Abstract
Membrane proteins and lipids have the capacity to associate into lateral domains in cell membranes through mutual or collective interactions. Lipid rafts are functional lateral domains that are formed through collective interactions of certain lipids and which can include or exclude proteins. These domains have been implicated in cell signaling and protein trafficking and seem to be of importance for virus–host interactions. We therefore want to investigate if raft and viral membrane proteins present similar structural features, and how these features are distributed throughout viruses. For this purpose, we performed a bioinformatics analysis of raft and viral membrane proteins from available online databases and compared them to nonraft proteins. In general, transmembrane proteins of rafts and viruses had higher proportions of palmitoyl and phosphoryl residues compared to nonraft proteins. They differed in terms of transmembrane domain length and thickness, with viral proteins being generally shorter and having a smaller accessible surface area per residue. Nontransmembrane raft proteins had increased amounts of palmitoyl, prenyl, and phosphoryl moieties while their viral counterparts were largely myristoylated and phosphorylated. Several of these structural determinants such as phosphorylation are new to the raft field and are extensively discussed in terms of raft functionality and phase separation. Surprisingly, the proportion of palmitoylated viral transmembrane proteins was inversely correlated to the virus size which indicated the implication of palmitoylation in virus membrane curvature and possibly budding. The current results provide new insights into the raft–virus interplay and unveil possible targets for antiviral compounds.
Introduction
Cell membrane lipids and proteins present a large degree of spatial heterogeneity in the lateral plane.1−5 The formation of lateral domains facilitates or hinders interactions between membrane components guiding signal transduction6 and intraorganellar transport,7,8 among other cellular functions.9 Some types of membrane domains are formed mainly from collective lipid interactions as is the case for lipid membrane rafts.10,11 In rafts, favorable interactions between cholesterol, sphingolipids, saturated acyl chains, glycolipids, and also membrane proteins drive the formation of transient domains involved in signaling, as well as lipid and protein transport.11
Curiously, some types of viruses have been linked to raft formation as they require cholesterol, sphingomyelin and membrane lipid heterogeneity for fusion with the cell membrane.12,13Influenza and coronavirus association and binding to host cells is facilitated by raft formation,14,15 while some viruses such as human immunodeficiency viruses (HIV) assemble into raft domains before maturation and budding.16−22 The importance of lipid rafts for viral assembly is further underlined by the fact that viral lipidomes are essentially composed of raft lipids.23,24 Furthermore, it seems that raft localization of certain viral proteins is essential for membrane dependent viral assembly, budding, and fission.17,18,22,25,26
In light of these implications for rafts in cellular and viral functions, it is essential to know which structural parameters are typical for proteins in raft domains and if these are similar in viral proteins. It has been shown that certain lipid modifications, and structural parameters of transmembrane domains (TMDs) play important roles in raft partitioning of proteins.27−29 A predictive model for raft partitioning for single-pass transmembrane domains was created from energy contributions of palmitoyl residues, hydrophobic matching, and TMD/membrane interfacial energy.29 This model is not applicable for multipass transmembrane domains or membrane attached (peripheral) proteins and also ignores other possible structural properties, capable of interacting with membranes and raft domains. These knowledge gaps on structural information are aggravated by contradictory results on some protein modifications such as prenylations, which have been listed as raft preferring30 or raft avoiding.31
We therefore use a bioinformatics approach to compare various structural properties of raft, viral, and nonraft membrane proteins which are based on large available data sets. This approach should allow us to gather statistical relevant data and determine the importance of structural properties for raft partitioning and examine the occurrence of those in viral proteins. We especially focus on post-translational modifications which have been shown to influence raft partitioning in the past and investigate new parameters such as phosphorylations, disulfide bonds, and glycosylations, which could potentially influence raft localization.
Besides the relevance of structural information for raft partitioning, we will investigate if these structural properties are conserved in different viral families and if they are involved in the generation of membrane curvature during the maturation process. The reason for this assumption is the fact that palmitoylated proteins were able to regulate curvature in influenza viruses.32 We therefore explore if virus size or curvature can be linked to the explored structural parameters and their variability upon viral families.
Methods
Data Mining
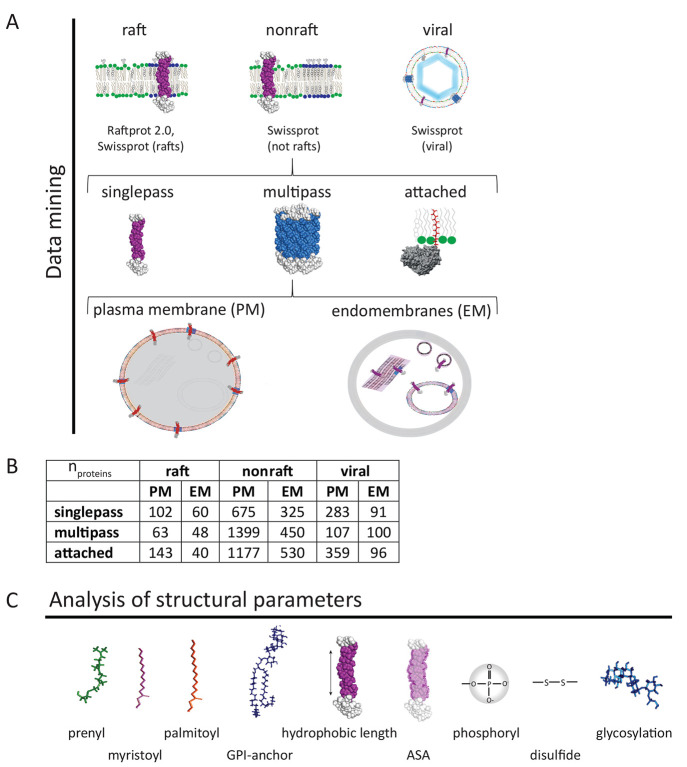
To build a raft protein database, we combine human raft proteins from the Swiss-prot server, and those from the RaftProt 2.0 database in which raft partitioning has been determined by at least 2 different methods (Figure 1A).33 We further create a second group of nonraft proteins, including all human membrane proteins from the Swiss-prot database which are not in the established raft database. A third group contains all human viral membrane proteins in the Swiss-Prot database. Those three membrane protein groups are further split upon plasma membrane (PM) or endomembrane (EM) localization in the UniProt/Swiss-Prot database. Endomembranes are defined here as all internal membranes except mitochondria. This approach takes into account the fact that proteins possess some structural organellar identity, rendering comparison of raft versus nonraft proteins from different organelles difficult.34 Proteins are further grouped into single-pass, multipass, and membrane attached (peripheral) proteins because they interact quite differently with membranes. To avoid irrelevant structural information we only take peer reviewed proteins into account. The exact number of proteins in each group is shown in (Figure 1B).
Figure 1.
Data mining process and distribution of membrane proteins into different categories upon type (singlepass, multipass, and attached) and localization (plasma membrane or endomembrane system) (A). Number of proteins in each group (B). The analysis of displayed structural parameters is performed on all proteins except for hydrophobic length and ASA (accessible surface area) on attached membrane proteins. We determine the fraction of proteins which contain a certain modification (Figure 2) and the average number of modifications per protein with its distribution (Figure 3).
Analysis of Structural Parameters
For all subgroups, we analyze various structural parameters which could be relevant for raft partitioning (Figure 1C).27,29 We are mainly interested in post-translational lipid modifications of membrane proteins such as prenylation, myristoylation, palmitoylation, and GPI-anchors since they have previously shown to be involved in raft localization of membrane proteins.27 We further investigate other post-translational modifications which could potentially influence raft partitioning. Glycosylation was analyzed since it may influence raft partitioning through hydrogen bonding with glycolipids or other glycoproteins.11 Disulfide bonds and phosphorylation are also investigated since they can induce oligomerization of proteins which might help in raft assembly or partitioning to preformed domains.5,6 The presence of all post-translational modifications in proteins is determined via the correspondent annotations in the Swiss-Prot/UniProtKB database.35 In detail, we count the numbers of modified proteins in each group which is determined as fraction of, i.e., prenylated proteins (Fprenylated, Figure 2). The number of modifications per proteins (Figure 3) is determined according to annotations in the Swiss-Prot databank. These can be either published modifications, predicted modifications by certain algorithms (unirule, prosite–prorule), or manual assertions according to sequence analysis and similarity.
