Abstract
Modern combination antiretroviral therapy (cART) can bring HIV-1 in blood plasma to level undetectable by standard tests, prevent the onset of acquired immune deficiency syndrome (AIDS), and allow a near-normal life expectancy for HIV-infected individuals. Unfortunately, cART is not curative, as within a few weeks of treatment cessation, HIV viremia in most patients rebounds to pre-cART levels. The primary source of this rebound, and the principal barrier to a cure, is the highly stable reservoir of latent yet replication-competent HIV-1 proviruses integrated into the genomic DNA of resting memory CD4+ T cells. In this review, prevailing models for how the latent reservoir is established and maintained, residual viremia and viremic rebound upon withdrawal of cART, and the types and characteristics of cells harboring latent HIV-1 will be discussed. Selected technologies currently being used to advance our understanding of HIV latency will also be presented, as will a perspective on which areas of advancement are most essential for producing the next generation of HIV-1 therapeutics.
Keywords: HIV, latency, provirus, integration, residual viremia, antiretroviral, T cell, CD4, TCR, clonal expansion, homeostatic maintenance, multiple displacement amplification, MDA
1. INTRODUCTION
Combination antiretroviral therapy (cART) against HIV-1 infection is a truly remarkable achievement of modern medicine, capable of reducing viral load to <50 particles/mL and halting disease progression [1-3]. Moreover, individuals whose viral load is stably suppressed cannot sexually transmit the virus [4]. Typically comprised of two nucleoside reverse transcriptase inhibitors (NRTIs) and an integrase strand transfer inhibitor (INSTI), a non-nucleoside reverse transcriptase inhibitor (NNRTI), or a protease inhibitor (PI) with a pharmacokinetic (PK) enhancer such as cobicistat or ritonavir [5], modern cART is better tolerated than previous combinations of antiretroviral drugs. However, the cost of cART treatment is high, long term cardiac and hepatic toxicity remains a concern [6-9], and because low-level viremia persists on cART and quickly rebounds upon cART cessation [10, 11], lifelong treatment is required.
The principal barrier to a cure for HIV-1 infection is the stable reservoir of latent virus in resting memory CD4+ T cells [12-18] maintained even in patients whose viremia is suppressed on long-term cART [19-24]. With a t½ alternatively measured at 3.7 [21, 23] or 3.6 years [20], the slow decay rate of the latent reservoir is prohibitive to cure by lifelong cART alone. Hence, the next generation of antiretroviral therapeutics must target the latent viral reservoir for reduction and eventual elimination, and for this, a better understanding of HIV-1 latency and the processes that promote, maintain, and reverse it, is required.
Prolonged, reversibly quiescent infection is the essence of viral latency [25]. This requires that (i) the infecting virus does not elicit an immune response that kills the host cell, (ii) expression of viral genes is highly restricted yet can be activated under favorable intracellular conditions [25], and (iii) infected cells and their progeny can persist for an extended period of time. In the case of HIV-1, the first condition is met by integration of viral DNA into the genome of the infected cell [26, 27], creating a provirus essentially invisible to host antiviral surveillance. How HIV-1 meets the second condition is less clear, although evidence that the latent reservoir is maintained in CD4+ resting memory T cells that have an intracellular environment unfavorable for viral RNA transcription will be discussed. Finally, resting memory T cells are long lived and maintained by homeostatic proliferation, thus satisfying the third condition.
The reservoir of latent virus is established early in infection, even among those rare individuals who spontaneously control HIV-1 infection without cART [28, 29]. Hence, early administration of cART, while reducing the size and diversity of the latent reservoir relative to those treated during later stages of infection [17, 30], does not prevent its establishment [29, 31]. This result and conclusion are supported by experiments with rhesus macaques infected with SIV, which also establishes a latent reservoir in resting CD4+ T cells [32, 33]. Specifically, initiation of cART in these animals three days after infection effectively suppressed viremia but did not prevent establishment of the latent reservoir [34]. Taken together, these observations demonstrate that despite continuous cART, the latent reservoir of HIV-1 in resting T cells is established within a few days of infection and is large and stable enough to support continual low-level viremia and viral rebound if treatment is withdrawn.
2. A GENERALIZED MODEL OF HIV-1 LATENCY
There is ample evidence that the latent HIV-1 reservoir is comprised primarily of resting memory CD4+ T cells. For instance, by activating cells to reverse latency, and regardless of the duration of ART, replication-competent HIV-1 can be recovered from highly purified resting CD4+ T cells isolated from infected individuals [14, 19-22, 35]. Indeed, resting CD4+ T cells are the only undisputed source of persistent, replication-competent HIV-1 when cART is optimal [36]. Other cell types, including macrophages, may contribute to HIV persistence [37-48], although this is not easy to confirm, since tissue macrophages, especially those that are CNS resident, can be difficult to sample [42]. Latent HIV-1 has also been found in naïve CD4+ T cells [12-14, 16-18]; however, these cells constitute only a small fraction of the latent reservoir.
Any theory explaining the mechanisms of HIV-1 latency must address how HIV-1, while demonstrating a strong propensity to infect activated CD4+ T cells [49, 50], establishes a latent reservoir primarily in resting memory CD4+ T cells [12-14, 16-18]. Actively infected T cells are short lived (t½ of 1-2 days [51, 52]), yet each is capable of producing on the order of 2000 infectious particles during its lifespan [53]. In contrast, the latent reservoir of infected memory T cells is largely inert with respect to virus production and has a collective t½ of 3.6-3.7 years [20, 21, 23]. Arguably, the most straightforward explanation for this dichotomy is that HIV-1 primarily infects activated T cells, some fraction of which is then transformed into memory cells that enter a quiescent, resting state conducive to viral latency [54].
There are several differences in the cellular microenvironments of activated and resting memory CD4+ T cells that may account for their differing permissiveness with respect to HIV-1 fusion with the cell membrane, reverse transcription, integration, and gene expression. For instance, CCR5, a critical co-receptor for entry of the commonly transmitted forms of HIV-1 [55-60], is upregulated upon T cell activation [61], thus facilitating viral entry. Subsequent reverse transcription of viral RNA and integration of viral DNA into the infected cell genome occurs within hours [62], in part due to the relatively fluid cytoskeleton of the activated T cell and ready availability of cellular factors required for nuclear entry and chromatin binding. Active nuclear forms of key host factors such as NF-kB, NFAT, and pTEFb, which promote and facilitate transcription of viral RNA from the integrated provirus, are also present in abundance in activated cells [63-70]. Collectively, these cellular characteristics demonstrate why infection and virus replication are favored in activated T cells at the expense of latency.
Activated T cells are also short-lived, undergoing programmed cell death as part of the contraction phase of the T cell response [71], and their lifespan may be shortened further by infection with HIV-1. Studies of viral dynamics after initiation of cART demonstrate a rapid, almost immediate decline in viremia, a function of both the short life span of actively infected T cells (t½ of ~ 1 day) and the decay of plasma virions (t½ of minutes) [3, 52, 72, 73]. In addition to contraction-phase apoptosis, cell death can result from the production of toxic viral proteins, integration of the HIV-1 provirus into a metabolically critical segment of the host cell genome [74, 75], or a CTL response [76-79]. The contribution of the last of these possibilities is apparently minor however, as the CTL response does not appear to shorten the t½ of productively infected cells in aggregate [80, 81].
In contrast to activated CD4+ T cells, resting memory T cells generally do not express the CCR5 HIV-1 co-receptor [61], thus impeding cellular entry, and the static nature of the actin cytoskeleton inhibits delivery of the reverse transcription complex to the nucleus [82]. In addition, SAMHD1, a cellular restriction factor expressed at high levels in myeloid cells and resting CD4+ T cells [55-60], depletes cytoplasmic dNTP levels, thereby inhibiting reverse transcription directly [83-85]. As a consequence, reverse transcription of HIV-1 RNA in resting T cells can take as long as three days [86-88]. Prolonged residency of the reverse transcription complex in the cytoplasm increases the likelihood that viral DNA intermediates are recognized by the intracellular DNA sensor IFI16, which leads to activation of caspase-1 and a pro-inflammatory form of cell death known as pyroptosis [89-91]. Together, these aspects of the cellular microenvironment serve to inhibit all stages of the HIV-1 life cycle up to and including integration. However, if viral DNA integration is successful, transcription of viral RNA is hindered by reduced levels of the active forms of NF-kB, NFAT, and pTEFb characteristic of resting memory T cells [63-65, 67, 68, 70]. Epigenetic modifications to the host cell genome, which contributes to the quiescence of memory T cells in the resting state, may likewise contribute to longer-term and more complete silencing of transcription from the HIV-1 provirus [92-94]. Hence, both the dearth of important transcription factors and epigenetic modification of host cell DNA in resting memory T cells are likely to play important roles in establishing and maintaining the latency of an otherwise transcriptionally prolific virus.
3. DIFFERENTIATION OF CD4+ MEMORY T CELLS
At least three models have been proposed to explain transition and differentiation among naïve and memory T cell subsets, namely, linear differentiation (LD), decreasing potential/progressive differentiation (DP/PD), and divergent differentiation/disparate fate (DD/DF) [95]. In the LD model, naïve CD4+ T cells challenged with antigen become activated and progressively differentiate along a linear path to become effector cells, some of which then survive the contraction phase and persist as quiescent (resting) memory cells [71, 96-101]. Primary effector cells also become progressively differentiated into effector cells in the DP/PD model; however, their progression down this path, and their capacity for acquiring memory function, respectively, are directly and inversely related to the strength and duration of TCR and costimulatory signaling [97, 102]. In other words, in the DP/PD model, early CD4+ effector intermediates can ultimately transition into memory cells, but the most activated, terminally differentiated effector T cells cannot. Finally, according to the DD/DF model, initial proliferation of activated cells during the expansion phase yields heterogenous progeny with different capacities for differentiation to either effector or memory cells. This fascinating model invokes asymmetric cell division, in which the distribution of activation-associated signaling receptors between progeny is uneven [103, 104]. The DD/DF model also supports the existence of terminal effector and memory precursor CD4+ T cells subsets homologous to subsets in the CD8+ T cell lineage [98]. Since none of these models alone can fully explain the observations of CD4+ T cell differentiation reported in the literature, it is likely that memory is generated through multiple pathways, and the range of phenotypes and capacity for plasticity among effector and memory cells is extensive.
