Abstract
Autism spectrum disorder is a highly prevalent and highly heritable neurodevelopmental condition, but studies have mostly taken traditional categorical diagnosis approach (yes/no for autism spectrum disorder). In contrast, an emerging notion suggests a continuum model of autism spectrum disorder with a normal distribution of autistic tendencies in the general population, where a full diagnosis is at the severe tail of the distribution. We set out to investigate such a viewpoint by investigating the interaction of polygenic risk scores for autism spectrum disorder and Age2 on neuroimaging measures (cortical thickness and white matter connectivity) in a general population (n = 391, with age ranging from 3 to 21 years from the Pediatric Imaging, Neurocognition and Genetics study). We observed that children with higher polygenic risk for autism spectrum disorder exhibited greater cortical thickness for a large age span starting from 3 years up to ∼14 years in several cortical regions localized in bilateral precentral gyri and the left hemispheric postcentral gyrus and precuneus. In an independent case–control dataset from the Autism Brain Imaging Data Exchange (n = 560), we observed a similar pattern: children with autism spectrum disorder exhibited greater cortical thickness starting from 6 years onwards till ∼14 years in wide-spread cortical regions including (the ones identified using the general population). We also observed statistically significant regional overlap between the two maps, suggesting that some of the cortical abnormalities associated with autism spectrum disorder overlapped with brain changes associated with genetic vulnerability for autism spectrum disorder in healthy individuals. Lastly, we observed that white matter connectivity between the frontal and parietal regions showed significant association with polygenic risk for autism spectrum disorder, indicating that not only the brain structure, but the white matter connectivity might also show a predisposition for the risk of autism spectrum disorder. Our findings showed that the fronto-parietal thickness and connectivity are dimensionally related to genetic risk for autism spectrum disorder in general population and are also part of the cortical abnormalities associated with autism spectrum disorder. This highlights the necessity of considering continuum models in studying the aetiology of autism spectrum disorder using polygenic risk scores and multimodal neuroimaging.
Keywords: autism spectrum disorders, polygenic risk score, cortical thickness, structural connectivity, genetics
Cortical thickness of the frontal and parietal regions is dimensionally related to genetic risk for autism in general population and is also part of the cortical abnormalities associated with autism. Our study highlights the necessity of considering continuum models in studying aetiology of autism using polygenic risk scores and neuroimaging.
Graphical Abstract
Graphical Abstract.
Introduction
Autism spectrum disorders (ASD) comprise a collection of neurodevelopmental conditions characterized by difficulties with social interactions, verbal and non-verbal communication and repetitive behaviours. ASD is highly prevalent (∼1 in 59 individuals) and highly heritable (Gaugler et al., 2014; Iossifov et al., 2014). ASD is conceptualized as a brain disorder, with several neuroimaging studies showing cortical alterations in individuals with ASD compared to healthy individuals (read as controls, CTL) (Hadjikhani et al., 2006; Hyde et al., 2010; Wallace et al., 2010; Zielinski et al., 2014; Lange et al., 2015; Khundrakpam et al., 2017). For instance, a recent study with the largest sample size (n = 3222: 1571 ASD, 1651 CTL with age 2–64 years) observed increased cortical thickness in the frontal cortex and decreased thickness in the temporal cortex with effect size ranging from −0.21 to 0.20 (van Rooij et al., 2017). However, these studies so far have, as with other traditional approaches in psychiatric imaging research, compare cases to CTL (yes/no for ASD) and ignore the possibility of intermediate outcomes.
In contrast, an emerging viewpoint suggests ASD as a continuum with a normal distribution of autistic tendencies in the general population, where a full diagnosis is at the severe tail of the distribution (Constantino and Todd, 2003; Wakabayashi et al., 2006; Plomin et al., 2009; Robinson et al., 2011, 2016). For instance, considerable variability in social communication and social interaction capabilities has been observed in the general population (Plomin et al., 2009). In addition, subthreshold autistic traits have been observed in unaffected siblings and family members of people with ASD (Constantino et al., 2010; Hazlett et al., 2012, 2017; Elison et al., 2013). The idea of continuum models for phenotypes is not new and has been shown for phenotypes that are easily quantifiable (such as intellectual disability and intelligence). For instance, in individuals with de novo deletions of chromosome 16p11.2, a quantitative reduction (∼2 standard deviation) in intelligence (from that of their non-carrier parents’ mean IQ) was observed as opposed to a traditional categorical diagnosis (yes/no for intellectual disability) (Moreno-De-Luca et al., 2015). Treating ASD as continuum will enable the study of intermediate levels of ASD in larger more accessible samples. Furthermore, this will allow investigation of the underlying mechanisms of ASD without the potential confounding effect of clinical state. Such approach has been beneficial for depression, where studying subclinical levels of depression largely highlights similar genetic mechanisms (McIntosh et al., 2019). We propose using a similar approach in ASD. However, traditionally such approach is limited by the fact that not many biological cohorts have included a continuum measure of ASD.
We aimed to resolve this question by studying the genetic potential for ASD in general (paediatric) population using polygenic risk scores (PRS). The polygenic form of autism is made up of the additive effects of individual single-nucleotide polymorphisms (SNPs), which collectively capture the variance explained by common alleles (Plomin, 2013; Wray et al., 2014). This way, the brain patterns of autistic-like traits can be studied in any cohort, with genetic and brain information available. This approach has already been used to show that PRS for ASD correlates with cognitive abilities (including educational attainment, intelligence quotient, logical memory and executive function) in general population (Clarke et al., 2016; Grove et al., 2019; Schork et al., 2019). To the best of our knowledge, no study has yet explored polygenic risk profiling along with neuroimaging data to shed light on identifying brain changes/alterations in people (among the general population) who are at a high risk for ASD.
We, therefore, set out to investigate the association between PRS for ASD (based on the most recent and largest genetic training dataset available; Grove et al., 2019) and neuroimaging measures (cortical thickness and white matter connectivity) in a general (paediatric) population. We chose cortical thickness and white matter connectivity because they are highly heritable and have been shown to be suitable neuroimaging measures for imaging genetics (Meyer-Lindenberg and Weinberger, 2006; Meyer-Lindenberg, 2009; Winkler et al., 2010). Based on findings from recent large-scale MRI studies (Khundrakpam et al., 2017; van Rooij et al., 2017), we hypothesized age-by-PRS interactions on regional cortical thickness. Such a hypothesis was also supported by recent reports of non-uniform expansion of regional surface area during development (Reardon et al., 2018). In addition, based on findings of previous studies (Catani et al., 2016), we also hypothesized that white matter fibres of the frontal regions would show decreased connectivity for individuals with higher PRS for ASD. Lastly, in an independent dataset comprising healthy CTL and subjects with ASD, we set out to investigate whether brain regions with high PRS for ASD also demonstrate significant group difference (ASD–CTL) in cortical thickness.
Materials and methods
Subjects
The data for the study were obtained from the Pediatric Imaging, Neurocognition and Genetics (PING) study (Jernigan et al., 2016). The PING study is a wide-ranging, publicly shared data resource for investigating neuroimaging, cognition and genetics in normally developing children and adolescents, comprising cross-sectional data from 1493 subjects collected from 10 different sites across the USA. Details of the study are described elsewhere (Jernigan et al., 2016).