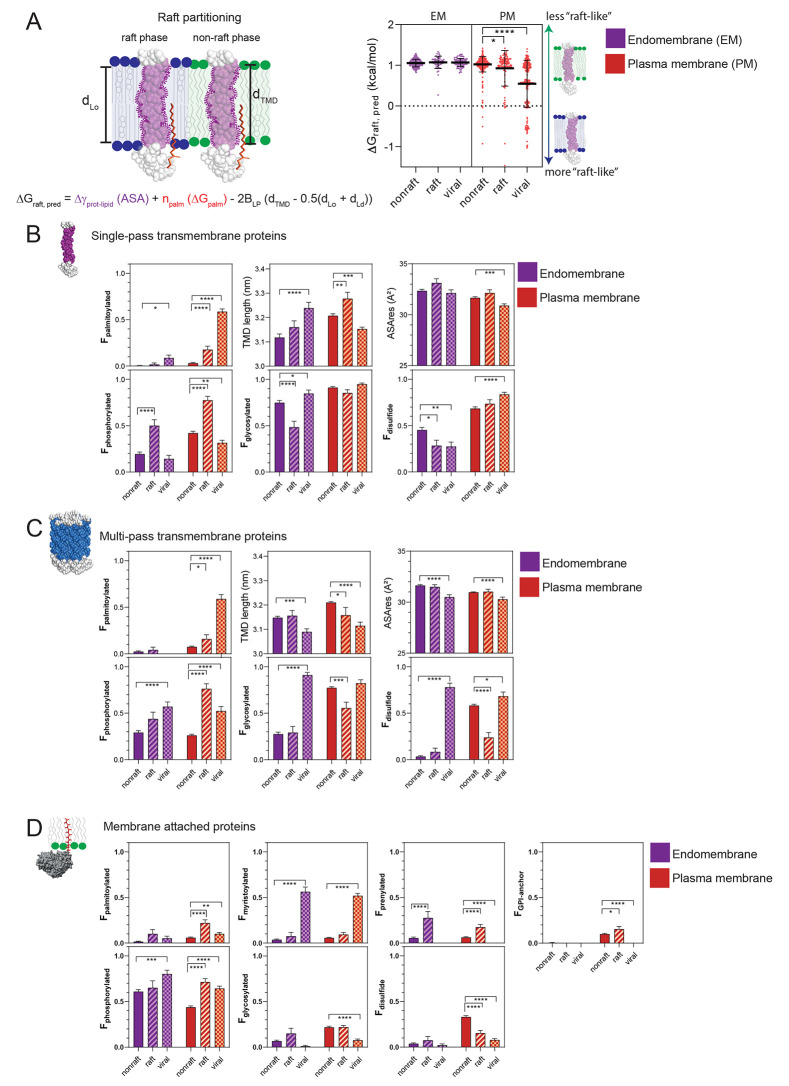
Figure 2.
Schematic of structural determinants and associated formula to predict the free energy (ΔGraft,pred) of raft partitioning according to Lorent el al.29 (A, left). Predictions have been made for single-pass transmembrane proteins in the nonraft, raft and viral database upon cellular localization (A, right). Membrane protein fractions (F) ± SEM containing structural modifications in the nonraft (full bars), raft (striped bars), and viral protein (squared bars) database upon localization in endomembranes (purple color) or the plasma membrane (orange color) (B–D). In addition, TMD length and accessible surface area/residue (ASAres) are determined for transmembrane proteins (B, C). Proteins are grouped upon single-pass transmembrane (B), multipass transmembrane (C), and membrane attached proteins (D). A two-way ANOVA test has been performed to compare the raft and viral proteins versus non raft data sets (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
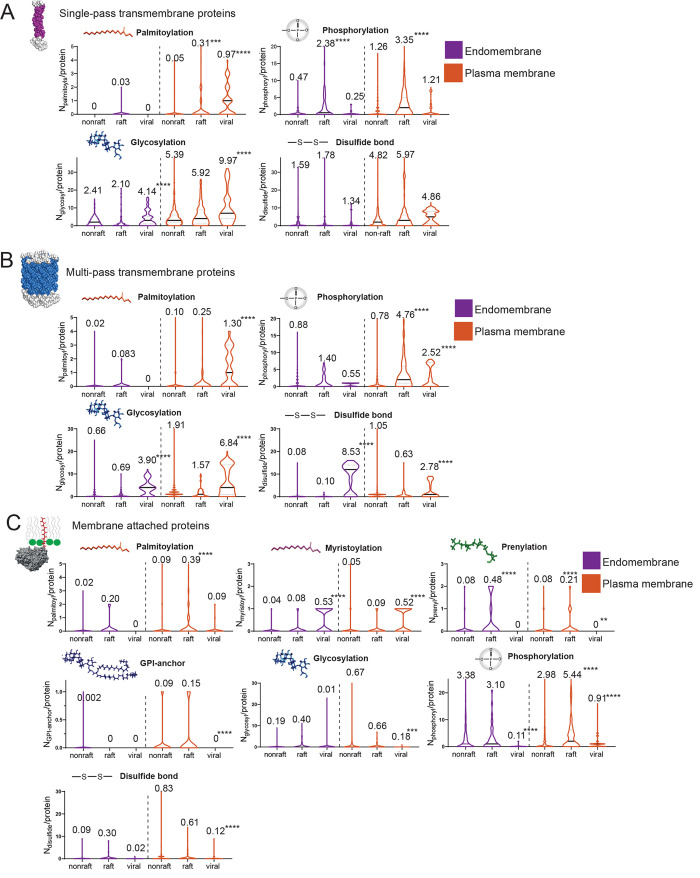
Figure 3.
Distributions of the amount of modifications per protein (Nx/protein) shown as violin shaped plots. The straight black line reflects the median of the distribution and the dotted lines the quartiles. The number on top of the violin plot is the average number of modifications per protein including the statistical difference of a one-way ANOVA versus the nonraft phase (**p < 0.01, ***p < 0.001, ****p < 0.0001). Plots are given for single-pass (A), multipass (B), and attached membrane proteins (C) in endomembranes (purple) and cell membranes (orange) for nonraft, raft, and viral proteins.
Prediction of Raft-Partitioning of Single-Pass Transmembrane Proteins
For transmembrane proteins, we additionally calculate transmembrane domain (TMD) length, TMD accessible surface area per residue (ASAres), and the free energy of raft partitioning (ΔGraft,pred) as predicted for single-pass transmembrane proteins.29 Briefly, the transmembrane domain for single pass transmembrane proteins is determined from annotations in the Uniprot databank which is mainly based on predictions by the TMHMM algorithm.36 We further multiply the number of amino acids in a TMD by 0.15 nm which is the statistical length of a residue in an α-helix. For multipass transmembrane domains, the TMD length is considered as the mean of all individual TMD lengths. The accessible surface area per residue in a TMD (ASAres) was determined by summing up ASA values for all individual residues in TMDs as described by Yuan37 and further normalizing to the length of the TMD.