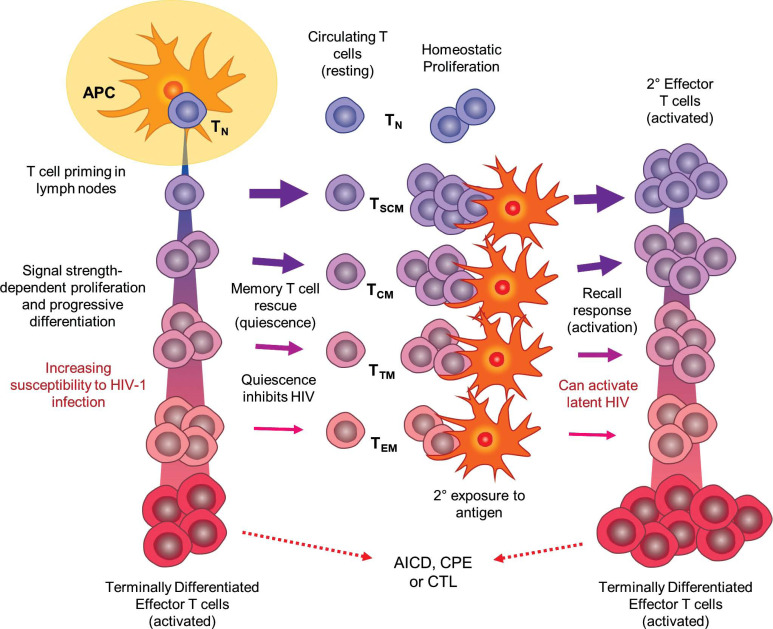
In Fig. (1), proposed mechanisms for HIV-1 infection, latency, and emergence from latency are superimposed on the DP/PD model of CD4+ T-cell differentiation and memory response. However, regardless of which specific T cell differentiation pathways are applicable, the simplest explanation for establishing HIV-1 latency involves infection of T cells of sufficient activated character to permit progression through stages of the virus life cycle up to and including integration, but which assume a resting state quickly thereafter so as to be non-permissive to high-level transcription of viral RNA and virus replication [15]. Such a mechanism would not only explain most physiological and clinical findings related to HIV persistence, but would do so without requiring the evolution of special viral mechanisms of latency. One notable consequence of the stringent timing requirement suggested by this model is that establishment of latent infection would be expected to be rare, and indeed, only approximately 1 in 106 resting CD4+ T cells are latently infected in HIV-infected individuals on cART [14, 20-23]. Moreover, the feasibility of this model is supported by experiments in which induction of and emergence from HIV-1 latency have been successfully recapitulated in primary T cells in vitro; i.e., isolated primary T cells are activated, infected, and then cultured under conditions that promote reversion to a resting state [105-109]. Re-stimulation of these cells through the T cell receptor (TCR) leads to HIV-1 gene expression.
Fig. (1).
Depiction of the DP/PD model of CD4+ T cell differentiation and immune response as a framework for HIV-1 infection, latency, and re-activation (see text for details). Upon initial antigen exposure, naïve T cells (TN) are primed by antigen-presenting cells (APCs) in secondary lymphoid organs. Depending on the strength and quality of stimulatory signals, CD4+ T cells progress along an activation and differentiation pathway, losing naïve and acquiring effector properties. It is postulated that less differentiated cells are rescued into memory with greater frequency, while highly differentiated effectors are more susceptible to HIV-1 infection and support virus replication. All terminally differentiated and most other effector cells are not rescued into memory, but are instead subject to programmed, activation-induced cell death (AICD), are killed by cytotoxic T-lymphcytes (CTL), or if infected with HIV-1, are killed by the cytopathic effects of active virus. Latency is established when infected effector cells are rescued into memory T cells in the resting state after the stimulating antigen is removed. Differentiation of memory cell subsets (TSCM, TCM, TTM, TEM) parallels effector cell differentiation, with less specialized cells tending to be longer-lived and having a greater capacity for homeostatic proliferation. Response to subsequent antigen exposure is faster and more potent as memory cells re-enter the effector differentiation pathway; however, this process may also cause quiescent HIV-1 to emerge from latency. As with initial exposure, some secondary effector cells are rescued into memory after the antigen has been removed, thus perpetuating the cycle of T-cell maintenance and response, and potentially HIV-1 latency.
4. T CELL SUBTYPES OF THE LATENT RESERVOIR
CD4+ memory T cell pools possess vast functional and phenotypic heterogeneity. Hence, identifying and characterizing the subsets that host long-term viral persistence during cART represents an area of active investigation and may have consequences for designing next-generation strategies to reduce and eliminate the latent reservoir. CD4+ memory T cells populate a differentiation hierarchy that includes naïve, T stem cell memory (TSCM), T central memory (TCM), T transitional-memory (TTM), T effector-memory (TEM) and terminally differentiated helper T cells (Fig. 1) [110]. The more primitive T cells in this progression express a mix of naïve and memory cell markers, tend to be longer lived, and are more capable of homeostatic self-renewal, while the more differentiated cells are increasingly committed to specialized effector functions, are short lived, and are programmed to perish soon after the activating antigen is no longer present [111, 112]. Memory T cells may also be classified into subpopulations by function, each defined by specific antimicrobial properties, cytokine secretion patterns, and expression of signature transcription factors [113]. These include Th1 (antibacterial immune defense), Th2 (protection against helminths and other parasites), Th17 (protection against extracellular bacterial and fungal infections), Th9 (regulation of allergic inflammation, anti-tumor and anti-parasitic immunity), regulator T cells (regulation of antimicrobial immunity), and follicular helper cells (TFH; support of humoral immunity). Tissue-resident memory cells represent another distinct subset, localizing to barrier tissues at interfaces with the environment, but also within the brain, kidney, and joints rather than the secondary lymphoid organs [114], as do γδ T cells, which expressing a T cell receptors (TCR) produced from recombinant γ and δ genes instead of the typical α and β [115]. The γδ cells play important functional roles in recognition of lipid antigens, and do not require antigen presentation by MHC complexes [116].
TCM cells generally constitute the largest fraction of the memory CD4+ T cell pool. In one study, among the cell subsets classified, TCM cells made the greatest fractional contribution to the viral reservoir in individuals on cART (i.e., TCM, TTM, TEM, terminally differentiated, and naïve CD4+ cells contributed an average of 51.7, 34.3, 13.9, 1.9 and 0.3% to the infected cell populations, respectively) [13]. Persistence of HIV-1 infection in this cell subset is likely facilitated by the relatively long half-life of these cells, greatly exceeding that of EM cells and approximating that of quiescent naïve T cells [117]. The preferential persistence of HIV-1 in TCM cells is consistent with the increased propensity of these cells toward homeostatic proliferation relative to TTM and TEM [118, 119]. When driven by IL-7, this maintenance mechanism is not associated with viral reactivation or expression of viral antigens, and therefore is unlikely to be restricted by antiviral immune mechanisms [120]. Some rare individuals, who after an initial period of suppressive therapy spontaneously maintained undetectable levels of HIV-1 replication in the absence of cART, were demonstrated to have very low residual levels of HIV-1 infected TCM cells [121], thus emphasizing the importance of these cells in maintaining the latent reservoir and contributing to viral rebound.
TSCM cells likewise make a significant contribution to the latent reservoir of patients on cART. Although these cells represent a minor fraction of total memory CD4+ T cells, and their contribution to the proviral reservoir has been measured as ~8%, the stem cell-like properties of TSCM cells confer greater capacity to proliferate and survive than even TCM cells [122]. TSCM cells appear to occupy a stage of differentiation between naïve cells and TCM cells, having a t½ of 277 months (compared to 144, 133, and 88 months for TCM, TTM, and TEM cells, respectively) and a strong propensity for homeostatic proliferation [16, 122]. Among patients on cART, the longitudinal decline in HIV-1 DNA associated with TSCM cells was less steep than in the TEM and terminally differentiated subsets [122]. Hence, HIV-1 infected TSCM cells are likely to be overrepresented in the latent viral reservoirs of many patients on long term cART [16, 122], and this fractional contribution is likely to increase with the duration of therapy.
Functional polarization of T cell subsets is associated with developmental and maturational properties that influence cellular longevity and long-term survival [123], and the process is regulated by specialized transcription factors that determine characteristic gene expression changes in each polarized cell population. These properties are also likely to influence the capacity of functionally polarized T cells to host latent HIV-1 infection. For example, while Bcl-6 [124], FoxP3 [125] and Gata3 [126] transcription factors have been shown to contribute to functional polarization, they can also bind to the HIV-1 promoter, thus potentially activating HIV-1 gene expression and reversing latency. Th17 cells in particular exhibit common characteristics among cells known to contribute to the latent reservoir, including longevity and propensity for homeostatic proliferation [123, 127, 128], and were found to harbor high levels of HIV-1 DNA with greater longitudinal stability than Th1 cells in individuals on long term cART [129]. Moreover, some Th17-specific gene expression signatures produce a cellular microenvironment favorable for HIV-1 replication [130], and cells of this subset also seem to be intrinsically susceptible to HIV-1 infection in ex vivo assays [131].
HIV-1 can also infect TFH cells [132-135], which represent a major site for HIV-1 replication and production during untreated HIV-1 infection [135]. These cells are characterized by surface expression of CXCR5 and PD-1, reside in lymph node follicles in immediate anatomical proximity to B cells, and support the germinal center reaction essential for generation of effective humoral immunity [136]. This tissue residency may render HIV-infected TFH cells especially difficult to eradicate by immunotherapy since CD8+ CTL lack chemokine receptors needed for migrating into B cell follicles [133]. This problem is exemplified in rhesus macaques that spontaneously control SIV, and where viral replication is restricted to TFH, presumably because CD8+ CTL lyse infected cells elsewhere in the lymph nodes [133]. Hence, if latently infected TFH cells persist, HIV-1 eradication strategies that disrupt B cell follicles to permit access by CTL may need to be considered [133]. Currently, however, the extent to which TFH serve as a long-term reservoir for HIV-1 in the setting of optimal ART remains to be determined, although one study revealed a sharp decline of HIV-1 DNA levels in TFH within 1-2 years of suppressive cART [135]. Relatedly, HIV-1 DNA has been detected in PD-1 and CXCR5-expressing TFH-like cells circulating in peripheral blood in individuals on cART [129], although the contribution of these cells to HIV-1 persistence is unknown.
The contribution of regulatory T cells to the latent reservoir has likewise not been determined. These cells express FoxP3 as a master transcription factor and play a dual role in HIV-1 pathogenesis, both reducing pathologic immune activation and inhibiting beneficial antiviral immunity. Regulatory CD4+ T cells from patients on cART have been shown to harbor abundant levels of HIV-1 DNA, with an infected cell t½ of 20 months [137].
Constitutively expressing low levels of CD4 receptor, γδ T cells may be thought to represent less preferred target cells for HIV-1 infection. However, HIV-1 DNA has previously not only been detected in γδ T cells in patents on cART at levels exceeding those in resting CD4 T cells, but in most such instances, virus retrieved from these samples was found by the virus outgrowth assay to be replication competent [138]. Finally, tissue-resident memory cells represent a subset of CD4+ T cells that includes lymphocyte populations in peripheral mucosal tissues, barrier surfaces, and in other non-lymphoid and lymphoid sites, and, at least in adipose tissues, may support HIV-1 infection [139, 140]. However, whether either γδ T cells or tissue resident memory cells contribute significantly to HIV-1 reservoirs in individuals on cART remains unclear.
5. RESIDUAL VIREMIA AND HIV PERSISTENCE
Analyzing the evolution of viremia and HIV-1 DNA associated with peripheral blood mononuclear cells (PBMC) throughout the course of cART can provide insight into the mechanisms of HIV-1 persistence. For instance, levels of plasma HIV-1 RNA have been found to decrease by 4-5 orders of magnitude during of the first year of therapy, suggesting that pre-treatment viremia is almost exclusively produced by short-lived infected cells [141-143]. In contrast, levels of PBMC-associated HIV-1 DNA in these individuals was reduced no more than 10-fold over the same period; hence, infected cells that persist despite cART harbor HIV-1 DNA but do not produce virus at high levels.
