Abstract
Telomeres are structurally nucleoprotein complexes at termini of linear chromosomes and essential to chromosome stability/integrity. In normal human cells, telomere length erodes progressively with each round of cell divisions, which serves as an important barrier to uncontrolled proliferation and malignant transformation. In sharp contrast, telomere maintenance is a key feature of human malignant cells and required for their infinite proliferation and maintenance of other cancer hallmarks as well. Thus, a telomere-based anti-cancer strategy has long been suggested. However, clinically efficient and specific drugs targeting cancer telomere-maintenance have still been in their infancy thus far. To achieve this goal, it is highly necessary to elucidate how exactly cancer cells maintain functional telomeres. In the last two decades, numerous studies have provided profound mechanistic insights, and the identified mechanisms include the aberrant activation of telomerase or the alternative lengthening of telomere pathway responsible for telomere elongation, dysregulation and mutation of telomere-associated factors, and other telomere homeostasis-related signaling nodes. In the present review, these various strategies employed by malignant cells to regulate their telomere length, structure and function have been summarized, and potential implications of these findings in the rational development of telomere-based cancer therapy and other clinical applications for precision oncology have been discussed.
Keywords: Cancer therapy, Gene transcription, TERC, TERRA, TERT, TERT promoter mutation, Telomerase, Telomere
1. INTRODUCTION
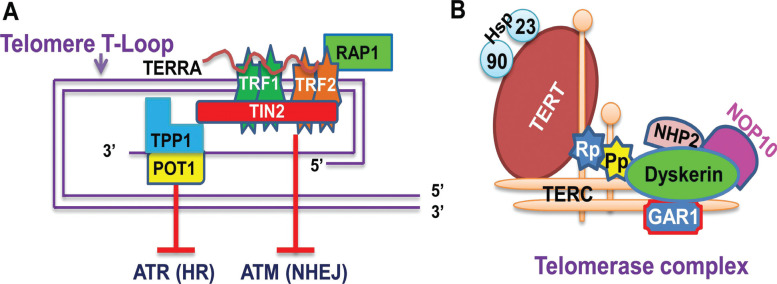
Human linear chromosomes terminate with arrays of TTAGGG sequences lasting 6 – 20 kb long so-called telomeric DNA, and these telomeric repeats normally harbor 3’-signle-stranded DNA overhang that invades into the duplexed region to generate T-loops [1, 2]. Such a DNA structure is further bound by a six protein-containing shelterin (TRF1, TRF2, RAP1, TIN2, TPP1 and POT1) to form a special nucleoprotein complex of telomeres [1, 2] (Fig. 1). The nucleoprotein structure of telomeres functions as a protective cap at the end of chromosomes to maintain genomic stability and integrity, which is primarily achieved by inhibiting the DNA damage response through the following mechanisms: First, the telomere structure hides the chromosome end being recognized as DNA breaks, and second, TRF2 retards Ataxia Telangiectasia-mutated (ATM) Signaling/Non-homologous End Joining (NHEJ), whereas POT1 inhibits Rad3-related (ATR) Signaling/Homologous Recombination (HR) [1, 2]. In addition, the telomere complex also contains long non-coding RNA transcribed from telomeric repetitive DNA (TERRA), a recently identified component required for the maintenance of functional telomere structure [3] (Fig. 1).
Fig. (1).
The telomere structure/shelterin association and telomerase complex. (A) Telomeric DNA association with the shelterin complex. Telomeric DNA is 8 – 20 kb long double-stranded TTAGGG repeats with a shorter G-rich, single stranded overhang. The overhang invades into the duplexed region to form T-loops. The shelterin complex is composed of six different factors, including TRF1 (telomeric repeat-binding factor 1), TRF2 (telomeric repeat-binding factor 2), RAP1 (repressor and activator protein 1), TIN2 (TRF1-interacting nuclear protein 2), POT1 (protection of telomeres 1) and TPP1 (TINT1/PTOP/PIT1) or ACD (adrendocortical dysplasia homolog). They bind to either double- (TRF1, TRF2, RAP1 and TIN2) or single-stranded telomeric DNA (TPP1 and POT1). TRF2 and POT1 inhibit ataxia telangiectasia-mutated (ATM) Signaling/Non-homologous End Joining (NHEJ) and Rad3-related (ATR) signaling, respectively, thereby preventing DNA damage response. TPP1/POT1 regulates telomerase accessibility to telomeres for telomeric DNA synthesis in telomerase-proficient cells. TERRA interacts with TRF1/2 and other factors associated with telomere chromatin, and it is required for the maintenance of normal telomere structure and function. (B) The schematic of the telomerase complex. TERT and TERC are the core of the enzyme and other components include dyskerin, NOP10, NHP2, GAR, repotin (RP), pontin (Pp), heat shock proteins (hsp) 90 and 23.
One essential hallmark of telomeres is their progressive attrition with successive divisions of human somatic cells due to the end-replication problem plus the lack or insufficiency of telomere-lengthening activities [1, 2]. When telomeres shorten to a threshold length to disrupt their capping function, the DNA damage response pathway is activated and cells are subsequently induced to enter a stable growth arrest status named replicative senescence or M1 stage [1, 4, 5]. Thus, telomere shortening acts as a mitotic O’clock, counting dividing times of cells and conferring them a limited lifespan. In sharp contrast, however, infinite proliferation is one of key hallmarks for cancer cells [6]. To acquire an immortal phenotype, malignant cells must overcome the senescence barrier by stabilizing their telomere length, and two major mechanisms have so far been identified to actively lengthen telomeres in cancer cells: activation of telomerase and Alternative-lengthening of Telomere (ALT) [1, 6-8]. The telomerase and ALT pathways are responsible for telomere length maintenance of ~ 90% and 5% - 10% of human cancers, respectively [7-9].
As described above, telomerase activation is the predominant strategy utilized by cancer cells to maintain their telomere length, and therefore has been one of the extensively studied areas in cancer research [1, 5, 7, 8]. Telomerase is an RNA-dependent DNA polymerase adding telomeric DNA onto chromosome ends [1, 7]. Although as a multi-unit complex, its core components only include the catalytic Subunit Telomerase Reverse Transcriptase (TERT) and a RNA component (TERC) as a template [1] (Fig. 1). TERC is ubiquitously expressed in human somatic cells, while the TERT gene is tightly repressed at the transcriptional level in the vast majority of normal human cells [1, 8, 10], which results in telomerase silence in these cells. During the tumorigenic process, the de-repression of the TERT gene and induction of TERT expression are required for telomerase activation [1, 8, 10]. Thus, TERT is a rate-limiting element to control telomerase activity. Moreover, in addition to its canonical telomere-lengthening function, TERT has recently been shown to exhibit multi-extratelomeric activities, which include its roles in DNA damage repair, transcription regulation, mitochondrial function, stem cell biology and among others [11-21]. All these telomere lengthening-dependent and independent functions significantly contribute to cancer hallmarks, and promote cancer initiation and progression.
In addition to the telomere lengthening pathways that actively elongate telomeric repeats, the shelterin proteins, TERRA, and other telomere-associated components also play an important part in the regulation of telomere length and function [2, 22]. For instance, manipulating TRF1 or TRF2 expression leads to significant changes in telomere length and structure without affecting telomerase activity [22, 23]. Moreover, aberrant alterations in the genetics and expression of shelterin factors have been observed in human cancer [2, 24, 25]. These cancer-related changes can remodel telomere chromatin, promote telomerase recruitment and accessibility to telomeres, increase genomic instability, and even exert extratelomeric activities, eventually facilitating cancer formation and/or progression.
Collectively, the last two decades have evidenced tremendous progress in cancer telomere biology. In the present article, the key findings in the cancer-associated telomere-maintenance research are summarized and how the current knowledge can be translated into the rational development of the telomere-based anti-cancer strategy and other potential applications for precision oncology is discussed.
2. TELOMERE DYNAMICS IN CANCER CELLS: SHORTER BUT STABILIZED
Cancer cells are known to maintain their telomere length via the activation of either telomerase or ALT for their infinite proliferation, but their telomeres are in general shorter than those in their normal counterparts [1, 5] (Fig. 2A). The co-existence of shorter telomeres and telomerase activity in cancer cells, seemingly paradoxical, is primarily attributable to late activation of telomerase during the oncogenic process [1, 5]. Another potential explanation is that individuals with shorter telomeres had increased cancer risk [26].
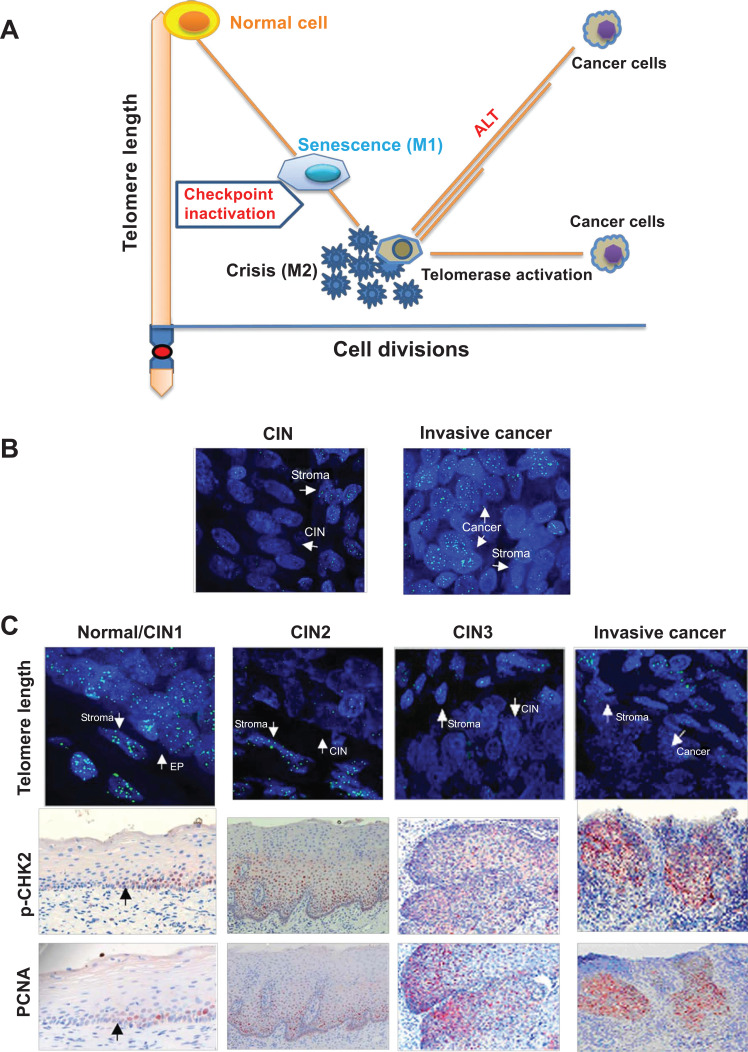
Fig. (2).
The mechanism underlying shorter but stabilized telomeres in human cancer. (A) Telomere dynamics during cellular proliferation, senescence, crisis and malignant transformation. Normal human somatic cells lose their telomeric DNA progressively with each round of cell divisions and critical short telomeres trigger senescence if the checkpoint signaling (TP53 and pRB) is intact. However, their inactivation induces cells to bypass the senescence barrier and to continue proliferation until the M2 crisis stage where genomic catastrophes occur. Most cells undergo apoptosis, while rare cells survive and acquire immortal phenotype through telomerase activation. The presence of telomerase activity stabilizes cell telomeres at a short balance even after their transformation. Under rare circumstances, the alternative lengthening of telomere (ALT) pathway may be activated and the cells have heterogeneous telomere length (Shown as different lengths of orange lines). (B) The ALT activation induces longer telomere in cervical tumors. The CIN and invasive cancer samples from the same patient are analyzed for telomere length using FISH. Telomere signals (length) are very weak in CIN, but became significantly stronger in fully transformed tumor derived from that same patient. The patient was confirmed with ALT in her tumor. (C) Significantly telomere shortening occurs already in precursors during the carcinogenesis of uterine cervix. Telomere length assessment using quantitative fluorescence in situ hybridization (Q-FISH) shows that telomere length (green signals) is comparable between normal cervical epithelial (EP) and stromal cells, but greatly reduced telomere signals are seen at CIN2 and 3, and telomerase-positive invasive cancer cells as well. Telomere shortening is coupled with the activation of DNA damage response and increased proliferation as indicated by positive staining of the phosphorylated CHK2 (pCHK2) and PCNA, respectively. Left: The normal/CIN1 and CIN2 samples were derived from the same patient and the black arrowheads indicate a transition site between normal and CIN1 tissues in the patient sample. Right: The CIN3 and cancer samples were derived from the same patient. The images in (B) and (C) are adapted from Oncogene [27] with permission from Springer/Nature.
As stated above, dividing normal human cells lose their telomeric repeats progressively due to the lack or insufficiency of telomere-maintaining activity and critical shortened (dysfunctional) telomeres activate the DNA damage response signal through which tumor suppressive/growth-inhibiting (checkpoint) genes are induced and cellular senescence subsequently occurs [1, 5]. In this process, the TP53-CDKN1A and CDKN2A-pRB checkpoint signalings are the major players to initiate stable growth arrest. However, proliferating cells frequently accumulate DNA mutations resulting from genetic insults and some of them may acquire the loss-of-function mutations of genes responsible for the DNA damage response, including TP53 and pRB [1, 5]. In that case, cells are capable of bypassing senescence and continuing to proliferate even if telomeres become dysfunctional until a mitotic catastrophe or M2 stage crisis occurs [1, 5]. Cells at M2 crisis are characterized by a massive genomic instability including loss of telomere capping, breakage fusion-bridge cycles, end-end fusions, rearrangements of chromosomes, and many other chromosome aberrations. The vast majority of these cells undergoes apoptosis, while few of them do survive genomic catastrophes due to TERT induction/telomerase activation. Activated telomerase stabilizes telomeres at a short balance and minimizes genomic instability to a tolerable level, thereby conferring cells the ability to proliferate infinitely, but usually insufficient to further increase telomere length to the pre-senescent level [1, 5] (Fig. 2A). In addition, the activation of the ALT pathway instead of telomerase may happen, although its frequency is much lower [1, 5, 9] (Fig. 2A and B). Nevertheless, the stabilization of telomere length and acquisition of immortality create a prerequisite for cells to undergo malignant transformation.
The M1 senescence and M2 crisis cell model above is not only demonstrated by in vitro cellular experiments but also supported by the findings from analyses of human tissues during a stepwise carcinogenesis process [27, 28]. In a previous study, telomere length, DNA damage response and proliferation during the evolution from precursor lesions (cervical intraepithelial neoplasia, CIN) have been examined to invasive cancers of the uterine cervix in sequential patient samples. Significantly shorter telomeres, the activated DNA damage response signaling and increased cellular proliferation were readily present in CIN tissues (Fig. 2C), while TERT induction occurred preferably at stages of CIN3 and in invasive cancers [27]. Shorter telomeres were coupled with the activation of DNA damage response and increased proliferation in both CINs and invasive cervical cancers (Fig. 2C). Such telomere erosions were similarly observed in other types of pre-malignant lesions and carcinomas [1, 5]. Based on the analysis of 18 430 cancerous materials derived from 31 different cancer types in The Cancer Genome Atlas (TCGA) dataset, Barthel et al. [7] further showed that telomerase-positive tumors had shorter telomeres than did their normal adjacent tissues, whereas those ALT-positive tumors carried heterogeneous lengths of telomeres, either longer, shorter or unchanged.
All the above in vitro and in vivo findings demonstrate that shorter telomeres coupled with telomerase activation are widely present in human cancers, which provide great advantages for the application of telomerase-based therapies in cancer patients. First, telomerase is silent in most human somatic cells, and targeting telomerase should thus be specific to cancer cells with minimal side-effects on normal ones. Second, tumor cells should be more sensitive to telomerase inhibition [5]. It is known that small subsets of normal human cells, including stem/progenitor cells, activated lymphocytes and other highly proliferative cells exhibit telomerase activity, and they may be potentially affected by telomerase inhibition. However, these cells carry significantly longer telomeres than do cancer cells, which offer a valuable therapeutic window: Telomere stabilization can be readily disrupted in tumor cells by telomerase perturbation long before detrimental side-effects occur to normal telomerase-positive human cells. Moreover, it was shown that the level of telomerase activity was only sufficient to maintain the shortest telomeres in most cancer cells and these cells might be even super-sensitive to telomerase inhibition [1]. However, this also indicates that minimal residual levels of telomerase activity may compromise the efficacy of telomerase-targeted therapies if the enzyme function is not fully abolished.
3. THE ACTIVATION OF TELOMERASE OR ALT IN CANCER: MECHANISTIC INSIGHTS
3.1. Telomerase Activation Through Multiple Mechanisms in Cancer
Present as a multi component-containing complex (Fig. 1), telomerase activity is theoretically subject to being regulated at numerous different levels. However, TERC and TERT are two essential subunits of the telomerase holy-enzyme, and have been shown as master determinants to govern telomerase activation in cancer. Therefore, most studies have been focused on these two telomerase components, especially the catalytic subunit TERT.
3.1.1. TERT Expression and Regulation in Normal Human Cells
Profund insights into the controlling mechanism for the TERT transcription in a physiological setting are very helpful to better understandings of the de-repression of the TERT gene during oncogenesis. The TERT gene is transcriptionally repressed in most differentiated human cells, which results in telomerase silencing [1, 8]. However, TERT is expressed in subsets of cells, including stem cells, activated lymphocytes and other cells with highly proliferative potentials [1, 8, 29]. The MYC network family transcription factors and WNT/β-catenin signaling have been shown as the key players to activate the TERT gene transcription and telomerase in these cells [30-34]. In addition, the TERT promoter bears estrogen response elements, and estrogen is capable of inducing TERT expression in certain types of estrogen receptor-expressing cells, such as lymphocytes, endometrial epithelial cells and endothelial cells [35-38]. Such TERT expression/telomerase activation is essential to maintain physiological activities and functions of these normal cells, while impaired TERT induction results in defective cell proliferation and/or differentiation, eventually leading to organ or tissue dysfunction [29, 32, 39].