The free energy of raft partitioning (ΔGraft,pred), which gives an indication on how the protein would preferentially partition into the raft phase in giant plasma membrane vesicles (GPMVs) is predicted by the formula in Figure 2A for single pass transmembrane proteins. The three individual terms arise from the difference of interfacial energy between the protein in the raft and nonraft phase (ΔγTMD,Lo-Ld = 1.1 pN/nm), the free energy contribution from palmitoyl residues (ΔGpalm=-0.48 kcal/mol per palmitoyl residue) and the free energy arising from hydrophobic matching as described in the mattress model (−2BLP(dTMD – 0.5(dLo + dLd))) in which BLP = 7.5 × 10–2 kcal/mol·nm, dLo = 3.9 nm, and dLd = 3.6 nm.29,38
Structural Parameters upon Viral Families
We further investigate if post-translational modifications including TMD length and accessible surface area are similarly distributed in single-pass transmembrane proteins of different enveloped viruses in the host plasma membrane. To do so, we study the coefficient of variation of given structural parameters upon viral families (Table 1). We also cross-correlate the fractions of proteins containing certain modifications with each other upon different virus families. This allows us to investigate if structural properties might develop in parallel (Figure 6). For example, the fraction of palmitoylated proteins might increase in parallel with the TMD length and decrease with the accessible surface area from one species to the other because proteins of certain viral species prefer to partition into lipid rafts. We include into these correlations membrane curvature or virus size because palmitoylation has recently been linked to membrane curvature in HIV which might result from the participation of palmitoylation in virus budding.32,39
Table 1. Variation of Selected Properties in Viral Single-Pass Transmembrane Proteins Summarized in the Coefficient of Variation (C.V.)a.
| species | MVS (L.L-U.L.) in nm | Fpalmitoyls | FPO4 | Fglycosylation | Fdisulfide | length (nm) | ASAres (A2) | Nproteins |
|---|---|---|---|---|---|---|---|---|
| Coronaviridae | 140 (120–160) | 0.54 | 0.00 | 1.00 | 1.00 | 3.15 | 36.38 | 24 |
| Flaviviridae | 47.5 (45–50) | 0.97 | 1.00 | 1.00 | 0.88 | 3.15 | 30.61 | 33 |
| Herpesvirales | 230 (160–300) | 0.00 | 0.24 | 0.90 | 0.48 | 3.13 | 30.43 | 82 |
| Orthomyxoviridae | 100 (80–120) | 0.50 | 0.23 | 0.93 | 0.96 | 3.19 | 30.15 | 139 |
| Paramyxoviridae | 225 (150–300) | 0.10 | 0.00 | 0.98 | 0.48 | 3.16 | 32.07 | 61 |
| Retroviridae | 105 (90–120) | 0.72 | 0.01 | 1.00 | 0.98 | 3.15 | 32.56 | 103 |
| Togaviridae | 70 (70–70) | 1.00 | 0.38 | 1.00 | 1.00 | 3.13 | 27.33 | 32 |
| C.V. (%) | 54.9 | 71.4 | 134.0 | 4.20 | 29.3 | 0.6 | 8.9 |
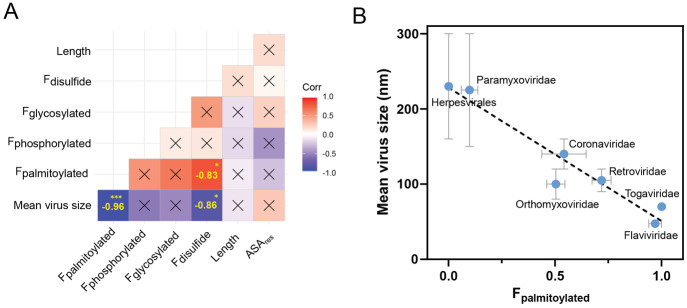
Figure 6.
Correlation matrix (Pearson coefficient) of viral single-pass transmembrane protein properties with correspondent significance. *p < 0.05, ***p < 0.001 (A); Virus size or curvature is strongly correlated to the fraction of palmitoylated proteins (B).
Results
Prediction of Raft Partitioning of Single-Pass Transmembrane Proteins from the Databases
It was shown in giant plasma membrane vesicles (GPMVs) that raft partitioning of single-pass transmembrane proteins depends first on the differential interfacial energy created between a protein’s transmembrane domain and the lipid environment in the tightly packed raft phase versus the nonraft phase, second the amount of palmitoyl residues and third the length of the transmembrane domain. From these parameters, a quantitative law was established to predict the free energy (ΔGraft,pred) of raft partitioning in giant plasma membrane vesicles (see prediction of raft partitioning in methods and Figure 2A for details).29 We hence predicted raft partitioning for single-pass transmembrane proteins in the generated nonraft, raft, and viral databases upon cellular localization (Figure 2A). We observed that proteins from the raft and viral data set would preferentially partition into the raft phase in GPMVs compared to proteins from the nonraft database (Figure 2A). It seems that viral proteins would have on average a lower ΔGraft,pred corresponding to a higher raft affinity than proteins from the raft database. Those values seem to concur with the general assumption that some viral proteins partition into lipid rafts.
Structural Peculiarities of Proteins from the Raft Database
The lower ΔGraft,pred values of single-pass transmembrane proteins from the raft-phase in plasma membranes can mainly be explained by the higher fraction of palmitoylated proteins (Figure 2B, Fpalmitoylation) and the higher amount of palmitoyl residues/protein (Figure 3A, Npalmitoyl/protein). Palmitoyl residues have a high energetic contribution of −0.48 kcal/nmol toward raft partitioning, “dragging” large proteins into the raft phase.29,40 In addition, plasma membrane raft proteins had longer transmembrane domains (Figure 2B, TMDlength) but no significant difference in accessible surface area per TMD residue (Figure 2B, ASA/residue). The absence of difference in ASA was surprising because proteins with low ASA would prefer the raft phase. Those results are later discussed.
Besides the “classic” protein modifications involved in raft partitioning (see above and Methods), we also investigated protein glycosylations, phosphorylations, and disulfide bonds. Glycosyl residues of proteins can potentially interact through hydrogen bonds or other polar interactions with glycosphingolipids, which are largely enriched in lipid rafts.41,42 Unexpectedly, there was either a decrease of glycosylated proteins in endomembrane raft versus nonraft proteins or no significant change in both plasma membrane groups (Figures 2B and 3A). This might indicate that glycosyl residues do not contribute to raft partitioning in single-pass transmembrane proteins. Protein phosphorylation is a post-translational modification which is essential in signal transduction and protein-induced phase separation. Phosphorylation of multiple protein residues can lead to the formation of protein phase separation or aggregation at the inner cell membrane interface as shown for phosphorylation of Linker for activation of T-cells (LAT).5,6 It had not been clear yet if protein phase separation is linked to raft formation but we observed a large increase in the fraction of proteins being phosphorylated (Figure 2B, Fphosphorylation) and the number of phosphorylation sites in annotated raft proteins compared to nonrafts (Figure 3A, Nphosphoryl/protein). This might hint at the importance of phosphorylation for raft formation, stabilization, or raft functionality, since protein phosphorylation events are important for signal transmission. Intermolecular disulfide bonds could potentially enhance or stabilize a raft phase by forming homo- or hetero-oligomers. However, while the fraction of proteins with disulfide bonds in the raft phase did not increase (Figure 2B), the average number of disulfide bonds/protein increased (Figure 3A). These results indicate that only a small number of proteins in the raft database possesses higher amounts of disulfide bonds compared to nonraft proteins.
Multipass transmembrane proteins interact differently with the membrane because their individual transmembrane domains interact closely with each other and thereby exclude protein–lipid interactions on a large scale. It makes sense that they do not exactly follow the same trends as single-pass transmembrane proteins, but we similarly observed an increase in fractions of palmitoylated and phosphorylated proteins and numbers of those modifications in plasma membrane raft proteins (Figures 2C and 3B). TMD length, glycosylations, and disulfide bonds were reduced in plasma membrane raft proteins and there were no obvious changes observed for endomembrane proteins. Other lipid modifications like myristoylations or prenylations were only present in infinitesimal amounts and were therefore not considered important for raft partitioning of transmembrane proteins.
Membrane-attached proteins (Figures 2D and 3C) were distinct in that they possessed other lipid modifications than transmembrane proteins. We observed a general increase of prenylated proteins in the raft database, while the fraction of palmitoylated and GPI-modified proteins increased only in raft proteins of the plasma membrane. Curiously, prenylations have been considered nonraft signals because of their rather bulky nature, and their presence here is rather surprising considering that the raft phase is tightly packed.31,40 GPI-anchors have always been considered raft signals, and the present result concurs with previous findings.8,42−44 Regarding nonlipid modifications, we observed once more an increase of phosphorylated proteins in raft proteins of the plasma membrane, emphasizing the importance of phosphorylations for raft proteins.
