COVID-19 is caused by SARS-CoV-2.1,2 By July 25, 2020, globally, 15,672,841 diagnosed cases and 638,352 deaths were reported (https://coronavirus.jhu.edu/map.html).3 High titers of Spike protein (S protein)-specific antibodies are found in the blood of COVID-19 patients, especially IgG for both SARS-CoV4 and SARS-CoV-2.5,6 Because of the central role that S protein plays in the entry of the virus into the host cell, S1 and, more specifically, the RBD (receptor-binding domain) is the most targeted region for the development of COVID-19 therapeutic antibodies7,8 and vaccines.9 It is known that in addition to the RBD, other areas/epitopes of S protein may also elicit neutralizing antibodies.10 However, antibody responses to full-length S protein have not been investigated at epitope resolution, and the capability of linear epitopes to elicit neutralizing antibodies has still not been explored.
To precisely decipher the B-cell linear epitopes of the S protein, we constructed a peptide microarray. A total of 211 peptides (Supplementary Table 1) were synthesized and conjugated to BSA (Supplementary Fig. 1a–c). The conjugates along with control proteins were prepared in triplicate at three dilutions. High reproducibility among triplicate spots or repeated arrays for serum profiling was achieved (Supplementary Fig. 1d, e). Peptides with variable concentrations may enable dynamic detection of antibody responses and indicate that antibodies against different epitopes may have different kinetic characteristics (Supplementary Figs. 1f and 2a). Moreover, an inhibitory assay using free peptides verified the specificity of the signals generated against the peptides (Supplementary Fig. 2b).
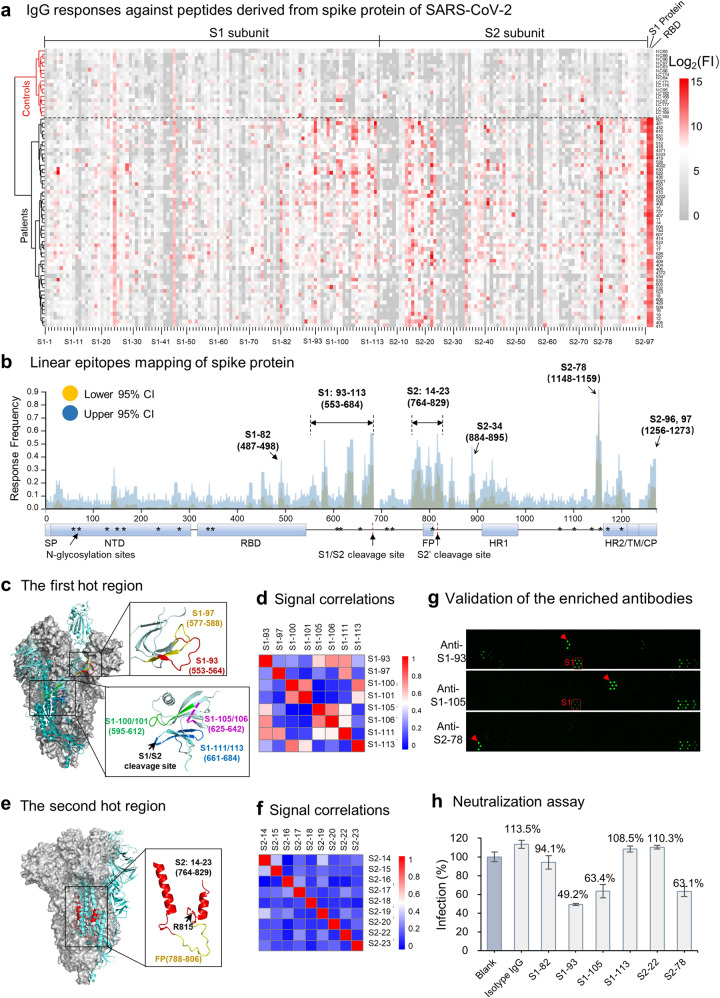
Fifty-five sera from convalescent COVID-19 patients and 18 control sera (Supplementary Table 2) were screened on the peptide microarray for both IgG and IgM responses (Fig. 1a and Supplementary Fig. 3). For IgG, COVID-19 patients were completely separated from controls, and distinct and specific signals were shown for some peptides. In contrast, the assay results were not distinct enough for IgM responses. We then focused on IgG for further analysis. Epitope maps of S protein were generated based on the response frequency (Fig. 1b).
Fig. 1.