Genomic data
A total of 550 000 SNPs were genotyped from saliva samples using the Illumina Human660W-Quad BeadChip. The data were prepared for imputation using the ‘imputePrepSanger’ pipeline (https://hub.docker.com/r/eauforest/imputeprepsanger/), implemented on CBRAIN (Sherif et al., 2014), using Human660W-Quad_v1_A-b37-strand chip as reference. Namely, the pipeline took Plink genotype files, adjusted the strand, the positions, the reference alleles to match Haplotype Reference Consortium panel and performed quality control (QC) steps and output a vcf file. The Plink QC steps included ensuring that people and SNPs have enough data available (–mind 0.1, –geno 0.1), keeping only common SNPs (–maf 0.05) that pass the Hardy–Weinberg equilibrium test (–hwe 5e−8). The pipeline also removed indels, palindromic SNPs, SNPs with differing alleles, SNPs with no match to the reference panel, SNPs with >0.2 allele frequency difference to the reference and duplicates. The data were imputated with Sanger Imputation Service (McCarthy et al., 2016), using default settings and the Haplotype Reference Consortium (http://www.haplotype-reference-consortium.org/) as the reference panel.
The imputed SNPs were then filtered in Plink 1.9 (Chang et al., 2015), keeping SNPs that had (i) unique names, (ii) only ACTG and (iii) MAF >0.05. We verified that all SNPs had INFO scores R2 > 0.9 with Plink 2.0. Polygenic score software PRSice 2.1.2 (Euesden et al., 2015) excluded further ambiguous variants, resulting in 4 696 385 variants being available for polygenic scoring. Participants were filtered to have 0.95 loadings to the European principal component (Genetic ancestry factor_Europe variable provided with the PING data), resulting in 526 participants. The same participants were used to calculate the 20 principal components (PC) with Plink 1.9.
The polygenic risk score for ASD was based on ASD genome-wide association study trained on 18 381 individuals with ASD and 27 969 CTL (Grove et al., 2019). We clumped the data as per PRSice default settings (clumping distance = 250 kb, threshold r2 = 0.1), using P = 0.001 cut-off criterion. After matching with available variants in the data, the autism polygenic score was based on 1245 variants.
Image acquisition and pre-processing
Each site administered a standardized structural MRI protocol. Steps, detailed elsewhere (Jernigan et al., 2016), included: (i) a 3D T1-weighted inversion prepared RF-spoiled gradient echo scan using prospective motion correction, for cortical and subcortical segmentation; (ii) a 3D T2-weighted variable flip angle fast spin echo scan, also using prospective motion correction, for the detection and quantification of white matter lesions and segmentation of CSF; and (iii) a high angular resolution diffusion imaging scan, with integrated B0 distortion correction for the segmentation of white matter tracts and the measurement of diffusion parameters.
The CIVET processing pipeline (http://www.bic.mni.mcgill.ca/ServicesSoftware/CIVET) developed at the Montreal Neurological Institute (MNI) was used to compute cortical thickness measurements at 81 924 regions covering the entire cortex. The pipeline includes the following steps: the T1-weighted image is first non-uniformity corrected and then linearly registered to the Talairach-like MNI152 template. The non-uniformity correction is then repeated using the template mask. The non-linear registration from the resultant volume to the MNI152 template is then computed, and the transform used to provide priors to segment the image into grey matter, white matter and cerebrospinal fluid. Inner and outer GM surfaces are then extracted using the Constrained Laplacian-based Automated Segmentation with Proximities algorithm, and cortical thickness is measured in native space using the linked distance between the two surfaces at 81 924 vertices. Each subject’s cortical thickness map was blurred using a 30-mm full width at half maximum surface-based diffusion smoothing kernel to impose a normal distribution on the corticometric data and to increase the signal-to-noise ratio. QC of these data was performed by two independent reviewers: only scans with consensus of the two reviewers were used. As a result of this process, data with motion artefacts, a low signal-to-noise ratio, artefacts due to hyperintensities from blood vessels, surface–surface intersections or poor placement of the grey or white matter surface for any reason were excluded.
Diffusion MRI data were pre-processed using the FSL pipeline (FMRIB Software Library v5.0.9) (Jenkinson et al., 2012). Steps include: (i) correction of the effects of distortion induced by eddy currents, inter-volume movements and susceptibility of the diffusion data; (ii) rigid alignment of the individual unweighted image with the structural image using flirt; (iii) non-linear registration to transform individual structural image to an MNI152 standard T1-weighted template using fnirt; and (iv) computing the forward and backward warp field images between individual diffusion MRI and MNI T1 spaces by concatenating (or inverting) the rigid transformation matrix and the warp field image. Diffusion parameters at each voxel were estimated by using Markov Chain Monte Carlo sampling. In this step, up to two possible fibre populations were modelled for each voxel after 2000 iterations. For QC, we checked the structural image and the average of the non-diffusion-weighted images for each participant. For example, a subject was excluded from further analysis if the signal–noise rate of structural image or unweighted-diffusion image was lower than 800. Also, the results of registration were evaluated by visual inspection. Furthermore, subjects who had >2-mm frame-wise displacements of the diffusion MRI were excluded from further analyses.
Of the total 1493 subjects, filtering for subjects with 0.95 loadings to the European principal component (Genetic ancestry factor_Europe variable provided with the PING data) resulted in 526 participants. Of these, 95 subjects did not have MRI data and 2 subjects did not have information about age, resulting to 429 subjects. Next, 13 subjects were excluded before any processing (raw data) due to severe motion and slicing artefacts. A subsequent 25 subjects failed CIVET pipeline (for a number of reasons including the presence of bright blood vessels and poor contrast) and were excluded in further analysis. Thus, the final sample comprised 391 participants (males/females = 207/184, age = 12.1 ± 4.7 years). The demographics of the resulting participants from the PING dataset, which were used in the study, are given in Table 1A.
Table 1.
Demographics of the subjects used in the study: (A) sample from PING dataset comprising healthy individuals and (B) sample from ABIDE dataset comprising healthy individuals and individuals with ASD
| A. PING dataset |
| Total number of subjects, N = 391 |
| Males/females = 207/184 |
| Age = 3.1–21.0 (12.1 ± 4.7) years |
| Ethnicity = European (86.70%), mixed (13.30%) |
| Handedness = Right-handed (86.71%), left-handed (9.97%), mixed (3.32%) |
| B. ABIDE dataset |
| Total number of subjects, N = 560 |
| Males/females = 560/0 |
| ASD/CTL = 266/294 |
| Age = ASD: 7.1–35.0 (17.2 ± 6.4) years |
| CTL: 6.5–35.0 (17.0 ± 6.4) years |
| ASD (with ADOS), N = 218 |
| ADOS Soc = 0–14 (6.9 ± 3.6) |
| ADOS Comm = 0–8 (3.3 ± 1.8) |
| ADOS severity = 1–9 (5.4 ± 2.3) |
Note that ASD refers to total number of subjects categorized as autistic, while ASD with ADOS refers to a subset of the ASD subjects with concurrent measures of ADOS Soc and ADOS Comm. Means, with standard deviation, are given in parentheses.
CTL = healthy controls; ADOS Soc = Autism diagnostic observation schedule Social; ADOS Comm = ADOS Communication.
Features of the polygenic risk scores for autism spectrum disorders
As a first step, we investigated whether the PRS for ASD for the whole data sample was normally distributed. Next, using general linear model (GLM), we investigated whether there was any effect of age on the PRS. Since scanner is categorical variable, we used ANOVA to test whether scanner has an effect on PRS. Lastly, using GLM, we also investigated whether there was any association of PRS and language measures. For the language measures, we used the scores of the Picture Vocabulary and the Oral Reading Recognition tests available with the PING dataset.