Structural Properties of Proteins from the Virus Database
As we have shown, single-pass transmembrane viral proteins localized at the host cell membrane were predicted to partition into the raft phase compared to nonraft proteins (Figure 2A, ΔGraft,pred). This seems to be largely due to the great amount of palmitoyl residues (0.97 palmitoyl residues/protein) compared to only 0.05 palmitoyl residues/protein in nonraft proteins (Figure 3A, orange violin plots). Multipass transmembrane proteins in viruses were roughly 60% being palmitoylated (Figure 2B) which corresponded to 1.3 palmitoyl residues/protein compared to 0.1 palmitoyl residues/protein in the nonraft phase (Figure 3B). Interestingly, transmembrane viral proteins had largely higher amounts of palmitoyl residues than proteins from the raft database (Figure 2 and 3). This means either that viral proteins partition extremely well into the raft phase, which would correspond to the low predicted free energy of raft partitioning (ΔGraft,pred) (Figure 1A), or that palmitoylation has additional functions in viral proteins. Regarding transmembrane domain properties, it seems that viral multipass TMDs were generally shorter, which is counterintuitive if we assume that viral TMDs have to “match” the thicker raft phase. The ASAres decreased in virus proteins except for single pass TMDs in endomembranes. The fraction of glycosylated transmembrane proteins was overall high (85–95%) in nonraft, raft, and viral proteins of the cell membrane (Figure 2B,C, purple color). Remarkably, the number of glycosylations per protein jumped from 5 to 6 in nonraft and raft, respectively, to 10 in viral single pass proteins. In multipass proteins, the numbers increased from 2 to 3 glycosyls/protein to 6. Glycosylation has a functional role in virus proteins since it can prevent recognition by the immune system.45,46 This might explain the higher number of glycosyl residues in virus encoded proteins and might hence be unrelated to the formation of a raft or raft-like phase. The fraction of phosphorylated proteins and the number of phosphoryl residues/protein increased significantly in plasma membrane multipass transmembrane virus proteins compared to nonraft proteins. Although the number was lower than in raft proteins and seemed to be of less importance in single pass transmembrane proteins or endomembrane proteins. Disulfide bonds presented an hourglass shape distribution (bimodal) in viral plasma membrane transmembrane proteins. The proportion of disulfide bonds was also higher in the plasma membrane compared to the endomembrane system. It might be that several viruses combine plasma membrane specific signals with raft signals.
Membrane-attached viral proteins varied only slightly for palmitoylation but around half of their proteins were myristoylated compared to 4–5% of nonviral proteins (Figure 2D). The distribution of myristoyl residues revealed that about half of the proteins contained 1 myristyol residue/protein while the other half contained none (Figure 3C). The role of myristoylation for raft partitioning in literature is not clear and it seems that a single myristoyl residue is not enough for raft partitioning.27 We did not observe a clear increase for this lipid modification in the raft database, so it seems that this modification is typical for viral proteins. There was no change observed for prenylations and GPI-anchors conversely to the raft database. Regarding nonlipid modifications, an increase of phosphorylated proteins was again observed while the numbers of phosphoryl residues remained low compared to nonraft proteins. Glycosylations and disulfide bonds were almost absent in this protein group, maybe because of their preferred localization on the inner membrane leaflet.
Principal Component Analysis of Proteins Reveals the Most Important Structural Determinants
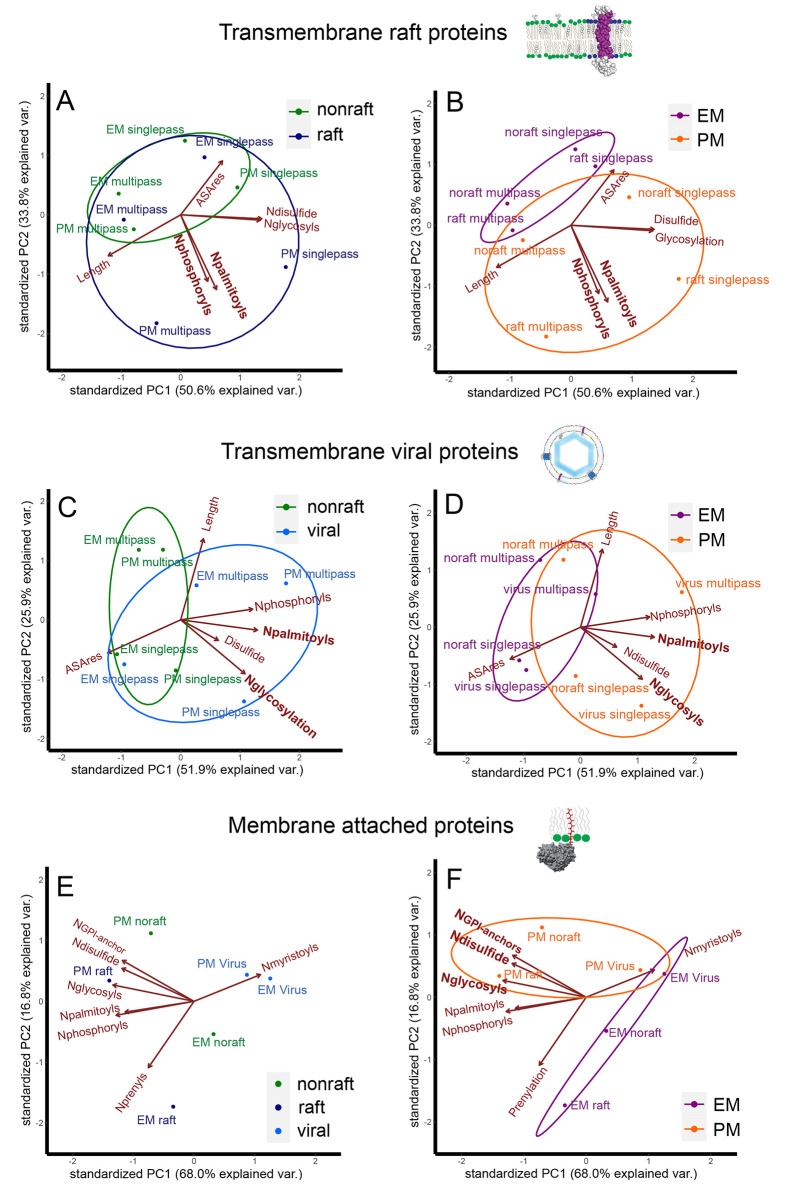
To gain a clearer picture of significant modifications in nonraft, raft, and viral proteins, we performed a principal component analysis (PCA) upon the average number of modifications per protein (Figure 4). After analysis, we regrouped proteins either upon nonraft, raft, and viral proteins (Figure 4, left column) or upon cellular localization (Figure 4, right column). This should enable us to distinguish between raft and virus or localization dependent properties, respectively. We combined single and multipass transmembrane proteins in the analysis to determine common properties for all transmembrane proteins in the three databases (Figure 4A–D). A first PCA of nonraft versus raft transmembrane proteins revealed that the chosen structural properties partially overlapped (Figure 4A), although a certain trend appeared. A smooth transition from nonraft to raft proteins had been observed in GPMVs in which many proteins presented an intermediate raft partitioning coefficient.29 We can therefore assume that many proteins are able to partition in between raft and nonraft phases which makes it more complicated to find strict raft determining parameters. However, palmitoylation and phosphorylation contributed most to the distinction between raft and nonraft proteins. Similar conclusions could be drawn from a PCA of the fraction of palmitoylated proteins (Figure S1A).
Figure 4.
Principal component analysis of the average number of modifications per protein (Nx/protein) in transmembrane (A–D) and attached membrane proteins (E, F), in raft versus nonraft (A, B) and virus versus nonraft (C, D) proteins. After PCA, proteins are colored upon protein type in the left column (green = nonraft, dark blue = raft, and light blue = viral) or cellular localization in the right column (orange = plasma membrane and purple = endomembranes).