Viremia declines steadily for approximately four years after treatment initiation [144-146], after which trace levels of free virus on the order of 1 copy/mL of blood plasma can be detected in individuals on effective cART. Such residual viremia reflects the nature of cART, which inhibits attachment and fusion, reverse transcription, integration, and/or particle maturation after release but does not prevent virus production or release from the infected cell. Hence, virus generated despite effective cART is unlikely to be fully mature, and new infections originating from these viruses should be strongly inhibited at multiple stages. These postulates are supported by data indicating that residual viremia in individuals on effective cART does not produce new infections [147-149]. Moreover, when administered early, cART seems to be effective in arresting expansion and diversification of the HIV-1 reservoir, as evidenced by the strong correlation among effectively treated patients between levels of PBMC-associated HIV-1 DNA and the state of infection at the time therapy was initiated [142, 143, 150]. These data also demonstrate that early treatment is critical to minimizing the size and diversity of the latent reservoir.
Residual viremia in well controlled patients does not appear to be affected by treatment intensification, suggesting that there is little or no background virus replication to suppress by supplementing cART with additional classes of antiviral drugs [147-149, 151]. Longitudinal studies of HIV-1 sequence diversity in individuals on cART support this postulate as well. Specifically, HIV-1 RNA sequences obtained from patient blood samples after prolonged cART resemble those present earlier in infection and generally do not show evidence of ongoing evolution [152-156]. In fact, HIV-1 sequences obtained at different times during treatment are often identical, and the fraction of identical sequences within samples increases with time [152, 157, 158]. Together, these observations controvert the notion of HIV-1 persistence being driven by ongoing replication and multiple rounds of infection, and instead support the idea of rare, sporadic viral activation from a reservoir of latently infected resting memory T cells maintained by clonal expansion [159].
When cART is interrupted, viral rebound is usually observed within two weeks [10, 11]. It is postulated that this is the time required for systemic clearance of antiviral drugs and stochastic activation of sufficient latent virus to seed new infections systemically. Despite an estimated 2 log variation in reservoir size, variation in the interval between treatment withdrawal and viral rebound is limited [160], and there is evidence to suggest that rebound is seeded by spontaneous activation of multiple cells per day from multiple anatomical sites (e.g., lymph node, ileum, and rectum) [11]. Moreover, as with residual viremia, sequences from early rebound viruses resemble those of viruses before treatment withdrawal, suggesting that rebound, like residual viremia, originates from a stable latent reservoir established before initiation of cART rather than an immune-privileged pool of perpetually replicating virus [161].
There have been at least three notable cases of “near cure” in which the interval between treatment withdrawal and HIV-1 rebound greatly exceeded two weeks. Specifically, virus rebound in the two “Boston patients” did not occur until 3 and 8 months after cART withdrawal [162, 163]. Prior to cART cessation, both individuals received stem cell bone marrow transplants to treat lymphomas not responding to chemotherapy, and this was preceded by “conditioning” therapy in which the patients’ existing T cells were destroyed by chemotherapy and radiation. In the case of the “Mississippi baby”, an infant was born to a mother known to be HIV-1 positive and was treated with cART almost immediately after birth [30]. Treatment was withdrawn after 15 to 18 months against medical advice, after which the child had undetectable levels of viremia for approximately 3 years prior to virus rebound. An important shared feature of these cases is that the reservoir of infected T cells was probably extremely small when antiviral treatment was withdrawn, being either diminished by pre-transplant conditioning or prevented from becoming established by early administration of cART. This marked reduction in reservoir size might, in turn, be expected to delay virus rebound, especially if rebound requires rare, spontaneous activation of latent HIV-1 harbored in quiescent CD4+ T cells. Conversely, it is difficult to imagine why rebound seeded by even a few T cells perpetually generating large amounts of virus would be delayed at all, since infection would be expected to expand exponentially in this case. Such exponential growth is actually typical of new infections, as indicated in phylogenetic studies of HIV-1 demonstrating that nascent infections are most often seeded by a single “founder” virus yet produce extreme levels of viremia over the course of a few weeks [164, 165].
Collectively, these data support the idea that residual viremia and viral rebound are seeded by a stable reservoir of latently infected cells that infrequently become activated to produce low levels of virus despite cART [166]. Nevertheless, there remains a contention that residual viremia and the persistent HIV-1 reservoir are in part maintained by ongoing virus replication in sanctuary sites such as lymph nodes [167-171] with subsequent trafficking of recently infected cells into the blood [171, 172]. Proponents of this argument cite evidence of HIV-1 evolution in lymph node and blood samples taken from three patients on cART [171], as well as reports of poor drug penetration in lymph nodes and mucosa-associated lymphoid tissue (MALT) [169]. However, it must be noted that these findings have not been reproduced by other investigators applying different models of evolution to the same set of HIV-1 sequences [173], and most other studies of patients on long-term suppressive cART have not found evidence of sequence diversification from pre-therapy in blood or tissues [17, 152, 159, 174-176].
6. MAINTAINING THE HIV-1 RESERVOIR BY CLONAL EXPANSION OF CD4+ MEMORY T CELLS
Numerous lines of evidence indicate that HIV-1 persists in individuals on effective cART within a reservoir of long-lived, latently infected CD4+ resting memory T cells. As depicted in (Fig. 1), such cells are natively produced by differentiation from naïve T cells primed by their cognate antigen for the purpose of establishing a capacity for memory response to a subsequent exposure. Memory responses can provide lifelong immunity, as exemplified in individuals who received the smallpox vaccine or have cleared hepatitis C infection [177, 178]. However, this persistence cannot be explained solely on the basis of memory T cell longevity, as the average t½ of memory T cells (approximately 22 weeks; [179-182]) is substantially shorter than that of naïve T cells (1-8 years), the functional memory T cells response (8-12 years), and even the HIV-1 reservoir (3.7 years). Hence, it stands to reason that homeostatic proliferation of memory T cells must also contribute to persistence of both the functional memory T cell response and the reservoir of HIV-infected T cells in patients on effective cART.
The cytokines thought to drive homeostatic clonal expansion of CD4+ memory T cells are IL-7 and IL-15 [183]. Indeed, in vitro studies in a primary cell model of HIV-1 latency confirm that latently infected cells can proliferate in response to IL-7 (and IL-2) without upregulation of HIV-1 gene expression [120], which would be detrimental to cell survival [120, 184-186]. These results are supported by clinical studies of patients on cART, where infusion of IL-7 leads to the proliferation of latently infected memory CD4+ T cells with almost no induction of HIV-1 gene expression [119, 187]. Like IL-7, IL-15 has also been shown to be important for homeostatic proliferation of CD4+ memory T cells [188].
Clonal expansion of CD4+ memory T cells can also be driven by recognition of cognate antigens presented on MHC class II receptors by antigen-presenting cells, as well as by cytokines produced as part of the normal course of an immune response. The antigen specificity of HIV-1 infected T cells has not been well classified, although some may be HIV-1 specific [189]. Unlike homeostatic proliferation, antigen-driven proliferation acting through the T cell receptor (TCR) is likely to both activate infected memory T cells and upregulate expression of HIV-1. Such activation, coupled with proliferation and subsequent reversion of a small number of activated cells to the resting state, may contribute to the maintenance of the HIV-1 reservoir, persistence of residual viremia of individuals on effective cART, and virus rebound upon cessation of treatment.
Precise identification of HIV-1 integration sites within T cell genomic DNA has become an important tool for characterizing and quantifying clonally expanded infected T cell populations. Although HIV-1 integration is not random, demonstrating a local sequence selectivity and favoring active transcriptional units [190, 191], sites of integration are of sufficient diversity that the probability of independent infections producing proviruses integrated at precisely the same location in the respective host cell genomes is exceedingly low. Instead, infected cells harboring proviruses that share a common integration site must almost certainly be the product of clonal expansion from a common parent cell. Such determinations can now be made relatively routinely, due to development of novel integration site mapping techniques reliant on semi-nested PCR and next-generation sequencing [184, 192]. Moreover, the application of such methods to DNA extracted from PBMC patient samples has provided convincing evidence for the clonal expansion of HIV-1 infected cells [184, 186, 193].
Results from a seminal study in this area showed that of 2410 integration sites identified in CD4+ T cells from five HIV-infected individuals, 43% were from clonally expanded populations [184], thus conclusively demonstrating that populations of infected cells are maintained by clonal expansion. Intriguingly, some of the integration sites identified in the clonally expanded populations were within known human proto-oncogenes associated with cell growth [194-196] and demonstrated in other in vivo and in vitro studies to harbor HIV-1 integrants [184, 186, 190, 193, 197]. These genes include (i) myocardin-like protein 2 (MKL2), a transcription factor, and (ii) basic leucine zipper transcription factor 2 (BACH2), a transcription regulator affecting lymphocyte growth, activation, senescence, and cytokine homeostasis. Of integration sites in these genes detected in patient samples, all were localized to specific intronic regions and found to be in the same transcriptional orientation as the gene in question, although a similar pattern was not observed in DNA acquired from in vitro infections [184, 186, 191]. Insertion of the HIV-1 promoter into a non-coding region of a growth-related gene might be expected to increase transcription levels, thereby conferring a selective advantage favoring proliferation and/or survival of these T cell clones in vivo. Indeed, enrichment for BACH2 integrants was confirmed in a recent study of HIV-1 integration sites in T cells both in vitro and from patient samples [198]. The same study also shows disproportional integration within FOXP1 and STAT5B, two genes involved in T cell differentiation and activity. Together, these data suggest that HIV-1 integration at specific sites and in a favorable orientation within select host genes may increase the propensity of the infected cells toward homeostatic proliferation, prolonged survival, or both.
While the notion that HIV-1 integration can confer a selective advantage to latently infected cells is intriguing, it is important to note that all MKL2 and BACH2 proviruses in clinical isolates analyzed to date have been defective [199], and thus are not truly part of the latent HIV-1 reservoir. This is consistent with findings that among individuals on long term cART, approximately 98% of proviruses in long-lived cells are not intact [200, 201]. This fraction is significantly higher than has been observed with SIV infection [202], and may even constitute an underrepresentation, as HIV-1 integrants having extensive deletions or hypermutations mediated by host cytidine deaminases of the APOBEC3 family [203] are difficult to detect by PCR. Moreover, there is probably selection against integration of a replication-competent provirus into a growth-related gene, since it is hard to imagine how HIV-1 promoter activity could increase expression of the host gene without also activating viral gene synthesis and bringing the quiescent infection out of latency. Nevertheless, with respect to a general characterization of the latent reservoir, the question of whether cells with a high propensity for clonal expansion are capable of both harboring and maintaining the latency of a replication-competent provirus is highly salient.