Linear epitope mapping of SARS-CoV-2 S protein and neutralizing activities of the elicited antibodies. a Heatmap of IgG antibody responses of 55 sera from COVID-19 convalescent patients and controls (healthy donors and lung cancer patients). FI fluorescence intensity. b Epitope mapping according to the response frequency. CI confidence interval. c Detailed structural information of the epitopes of the first hot areas on S protein (PDB: 6VYB). d Correlations of the antibody responses among the peptides for the first hot areas. e Detailed structural information of the epitopes of the second hot areas on S protein (PDB: 6VYB). f Correlations of the antibody responses among the peptides for the second hot areas. g Peptide microarray results for the enriched epitope-specific antibodies. h Neutralization assay with epitope-specific antibodies. Infection rates for each sample relative to that of the blank control are indicated. Triplicate experiments were performed, and the error bars indicate the SEM (standard error of the mean) value
Primarily, there are three hot epitope areas across S protein. The first is on the CTD (C terminal domain) that immediately follows the RBD, i.e., from S1–93 to S1–113. Interestingly, the identified epitopes, S1–93, 97, 100/101, 105/106, 111, and 113, are located predominantly in flexible loops (Fig. 1c). In addition, the signals of some epitopes had moderate correlations with others (Fig. 1d), and most of these epitopes were positively correlated with S1 (Supplementary Fig. 4c–f). The second hot area is from S2–14 to S2–23, including the FP (fusion peptide, aa 788–806) region and the S2′ cleavage site (R815) (Fig. 1e). In contrast to those for the first hot region, the antibody responses against epitopes of this region had poor correlations with each other (Fig. 1f), possibly due to the capability of this region to generate continuous but competitive epitopes. Moreover, part of this area is shielded by other parts in trimeric S (Fig. 1e), suggesting that this part would be easily accessed by the immune system after the departure of S2 from S1. The third hot area is S2–78 or aa 1148–1159, connecting HR1 (heptad repeat 1) and HR2 (heptad repeat 2) on the S2 subunit. IgG antibodies against this epitope can be detected in ~90% of COVID-19 patients, indicating that it is an extremely dominant epitope. In addition to these three areas, S2–34 (aa 884–895) and S2–96/97 (aa 1256–1273) also elicited antibodies in some patients.
The RBD can elicit a high titer of antibodies and highly correlates with the S1 protein6 (Supplementary Fig. 4a), suggesting that the RBD is a dominant region. A peptide, S1–82, located exactly on the surface of the RBM (receptor-binding motif) (Supplementary Fig. 5a), was identified as an epitope. However, further analysis demonstrated that the epitope had poor specificity (Supplementary Fig. 5b), probably due to sequence similarity (Supplementary Fig. 5c–g). Thus, the validity of epitope S1–82 may need further investigation. Other than S1–82, no significant binding was observed for the rest of the peptides located at the RBD, suggesting that conformational epitopes are dominant for the RBD.
In addition to the RBD, other areas/epitopes of S protein may also elicit neutralizing antibodies.10 To explore this possibility, we chose six representative epitopes, i.e., S1–82, S1–93, S1–105, S1–113, S2–22, and S2–78, to test. Antibodies that specifically bind these epitopes were separately enriched from five sera. High specificity of these antibodies, except for S1–82, was demonstrated (Fig. 1g, Supplementary Fig. 6). A pseudotype virus neutralization assay with the enriched antibodies was then performed. Because of the limited amount of the enriched epitope-specific antibodies, the assay was performed for only a single point with the highest antibody concentration that was applicable for each epitope-specific antibody. Surprisingly, the antibodies against three epitopes, i.e., S1–93, S1–105, and S2–78, exhibited potent neutralizing activity, with virus infection inhibitory efficiencies of 51%, 35%, and 35% at 8.3, 10.4, and 21 μg/mL, respectively (Fig. 1h). However, S1–82, S1–113, and S2–22 at 2.6, 7.6, and 21 μg/mL, respectively, were not effective.
S1–93 is located at the CTD of S1. The antibody against this epitope has neutralizing activity, which is consistent with a recent study.11 Antibody binding to this epitope may affect the conformational change of S for ACE2 binding. S1–105 also belongs to the CTD but is close to the S1/S2 cleavage site. The antibodies may block effective protease cleavage at this site, which is critical for the entry of the virus. S2–78 is located adjacent to HR2, and antibodies that bind to this epitope may interfere with the formation of 6-HB (helical bundle), an essential structure for cell membrane fusion.12
Potent neutralizing antibodies could provide therapeutic and prophylactic reagents to fight against the COVID-19 pandemic.13 However, it is risky to focus only on the RBD region, and evolutionary pressure on this “hot” area may cause potential mutations in this region. This may overcome or diminish the effectiveness of the RBD region-centered therapeutic antibodies and vaccines in the development pipeline.14 Thus, identification of other domains or epitopes that can elicit neutralizing antibodies is essential as well. The combination of the potent antigenicity of these peptides and neutralizing activity of the corresponding antibodies makes these epitopes potential candidates for both therapeutic antibodies and vaccine development.