Interestingly, the same PCA can better explain differences between plasma membrane and endomembrane proteins since both groups only slightly overlap (Figures 4B and S1B). The most important parameters to explain the differences seem again to be palmitoylation and phosphorylation. It appears though that raft partitioning follows similar structural rules as plasma membrane localization, but that the difference between protein structures is somehow larger (Figure 4A,B).
The chosen structural parameters can also not completely explain differences between transmembrane viral and nonviral proteins (Figures 4C and S1C). Differences between viral and nonviral transmembrane proteins were best explained by an increased amount of palmitoylations and glycosylations, and to a lesser extent by disulfide bonds and phosphorylations (Figure 4C). Although it seemed that a higher percentage of proteins had disulfide bonds even if the number of modifications was less important (Figures 2, 3, and S1). Plasma membrane localization of viral proteins depended on similar parameters but differences between both groups seemed to be less important (Figure 4D).
Membrane attached proteins showed different properties for raft, nonraft, and viral proteins. (Figure 4E). While raft proteins displayed higher amounts of phosphorylation, palmitoylation, glycosylation, and prenylations compared to nonraft proteins, viral proteins were rather myristoylated. These differences were rather striking and created different clusters. Interestingly, if the PCA was performed on the fraction of proteins possessing a certain modification, phosphorylation was also a defining factor for viral proteins (Figure S1) which is expected from (Figure 2D). Cell membrane localization of attached membrane proteins was best explained by glycosylation, GPI-anchors and disulfide bonds (Figures 4F and S1F). Those modifications are expected of peripheral plasma membrane proteins present on the exoplasmic leaflet.47
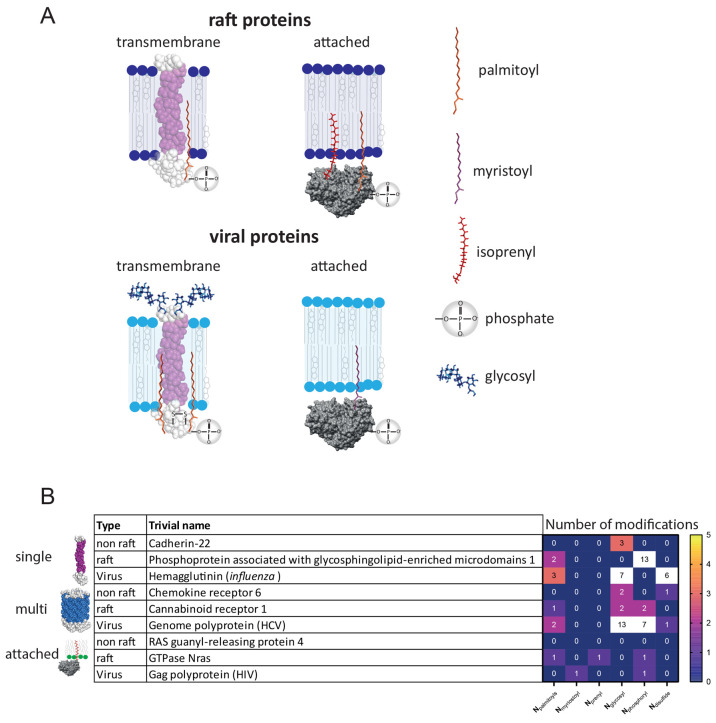
We graphically summarized the decisive structural peculiarities which distinguished raft and viral proteins from nonraft proteins (Figure 5A) and gave some examples of common proteins with typical modifications for each class (Figure 5B).
Figure 5.
Schematic of typical modifications in raft and viral plasma membrane proteins (A). Examples of typical nonraft, raft, and viral plasma membrane proteins with numbers of structural modifications (B).
Variability and Correlation of Structural Parameters in Viruses
Since viral proteins presented peculiar structural parameters compared to nonraft and raft proteins (Figure 5), we wanted to investigate the interspecies variability of those parameters. To avoid localization dependent biases, we only analyzed single-pass transmembrane proteins of the host plasma membrane (Table 1). Variations were very high in the fractions of palmitoylated and phosphorylated proteins (>70%) contrarily to other parameters in which variability remained relatively low (<30%).
Next, we created a correlation matrix to determine if different properties are inherently correlated between viral families (Figure 6A and Table S1). Astonishingly, we found that the mean virus size (MVS) is negatively correlated to the fraction of palmitoylated proteins (Figure 6A,B). Very small and therefore highly curved viruses like Togaviridae or Flaviviridae presented very high fractions of palmitoylated proteins (around 100%) while in larger viruses the amount remained low (Table 1 and Figure 6B). It had previously been shown that palmitoyl residues on the internal leaflet of the envelope in influenza virus like particles (Orthomyxoviridae) increased the negative curvature strain and thereby promoted formation of small virions.32 This observation seems to hold for other viral families since the amount of curvature and palmitoylation is correlated upon families. Two other weaker but significant positive correlations have been found, one between disulfide bonds and palmitoyl residues, and another between disulfide bonds and the mean virus size. Since both modifications are related to virus size, they both might be related to curvature. Maybe some conformational changes in proteins related to the formation of disulfide bridges can increase the intrinsic curvature of membrane proteins and thereby, in addition to palmitoyl residues, help with virus budding.
Discussion
The analyzed structural parameters for protein partitioning into raft domains differed upon their cellular localization and protein type (single, multiple, and attached membrane proteins). Interestingly, the most important features to discern nonraft from raft proteins were palmitoylation and phosphorylation while prenylation was also a determinant for raft partitioning of attached membrane proteins (Figure 4). Palmitoylation has been shown to be of importance for raft partitioning in numerous models in accordance with the analyzed proteins from the raft database.27,28,49−51 Prenylation is supposed to be a decreasing factor for raft partitioning,31,40 and its higher presence in the raft database is rather surprising since it is difficult to imagine very bulky isoprenyl tails inserting into the very packed raft environment. Some authors proposed that isoprenyls are found on the interface between raft and nonraft phase which might explain differing results since partitioning might depend on the detergent resistant membrane (DRM) isolation methods used.52,53
The observed presence of phosphorylation residues is to our knowledge an unknown raft directing factor but has been associated with raft partitioning.54−56 Multiple protein phosphorylations can induce the formation of a large protein phase separation at the cytosolic membrane interface as it has been shown for LAT.5 LAT has also been shown to be associated with the raft phase independently of phosphorylation sites in GPMVs, which means that phosphorylation is most probably not a prerequisite for raft association but maybe a functional aspect or stabilizing factor for rafts. Raft association of phosphorylatable proteins could lead to their close association, and further phosphorylation could lead to a large protein-raft phase implicated in signal transduction or protein trafficking. Another explanation for the high presence of phosphorylations might be their implication in signaling. Protein signaling is often regulated through the presence of phosphorylated residues by kinases and phosphatases. Since rafts are signaling domains, the increased presence of phosphorylated proteins would reflect their functional aspect.
Parameters such as transmembrane domain length and accessible surface area were important for raft partitioning of single pass transmembrane proteins in GPMVs. Single-pass transmembrane proteins from the raft database were also longer but had an increased surface area per residue. Multipass transmembrane domains were surprisingly shorter in the raft database but plasma membrane proteins were generally longer than endomembrane proteins. The differences from experimental GPMV data and the raft database might arise from the fact that the RaftProt 2.0 database is 86% based on DRM experiments.33 DRMs suffer from several artifacts such as the induction of a raft phase by the detergent itself.57 This might favor effects from lipid modifications over transmembrane domains since the detergent might intercalate in between the lipids and the transmembrane domain and hence reduce effects due to interfacial tension (Figure 2A). The results concerning TMD-length and TMD accessible surface area are therefore to be interpreted cautiously.