3.1.2. Cancer-specific TERT Induction via Transcriptional De-repression
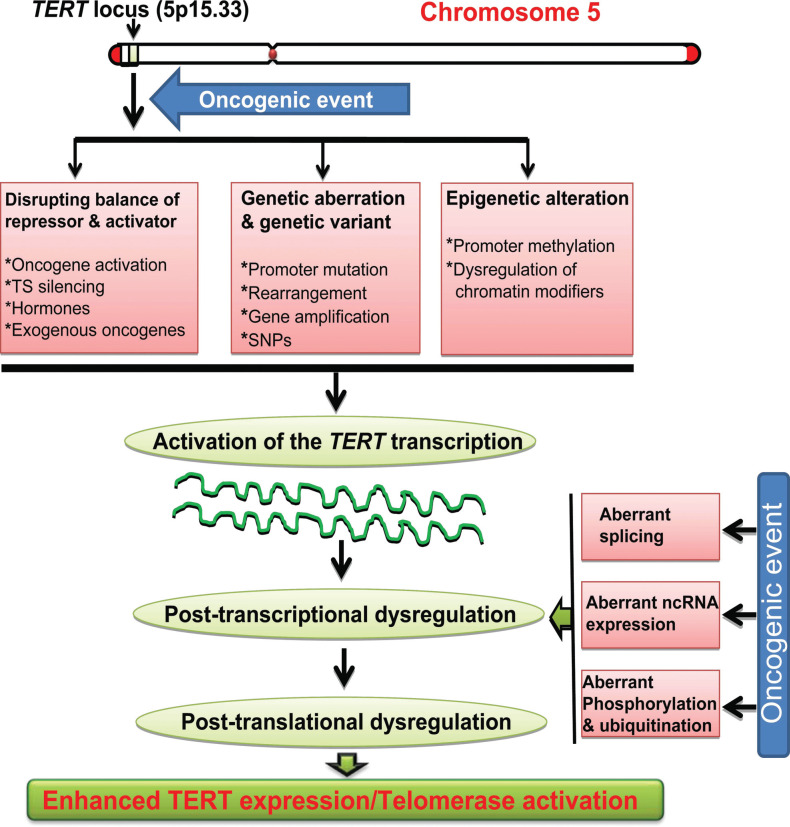
The transcriptional regulation of the TERT gene represents a fundamental mechanism to suppress telomerase activity in normal cells [19, 40, 41]. Induction of TERT expression by de-repressing this gene is an essential step to activate telomerase during oncogenesis [8, 19]. It has now been clear that various strategies are employed by malignant cells to stimulate TERT transcription and expression, which include the gain of oncogenic signals and/or loss of tumor suppression function, tumor virus-derived exogenous oncoproteins, and epigenetic and/or genetic alterations such as the promoter hypermethylation or mutation, gene rearrangements and amplification [8, 19] (Fig. 3).
Fig. (3).
The activation of the TERT gene transcription and telomerase mediated by oncogenic events during oncogenesis. Oncogenic factors disrupt the balance between TERT repressors and activators, and/or induce genetic aberrations, and epigenetic alterations at the TERT locus, thereby leading to the trans-activation of the TERT gene. These oncogenic factors may also promote aberrant mRNA splicing, dysregulation of non-coding RNAs (ncRNAs) and/or phosphorylation ubiqitination, through which TERT expression is further amplified at post-transcriptional and post-translational levels. Eventually, TERT expression and telomerase activity is induced in transformed cells. TS: Tumor suppressor.
3.1.2.1. Cellular or Viral Oncogenes and Tumor Suppressors
Given the critical role of TERT and telomerase in malignant transformation, it is natural that oncogenic factors target the TERT gene for transcription and telomerase activation. Indeed, MYC was the first identified cellular oncogene to possess such a function [30]. Consistently, the cloned TERT promoter did reveal the presence of two E-boxes (MYC binding motifs) in the proximal region required for the promoter activity [40, 41]. Further analyses showed that the MYC family network proteins (MYC/MAX/MAD1) were heavily involved in the regulation of the TERT transcription in both normal and cancerous cells [31, 34, 42]. Using a genetic screening system, Lin et al. identified the MYC antagonist MAD1 as one key negative regulator for TERT transcription in normal human cells [34]. It is well characterized that the MYC/MAX and MAX/MAD1 heterodimers compete to bind the E-box on their target gene promoters dependent on the abundance of MYC and MAD1 expression [34]. Using a differentiation model of human leukemic cells (HL60 cells), the dynamic changes in the MYC/MAX and MAD1/MAX occupancy on the TERT promoter and TERT expression during cellular terminal differentiation have been analyzed [42]. The MYC/MAX association with the promoter was coupled with high levels of MYC and TERT expression in undifferentiated HL60 cells, while MAD1 expression was up-regulated and the MAD1/MAX complex replaced MYC/MAX to occupy the TERT promoter accompanied by the diminished TERT expression upon terminal differentiation of these leukemia cells [42]. In contrast, the totally opposite scenario occurred in the in vitro conversion of normal human cells to malignant ones [43]. Clinically, MYC is overexpressed in almost half of the human malignancies, and moreover, certain factors may indirectly regulate the TERT transcription through the MYC/MAX/MAD1 network family, which collectively suggest the importance of the MYC-mediated TERT transcription in human carcinogenesis.
The Wnt/β-Catenin signaling is another good example. The dysregulated β-Catenin expression and its activating mutations are frequently observed in human cancer and function as a driver for malignant transformation and progression. In 2012, two groups showed that β-Catenin directly regulates the TERT transcription by binding to the TCF site in the TERT promoter [32, 33]. Upon β-Catenin binding, the lysine methyltransferase Setd1a is recruited to the promoter, catalyzing histone H3K4 trimethylation locally and initiating transcription [32]. TCF1, TCF4 and KLF4 may be also involved in β-Catenin-mediated TERT transcription [32, 33].
In addition to the MYC/MAX/MAD family and Wnt/β-Catenin pathways, other cancer-promoting factors, which include estrogen, HIFs, Ets, NFX1, Tax, DJ-1, E2F, COX2, survivin, FoxM1, Reptin, mutant FLT3, seminal fluids, growth factors and cytokines, directly or indirectly regulate the transcription of the TERT gene, as well [8, 21, 44-50]. On the other hand, it should be pointed out that tumor suppressors may transcriptionally inhibit TERT expression under physiological settings. Menin, wt p53, WT1, TGF-β and MAD1 act as strong repressors for the TERT gene in normal cells, while their inhibition or inactivation can lead to TERT re-expression [34, 51]. Indeed, during oncogenesis, the inactivation of tumor suppressors is widespread. A smarter strategy is even employed in renal cell carcinoma where the mutation of E-box on the TERT promoter erases the MAD1 binding and its inhibitory action [52]. Taken together, oncogene activation and/or loss of tumor suppression function coordinate to induce TERT expression in human cancer.
Intriguingly, cellular oncogenes may be hijacked by exogenous viral onco-proteins to induce TERT expression and to amplify their oncogenic effects. Infection with tumor viruses is believed to be related to 15% of human cancer, and these viruses include Human Papillomavirus (HPV), Hepatitis B Virus (HBV), Hepatitis C Virus (HCV), Epstein-Barr Virus (EBV), Cytomegalovirus (CMV), Kaposi sarcoma-associated Herpesvirus (KSHV), human T-cell Leukemia Virus-1 (HTLV-1) and others [53-55]. Among all these tumor viruses, the HPV-encoded E6 is a well-characterized viral oncogene to activate telomerase by inducing the TERT transcription. E6 and E6-associated protein interact with c-MYC to transcriptionally activate the TERT gene through the E-box-binding in the TERT core promoter [44, 55]. The CMV immediate-early protein 72 was shown to cooperate with the host transcription factor Sp1 to robustly promote TERT expression at the transcriptional level in glioblastoma cells [54]. Such targeted activation of TERT transcription might be one of the key strategies for virus-mediated carcinogenesis.
3.1.2.2. Epigenetic Alterations
The presence and recruitment/assembling of transcription factors to the TERT promoter are required for its transcription, but their access and binding to the promoter is tightly controlled by an epigenetic mechanism involved in DNA methylation, and histone acetylation, methylation and phosphorylation [8, 19, 31, 42, 56-60]. The TERT promoter is embedded in a CpG island and in general, unmethylated in normal human cells [8, 57, 58]. It is believed that the hypomethylated TERT promoter allows the binding of repressors to maintain the closed chromatin structure in those cells [58]. In sharp contrast, the TERT promoter is hypermethylated in cancer cells, and sequencing-based high-resolution mapping has recently identified the TERT Hypermethylated Oncological Region (THOR) as a key epigenetic mechanism to activate TERT expression [58]. THOR is a 433-bp genomic region covering 52 CpGs located immediately upstream of the TERT core promoter and responsible for telomerase activation in 90% of human malignancies [58].
However, such TERT promoter hypermethylation or THOR is only seen in cancer cells [58, 61]. Normal lymphocytes, when activated, express high levels of TERT mRNA and telomerase activity but lack the promoter hypermethylation [57, 61]. It is currently unclear how the THOR hypermethylation occurs in oncogenesis and why it is not required for the TERT trans-activation in normal human cells. Elucidating this issue may provide new strategies for telomerase-based cancer therapy.
The chromatin configuration is further governed by histone modifications. Histone acetylation/deacetylation, methylation/demethylation and phosphorylation/de-phosphorylation are all involved in the regulation of TERT transcription [8, 19, 31, 42, 56, 59, 60, 62]. It was previously shown that inhibiting histone deacetylases (HDACs) increased histone acetylation at the TERT promoter and thereby directly induced TERT expression in normal human cells lacking telomerase activity [42, 62]. The tumor suppressor MAD1 occupies the E-box in the TERT promoter and recruits HDACs to catalyze histone deacetylation through which repressive chromatin is maintained, while the oncogene MYC attracts histone acetyltransferases to acetylate histones associated with the TERT promoter [42, 62]. Therefore, histone acetylation/deacetylation is a common underlying feature of TERT transactivation/ repression in human cells. In addition, the histone methytransferase SMYD3 directly binds to (CCCTCC) motifs within the TERT proximal promoter and specifically catalyzes histone H3-K4 tri-methylation, thereby activating TERT transcription [60]. It has also been shown that SMYD3-mediated H3-K4 tri-methylation licenses the MYC and Sp1 binding to the TERT promoter [60]. Taken together, epigenetic regulators intimately interact with transcription factors to exert their regulatory effects on the TERT transcription.
The epigenetic mechanism is also involved in the TERT regulation via Telomere Position Effect-over Long Distances (TPE-OLD), a TPE-like mechanism [63]. Long telomeres, present in early passages of normal human cells, form a telomere-loop structure in the region near the TERT locus, which mimics the compacted chromatin and leads to a repressed TERT epigenetic state, however, the replication-induced telomere shortening in aged cells disrupt the repressive loop, which in turn re-configures the chromatin structure characterized by increased H3-K4 acetylation and methylation associated with the TERT promoter [63]. Significantly altered DNA methylation in the TERT promoter was also observed in cells carrying very short telomeres. These permissive changes in the TERT chromatin favor the activation of the TERT transcription. As very short telomeres are widespread in human cancer, TPE-OLD may contribute to TERT/telomerase activation in cancer cells [63]. Indeed, the effect of TPE-OLD on the TERT expression was observed in HeLa cells when their telomere length was experimentally manipulated.
3.1.2.3. Genetic Aberrations and Germline Variants
The comprehensive characterization of human cancer genomics by high-throughput sequencing technologies has unraveled novel mechanisms to activate TERT transcription and telomerase. Recurrent TERT promoter mutations, focal rearrangements/amplification of the TERT locus, and onco-viral DNA insertion have been shown to play a key role in the cancer-specific TERT induction [8]. Additionally, certain Single Nucleotide Polymorphisms (SNPs) in the TERT gene are involved in the regulation of TERT transcription and telomere length, thereby contributing to the susceptibility or initiation and even progression of cancer [64-66].
3.1.2.3.1. TERT Promoter Mutations
Two hotspot mutations with a cytidine-to-thymidine (C>T) dipyrimidine transition at the proximal region of the TERT promoter (−124 and −146 bp from the ATG), so-named C228T and C250T, respectively, were originally identified in sporadic and familiar malignant melanomas six years ago [67, 68]. Since then, almost all human cancer types have been analyzed and the mutations frequently observed [69]. Based on the analysis of all TCGA cohorts with available mutation data, Barthel et al. [7] showed that average 27% of tumors carried either C228T or C250T mutation, being the most common mutations of non-coding regulatory regions in human cancer. The highest mutation rate (up to 80 – 90%) occurs in malignant melanoma, glioblastoma, urothelial bladder cancer, myxoid liposarcoma, certain types of skin cancer and medulloblastoma [7, 67-71]. The intermediate level of the mutation frequency (15 – 50%) is observed in Upper Tract Urinary Carcinoma (UTUC), thyroid cancer, hepatocellular carcinoma (HCC), head and neck cancer, ovarian clear cell carcinoma and among others. Malignancies derived from lung, breast, gastrointestinal, prostate and kidney, and blood or bone marrow either lack the mutations or have a low mutation rate (<10%) [7, 69, 70, 72-74]. Importantly, the TERT promoter mutation is not found in adjacent non-tumorous tissues, or other types of normal cells from sporadic cancer patients. When the C228T mutation was introduced into normal human bladder stem cells (SCs), this mutation alone was sufficient to promote transformation of these SCs by up-regulating TERT expression and activating telomerase [75], indicating its functional importance in tumorigenesis.
Primary tumors harboring the TERT promoter mutations in general express high levels of TERT and telomerase activity [69]. Mechanistically, either C228T or C250T mutation creates a de novo binding motif for ETS transcription factors, thereby up-regulating TERT expression [67, 76]. It was soon demonstrated that GA-binding proteins (GABPA and GABPB1), the members of the ETS family transcription factors, were specifically recruited to the mutant TERT promoter in cancer cells through which the TERT gene is transcriptionally induced [76]. In accordance with these observations, inhibiting the expression of GABPA or GABPB1 led to the down-regulation of TERT expression in cancer cells bearing a mutant TERT promoter [76-78]. In mutant TERT promoter-harboring glioblastoma cells, GABPB1 knockdown inhibited TERT expression, induced telomere shortening/dysfunction, impaired proliferation/ survival, and eventually attenuated tumorigenic ability [78]. Given these findings, GABPA/B1 was thus suggested as a therapeutic target for mutant TERT promoter-carrying malignancies.
However, recent findings raised a number of questions regarding the relationship between GABPA and TERT promoter mutations [79, 80]. In Thyroid Cancer (TC) cells, GABPA knocking-down inhibited TERT expression in both wt and mutant TERT promoter-bearing cells [79]. This indicates that GABPA regulates TERT expression independently of the TERT promoter mutation in TC cells. Moreover, GABPA depletion facilitated the invasion of TC cells, although TERT expression was down-regulated. Consistently, higher GABPA expression was inversely associated with metastatic disease and poor outcomes in TC patients [79]. Mechanistically, DICER1, a component of the microRNA machinery to inhibit cancer metastasis, was identified as a direct target gene for GABPA [79]. By promoting DICER1 expression, GABPA inhibits the invasive phenotype of TC cells [79]. Furthermore, GABPA was shown to suppress the aggressive phenotype of bladder cancer (BC) by activating the transcription of FoxA1 and GATA3, two transcription factors responsible for luminal differentiation of urothelial cells [80]. The TERT promoter mutations occur in up to 85% of BC tumors, while the GABPA inhibition indeed leads to the down-regulation of TERT expression, but promotes proliferation, stemness, invasion and drug resistance of BC cells [80]. In the clinical setting, the aberrant methylation and deletion of the GABPA gene are widespread in primary BC tumors [80]. Taken together, the GABPA effect is context-dependent in oncogenesis and it may act as a tumor suppressor rather than the oncogenic driver in the TC and BC pathogenesis despite its stimulatory effect on TERT expression [79, 80]. Importantly, because it takes time for cancer cells to undergo senescence/apoptosis resulting from telomerase inhibition mediated by GABP factor depletion [78], while GABPA knockdown-induced invasiveness may occur immediately, GABPA inactivation will likely lead to rapid TC and BC tumor disseminating [79, 80]. Thus, it should be very cautious to target GABP factors for telomerase-based cancer therapeutics.
3.1.2.3.2. TERT Gene Rearrangements
The rearrangement involved in the TERT locus has long been noticed in cancer, however, its functional significance has not yet been well addressed until recently. Several lines of evidence demonstrate that the rearrangement alters the genomic structure of the TERT locus, thereby leading to intrinsic enhancer hijacking to activate TERT transcription [53, 81].
The TCGA cohort patient analyses revealed that the TERT structural variants/rearrangements were observed in more than 15 cancer types and the highest frequency occurred in sarcoma (22%), however, the detailed dissection of this genetic alteration was not performed. Neuroblastoma is a tumor where The TERT gene rearrangement has been most comprehensively analyzed so far [7]. Peifer et al. showed that 23% of neuroblastoma patients (39/169) at advanced stages exhibited recurrent rearrangements in a 50 kb region proximal of the TERT gene [82]. Similar findings were reported by the other two groups [81, 83]. Based on these findings, Valentijn et al. further reviewed NIH Epigenomics Roadmap data (containing 127 human tissues), and they consistently observed a highly repressed region within 20-kbs upstream of TERT characterized by a Polycomb-modified chromatin signature, whereas super-enhancers were readily found beyond this repressive region [81]. The majority of the TERT rearrangements lead to the disruption of the Polycomb-silenced region and the direct overlapping between super-enhancers and juxtaposed TERT coding sequence. Such overlapping results in enhancer-hijacking through which massive chromatin remodeling and transcriptional activation of the TERT gene are achieved [81]. All the findings above indicate that TERT rearrangements are not random events, and it was observed that the TERT rearrangement induced TERT expression much more efficiently than did TERT promoter mutations [7].