Statistical analyses
GLMs were used to investigate the age-by-PRS interactions on cortical thickness. Motivated by earlier studies (Shaw et al., 2006, 2007, 2008; Nguyen et al., 2013; Khundrakpam et al., 2017), we first explored models with linear, quadratic and cubic age effects. For this, the best-fit model was selected using Akaike information criterion (Akaike, 1974). Akaike information criterion was used to penalize the added parameters, and the model with the lowest Akaike information criterion value was selected. Models with quadratic age terms, which had the lowest Akaike information criterion values, were, therefore, selected.
Cortical thickness was modelled as:
where i is a vertex, Age is mean centred, ε is the residual error, PRS is the polygenic risk score and the intercept and the β terms are the fixed effects. To minimize the chance of population structure explaining the polygenic score results, we extracted 20 first PC (PC20) and used them as covariates. Without controlling for those PC, random differences in population genomic signature can explain outcomes, if different populations also happen to differ in the outcome (Hamer and Sirota, 2000). Since there were 9 sites but 13 scanners, device serial number (unique for each scanner, provided in PING) was put as covariate in the analyses. All GLM analyses were done using the SurfStat toolbox (http://www.math.mcgill.ca/keith/surfstat/).
Note that the aim of our model was to compute the significance of β8 (i.e. the influence of Age2 × PRS) on cortical thickness. However, the interpretation could be challenging since the interaction terms and power of age might lead to high correlation between the predictors. To substantiate the effect, we compared the above model using a likelihood ratio test to
| (A) |
| (B) |
The comparison of original model with model (A) would study whether PRS and its interactions with Age and Age2 jointly have a significant impact, whereas the comparison of original model with model (B) would test whether the interactions of PRS with Age and Age2 jointly have a significant impact.
At every cortical point, the t-statistic for the interaction of Age2 × PRS on cortical thickness (at 81 924 vertices) was mapped onto a standard surface. Correction for multiple comparisons, using random field theory was then applied to the resultant map to determine the regions of cortex showing statistically significant association between PRS-ASD and cortical thickness (Worsley et al., 2004). Next, the median value of the PRS was used to divide the whole data sample to two groups: low PRS (up to median value) and high PRS (remaining data). At each of the identified significant regions, we fit quadratic polynomial curves to the thickness data (after regressing for covariates) for both the groups separately.
To better understand the impact of sex on the interaction of Age2 × PRS on cortical thickness, similar analysis was performed for male- and female-only groups by splitting the data sample.
Age3 × Group interaction on cortical thickness in independent dataset
Next, we set out to investigate whether the cortical regions [that showed significant association of Age2 × PRS on cortical thickness] would also show Age3 × Group on cortical thickness (ASD and CTL groups) in cortical thickness in an independent dataset (cubic model of age was based on our previous study; Khundrakpam et al., 2017). For this, we used the Autism Brain Imaging Data Exchange (ABIDE) dataset, an agglomerative dataset of MRI scans of normal subjects and subjects with ASD (Di Martino et al., 2014). We leveraged the ABIDE data (Table 1B) used in our recent publication where details of the pre-processing and exclusion criteria are described (Khundrakpam et al., 2017). The pre-processing and QC procedure were similar as that of the PING dataset described above. Apart from QC-failed subjects, exclusion criteria included insufficient number of subjects in each category (ASD and CTL) to determine group difference, too few females resulting to excluding all females, and excluding subjects over 35 years of age due to insufficient numbers. The final sample comprised 560 male subjects, of whom 294 were controls (17 ± 6.4 years) and 266 were individuals with ASD (17.2 ± 6.4 years) (Table 1B). For ASD diagnosis, similar to the previous study (Di Martino et al., 2014), ASD diagnoses were reached by combining clinical judgement and diagnostic instruments—Autism Diagnostic Observation Schedule and/or Autism Diagnostic Interview-Revised. Some sites had missing information for the Autism Diagnostic Observation Schedule scores, and therefore, a subset (n = 218) of the total data (n = 266) used for the group difference analysis had information for Autism Diagnostic Observation Schedule Social ranging from 0 to 14 (6.9 ± 3.6), Autism Diagnostic Observation Schedule Communication ranging from 0 to 8 (3.3 ± 1.8) and Autism Diagnostic Observation Schedule severity ranging from 1 to 9 (5.4 ± 2.3) (Table 1B). As has been done in our previous study (Khundrakpam et al., 2017), GLMs (with cubic age models) were built on these data to compute Age3 × Group on cortical thickness (group = ASD–CTL) in cortical thickness for all vertices covering the entire cortex.
where i is a vertex, Age is mean centred, ε is the residual error, and the intercept and the β terms are the fixed effects.
Overlap of the map of Age2 × polygenic risk scores on cortical thickness in Pediatric Imaging, Neurocognition and Genetics and the map of Age3 × Group on cortical thickness in Autism Brain Imaging Data Exchange
To statistically check whether there was regional overlap between ‘map of the Age2 × PRS on cortical thickness using the PING dataset’ and ‘map of the Age3 × Group on cortical thickness using the ABIDE dataset’, we used the spin test developed by (Alexander-Bloch et al., 2018). In short, the method, using a spatial permutation framework, generates null models of overlap by applying random rotations to spherical representations of the cortical surface. As in previous studies (Reardon et al., 2018), 1000 surface rotations of the PING map were generated and the statistical overlap was checked by comparing whether the observed cross-vertex correlation between PING and ABIDE maps was statistically greater (P < 0.05) than those with 1000 rotations.
Association of structural connectivity between the identified significant cortical regions and polygenic risk scores-autism spectrum disorders
Lastly, we set out to study whether the structural connectivity between the identified significant cortical regions associates with the PRS for ASD. For this, we first re-ran the GLM analysis for the association of Age2 × PRS on cortical thickness at the region level with 64 brain regions based on the Desikan–Killany–Tourville atlas (Desikan et al., 2006). Next, structural connectivity (measured as streamline count) between these identified significant Desikan–Killany–Tourville regions was computed. Lastly, we ran GLM to assess the association of the structural connectivity between the significant Desikan–Killany–Tourville regions and the PRS for ASD. Age, sex, scanner and the first 20 PC were put as covariates. Correction for multiple comparisons was performed using false discovery rate at q = 0.05 (Genovese et al., 2002).
Data availability
The PING data used in the study are available at https://nda.nih.gov/ after providing the necessary data user agreement. The raw data of ABIDE are available at http://fcon_1000.projects.nitrc.org/indi/abide/, and its pre-processed data are available at http://preprocessed-connectomes-project.org/abide/.
Results
Features of the polygenic risk scores for autism spectrum disorders
Since our data sample (PING) did not have ASD diagnosis, we could not validate the PRS. However, previous research has shown that polygenic score for ASD can predict ASD diagnosis with mean added R2 = 0.0245 (Grove et al., 2019).