We suspected virus proteins to be similar to raft proteins because many papers published the importance for rafts in virus binding and assembly in the host cell.12,14,15,17,20,21,58 Virus transmembrane proteins were, like raft proteins, mainly enriched in palmitoyl residues and phosphorylation sites, but they presented more glycosylations and had a slightly shorter and thinner TMD (lower ASA). Attached viral proteins presented higher proportions of myristoyl residues compared to high amounts of palmitoylation and prenylation sites in raft proteins. An increase of phosphorylation sites seemed also to be a common structural feature of nontransmembrane viral proteins (Figure 4).
Palmitoylation was a common feature of raft and viral proteins, but the average amount of palmitoylated proteins was much higher in viruses compared to the raft database. A large interspecies variation in the proportion of palmitoylated virus proteins could specifically be related to the virus size. A high amount of palmitoylated proteins in a certain species reflected a small virus size and vice versa (Figure 5). This result revealed that palmitoyl residues in viruses might not only be involved in raft association but also in the induction of curvature. It was shown that palmitoyl residues are able to increase the negative curvature of the inner leaflet and hence induce the formation of smaller virus-like particles.32 Palmitoylation might therefore fulfill a double function in viral proteins which is raft association and the induction of negative curvature during budding. The very low amount of palmitoylated proteins (almost 0) in certain viruses further suggests that their proteins most probably do not associate with lipid rafts. Interestingly, some viruses are able to change the amount of palmitoylation and myristoylation in the host cells, which could also potentially change raft partitioning of host proteins.59 Inhibition of palmitoylation could therefore be a very interesting antiviral drug target. The increase of phosphorylation in viral proteins could be related to their functional aspects. Viruses are able to produce their own kinases but also recruit host kinases for proliferation which makes them interesting targets for antiviral therapy.60,61 It is not clear to what extend phosphorylation in viruses is involved in raft partitioning or protein phase separation, but it is definitely worth further experimental investigation.5,6 The reduction in length and thickness of TMDs in viral transmembrane proteins could biophysically reflect a thinner and more tightly packed membrane, respectively.29,38 Alternatively, TMD length seems to be related to the viral entry mechanism, and it is usually higher for proteins which enter through the plasma membrane but shorter for proteins, which are guided through endocytosis.62 Protein myristoylation was increased in viral attached proteins but not in raft proteins. It is not clear if myristoyl residues would contribute to raft association since the literature on myristoylation is controversial.27,40 Biophysically, the C14 myristoyl tail should be easier to integrate into a thinner membrane compared to the longer C16 palmitoyl tail. However, myristoylation of viral proteins seems to be an interesting drug target as it was shown for HIV.63 The high amount of glycosylation in viral proteins has been discussed above, and it remains unclear if glycosylation is a raft-dependent structural parameter.8,64,65
Recently, a mechanism that describes raft-dependent association of HIV proteins fits very well with our observations.22 In their model, virus and raft proteins assemble first through a raft-dependent mechanism. Later, the raft domains increase in curvature and bud out of the membrane.22 With respect to our data, the higher proportion of palmitoyl residues, the lower accessible surface area of TMDs, and increased amounts of myristoylated-attached proteins could first drive raft partitioning of virus proteins.29 Furthermore, multiple phosphorylations might guide protein assembly at the membrane or induce signaling,66 and high amounts of palmitoylated transmembrane proteins could help increase the curvature of domains.32 A high curvature could further favor assembly of curved raft-like lipids into the final virion, which has previously been observed.23,24
Summary/Conclusion
From the current data, it emerges that raft and virus membrane proteins have similar and differentiating features in terms of studied modifications and TMD properties. On average, virus and raft transmembrane proteins were enriched in palmitoylated and phosphorylated residues, but the amounts were very variable upon viral species. While palmitoylation is a known feature for raft partitioning, phosphorylation is new and could either be related to raft functionality and signaling, to protein phase separation and the stabilization of a raft phase, or both. Palmitoylation and disulfide bonds could be correlated to virus size, indicating that they are possible drivers of membrane curvature. Membrane-attached raft proteins were similarly enriched in palmitoyl and phosphoryl residues but surprisingly possessed isoprenyl residues that had been labeled as nonraft signals in the past. Attached viral proteins possessed shorter myristoyl residues and phosphorylation was higher in terms of protein fraction. We have to keep in mind that most of the analyzed raft data were based on DRM experiments which have their caveats, and further experimental data are certainly necessary to confirm these features in terms of raft partitioning and curvature. However, other structural features which might influence raft partitioning or curvature but were not analyzed in this study should also not be dismissed, i.e., the formation of dimers and oligomers.67
Acknowledgments
We thank Dr. Ilya Levental and Dr. Can Kesmir for suggestions in the accomplishment of this work and Adrian Kopf for a review of the manuscript. We also would like to thank Dr. Maria Sperotto for fruitful discussions on features that discriminate raft proteins from nonraft proteins.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jpcb.0c03435.
Principal component analysis and correlation matrix of viral protein modifications (PDF)
The authors declare no competing financial interest.
Special Issue
Published as part of The Journal of Physical Chemistry virtual special issue “Computational and Experimental Advances in Biomembranes”.
Supplementary Material
References
- Goswami D.; Gowrishankar K.; Bilgrami S.; Ghosh S.; Raghupathy R.; Chadda R.; Vishwakarma R.; Rao M.; Mayor S. Nanoclusters of GPI-anchored proteins are formed by cortical actin-driven activity. Cell 2008, 135, 1085–97. 10.1016/j.cell.2008.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowrishankar K.; Ghosh S.; Saha S.; C. R.; Mayor S.; Rao M. Active remodeling of cortical actin regulates spatiotemporal organization of cell surface molecules. Cell 2012, 149, 1353–1367. 10.1016/j.cell.2012.05.008. [DOI] [PubMed] [Google Scholar]
- Scolari S.; Engel S.; Krebs N.; Plazzo A. P.; De Almeida R. F.; Prieto M.; Veit M.; Herrmann A. Lateral distribution of the transmembrane domain of influenza virus hemagglutinin revealed by time-resolved fluorescence imaging. J. Biol. Chem. 2009, 284, 15708–16. 10.1074/jbc.M900437200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggeling C.; Ringemann C.; Medda R.; Schwarzmann G.; Sandhoff K.; Polyakova S.; Belov V. N.; Hein B.; von Middendorff C.; Schonle A.; Hell S. W. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature 2009, 457, 1159–62. 10.1038/nature07596. [DOI] [PubMed] [Google Scholar]
