies. Common treatments, such as tyrosine kinase inhibitor sunitinib and VEGF inhibitor bevacizumab, have shown limited benefits in treating KIRC (5). In summary, identifying biomarkers and potential targeted drugs will be critical in the development of individualized therapy in successfully determining the progression and prognosis of KIRC.
BAP1 is a protein-encoding, tumor suppressor gene. Germline mutations of BAP1 has shown to increase the susceptibility to malignant tumors such as melanoma of the skin and uveal, malignant mesothelioma, KIRC, cholangiocarcinoma, and basal cell carcinoma (6). Additionally, changes in BAP1 expression may lead to the formation of RCC, which is of great significance for the clinical prognosis and development of precision therapy. BAP1 can deubiquitylate Ub-H2AK119 at the site of UV-induced DNA damage, and promote DNA repair by regulating the accumulation of repair proteins (BRCA1, RAD51, and RPA) (7). BAP1 mutations most often affect its catalytic activity by regulating methylated histone H3 lysine 27 (H3K27) and polycomb repressive complex 2 (PRC2) (8). Also, BAP1 mutation has shown to be related to aerobic glycolysis, where significant differences were observed in the characteristics of glycolytic metabolites between mutated and wild-type BAP1 fibroblast cell cultures (9,10). In other reports, BAP1 targets type 3 inositol 1,4,5 triphosphate receptor (IP3R3) located in the endoplasmic reticulum, and subsequently regulates Ca2+ release to promote apoptosis (9). In summary, BAP1 mutations can affect the treatment choice and serve as a predictive tool for the progression and prognosis of cancer. Therefore, the response to treatments of patients with KIRC may alter during the development and progression of the disease. Thus, our exploration of the potential relationship between BAP1 mutations and related signal pathways will contribute to the understanding of BAP1 mutations and their significance in the incidence and progression of KIRC. Additionally, this investigation may further provide a theoretical basis for the development of targeted KIRC therapies.
In conclusion, BaP1 mutation plays an important role in cancer progression. However, as far as we know, studies based on BaP1 mutation in drug treatment selection have not been found, which is crucial. In this study, our objective was to determine the potential signal pathways and genes related to BAP1 mutations through analyzing the RNA-Seq dataset and Genomics of Drug Sensitivity in Cancer (GDSC) database of KIRC by bioinformatics analysis. We also evaluated their significance in drug selection to potentially identify the crucial role of BAP1 mutations in individualized therapy. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tau-20-1079).
Methods
RNA-Seq data
The RNA-Seq dataset and corresponding clinical data of renal clear cell carcinoma from the TCGA database was downloaded and reviewed for relevant data of BAP1 mutated patients on the cBioPortal (11).
Data analysis and mining of the GDSC database
Compounds with significant selectivity to BAP1 mutations was initially identified on the GDSC (12). Subsequently, the sensitivity was isolated to KIRC cells before narrowing the hits. Volcanic maps and scatter maps were calculated and generated by GDSC.
Gene set enrichment analysis (GSEA)
It was important to identify the effect of BAP1 mutation in KIRC patients. We therefore identified the differences in mRNA expression levels of biological function annotation and pathways between mutated and wild type BAP1 patients by GSEA analysis.
DEGs
The differential expression of RNA-Seq count data to screen the differential expression of genes between BAP1 mutated and wild typed in KIRC patients were detected by EdgeR (13). |Fold change (FC)| ≥2 was the criterion, as well as the P value and FDR <0.05.
Functional annotation and pathway enrichment analysis of DEGs
Enrichment analysis was performed through uploading GO and KEGG analysis to the Database for Annotation, Visualization and Integrated Discovery (DAVID), while statistical significance was set as P value <0.05 (14).