This question was answered in studies involving an HIV-infected individual who also had squamous cell carcinoma [184, 185]. A single, highly-expanded clone was identified in samples obtained from this individual, comprising more than 50% of the patient’s HIV-infected cells and approximately 108 cells in total. The integrant was designated AMBI-1 (ambiguous) to indicate that the integration site of the provirus could not be uniquely determined. Cells harboring the AMBI-1 integrant were found to be widely anatomically distributed but enriched in cancer metastases, suggesting that the clone expanded in response to cancer antigen. More importantly, the AMBI-1 provirus was found to be intact and replication-competent, although only a small fraction of the highly expanded clone produced detectable levels of HIV-1 RNA at any given time. This highly expanded clone is the first shown to be capable of harboring a largely latent, replication-competent HIV-1 provirus despite cART.
CONCLUSION AND PERSPECTIVE
The principle barrier to a cure for HIV-infection in individuals on effective cART is the reservoir consisting primarily of long-lived, latently-infected, CD4+ resting memory T cells maintained by clonal expansion. Residual viremia in these patients is supported by infrequent, stochastic activation of a few quiescent cells from this reservoir each day, as is virus rebound if cART is discontinued. The capacity to rapidly and thoroughly characterize these viral reservoirs is of paramount importance for developing the next generation of antiretroviral therapeutics. However, progress in this area has been slow, in large part, because the methodologies used for this purpose are often labor intensive, of low throughput, or incompletely characterize HIV-1 proviruses and infected cells.
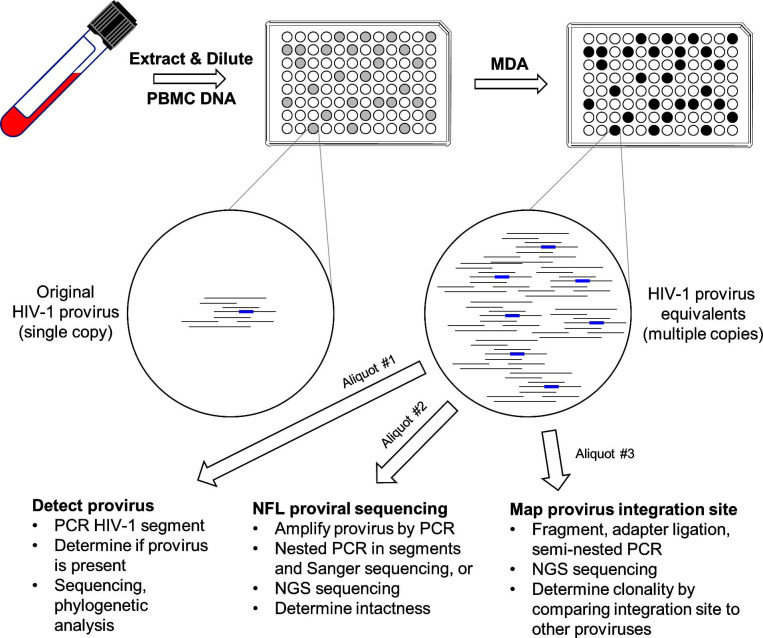
Fortunately, methods are improving. Tens of thousands of viral RNA molecules can now be independently sequenced in a single experiment [204], and sequences of proviruses can be obtained almost in their entirety by nested PCR combined with Sanger or next-generation sequencing (NGS) [205]. Moreover, using NGS, hundreds of integration sites can be sequenced in parallel after implementing a multistep preparatory method involving DNA fragmentation, adapter ligation, and PCR [184]. More recently, an exciting technology was developed that combines the properties of the last two of these assays [206, 207]. In brief, PBMC DNA samples are diluted and partitioned into aliquots of which approximately 30% contain a single HIV-1 provirus. Each aliquot is then subjected to multiple displacement amplification (MDA; [208]), which uniformly amplifies both host and proviral DNA on the order of 1000-fold. Finally, by applying different assays to aliquots of each MDA reaction, both the sequences and integration sites of individual proviruses can be determined, thus significantly streamlining the characterization of proviral intactness and clonality (Fig. 2).
Fig. (2).
Multiple displacement amplification (MDA) – single HIV-1 genome sequencing (SGS) workflow. Genomic PBMC DNA from an HIV-infected individual is diluted and dispensed into 96-well plates to maximize the number of wells containing exactly one provirus. MDA is then used to amplify DNA in each of these wells on the order of 1000-fold. Reaction products from wells originally containing a single HIV-1 provirus may thus be partitioned for downstream assays, including PCR-amplification and sequencing of segments of the virus genome (e.g., the RT gene), amplification and sequencing of near full length (NFL) provirus, and integration site determination. Further and more complete characterization of the latent HIV-1 reservoir will depend upon continued development of techniques such as these that permit examination of multiple facets of individual HIV-infected cells.
Future efforts to characterize the latent HIV-1 reservoir will benefit broadly from continued development of methodologies that combine the advantages of existing approaches. Taken to the extreme, this could entail combining several assays into one and reducing reaction volumes to the nanoliter and single-cell scale. This might allow researchers to sequence and determine the integration sites of thousands of individual replication-competent proviruses in a single experiment, as well as examine the transcriptional profiles and sequence the TCR mRNA of infected cells.
The last of these possibilities is particularly intriguing, given the importance of clonal expansion in maintaining the latent reservoir. For instance, one of the greatest limitations of the otherwise promising ‘shock and kill’ strategy for eradicating the latent reservoir is that it is not selective; i.e., there are no obvious means of activating only the latently infected resting memory T cells. Consequently, clinicians must either use an approach that activates only a small fraction of an individual’s CD4+ T cells, leaving most of the reservoir unaffected, or activate them all, the immunological consequences of which would likely be severe. However, because a recombinant TCR is unique, yet common to all cells in an expanded clone [209], a TCR sequence determined for a single infected cell will be shared by all cells in the clonal population. Moreover, if the TCR sequences of multiple latently infected clones are known, this information could theoretically be used to segregate, activate, or even eliminate the respective clonal populations while leaving the unspecified T cells unaffected. Based on the promise of ideas such as these, the continued development of methods that combine the strengths of established approaches will advance our understanding of the latent HIV-1 reservoir and further the development of the next generation of antiretro- viral therapeutics.
ACKNOWLEDGEMENTS
The authors wish to acknowledge and thank all the patient volunteers who participated in the clinical studies cited in this article.
LIST OF ABBREVIATIONS
- cART
Combination Antiretroviral Therapy
- MDA
Multiple Displacement Amplification
- PCR
Polymerase Chain Reaction
- TCR
T Cell Receptor
- TEM
Effector Memory T cell
- TFH
Follicular Helper T cell
- TSCM
Stem Cell-Like Memory T Cell
- TTM
Transitional Memory T Cell
- TCM
Central Memory T Cell
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
Intramural Research Program of the National Cancer Institute, National Institutes of Health, Department of Health and Human Services [ZIA BC 010493 to S.F.J.L.G.]. Funding for open access charge: Intramural Research Program of the National Cancer Institute, National Institutes of Health, Department of Health and Human Services [ZIA BC 010493].
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Gulick R.M., Mellors J.W., Havlir D., et al. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N. Engl. J. Med. 1997;337(11):734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 2.Hammer S.M., Squires K.E., Hughes M.D., et al. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. AIDS Clinical Trials Group 320 Study Team. N. Engl. J. Med. 1997;337(11):725–733. doi: 10.1056/NEJM199709113371101. [DOI] [PubMed] [Google Scholar]
- 3.Perelson A.S., Essunger P., Cao Y., et al. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature. 1997;387(6629):188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 4.Calabrese S.K., Mayer K.H. Providers should discuss U=U with all patients living with HIV. Lancet HIV. 2019;6(4):e211–e213. doi: 10.1016/S2352-3018(19)30030-X. [DOI] [PubMed] [Google Scholar]
- 5.Günthard H.F., Aberg J.A., Eron J.J., et al. International Antiviral Society-USA Panel. Antiretroviral treatment of adult HIV infection: 2014 recommendations of the International Antiviral Society-USA Panel. JAMA. 2014;312(4):410–425. doi: 10.1001/jama.2014.8722. [DOI] [PubMed] [Google Scholar]
- 6.Esser S., Helbig D., Hillen U., Dissemond J., Grabbe S. Side effects of HIV therapy. J. Dtsch. Dermatol. Ges. 2007;5(9):745–754. doi: 10.1111/j.1610-0387.2007.06322.x. [DOI] [PubMed] [Google Scholar]
- 7.Hemkens L.G., Bucher H.C. HIV infection and cardiovascular disease. Eur. Heart J. 2014;35(21):1373–1381. doi: 10.1093/eurheartj/eht528. [DOI] [PubMed] [Google Scholar]
- 8.Kovari H., Sabin C.A., Ledergerber B., et al. Antiretroviral drug-related liver mortality among HIV-positive persons in the absence of hepatitis B or C virus coinfection: the data collection on adverse events of anti-HIV drugs study. Clin. Infect. Dis. 2013;56(6):870–879. doi: 10.1093/cid/cis919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lugassy D.M., Farmer B.M., Nelson L.S. Metabolic and hepatobiliary side effects of antiretroviral therapy (ART). Emerg. Med. Clin. North Am. 2010;28(2):409–419. doi: 10.1016/j.emc.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 10.Davey R.T., Jr, Bhat N., Yoder C., et al. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc. Natl. Acad. Sci. USA. 1999;96(26):15109–15114. doi: 10.1073/pnas.96.26.15109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rothenberger M.K., Keele B.F., Wietgrefe S.W., et al. Large number of rebounding/founder HIV variants emerge from multifocal infection in lymphatic tissues after treatment interruption. Proc. Natl. Acad. Sci. USA. 2015;112(10):E1126–E1134. doi: 10.1073/pnas.1414926112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brenchley J.M., Hill B.J., Ambrozak D.R., et al. T-cell subsets that harbor human immunodeficiency virus (HIV) in vivo: implications for HIV pathogenesis. J. Virol. 2004;78(3):1160–1168. doi: 10.1128/JVI.78.3.1160-1168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chomont N., El-Far M., Ancuta P., et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat. Med. 2009;15(8):893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chun T.W., Carruth L., Finzi D., et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387(6629):183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 15.Chun T.W., Finzi D., Margolick J., Chadwick K., Schwartz D., Siliciano R.F. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat. Med. 1995;1(12):1284–1290. doi: 10.1038/nm1295-1284. [DOI] [PubMed] [Google Scholar]