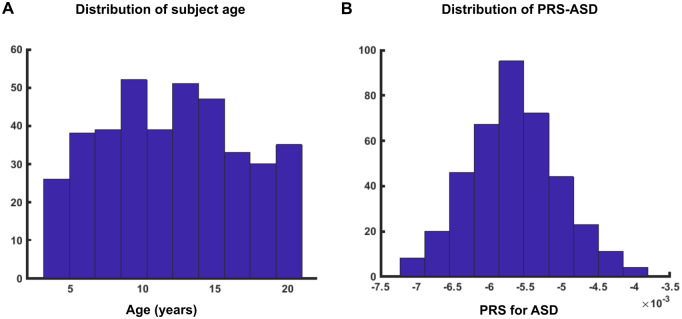
As shown in Fig. 1A, subjects were properly distributed for the whole age range. The PRS for ASD were normally distributed for our data sample (Fig. 1B). There was no significant association of PRS-ASD with age (T = 1.45, P = 0.14, Supplementary Fig. 1A). Scanner had no significant effect on the PRS (F = 1.27, P = 0.23, Supplementary Fig. 1B). Thus, the data sample characteristics such as age and scanner did not have a significant impact on the PRS for ASD.
Figure 1.
Distribution of age and PRS for ASD. (A) Subjects were properly distributed for the whole age range (3–21 years). (B) PRS for ASD were normally distributed for the data sample.
Age2 × polygenic risk scores interaction on cortical thickness
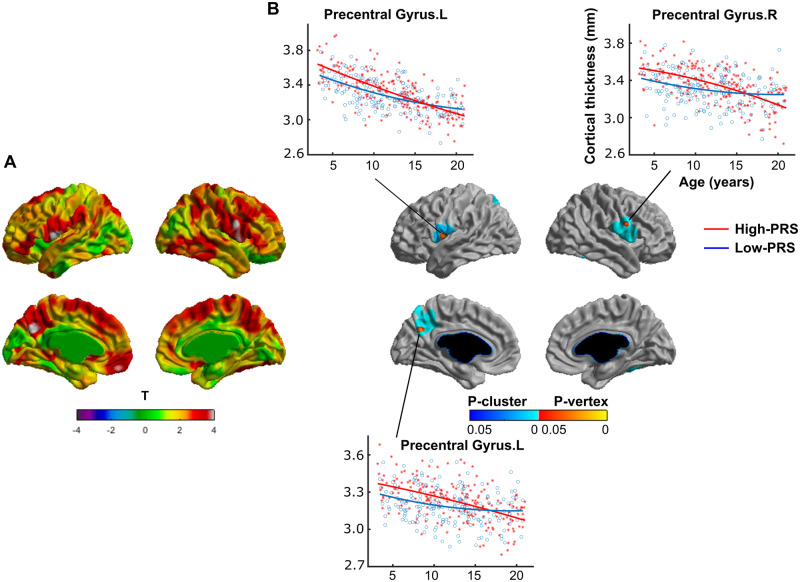
Significant positive interaction (P < 0.05, random field theory corrected) of Age2 × PRS on cortical thickness was observed in several cortical regions located in the bilateral precentral gyri and the left hemispheric postcentral gyrus and precuneus (Fig. 2). To be precise, the four peak vertices that survived random field theory correction were localized at: ‘the left precentral gyrus’ (t = 4.30, P = 0.015, MNI coordinates: X = −54.8, Y = −1.7, Z = 8.4), ‘the left precuneus’ (t = 4.29, P = 0.016, MNI coordinates: X = −3.4, Y = −63.2, Z = 31.8), ‘the right precentral gyrus’ (t = 4.22, P = 0.021, MNI coordinates: X = 61.4, Y = 4.1, Z = 24.8) and ‘the left postcentral gyrus’ (t = 4.11, P = 0.031, MNI coordinates: X = −59.7, Y = −4.0, Z = 11.2) (Table 2). At these peaks, scatter plots of the cortical thickness were drawn for the low-PRS and high-PRS groups (see ‘Materials and methods’ section for the groups). For all the peak vertices, the high-PRS group exhibited greater thickness for a large age span starting from 3 years lasting up to ∼14 years (Fig. 2B). Note that, for better visualization purpose, scatter plots were shown for only three peaks.
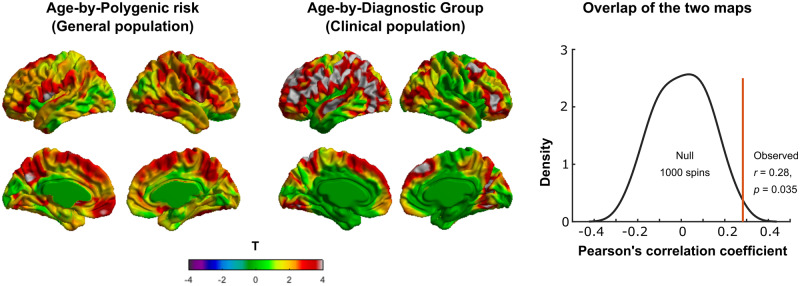
Figure 2.
Positive interaction of (Age2 × PRS) on cortical thickness. (A) The t-statistics across vertices on the surface and (B) multiple comparison-corrected P-statistics (P < 0.05, RFT corrected for multiple comparisons, see ‘Materials and methods’ section) for the interaction of (Age2 × PRS) on cortical thickness. Note that there is significant positive interaction (P < 0.05, RFT corrected) of Age2 × PRS on cortical thickness in several cortical regions located in the bilateral precentral gyri and the left hemispheric postcentral gyrus and precuneus (for peak vertices, see Table 2). At these peaks, scatter plots of the cortical thickness are shown for the low-PRS and high-PRS groups (see ‘Materials and methods’ section for the groups). Note that, for all the peak vertices, the high-PRS group exhibits greater thickness for a large age span starting from 3 years lasting up to ∼14 years. Note that, for better visualization purpose, scatter plots were shown for only three peaks. RFT = random field theory, L = left hemisphere, R = right hemisphere.
Table 2.
Brain regions with significant interaction of Age2 × PRS on cortical thickness
| X | Y | Z | Cluster ID | t | P-value | DKT label |
|---|---|---|---|---|---|---|
| −54.8 | −1.7 | 8.4 | 3 | 4.30 | 0.015 | ‘Precentral Gyrus.L’ |
| −3.4 | −63.2 | 31.8 | 2 | 4.29 | 0.016 | ‘Precuneus.L’ |
| 61.4 | 4.1 | 24.8 | 1 | 4.22 | 0.021 | ‘Precentral Gyrus.R’ |
| −59.7 | −4.0 | 11.2 | 3 | 4.11 | 0.031 | ‘Postcentral Gyrus.L’ |
Peak vertices (maximum t-statistics) are shown for significant interaction of Age2 × PRS on cortical thickness (for details, see ‘Materials and methods’ section). X, Y and Z denote MNI coordinates.
DKT = Desikan–Killany–Tourville atlas.
Comparison of the original GLM model with model (A) (see ‘Materials and methods’ section) using likelihood ratio test revealed: for peak at Precentral Gyrus.L, likelihood ratio statistics, LRStat = 17.41, P = 0.0005; for peak at Precuneus.L, LRStat = 16.59, P = 0.0008; for peak at Precentral Gyrus.R, LRStat = 15.28, P = 0.0015; and for peak at Postcentral Gyrus.L, LRStat = 16.45, P = 0.0009. These results indicated that PRS and its interactions with Age and Age2 jointly have a significant impact. Next, comparison of the original GLM model with model (B) (see ‘Materials and methods’ section) using likelihood ratio test revealed: for peak at Precentral Gyrus.L, LRStat = 9.93, P = 0.006; for peak at Precuneus.L, LRStat = 9.75, P = 0.008; for peak at Precentral Gyrus.R, LRStat = 10.03, P = 0.006; and for peak at Postcentral Gyrus.L, LRStat = 9.64, P = 0.007. These results indicated that the interactions of PRS with Age and Age2 jointly have a significant impact.