PPI network and module analysis
The STRING (version 9.0) database is a tool for developing DEGs-encoded protein and PPI network (15), covering 5,214,234 proteins from 1,133 organisms. In this study, we tried to screen the STRING database to investigate the interaction and hub genes of DEGs. Cytoscape software was used to conduct a PPI network for the interaction between proteins encoded by candidate DEGs (score >0.4). The module selection by MCODE in Cytoscape (scores >3 and nodes >4) (16) was made, based on the PPI network.
Statistical analysis
The student’s t-test was used to detect the difference in BAP1 mRNA expression between BAP1 mutated and wild type KIRC. Subsequently, the clinical efficacy of BAP1 mutation and wild type, determined by Graphpad, was calculated with the Kaplan-Meier method combined with the logrank test. We adjusted FDR in edgeR and GSEA, respectively, by multiple testing via the Benjamini-Hochberg procedure (17,18). A P<0.05 was considered statistically significant. Graphpad and R 3.6.3 were used to perform the statistical analyses.
Results
BAP1 mutation is more common in KIRC
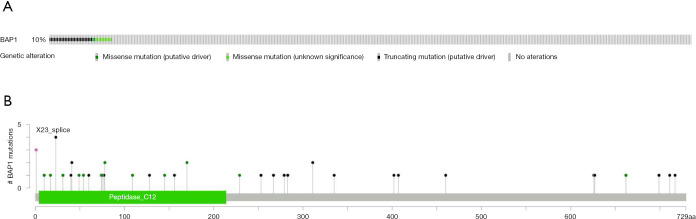
Information of 530 patients with KIRC, corresponding RNA-Seq datasets of cancer tissues and completed follow-up data were downloaded from the TCGA database. Data pertaining to BAP1 mutations in KIRC patients was also obtained from the cBioPortal. Forty-one KIRC patients (10%) were shown to carry BAP1 mutations (Figure 1A). Mutation types including missense, and truncating mutations are displayed in Figure 1B.
Figure 1.
Forty-one KIRC patients (10%) carried BAP1 mutations (A), while the mutation types including missense, and truncating mutations (B) had been found in KIRC reproduced from the TCGA database.
Correlation between BAP1 mutation and overall survival in KIRC
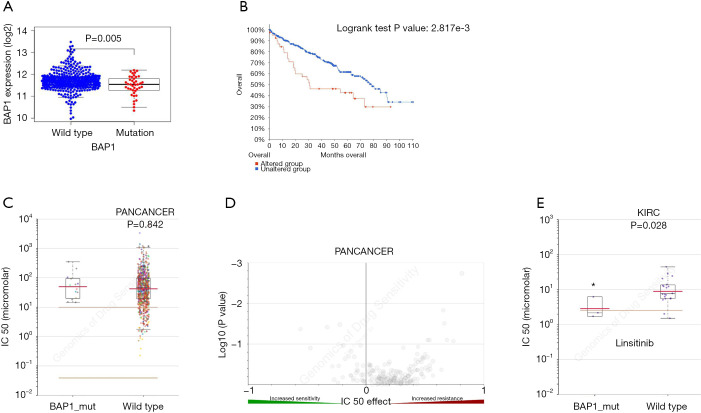
Based on the clinical data obtained from TCGA, we investigated the effect of BAP1 mutation on the progression and prognosis of KIRC. Patients were divided into two groups, based on their clinical characteristics (Table 1). mRNA expression levels were examined across the mutant and wildtype groups. Analysis indicated a down-regulation of BAP1 levels in the tumor tissues of KIRC patients (Figure 2A). The overall survival of patients with BAP1 mutation was shown to be poor (Figure 2B), suggesting that BAP1 mutation may contribute to the progression of KIRC. The results indicate that early genetic mutation and expression analysis may be beneficial to the prognosis of patients with BAP1 mutation.