- 16.Jaafoura S., de Goër de Herve M.G., Hernandez-Vargas E.A., et al. Progressive contraction of the latent HIV reservoir around a core of less-differentiated CD4+ memory T Cells. Nat. Commun. 2014;5:5407. doi: 10.1038/ncomms6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Josefsson L., von Stockenstrom S., Faria N.R., et al. The HIV-1 reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. Proc. Natl. Acad. Sci. USA. 2013;110(51):E4987–E4996. doi: 10.1073/pnas.1308313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pierson T., Hoffman T.L., Blankson J., et al. Characterization of chemokine receptor utilization of viruses in the latent reservoir for human immunodeficiency virus type 1. J. Virol. 2000;74(17):7824–7833. doi: 10.1128/JVI.74.17.7824-7833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chun T.W., Stuyver L., Mizell S.B., et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA. 1997;94(24):13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crooks A.M., Bateson R., Cope A.B., et al. Precise Quantitation of the Latent HIV-1 Reservoir: Implications for Eradication Strategies. J. Infect. Dis. 2015;212(9):1361–1365. doi: 10.1093/infdis/jiv218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finzi D., Blankson J., Siliciano J.D., et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 1999;5(5):512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 22.Finzi D., Hermankova M., Pierson T., et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278(5341):1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 23.Siliciano J.D., Kajdas J., Finzi D., et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 2003;9(6):727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 24.Wong J.K., Hezareh M., Günthard H.F., et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278(5341):1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 25.Speck S.H., Ganem D. Viral latency and its regulation: lessons from the gamma-herpesviruses. Cell Host Microbe. 2010;8(1):100–115. doi: 10.1016/j.chom.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson E.M., Maldarelli F. The role of integration and clonal expansion in HIV infection: live long and prosper. Retrovirology. 2018;15(1):71. doi: 10.1186/s12977-018-0448-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hughes S.H., Coffin J.M. What Integration Sites Tell Us about HIV Persistence. Cell Host Microbe. 2016;19(5):588–598. doi: 10.1016/j.chom.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buckheit R.W., III, Salgado M., Martins K.O., Blankson J.N. The implications of viral reservoirs on the elite control of HIV-1 infection. Cell. Mol. Life Sci. 2013;70(6):1009–1019. doi: 10.1007/s00018-012-1101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chun T.W., Engel D., Berrey M.M., Shea T., Corey L., Fauci A.S. Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proc. Natl. Acad. Sci. USA. 1998;95(15):8869–8873. doi: 10.1073/pnas.95.15.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Persaud D., Gay H., Ziemniak C., et al. Absence of detectable HIV-1 viremia after treatment cessation in an infant. N. Engl. J. Med. 2013;369(19):1828–1835. doi: 10.1056/NEJMoa1302976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colby D.J., Trautmann L., Pinyakorn S., et al. RV411 study group. Rapid HIV RNA rebound after antiretroviral treatment interruption in persons durably suppressed in Fiebig I acute HIV infection. Nat. Med. 2018;24(7):923–926. doi: 10.1038/s41591-018-0026-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dinoso J.B., Rabi S.A., Blankson J.N., et al. A simian immunodeficiency virus-infected macaque model to study viral reservoirs that persist during highly active antiretroviral therapy. J. Virol. 2009;83(18):9247–9257. doi: 10.1128/JVI.00840-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen A., Zink M.C., Mankowski J.L., et al. Resting CD4+ T lymphocytes but not thymocytes provide a latent viral reservoir in a simian immunodeficiency virus-Macaca nemestrina model of human immunodeficiency virus type 1-infected patients on highly active antiretroviral therapy. J. Virol. 2003;77(8):4938–4949. doi: 10.1128/JVI.77.8.4938-4949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitney J.B., Hill A.L., Sanisetty S., et al. Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature. 2014;512(7512):74–77. doi: 10.1038/nature13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eriksson S., Graf E.H., Dahl V., et al. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog. 2013;9(2):: e1003174. doi: 10.1371/journal.ppat.1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eisele E., Siliciano R.F. Redefining the viral reservoirs that prevent HIV-1 eradication. Immunity. 2012;37(3):377–388. doi: 10.1016/j.immuni.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arfi V., Rivière L., Jarrosson-Wuillème L., et al. Characterization of the early steps of infection of primary blood monocytes by human immunodeficiency virus type 1. J. Virol. 2008;82(13):6557–6565. doi: 10.1128/JVI.02321-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Babas T., Muñoz D., Mankowski J.L., Tarwater P.M., Clements J.E., Zink M.C. Role of microglial cells in selective replication of simian immunodeficiency virus genotypes in the brain. J. Virol. 2003;77(1):208–216. doi: 10.1128/JVI.77.1.208-216.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cribbs S.K., Lennox J., Caliendo A.M., Brown L.A., Guidot D.M. Healthy HIV-1-infected individuals on highly active antiretroviral therapy harbor HIV-1 in their alveolar macrophages. AIDS Res. Hum. Retroviruses. 2015;31(1):64–70. doi: 10.1089/aid.2014.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gartner S., Markovits P., Markovitz D.M., Kaplan M.H., Gallo R.C., Popovic M. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science. 1986;233(4760):215–219. doi: 10.1126/science.3014648. [DOI] [PubMed] [Google Scholar]
- 41.González-Scarano F., Martín-García J. The neuropathogenesis of AIDS. Nat. Rev. Immunol. 2005;5(1):69–81. doi: 10.1038/nri1527. [DOI] [PubMed] [Google Scholar]
- 42.Honeycutt J.B., Wahl A., Baker C., et al. Macrophages sustain HIV replication in vivo independently of T cells. J. Clin. Invest. 2016;126(4):1353–1366. doi: 10.1172/JCI84456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Igarashi T., Brown C.R., Endo Y., et al. Macrophage are the principal reservoir and sustain high virus loads in rhesus macaques after the depletion of CD4+ T cells by a highly pathogenic simian immunodeficiency virus/HIV type 1 chimera (SHIV): Implications for HIV-1 infections of humans. Proc. Natl. Acad. Sci. USA. 2001;98(2):658–663. doi: 10.1073/pnas.98.2.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koenig S., Gendelman H.E., Orenstein J.M., et al. Detection of AIDS virus in macrophages in brain tissue from AIDS patients with encephalopathy. Science. 1986;233(4768):1089–1093. doi: 10.1126/science.3016903. [DOI] [PubMed] [Google Scholar]
- 45.Peng G., Greenwell-Wild T., Nares S., et al. Myeloid differentiation and susceptibility to HIV-1 are linked to APOBEC3 expression. Blood. 2007;110(1):393–400. doi: 10.1182/blood-2006-10-051763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Redel L., Le Douce V., Cherrier T., et al. HIV-1 regulation of latency in the monocyte-macrophage lineage and in CD4+ T lymphocytes. J. Leukoc. Biol. 2010;87(4):575–588. doi: 10.1189/jlb.0409264. [DOI] [PubMed] [Google Scholar]
- 47.Schnell G., Joseph S., Spudich S., Price R.W., Swanstrom R. HIV-1 replication in the central nervous system occurs in two distinct cell types. PLoS Pathog. 2011;7(10):: e1002286. doi: 10.1371/journal.ppat.1002286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schnell G., Spudich S., Harrington P., Price R.W., Swanstrom R. Compartmentalized human immunodeficiency virus type 1 originates from long-lived cells in some subjects with HIV-1-associated dementia. PLoS Pathog. 2009;5(4):: e1000395. doi: 10.1371/journal.ppat.1000395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Margolick J.B., Volkman D.J., Folks T.M., Fauci A.S. Amplification of HTLV-III/LAV infection by antigen-induced activation of T cells and direct suppression by virus of lymphocyte blastogenic responses. J. Immunol. 1987;138(6):1719–1723. [PubMed] [Google Scholar]
- 50.Zhang Z., Schuler T., Zupancic M., et al. Sexual transmission and propagation of SIV and HIV in resting and activated CD4+ T cells. Science. 1999;286(5443):1353–1357. doi: 10.1126/science.286.5443.1353. [DOI] [PubMed] [Google Scholar]
- 51.Coffin J., Swanstrom R. HIV pathogenesis: dynamics and genetics of viral populations and infected cells. Cold Spring Harb. Perspect. Med. 2013;3(1):: a012526. doi: 10.1101/cshperspect.a012526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perelson A.S., Neumann A.U., Markowitz M., Leonard J.M., Ho D.D. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271(5255):1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 53.Eckstein D.A., Penn M.L., Korin Y.D., et al. HIV-1 actively replicates in naive CD4(+) T cells residing within human lymphoid tissues. Immunity. 2001;15(4):671–682. doi: 10.1016/S1074-7613(01)00217-5. [DOI] [PubMed] [Google Scholar]
- 54.Murray A.J., Kwon K.J., Farber D.L., Siliciano R.F. The Latent Reservoir for HIV-1: How Immunologic Memory and Clonal Expansion Contribute to HIV-1 Persistence. J. Immunol. 2016;197(2):407–417. doi: 10.4049/jimmunol.1600343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alkhatib G., Combadiere C., Broder C.C., et al. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272(5270):1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 56.Choe H., Farzan M., Sun Y., et al. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85(7):1135–1148. doi: 10.1016/S0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 57.Deng H., Liu R., Ellmeier W., et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381(6584):661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 58.Dragic T., Litwin V., Allaway G.P., et al. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381(6584):667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 59.Trkola A., Dragic T., Arthos J., et al. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384(6605):184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 60.Wu L., Gerard N.P., Wyatt R., et al. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384(6605):179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 61.Bleul C.C., Wu L., Hoxie J.A., Springer T.A., Mackay C.R. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc. Natl. Acad. Sci. USA. 1997;94(5):1925–1930. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mohammadi P., Desfarges S., Bartha I., et al. 24 hours in the life of HIV-1 in a T cell line. PLoS Pathog. 2013;9(1):: e1003161. doi: 10.1371/journal.ppat.1003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adams M., Sharmeen L., Kimpton J., et al. Cellular latency in human immunodeficiency virus-infected individuals with high CD4 levels can be detected by the presence of promoter-proximal transcripts. Proc. Natl. Acad. Sci. USA. 1994;91(9):3862–3866. doi: 10.1073/pnas.91.9.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Böhnlein E., Lowenthal J.W., Siekevitz M., Ballard D.W., Franza B.R., Greene W.C. The same inducible nuclear proteins regulates mitogen activation of both the interleukin-2 receptor-alpha gene and type 1 HIV. Cell. 1988;53(5):827–836. doi: 10.1016/0092-8674(88)90099-2. [DOI] [PubMed] [Google Scholar]
- 65.Duh E.J., Maury W.J., Folks T.M., Fauci A.S., Rabson A.B. Tumor necrosis factor alpha activates human immunodeficiency virus type 1 through induction of nuclear factor binding to the NF-kappa B sites in the long terminal repeat. Proc. Natl. Acad. Sci. USA. 1989;86(15):5974–5978. doi: 10.1073/pnas.86.15.5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kinoshita S., Chen B.K., Kaneshima H., Nolan G.P. Host control of HIV-1 parasitism in T cells by the nuclear factor of activated T cells. Cell. 1998;95(5):595–604. doi: 10.1016/S0092-8674(00)81630-X. [DOI] [PubMed] [Google Scholar]
- 67.Lin X., Irwin D., Kanazawa S., et al. Transcriptional profiles of latent human immunodeficiency virus in infected individuals: effects of Tat on the host and reservoir. J. Virol. 2003;77(15):8227–8236. doi: 10.1128/JVI.77.15.8227-8236.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nabel G., Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature. 1987;326(6114):711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- 69.Pessler F., Cron R.Q. Reciprocal regulation of the nuclear factor of activated T cells and HIV-1. Genes Immun. 2004;5(3):158–167. doi: 10.1038/sj.gene.6364047. [DOI] [PubMed] [Google Scholar]