In neuroblastoma, the TERT gene rearrangement is mutually exclusive with MYCN amplification and the mutation of the α-thalassemia/mental retardation X-linked (ATRX) gene, encoding a SWI/SNF chromatin-remodeling ATP-dependent helicase [81, 83]. Amplification of the MYCN gene, the most frequent genomic event in neuro-blastoma, leads to its over-expression, thereby promoting TERT transcription [81, 83]. The ATRX mutation is associated with the acquisition of ALT activity, a recombination-based mechanism for telomere maintenance [84] (See the ALT part below for details).
3.1.2.3.3. Tumor Viral DNA Insertions at the TERT Locus
In tumor virus-associated cancer, the TERT transcription is activated not only by viral oncoproteins as described above but also by the host genome integration of the viral DNA. This later mechanism is especially important in HBV-related pathogenesis of HCC. Horikawa et al. first demonstrated the presence of the HBV enhancer-containing DNA in the TERT promoter region and the exogenous viral enhancer-driven TERT up-regulation in HCC cells [85]. Using the next generation sequencing technology to analyze 81 HBV-positive HCC tumors, Sung et al. showed that the TERT locus was the most frequent target as HBV integration breakpoints; and they identified the insertion of HBV DNA there in 18 of examined HCC tumors [86]. The HBV DNA insertion is not random, because it occurred predominantly in the TERT promoter region, and almost all the inserted sequences contained at least one viral enhancer or promoter [53]. These exogenous enhancers or promoters are hijacked by HCC cells to promote the TERT transcription and telomerase activation. Higher levels of TERT expression were readily observed in these tumors [86].
HBV-related HCC is widespread in East Asian, especially in China, but HBV-negative HCC is predominant in western countries. In these HCCs, the integration of viral DNA derived from adeno-associated virus type 2 (AAV2) was observed in 11 of 193 tumors and all the insertion breakpoints were localized in the regions of cancer-related genes including TERT [87]. Interestingly, the AAV2 DNA fragment cloned from the HCC patient tumor substantially augmented the TERT promoter activity [87]. Likely, a similar hijacking mechanism is employed by AAV2 to activate telomerase in the HCC pathogenesis.
Hijacking inserted HBV promoters or enhancers to activate the TERT gene is well documented in HCC, however, it has remained elusive whether the TERT locus is perturbed by other onco-viral insertions. Analyzing the DNA integration from HPV, EBV, and BKV in human malignancies, Chen et al. observed the breakpoints and integrating events at a number of cancer-driving genes, but the TERT locus was not the target for insertions by any of these viruses [53]. A recent preliminary analysis on cells and tumors derived from head and neck cancer showed that HPV DNA was inserted into the TERT locus [88], but it is unclear if this finding is representative, and functionally important. As HPV infection is intimately associated with the vast majority of cervical cancer [27], more detailed and comprehensive analyses of this cancer type should help to answer the question above.
3.1.2.3.4. TERT Gene Amplification
Chromosome 5p gains in various kinds of human cancer have long been documented, for instance, this genomic event was observed in 70% of tumors derived from patients with lung adenocarcinoma [89], however, its functional significance had been elusive until the TERT gene was cloned in 1997 [90, 91]. Mapping the TERT gene within a ~40 kb gene body on the short arm of the chromosome 5 (5p.15:33) immediately attracted our attention: the TERT locus might be a target for the amplification in carcinogenesis. Using Fluorescence In Situ Hybridization (FISH), it has been demonstrated that the TERT amplification was widespread in human cancer [92]. Focal copy gains or amplification at the TERT locus were frequently detected in most cancer types, while the TERT gene was typically amplified in double-minuses in neuroblastoma-derived cells, and each cell carried more than 100 TERT copies [92]. During last 20 years, numerous conventional FISH or qPCR studies and recent next-generation sequencing analyses have revealed the prevalence of TERT amplification in multiple types of malignancies [8]. Moreover, the examination of the TCGA cohort (6835 patients from 31 tumor types) showed that TERT-amplified tumors expressed the highest levels of TERT mRNA and telomerase activity [7], which suggests a critical role of the TERT amplification in telomerase activation for cancer initiation and/or progression.
3.1.2.3.5. SNPs at the TERT Locus
The TERT gene carries multiple SNPs or genetic variants and a panel of them has been shown to regulate the TERT transcription and telomerase activity, thereby contributing to cancer susceptibility, cancer treatment resistance, progression and patient survival [66]. In all the TERT SNPs, rs2736100 (located in TERT intron 2) is most investigated, and the variants are associated with the risk of multiple types of cancer, as shown by published observations [66]. In myeloproliferative neoplasia (MPN), MPN cells derived from patients with rs2736100-CC genotype were found to express the highest levels of TERT mRNA compared with those from AC- and AA-carriers (CC>AC>AA) [93]. Consistently, the luciferase reporter assay showed that the reporter driven by the rs2736100-CC-containing sequences exhibited significantly higher luciferase activity than that by the AA allele-carrying fragments [94]. Higher TERT mRNA levels coupled with longer telomere were observed in lung cancer patients harboring CC genotypes [94]. Taken together, the rs2736100-CC genotype may facilitate TERT transcription and expression, which is consistent with increased cancer risk in individuals carrying this variant [94, 95].
In addition to its direct effect on TERT expression, the rs2736100 variant was also shown to affect the occurrence of TERT promoter mutations in HCC [65]. The rs2736100-CC-bearing HCC patients exhibited significantly lower rates of TERT promoter mutations than those with the AA genotype [65]. This scenario is likely due to the presence of shorter telomeres in patients carrying rs2376100-AA, because shorter telomere length is a driver for TERT promoter mutations [72]. Another TERT variant rs2853669, located in the TERT core promoter, also acts as a modifier of the effect of the TERT promoter mutations on survival and tumor recurrence in different cancer types [96]. These data collectively suggest that germline TERT variants cross-talk with somatic genetic alterations to coordinately regulate TERT expression and oncogenesis. In addition, many other TERT SNPs may be associated with TERT regulation and cancer risk [66]. The rs2736098 is an SNP in the second exon of the TERT gene, and its G-allele associated with the risk of multiple cancer types [66]. The AA genotype of this SNP is also involved in the regulation of the TERT promotor mutation frequency in HCC [65]. Moreover, individuals carrying rs2736098-AA exhibit a lower risk for HBV infection [97], an onco-virus to drive the HCC pathogenesis.
3.1.2.4. TERT Regulation at Post-transcriptional and Translational Levels
3.1.2.4.1. The Post-transcriptional Regulation of TERT Expression
The TERT is a single copy gene consisting of 16 exons and 15 introns. With a single transcription start site, the TERT gene is subject to regulation by an alternative splicing mechanism [41, 98]. The major splicing sites within a TERT pre-mRNA include three deletion (α, β, and γ) and four insertion sites. Splicing at one of these sites alone or different combinations generates various types of spliced TERT mRNA, and so far, more than 20 splicing variants have been identified [98, 99]. As the functional reverse transcriptase and telomerase-specific T motifs of the TERT protein are scattered on separate individual exons, the spliced variants such as α-, β- and γ-variants miss parts of sequences encoding the reverse transcriptase domain, and may exert a dominant-negative effect [99]. Some other variants have defective termination Condon or translation [98, 99]. Therefore, despite the presence of many transcript variants, the full-length TERT mRNA is the only transcript that is translated into a functional protein for telomerase activity.
Intriguingly, certain normal human cells express non-functional or dominant-negative TERT transcripts, but the underlying physiological significance is unclear [100]. Likely, residual levels of TERT promoter activity remain in these cells, and the splicing mechanism is required to totally inhibit telomerase activity by switching-off expression of the full-length TERT mRNA [99]. Conceivably, a TERT splicing switch to a functional TERT mRNA is necessary for tumorigenesis. It has been previously observed that MYC expression was closely correlated with the presence of the full-length TERT mRNA in renal cell carcinoma and activated lymphocytes [101]. Turning on and off MYC expression led to corresponding changes in expression of full-length TERT mRNA in lymphoid cells transfected with an inducible MYC expression vector. Thus, MYC is a key driver for expression of the functional TERT transcript. The oncogenic fusion protein EWS-FLI1 was also observed to promote the full-length TERT splicing switch [102]. In addition, BRG1 and BRM, the catalytic subunits of the chromatin-remodeling SWI/SNF complex, are actively involved in the TERT splicing regulation. They interact with the splicing factors p54(nrb) and PSF, and are specifically co-localized with phosphorylated RNA polymerase II in a region incorporating an alternative splicing acceptor site of TERT exon 7, which contributes to splicing switch toward full-length TERT transcripts. Whereas BRM depletion led to down-regulation of TERT expression and enhancement of ratios of exon-7-and-8-excluded TERT mRNA that encodes a β-site-deleted inactive protein [103]. These cells underwent telomere shortening and eventual replicative senescence within a period. More recently, a number of splicing factors and RNA-binding proteins were shown to directly regulate TERT pre-mRNA splicing. The over-expression of splicing proteins SRSF11, hnRNPL and hnRNPH2 contribute to TERT minus beta splicing choice [104]. Sayed et al. identified that Neuro-oncological Ventral Antigen 1 (NOVA1), an RNA-binding factor, directly interacted with TERT pre-mRNA and promoted the inclusion of exons in the reverse transcriptase domain of TERT through which splicing was switched to the production of active TERT transcripts [105]. Stably inhibiting NOVA1 expression in cancer cells leads to splicing away from full-length TERT transcripts, thereby inactivating telomerase, inducing telomere dysfunction and eventually triggering cell growth arrest and apoptosis [105]. These data suggest that the splicing regulation of TERT transcripts may be targeted for cancer therapy.
3.1.2.4.2. The Post-translational Regulation of TERT Expression/Localization via Phosphorylation and Ubiquitination
TERT expression is subject to the post-translational regulation by phosphorylation and ubiquitination. Liu’s group first documented that protein phosphorylation was involved in the regulation of telomerase activity [106]. It was soon identified that AKT kinase phosphorylated TERT protein at Ser227 and Ser824 sites, and thereby increased telomerase activity [107]. Many studies have so far shown that TERT protein can be phosphorylated by a number of protein kinases, including AKT, PKCs, SRC and c-ABL [107-110], but the functional consequence of TERT phosphorylation is different, dependent on the phosphorylated sites. In general, AKT and PKCs promote while SRC and c-ABL inhibit TERT activity.
Phosphorylated TERT exhibits a direct effect on the regulation of telomerase activity, but its subcellular re-distribution may be equally important. AKT-mediated TERT phosphorylation at Ser227 enhances the interaction between TERT and importin-α, a nuclear import receptor, and increases TERT retention and availability in nuclei, and this phosphorylation event is required for telomere stabilization and cellular immortalization [111]. In contrast, the TERT Tyr707 phosphorylated by Src kinase promotes its cytoplasmic translocation from nuclei via the export receptor CRM1 when cells suffer from oxidative insults [109, 112]. In that case, TERT is no longer available for telomere lengthening or other activities and undergoes a rapid degradation in the cytoplasm, which subsequently compromises cell survival. It is thus evident from these studies that TERT phosphorylation mediated by protein kinases is functionally important in tumorigenesis.
The ubiquitination pathway also participates in the regulation of TERT expression in cancer. It was previously identified that the chaperones Hsp23 and Hsp90 interacted with TERT and were required for the active telomerase complex assembling [113]. Further studies showed that Hsp23 and Hsp90 protected TERT from degradation through ubiquitin-dependent proteolysis. When the interaction between Hsp90 and TERT is disrupted, TERT undergoes ubiquitination modification catalyzed by the E3 ligase MKRN1 and subsequent proteasome-mediated degradation [114]. Immunophilin FK506-binding protein (FKBP)52, an Hsp90-binding factor, is also within the TERT-Hsp90 complex, and FKBP52 inhibition mimics Hsp90-TERT disassociation, abolishes the nuclear translocation of TERT, resulting in cytoplasmic accumulation where the proteasome-dependent degradation of TERT occurs rapidly [115]. On the other hand, C terminus of Hsc70-interacting protein sequesters Hsp23 from the TERT complex, thereby leading to the cytoplasmic retention and degradation of TERT [116]. In addition, another E3-ligase HDM2 was shown to directly interact with and polyubiquitinate TERT for its degradation independently of Hsp90 or Hsp23 [117].
3.1.2.4.3. MicroRNA and Long Non-Coding RNA Regulation of TERT Expression
The dysregulation of microRNAs (miRNAs), widespread in human cancer, plays a key role in oncogenesis [118]. Given the critical role of telomerase activation in cancer formation or progression, TERT could be a key target for cancer-related miRNAs. Indeed, a panel of such miRNAs has been identified to regulate TERT expression in various types of cancer, and interestingly, oncogenic and tumor-suppressive miRNAs in general exhibit opposite effects on TERT expression. Tumor suppressive miRNAs include miRNA-34, 133, 138, 299, 422, 491, 498, 512, 532, 615, 661, 1182, 1266, etc.; and they directly bind to 3’-UTR of TERT mRNA, inhibiting its translation and/or promoting mRNA degradation [118]. Oncogenic miRNAs such as miRNA-19b, 21, and 202 mainly enhance TERT expression by targeting TERT regulators in an indirect manner [118].
Long non-coding RNAs (lncRNAs) have recently emerged as players in tumorigenesis, but their regulatory effect on telomere homeostasis and telomerase activation not well explored except telomerase-associated TERC and TERRA (See the discussion below for details). BC032469, a lncRNA over-expressed in gastric and esophageal cancer, was observed to up-regulate TERT expression by sponging miR-1207-5p, a negative TERT regulator [119]. Estrogen receptor regulates and cooperates with lncRNAs HOTAIR and MALAT1 to control the TERT transcription by binding to the estrogen response element on the TERT promoter in prostate cancer cells [120]. Compared to miRNAs, lncRNAs seem to employ diverse strategies to regulate TERT expression.
3.1.2.5. Dysregulation of TERC and Other Telomerase Components in Human Cancer
TERC, a 451 nucleotide-long lncRNA, functions as an intrinsic template for telomerase-mediated telomere lengthening, and it together with TERT forms the core of the telomerase holoenzyme. Unlike TERT, TERC is ubiquitously expressed in human somatic cells, but its up-regulation still occurs widely in human cancer, which indicates the importance of TERC over-expression in cancer formation and progression [10]. The cancer-promoting effect of TERC is well exemplified by Marek's disease herpesvirus (MDV)-mediated malignant T cell lymphoma in chickens [121]. MDV encodes a viral TERC (vTERC) sharing 88% sequence identity with chicken TERC (chTERC). MDV infection of one-day-old chickens drives tumor formation via vTERC expression. Moreover, the replacement of vTERC by chTERC in the MDV genome retains a strong oncogenic activity, suggesting a similar oncogenic effect of cellular TERC [122]. In TERC-/- mice, when the tumor suppressor TP53 is functional, the tumor incidence declined substantially [123]. Papilloma frequency induced by carcinogen treatment was 20-fold lower in TERC−/− mice compared with their wt counterparts [124]. In humans, individuals with defective TERC have very low cancer incidences despite an increased hematological precursor lesion such as myelodysplastic syndrome [125].
All the data above collectively support the cancer-promoting activity of TERC, which may be telomere-lengthening dependent or independent. Like TERT, TERC is not only required for telomere extension, but also exhibits extra-telomeric functions. For example, TERC interacts with ATR and inhibits the ATR-mediated DNA damage response, thereby suppressing the activation of the p53/CHK1-dependent signaling in response to genotoxic insults [126], while TERC inhibition results in cell cycle arrest by activating p53 and CHK1. More recently, TERC was shown to function as a transcription factor to stimulate expression of inflammatory mediators, including LIN37, TPRG1L, TYROBP, USP16 and others, by forming RNA-DNA triplexes on the promoters of these genes [127]. This novel finding raises the possibility of the direct activation of oncogenic factors by TERC.
The TERC gene is localized at chromosome 3q26 and its expression in cancer is regulated by many different mechanisms, including aberrant alterations in transcriptional and epigenetic statuses, oncogenic signalings, gene amplification, and among others, which depend on cancer types [10]. The TERC promoter is rich with binding sites for numbers of hormone receptors, and transcription factors Sp1, AP1, ETS, and HIF-1 [10]. In addition, three non-canonical E-boxes are present on the promoter region and MYC strongly activates the TERC transcription in prostate, breast, lung and colorectal cancer-derived cells [128]. Therefore, the transcriptional controlling of TERC expression is one of the key regulatory mechanisms in these tumors. While in cervical cancer, the TERC amplification is a predominant approach to up-regulate TERC expression and observed in the vast majority of patients [10].
A key feature of TERC RNA is its unusual long half-life, lasting more than 2 weeks in TERT-positive cancer cells [129], however, its maturation and stability may decline dramatically under certain pathological conditions. Dyskerin, encoded by dyskeratosis congenita 1 (DKC1) gene, is a known component in the telomerase complex and stabilizes TERC RNA, while its mutation accelerates TERC RNA degradation through which telomerase activity is diminished, telomere homeostasis is impaired and the onset of dyskeratosis congenita is triggered [130]. In addition, TERC RNA maturation was recently shown to be governed by Poly(A)-specific Ribonuclease (PARN) at a posttranscriptional level, and the inactivating PARN mutations result in incomplete 3′ end processing and increased destruction of nascent TERC transcripts [131]. Therefore, telomerase deficiency and related disorders are observed in PARN-mutation-bearing individuals [131, 132].
In addition to TERT and TERC, two essential subunits for telomerase activity, alterations of other known components in the telomerase complex may occur in tumorigenesis, too. The augmented DKC1 expression mediated by GATA1 robustly increases telomerase activity in human erythroblasts [133]. Hsp90 was shown to be up-regulated during prostate cancer progression, through which telomerase activity was enhanced [134]. The AAA+ superfamily ATPase Reptin is not only associated with the telomerase complex [135], but also acts as a transcription factor to promote TERT expression [136], while it, together with its partner Pontin, is widely overexpressed in human malignancies [135, 136]. All these factors in the telomerase complex are required for telomerase activation/telomere lengthening on the one hand, and may also display other biological activities to promote tumorigenesis independent of their stimulatory effect on telomerase on the other.