Table 1. The clinical characteristics of KIRC patients in TCGA.
| Characteristics | BAP1 status | |
|---|---|---|
| Wild-type | Mutated | |
| Age [years] | 60.7 [26–90] | 62.1 [41–85] |
| Gender | ||
| Male | 257 | 17 |
| Female | 120 | 24 |
| Tumor stage | ||
| T1 | 194 | 8 |
| T2 | 40 | 10 |
| T3 | 137 | 23 |
| T4 | 6 | 0 |
| N state | ||
| N0 | 166 | 26 |
| N1 | 10 | 2 |
| NX | 201 | 13 |
| AJCC stage | ||
| Stage I | 190 | 7 |
| Stage II | 34 | 7 |
| Stage III | 96 | 15 |
| Stage IV | 57 | 12 |
| Histologic grade | ||
| G1 | 7 | 0 |
| G2 | 162 | 12 |
| G3 | 154 | 15 |
| G4 | 54 | 13 |
| GX | 0 | 1 |
KIRC, kidney renal clear cell carcinoma.
Figure 2.
Mutations of BAP1 are associated with KIRC prognosis and drug selection. (A) Correlation between the BAP1 mutation and mRNA expression. (B) Kaplan-Meier survival curves for KIRC patients stratified by the BAP1 mutation. (C,D) Scattered plot and volcano plot show that multiple cancer cell types with the BAP1 mutation were not significantly inhibited by linsitinib. (E) Reproduction of the GDSC database by excluding cancer of other types showed that KIRC cells with the BAP1 mutation were significantly inhibited by linsitinib. *, P<0.05. GDSC, Genomics of Drug Sensitivity in Cancer; KIRC, kidney renal clear cell carcinoma.
KIRC cells with BAP1 mutation are sensitive to linsitinib
The benefits of current targeted therapies in treating metastatic KIRC is limited and highly susceptible to drug resistance. Therefore, we further investigated the influence of BAP1 on treatment response. To develop more effective and targeted therapeutics, we investigated whether KIRC patients carrying the BAP1 mutation showed potentially better selective drugs by searching the GDSC database. The investigation initially revealed that linsitinib showed no significant difference in IC50 values in various cancers between BAP1 mutated and wild-type groups (P=0.842) (Figure 2C,D). However, linsitinib specifically benefitted KIRC patients with BAP1 mutation (P=0.028) (Figure 2E). Above all, the results demonstrated that linsitinib, a dual inhibitor of the IGF-1 receptor and insulin receptor, was a potential targeted antineoplastic drug for KIRC patients with BAP1 mutation.
Results of GSEA
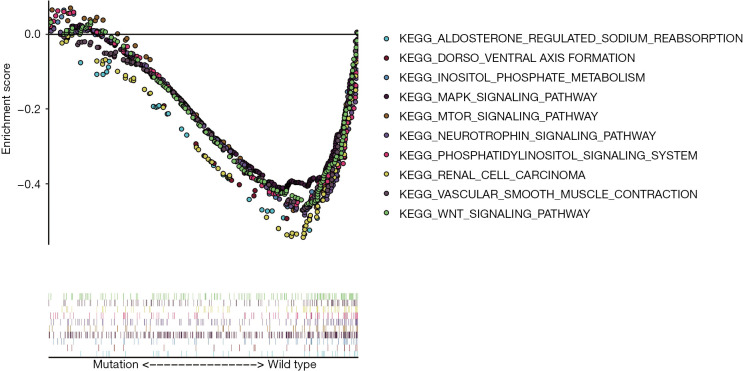
To further explore and demonstrate the potential mechanism that link the occurrence, progression, prognosis, and drug selection of KIRC to BAP1 mutation, the effect of BAP1 mutations on cellular processes was analyzed. We first examined various biological functional gene sets by GSEA. The result indicated aldosterone regulates sodium reabsorption, dorsal-ventral axis formation, inositol phosphate metabolism, MAPK signal pathway, mTOR signal pathway, neurotrophin signal pathway, phosphatidylinositol signal system, RCC, vascular smooth muscle contraction and WNT signal pathway were significantly enriched and more active in patients with BAP1 wild-type (Figure 3). The result suggests BAP1 mutation may contribute to the progress of KIRC by affecting tumor formation, metabolism, invasion and metastasis, growth and differentiation, apoptosis and DNA repair.