Separate analysis for the male and female groups revealed trend-level interaction of Age2 × PRS on cortical thickness for the male-only group, similar to the interaction pattern that was observed for the whole data sample (Supplementary Fig. 3). None of the brain regions showed significant interaction possibly due to the considerable reduction in sample size (N = 207 for males and N = 184 for females).
Age3 × Group interaction on cortical thickness in the Autism Brain Imaging Data Exchange dataset
As shown in our previous study (Khundrakpam et al., 2017), in the ABIDE dataset, we observed significant (Age3 × Group) interaction on cortical thickness, such that ASD group exhibited greater cortical thickness (compared to controls) for a large age span starting from 6 up to ∼14 years [Supplementary Fig. 4, modified from Figure 4 of Khundrakpam et al. (2017)]. This significant association was observed at several cortical regions located predominantly in the left hemisphere, in the inferior and precentral gyri of the frontal lobe, postcentral and supramarginal gyri of the parietal lobe, middle and superior temporal and fusiform gyri of the temporal lobe and the anterior of the inferior and middle occipital gyri of the occipital lobe. Right hemispheric regions included the superior and inferior frontal gyri, medial prefrontal gyrus, precuneus and fusiform gyrus.
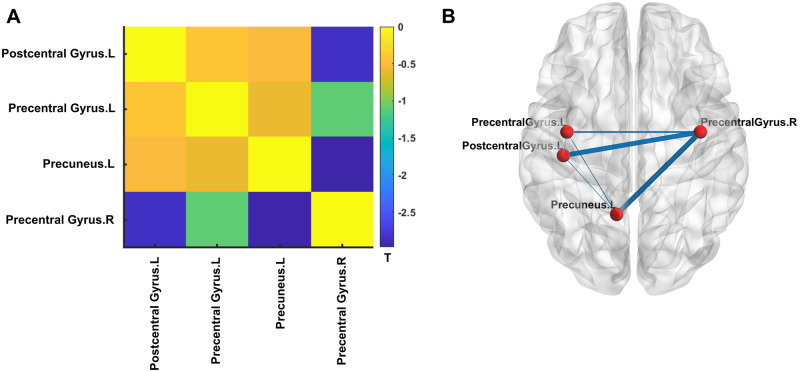
Figure 4.
Association of PRS for ASD and structural connectivity. (A) Matrix of the t-statistics for the association of PRS-ASD and structural connectivity for the four DKT brain regions namely Postcentral Gyrus.L, Precentral Gyrus.L, Precuneus.L and Precentral Gyrus.R (see ‘Materials and methods’ section). Note that all connections show negative correlation with PRS. Of the total six connections (corresponding to four regions), only two (namely Postcentral Gyrus.L—Precentral Gyrus.R and Precuneus.L—Precentral Gyrus.R) survived correction for multiple comparisons using false discovery rate at q = 0.05. (B) For better visualization, the t-statistics of the correlation of PRS for ASD and structural connectivity were visualized as edge weights between the four cortical regions (represented by the nodes) on a standard surface. DKT = Desikan–Killany–Tourville; L = left hemisphere; R = right hemisphere.
Overlap of the map of Age2 × polygenic risk scores on cortical thickness in Pediatric Imaging, Neurocognition and Genetics and the map of Age3 × Group on cortical thickness in Autism Brain Imaging Data Exchange
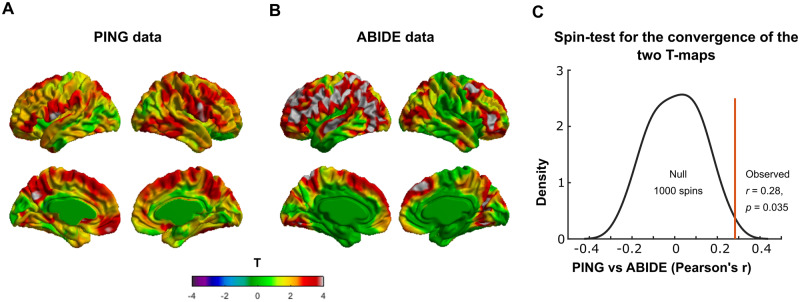
Using 1000 surface-based rotations of the PING map, we found that the observed cross-vertex correlation of the PING and ABIDE maps (r = 0.28) was statistically greater (P = 0.035) than those with the 1000 rotations (Fig. 3). Thus, there was regional overlap between the ‘map of Age2 × PRS on cortical thickness in the PING dataset’ and the ‘map of Age3 × Group on cortical thickness in the ABIDE dataset’. Note that the interactive effect of Age3 and group (ASD–CTL) on cortical thickness was much more wide spread, and although the overlap between the PING and ABIDE maps was significant at P = 0.035, one should keep in mind that the convergence was not very strong.
Figure 3.
Overlap of the ‘map of Age2 × PRS on cortical thickness in PING’ and the ‘map of Age3 × Group on cortical thickness in ABIDE’. (A) T-map of Age2 × PRS interaction on cortical thickness in the PING dataset. (B) T-map of Age3 × Group interaction on cortical thickness (groups: ASD and CTL) in the ABIDE dataset. (C) Spin test for statistically checking the regional overlap between the two maps using PING and ABIDE datasets. Note that using 1000 surface-based rotations of the map using PING, we found that the observed cross-vertex correlation of the PING and ABIDE maps (r = 0.29) was statistically greater (P = 0.03) that those with the 1000 rotations. Diagnostic groups = ASD and CTL; CTL = healthy controls.
Similar analysis for the male-only group from the PING dataset with that of ABIDE (comprising of males only) revealed significant overlap with r = 0.14, P = 0.03 (Supplementary Fig. 5). Thus, compared to the whole data sample of PING (combining males and females), we observed reduced overlap of the male-only PING with that of the male-only ABIDE data, which might be due to the considerable reduction in sample size.
Association of structural connectivity and polygenic risk scores-autism spectrum disorders
At the Desikan–Killany–Tourville atlas level, four cortical regions showed significant association of PRS for ASD and cortical thickness (Supplementary Fig. 6). The structural connectivity between these regions showed negative correlation with PRS for ASD, with two connections namely Postcentral Gyrus.L—Precentral Gyrus.R and Precuneus.L—Precentral Gyrus.R, showing significant correlation (Fig. 4). To better illustrate the results, the t-statistics of the correlation of PRS for ASD and structural connectivity were visualized as edge weights between the four cortical regions (represented by the nodes) on a standard surface (Fig. 4B).
Discussion
Using MRI scans from a large population of typically developing children and adolescents (n = 391), we studied the interactive effect of Age2 × PRS on cortical thickness. We observed significant positive interaction such that children with higher PRS for ASD exhibited greater cortical thickness for a large age span starting from 3 years up to ∼14 years in several cortical regions localized in bilateral precentral gyri and the left hemispheric postcentral gyrus and precuneus. Critically, we observed similar pattern of interactive effect of Age3 × Group on cortical thickness in the ABIDE dataset such that children with ASD exhibited greater cortical thickness starting from 6 years onwards till ∼14 years in wide-spread cortical regions including (the ones identified using PING dataset). We observed statistically significant regional overlap between the maps using PING and ABIDE datasets, suggesting that some of the cortical abnormalities associated with ASD overlapped with brain changes associated with genetic vulnerability for ASD in healthy individuals. Lastly, we observed that structural connectivity (assessed from diffusion tensor imaging scans) between the right precentral gyrus and left postcentral gyrus and between the right precentral gyrus and left precuneus showed significant negative association with PRS for ASD, indicating that not only the brain structure, but the structural brain connectivity might also show predisposition for risk of ASD.