3.2. The ALT Pathway Activation in Human Cancer
Different from telomerase-mediated telomere synthesis, the ALT pathway employs HR and homology-based telomere lengthening strategies to maintain telomeres [9]. According to the analysis of TCGA cohorts of cancer patients, Berthal et al. estimated that the ALT activation occurred in 5% of all human tumors, lower than previously expected [7], however, a high frequency (up to 50% or even higher) of ALT is seen in malignancies derived from mesenchymal and neural epithelial origins, including various sarcomas, gliomas, neuroblastomas, medullary thyroid carcinoma, pancreatic neuroendocrine tumors and among others [7, 9]. The ALT-positive cancer cells are characterized by the following features [7, 9]: (1) Lack of TERT expression and telomerase activity; (2) Heterogeneous telomere length coupled with frequent telomeric-sister chromatid exchange and extrachro-mosomal linear/circular DNA of telomeric repeats; and (3) Presence of ALT-associated promyelocytic leukemia bodies (APBs).
It remains elusive why the ALT pathway is preferably activated in mesenchymal/neural epithelial tissue-originated malignancies. One potential explanation is the absence or trace of TERT and telomerase activity in normal human mesenchymal stem cells [7, 9]. Genetically, the inactivation of the ATP-dependent chromatin remodelers, ATRX and/or death associated protein-6 (DAXX) promotes ALT and is frequently observed in ALT-positive tumors [9, 84, 137]. ATRX and DAXX cooperate to deposit histone H3.3 into telomeric and pericentromeric chromatin to prevent the formation of a G-quadruplex DNA structure facilitating HR, thereby leading to telomere shortening. Therefore, the loss-of-function of ATRX or DAXX gene mutations/deletions contributes to the ALT activation by destabilizing repressive heterochromatin, accumulating G-quadruplex structures of telomeric DNA and inducing HR [84, 137]. In line with the inhibitory effect of ATRX and DAXX on ALT, the re-introduction of ATRX and DAXX into ALT-positive, ATRX and DAXX-negative cells, respectively, has been shown to abolish the ALT phenotype [137, 138]. In addition, lncRNA TERRA plays a part in the ALT activation. High levels of TERRA are observed in ALT-positive cells and present in APBs [139]. TERRA may form RNA/DNA hybrids with the telomeric C-rich strand, which creates a permissive environment for the recombination of telomeric DNA [139].
The ALT pathway is only seen in 5% of human cancer, but has important significances in telomere-based cancer therapy. If the telomere-lengthening mechanism by telomerase and ATL is switchable, targeting telomerase in telomerase-proficient cancer cells may induce treatment resistance through the ALT activation. Indeed, Hu et al. [140] showed the ALT switch in telomerase-positive cells when telomerase was inhibited, combined with the disruption of ATRX/DAXX complex and induction of telomeric DNA damage. In addition, the depletion of histone chaperons ASF1-A/B alone was reported to be sufficient for the ALT induction, coupled with TERT inhibition [141], in telomerase-positive cancer cells, suggesting a key role of ASF1 in suppressing ALT. However, we failed to observe significantly inhibitory effects of ASF1 knocking-down on TERT expression in different cancer cells; instead, these cells underwent senescence due to DNA damage response provoked by ASF1 inhibition [142, 143]. Because the forced TERT expression in ALT-positive cells constituted telomerase activity and both mechanisms are active for telomere lengthening [41], the additional issue is whether telomerase and ALT can be co-present in the same tumor cell or in different clones in same tumor tissues. If it is the case, targeting telomerase-positive clones will most likely fuel outgrowth of ALT-positive clones. Further investigations are required to answer these questions.
4. THE ABERRANT ALTERATIONS IN TELOMERE-ASSOCIATED SHELTERIN FACTORS AND TERRA IN CANCER
The physiological function of telomeres is dependent on its intact structure formed by telomeric DNA, associated factors (mainly the shelterin complex) and TERRA. Telomerase or ALT activation stabilizes telomere length in cancer cells, however, the aberrant alterations in shelterin factors and TERRA expression also occur frequently during oncogenesis, which directly or indirectly shapes telomere chromatins favoring tumor formation and/or progression.
4.1. The Genetic Aberrations and Dysregulation of Shelterin Factors in Cancer
4.1.1. Mutations of the Shelterin Factor-encoded Genes in Human Cancer
Cancer-associated shelterin mutations occur in the POT1 and TPP1-encoded ACD genes. Somatic POT1 mutations were initially identified in Chronic Lymphocytic Leukemia (CLL), and more frequently seen (up to 9%) in patients bearing a non-mutated Immunoglobulin Heavy Chain (IGHV) gene [25, 144]. The mutations are predominantly within the region encoding the domain harboring oligosaccharide-oligonucleotide (OB) folds, which impairs POT1 binding to telomeric DNA and thereby leads to telomere dysfunction and genomic instability. Consistently, CLL cells from these POT1 mutation-harboring patients do have numerous telomeric and chromosomal abnormalities [25]. The disruption of the interaction between POT1 and telomeric DNA increases the accessibility of telomerase to telomeres for their extension. Longer telomeres render CLL cells more malignant characters. It is known that CLL without IGHV mutations is aggressive [28], in which POT1 mutation-mediated chromosome instability likely drives the disease, even more, exacerbated [25, 144]. Thus, the analysis of POT1 mutations in CLL should help improve disease prognostication and management.
In addition to the somatic POT1 mutations in sporadic CLL, Speedy et al. further identified its germline mutations in familiar CLL (4/66 families, 6%) [145]. The mutations either impair the POT1 association with telomeric DNA or disrupt the interaction between POT1 and TPP1. Telomeric and cytogenetic aberrations in these patients are similar to those seen in POT1-mutated sporadic CLLs. Moreover, the POT1 germline mutation has also been found in other familiar malignancies, including Hodgkin lymphoma, colorectal cancer, melanoma, cardiac angiosarcoma and glioma [146-149]. Each of these different familiar malignancies with POT1 mutations, in general, does not cross with each other, suggesting the modest penetrance of this genetic event [145].
Searching for potential mutational-drivers in 3 childhood patients with acute lymphoblastic leukemia (ALL), Spinella et al. [150] identified the somatic mutation of ACD, a gene encoding TPP1 shelterin protein in one patient. The mutation (ACD p.G223V) is adjacent to the TEL patch in the OB-fold domain of TPP1. The introduction of this mutant form of TPP1 into leukemia cells promotes telomere lengthening and resistance to apoptosis induced by chemotherapeutic drugs. Based on the analyses of the public cancer database COSMIC (version 71), missense or frameshift mutations in the ACD gene are found in various cancer types, and many of these mutations are located in the OB-fold, adjacent to the TEL patch, and in the POT1 interaction domain. In addition, the ACD mutations have also been observed in familiar CLL and melanoma [145, 146]. Thus, the ACD mutation may not be a rare genetic event in human cancer.
4.1.2. Dysregulation of Shelterin Factor Expression in Cancer
The regulation of shelterin factor expression in cancer cells may occur at multiple levels, including transcriptional, post-transcriptional and post-translational. TRF1 mRNA expression in breast cancer is in general down-regulated, likely due to the promoter hypermethylation and miRNA-mediated degradation [151-153]. However, contradictory results obtained from TRF1 mRNA and protein analyses, either high or low, were reported in gastric, lung, liver, prostate and other cancers [24, 154-159]. Nevertheless, Bejarano et al. showed that TRF1 was over-expressed in Glioblastoma Multiforme (GBM) cells, especially GBM stem cells (GSCs), independently of telomere length. TRF1 deletion induced telomeric DNA damage and reduced proliferation and GSC phenotypes. TRF1 chemical inhibitors recapitulated these effects in human GBM cells, and inhibited GSC self-renewal and tumor growth in xenografts from patient-derived primary GSCs. Interestingly, BRAF and ERK2 kinases, in the downstream of the RAS pathway, phosphorylate TRF1 protein [160, 161], thereby increasing its stability and binding to telomeres. Inhibition of these kinases mimicked the effects of TRF1 deletion on telomere damage and GSC phenotypes in GBM cells, exhibiting good therapeutic efficacy both in vitro and in vivo. One critical issue is whether TRF1 inhibitors are useful for tumors with normal and even lower levels of TRF1 expression, or whether low TRF1-expressing tumors are more sensitive to TRF1 inhibition. Further investigations are required to determine the effect of TRF1 inhibitors on tumor cells expressing different levels of TRF1. In addition, drugs specifically targeting BRAF and MEK have been applied in malignancies carrying BRAF or RAS mutations, and it may be worthy of assessing whether TRF1 is able to serve as a variable to predict treatment response.
Unlike TRF1, most studies revealed that TRF2 was over-expressed in different types of human cancer and promoted tumorigenesis through telomere protection-dependent or independent manners [24, 158, 159, 162-164]. Interestingly, TRF2 is also a direct target gene of β-Catenin, an oncogenic factor frequently dysregulated in human malignancies [163]. In addition, higher POT1 expression was observed in several cancer types and hematological neoplasia [24, 165-168]. Increased POT1 expression was suggested as a driver during the evolution from a precursor lesion monoclonal gammopathy of undetermined significance to multiple myeloma, a fully transformed malignancy [165]. However, the down-regulation of POT1 occurred in ALL and was associated with chromosomal instability [125, 169]. Three other shelterin proteins (TPP1, TIN2 and RAP1) in cancer have been so far rarely explored and will not be discussed here.
4.1.3. TERRA Expression in Cancer
LncRNA TERRA is transcribed by RNA Pol II from the subtelomere C-rich strand towards the telomere, and its expression is transcriptionally activated by the chromatin organizing factor CTCF and the cohesin Rad21 [3, 170, 171]. In addition, TRF1 interacts directly with Pol II to promote TERRA transcription. It has also been shown that a compacted chromatin status mediated by H3K9 and H4K20 trimethylation and telomeric DNA methylation represses TERRA expression, which results in differential expression levels of TERRA between normal and telomerase-positive malignant cells. The hypermethylation of subtelomere regions coupled with reduced TERRA expression was observed in telomerase-proficient tumors compared to those in normal cells [172]. ALT-activated tumor cells, in general, express the highest TERRA, likely due to open chromatin conformation at subtelomeres and abnormally long telomeres resulting from the mutation of chromatin remodeling factors ATRX or DAXX [9].
TERRA plays an important role in the regulation of telomere structure and function. First, it interacts with TRF1 and TRF2, directing the proper assembly of the shelterin factors at telomeres. Second, it facilitates telomeric DNA replication by recruiting origin replication complex 1, or forming R-loops at chromosome ends [173]. Third, it promotes DNA damage response and repair at dysfunctional telomeres. Finally, it is actively involved in the formation of heterochromatin at subtelomere and telomere regions. In addition, TERRA may bind to TERC or telomeric repeats to interfere with telomerase action in telomerase-activated cells, while promote HR-mediated telomere lengthening through R-loop formation in ATL-positive cancer cells. Intriguingly, the deletion of the major TERRA locus induces dramatic loss of telomeric repeats and a massive DNA damage response in cancer cells regardless activation of telomerase or ALT [174]. These findings suggest that targeting TERRA may be a potential strategy for cancer therapy.
5. IMPLICATIONS OF CANCER-RELATED TELOMERE MAINTENANCE IN PRECISION ONCOLOGY
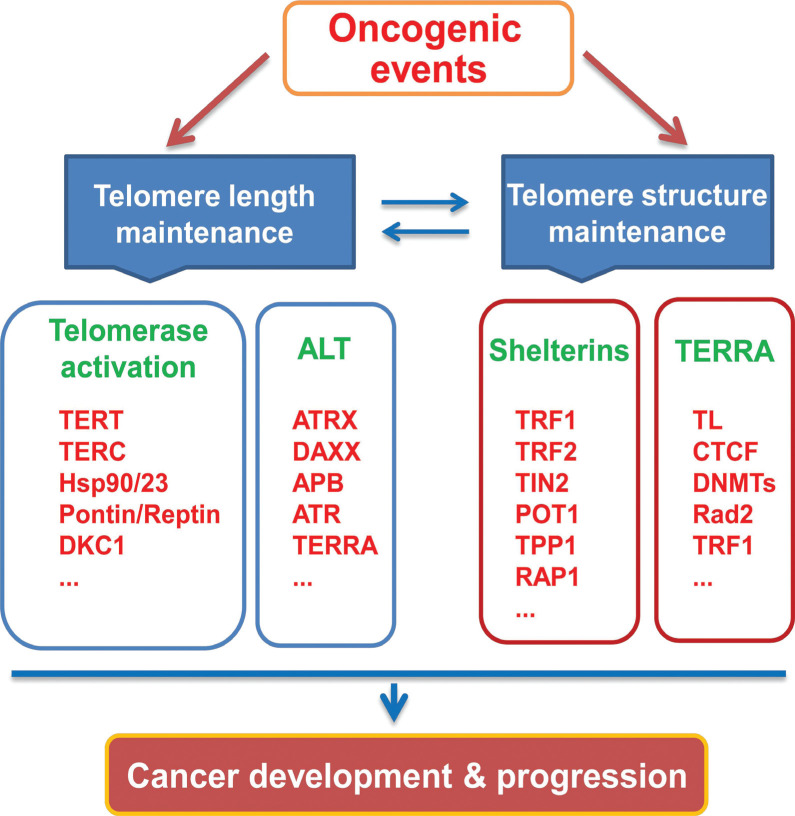
Telomere maintenance is essential to cancer development and progression, and malignant cells have evolved multiple specific strategies to achieve this goal. These strategies or mechanisms involve featured alterations in transcriptomes, genetics and epigenetics of telomere-related factors (Fig. 4), which have aroused great enthusiasm for their potential clinical applications for precision oncology including cancer intervention, diagnosis and surveillance and prognostication.
Fig. (4).
Multiple telomere-related factors are targeted by oncogenic events to maintain telomere stabilization/protection, unlimited proliferation and other hallmarks of cancer cells. Telomere length and structure is interdependent with each other. The oncogenic event targets one or more components in telomere length maintenance and/or telomere chromatin for telomere stabilization and protection during the malignant transformation and cancer progression.
5.1. Targeting Telomere Maintenance for Cancer Therapy
As discussed above, the unique features of telomere maintenance in cancer make it an ideal target for cancer therapy: (1) The vast majority of cancer cells carry significantly shorter telomeres than do normal cells; and (2) Telomerase is activated in up to 90% of human cancer. These scenarios thus provide a valuable therapeutic window for specifically killing cancer cells with minimal side-effects. Indeed, various types of small-molecules compounds inhibiting telomerase activity through different mechanisms have been developed [5, 175]. Imetelstat or GRN163L, a synthetic lipid-conjugated 13-mer N3′→P5′ thio-phosphoramidate deoxyribo-oligonucleotide that interferes with TERC function, has been shown to efficiently inhibit telomerase activity in cancer cells [5, 175]. Imetelstat bears a complementary sequence to TERC, antagonizing a 13-mer template within TERC for TTAGGG repeat synthesis. Thus, Imetelstat treatment of cancer cells leads to telomerase inhibition accompanied by progressive telomere erosions, eventually inducing cellular senescence or apoptosis. More importantly, the Imetelstat application in Myeloproliferative Neoplasia (MPN) has proven highly effective, and most patients underwent rapid and durable hematologic and molecular remissions following the drug administration [176-178]. However, the therapeutic benefit of Imetelstat to patients with solid tumors is minimal and severe side-effects have been documented [179]. Imetelstat seems suitable for slowly developed malignancies rather than aggressive tumors [179]. Its combination with other therapeutic strategies may be required to treat solid cancers.
Theoretically, TERT should be the most perfect target for telomerase inhibition in cancer. However, despite tremendous efforts, the development of the catalytic TERT inhibitors has so far not been very successful. BIBR1532, a potent and selective telomerase inhibitor, is a non-competitive small molecule targeting the superficial region of TERT [180, 181]. It was shown to inhibit TERT activity and reduce telomere length in cultured cancer cells without causing acute cytotoxicity, however, its efficacy is highly telomere length-dependent, so that it may not be an ideal single therapy agent for most cancers [5, 180]. In addition, MST-312, the synthesized tea catechin EGCG analogue, was observed to potently inhibit telomerase activity in different types of malignant cells [182, 183]. An epigenetic mechanism may be involved in the down-regulation of TERT expression mediated by EGCG and MST-312 [184].
For ALT-activated cancer cells, ATR inhibitors VE-821 and KU-55933 were found to induce chromosome fragmentation and cell apoptosis [137]. Mechanistically, these molecules inhibit telomere lengthening by interfering with the HR activity mediated by ATR kinase [137]. Several ATR inhibitors have been under clinical trials for cancer treatment and it is important to evaluate their efficacy on ALT-positive tumors in clinical studies.
A number of factors maintaining telomere structure such as TRF1, TERRA and others have been experimentally tested for their potential values in anti-cancer therapies, and obtained results did show that their inhibition led to telomere dysfunction and reduced tumor growth [23, 174]. It should be highlighted, however, that highly selective killing of tumor cells is of great importance for the development of anti-cancer drugs. Telomere structure maintenance is similarly essential to normal human cells, and it is currently unclear whether these same treatments are deleterious to them, especially to normal stem/progenitor cells, immune cells and other highly proliferative cells. If it is the case, severe side-effects might be expected, which will certainly prevent their potential clinical applications. Therefore, it is important to assess whether targeting these factors is highly selective to tumor cells and how wide a therapeutic window is.
5.2. Telomere-related Biomarkers for Cancer Diagnosis, Disease Monitoring and Prognostication
Telomere-related biomarkers specific to cancer are very useful for cancer diagnosis, progression surveillance and outcome predictions. Telomerase activity, TERT mRNA, TERT and TERC gene copy numbers and TERT promoter mutation or hypermethylation have been extensively tested for their application in cancer diagnostics. The non-invasive detection of these biomarkers in circulation or plasma, urine, stool, and cerebrospinal fluid for the diagnosis/monitoring of different kinds of cancer are especially attractive [57, 69, 71, 73, 185, 186]. For instance, a very high frequency of the TERT promoter mutation occurs in bladder cancer and UTUC, and the urinary assessment of the mutant TERT promoter has proven valuable in the disease diagnosis and relapse prediction [71, 73, 185, 187]. The TERC copy number assay showed high sensitivity and specificity for CINs and cervical cancer [188].