- 70.Rice A.P., Herrmann C.H. Regulation of TAK/P-TEFb in CD4+ T lymphocytes and macrophages. Curr. HIV Res. 2003;1(4):395–404. doi: 10.2174/1570162033485159. [DOI] [PubMed] [Google Scholar]
- 71.Ahmed R., Gray D. Immunological memory and protective immunity: understanding their relation. Science. 1996;272(5258):54–60. doi: 10.1126/science.272.5258.54. [DOI] [PubMed] [Google Scholar]
- 72.Ho D.D., Neumann A.U., Perelson A.S., Chen W., Leonard J.M., Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373(6510):123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 73.Wei X., Ghosh S.K., Taylor M.E., et al. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373(6510):117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 74.Cooper A., García M., Petrovas C., Yamamoto T., Koup R.A., Nabel G.J. HIV-1 causes CD4 cell death through DNA-dependent protein kinase during viral integration. Nature. 2013;498(7454):376–379. doi: 10.1038/nature12274. [DOI] [PubMed] [Google Scholar]
- 75.Sakai K., Dimas J., Lenardo M.J. The Vif and Vpr accessory proteins independently cause HIV-1-induced T cell cytopathicity and cell cycle arrest. Proc. Natl. Acad. Sci. USA. 2006;103(9):3369–3374. doi: 10.1073/pnas.0509417103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Borrow P., Lewicki H., Wei X., et al. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 1997;3(2):205–211. doi: 10.1038/nm0297-205. [DOI] [PubMed] [Google Scholar]
- 77.Koup R.A., Safrit J.T., Cao Y., et al. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 1994;68(7):4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schmitz J.E., Kuroda M.J., Santra S., et al. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283(5403):857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 79.Walker B.D., Chakrabarti S., Moss B., et al. HIV-specific cytotoxic T lymphocytes in seropositive individuals. Nature. 1987;328(6128):345–348. doi: 10.1038/328345a0. [DOI] [PubMed] [Google Scholar]
- 80.Klatt N.R., Shudo E., Ortiz A.M., et al. CD8+ lymphocytes control viral replication in SIVmac239-infected rhesus macaques without decreasing the lifespan of productively infected cells. PLoS Pathog. 2010;6(1):: e1000747. doi: 10.1371/journal.ppat.1000747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wong J.K., Strain M.C., Porrata R., et al. In vivo CD8+ T-cell suppression of siv viremia is not mediated by CTL clearance of productively infected cells. PLoS Pathog. 2010;6(1):: e1000748. doi: 10.1371/journal.ppat.1000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yoder A., Yu D., Dong L., et al. HIV envelope-CXCR4 signaling activates cofilin to overcome cortical actin restriction in resting CD4 T cells. Cell. 2008;134(5):782–792. doi: 10.1016/j.cell.2008.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Baldauf H.M., Pan X., Erikson E., et al. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat. Med. 2012;18(11):1682–1687. doi: 10.1038/nm.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Berger A., Sommer A.F., Zwarg J., et al. SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutières syndrome are highly susceptible to HIV-1 infection. PLoS Pathog. 2011;7(12):: e1002425. doi: 10.1371/journal.ppat.1002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Laguette N., Sobhian B., Casartelli N., et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474(7353):654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pierson T.C., Zhou Y., Kieffer T.L., Ruff C.T., Buck C., Siliciano R.F. Molecular characterization of preintegration latency in human immunodeficiency virus type 1 infection. J. Virol. 2002;76(17):8518–8531. doi: 10.1128/JVI.76.17.8518-8513.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Taylor H.E., Simmons G.E., Jr, Mathews T.P., et al. Phospholipase D1 Couples CD4+ T Cell Activation to c-Myc-Dependent Deoxyribonucleotide Pool Expansion and HIV-1 Replication. PLoS Pathog. 2015;11(5):: e1004864. doi: 10.1371/journal.ppat.1004864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zack J.A., Arrigo S.J., Weitsman S.R., Go A.S., Haislip A., Chen I.S. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61(2):213–222. doi: 10.1016/0092-8674(90)90802-L. [DOI] [PubMed] [Google Scholar]
- 89.Doitsh G., Galloway N.L., Geng X., et al. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature. 2014;505(7484):509–514. doi: 10.1038/nature12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Monroe K.M., Yang Z., Johnson J.R., et al. IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science. 2014;343(6169):428–432. doi: 10.1126/science.1243640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Muñoz-Arias I., Doitsh G., Yang Z., Sowinski S., Ruelas D., Greene W.C. Blood-Derived CD4 T Cells Naturally Resist Pyroptosis during Abortive HIV-1 Infection. Cell Host Microbe. 2015;18(4):463–470. doi: 10.1016/j.chom.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.He G., Ylisastigui L., Margolis D.M. The regulation of HIV-1 gene expression: the emerging role of chromatin. DNA Cell Biol. 2002;21(10):697–705. doi: 10.1089/104454902760599672. [DOI] [PubMed] [Google Scholar]
- 93.West M.J., Lowe A.D., Karn J. Activation of human immunodeficiency virus transcription in T cells revisited: NF-kappaB p65 stimulates transcriptional elongation. J. Virol. 2001;75(18):8524–8537. doi: 10.1128/JVI.75.18.8524-8537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ylisastigui L., Archin N.M., Lehrman G., Bosch R.J., Margolis D.M. Coaxing HIV-1 from resting CD4 T cells: histone deacetylase inhibition allows latent viral expression. AIDS. 2004;18(8):1101–1108. doi: 10.1097/00002030-200405210-00003. [DOI] [PubMed] [Google Scholar]
- 95.Gasper D.J., Tejera M.M., Suresh M. CD4 T-cell memory generation and maintenance. Crit. Rev. Immunol. 2014;34(2):121–146. doi: 10.1615/CritRevImmunol.2014010373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Harrington L.E., Janowski K.M., Oliver J.R., Zajac A.J., Weaver C.T. Memory CD4 T cells emerge from effector T-cell progenitors. Nature. 2008;452(7185):356–360. doi: 10.1038/nature06672. [DOI] [PubMed] [Google Scholar]
- 97.Kaech S.M., Wherry E.J., Ahmed R. Effector and memory T-cell differentiation: implications for vaccine development. Nat. Rev. Immunol. 2002;2(4):251–262. doi: 10.1038/nri778. [DOI] [PubMed] [Google Scholar]
- 98.Marshall H.D., Chandele A., Jung Y.W., et al. Differential expression of Ly6C and T-bet distinguish effector and memory Th1 CD4(+) cell properties during viral infection. Immunity. 2011;35(4):633–646. doi: 10.1016/j.immuni.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pepper M., Jenkins M.K. Origins of CD4(+) effector and central memory T cells. Nat. Immunol. 2011;12(6):467–471. doi: 10.1038/ni.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pepper M., Linehan J.L., Pagán A.J., et al. Different routes of bacterial infection induce long-lived TH1 memory cells and short-lived TH17 cells. Nat. Immunol. 2010;11(1):83–89. doi: 10.1038/ni.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Topham D.J., Doherty P.C. Longitudinal analysis of the acute Sendai virus-specific CD4+ T cell response and memory. J. Immunol. 1998;161(9):4530–4535. [PubMed] [Google Scholar]
- 102.Lanzavecchia A., Sallusto F. Progressive differentiation and selection of the fittest in the immune response. Nat. Rev. Immunol. 2002;2(12):982–987. doi: 10.1038/nri959. [DOI] [PubMed] [Google Scholar]
- 103.Chang J.T. Polarity and lymphocyte fate determination. Curr. Opin. Cell Biol. 2012;24(4):526–533. doi: 10.1016/j.ceb.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Choi Y.S., Kageyama R., Eto D., et al. ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity. 2011;34(6):932–946. doi: 10.1016/j.immuni.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bosque A., Planelles V. Induction of HIV-1 latency and reactivation in primary memory CD4+ T cells. Blood. 2009;113(1):58–65. doi: 10.1182/blood-2008-07-168393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sahu G.K., Lee K., Ji J., Braciale V., Baron S., Cloyd M.W. A novel in vitro system to generate and study latently HIV-infected long-lived normal CD4+ T-lymphocytes. Virology. 2006;355(2):127–137. doi: 10.1016/j.virol.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 107.Saleh S., Solomon A., Wightman F., Xhilaga M., Cameron P.U., Lewin S.R. CCR7 ligands CCL19 and CCL21 increase permissiveness of resting memory CD4+ T cells to HIV-1 infection: a novel model of HIV-1 latency. Blood. 2007;110(13):4161–4164. doi: 10.1182/blood-2007-06-097907. [DOI] [PubMed] [Google Scholar]
- 108.Tyagi M., Pearson R.J., Karn J. Establishment of HIV latency in primary CD4+ cells is due to epigenetic transcriptional silencing and P-TEFb restriction. J. Virol. 2010;84(13):6425–6437. doi: 10.1128/JVI.01519-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yang H.C., Xing S., Shan L., et al. Small-molecule screening using a human primary cell model of HIV latency identifies compounds that reverse latency without cellular activation. J. Clin. Invest. 2009;119(11):3473–3486. doi: 10.1172/JCI39199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Restifo N.P., Gattinoni L. Lineage relationship of effector and memory T cells. Curr. Opin. Immunol. 2013;25(5):556–563. doi: 10.1016/j.coi.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gattinoni L., Lugli E., Ji Y., et al. A human memory T cell subset with stem cell-like properties. Nat. Med. 2011;17(10):1290–1297. doi: 10.1038/nm.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sallusto F., Lanzavecchia A. Memory in disguise. Nat. Med. 2011;17(10):1182–1183. doi: 10.1038/nm.2502. [DOI] [PubMed] [Google Scholar]
- 113.Sallusto F., Lanzavecchia A. Heterogeneity of CD4+ memory T cells: functional modules for tailored immunity. Eur. J. Immunol. 2009;39(8):2076–2082. doi: 10.1002/eji.200939722. [DOI] [PubMed] [Google Scholar]
- 114.Carbone F.R. Tissue-resident memory t cells and fixed immune surveillance in nonlymphoid organs. J. Immunol. 2015;195(1):17–22. doi: 10.4049/jimmunol.1500515. [DOI] [PubMed] [Google Scholar]