Increased cortical thickness during early brain development has been observed in ASD (Zielinski et al., 2014; Khundrakpam et al., 2017), consistent with the observation of brain overgrowth in ASD (Kemper and Bauman, 1998; Courchesne and Pierce, 2005; Hazlett et al., 2005). Several studies suggest that the abnormal brain growth pattern starts at the first year of life with a period of accelerated growth, which continues during early childhood, achieving near-adult brain size earlier than in typical development (Courchesne et al., 2003; Hazlett et al., 2005). This increased brain size relative to typically developing children persists into adolescence (Redcay and Courchesne, 2005). Our findings contribute to the existing evidence and suggest that increased cortical thickness can even be observed in individuals with genetic risk for ASD. This is not surprising because PRS/genetic risk variants for neurological disorders have been shown to be associated with brain measures across disorder-vulnerable regions of interest in healthy subjects. For example, higher PRS for Alzheimer’s disease is associated with lower left hippocampal volume (a key brain region implicated in Alzheimer’s disease) in young healthy subjects (Foley et al., 2017). Similarly, higher PRS for major depressive disorder is associated with decreased white matter integrity in right superior longitudinal fasciculus (Whalley et al., 2013).
Although findings of structural connectivity in subjects with ASD have been inconsistent, several studies indicate an overall decreased fractional anisotropy and increased mean diffusivity in corpus callosum, superior longitudinal fasciculus and occipito-frontal fasciculus [for a detailed review, see Rane et al. (2015)]. In particular, converging evidence suggests that ASD is associated with altered structural connectivity (decreased fractional anisotropy, increased mean diffusivity and reduced number of fibres) of the frontal lobes (Catani et al., 2016). Consistent with this, we observed decreased number of fibres for the fronto-parietal connections. Although diffusion tensor imaging metrics (fractional anisotropy, mean diffusivity and fibre count) represent different biological processes, it seems likely that ASD is characterized by altered long-range connectivity between the frontal and parietal regions. Our findings, performed on general population, add to this extant literature: abnormal structural connectivity between the frontal and parietal regions may be particularly susceptible to genetic predisposition for the risk of ASD. Indeed, a recent longitudinal study showed abnormal structural connectivity (increased fractional anisotropy) in several white matter tracts including those reported in our paper in high-risk infants who later developed ASD (Wolff et al., 2012). It may be noted that, in our study, we did not observe significant association of PRS for ASD and language measures such as vocabulary (Supplementary Fig. 2A) and reading (Supplementary Fig. 2B). One possible reason could be that, although the genetic predisposition of ASD had a significant impact on brain structure, its impact on cognition (such as language) was not strong enough to be captured in the data.
Our findings of increased cortical thickness with increased PRS for ASD were based on healthy individuals (from the PING dataset); as such, the underlying biological mechanisms are not known. However, if we look at these findings along with the observation of increased cortical thickness in individuals with ASD (in the ABIDE dataset), we could speculate ASD as a continuum with a normal distribution of autistic tendencies in the general population, where a full diagnosis is at the severe tail of the distribution (see ‘Conclusion’ section for more discussion on this). In light of this viewpoint, we can leverage previous studies on ASD research in speculating the possible biological mechanisms behind our findings. Increased cortical thickness in ASD is thought to reflect increased number of synaptic spines and reduced developmental synaptic pruning (Tang et al., 2014), increased number of neurons (Courchesne et al., 2011) and greater microglial cell density and somal volume (Morgan et al., 2010). At the same time, under-connectivity of long-range white matter fibres (including fibres observed in our study) in ASD could arise from decreased myelin thickness, axons with larger diameter (Zikopoulos and Barbas, 2010), increased oedema from inflammation (Vargas et al., 2005) and increased packing density (Bauman and Kemper, 2005). Since post mortem studies are limited, advancements in the field of in vivo neuroimaging, particularly in the field of diffusion tensor imaging such as neurite orientation dispersion and density mapping, NODDI (Zhang et al., 2012; Sato et al., 2017), could provide more clues about the underlying biological processes in the pathology of ASD.
It may be mentioned that our findings of the group difference (ASD–CTL) in cortical thickness in the ABIDE dataset were based on male participants only, whereas the findings of association of PRS for ASD and cortical thickness in the PING dataset were based on both male and female participants. However, similar analysis separately for male and female participants (in the PING dataset) yielded similar patterns of association (albeit with lower statistical power). Given the lack of enough male subjects, and the fact that sex did not have an impact on the association of PRS for ASD and cortical thickness, we therefore compared the findings in the two datasets. Future studies using larger datasets such as the Adolescent Brain Cognitive Development dataset (Casey et al., 2018) should seek to identify the association of PRS for ASD and cortical thickness separately for males and females.
Conclusion
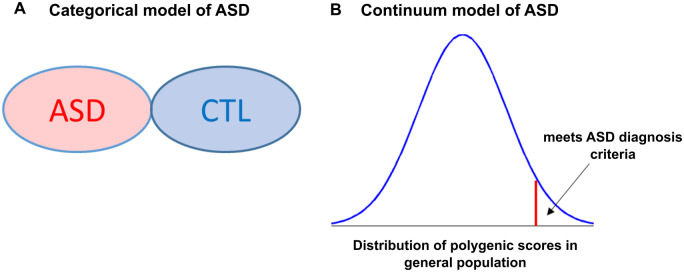
In conclusion, our findings support the notion of a continuum model of ASD as opposed to traditional categorical psychiatric diagnoses (Fig. 5). As shown in Fig. 5A, conventional approach towards psychiatric neuroimaging involves categorizing population sample in two groups: ‘individuals with ASD’ and ‘healthy controls’. Accordingly, much of the neuroimaging studies on ASD have focused on finding cortical differences between cases and CTL and ignore the possibility of intermediate outcomes. On the other hand, our findings add to the emerging viewpoint suggesting ASD as a continuum with a normal distribution of autistic tendencies in the general population, where a full diagnosis is at the severe tail of the distribution (reflected in Fig. 5B) (Constantino and Todd, 2003; Wakabayashi et al., 2006; Plomin et al., 2009; Robinson et al., 2011, 2016). Our main motivation here is to highlight the categorical vs continuum model of ASD in an easily understandable way. However, it may be noted that our figure for continuum model (Fig. 5B) is likely too simplistic, and one should keep in mind the multi-dimensional relationship between genetics, epigenetics and brain development. Our findings further indicate the added value of investigating genetic risk rather than diagnostic categories. For instance, our observations suggest that individuals (in the general population) with high risk for ASD have brain alterations that overlap with the brain abnormalities seen in subjects with ASD (Fig. 3), indicating that although predisposed for genetic risk, there may be compensatory factors that protect their brains. Our findings also highlight the critical role of the fronto-parietal thickness and connectivity in a dimensional disease model of ASD spanning from healthy individuals with genetic risk to patients with ASD. Thus, PRS and multimodal neuroimaging should be considered as useful approaches to identify the underlying brain alterations in ASD.
Figure 5.
Categorical vs continuum model of ASD. (A) Categorical model of ASD treats a population as group with ASD and CTL. (B) Continuum model of ASD treats ASD as a continuum with a normal distribution of polygenic scores in the general population, where the severe tail of the distribution meets ASD diagnosis criteria. Note that our figure for continuum model (B) is likely too simplistic and one should keep in mind the multi-dimensional relationship between genetics, epigenetics and brain development. CTL = controls.