- Banjade S.; Rosen M. K. Phase transitions of multivalent proteins can promote clustering of membrane receptors. eLife 2014, 3, 04123. 10.7554/eLife.04123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X.; Ditlev J. A.; Hui E.; Xing W.; Banjade S.; Okrut J.; King D. S.; Taunton J.; Rosen M. K.; Vale R. D. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 2016, 352, 595–9. 10.1126/science.aad9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Rohrer B. B.; Levental K. R.; Simons K.; Levental I. Membrane raft association is a determinant of plasma membrane localization. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 8500–5. 10.1073/pnas.1404582111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. A.; Rose J. K. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell 1992, 68, 533–44. 10.1016/0092-8674(92)90189-J. [DOI] [PubMed] [Google Scholar]
- Sezgin E.; Levental I.; Mayor S.; Eggeling C. The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 2017, 18, 361–374. 10.1038/nrm.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood D.; Ries J.; Schwille P.; Simons K. Plasma membranes are poised for activation of raft phase coalescence at physiological temperature. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 10005–10. 10.1073/pnas.0804374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood D.; Simons K. Lipid rafts as a membrane-organizing principle. Science 2010, 327, 46–50. 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- Yang S. T.; Kreutzberger A. J. B.; Kiessling V.; Ganser-Pornillos B. K.; White J. M.; Tamm L. K. HIV virions sense plasma membrane heterogeneity for cell entry. Sci. Adv. 2017, 3, e1700338. 10.1126/sciadv.1700338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. T.; Kreutzberger A. J. B.; Lee J.; Kiessling V.; Tamm L. K. The role of cholesterol in membrane fusion. Chem. Phys. Lipids 2016, 199, 136–143. 10.1016/j.chemphyslip.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma D. K.; Gupta D.; Lal S. K. Host lipid rafts play a major role in binding and endocytosis of influenza A virus. Viruses 2018, 10, 650. 10.3390/v10110650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H.; Huang M.; Yuan Q.; Wei Y.; Gao Y.; Mao L.; Gu L.; Tan Y. W.; Zhong Y.; Liu D.; Sun S. The Important Role of Lipid Raft-Mediated Attachment in the Infection of Cultured Cells by Coronavirus Infectious Bronchitis Virus Beaudette Strain. PLoS One 2017, 12, e0170123. 10.1371/journal.pone.0170123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel S.; Scolari S.; Thaa B.; Krebs N.; Korte T.; Herrmann A.; Veit M. FLIM-FRET and FRAP reveal association of influenza virus haemagglutinin with membrane rafts. Biochem. J. 2010, 425, 567–73. 10.1042/BJ20091388. [DOI] [PubMed] [Google Scholar]
- Giese S. I.; Woerz I.; Homann S.; Tibroni N.; Geyer M.; Fackler O. T. Specific and distinct determinants mediate membrane binding and lipid raft incorporation of HIV-1(SF2) Nef. Virology 2006, 355, 175–91. 10.1016/j.virol.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Koshizuka T.; Kawaguchi Y.; Nozawa N.; Mori I.; Nishiyama Y. Herpes simplex virus protein UL11 but not UL51 is associated with lipid rafts. Virus Genes 2007, 35, 571–5. 10.1007/s11262-007-0156-2. [DOI] [PubMed] [Google Scholar]
- Ohkura T.; Momose F.; Ichikawa R.; Takeuchi K.; Morikawa Y. Influenza A virus hemagglutinin and neuraminidase mutually accelerate their apical targeting through clustering of lipid rafts. J. Virol. 2014, 88, 10039–55. 10.1128/JVI.00586-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiffele P.; Roth M. G.; Simons K. Interaction of influenza virus haemagglutinin with sphingolipid-cholesterol membrane domains via its transmembrane domain. EMBO J. 1997, 16, 5501–8. 10.1093/emboj/16.18.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaa B.; Levental I.; Herrmann A.; Veit M. Intrinsic membrane association of the cytoplasmic tail of influenza virus M2 protein and lateral membrane sorting regulated by cholesterol binding and palmitoylation. Biochem. J. 2011, 437, 389–97. 10.1042/BJ20110706. [DOI] [PubMed] [Google Scholar]
- Sengupta P.; Seo A. Y.; Pasolli H. A.; Song Y. E.; Johnson M. C.; Lippincott-Schwartz J. A lipid-based partitioning mechanism for selective incorporation of proteins into membranes of HIV particles. Nat. Cell Biol. 2019, 21, 452–461. 10.1038/s41556-019-0300-y. [DOI] [PubMed] [Google Scholar]
- Ivanova P. T.; Myers D. S.; Milne S. B.; McClaren J. L.; Thomas P. G.; Brown H. A. Lipid composition of viral envelope of three strains of influenza virus - not all viruses are created equal. ACS Infect. Dis. 2015, 1, 435. 10.1021/acsinfecdis.5b00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Genderen I. L.; Brandimarti R.; Torrisi M. R.; Campadelli G.; van Meer G. The phospholipid composition of extracellular herpes simplex virions differs from that of host cell nuclei. Virology 1994, 200, 831–6. 10.1006/viro.1994.1252. [DOI] [PubMed] [Google Scholar]
- Djordjevic J. T.; Schibeci S. D.; Stewart G. J.; Williamson P. HIV type 1 Nef increases the association of T cell receptor (TCR)-signaling molecules with T cell rafts and promotes activation-induced raft fusion. AIDS Res. Hum. Retroviruses 2004, 20, 547–55. 10.1089/088922204323087804. [DOI] [PubMed] [Google Scholar]
- Nikolaus J.; Scolari S.; Bayraktarov E.; Jungnick N.; Engel S.; Plazzo A. P.; Stockl M.; Volkmer R.; Veit M.; Herrmann A. Hemagglutinin of influenza virus partitions into the nonraft domain of model membranes. Biophys. J. 2010, 99, 489–498. 10.1016/j.bpj.2010.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorent J. H.; Levental I. Structural determinants of protein partitioning into ordered membrane domains and lipid rafts. Chem. Phys. Lipids 2015, 192, 23–32. 10.1016/j.chemphyslip.2015.07.022. [DOI] [PubMed] [Google Scholar]
- Levental I.; Lingwood D.; Grzybek M.; Coskun U.; Simons K. Palmitoylation regulates raft affinity for the majority of integral raft proteins. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 22050–4. 10.1073/pnas.1016184107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorent J. H.; Diaz-Rohrer B.; Lin X.; Spring K.; Gorfe A. A.; Levental K. R.; Levental I. Structural determinants and functional consequences of protein affinity for membrane rafts. Nat. Commun. 2017, 8, 1219. 10.1038/s41467-017-01328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D.; Kolter R. Functional microdomains in bacterial membranes. Genes Dev. 2010, 24, 1893–902. 10.1101/gad.1945010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkonian K. A.; Ostermeyer A. G.; Chen J. Z.; Roth M. G.; Brown D. A. Role of lipid modifications in targeting proteins to detergent-resistant membrane rafts. Many raft proteins are acylated, while few are prenylated. J. Biol. Chem. 1999, 274, 3910–7. 10.1074/jbc.274.6.3910. [DOI] [PubMed] [Google Scholar]
- Chlanda P.; Mekhedov E.; Waters H.; Sodt A.; Schwartz C.; Nair V.; Blank P. S.; Zimmerberg J. Palmitoylation contributes to membrane curvature in influenza A virus assembly and hemagglutinin-mediated membrane fusion. J. Virol. 2017, 91, e00947-17. 10.1128/JVI.00947-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed A.; Shah A. D.; Chen D.; Hill M. M. RaftProt V2: understanding membrane microdomain function through lipid raft proteomes. Nucleic Acids Res. 2019, 47, D459–D463. 10.1093/nar/gky948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe H. J.; Stevens T. J.; Munro S. A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell 2010, 142, 158–69. 10.1016/j.cell.2010.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A.; Apweiler R. The SWISS-PROT Protein Sequence Data Bank and Its New Supplement TREMBL. Nucleic Acids Res. 1996, 24, 21–25. 10.1093/nar/24.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A.; Larsson B.; von Heijne G.; Sonnhammer E. L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001, 305, 567–80. 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Yuan Z.; Zhang F.; Davis M. J.; Boden M.; Teasdale R. D. Predicting the solvent accessibility of transmembrane residues from protein sequence. J. Proteome Res. 2006, 5, 1063–70. 10.1021/pr050397b. [DOI] [PubMed] [Google Scholar]