- 115.Ebert A., Hill L., Busslinger M. Spatial Regulation of V-(D)J Recombination at Antigen Receptor Loci. Adv. Immunol. 2015;128:93–121. doi: 10.1016/bs.ai.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 116.Kalyan S., Kabelitz D. Defining the nature of human γδ T cells: a biographical sketch of the highly empathetic. Cell. Mol. Immunol. 2013;10(1):21–29. doi: 10.1038/cmi.2012.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kovacs J.A., Lempicki R.A., Sidorov I.A., et al. Induction of prolonged survival of CD4+ T lymphocytes by intermittent IL-2 therapy in HIV-infected patients. J. Clin. Invest. 2005;115(8):2139–2148. doi: 10.1172/JCI23196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Soriano-Sarabia N., Bateson R.E., Dahl N.P., et al. Quantitation of replication-competent HIV-1 in populations of resting CD4+ T cells. J. Virol. 2014;88(24):14070–14077. doi: 10.1128/JVI.01900-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Vandergeeten C., Fromentin R., DaFonseca S., et al. Interleukin-7 promotes HIV persistence during antiretroviral therapy. Blood. 2013;121(21):4321–4329. doi: 10.1182/blood-2012-11-465625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bosque A., Famiglietti M., Weyrich A.S., Goulston C., Planelles V. Homeostatic proliferation fails to efficiently reactivate HIV-1 latently infected central memory CD4+ T cells. PLoS Pathog. 2011;7(10):: e1002288. doi: 10.1371/journal.ppat.1002288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sáez-Cirión A., Bacchus C., Hocqueloux L., et al. Post-treatment HIV-1 controllers with a long-term virological remission after the interruption of early initiated antiretroviral therapy ANRS VISCONTI Study. PLoS Pathog. 2013;9(3):: e1003211. doi: 10.1371/journal.ppat.1003211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Buzon M.J., Sun H., Li C., et al. HIV-1 persistence in CD4+ T cells with stem cell-like properties. Nat. Med. 2014;20(2):139–142. doi: 10.1038/nm.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Muranski P., Restifo N.P. Essentials of Th17 cell commitment and plasticity. Blood. 2013;121(13):2402–2414. doi: 10.1182/blood-2012-09-378653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Baron B.W., Desai M., Baber L.J., et al. BCL6 can repress transcription from the human immunodeficiency virus type I promoter/enhancer region. Genes Chromosomes Cancer. 1997;19(1):14–21. doi: 10.1002/(SICI)1098-2264(199705)19:1<14:AID-GCC3>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 125.Holmes D., Knudsen G., Mackey-Cushman S., Su L. FoxP3 enhances HIV-1 gene expression by modulating NFkappaB occupancy at the long terminal repeat in human T cells. J. Biol. Chem. 2007;282(22):15973–15980. doi: 10.1074/jbc.M702051200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yang Z., Engel J.D. Human T cell transcription factor GATA-3 stimulates HIV-1 expression. Nucleic Acids Res. 1993;21(12):2831–2836. doi: 10.1093/nar/21.12.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Muranski P., Borman Z.A., Kerkar S.P., et al. Th17 cells are long lived and retain a stem cell-like molecular signature. Immunity. 2011;35(6):972–985. doi: 10.1016/j.immuni.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wei S., Zhao E., Kryczek I., Zou W. Th17 cells have stem cell-like features and promote long-term immunity. OncoImmunology. 2012;1(4):516–519. doi: 10.4161/onci.19440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sun H., Kim D., Li X., et al. Th1/17 Polarization of CD4 T Cells Supports HIV-1 Persistence during Antiretroviral Therapy. J. Virol. 2015;89(22):11284–11293. doi: 10.1128/JVI.01595-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cleret-Buhot A., Zhang Y., Planas D., et al. Identification of novel HIV-1 dependency factors in primary CCR4(+)CCR6(+)Th17 cells via a genome-wide transcriptional approach. Retrovirology. 2015;12:102. doi: 10.1186/s12977-015-0226-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gosselin A., Monteiro P., Chomont N., et al. Peripheral blood CCR4+CCR6+ and CXCR3+CCR6+CD4+ T cells are highly permissive to HIV-1 infection. J. Immunol. 2010;184(3):1604–1616. doi: 10.4049/jimmunol.0903058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Connick E., Folkvord J.M., Lind K.T., et al. Compartmentalization of simian immunodeficiency virus replication within secondary lymphoid tissues of rhesus macaques is linked to disease stage and inversely related to localization of virus-specific CTL. J. Immunol. 2014;193(11):5613–5625. doi: 10.4049/jimmunol.1401161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fukazawa Y., Lum R., Okoye A.A., et al. B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers. Nat. Med. 2015;21(2):132–139. doi: 10.1038/nm.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hong J.J., Amancha P.K., Rogers K., Ansari A.A., Villinger F. Spatial alterations between CD4(+) T follicular helper, B, and CD8(+) T cells during simian immunodeficiency virus infection: T/B cell homeostasis, activation, and potential mechanism for viral escape. J. Immunol. 2012;188(7):3247–3256. doi: 10.4049/jimmunol.1103138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Perreau M., Savoye A.L., De Crignis E., et al. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J. Exp. Med. 2013;210(1):143–156. doi: 10.1084/jem.20121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41(4):529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tran T.A., de Goër de Herve M.G., Hendel-Chavez H., et al. Resting regulatory CD4 T cells: a site of HIV persistence in patients on long-term effective antiretroviral therapy. PLoS One. 2008;3(10):: e3305. doi: 10.1371/journal.pone.0003305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Soriano-Sarabia N., Archin N.M., Bateson R., et al. Peripheral Vγ9Vδ2 T Cells Are a Novel Reservoir of Latent HIV Infection. PLoS Pathog. 2015;11(10):: e1005201. doi: 10.1371/journal.ppat.1005201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Couturier J., Suliburk J.W., Brown J.M., et al. Human adipose tissue as a reservoir for memory CD4+ T cells and HIV. AIDS. 2015;29(6):667–674. doi: 10.1097/QAD.0000000000000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Damouche A., Lazure T., Avettand-Fènoël V., et al. Adipose Tissue Is a Neglected Viral Reservoir and an Inflammatory Site during Chronic HIV and SIV Infection. PLoS Pathog. 2015;11(9):: e1005153. doi: 10.1371/journal.ppat.1005153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Furtado M.R., Callaway D.S., Phair J.P., et al. Persistence of HIV-1 transcription in peripheral-blood mononuclear cells in patients receiving potent antiretroviral therapy. N. Engl. J. Med. 1999;340(21):1614–1622. doi: 10.1056/NEJM199905273402102. [DOI] [PubMed] [Google Scholar]
- 142.Koelsch K.K., Liu L., Haubrich R., et al. Dynamics of total, linear nonintegrated, and integrated HIV-1 DNA in vivo and in vitro. J. Infect. Dis. 2008;197(3):411–419. doi: 10.1086/525283. [DOI] [PubMed] [Google Scholar]
- 143.Morand-Joubert L., Marcellin F., Launay O., et al. Contribution of cellular HIV-1 DNA quantification to the efficacy analysis of antiretroviral therapy: a randomized comparison of 2 regimens, including 3 drugs from 2 or 3 classes (TRIANON, ANRS 081). J. Acquir. Immune Defic. Syndr. 2005;38(3):268–276. [PubMed] [Google Scholar]
- 144.Dornadula G., Zhang H., VanUitert B., et al. Residual HIV-1 RNA in blood plasma of patients taking suppressive highly active antiretroviral therapy. JAMA. 1999;282(17):1627–1632. doi: 10.1001/jama.282.17.1627. [DOI] [PubMed] [Google Scholar]
- 145.Maldarelli F., Palmer S., King M.S., et al. ART suppresses plasma HIV-1 RNA to a stable set point predicted by pretherapy viremia. PLoS Pathog. 2007;3(4):: e46. doi: 10.1371/journal.ppat.0030046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Palmer S., Wiegand A.P., Maldarelli F., et al. New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 2003;41(10):4531–4536. doi: 10.1128/JCM.41.10.4531-4536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Dinoso J.B., Kim S.Y., Wiegand A.M., et al. Treatment intensification does not reduce residual HIV-1 viremia in patients on highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA. 2009;106(23):9403–9408. doi: 10.1073/pnas.0903107106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gandhi R.T., Zheng L., Bosch R.J., et al. The effect of raltegravir intensification on low-level residual viremia in HIV-infected patients on antiretroviral therapy: a randomized controlled trial. PLoS Med. 2010;7(8):: e1000321. doi: 10.1371/journal.pmed.1000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.McMahon D., Jones J., Wiegand A., et al. Short-course raltegravir intensification does not reduce persistent low-level viremia in patients with HIV-1 suppression during receipt of combination antiretroviral therapy. Clin. Infect. Dis. 2010;50(6):912–919. doi: 10.1086/650749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Besson G.J., Lalama C.M., Bosch R.J., et al. HIV-1 DNA decay dynamics in blood during more than a decade of suppressive antiretroviral therapy. Clin. Infect. Dis. 2014;59(9):1312–1321. doi: 10.1093/cid/ciu585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang X., Mink G., Lin D., Song X., Rong L. Influence of raltegravir intensification on viral load and 2-LTR dynamics in HIV patients on suppressive antiretroviral therapy. J. Theor. Biol. 2017;416:16–27. doi: 10.1016/j.jtbi.2016.12.015. [DOI] [PubMed] [Google Scholar]
- 152.Bailey J.R., Sedaghat A.R., Kieffer T., et al. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J. Virol. 2006;80(13):6441–6457. doi: 10.1128/JVI.00591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hermankova M., Ray S.C., Ruff C., et al. HIV-1 drug resistance profiles in children and adults with viral load of <50 copies/ml receiving combination therapy. JAMA. 2001;286(2):196–207. doi: 10.1001/jama.286.2.196. [DOI] [PubMed] [Google Scholar]
- 154.Kieffer T.L., Finucane M.M., Nettles R.E., et al. Genotypic analysis of HIV-1 drug resistance at the limit of detection: virus production without evolution in treated adults with undetectable HIV loads. J. Infect. Dis. 2004;189(8):1452–1465. doi: 10.1086/382488. [DOI] [PubMed] [Google Scholar]
- 155.Nettles R.E., Kieffer T.L., Kwon P., et al. Intermittent HIV-1 viremia (Blips) and drug resistance in patients receiving HAART. JAMA. 2005;293(7):817–829. doi: 10.1001/jama.293.7.817. [DOI] [PubMed] [Google Scholar]
- 156.Persaud D., Siberry G.K., Ahonkhai A., et al. Continued production of drug-sensitive human immunodeficiency virus type 1 in children on combination antiretroviral therapy who have undetectable viral loads. J. Virol. 2004;78(2):968–979. doi: 10.1128/JVI.78.2.968-979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Anderson J.A., Archin N.M., Ince W., et al. Clonal sequences recovered from plasma from patients with residual HIV-1 viremia and on intensified antiretroviral therapy are identical to replicating viral RNAs recovered from circulating resting CD4+ T cells. J. Virol. 2011;85(10):5220–5223. doi: 10.1128/JVI.00284-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Tobin N.H., Learn G.H., Holte S.E., et al. Evidence that low-level viremias during effective highly active antiretroviral therapy result from two processes: expression of archival virus and replication of virus. J. Virol. 2005;79(15):9625–9634. doi: 10.1128/JVI.79.15.9625-9634.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wagner T.A., McKernan J.L., Tobin N.H., Tapia K.A., Mullins J.I., Frenkel L.M. An increasing proportion of monotypic HIV-1 DNA sequences during antiretroviral treatment suggests proliferation of HIV-infected cells. J. Virol. 2013;87(3):1770–1778. doi: 10.1128/JVI.01985-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hill A.L., Rosenbloom D.I., Fu F., Nowak M.A., Siliciano R.F. Predicting the outcomes of treatment to eradicate the latent reservoir for HIV-1. Proc. Natl. Acad. Sci. USA. 2014;111(37):13475–13480. doi: 10.1073/pnas.1406663111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Joos B., Fischer M., Kuster H., et al. HIV rebounds from latently infected cells, rather than from continuing low-level replication. Proc. Natl. Acad. Sci. USA. 2008;105(43):16725–16730. doi: 10.1073/pnas.0804192105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Henrich T.J., Hanhauser E., Marty F.M., et al. Antiretroviral-free HIV-1 remission and viral rebound after allogeneic stem cell transplantation: report of 2 cases. Ann. Intern. Med. 2014;161(5):319–327. doi: 10.7326/M14-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Henrich T.J., Hu Z., Li J.Z., et al. Long-term reduction in peripheral blood HIV type 1 reservoirs following reduced-intensity conditioning allogeneic stem cell transplantation. J. Infect. Dis. 2013;207(11):1694–1702. doi: 10.1093/infdis/jit086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kearney M., Maldarelli F., Shao W., et al. Human immunodeficiency virus type 1 population genetics and adaptation in newly infected individuals. J. Virol. 