There are some limitations in this study. Because a very limited amount of purified antibodies could be enriched from valuable serum samples, we could not perform a full set of neutralization assays by serial dilution to obtain the exact IC50 values. The IC50 values estimated based on the present data were 5–20 μg/mL for the antibodies. However, monoclonal antibodies presumably have better neutralization activity, so it is an option to acquire reactive B-cell clones and express recombinant monoclonal antibodies.2,7 Because of the poor availability of convalescent sera, we did not thoroughly examine all the peptides that may potentially raise neutralizing antibodies. The other epitopes that raised antibodies may also have neutralizing activities, such as S1–97, which is very close to S1–93, or the peptides derived from the FP region, such as S2–18, –19, and –20, which are worth further investigation.
Supplementary information
Acknowledgements
This work was partially supported by the National Key Research and Development Program of China Grant (No. 2016YFA0500600), National Natural Science Foundation of China (Nos. 31970130, 31600672, 31670831, and 31370813), Shenzhen Key Laboratory of Synthetic Genomics (ZDSYS201802061806209), Shenzhen Science and Technology Program (KQTD20180413181837372), Guangdong Provincial Key Laboratory of Synthetic Genomics (2019B030301006), and Foshan Scientific and Technological Key Project for COVID-19 (No. 2020001000430).
Author contributions
S.-C.T. developed the conceptual ideas and designed the study. J.Z., W.W., and X.Y. collected the serum samples and provided key reagents. Y.L., D.-Y.L., H.-N.Z., X.-L.T., H.-W.J., M.-L.M., H.Q., Q.-F.M. and S.-J.G. performed the experiments. T.-L.Y. and Y.L.-W. provided critical suggestions for the neutralization assay. S.-C.T. and Y.L. wrote the paper with suggestions from the other authors. D.-W.S. and J.-B.D. provided key reagents or data analysis.
Data availability
The peptide microarray data are deposited in the Protein Microarray Database (http://www.proteinmicroarray.cn) under the accession number PMDE242. Additional data related to this paper may be requested from the authors.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Yang Li, Dan-yun Lai, Hai-nan Zhang, He-wei Jiang, Xiaolong Tian
Contributor Information
Tianlei Ying, Email: tlying@fudan.edu.cn.
Jie Zhou, Email: zjet65@163.com.
Sheng-ce Tao, Email: taosc@sjtu.edu.cn.
Supplementary information
The online version of this article (10.1038/s41423-020-00523-5) contains supplementary material.
References
- 1.Zhou P, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li G, Chen X, Xu A. Profile of specific antibodies to the SARS-associated coronavirus. N. Engl. J. Med. 2003;349:508–509. doi: 10.1056/NEJM200307313490520. [DOI] [PubMed] [Google Scholar]
- 5.Long QX, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 6.Jiang H, et al. SARS-CoV-2 proteome microarray for global profiling of COVID-19 specific IgG and IgM responses. Nat. Commun. 2020;11:3581. doi: 10.1038/s41467-020-17488-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao Y, et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. 2020;182:73–84.e.16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Y, et al. Identification of human single-domain antibodies against SARS-CoV-2. Cell Host Microbe. 2020;27:891–898. doi: 10.1016/j.chom.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amanat F, Krammer F. SARS-CoV-2 vaccines: status report. Immunity. 2020;52:583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chi X, et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poh CM, et al. Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients. Nat. Commun. 2020;11:2806. doi: 10.1038/s41467-020-16638-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia S, et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grifoni A, et al. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671–680.e2. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, et al. Importance of neutralizing monoclonal antibodies targeting multiple antigenic sites on the middle east respiratory syndrome coronavirus spike glycoprotein to avoid neutralization escape. J. Virol. 2018;92:e02002–e02017. doi: 10.1128/JVI.02002-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The peptide microarray data are deposited in the Protein Microarray Database (http://www.proteinmicroarray.cn) under the accession number PMDE242. Additional data related to this paper may be requested from the authors.