Telomerase/telomere-related alterations as prognostic factors for cancer patients have been evaluated in many clinical observations, but most of them are focused on TERT. In most analyzed human tumors, higher telomerase activity and/or TERT expression are closely associated with poor patient outcomes [7, 64, 69, 79, 189]. GBM patients with ALT have better outcomes than those with telomerase activation/TERT expression. In neuroblastoma, tumors without detectable TERT mRNA frequently undergo spontaneous regression [190], whereas TERT expression, and/or its gene amplification or rearrangements are the driving-force for the aggressive disease and associated with significantly shorter patient survival [81-83]. The presence of TERT promoter mutations has recently been shown as an unfavorable prognostic factor in a number of cancer types including papillary thyroid carcinoma, glioblastoma, bladder cancer, and others [69, 79, 191-193]. In addition, the association between TERT promoter hypermehtylation and poor outcomes or progression was reported in brain tumors and adrenocortical carcinoma [58, 61, 194].
A very few of studies have explored the association between alterations in cancer-related shelterin factors and prognosis. TRF2 over-expression was shown to predict shorter survival in oral carcinoma and HCC [24, 195]. POT1 mutations were associated with aggressive diseases in CLL patients [144].
CONCLUSION
Telomere stabilization through activation of either telomerase or ALT pathway is required for malignant transformation of human somatic cells and cancer progression. In the meanwhile, many other telomere-related elements are aberrantly altered in oncogenesis. These cancer-associated changes have been extensively investigated and the obtained results have so far provided profound insights into cancer telomere biology, which is expected to path the way for the application of telomere-based strategies in precision oncology. It should also be highlighted that the telomere stabilization is only one part of the whole oncogenic program. Thus, a better understanding of the oncogenic program and targeting upstream events may help efficiently block telomere-related aberrations in cancer.
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
This work was supported by grants from China Postdoctoral Science Foundation Grant (2019M652404), the Swedish Cancer Society (19 0018 Pj), Swedish Research Council (2018-02993), the Cancer Society in Stockholm (171223), Karolinska Institutet Foundation (2018-01524), Swedish Foundation for International Cooperation in Research and Higher Education (STINT, CH2016-6632), Ruth and Richard Julin Foundation and Nanna Svartz´ Foundation(2018-00183).
ACKNOWLEDGEMENTS
Declared none.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Shay J.W., Wright W.E. Telomeres and telomerase: three decades of progress. Nat. Rev. Genet. 2019;20(5):299–309. doi: 10.1038/s41576-019-0099-1. [DOI] [PubMed] [Google Scholar]
- 2.Veverka P., Janovič T., Hofr C. Quantitative biology of human shelterin and telomerase: searching for the weakest point. Int. J. Mol. Sci. 2019;20(13):, E3186. doi: 10.3390/ijms20133186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azzalin C.M., Lingner J. Telomere functions grounding on TERRA firma. Trends Cell Biol. 2015;25(1):29–36. doi: 10.1016/j.tcb.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 4.Hayflick L., Moorhead P.S. The serial cultivation of human diploid cell strains. Exp. Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 5.Harley C.B. Telomerase and cancer therapeutics. Nat. Rev. Cancer. 2008;8(3):167–179. doi: 10.1038/nrc2275. [DOI] [PubMed] [Google Scholar]
- 6.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Barthel F.P., Wei W., Tang M., Martinez-Ledesma E., Hu X., Amin S.B., Akdemir K.C., Seth S., Song X., Wang Q., Lichtenberg T., Hu J., Zhang J., Zheng S., Verhaak R.G. Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat. Genet. 2017;49(3):349–357. doi: 10.1038/ng.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan X., Larsson C., Xu D. Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: old actors and new players. Oncogene. 2019;38(34):6172–6183. doi: 10.1038/s41388-019-0872-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pickett H.A., Reddel R.R. Molecular mechanisms of activity and derepression of alternative lengthening of telomeres. Nat. Struct. Mol. Biol. 2015;22(11):875–880. doi: 10.1038/nsmb.3106. [DOI] [PubMed] [Google Scholar]
- 10.Cairney C.J., Keith W.N. Telomerase redefined: integrated regulation of hTR and hTERT for telomere maintenance and telomerase activity. Biochimie. 2008;90(1):13–23. doi: 10.1016/j.biochi.2007.07.025. [DOI] [PubMed] [Google Scholar]
- 11.Ding D., Xi P., Zhou J., Wang M., Cong Y.S. Human telomerase reverse transcriptase regulates MMP expression independently of telomerase activity via NF-κB-dependent transcription. FASEB J. 2013;27(11):4375–4383. doi: 10.1096/fj.13-230904. [DOI] [PubMed] [Google Scholar]
- 12.Luo Z., Wang W., Li F., Songyang Z., Feng X., Xin C., Dai Z., Xiong Y. Pan-cancer analysis identifies telomerase-associated signatures and cancer subtypes. Mol. Cancer. 2019;18(1):106. doi: 10.1186/s12943-019-1035-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Im E., Yoon J.B., Lee H.W., Chung K.C. Human Telomerase reverse transcriptase (hTERT) positively regulates 26S proteasome activity. J. Cell. Physiol. 2017;232(8):2083–2093. doi: 10.1002/jcp.25607. [DOI] [PubMed] [Google Scholar]
- 14.Park J.I., Venteicher A.S., Hong J.Y., Choi J., Jun S., Shkreli M., Chang W., Meng Z., Cheung P., Ji H., McLaughlin M., Veenstra T.D., Nusse R., McCrea P.D., Artandi S.E. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature. 2009;460(7251):66–72. doi: 10.1038/nature08137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saretzki G. Extra-telomeric functions of human telomerase: cancer, mitochondria and oxidative stress. Curr. Pharm. Des. 2014;20(41):6386–6403. doi: 10.2174/1381612820666140630095606. [DOI] [PubMed] [Google Scholar]
- 16.Liu Z., Li Q., Li K., Chen L., Li W., Hou M., Liu T., Yang J., Lindvall C., Björkholm M., Jia J., Xu D. Telomerase reverse transcriptase promotes epithelial-mesenchymal transition and stem cell-like traits in cancer cells. Oncogene. 2013;32(36):4203–4213. doi: 10.1038/onc.2012.441. [DOI] [PubMed] [Google Scholar]
- 17.Masutomi K., Possemato R., Wong J.M., Currier J.L., Tothova Z., Manola J.B., Ganesan S., Lansdorp P.M., Collins K., Hahn W.C. The telomerase reverse transcriptase regulates chromatin state and DNA damage responses. Proc. Natl. Acad. Sci. USA. 2005;102(23):8222–8227. doi: 10.1073/pnas.0503095102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu J., Yuan X., Sjöholm L., Liu T., Kong F., Ekström T.J., Björkholm M., Xu D. Telomerase reverse transcriptase regulates DNMT3B expression/aberrant DNA methylation phenotype and AKT activation in hepatocellular carcinoma. Cancer Lett. 2018;434:33–41. doi: 10.1016/j.canlet.2018.07.013. [DOI] [PubMed] [Google Scholar]
- 19.Yuan X., Xu D. Telomerase reverse transcriptase (TERT) in action: Cross-talking with epigenetics. Int. J. Mol. Sci. 2019;20(13):, E3338. doi: 10.3390/ijms20133338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang K., Guo Y., Wang X., Zhao H., Ji Z., Cheng C., Li L., Fang Y., Xu D., Zhu H.H., Gao W.Q. WNT/β-catenin directs self-renewal symmetric cell division of hTERThigh prostate cancer stem cells. Cancer Res. 2017;77(9):2534–2547. doi: 10.1158/0008-5472.CAN-16-1887. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X., Li B., Yu J., Dahlström J., Tran A.N., Björkholm M., Xu D. MYC-dependent downregulation of telomerase by FLT3 inhibitors is required for their therapeutic efficacy on acute myeloid leukemia. Ann. Hematol. 2018;97(1):63–72. doi: 10.1007/s00277-017-3158-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Steensel B., Smogorzewska A., de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92(3):401–413. doi: 10.1016/S0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 23.Bejarano L., Schuhmacher A.J. Mendez, M Inhibition of TRF1 telomere protein impairs tumor initiation and progression in glioblastoma mouse models and patient-derived xenografts. Cancer Cell. 2017;32:590-607. doi: 10.1016/j.ccell.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 24.Kim H., Yoo J.E., Cho J.Y., Oh B.K., Yoon Y.S., Han H.S., Lee H.S., Jang J.J., Jeong S.H., Kim J.W., Park Y.N. Telomere length, TERT and shelterin complex proteins in hepatocellular carcinomas expressing “stemness”-related markers. J. Hepatol. 2013;59(4):746–752. doi: 10.1016/j.jhep.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 25.Ramsay A.J., Quesada V., Foronda M., Conde L., Martínez-Trillos A., Villamor N., Rodríguez D., Kwarciak A., Garabaya C., Gallardo M., López-Guerra M., López-Guillermo A., Puente X.S., Blasco M.A., Campo E., López-Otín C. POT1 mutations cause telomere dysfunction in chronic lymphocytic leukemia. Nat. Genet. 2013;45(5):526–530. doi: 10.1038/ng.2584. [DOI] [PubMed] [Google Scholar]
- 26.Yuan X., Kronström M., Hellenius M.L., Cederholm T., Xu D., Sjögren P. Longitudinal changes in leukocyte telomere length and mortality in elderly Swedish men. Aging (Albany NY) 2018;10(10):3005–3016. doi: 10.18632/aging.101611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang A., Wang J., Zheng B., Fang X., Angström T., Liu C., Li X., Erlandsson F., Björkholm M., Nordenskjörd M., Gruber A., Wallin K.L., Xu D. Telomere attrition predominantly occurs in precursor lesions during in vivo carcinogenic process of the uterine cervix. Oncogene. 2004;23(44):7441–7447. doi: 10.1038/sj.onc.1207527. [DOI] [PubMed] [Google Scholar]
- 28.Augereau A., T’kint de Roodenbeke C., Simonet T., Bauwens S., Horard B., Callanan M., Leroux D., Jallades L., Salles G., Gilson E., Poncet D. Telomeric damage in early stage of chronic lymphocytic leukemia correlates with shelterin dysregulation. Blood. 2011;118(5):1316–1322. doi: 10.1182/blood-2010-07-295774. [DOI] [PubMed] [Google Scholar]
- 29.Kong F., Zheng C., Xu D. Telomerase as a “stemness” enzyme. Sci. China Life Sci. 2014;57(6):564–570. doi: 10.1007/s11427-014-4666-6. [DOI] [PubMed] [Google Scholar]
- 30.Wang J., Xie L.Y., Allan S., Beach D., Hannon G.J. Myc activates telomerase. Genes Dev. 1998;12(12):1769–1774. doi: 10.1101/gad.12.12.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ge Z., Liu C., Björkholm M., Gruber A., Xu D. Mitogen-activated protein kinase cascade-mediated histone H3 phosphorylation is critical for telomerase reverse transcriptase expression/telomerase activation induced by proliferation. Mol. Cell. Biol. 2006;26(1):230–237. doi: 10.1128/MCB.26.1.230-237.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffmeyer K., Raggioli A., Rudloff S., Anton R., Hierholzer A., Del Valle I., Hein K., Vogt R., Kemler R. Wnt/β-catenin signaling regulates telomerase in stem cells and cancer cells. Science. 2012;336(6088):1549–1554. doi: 10.1126/science.1218370. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y., Toh L., Lau P., Wang X. Human telomerase reverse transcriptase (hTERT) is a novel target of the Wnt/β-catenin pathway in human cancer. J. Biol. Chem. 2012;287(39):32494–32511. doi: 10.1074/jbc.M112.368282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin S.Y., Elledge S.J. Multiple tumor suppressor pathways negatively regulate telomerase. Cell. 2003;113(7):881–889. doi: 10.1016/S0092-8674(03)00430-6. [DOI] [PubMed] [Google Scholar]
- 35.Grasselli A., Nanni S., Colussi C., Aiello A., Benvenuti V., Ragone G., Moretti F., Sacchi A., Bacchetti S., Gaetano C., Capogrossi M.C., Pontecorvi A., Farsetti A. Estrogen receptor-alpha and endothelial nitric oxide synthase nuclear complex regulates transcription of human telomerase. Circ. Res. 2008;103(1):34–42. doi: 10.1161/CIRCRESAHA.107.169037. [DOI] [PubMed] [Google Scholar]
- 36.Kyo S., Takakura M., Kanaya T., Zhuo W., Fujimoto K., Nishio Y., Orimo A., Inoue M. Estrogen activates telomerase. Cancer Res. 1999;59(23):5917–5921. [PubMed] [Google Scholar]
- 37.Willing C., Peich M., Danescu A., Kehlen A., Fowler P.A., Hombach-Klonisch S. Estrogen-independent actions of environmentally relevant AhR-agonists in human endometrial epithelial cells. Mol. Hum. Reprod. 2011;17(2):115–126. doi: 10.1093/molehr/gaq081. [DOI] [PubMed] [Google Scholar]
- 38.Benko A.L., Olsen N.J., Kovacs W.J. Estrogen and telomerase in human peripheral blood mononuclear cells. Mol. Cell. Endocrinol. 2012;364(1-2):83–88. doi: 10.1016/j.mce.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang A., Zheng C., Hou M., Lindvall C., Li K.J., Erlandsson F., Björkholm M., Gruber A., Blennow E., Xu D. Deletion of the telomerase reverse transcriptase gene and haploinsufficiency of telomere maintenance in Cri du chat syndrome. Am. J. Hum. Genet. 2003;72(4):940–948. doi: 10.1086/374565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takakura M., Kyo S., Kanaya T., Hirano H., Takeda J., Yutsudo M., Inoue M. Cloning of human telomerase catalytic subunit (hTERT) gene promoter and identification of proximal core promoter sequences essential for transcriptional activation in immortalized and cancer cells. Cancer Res. 1999;59(3):551–557. [PubMed] [Google Scholar]
- 41.Cong Y.S., Wen J., Bacchetti S. The human telomerase catalytic subunit hTERT: organization of the gene and characterization of the promoter. Hum. Mol. Genet. 1999;8(1):137–142. doi: 10.1093/hmg/8.1.137. [DOI] [PubMed] [Google Scholar]
- 42.Xu D., Popov N., Hou M., Wang Q., Björkholm M., Gruber A., Menkel A.R., Henriksson M. Switch from Myc/Max to Mad1/Max binding and decrease in histone acetylation at the telomerase reverse transcriptase promoter during differentiation of HL60 cells. Proc. Natl. Acad. Sci. USA. 2001;98(7):3826–3831. doi: 10.1073/pnas.071043198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Casillas M.A., Brotherton S.L., Andrews L.G., Ruppert J.M., Tollefsbol T.O. Induction of endogenous telomerase (hTERT) by c-Myc in WI-38 fibroblasts transformed with specific genetic elements. Gene. 2003;316:57–65. doi: 10.1016/S0378-1119(03)00739-X. [DOI] [PubMed] [Google Scholar]
- 44.Gewin L., Myers H., Kiyono T., Galloway D.A. Identification of a novel telomerase repressor that interacts with the human papillomavirus type-16 E6/E6-AP complex. Genes Dev. 2004;18(18):2269–2282. doi: 10.1101/gad.1214704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koshiji M., Kageyama Y., Pete E.A., Horikawa I., Barrett J.C., Huang L.E. HIF-1alpha induces cell cycle arrest by functionally counteracting Myc. EMBO J. 2004;23(9):1949–1956. doi: 10.1038/sj.emboj.7600196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y., Liu L., Tollefsbol T.O. Glucose restriction can extend normal cell lifespan and impair precancerous cell growth through epigenetic control of hTERT and p16 expression. FASEB J. 2010;24(5):1442–1453. doi: 10.1096/fj.09-149328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu D., Dwyer J., Li H., Duan W., Liu J.P. Ets2 maintains hTERT gene expression and breast cancer cell proliferation by interacting with c-Myc. J. Biol. Chem. 2008;283(35):23567–23580. doi: 10.1074/jbc.M800790200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chou W.C., Hawkins A.L., Barrett J.F., Griffin C.A., Dang C.V. Arsenic inhibition of telomerase transcription leads to genetic instability. J. Clin. Invest. 2001;108(10):1541–1547. doi: 10.1172/JCI14064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu L., Liu C., Lou F., Zhang G., Wang X., Fan Y., Yan K., Wang K., Xu Z., Hu S., Björkholm M., Xu D. Activation of telomerase by seminal plasma in malignant and normal cervical epithelial cells. J. Pathol. 2011;225(2):203–211. doi: 10.1002/path.2914. [DOI] [PubMed] [Google Scholar]
- 50.Sitaram R.T., Cairney C.J., Grabowski P., Keith W.N., Hallberg B., Ljungberg B., Roos G. The PTEN regulator DJ-1 is associated with hTERT expression in clear cell renal cell carcinoma. Int. J. Cancer. 2009;125(4):783–790. doi: 10.1002/ijc.24335. [DOI] [PubMed] [Google Scholar]