Supplementary Material
Acknowledgements
Data used in preparation of this article were obtained from the Pediatric Imaging, Neurocognition and Genetics Study (PING) database (http://ping.chd.ucsd.edu). As such, the investigators within PING contributed to the design and implementation of PING and/or provided data but did not participate in analysis or writing of this report.
Funding
A.E. was supported by the Azrieli Neurodevelopmental Research Program in partnership with Brain Canada Multi-Investigator Research Initiative (MIRI) grant. U.V. was supported by Estonian Research Council’s grants PUTJD654 and MOBTP94.
Competing interests
The authors report no competing interests.
Glossary
- ABIDE =
Autism Brain Imaging Data Exchange
- ASD =
autism spectrum disorders
- CTL =
controls
- GLM =
general linear model
- MNI =
Montreal Neurological Institute
- PC =
principal components
- PING =
Pediatric Imaging, Neurocognition and Genetics
- PRS =
polygenic risk scores
- QC =
quality control
- SNPs =
single-nucleotide polymorphisms
References
- Akaike H. A new look at the statistical model identification. IEEE Trans Automat Contr 1974; 19: 716–23. [Google Scholar]
- Alexander-Bloch AF, Shou H, Liu S, Satterthwaite TD, Glahn DC, Shinohara RT, et al. On testing for spatial correspondence between maps of human brain structure and function. Neuroimage 2018; 178: 540–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauman ML, Kemper TL.. Neuroanatomic observations of the brain in autism: a review and future directions. Int J Dev Neurosci 2005; 23: 183–7. [DOI] [PubMed] [Google Scholar]
- Casey BJ, Cannonier T, Conley MI, Cohen AO, Barch DM, Heitzeg MM, et al. The Adolescent Brain Cognitive Development (ABCD) study: imaging acquisition across 21 sites. Dev Cogn Neurosci 2018; 32: 43–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catani M, Dell’Acqua F, Budisavljevic S, Howells H, Thiebaut De Schotten M, Froudist-Walsh S, et al. Frontal networks in adults with autism spectrum disorder. Brain 2016; 139: 616–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ.. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaSci 2015; 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke T-K, Lupton MK, Fernandez-Pujals AM, Starr J, Davies G, Cox S, et al. Common polygenic risk for autism spectrum disorder (ASD) is associated with cognitive ability in the general population. Mol Psychiatry 2016; 21: 419–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantino JN, Todd RD.. Autistic traits in the general population. Arch Gen Psychiatry 2003; 60: 524. [DOI] [PubMed] [Google Scholar]
- Constantino JN, Zhang Y, Frazier T, Abbacchi AM, Law P.. Sibling recurrence and the genetic epidemiology of autism. Am J Psychiatry 2010; 167: 1349–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne E, Carper R, Akshoomoff N.. Evidence of brain overgrowth in the first year of life in autism. JAMA 2003; 290: 337. [DOI] [PubMed] [Google Scholar]
- Courchesne E, Mouton PR, Calhoun ME, Semendeferi K, Ahrens-Barbeau C, Hallet MJ, et al. Neuron number and size in prefrontal cortex of children with autism. JAMA 2011; 306: 2001–10. [DOI] [PubMed] [Google Scholar]
- Courchesne E, Pierce K.. Brain overgrowth in autism during a critical time in development: implications for frontal pyramidal neuron and interneuron development and connectivity. Int J Dev Neurosci 2005; 23: 153–70. [DOI] [PubMed] [Google Scholar]
- Di Martino A, Yan CG, Li Q, Denio E, Castellanos FX, Alaerts K, et al. The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol Psychiatry 2014; 19: 659–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 2006; 31: 968–80. [DOI] [PubMed] [Google Scholar]
- Elison JT, Paterson SJ, Wolff JJ, Reznick JS, Sasson NJ, Gu H, et al. ; for the IBIS Network. White matter microstructure and atypical visual orienting in 7-month-olds at risk for autism. Am J Psychiatry 2013; 170: 899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euesden J, Lewis CM, O’Reilly PF.. PRSice: polygenic risk score software. Bioinformatics 2015; 31: 1466–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley SF, Tansey KE, Caseras X, Lancaster T, Bracht T, Parker G, et al. Multimodal brain imaging reveals structural differences in Alzheimer’s disease polygenic risk carriers: a study in healthy young adults. Biol Psychiatry 2017; 81: 154–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaugler T, Klei L, Sanders SJ, Bodea CA, Goldberg AP, Lee AB, et al. Most genetic risk for autism resides with common variation. Nat Genet 2014; 46: 881–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese CR, Lazar NA, Nichols T.. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage 2002; 15: 870–8. [DOI] [PubMed] [Google Scholar]
- Grove J, Ripke S, Als TD, Mattheisen M, Walters RK, Won H, et al. ; Autism Spectrum Disorder Working Group of the Psychiatric Genomics Consortium. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet 2019; 51: 431–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjikhani N, Joseph RM, Snyder J, Tager-Flusberg H.. Anatomical differences in the mirror neuron system and social cognition network in autism. Cereb Cortex 2006; 16: 1276–82. [DOI] [PubMed] [Google Scholar]
- Hamer D, Sirota L.. Beware the chopsticks gene. Mol Psychiatry 2000; 5: 11–3. [DOI] [PubMed] [Google Scholar]
- Hazlett HC, Gu H, McKinstry RC, Shaw DWW, Botteron KN, Dager SR, et al. ; the IBIS Network. Brain volume findings in 6-month-old infants at high familial risk for autism. Am J Psychiatry 2012; 169: 601–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazlett HC, Gu H, Munsell BC, Kim SH, Styner M, Wolff JJ, et al. ; the IBIS Network. Early brain development in infants at high risk for autism spectrum disorder. Nature 2017; 542: 348–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazlett HC, Poe M, Gerig G, Smith RG, Provenzale J, Ross A, et al. Magnetic resonance imaging and head circumference study of brain size in autism: birth through age 2 years. Arch Gen Psychiatry 2005; 62: 1366–76. [DOI] [PubMed] [Google Scholar]
- Hyde KL, Samson F, Evans AC, Mottron L.. Neuroanatomical differences in brain areas implicated in perceptual and other core features of autism revealed by cortical thickness analysis and voxel-based morphometry. Hum Brain Mapp 2010; 31: 556–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 2014; 515: 216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM.. FSL. Neuroimage 2012; 62: 782–90. [DOI] [PubMed] [Google Scholar]
- Jernigan TL, Brown TT, Hagler DJ, Akshoomoff N, Bartsch H, Newman E, et al. The Pediatric Imaging, Neurocognition, and Genetics (PING) data repository. Neuroimage 2016; 124: 1149–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper TL, Bauman M.. Neuropathology of infantile autism. J Neuropathol Exp Neurol 1998; 57: 645–52. [DOI] [PubMed] [Google Scholar]
- Khundrakpam BS, Lewis JD, Kostopoulos P, Carbonell F, Evans AC.. Cortical thickness abnormalities in autism spectrum disorders through late childhood, adolescence, and adulthood: a large-scale MRI study. Cereb Cortex 2017; 27: 1–11. [DOI] [PubMed] [Google Scholar]