- Mouritsen O. G.; Bloom M. Mattress model of lipid-protein interactions in membranes. Biophys. J. 1984, 46, 141–53. 10.1016/S0006-3495(84)84007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelderblom H. R.Structure and Classification of Viruses. In Medical Microbiology, 4th edition, Baron S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, 1996. [PubMed] [Google Scholar]
- Levental I.; Grzybek M.; Simons K. Greasing their way: lipid modifications determine protein association with membrane rafts. Biochemistry 2010, 49, 6305–16. 10.1021/bi100882y. [DOI] [PubMed] [Google Scholar]
- Rog T.; Vattulainen I. Cholesterol, sphingolipids, and glycolipids: What do we know about their role in raft-like membranes?. Chem. Phys. Lipids 2014, 184C, 82–104. 10.1016/j.chemphyslip.2014.10.004. [DOI] [PubMed] [Google Scholar]
- Komura N.; Suzuki K. G.; Ando H.; Konishi M.; Koikeda M.; Imamura A.; Chadda R.; Fujiwara T. K.; Tsuboi H.; Sheng R.; Cho W.; Furukawa K.; Furukawa K.; Yamauchi Y.; Ishida H.; Kusumi A.; Kiso M. Raft-based interactions of gangliosides with a GPI-anchored receptor. Nat. Chem. Biol. 2016, 12, 402–10. 10.1038/nchembio.2059. [DOI] [PubMed] [Google Scholar]
- Garner A. E.; Smith D. A.; Hooper N. M. Sphingomyelin chain length influences the distribution of GPI-anchored proteins in rafts in supported lipid bilayers. Mol. Membr. Biol. 2007, 24, 233–242. 10.1080/09687860601127770. [DOI] [PubMed] [Google Scholar]
- Reeves V. L.; Thomas C. M.; Smart E. J. Lipid rafts, caveolae and GPI-linked proteins. Adv. Exp. Med. Biol. 2012, 729, 3–13. 10.1007/978-1-4614-1222-9_1. [DOI] [PubMed] [Google Scholar]
- Iraqi M.; Edri A.; Greenshpan Y.; Kundu K.; Bolel P.; Cahana A.; Ottolenghi A.; Gazit R.; Lobel L.; Braiman A.; Porgador A. N-Glycans mediate the ebola virus-GP1 shielding of ligands to immune receptors and immune evasion. Front. Cell. Infect. Microbiol. 2020, 10, 48. 10.3389/fcimb.2020.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver Z. A.; Antonopoulos A.; Haslam S. M.; Dell A.; Dickinson G. M.; Seaman M. S.; Desrosiers R. C. Discovery of O-linked carbohydrate on HIV-1 envelope and its role in shielding against one category of broadly neutralizing antibodies. Cell Rep. 2020, 30, 1862–1869. 10.1016/j.celrep.2020.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler K. B.; Costello C. E. Glycomics and glycoproteomics of membrane proteins and cell-surface receptors: Present trends and future opportunities. Electrophoresis 2016, 37, 1407–19. 10.1002/elps.201500552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A. M. Q.Virus Taxonomy, Ninth report of the international Committee on Taxonomy of Viruses, Part I. Introduction. In Virus Taxonomy; King A. M. Q., Adams M. J.; Carstens E. B., Lefkowitz E. J., Eds.; Elsevier: San Diego, CA, 2012; pp 1–20. [Google Scholar]
- Neumann-Giesen C.; Falkenbach B.; Beicht P.; Claasen S.; Lüers G.; Stuermer C. A.; Herzog V.; Tikkanen R. Membrane and raft association of reggie-1/flotillin-2: role of myristoylation, palmitoylation and oligomerization and induction of filopodia by overexpression. Biochem. J. 2004, 378, 509–518. 10.1042/bj20031100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogomori H.; Hammond A. T.; Ostermeyer-Fay A. G.; Barr D. J.; Feigenson G. W.; London E.; Brown D. A. Palmitoylation and intracellular domain interactions both contribute to raft targeting of linker for activation of T cells. J. Biol. Chem. 2005, 280, 18931–42. 10.1074/jbc.M500247200. [DOI] [PubMed] [Google Scholar]
- Wang X.; Tian Q. B.; Okano A.; Sakagami H.; Moon I. S.; Kondo H.; Endo S.; Suzuki T. BAALC 1–6-8 protein is targeted to postsynaptic lipid rafts by its N-terminal myristoylation and palmitoylation, and interacts with alpha, but not beta, subunit of Ca/calmodulin-dependent protein kinase II. J. Neurochem. 2005, 92, 647–59. 10.1111/j.1471-4159.2004.02902.x. [DOI] [PubMed] [Google Scholar]
- Trabelsi S.; Zhang S.; Lee T. R.; Schwartz D. K. Linactants: surfactant analogues in two dimensions. Phys. Rev. Lett. 2008, 100, 037802. 10.1103/PhysRevLett.100.037802. [DOI] [PubMed] [Google Scholar]
- Munro S. Lipid rafts: elusive or illusive?. Cell 2003, 115, 377–88. 10.1016/S0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- Bryant M. R.; Marta C. B.; Kim F. S.; Bansal R. Phosphorylation and lipid raft association of fibroblast growth factor receptor-2 in oligodendrocytes. Glia 2009, 57, 935–46. 10.1002/glia.20818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson D.; Bakinowski M.; Thomas M. L.; Horejsi V.; Veillette A. Phosphorylation-dependent regulation of T-cell activation by PAG/Cbp, a lipid raft-associated transmembrane adaptor. Mol. Cell. Biol. 2003, 23, 2017–28. 10.1128/MCB.23.6.2017-2028.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.; Trible R. P.; Samelson L. E. LAT palmitoylation: its essential role in membrane microdomain targeting and tyrosine phosphorylation during T cell activation. Immunity 1998, 9, 239–46. 10.1016/S1074-7613(00)80606-8. [DOI] [PubMed] [Google Scholar]
- Brown D. A. Lipid rafts, detergent-resistant membranes, and raft targeting signals. Physiology 2006, 21, 430–9. 10.1152/physiol.00032.2006. [DOI] [PubMed] [Google Scholar]
- Chen B. J.; Takeda M.; Lamb R. A. Influenza virus hemagglutinin (H3 subtype) requires palmitoylation of its cytoplasmic tail for assembly: M1 proteins of two subtypes differ in their ability to support assembly. J. Virol. 2005, 79, 13673–84. 10.1128/JVI.79.21.13673-13684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colquhoun D. R.; Lyashkov A. E.; Mohien C. U.; Aquino V. N.; Bullock B. T.; Dinglasan R. R.; Agnew B. J.; Graham D. R. Bioorthogonal mimetics of palmitoyl-CoA and myristoyl-CoA and their subsequent isolation by click chemistry and characterization by mass spectrometry reveal novel acylated host-proteins modified by HIV-1 infection. Proteomics 2015, 15, 2066–2077. 10.1002/pmic.201500063. [DOI] [PubMed] [Google Scholar]
- Keating J. A.; Striker R. Phosphorylation events during viral infections provide potential therapeutic targets. Rev. Med. Virol. 2012, 22, 166–81. 10.1002/rmv.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yángüez E.; Hunziker A.; Dobay M. P.; Yildiz S.; Schading S.; Elshina E.; Karakus U.; Gehrig P.; Grossmann J.; Dijkman R.; Schmolke M.; Stertz S. Phosphoproteomic-based kinase profiling early in influenza virus infection identifies GRK2 as antiviral drug target. Nat. Commun. 2018, 9, 3679. 10.1038/s41467-018-06119-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S.; Mittal A. Transmembrane domain lengths serve as signatures of organismal complexity and viral transport mechanisms. Sci. Rep. 2016, 6, 22352. 10.1038/srep22352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindwasser O. W.; Resh M. D. Myristoylation as a target for inhibiting HIV assembly: unsaturated fatty acids block viral budding. Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 13037–42. 10.1073/pnas.212409999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantini J. Interaction of proteins with lipid rafts through glycolipid-binding domains: biochemical background and potential therapeutic applications. Curr. Med. Chem. 2007, 14, 2911–7. 10.2174/092986707782360033. [DOI] [PubMed] [Google Scholar]
- Lorent J.; Lins L.; Domenech O.; Quetin-Leclercq J.; Brasseur R.; Mingeot-Leclercq M. P. Domain formation and permeabilization induced by the saponin alpha-hederin and its aglycone hederagenin in a cholesterol-containing bilayer. Langmuir 2014, 30, 4556–69. 10.1021/la4049902. [DOI] [PubMed] [Google Scholar]
- Yamauchi S.; Takeuchi K.; Chihara K.; Sun X.; Honjoh C.; Yoshiki H.; Hotta H.; Sada K. Hepatitis C Virus Particle Assembly Involves Phosphorylation of NS5A by the c-Abl Tyrosine Kinase. J. Biol. Chem. 2015, 290, 21857–64. 10.1074/jbc.M115.666859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paladino S.; Sarnataro D.; Pillich R.; Tivodar S.; Nitsch L.; Zurzolo C. Protein oligomerization modulates raft partitioning and apical sorting of GPI-anchored proteins. J. Cell Biol. 2004, 167, 699–709. 10.1083/jcb.200407094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.