- 51.Xu D., Wang Q., Gruber A., Björkholm M., Chen Z., Zaid A., Selivanova G., Peterson C., Wiman K.G., Pisa P. Downregulation of telomerase reverse transcriptase mRNA expression by wild type p53 in human tumor cells. Oncogene. 2000;19(45):5123–5133. doi: 10.1038/sj.onc.1203890. [DOI] [PubMed] [Google Scholar]
- 52.Mitchell T.J., Turajlic S., Rowan A. Timing the landmark events in the evolution of clear cell renal cell cancer: TRACERx Renal. Cell. 2018;173:611-623. doi: 10.1016/j.cell.2018.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen X., Kost J., Sulovari A., Wong N., Liang W.S., Cao J., Li D. A virome-wide clonal integration analysis platform for discovering cancer viral etiology. Genome Res. 2019;29(5):819–830. doi: 10.1101/gr.242529.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strååt K., Liu C., Rahbar A., Zhu Q., Liu L., Wolmer-Solberg N., Lou F., Liu Z., Shen J., Jia J., Kyo S., Björkholm M., Sjöberg J., Söderberg-Nauclér C., Xu D. Activation of telomerase by human cytomegalovirus. J. Natl. Cancer Inst. 2009;101(7):488–497. doi: 10.1093/jnci/djp031. [DOI] [PubMed] [Google Scholar]
- 55.Bellon M., Nicot C. Regulation of telomerase and telomeres: human tumor viruses take control. J. Natl. Cancer Inst. 2008;100(2):98–108. doi: 10.1093/jnci/djm269. [DOI] [PubMed] [Google Scholar]
- 56.Lewis K.A., Tollefsbol T.O. Regulation of the telomerase Reverse transcriptase subunit through epigenetic mechanisms. Front. Genet. 2016;7:83. doi: 10.3389/fgene.2016.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu L., Liu C., Fotouhi O., Fan Y., Wang K., Xia C., Shi B., Zhang G., Wang K., Kong F., Larsson C., Hu S., Xu D. TERT promoter hypermethylation in gastrointestinal cancer: a potential stool biomarker. Oncologist. 2017;22(10):1178–1188. doi: 10.1634/theoncologist.2017-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee D.D., Leão R., Komosa M., Gallo M., Zhang C.H., Lipman T., Remke M., Heidari A., Nunes N.M., Apolónio J.D., Price A.J., De Mello R.A., Dias J.S., Huntsman D., Hermanns T., Wild P.J., Vanner R., Zadeh G., Karamchandani J., Das S., Taylor M.D., Hawkins C.E., Wasserman J.D., Figueiredo A., Hamilton R.J., Minden M.D., Wani K., Diplas B., Yan H., Aldape K., Akbari M.R., Danesh A., Pugh T.J., Dirks P.B., Castelo-Branco P., Tabori U. DNA hypermethylation within TERT promoter upregulates TERT expression in cancer. J. Clin. Invest. 2019;129(1):223–229. doi: 10.1172/JCI121303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ge Z., Li W., Wang N., Liu C., Zhu Q., Björkholm M., Gruber A., Xu D. Chromatin remodeling: recruitment of histone demethylase RBP2 by Mad1 for transcriptional repression of a Myc target gene, telomerase reverse transcriptase. FASEB J. 2010;24(2):579–586. doi: 10.1096/fj.09-140087. [DOI] [PubMed] [Google Scholar]
- 60.Liu C., Fang X., Ge Z., Jalink M., Kyo S., Björkholm M., Gruber A., Sjöberg J., Xu D. The telomerase reverse transcriptase (hTERT) gene is a direct target of the histone methyltransferase SMYD3. Cancer Res. 2007;67(6):2626–2631. doi: 10.1158/0008-5472.CAN-06-4126. [DOI] [PubMed] [Google Scholar]
- 61.Castelo-Branco P., Choufani S., Mack S., Gallagher D., Zhang C., Lipman T., Zhukova N., Walker E.J., Martin D., Merino D., Wasserman J.D., Elizabeth C., Alon N., Zhang L., Hovestadt V., Kool M., Jones D.T., Zadeh G., Croul S., Hawkins C., Hitzler J., Wang J.C., Baruchel S., Dirks P.B., Malkin D., Pfister S., Taylor M.D., Weksberg R., Tabori U. Methylation of the TERT promoter and risk stratification of childhood brain tumours: an integrative genomic and molecular study. Lancet Oncol. 2013;14(6):534–542. doi: 10.1016/S1470-2045(13)70110-4. [DOI] [PubMed] [Google Scholar]
- 62.Hou M., Wang X., Popov N., Zhang A., Zhao X., Zhou R., Zetterberg A., Björkholm M., Henriksson M., Gruber A., Xu D. The histone deacetylase inhibitor trichostatin A derepresses the telomerase reverse transcriptase (hTERT) gene in human cells. Exp. Cell Res. 2002;274(1):25–34. doi: 10.1006/excr.2001.5462. [DOI] [PubMed] [Google Scholar]
- 63.Kim W., Ludlow A.T., Min J., Robin J.D., Stadler G., Mender I., Lai T.P., Zhang N., Wright W.E., Shay J.W. Regulation of the human telomerase gene tert by telomere position effect-over long distances (TPE-OLD): implications for aging and cancer. PLoS Biol. 2016;14(12):, e2000016. doi: 10.1371/journal.pbio.2000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ma R, Liu C, Lu M. The TERT locus genotypes of rs2736100- CC/CA and rs2736098-AA predict shorter survival in renal cell carcinoma. Urol. Oncol. 2019;37(301):e1-301-e10. doi: 10.1016/j.urolonc.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 65.Yuan X., Cheng G., Yu J., Zheng S., Sun C., Sun Q., Li K., Lin Z., Liu T., Li P., Xu Y., Kong F., Bjorkholm M., Xu D. The TERT promoter mutation incidence is modified by germline TERT rs2736098 and rs2736100 polymorphisms in hepatocellular carcinoma. Oncotarget. 2017;8(14):23120–23129. doi: 10.18632/oncotarget.15498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mocellin S., Verdi D., Pooley K.A., Landi M.T., Egan K.M., Baird D.M., Prescott J., De Vivo I., Nitti D. Telomerase reverse transcriptase locus polymorphisms and cancer risk: a field synopsis and meta-analysis. J. Natl. Cancer Inst. 2012;104(11):840–854. doi: 10.1093/jnci/djs222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang F.W., Hodis E., Xu M.J., Kryukov G.V., Chin L., Garraway L.A. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339(6122):957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Horn S., Figl A., Rachakonda P.S., Fischer C., Sucker A., Gast A., Kadel S., Moll I., Nagore E., Hemminki K., Schadendorf D., Kumar R. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339(6122):959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 69.Liu T., Yuan X., Xu D. Cancer-Specific telomerase reverse transcriptase (tert) promoter mutations: biological and clinical implications. Genes (Basel) 2016;7(7):, E38. doi: 10.3390/genes7070038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Killela P.J., Reitman Z.J., Jiao Y., Bettegowda C., Agrawal N., Diaz L.A., Jr, Friedman A.H., Friedman H., Gallia G.L., Giovanella B.C., Grollman A.P., He T.C., He Y., Hruban R.H., Jallo G.I., Mandahl N., Meeker A.K., Mertens F., Netto G.J., Rasheed B.A., Riggins G.J., Rosenquist T.A., Schiffman M., Shih IeM., Theodorescu D., Torbenson M.S., Velculescu V.E., Wang T.L., Wentzensen N., Wood L.D., Zhang M., McLendon R.E., Bigner D.D., Kinzler K.W., Vogelstein B., Papadopoulos N., Yan H. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc. Natl. Acad. Sci. USA. 2013;110(15):6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang K., Liu T., Liu C., Meng Y., Yuan X., Liu L., Ge N., Liu J., Wang C., Ren H., Yan K., Hu S., Xu Z., Fan Y., Xu D. TERT promoter mutations and TERT mRNA but not FGFR3 mutations are urinary biomarkers in Han Chinese patients with urothelial bladder cancer. Oncologist. 2015;20(3):263–269. doi: 10.1634/theoncologist.2014-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu T., Wang N., Cao J., Sofiadis A., Dinets A., Zedenius J., Larsson C., Xu D. The age- and shorter telomere-dependent TERT promoter mutation in follicular thyroid cell-derived carcinomas. Oncogene. 2014;33(42):4978–4984. doi: 10.1038/onc.2013.446. [DOI] [PubMed] [Google Scholar]
- 73.Wang K., Liu T., Liu L., Liu J., Liu C., Wang C., Ge N., Ren H., Yan K., Hu S., Björkholm M., Fan Y., Xu D. TERT promoter mutations in renal cell carcinomas and upper tract urothelial carcinomas. Oncotarget. 2014;5(7):1829–1836. doi: 10.18632/oncotarget.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang N., Liu T., Sofiadis A., Juhlin C.C., Zedenius J., Höög A., Larsson C., Xu D. TERT promoter mutation as an early genetic event activating telomerase in follicular thyroid adenoma (FTA) and atypical FTA. Cancer. 2014;120(19):2965–2979. doi: 10.1002/cncr.28800. [DOI] [PubMed] [Google Scholar]
- 75.Li C., Wu S., Wang H., Bi X., Yang Z., Du Y., He L., Cai Z., Wang J., Fan Z. The C228T mutation of TERT promoter frequently occurs in bladder cancer stem cells and contributes to tumorigenesis of bladder cancer. Oncotarget. 2015;6(23):19542–19551. doi: 10.18632/oncotarget.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bell R.J., Rube H.T., Kreig A., Mancini A., Fouse S.D., Nagarajan R.P., Choi S., Hong C., He D., Pekmezci M., Wiencke J.K., Wrensch M.R., Chang S.M., Walsh K.M., Myong S., Song J.S., Costello J.F. Cancer. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science. 2015;348(6238):1036–1039. doi: 10.1126/science.aab0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stern J.L., Theodorescu D., Vogelstein B., Papadopoulos N., Cech T.R. Mutation of the TERT promoter, switch to active chromatin, and monoallelic TERT expression in multiple cancers. Genes Dev. 2015;29(21):2219–2224. doi: 10.1101/gad.269498.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mancini A., Xavier-Magalhães A., Woods W.S. Disruption of the beta1L isoform of GABP reverses glioblastoma replicative immortality in a tert promoter mutation-dependent manner. Cancer Cell. 2018;34:513-528. doi: 10.1016/j.ccell.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yuan X., Mu N., Wang N., Strååt K., Sofiadis A., Guo Y., Stenman A., Li K., Cheng G., Zhang L., Kong F., Ekblad L., Wennerberg J., Nilsson I.L., Juhlin C.C., Larsson C., Xu D. GABPA inhibits invasion/metastasis in papillary thyroid carcinoma by regulating DICER1 expression. Oncogene. 2019;38(7):965–979. doi: 10.1038/s41388-018-0483-x. [DOI] [PubMed] [Google Scholar]
- 80.Guo Y., Yuan X., Li K., Dai M., Zhang L., Wu Y., Sun C., Chen Y., Cheng G., Liu C., Strååt K., Kong F., Zhao S., Bjorkhölm M., Xu D. GABPA is a master regulator of luminal identity and restrains aggressive diseases in bladder cancer. Cell Death Differ. 2019 doi: 10.1038/s41418-019-0466-7. Epub ahead of Print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Valentijn L.J., Koster J., Zwijnenburg D.A., Hasselt N.E., van Sluis P., Volckmann R., van Noesel M.M., George R.E., Tytgat G.A., Molenaar J.J., Versteeg R. TERT rearrangements are frequent in neuroblastoma and identify aggressive tumors. Nat. Genet. 2015;47(12):1411–1414. doi: 10.1038/ng.3438. [DOI] [PubMed] [Google Scholar]
- 82.Peifer M., Hertwig F., Roels F., Dreidax D., Gartlgruber M., Menon R., Krämer A., Roncaioli J.L., Sand F., Heuckmann J.M., Ikram F., Schmidt R., Ackermann S., Engesser A., Kahlert Y., Vogel W., Altmüller J., Nürnberg P., Thierry-Mieg J., Thierry-Mieg D., Mariappan A., Heynck S., Mariotti E., Henrich K.O., Gloeckner C., Bosco G., Leuschner I., Schweiger M.R., Savelyeva L., Watkins S.C., Shao C., Bell E., Höfer T., Achter V., Lang U., Theissen J., Volland R., Saadati M., Eggert A., de Wilde B., Berthold F., Peng Z., Zhao C., Shi L., Ortmann M., Büttner R., Perner S., Hero B., Schramm A., Schulte J.H., Herrmann C., O’Sullivan R.J., Westermann F., Thomas R.K., Fischer M. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature. 2015;526(7575):700–704. doi: 10.1038/nature14980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ackermann S., Cartolano M., Hero B., Welte A., Kahlert Y., Roderwieser A., Bartenhagen C., Walter E., Gecht J., Kerschke L., Volland R., Menon R., Heuckmann J.M., Gartlgruber M., Hartlieb S., Henrich K.O., Okonechnikov K., Altmüller J., Nürnberg P., Lefever S., de Wilde B., Sand F., Ikram F., Rosswog C., Fischer J., Theissen J., Hertwig F., Singhi A.D., Simon T., Vogel W., Perner S., Krug B., Schmidt M., Rahmann S., Achter V., Lang U., Vokuhl C., Ortmann M., Büttner R., Eggert A., Speleman F., O’Sullivan R.J., Thomas R.K., Berthold F., Vandesompele J., Schramm A., Westermann F., Schulte J.H., Peifer M., Fischer M. A mechanistic classification of clinical phenotypes in neuroblastoma. Science. 2018;362(6419):1165–1170. doi: 10.1126/science.aat6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Heaphy C.M., de Wilde R.F., Jiao Y., Klein A.P., Edil B.H., Shi C., Bettegowda C., Rodriguez F.J., Eberhart C.G., Hebbar S., Offerhaus G.J., McLendon R., Rasheed B.A., He Y., Yan H., Bigner D.D., Oba-Shinjo S.M., Marie S.K., Riggins G.J., Kinzler K.W., Vogelstein B., Hruban R.H., Maitra A., Papadopoulos N., Meeker A.K. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333(6041):425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Horikawa I., Barrett J.C. cis-Activation of the human telomerase gene (hTERT) by the hepatitis B virus genome. J. Natl. Cancer Inst. 2001;93(15):1171–1173. doi: 10.1093/jnci/93.15.1171. [DOI] [PubMed] [Google Scholar]
- 86.Sung W.K., Zheng H., Li S., Chen R., Liu X., Li Y., Lee N.P., Lee W.H., Ariyaratne P.N., Tennakoon C., Mulawadi F.H., Wong K.F., Liu A.M., Poon R.T., Fan S.T., Chan K.L., Gong Z., Hu Y., Lin Z., Wang G., Zhang Q., Barber T.D., Chou W.C., Aggarwal A., Hao K., Zhou W., Zhang C., Hardwick J., Buser C., Xu J., Kan Z., Dai H., Mao M., Reinhard C., Wang J., Luk J.M. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat. Genet. 2012;44(7):765–769. doi: 10.1038/ng.2295. [DOI] [PubMed] [Google Scholar]
- 87.Nault J.C., Datta S., Imbeaud S., Franconi A., Mallet M., Couchy G., Letouzé E., Pilati C., Verret B., Blanc J.F., Balabaud C., Calderaro J., Laurent A., Letexier M., Bioulac-Sage P., Calvo F., Zucman-Rossi J. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nat. Genet. 2015;47(10):1187–1193. doi: 10.1038/ng.3389. [DOI] [PubMed] [Google Scholar]
- 88.Chang K.P., Wang C.I., Pickering C.R., Huang Y., Tsai C.N., Tsang N.M., Kao H.K., Cheng M.H., Myers J.N. Prevalence of promoter mutations in the TERT gene in oral cavity squamous cell carcinoma. Head Neck. 2017;39(6):1131–1137. doi: 10.1002/hed.24728. [DOI] [PubMed] [Google Scholar]
- 89.Balsara B.R., Sonoda G., du Manoir S., Siegfried J.M., Gabrielson E., Testa J.R. Comparative genomic hybridization analysis detects frequent, often high-level, overrepresentation of DNA sequences at 3q, 5p, 7p, and 8q in human non-small cell lung carcinomas. Cancer Res. 1997;57(11):2116–2120. [PubMed] [Google Scholar]
- 90.Meyerson M., Counter C.M., Eaton E.N., Ellisen L.W., Steiner P., Caddle S.D., Ziaugra L., Beijersbergen R.L., Davidoff M.J., Liu Q., Bacchetti S., Haber D.A., Weinberg R.A. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90(4):785–795. doi: 10.1016/S0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 91.Kilian A., Bowtell D.D., Abud H.E., Hime G.R., Venter D.J., Keese P.K., Duncan E.L., Reddel R.R., Jefferson R.A. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum. Mol. Genet. 1997;6(12):2011–2019. doi: 10.1093/hmg/6.12.2011. [DOI] [PubMed] [Google Scholar]
- 92.Zhang A., Zheng C., Lindvall C., Hou M., Ekedahl J., Lewensohn R., Yan Z., Yang X., Henriksson M., Blennow E., Nordenskjöld M., Zetterberg A., Björkholm M., Gruber A., Xu D. Frequent amplification of the telomerase reverse transcriptase gene in human tumors. Cancer Res. 2000;60(22):6230–6235. [PubMed] [Google Scholar]