Figure 3.
We further explored the differences in genes enrichment between BAP1 wild-type and BAP1 mutant patients by GSEA analysis. The result suggested that the signal pathways of gene enrichment are more active in patients with BAP1 wild-type. GSEA, gene set enrichment analysis; KIRC, kidney renal clear cell carcinoma.
Identification of DEGs
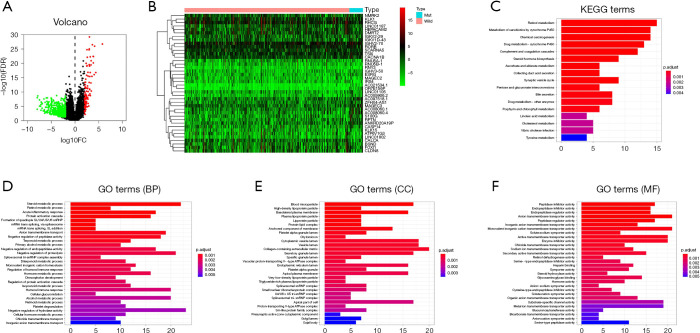
To explore the potential pathways and relevant genes involved with BAP1 mutation, the RNA-Seq data of 41 BAP1 mutant and wild-type KIRC patients were reviewed for DEG screening. A total of 730 genes were identified as DEGs, based on in-silico analysis with the |FC| ≥2.0 and P<0.05 criteria. The volcano plot of the DEGs is shown in Figure 4A. Subsequently, 617 genes were found to be down-regulated and 113 genes were up-regulated. Furthermore, we drew the heatmap according to the expression of the DEGs between KIRC patients with BAP1 mutated and BAP1 wild-type (Figure 4B).
Figure 4.
DAVID enrichment results of DEGs. (A) Volcano plot of DEGs. (B) Heatmap of DEGs. (C) KEGG pathway analysis of DEGs. (D,E,F) GO enrichment terms of DEGs. DAVID, Database for Annotation, Visualization and Integrated Discovery; DEGs, differentially expressed genes.
GO and KEGG analyses of DEGs
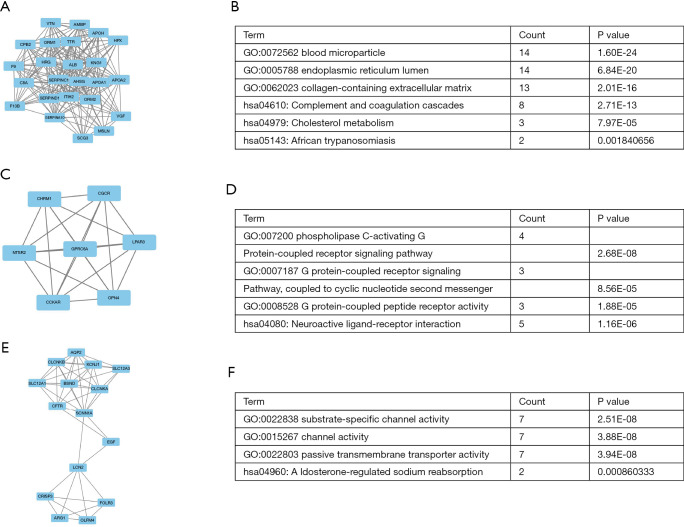
The DEGs underwent GO, KEGG pathways and DAVID analysis, before examination at a functional level. KEGG pathway analysis revealed that DEGs were significantly enriched in the retinol metabolism, metabolism of xenobiotics by cytochrome P450, chemical carcinogenesis, drug metabolism-cytochrome P450 (Figure 4C). The top three enriched GO terms for each category, BP, cell component (CC), and MF, were identified. It is noteworthy that the enriched BP terms included steroid metabolic process, negative regulation of hydrolase activity, and organic anion transport. The enriched CC terms included cytoplasmic vesicle lumen, vesicle lumen, and collagen-containing extracellular matrix. The enriched MF terms included enzyme inhibitor activity, anion transmembrane transporter activity, monovalent inorganic cation transmembrane transporter activity (Figure 4D,E,F).