2009;83(6):2715–2727. doi: 10.1128/JVI.01960-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Keele B.F., Giorgi E.E., Salazar-Gonzalez J.F., et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. USA. 2008;105(21):7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Nickle D.C., Jensen M.A., Shriner D., et al. Evolutionary indicators of human immunodeficiency virus type 1 reservoirs and compartments. J. Virol. 2003;77(9):5540–5546. doi: 10.1128/JVI.77.9.5540-5546.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chun T.W., Nickle D.C., Justement J.S., et al. HIV-infected individuals receiving effective antiviral therapy for extended periods of time continually replenish their viral reservoir. J. Clin. Invest. 2005;115(11):3250–3255. doi: 10.1172/JCI26197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Cory T.J., Schacker T.W., Stevenson M., Fletcher C.V. Overcoming pharmacologic sanctuaries. Curr. Opin. HIV AIDS. 2013;8(3):190–195. doi: 10.1097/COH.0b013e32835fc68a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Fletcher C.V., Staskus K., Wietgrefe S.W., et al. Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc. Natl. Acad. Sci. USA. 2014;111(6):2307–2312. doi: 10.1073/pnas.1318249111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Huang Y., Hoque M.T., Jenabian M.A., et al. Antiretroviral drug transporters and metabolic enzymes in human testicular tissue: potential contribution to HIV-1 sanctuary site. J. Antimicrob. Chemother. 2016;71(7):1954–1965. doi: 10.1093/jac/dkw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lorenzo-Redondo R., Fryer H.R., Bedford T., et al. Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature. 2016;530(7588):51–56. doi: 10.1038/nature16933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Boritz E.A., Darko S., Swaszek L., et al. Multiple Origins of Virus Persistence during Natural Control of HIV Infection. Cell. 2016;166(4):1004–1015. doi: 10.1016/j.cell.2016.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kearney M.F., Wiegand A., Shao W., et al. Ongoing HIV Replication During ART Reconsidered. Open Forum Infect. Dis. 2017;4(3):: ofx173. doi: 10.1093/ofid/ofx173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Brodin J., Zanini F., Thebo L., et al. Establishment and stability of the latent HIV-1 DNA reservoir. eLife. 2016;5:5. doi: 10.7554/eLife.18889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Evering T.H., Mehandru S., Racz P., et al. Absence of HIV-1 evolution in the gut-associated lymphoid tissue from patients on combination antiviral therapy initiated during primary infection. PLoS Pathog. 2012;8(2):: e1002506. doi: 10.1371/journal.ppat.1002506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kearney M.F., Spindler J., Shao W., et al. Lack of detectable HIV-1 molecular evolution during suppressive antiretroviral therapy. PLoS Pathog. 2014;10(3):: e1004010. doi: 10.1371/journal.ppat.1004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hammarlund E., Lewis M.W., Hansen S.G., et al. Duration of antiviral immunity after smallpox vaccination. Nat. Med. 2003;9(9):1131–1137. doi: 10.1038/nm917. [DOI] [PubMed] [Google Scholar]
- 178.Takaki A., Wiese M., Maertens G., et al. Cellular immune responses persist and humoral responses decrease two decades after recovery from a single-source outbreak of hepatitis C. Nat. Med. 2000;6(5):578–582. doi: 10.1038/75063. [DOI] [PubMed] [Google Scholar]
- 179.Hellerstein M.K., Hoh R.A., Hanley M.B., et al. Subpopulations of long-lived and short-lived T cells in advanced HIV-1 infection. J. Clin. Invest. 2003;112(6):956–966. doi: 10.1172/JCI200317533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Mclean A.R., Michie C.A. In vivo estimates of division and death rates of human T lymphocytes. Proc. Natl. Acad. Sci. USA. 1995;92(9):3707–3711. doi: 10.1073/pnas.92.9.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Michie C.A., McLean A., Alcock C., Beverley P.C. Lifespan of human lymphocyte subsets defined by CD45 isoforms. Nature. 1992;360(6401):264–265. doi: 10.1038/360264a0. [DOI] [PubMed] [Google Scholar]
- 182.Vrisekoop N., den Braber I., de Boer A.B., et al. Sparse production but preferential incorporation of recently produced naive T cells in the human peripheral pool. Proc. Natl. Acad. Sci. USA. 2008;105(16):6115–6120. doi: 10.1073/pnas.0709713105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Surh C.D., Sprent J. Homeostasis of naive and memory T cells. Immunity. 2008;29(6):848–862. doi: 10.1016/j.immuni.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 184.Maldarelli F., Wu X., Su L., et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345(6193):179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Simonetti F.R., Sobolewski M.D., Fyne E., et al. Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc. Natl. Acad. Sci. USA. 2016;113(7):1883–1888. doi: 10.1073/pnas.1522675113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wagner T.A., McLaughlin S., Garg K., et al. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014;345(6196):570–573. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Katlama C., Lambert-Niclot S., Assoumou L., et al. Treatment intensification followed by interleukin-7 reactivates HIV without reducing total HIV DNA: a randomized trial. AIDS. 2016;30(2):221–230. doi: 10.1097/QAD.0000000000000894. [DOI] [PubMed] [Google Scholar]
- 188.Purton J.F., Tan J.T., Rubinstein M.P., Kim D.M., Sprent J., Surh C.D. Antiviral CD4+ memory T cells are IL-15 dependent. J. Exp. Med. 2007;204(4):951–961. doi: 10.1084/jem.20061805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Brenchley J.M., Ruff L.E., Casazza J.P., Koup R.A., Price D.A., Douek D.C. Preferential infection shortens the life span of human immunodeficiency virus-specific CD4+ T cells in vivo. J. Virol. 2006;80(14):6801–6809. doi: 10.1128/JVI.00070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Han Y., Lassen K., Monie D., et al. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J. Virol. 2004;78(12):6122–6133. doi: 10.1128/JVI.78.12.6122-6133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Schröder A.R., Shinn P., Chen H., Berry C., Ecker J.R., Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110(4):521–529. doi: 10.1016/S0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 192.Berry C.C., Gillet N.A., Melamed A., Gormley N., Bangham C.R., Bushman F.D. Estimating abundances of retroviral insertion sites from DNA fragment length data. Bioinformatics. 2012;28(6):755–762. doi: 10.1093/bioinformatics/bts004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Cohn L.B., Silva I.T., Oliveira T.Y., et al. HIV-1 integration landscape during latent and active infection. Cell. 2015;160(3):420–432. doi: 10.1016/j.cell.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Flucke U., Tops B.B., de Saint Aubain Somerhausen N., et al. Presence of C11orf95-MKL2 fusion is a consistent finding in chondroid lipomas: a study of eight cases. Histopathology. 2013;62(6):925–930. doi: 10.1111/his.12100. [DOI] [PubMed] [Google Scholar]
- 195.Kobayashi S., Taki T., Chinen Y., et al. Identification of IGHCδ-BACH2 fusion transcripts resulting from cryptic chromosomal rearrangements of 14q32 with 6q15 in aggressive B-cell lymphoma/leukemia. Genes Chromosomes Cancer. 2011;50(4):207–216. doi: 10.1002/gcc.20845. [DOI] [PubMed] [Google Scholar]
- 196.Muehlich S., Hampl V., Khalid S., et al. The transcriptional coactivators megakaryoblastic leukemia 1/2 mediate the effects of loss of the tumor suppressor deleted in liver cancer 1. Oncogene. 2012;31(35):3913–3923. doi: 10.1038/onc.2011.560. [DOI] [PubMed] [Google Scholar]
- 197.Ikeda T., Shibata J., Yoshimura K., Koito A., Matsushita S. Recurrent HIV-1 integration at the BACH2 locus in resting CD4+ T cell populations during effective highly active antiretroviral therapy. J. Infect. Dis. 2007;195(5):716–725. doi: 10.1086/510915. [DOI] [PubMed] [Google Scholar]
- 198.Lucic B., Chen H.C., Kuzman M., et al. Spatially clustered loci with multiple enhancers are frequent targets of HIV-1 integration. Nat. Commun. 2019;10(1):4059. doi: 10.1038/s41467-019-12046-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Simonetti FR, Spindler J, Wu X, Hill S, Shao W, Mellors JW, et al. Analysis of HIV proviurses in clonally expanded cells in vivo. conference on retrovirues and opportunistic infections (Boston). 127
- 200.Bruner K.M., Murray A.J., Pollack R.A., et al. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat. Med. 2016;22(9):1043–1049. doi: 10.1038/nm.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ho Y.C., Shan L., Hosmane N.N., et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155(3):540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Bender A.M., Simonetti F.R., Kumar M.R., Fray E.J., Bruner K.M., Timmons A.E., et al. The Landscape of Persistent Viral Genomes in ART-Treated SIV, SHIV, and HIV-2 Infections. Cell Host Microbe. 2019;26(1):73–85. doi: 10.1016/j.chom.2019.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Harris R.S., Dudley J.P. APOBECs and virus restriction. Virology. 2015;479-480:131–145. doi: 10.1016/j.virol.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Boltz V.F., Rausch J., Shao W., et al. Ultrasensitive single-genome sequencing: accurate, targeted, next generation sequencing of HIV-1 RNA. Retrovirology. 2016;13(1):87. doi: 10.1186/s12977-016-0321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Hiener B., Eden J.S., Horsburgh B.A., Palmer S. Amplification of near full-length HIV-1 proviruses for next-generation sequencing. J. Vis. Exp. 2018;(140):: e58016. doi: 10.3791/58016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Einkauf K.B., Lee G.Q., Gao C., et al. Intact HIV-1 proviruses accumulate at distinct chromosomal positions during prolonged antiretroviral therapy. J. Clin. Invest. 2019;129(3):988–998. doi: 10.1172/JCI124291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Patro S.C., Brandt L.D., Bale M.J., Halvas E.K., Joseph K.W., Shao W., et al. Combined HIV-1 sequence and integration site analysis informs viral dynamics and allows reconstruction of replicating viral ancestors. Proceedings of the National Academy of Sciences of the United States of America. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Lasken R.S. Single-cell genomic sequencing using Multiple Displacement Amplification. Curr. Opin. Microbiol. 2007;10(5):510–516. doi: 10.1016/j.mib.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 209.De Simone M., Rossetti G., Pagani M. Single Cell T Cell Receptor Sequencing: Techniques and Future Challenges. Front. Immunol. 2018;9:1638. doi: 10.3389/fimmu.2018.01638. [DOI] [PMC free article] [PubMed] [Google Scholar]