- 93.Dahlström J., Liu T., Yuan X., Saft L., Ghaderi M., Wei Y.B., Lavebratt C., Li P., Zheng C., Björkholm M., Xu D. TERT rs2736100 genotypes are associated with differential risk of myeloproliferative neoplasms in Swedish and Chinese male patient populations. Ann. Hematol. 2016;95(11):1825–1832. doi: 10.1007/s00277-016-2787-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wei R., Cao L., Pu H., Wang H., Zheng Y., Niu X., Weng X., Zhang H., Favus M., Zhang L., Jia W., Zeng Y., Amos C.I., Lu S., Wang H.Y., Liu Y., Liu W. TERT Polymorphism rs2736100-C is associated with egfr mutation-positive non-small cell lung Cancer. Clin. Cancer Res. 2015;21(22):5173–5180. doi: 10.1158/1078-0432.CCR-15-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bojesen S.E., Pooley K.A., Johnatty S.E. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat. Genet. 2013;45:371-384. doi: 10.1038/ng.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rachakonda P.S., Hosen I., de Verdier P.J., Fallah M., Heidenreich B., Ryk C., Wiklund N.P., Steineck G., Schadendorf D., Hemminki K., Kumar R. TERT promoter mutations in bladder cancer affect patient survival and disease recurrence through modification by a common polymorphism. Proc. Natl. Acad. Sci. USA. 2013;110(43):17426–17431. doi: 10.1073/pnas.1310522110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cheng G., Yuan X., Wang F., Sun Q., Xin Q., Li K., Sun C., Lin Z., Luan Y., Xu Y., Li P., Kong F., Xu D. Association between the telomerase rs2736098_tt genotype and a lower risk of chronic hepatitis b and cirrhosis in chinese males. Clin. Transl. Gastroenterol. 2017;8(3):, e79. doi: 10.1038/ctg.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wong M.S., Wright W.E., Shay J.W. Alternative splicing regulation of telomerase: a new paradigm? Trends Genet. 2014;30(10):430–438. doi: 10.1016/j.tig.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Avin B.A., Umbricht C.B., Zeiger M.A. Human telomerase reverse transcriptase regulation by DNA methylation, transcription factor binding and alternative splicing (Review). Int. J. Oncol. 2016;49(6):2199–2205. doi: 10.3892/ijo.2016.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fan Y., Liu Z., Fang X., Ge Z., Ge N., Jia Y., Sun P., Lou F., Björkholm M., Gruber A., Ekman P., Xu D. Differential expression of full-length telomerase reverse transcriptase mRNA and telomerase activity between normal and malignant renal tissues. Clin. Cancer Res. 2005;11(12):4331–4337. doi: 10.1158/1078-0432.CCR-05-0099. [DOI] [PubMed] [Google Scholar]
- 101.Jalink M., Ge Z., Liu C., Björkholm M., Gruber A., Xu D. Human normal T lymphocytes and lymphoid cell lines do express alternative splicing variants of human telomerase reverse transcriptase (hTERT) mRNA. Biochem. Biophys. Res. Commun. 2007;353(4):999–1003. doi: 10.1016/j.bbrc.2006.12.149. [DOI] [PubMed] [Google Scholar]
- 102.Selvanathan S.P., Graham G.T., Erkizan H.V., Dirksen U., Natarajan T.G., Dakic A., Yu S., Liu X., Paulsen M.T., Ljungman M.E., Wu C.H., Lawlor E.R., Üren A., Toretsky J.A. Oncogenic fusion protein EWS-FLI1 is a network hub that regulates alternative splicing. Proc. Natl. Acad. Sci. USA. 2015;112(11):E1307–E1316. doi: 10.1073/pnas.1500536112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ito T., Watanabe H., Yamamichi N., Kondo S., Tando T., Haraguchi T., Mizutani T., Sakurai K., Fujita S., Izumi T., Isobe T., Iba H. Brm transactivates the telomerase reverse transcriptase (TERT) gene and modulates the splicing patterns of its transcripts in concert with p54(nrb). Biochem. J. 2008;411(1):201–209. doi: 10.1042/BJ20071075. [DOI] [PubMed] [Google Scholar]
- 104.Listerman I., Sun J., Gazzaniga F.S., Lukas J.L., Blackburn E.H. The major reverse transcriptase-incompetent splice variant of the human telomerase protein inhibits telomerase activity but protects from apoptosis. Cancer Res. 2013;73(9):2817–2828. doi: 10.1158/0008-5472.CAN-12-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sayed M.E., Yuan L., Robin J.D., Tedone E., Batten K., Dahlson N., Wright W.E., Shay J.W., Ludlow A.T. NOVA1 directs PTBP1 to hTERT pre-mRNA and promotes telomerase activity in cancer cells. Oncogene. 2019;38(16):2937–2952. doi: 10.1038/s41388-018-0639-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li H., Zhao L., Yang Z., Funder J.W., Liu J.P. Telomerase is controlled by protein kinase Calpha in human breast cancer cells. J. Biol. Chem. 1998;273(50):33436–33442. doi: 10.1074/jbc.273.50.33436. [DOI] [PubMed] [Google Scholar]
- 107.Kang S.S., Kwon T., Kwon D.Y., Do S.I. Akt protein kinase enhances human telomerase activity through phosphorylation of telomerase reverse transcriptase subunit. J. Biol. Chem. 1999;274(19):13085–13090. doi: 10.1074/jbc.274.19.13085. [DOI] [PubMed] [Google Scholar]
- 108.Chang J.T., Lu Y.C., Chen Y.J., Tseng C.P., Chen Y.L., Fang C.W., Cheng A.J. hTERT phosphorylation by PKC is essential for telomerase holoprotein integrity and enzyme activity in head neck cancer cells. Br. J. Cancer. 2006;94(6):870–878. doi: 10.1038/sj.bjc.6603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Haendeler J., Hoffmann J., Brandes R.P., Zeiher A.M., Dimmeler S. Hydrogen peroxide triggers nuclear export of telomerase reverse transcriptase via Src kinase family-dependent phosphorylation of tyrosine 707. Mol. Cell. Biol. 2003;23(13):4598–4610. doi: 10.1128/MCB.23.13.4598-4610.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kharbanda S., Kumar V., Dhar S., Pandey P., Chen C., Majumder P., Yuan Z.M., Whang Y., Strauss W., Pandita T.K., Weaver D., Kufe D. Regulation of the hTERT telomerase catalytic subunit by the c-Abl tyrosine kinase. Curr. Biol. 2000;10(10):568–575. doi: 10.1016/S0960-9822(00)00483-8. [DOI] [PubMed] [Google Scholar]
- 111.Jeong S.A., Kim K., Lee J.H., Cha J.S., Khadka P., Cho H.S., Chung I.K. Akt-mediated phosphorylation increases the binding affinity of hTERT for importin α to promote nuclear translocation. J. Cell Sci. 2015;128(15):2951. doi: 10.1242/jcs.176453. [DOI] [PubMed] [Google Scholar]
- 112.Miwa S., Czapiewski R., Wan T., Bell A., Hill K.N., von Zglinicki T., Saretzki G. Decreased mTOR signalling reduces mitochondrial ROS in brain via accumulation of the telomerase protein TERT within mitochondria. Aging (Albany NY) 2016;8(10):2551–2567. doi: 10.18632/aging.101089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Forsythe H.L., Jarvis J.L., Turner J.W., Elmore L.W., Holt S.E. Stable association of hsp90 and p23, but Not hsp70, with active human telomerase. J. Biol. Chem. 2001;276(19):15571–15574. doi: 10.1074/jbc.C100055200. [DOI] [PubMed] [Google Scholar]
- 114.Kim J.H., Park S.M., Kang M.R., Oh S.Y., Lee T.H., Muller M.T., Chung I.K. Ubiquitin ligase MKRN1 modulates telomere length homeostasis through a proteolysis of hTERT. Genes Dev. 2005;19(7):776–781. doi: 10.1101/gad.1289405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jeong Y.Y., Her J., Oh S.Y., Chung I.K. Hsp90-binding immunophilin FKBP52 modulates telomerase activity by promoting the cytoplasmic retrotransport of hTERT. Biochem. J. 2016;473(20):3517–3532. doi: 10.1042/BCJ20160344. [DOI] [PubMed] [Google Scholar]
- 116.Lee J.H., Khadka P., Baek S.H., Chung I.K. CHIP promotes human telomerase reverse transcriptase degradation and negatively regulates telomerase activity. J. Biol. Chem. 2010;285(53):42033–42045. doi: 10.1074/jbc.M110.149831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Oh W., Lee E.W., Lee D., Yang M.R., Ko A., Yoon C.H., Lee H.W., Bae Y.S., Choi C.Y., Song J. Hdm2 negatively regulates telomerase activity by functioning as an E3 ligase of hTERT. Oncogene. 2010;29(28):4101–4112. doi: 10.1038/onc.2010.160. [DOI] [PubMed] [Google Scholar]
- 118.Farooqi A.A., Mansoor Q., Alaaeddine N., Xu B. MicroRNA regulation of telomerase reverse transcriptase (tert): micro machines pull strings of papier-mâché Puppets. Int. J. Mol. Sci. 2018;19(4):, E1051. doi: 10.3390/ijms19041051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lü M.H., Tang B., Zeng S., Hu C.J., Xie R., Wu Y.Y., Wang S.M., He F.T., Yang S.M. Long noncoding RNA BC032469, a novel competing endogenous RNA, upregulates hTERT expression by sponging miR-1207-5p and promotes proliferation in gastric cancer. Oncogene. 2016;35(27):3524–3534. doi: 10.1038/onc.2015.413. [DOI] [PubMed] [Google Scholar]
- 120.Aiello A., Bacci L., Re A., Ripoli C., Pierconti F., Pinto F., Masetti R., Grassi C., Gaetano C., Bassi P.F., Pontecorvi A., Nanni S., Farsetti A. MALAT1 and HOTAIR long non-coding rnas play opposite role in estrogen-mediated transcriptional regulation in prostate cancer cells. Sci. Rep. 2016;6:38414. doi: 10.1038/srep38414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Trapp S., Parcells M.S., Kamil J.P., Schumacher D., Tischer B.K., Kumar P.M., Nair V.K., Osterrieder N. A virus-encoded telomerase RNA promotes malignant T cell lymphomagenesis. J. Exp. Med. 2006;203(5):1307–1317. doi: 10.1084/jem.20052240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kheimar A., Trimpert J., Groenke N., Kaufer B.B. Overexpression of cellular telomerase RNA enhances virus-induced cancer formation. Oncogene. 2019;38(10):1778–1786. doi: 10.1038/s41388-018-0544-1. [DOI] [PubMed] [Google Scholar]
- 123.Chin L., Artandi S.E., Shen Q., Tam A., Lee S.L., Gottlieb G.J., Greider C.W., DePinho R.A. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97(4):527–538. doi: 10.1016/S0092-8674(00)80762-X. [DOI] [PubMed] [Google Scholar]
- 124.González-Suárez E., Samper E., Flores J.M., Blasco M.A. Telomerase-deficient mice with short telomeres are resistant to skin tumorigenesis. Nat. Genet. 2000;26(1):114–117. doi: 10.1038/79089. [DOI] [PubMed] [Google Scholar]
- 125.Alter B.P., Giri N., Savage S.A., Rosenberg P.S. Cancer in dyskeratosis congenita. Blood. 2009;113(26):6549–6557. doi: 10.1182/blood-2008-12-192880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kedde M., le Sage C., Duursma A., Zlotorynski E., van Leeuwen B., Nijkamp W., Beijersbergen R., Agami R. Telomerase-independent regulation of ATR by human telomerase RNA. J. Biol. Chem. 2006;281(52):40503–40514. doi: 10.1074/jbc.M607676200. [DOI] [PubMed] [Google Scholar]
- 127.Liu H., Yang Y., Ge Y., Liu J., Zhao Y. TERC promotes cellular inflammatory response independent of telomerase. Nucleic Acids Res. 2019;47(15):8084–8095. doi: 10.1093/nar/gkz584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Baena-Del Valle J.A., Zheng Q., Esopi D.M., Rubenstein M., Hubbard G.K., Moncaliano M.C., Hruszkewycz A., Vaghasia A., Yegnasubramanian S., Wheelan S.J., Meeker A.K., Heaphy C.M., Graham M.K., De Marzo A.M. MYC drives overexpression of telomerase RNA (hTR/TERC) in prostate cancer. J. Pathol. 2018;244(1):11–24. doi: 10.1002/path.4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yi X., Tesmer V.M., Savre-Train I., Shay J.W., Wright W.E. Both transcriptional and posttranscriptional mechanisms regulate human telomerase template RNA levels. Mol. Cell. Biol. 1999;19(6):3989–3997. doi: 10.1128/MCB.19.6.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wong J.M., Collins K. Telomerase RNA level limits telomere maintenance in X-linked dyskeratosis congenita. Genes Dev. 2006;20(20):2848–2858. doi: 10.1101/gad.1476206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Boyraz B., Moon D.H., Segal M., Muosieyiri M.Z., Aykanat A., Tai A.K., Cahan P., Agarwal S. Posttranscriptional manipulation of TERC reverses molecular hallmarks of telomere disease. J. Clin. Invest. 2016;126(9):3377–3382. doi: 10.1172/JCI87547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tummala H., Walne A., Collopy L., Cardoso S., de la Fuente J., Lawson S., Powell J., Cooper N., Foster A., Mohammed S., Plagnol V., Vulliamy T., Dokal I. Poly(A)-specific ribonuclease deficiency impacts telomere biology and causes dyskeratosis congenita. J. Clin. Invest. 2015;125(5):2151–2160. doi: 10.1172/JCI78963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Richards LA, Kumari A, Knezevic K. DKC1 is a transcriptional target of GATA1 and drives upregulation of telomerase activity in normal human erythroblasts. Haematologica. 2019. p. Pii: haematol.2018.515699. [DOI] [PMC free article] [PubMed]
- 134.Akalin A., Elmore L.W., Forsythe H.L., Amaker B.A., McCollum E.D., Nelson P.S., Ware J.L., Holt S.E. A novel mechanism for chaperone-mediated telomerase regulation during prostate cancer progression. Cancer Res. 2001;61(12):4791–4796. [PubMed] [Google Scholar]
- 135.Venteicher A.S., Meng Z., Mason P.J., Veenstra T.D., Artandi S.E. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell. 2008;132(6):945–957. doi: 10.1016/j.cell.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Li W., Zeng J., Li Q., Zhao L., Liu T., Björkholm M., Jia J., Xu D. Reptin is required for the transcription of telomerase reverse transcriptase and over-expressed in gastric cancer. Mol. Cancer. 2010;9:132. doi: 10.1186/1476-4598-9-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Flynn R.L., Cox K.E., Jeitany M., Wakimoto H., Bryll A.R., Ganem N.J., Bersani F., Pineda J.R., Suvà M.L., Benes C.H., Haber D.A., Boussin F.D., Zou L. Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science. 2015;347(6219):273–277. doi: 10.1126/science.1257216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yost K.E., Clatterbuck Soper S.F., Walker R.L., Pineda M.A., Zhu Y.J., Ester C.D., Showman S., Roschke A.V., Waterfall J.J., Meltzer P.S. Rapid and reversible suppression of ALT by DAXX in osteosarcoma cells. Sci. Rep. 2019;9(1):4544. doi: 10.1038/s41598-019-41058-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Arora R., Lee Y., Wischnewski H., Brun C.M., Schwarz T., Azzalin C.M. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat. Commun. 2014;5:5220. doi: 10.1038/ncomms6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hu Y., Shi G., Zhang L., Li F., Jiang Y., Jiang S., Ma W., Zhao Y., Songyang Z., Huang J. Switch telomerase to ALT mechanism by inducing telomeric DNA damages and dysfunction of ATRX and DAXX. Sci. Rep. 2016;6:32280. doi: 10.1038/srep32280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.O’Sullivan R.J., Arnoult N., Lackner D.H., Oganesian L., Haggblom C., Corpet A., Almouzni G., Karlseder J. Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nat. Struct. Mol. Biol. 2014;21(2):167–174. doi: 10.1038/nsmb.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Liang X., Yuan X., Yu J., Wu Y., Li K., Sun C., Li S., Shen L., Kong F., Jia J., Björkholm M., Xu D. Histone Chaperone ASF1A predicts poor outcomes for patients with gastrointestinal cancer and drives cancer progression by stimulating transcription of β-catenin target genes. EBioMedicine. 2017;21:104–116. doi: 10.1016/j.ebiom.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wu Y., Li X., Yu J., Björkholm M., Xu D. ASF1a inhibition induces p53-dependent growth arrest and senescence of cancer cells. Cell Death Dis. 2019;10(2):76. doi: 10.1038/s41419-019-1357-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Herling C.D., Klaumünzer M., Rocha C.K., Altmüller J., Thiele H., Bahlo J., Kluth S., Crispatzu G., Herling M., Schiller J., Engelke A., Tausch E., Döhner H., Fischer K., Goede V., Nürnberg P., Reinhardt H.C., Stilgenbauer S., Hallek M., Kreuzer K.A. Complex karyotypes and KRAS and POT1 mutations impact outcome in CLL after chlorambucil-based chemotherapy or chemoimmunotherapy. Blood. 2016;128(3):395–404. doi: 10.1182/blood-2016-01-691550. [DOI] [PubMed] [Google Scholar]
- 145.Speedy H.E., Kinnersley B., Chubb D. Germ line mutations in shelterin complex genes are associated with familial chronic lymphocytic leukemia. Blood. 2016;128:2319–2326. doi: 10.1182/blood-2016-01-695692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Aoude L.G., Pritchard A.L., Robles-Espinoza C.D., Wadt K., Harland M., Choi J., Gartside M., Quesada V., Johansson P., Palmer J.M., Ramsay A.J., Zhang X., Jones K., Symmons J., Holland E.A., Schmid H., Bonazzi V., Woods S., Dutton-Regester K., Stark M.S., Snowden H., van Doorn R., Montgomery G.W., Martin N.G., Keane T.M., López-Otín C., Gerdes A.M., Olsson H., Ingvar C., Borg A., Gruis N.A., Trent J.M., Jönsson G., Bishop D.T., Mann G.J., Newton-Bishop J.A., Brown K.M., Adams D.J., Hayward N.K. Nonsense mutations in the shelterin complex genes ACD and TERF2IP in familial melanoma. J. Natl. Cancer Inst. 2014;107(2):, dju408. doi: 10.1093/jnci/dju408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Broderick P., Dobbins S.E., Chubb D. Validation of recently proposed colorectal cancer susceptibility gene variants in an analysis of families and patients-a systematic review. Gastroenterology. 2017;152:75-77. doi: 10.1053/j.gastro.2016.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bainbridge M.N., Armstrong G.N., Gramatges M.M., Bertuch A.A., Jhangiani S.N., Doddapaneni H., Lewis L., Tombrello J., Tsavachidis S., Liu Y., Jalali A., Plon S.E., Lau C.C., Parsons D.W., Claus E.B., Barnholtz-Sloan J., Il’yasova D., Schildkraut J., Ali-Osman F., Sadetzki S., Johansen C., Houlston R.S., Jenkins R.B., Lachance D., Olson S.H., Bernstein J.L., Merrell R.T., Wrensch M.R., Walsh K.M., Davis F.G., Lai R., Shete S., Aldape K., Amos C.I., Thompson P.A., Muzny D.M., Gibbs R.A., Melin B.S., Bondy M.L., Gliogene Consortium Germline mutations in shelterin complex genes are associated with familial glioma. J. Natl. Cancer Inst. 2014;107(1):384. doi: 10.1093/jnci/dju384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.McMaster M.L., Sun C., Landi M.T., Savage S.A., Rotunno M., Yang X.R., Jones K., Vogt A., Hutchinson A., Zhu B., Wang M., Hicks B., Thirunavukarason A., Stewart D.R., Koutros S., Goldstein A.M., Chanock S.J., Caporaso N.E., Tucker M.A., Goldin L.R., Liu Y. Germline mutations in Protection of Telomeres 1 in two families with Hodgkin lymphoma. Br. J. Haematol. 2018;181(3):372–377. doi: 10.1111/bjh.15203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Spinella J.F., Cassart P., Garnier N., Rousseau P., Drullion C., Richer C., Ouimet M., Saillour V., Healy J., Autexier C., Sinnett D. A novel somatic mutation in ACD induces telomere lengthening and apoptosis resistance in leukemia cells. BMC Cancer. 2015;15:621. doi: 10.1186/s12885-015-1639-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Poonepalli A., Banerjee B., Ramnarayanan K., Palanisamy N., Putti T.C., Hande M.P. Telomere-mediated genomic instability and the clinico-pathological parameters in breast cancer. Genes Chromosomes Cancer. 2008;47(12):1098–1109. doi: 10.1002/gcc.20608. [DOI] [PubMed] [Google Scholar]