Module screening from the PPI Network
We subsequently identified the top 10 hub genes ranked by degree, including ALB, KNG 1, AHSG, ORM1, APOA1, SERPINC 1, ORM2, ITIH2, EGF, C8A. The degree of ALB was shown to be the highest at 77. We identified the gene modules in the PPI network with the MCODE in Cytoscape and analyzed the enrichment of GO and KEGG pathways according to modules 1, 3, and 4 (Figure 5). The results indicated that the genes were mainly involved with blood microparticle, phospholipase C-activating G protein-coupled receptor signaling pathway, substrate-specific channel activity, complement and coagulation cascades, neuroactive ligand-receptor interaction, and aldosterone-regulated sodium reabsorption.
Figure 5.
Three modules from the PPI network. (A,B) PPI network and GO and KEGG analyses of module 1. (C,D) PPI network and GO and KEGG analyses of module 3. (E,F) PPI network and GO and KEGG analyses of module 4. PPI, protein-protein interaction.
Discussion
BAP1 is a tumor suppressor gene, where its mutation has been strongly associated with the formation of various malignant tumors. As a deubiquitinating enzyme, BAP1 contributes to gene transcription, cell differentiation, DNA damage repair, apoptosis, and cell metabolism in tumor inhibition. Recently, studies showed that aerobic glycolysis was more active in cells with BAP1 mutation (10). At the same time, BAP1 mutation promotes cell malignant transformation by altering cell metabolism and tumor microenvironment (10). Additionally, BAP1 promotes tumor cell apoptosis by regulating ER-mitochondrial calcium release (19). Besides, Guazzelli et al. pointed out that BAP1 plays an important role in cancer drug resistance, which may be associated with the sensitivity of BAP1 in different states to DNA damage (20). The potential mechanism may be related to ubiquitination during the DNA damage response regulated by BAP1and BRCA1-BARD1 complex (21). Additionally, Kumar et al. suggested BAP1 may modify response to vinorelbine by preventing mitotic microtubule formation, which is a potential predictive biomarker for chemotherapy sensitivity (22). As a deubiquitinating enzyme, BAP1 inhibits the expression of SLC7A11 by reducing the ubiquitination of H2A to inhibit the activity of SLC7A11 promoter (23).
Our study investigated the effect of BAP1 mutation on individualized treatment in KIRC patients. We analyzed 530 cases and identified approximately 10% of patients that carried truncated and missense BAP1 mutations. Survival analysis revealed significantly poor survival rates of KIRC patients with the BAP1 gene mutation. The above results show that the key role of BAP1 mutation in patients with KIRC is consistent with some recent findings. In clinical application, clinicians can determine the prognosis of patients, and select appropriate treatment plans, tailored to the individual’s genetic profiling. Comprehensive examinations and close follow-up in KIRC patients with BAP1 mutations require close attention to identify early tumor dissemination, or earlier use of selective drugs. More importantly, GDSC data reveals that KIRC patients with BAP1 mutation showed a higher sensitivity to linsitinib. Linsitinib could potentially be administered as a selective drug for individualized therapy in KIRC patients with BAP1 mutation in the future.