- Lange N, Travers BG, Bigler ED, Prigge MBD, Froehlich AL, Nielsen JA, et al. Longitudinal volumetric brain changes in autism spectrum disorder ages 6-35 years. Autism Res 2015; 8: 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, et al. ; Haplotype Reference Consortium. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 2016; 48: 1279–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh AM, Sullivan PF, Lewis CM.. Uncovering the genetic architecture of major depression. Neuron 2019; 102: 91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Lindenberg A. Neural connectivity as an intermediate phenotype: brain networks under genetic control. Hum Brain Mapp 2009; 30: 1938–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Weinberger DR.. Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nat Rev Neurosci 2006; 7: 818–27. [DOI] [PubMed] [Google Scholar]
- Moreno-De-Luca A, Evans DW, Boomer KB, Hanson E, Bernier R, Goin-Kochel RP, et al. The role of parental cognitive, behavioral, and motor profiles in clinical variability in individuals with chromosome 16p11.2 deletions. JAMA Psychiatry 2015; 72: 119. [DOI] [PubMed] [Google Scholar]
- Morgan JT, Chana G, Pardo CA, Achim C, Semendeferi K, Buckwalter J, et al. Microglial activation and increased microglial density observed in the dorsolateral prefrontal cortex in autism. Biol Psychiatry 2010; 68: 368–76. [DOI] [PubMed] [Google Scholar]
- Nguyen T-V, McCracken JT, Ducharme S, Cropp BF, Botteron KN, Evans AC, et al. Interactive effects of dehydroepiandrosterone and testosterone on cortical thickness during early brain development. J Neurosci 2013; 33: 10840–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomin R. Commentary: Missing heritability, polygenic scores, and gene-environment correlation. J Child Psychol Psychiatr 2013; 54: 1147–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomin R, Haworth CMA, Davis O.. Common disorders are quantitative traits. Nat Rev Genet 2009; 10: 872–8. [DOI] [PubMed] [Google Scholar]
- Rane P, Cochran D, Hodge SM, Haselgrove C, Kennedy DN, Frazier JA.. Connectivity in autism: a review of MRI connectivity studies. Harv Rev Psychiatry 2015; 23: 223–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon PK, Seidlitz J, Vandekar S, Liu S, Patel R, Park MTM, et al. Normative brain size variation and brain shape diversity in humans. Science 2018; 360: 1222–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redcay E, Courchesne E.. When is the brain enlarged in autism? A meta-analysis of all brain size reports. Biol Psychiatry 2005; 58: 1–9. [DOI] [PubMed] [Google Scholar]
- Robinson EB, Koenen KC, McCormick MC, Munir K, Hallett V, Happé F, et al. Evidence that autistic traits show the same etiology in the general population and at the quantitative extremes (5%, 2.5%, and 1%). Arch Gen Psychiatry 2011; 68: 1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson EB, St Pourcain B, Anttila V, Kosmicki JA, Bulik-Sullivan B, Grove J, et al. ; iPSYCH-SSI-Broad Autism Group. Genetic risk for autism spectrum disorders and neuropsychiatric variation in the general population. Nat Genet 2016; 48: 552–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Kerever A, Kamagata K, Tsuruta K, Irie R, Tagawa K, et al. Understanding microstructure of the brain by comparison of neurite orientation dispersion and density imaging (NODDI) with transparent mouse brain. Acta Radiol Open 2017; 6: 205846011770381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schork AJ, Brown TT, Hagler DJ, Thompson WK, Chen C-H, Dale AM, et al. ; for the Pediatric Imaging, Neurocognition and Genetics Study. Polygenic risk for psychiatric disorders correlates with executive function in typical development. Genes Brain Behav 2019; 18: e12480. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P, Eckstrand K, Sharp W, Blumenthal J, Lerch JP, Greenstein D, et al. Attention-deficit/hyperactivity disorder is characterized by a delay in cortical maturation. Proc Natl Acad Sci USA 2007; 104: 19649–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P, Greenstein D, Lerch J, Clasen L, Lenroot R, Gogtay N, et al. Intellectual ability and cortical development in children and adolescents. Nature 2006; 440: 676–9. [DOI] [PubMed] [Google Scholar]
- Shaw P, Kabani NJ, Lerch JP, Eckstrand K, Lenroot R, Gogtay N, et al. Neurodevelopmental trajectories of the human cerebral cortex. J Neurosci 2008; 28: 3586–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherif T, Rioux P, Rousseau M-E, Kassis N, Beck N, Adalat R, et al. CBRAIN: a web-based, distributed computing platform for collaborative neuroimaging research. Front Neuroinform 2014; 8: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Gudsnuk K, Kuo SH, Cotrina ML, Rosoklija G, Sosunov A, et al. Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron 2014; 83: 1131–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij D, Anagnostou E, Arango C, Auzias G, Behrmann M, Busatto GF, et al. Cortical and subcortical brain morphometry differences between patients with autism spectrum disorder and healthy individuals across the lifespan: results from the ENIGMA ASD Working Group. Am J Psychiatry 2017; 175: 359–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas DL, Nascimbene C, Krishnan C, Zimmerman AW, Pardo CA.. Neuroglial activation and neuroinflammation in the brain of patients with autism. Ann Neurol 2005; 57: 67–81. [DOI] [PubMed] [Google Scholar]
- Wakabayashi A, Baron-Cohen S, Wheelwright S.. Are autistic traits an independent personality dimension? A study of the Autism-Spectrum Quotient (AQ) and the NEO-PI-R. Pers Individ Dif 2006; 41: 873–83. [Google Scholar]
- Wallace GL, Dankner N, Kenworthy L, Giedd JN, Martin A.. Age-related temporal and parietal cortical thinning in autism spectrum disorders. Brain 2010; 133: 3745–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalley HC, Sprooten E, Hackett S, Hall L, Blackwood DH, Glahn DC, et al. Polygenic risk and white matter integrity in individuals at high risk of mood disorder. Biol Psychiatry 2013; 74: 280–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler AM, Kochunov P, Blangero J, Almasy L, Zilles K, Fox PT, et al. Cortical thickness or grey matter volume? The importance of selecting the phenotype for imaging genetics studies. Neuroimage 2010; 53: 1135–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff JJ, Gu H, Gerig G, Elison JT, Styner M, Gouttard S, et al. ; the IBIS Network. Differences in white matter fiber tract development present from 6 to 24 months in infants with autism. Am J Psychiatry 2012; 169: 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsley KJ, Taylor JE, Tomaiuolo F, Lerch J.. Unified univariate and multivariate random field theory. Neuroimage 2004; 23: S189–95. [DOI] [PubMed] [Google Scholar]
- Wray NR, Lee SH, Mehta D, Vinkhuyzen AAE, Dudbridge F, Middeldorp CM.. Research review: Polygenic methods and their application to psychiatric traits. J Child Psychol Psychiatr 2014; 55: 1068–87. [DOI] [PubMed] [Google Scholar]
- Zhang H, Schneider T, Wheeler-Kingshott CA, Alexander DC.. NODDI: Practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage 2012; 61: 1000–16. [DOI] [PubMed] [Google Scholar]
- Zielinski BA, Prigge MBD, Nielsen JA, Froehlich AL, Abildskov TJ, Anderson JS, et al. Longitudinal changes in cortical thickness in autism and typical development. Brain 2014; 137: 1799–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zikopoulos B, Barbas H.. Changes in prefrontal axons may disrupt the network in autism. J Neurosci 2010; 30: 14595–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The PING data used in the study are available at https://nda.nih.gov/ after providing the necessary data user agreement. The raw data of ABIDE are available at http://fcon_1000.projects.nitrc.org/indi/abide/, and its pre-processed data are available at http://preprocessed-connectomes-project.org/abide/.