- 152.Heng J., Zhang F., Guo X., Tang L., Peng L., Luo X., Xu X., Wang S., Dai L., Wang J. Integrated analysis of promoter methylation and expression of telomere related genes in breast cancer. Oncotarget. 2017;8(15):25442–25454. doi: 10.18632/oncotarget.16036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dinami R., Ercolani C., Petti E., Piazza S., Ciani Y., Sestito R., Sacconi A., Biagioni F., le Sage C., Agami R., Benetti R., Mottolese M., Schneider C., Blandino G., Schoeftner S. miR-155 drives telomere fragility in human breast cancer by targeting TRF1. Cancer Res. 2014;74(15):4145–4156. doi: 10.1158/0008-5472.CAN-13-2038. [DOI] [PubMed] [Google Scholar]
- 154.Miyachi K., Fujita M., Tanaka N., Sasaki K., Sunagawa M. Correlation between telomerase activity and telomeric-repeat binding factors in gastric cancer. J. Exp. Clin. Cancer Res. 2002;21(2):269–275. [PubMed] [Google Scholar]
- 155.Lin X., Gu J., Lu C., Spitz M.R., Wu X. Expression of telomere-associated genes as prognostic markers for overall survival in patients with non-small cell lung cancer. Clin. Cancer Res. 2006;12(19):5720–5725. doi: 10.1158/1078-0432.CCR-05-2809. [DOI] [PubMed] [Google Scholar]
- 156.Lantuejoul S., Raynaud C., Salameire D., Gazzeri S., Moro-Sibilot D., Soria J.C., Brambilla C., Brambilla E. Telomere maintenance and DNA damage responses during lung carcinogenesis. Clin. Cancer Res. 2010;16(11):2979–2988. doi: 10.1158/1078-0432.CCR-10-0142. [DOI] [PubMed] [Google Scholar]
- 157.Oh B.K., Kim Y.J., Park C., Park Y.N. Up-regulation of telomere-binding proteins, TRF1, TRF2, and TIN2 is related to telomere shortening during human multistep hepatocarcinogenesis. Am. J. Pathol. 2005;166(1):73–80. doi: 10.1016/S0002-9440(10)62233-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chen W., Wang Y., Li F., Lin W., Liang Y., Ma Z. Expression of telomere repeat binding factor 1 and TRF2 in prostate cancer and correlation with clinical parameters. BioMed Res. Int. 2017;2017:, 9764752. doi: 10.1155/2017/9764752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pal D., Sharma U., Singh S.K., Kakkar N., Prasad R. Over-expression of telomere binding factors (TRF1 & TRF2) in renal cell carcinoma and their inhibition by using SiRNA induce apoptosis, reduce cell proliferation and migration invitro. PLoS One. 2015;10(3):, e0115651. doi: 10.1371/journal.pone.0115651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Méndez-Pertuz M., Martínez P., Blanco-Aparicio C., Gómez-Casero E., Belen García A., Martínez-Torrecuadrada J., Palafox M., Cortés J., Serra V., Pastor J., Blasco M.A. Modulation of telomere protection by the PI3K/AKT pathway. Nat. Commun. 2017;8(1):1278. doi: 10.1038/s41467-017-01329-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tan R., Nakajima S., Wang Q. Nek7 Protects telomeres from oxidative DNA damage by phosphorylation and stabilization of TRF1. Mol. Cell. 2017;65:818-831. doi: 10.1016/j.molcel.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cherfils-Vicini J., Iltis C., Cervera L., Pisano S., Croce O., Sadouni N., Győrffy B., Collet R., Renault V.M., Rey-Millet M., Leonetti C., Zizza P., Allain F., Ghiringhelli F., Soubeiran N., Shkreli M., Vivier E., Biroccio A., Gilson E. Cancer cells induce immune escape via glycocalyx changes controlled by the telomeric protein TRF2. EMBO J. 2019;38(11):, e100012. doi: 10.15252/embj.2018100012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Diala I., Wagner N., Magdinier F., Shkreli M., Sirakov M., Bauwens S., Schluth-Bolard C., Simonet T., Renault V.M., Ye J., Djerbi A., Pineau P., Choi J., Artandi S., Dejean A., Plateroti M., Gilson E. Telomere protection and TRF2 expression are enhanced by the canonical Wnt signalling pathway. EMBO Rep. 2013;14(4):356–363. doi: 10.1038/embor.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Saha A., Roy S., Kar M., Roy S., Thakur S., Padhi S., Akhter Y., Banerjee B. Role of telomeric TRF2 in orosphere formation and csc phenotype maintenance through efficient dna repair pathway and its correlation with recurrence in OSCC. Stem Cell Rev Rep. 2018;14(6):871–887. doi: 10.1007/s12015-018-9823-z. [DOI] [PubMed] [Google Scholar]
- 165.Panero J., Stanganelli C., Arbelbide J., Fantl D.B., Kohan D., García Rivello H., Rabinovich G.A., Slavutsky I. Expression profile of shelterin components in plasma cell disorders. Clinical significance of POT1 overexpression. Blood Cells Mol. Dis. 2014;52(2-3):134–139. doi: 10.1016/j.bcmd.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 166.Pal D., Singh S.K., Kakkar N., Prasad R. Expression of telomere binding proteins (RAP1 and POT1) in renal cell carcinoma and their correlation with clinicopathological parameters. Indian J. Clin. Biochem. 2017;32(3):301–305. doi: 10.1007/s12291-016-0611-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wijaya A.B., Hidayatullah F., Setyabudhi V.V., Pahlevi F.R., Indra R., Nurseta T. Profile of POT1 as telomerase shelterin component discriminatesbetween cervical cancer and normal cervical cells. Turk. J. Med. Sci. 2017;47(2):417–423. doi: 10.3906/sag-1512-123. [DOI] [PubMed] [Google Scholar]
- 168.Dahlström J., Zhang X., Ghaderi M., Hultcrantz M., Björkholm M., Xu D. Dysregulation of shelterin factors coupled with telomere shortening in Philadelphia chromosome negative myeloproliferative neoplasms. Haematologica. 2015;100(10):e402–e405. doi: 10.3324/haematol.2015.125765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Poncet D., Belleville A., t’kint de Roodenbeke C., Roborel de Climens A., Ben Simon E., Merle-Beral H., Callet-Bauchu E., Salles G., Sabatier L., Delic J., Gilson E. Changes in the expression of telomere maintenance genes suggest global telomere dysfunction in B-chronic lymphocytic leukemia. Blood. 2008;111(4):2388–2391. doi: 10.1182/blood-2007-09-111245. [DOI] [PubMed] [Google Scholar]
- 170.Thijssen P.E., Tobi E.W., Balog J., Schouten S.G., Kremer D., El Bouazzaoui F., Henneman P., Putter H., Eline Slagboom P., Heijmans B.T., van der Maarel S.M. Chromatin remodeling of human subtelomeres and TERRA promoters upon cellular senescence: commonalities and differences between chromosomes. Epigenetics. 2013;8(5):512–521. doi: 10.4161/epi.24450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Schoeftner S., Blasco M.A. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 2008;10(2):228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 172.Le Berre G., Hossard V., Riou J.F., Guieysse-Peugeot A.L. Repression of TERRA expression by subtelomeric dna methylation is dependent on NRF1 Binding. Int. J. Mol. Sci. 2019;20(11):, E2791. doi: 10.3390/ijms20112791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Xu Y., Suzuki Y., Ishizuka T., Xiao C.D., Liu X., Hayashi T., Komiyama M. Finding a human telomere DNA-RNA hybrid G-quadruplex formed by human telomeric 6-mer RNA and 16-mer DNA using click chemistry: a protective structure for telomere end. Bioorg. Med. Chem. 2014;22(16):4419–4421. doi: 10.1016/j.bmc.2014.05.053. [DOI] [PubMed] [Google Scholar]
- 174.Montero J.J., López de Silanes I., Graña O., Blasco M.A. Telomeric RNAs are essential to maintain telomeres. Nat. Commun. 2016;7:12534. doi: 10.1038/ncomms12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Gomez D.L., Armando R.G., Cerrudo C.S., Ghiringhelli P.D., Gomez D.E. Telomerase as a cancer target. development of new molecules. Curr. Top. Med. Chem. 2016;16(22):2432–2440. doi: 10.2174/1568026616666160212122425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tefferi A. Telomerase Inhibitor Imetelstat in Essential Thrombocythemia and Myelofibrosis. N. Engl. J. Med. 2015;373(26):2580–2581. doi: 10.1056/NEJMc1512663. [DOI] [PubMed] [Google Scholar]
- 177.Tefferi A., Lasho T.L., Begna K.H., Patnaik M.M., Zblewski D.L., Finke C.M., Laborde R.R., Wassie E., Schimek L., Hanson C.A., Gangat N., Wang X., Pardanani A. A pilot study of the telomerase inhibitor imetelstat for myelofibrosis. N. Engl. J. Med. 2015;373(10):908–919. doi: 10.1056/NEJMoa1310523. [DOI] [PubMed] [Google Scholar]
- 178.Baerlocher G.M., Oppliger Leibundgut E., Ottmann O.G., Spitzer G., Odenike O., McDevitt M.A., Röth A., Daskalakis M., Burington B., Stuart M., Snyder D.S. Telomerase inhibitor imetelstat in patients with essential thrombocythemia. N. Engl. J. Med. 2015;373(10):920–928. doi: 10.1056/NEJMoa1503479. [DOI] [PubMed] [Google Scholar]
- 179.Thompson P.A., Drissi R., Muscal J.A., Panditharatna E., Fouladi M., Ingle A.M., Ahern C.H., Reid J.M., Lin T., Weigel B.J., Blaney S.M. A phase I trial of imetelstat in children with refractory or recurrent solid tumors: a Children’s Oncology Group Phase I Consortium Study (ADVL1112). Clin. Cancer Res. 2013;19(23):6578–6584. doi: 10.1158/1078-0432.CCR-13-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bryan C., Rice C., Hoffman H., Harkisheimer M., Sweeney M., Skordalakes E. Structural basis of telomerase inhibition by the highly Specific BIBR1532. Structure. 2015;23(10):1934–1942. doi: 10.1016/j.str.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Damm K., Hemmann U., Garin-Chesa P., Hauel N., Kauffmann I., Priepke H., Niestroj C., Daiber C., Enenkel B., Guilliard B., Lauritsch I., Müller E., Pascolo E., Sauter G., Pantic M., Martens U.M., Wenz C., Lingner J., Kraut N., Rettig W.J., Schnapp A. A highly selective telomerase inhibitor limiting human cancer cell proliferation. EMBO J. 2001;20(24):6958–6968. doi: 10.1093/emboj/20.24.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Morais K.D.S., Arcanjo D.D.S., de Faria Lopes G.P., da Silva G.G., da Mota T.H.A., Gabriel T.R., Rabello Ramos D.D.A., Silva F.P., de Oliveira D.M. Long-term in vitro treatment with telomerase inhibitor MST-312 induces resistance by selecting long telomeres cells. Cell Biochem. Funct. 2019;37(4):273–280. doi: 10.1002/cbf.3398. [DOI] [PubMed] [Google Scholar]
- 183.Seimiya H., Oh-hara T., Suzuki T., Naasani I., Shimazaki T., Tsuchiya K., Tsuruo T. Telomere shortening and growth inhibition of human cancer cells by novel synthetic telomerase inhibitors MST-312, MST-295, and MST-1991. Mol. Cancer Ther. 2002;1(9):657–665. [PubMed] [Google Scholar]
- 184.Berletch J.B., Liu C., Love W.K., Andrews L.G., Katiyar S.K., Tollefsbol T.O. Epigenetic and genetic mechanisms contribute to telomerase inhibition by EGCG. J. Cell. Biochem. 2008;103(2):509–519. doi: 10.1002/jcb.21417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hurst C.D., Platt F.M., Knowles M.A. Comprehensive mutation analysis of the TERT promoter in bladder cancer and detection of mutations in voided urine. Eur. Urol. 2014;65(2):367–369. doi: 10.1016/j.eururo.2013.08.057. [DOI] [PubMed] [Google Scholar]
- 186.Bougel S., Lhermitte B., Gallagher G., de Flaugergues J.C., Janzer R.C., Benhattar J. Methylation of the hTERT promoter: a novel cancer biomarker for leptomeningeal metastasis detection in cerebrospinal fluids. Clin. Cancer Res. 2013;19(8):2216–2223. doi: 10.1158/1078-0432.CCR-12-1246. [DOI] [PubMed] [Google Scholar]
- 187.Wang K., Liu T., Ge N., Liu L., Yuan X., Liu J., Kong F., Wang C., Ren H., Yan K., Hu S., Xu Z., Björkholm M., Fan Y., Zhao S., Liu C., Xu D. TERT promoter mutations are associated with distant metastases in upper tract urothelial carcinomas and serve as urinary biomarkers detected by a sensitive castPCR. Oncotarget. 2014;5(23):12428–12439. doi: 10.18632/oncotarget.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ji W., Lou W., Hong Z., Qiu L., Di W. Genomic amplification of HPV, h-TERC and c-MYC in liquid-based cytological specimens for screening of cervical intraepithelial neoplasia and cancer. Oncol. Lett. 2019;17(2):2099–2106. doi: 10.3892/ol.2018.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Xie H., Liu T., Wang N., Björnhagen V., Höög A., Larsson C., Lui W.O., Xu D. TERT promoter mutations and gene amplification: promoting TERT expression in Merkel cell carcinoma. Oncotarget. 2014;5(20):10048–10057. doi: 10.18632/oncotarget.2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Hiyama E., Hiyama K., Yokoyama T., Matsuura Y., Piatyszek M.A., Shay J.W. Correlating telomerase activity levels with human neuroblastoma outcomes. Nat. Med. 1995;1(3):249–255. doi: 10.1038/nm0395-249. [DOI] [PubMed] [Google Scholar]
- 191.Xing M., Liu R., Liu X., Murugan A.K., Zhu G., Zeiger M.A., Pai S., Bishop J. BRAF V600E and TERT promoter mutations cooperatively identify the most aggressive papillary thyroid cancer with highest recurrence. J. Clin. Oncol. 2014;32(25):2718–2726. doi: 10.1200/JCO.2014.55.5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Sahm F., Schrimpf D., Olar A., Koelsche C., Reuss D., Bissel J., Kratz A., Capper D., Schefzyk S., Hielscher T., Wang Q., Sulman E.P., Adeberg S., Koch A., Okuducu A.F., Brehmer S., Schittenhelm J., Becker A., Brokinkel B., Schmidt M., Ull T., Gousias K., Kessler A.F., Lamszus K., Debus J., Mawrin C., Kim Y.J., Simon M., Ketter R., Paulus W., Aldape K.D., Herold-Mende C., von Deimling A. TERT Promoter mutations and risk of recurrence in meningioma. J. Natl. Cancer Inst. 2015;108(5):, djv377. doi: 10.1093/jnci/djv377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Brennan C.W., Verhaak R.G., McKenna A., Campos B., Noushmehr H., Salama S.R., Zheng S., Chakravarty D., Sanborn J.Z., Berman S.H., Beroukhim R., Bernard B., Wu C.J., Genovese G., Shmulevich I., Barnholtz-Sloan J., Zou L., Vegesna R., Shukla S.A., Ciriello G., Yung W.K., Zhang W., Sougnez C., Mikkelsen T., Aldape K., Bigner D.D., Van Meir E.G., Prados M., Sloan A., Black K.L., Eschbacher J., Finocchiaro G., Friedman W., Andrews D.W., Guha A., Iacocca M., O’Neill B.P., Foltz G., Myers J., Weisenberger D.J., Penny R., Kucherlapati R., Perou C.M., Hayes D.N., Gibbs R., Marra M., Mills G.B., Lander E., Spellman P., Wilson R., Sander C., Weinstein J., Meyerson M., Gabriel S., Laird P.W., Haussler D., Getz G., Chin L., TCGA Research Network The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Svahn F., Juhlin C.C., Paulsson J.O., Fotouhi O., Zedenius J., Larsson C., Stenman A. Telomerase reverse transcriptase promoter hypermethylation is associated with metastatic disease in abdominal paraganglioma. Clin. Endocrinol. (Oxf.) 2018;88(2):343–345. doi: 10.1111/cen.13513. [DOI] [PubMed] [Google Scholar]
- 195.Benhamou Y., Picco V., Raybaud H., Sudaka A., Chamorey E., Brolih S., Monteverde M., Merlano M., Lo Nigro C., Ambrosetti D., Pagès G. Telomeric repeat-binding factor 2: a marker for survival and anti-EGFR efficacy in oral carcinoma. Oncotarget. 2016;7(28):44236–44251. doi: 10.18632/oncotarget.10005. [DOI] [PMC free article] [PubMed] [Google Scholar]