In our study, we first downloaded the RNA-Seq data set of KIRC from the TCGA database. Bioinformatics analysis was performed to determine the potential pathways and genes related to BAP1 mutation. We simultaneously performed GSEA analysis, revealing that BAP1 mutation was related to many cancer-related pathways. Interestingly, MAPK signal pathway, mTOR signal pathway, and WNT signal pathway were related to multiple drug resistance. Quispel-Janssen et al. (24) indicated BAP1 loss was associated with the increased expression of the receptors FGFR1/3 and ligands FGF9/18, which contributed to the sensitivity of FGFR inhibition. Besides, a study of families with BAP1 germline mutations shows that mutated BAP1 may promote oncogenesis and increase the risk of KIRC by 8-fold through altering cellular metabolism and the tumor microenvironment (25). BAP1 was also found to regulate Ca2+ release by binding to the IP3R3, contributing to apoptosis of DNA damaged cells. These cells were normally susceptible to cancer transformation due to decreased apoptosis with deficient BAP1 (19). Therefore, in the follow-up study, we can focus on the relationship between BAP1 mutation and MAPK, mTOR, and WNT signaling pathways in KIRC.
Furthermore, we identified 730 DEGs between the mutated and wild-types before undertaking GO and KEGG pathway analysis. In the meantime, DEGs in KIRC patients with BAP1 mutation were enriched in the biological processes of metabolism, drug metabolism-cytochrome P450, acute inflammatory response, chemical carcinogenesis, humoral immune response, and drug metabolism-other enzymes. Among them, drug metabolic enzymes play an important role in metabolic detoxification and activation. They elicit crucial drug effects and susceptibility to toxicity in tissues and organs. The main drug metabolic enzymes related to chemotherapeutic therapies are GST, CYP, UGT, TPMT, and DPD. These enzymes display genetic polymorphism and can be induced under certain conditions. The emergence of tumor drug resistance may be the result of joint actions from various molecular mechanisms, and the increase in the metabolic inactivation of drugs. The metabolic pathways of numerous enzymes and the complex interactions between phase I and phase II metabolic enzymes has not received sufficient attention in the study of tumor drug resistance. However, there is a lack of investigation on the possible mechanisms of the effect of BAP1 on drug metabolic enzymes in KIRC. Therefore, the potential relationship between the BAP1 mutation and the level of drug-related metabolic enzymes in KIRC may provide a potential therapeutic target.
Based on the degree of interaction, we identified the hub gene composed of 10 DEGs in the PPI network analysis. GO annotations related to this gene included blood microparticle, endoplasmic reticulum lumen, post-translational protein modification, vesicle lumen, cytoplasmic vesicle lumen, and secretory granule lumen.
The limitation of this paper is that the conclusion is based on pure bioinformatics analysis and lacks strong evidence. We will continue to improve the experiment in the future to further verify it.
Conclusions
In summary, our investigation identified the potential pathways and relevant genes related to the BAP1 mutation in KIRC, potentially contributing to the development of treatment strategies, predictive and prognostic tools for this special subtype of KIRC patients.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This study is funded by the High-level Hospital Construction Research Project of Maoming People’s Hospital, Guangdong Medical Science and Technology Research Fund (No. 2017101819316701), and Maoming Science and Technology Project (NO. 180324131700491).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tau-20-1079
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-1079). The authors have no conflicts of interest to declare.
(English Language Editor: E. Tan)
References
- 1.Liu F, Wu L, Wang A, et al. MicroRNA-138 attenuates epithelial-to-mesenchymal transition by targeting SOX4 in clear cell renal cell carcinoma. Am J Transl Res 2017;9:3611-22. [PMC free article] [PubMed] [Google Scholar]
- 2.He C, Zhao X, Jiang H, et al. Demethylation of miR-10b plays a suppressive role in ccRCC cells. Int J Clin Exp Pathol 2015;8:10595-604. [PMC free article] [PubMed] [Google Scholar]
- 3.Cimadamore A, Santoni M, Massari F, et al. Liquid biopsies in renal cell carcinoma with focus on epigenome analysis. Ann Transl Med 2019;7:S194. 10.21037/atm.2019.07.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsieh JJ, Le VH, Oyama T, et al. Chromosome 3p Loss-Orchestrated VHL, HIF, and Epigenetic Deregulation in Clear Cell Renal Cell Carcinoma. J Clin Oncol 2018;36:JCO2018792549. 10.1200/JCO.2018.79.2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heath EI. The Judgment of Paris: Treatment Dilemmas in Advanced Renal Cell Carcinoma. J Clin Oncol 2014;32:729-34. 10.1200/JCO.2013.53.6029 [DOI] [PubMed] [Google Scholar]
- 6.Machida YJ. A mechanism for the tissue specificity in BAP1 cancer syndrome. Transl Cancer Res 2019;8:S621-4. 10.21037/tcr.2019.06.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hendzel MJ. Germline Mutations in BAP1 Impair Its Function in DNA Double-Strand Break Repair. Cancer Res 2014;74:4282-94. 10.1158/0008-5472.CAN-13-3109 [DOI] [PubMed] [Google Scholar]
- 8.Murali R, Wiesner T, Scolyer RA. Tumours associated with BAP1 mutations. Pathology 2013;45:116-26. 10.1097/PAT.0b013e32835d0efb [DOI] [PubMed] [Google Scholar]
- 9.Bononi A, Yang H, Giorgi C, et al. Germline BAP1 mutations induce a Warburg effect. Cell Death Differ 2017;24:1694-704. 10.1038/cdd.2017.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warburg O. On the origin of cancer cells. Science 1956;123:309-14. 10.1126/science.123.3191.309 [DOI] [PubMed] [Google Scholar]
- 11.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang W, Soares J, Greninger P, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res 2013;41:D955-61. 10.1093/nar/gks1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139-40. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003;4:P3. 10.1186/gb-2003-4-5-p3 [DOI] [PubMed] [Google Scholar]
- 15.Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447-52. 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003;4:2. 10.1186/1471-2105-4-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benjamini Y, Drai D, Elmer G, et al. Controlling the false discovery rate in behavior genetics research. Behav Brain Res 2001;125:279-84. 10.1016/S0166-4328(01)00297-2 [DOI] [PubMed] [Google Scholar]
- 18.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 2003;19:368-75. 10.1093/bioinformatics/btf877 [DOI] [PubMed] [Google Scholar]
- 19.Bononi A, Giorgi C, Patergnani S, et al. BAP1 regulates IP3R3-mediated Ca(2+) flux to mitochondria suppressing cell transformation. Nature 2017;546:549-53. 10.1038/nature22798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guazzelli A, Meysami P, Bakker E, et al. BAP1 Status Determines the Sensitivity of Malignant Mesothelioma Cells to Gemcitabine Treatment. Int J Mol Sci 2019;20:429. 10.3390/ijms20020429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirosawa T, Ishida M, Ishii K, et al. Loss of BAP1 expression is associated with genetic mutation and can predict outcomes in gallbladder cancer. PLoS One 2018;13:e0206643. 10.1371/journal.pone.0206643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar N, Alrifai D, Kolluri KK, et al. Retrospective response analysis of BAP1 expression to predict the clinical activity of systemic cytotoxic chemotherapy in mesothelioma. Lung Cancer 2019;127:164-6. 10.1016/j.lungcan.2018.12.004 [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Koppula P, Gan B. Regulation of H2A ubiquitination and SLC7A11 expression by BAP1 and PRC1. Cell Cycle 2019;18:773-83. 10.1080/15384101.2019.1597506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quispel-Janssen JM, Badhai J, Schunselaar L, et al. Comprehensive Pharmacogenomic Profiling of Malignant Pleural Mesothelioma Identifies a Subgroup Sensitive to FGFR Inhibition. Clin Cancer Res 2018;24:84-94. 10.1158/1078-0432.CCR-17-1172 [DOI] [PubMed] [Google Scholar]
- 25.Popova T, Hebert L, Jacquemin V, et al. Germline BAP1 mutations predispose to renal cell carcinomas. Am J Hum Genet 2013;92:974-80. 10.1016/j.ajhg.2013.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as